Abstract
Artificial intelligence (AI) has been widely applied to medical imaging. The use of AI for emission computed tomography, particularly single-photon emission computed tomography (SPECT) emerged nearly 30 years ago but has been accelerated in recent years due to the development of AI technology. In this review, we will describe and discuss the progress of AI technology in SPECT imaging. The applications of AI are dispersed in disease prediction and diagnosis, post-reconstruction image denoising, attenuation map generation, and image reconstruction. These applications are relevant to many disease categories such as the neurological disorders, kidney failure, cancer, heart disease, etc. This review summarizes these applications so that SPECT researchers can have a reference overview of the role of AI in current SPECT studies. For each application, we followed the timeline to present the evolution of AI’s usage and offered insights on how AI was combined with the knowledge of underlying physics as well as traditional non-learning techniques. Ultimately, AI applications are critical to the progress of modern SPECT technology because they provide compensations for many deficiencies in conventional SPECT imaging methods and demonstrate unparalleled success. Nonetheless, AI also has its own challenges and limitations in the medical field, including SPECT imaging. These fundamental questions are discussed, and possible future directions and countermeasures are suggested.
Keywords: Artificial intelligence (AI), deep learning, machine learning, neural network, single-photon emission computed tomography (SPECT)
Introduction
As a functional medical imaging modality, single-photon emission computed tomography (SPECT) is an important tool for molecular imaging research and clinical studies. It can be used to evaluate normal physiology and monitor many diseases including cardiovascular diseases (1-4), disorders of the central nervous system (5-7), cancer (8-11), brain functions (12-14), etc. However, the drawbacks of SPECT imaging are also well known. Limited spatial resolution (1–2 cm full-width half maximum) and high noise are inherent to the current clinical SPECT systems due to the hardware design. The projection data is also affected by degrading factors such as attenuation, scatter, and collimator septal penetration. All those lead to challenges in image reconstruction, and consequently affect diagnostic accuracy of SPECT images.
Recently, many artificial intelligence (AI) algorithms have been proposed to compensate for such deficiencies. Studies include denoising and resolution improvement for SPECT images, attenuation map generation and correction, end-to-end image reconstruction that directly takes the hardware implications into account, and many others. These techniques have been widely applied to the diagnosis of different diseases and conditions. In this review, we discuss these AI systems and applications such that they can be shared among SPECT researchers so AI techniques can be better utilized to serve the SPECT academic community and clinical investigations.
This paper will not review how AI was used to perform SPECT image restoration, i.e., post-processing on existing images reconstructed by conventional approaches, because those essentially belong to the image processing ambit [for example in (13)]. Instead, it will concentrate on the application and integration of AI into SPECT imaging for the diagnosis and investigation of different diseases. Although most of the work on AI in SPECT emerged in the last few years, the history of AI in SPECT imaging can be traced back 30 years to the pioneers who deserve particular recognition (14-18). The field owes a great deal to those few teams who ventured into huge-data collection, validation, and clinical trials. We present the following article in accordance with the Narrative Review reporting checklist (available at http://dx.doi.org/10.21037/atm-20-5988).
Brain imaging
In the 1990s, AI entered its first wave of prosperity. The AI methodology quickly spread to many areas including medical imaging. C. E. Floyd, Jr. might be the first SPECT researcher in that age who investigated the feasibility of using an artificial neural network (ANN) for SPECT brain imaging (14). The network was trained with an ideal projection-image pair to learn the mapping rule, which was accomplished by minimizing the mean squared error for a sample projection-image pair in the absence of noise. The developed ANN was a two-layer linear feedforward network validated by successfully reconstructing the Hoffman brain phantom (15), which provided a reconstruction result similar to those provided by the filtered backprojection (FBP) algorithm (16).
Since attenuation, noise, scatter, and the geometric response function of the camera (all having an impact on the projection data) were not accounted for in the FBP algorithm, in 1994, Gopal and Hebert (17) developed a feedforward ANN to restore the projection data before the FBP reconstruction. This approach was not specifically proposed for restoring the brain SPECT data only, but more broadly for all SPECT data. Their validation was performed on Tc-99m methylene diphosphonate (MDP) bone SPECT studies and red blood cell (RBC) liver-spleen SPECT studies).
Fakhri et al. in 2001 developed an ANN method for scatter and crosstalk correction in simultaneous 123I/99mTc dual-isotope brain SPECT imaging (18). In this method, the measured energy spectrum in 26 energy channels from each projection bin was the input, and the ANN was trained to produce primary-to-total photon ratio of each radionuclide using a back-propagation algorithm. The estimated primary photons were then reconstructed using the ordered-subset expectation maximization (OSEM) algorithm with compensation for nonuniform attenuation and distance-dependent collimator response. The method was evaluated using Monte-Carlo (MC) simulations, and the accuracy and variability of both 99mTc and 123I activity estimates were very close to those obtained from data without any scatter. They concluded that the ANN method plus OSEM could be a promising approach for absolute activity quantitation in simultaneous 123I/99mTc SPECT.
Challenges
The pioneering research in the 1990s identified many challenges for AI in SPECT imaging. These challenges are fundamental: the architecture of ANNs was restricted to 2–3 feedforward layers that is too simple to solve complex problems in SPECT imaging, especially the nonlinear inverse problems. Furthermore, due to relatively insufficient computing power, it was challenging to develop advanced AI systems with a large training database, which significantly dampened the reliability.
Alzheimer’s disease (AD) diagnosis
Not until approximately 10 years ago, thanks to the substantial progress in computer technology, that more advanced AI algorithms and systems could be developed to solve more practical problems. One example was the AI-aided early detection of AD. The examination of the predictive value of SPECT imaging with respect to AD has been performed since functional SPECT imaging was found to be effective in providing information about regional cerebral blood flow (rCBF), which is valuable for early diagnosis of AD (19). The evaluation on SPECT of patients who may have AD is usually done through visual assessments performed by experts. In 2010, Illan et al. proposed an independent component analysis method for designing a computer-aided diagnosis system for AD (20). The method was based on a support vector machine (SVM), another typical machine learning (ML) algorithm differing from the NN approach. For training the diagnostic system, SPECT images (used for training) were labelled by physicians as one of four categories: normal (no symptoms of AD), possible (AD-1), probable AD (AD-2), and certain AD (AD-3) to distinguish between different levels of the typical characteristics of AD. The fully trained system was found to reach a 91.1% accuracy during the validation stage. Later, Rondina et al. developed another SVM approach to classify patients with mild AD versus age and gender (21). In contrast to using the whole-brain image, brain regions identified as the most relevant sources were used to discriminate the AD patients. The result was that such an approach contributed to the improvement of classification accuracy for 18F-FDG PET only, but not for rCBF SPECT.
In 2017, Höller et al. (22) combined quantitative markers from SPECT and electroencephalography (EGG) for differential diagnosis of disorders with amnestic symptoms. The classification was again accomplished by SVM algorithm. Training was performed on 39 patients examined with AD, 69 patients with depressive cognitive impairment (DCI), 71 patients with amnestic mild cognitive impairment (aMCI), and 41 patients with amnestic subjective cognitive complaints (aSCC). The conclusion was that quantitative analysis of EGG and SPECT imaging plus ML techniques can effectively differentiate AD, DCI, aMCI, and aSCC.
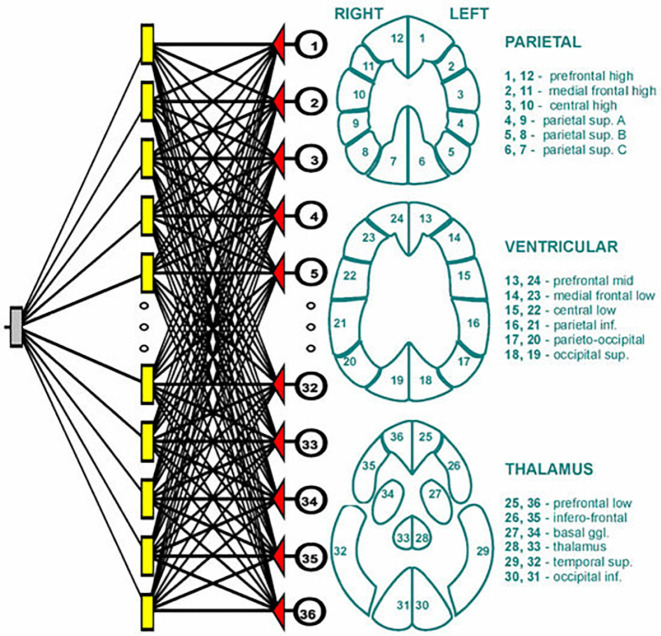
More recently, an ANN method was applied by Swietlik and Bialowas to identify AD using cerebral perfusion SPECT data (23). Data from 72 (43 female, 29 male) AD patients and 60 (44 female, 16 male) normal controls were involved in this study, where the training group contained 100 (55 AD, 45 normal) patients and the testing group had 32 (17 AD, 15 normal) cases. For each patient, the reconstructed brain profile was segmented into 36 fixed subregions, and then the total counts in each subregion were computed and used as an input signal into and NN, as shown in Figure 1. The NN architecture was quite simple, which consisted of three layers with 36 neurons in input layer, 21 neurons in the hidden layer, and 1 neuron in the output layer. A comparison of AD diagnosis by this NN and by conventional statistical methods did not show any statistically significant differences.
Figure 1.
The ANN and its input signal by Swietlik and Bialowas (23).
Parkinson’s disease (PD)
123I-Ioflupane (123I FP-CIT), also known as DaTscan, is an FDA approved (in 2011) radioactive tracer that binds to the dopamine transporter (DAT) on pre-synaptic neurons in the dopaminergic system. DAT SPECT has been rountinely used for the dignosis and monitoring of PD. There is an open database, Parkinson’s Progression Marker Initiave (PPMI, https://www.ppmi-info.org/), that contains thousands of DAT SPECT images and other relevant data, which has promoted more applications of AI in DAT SPECT for PD than many other SPECT studies. In addition to ML algorithms such as SVM (24-27), k-nearest neighbors (26,27), or deep learning NNs (28-33) that make diagnoses relying on reconstructed SPECT images, there are also many other interesting applications leveraging either the DAT SPECT images or DAT SPECT projection data.
Tang et al. developed an ML prediction model in 2019 (34), aiming to predict motor outcome from baseline (year 0) DAT SPECT imaging radiomic features and clinical measurements. Images fed to the NN were reconstructed by OSEM algorithm (35,36). The trained ANN was able to predict the unified PD rating scale (UPDRS) part III motor score in year 4 from 92 imaging features extracted on 12 different regions as well as 6 non-imaging measurements at baseline, with a prediction accuracy of 75%.
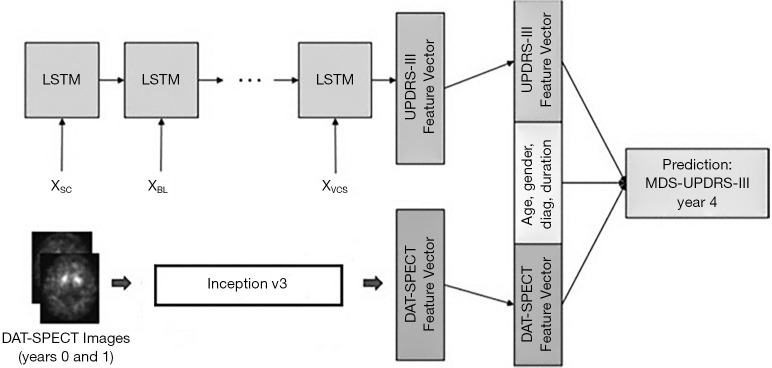
Similar work was conducted at Johns Hopkins University (JHU). In 2018, Leung et al. (37) designed a NN to predict outcome of patients of PD using longitudinal clinical data containing imaging and non-imaging information. Features were extracted from clinical data by the NN (imaging features were first extracted by an Inception V3 network which was then combined with non-imaging information, such as age, gender, and diagnostic duration, to be delivered to a long short-time memory (LSTM)-based network for making final processing, as illustrated in Figure 2) and then the motor performance (UPDRS part III) in year 4 can be outputted. The prediction result was compared with the true UPDRS part III in year 4. It was found that the accuracy of prediction given only the non-imaging data outperformed the approach given only the imaging data. But the performance was substantially improved when both imaging and non-imaging information were given to the NN. Recently, the same group has also worked on using NN approach to detect PD directly from DAT-SPECT projection data (38). Differing from other methods where reconstructed images were used (24-33), in this method the NN accepted SPECT projection data and a conclusion was made from the analysis in the projection-data space directly. The hypothesis was that all the necessary information for accurate diagnosis was already presented in the projection- data, and reconstruction into images was simply to allow easy interpretation by a human observer. A NN can retrieve such information directly from projection data, and thus eliminates the necessity of time-consuming image reconstruction processes. The team showed that the developed NN can yield an overall accuracy of 97%. Currently, they are working on comparing the performance of this approach with a NN that operates in the reconstructed DAT-SPECT image domain.
Figure 2.
A deep-learning architecture to predict UPDR-III scores in year 4 for PD patients, presented by the JHU group.
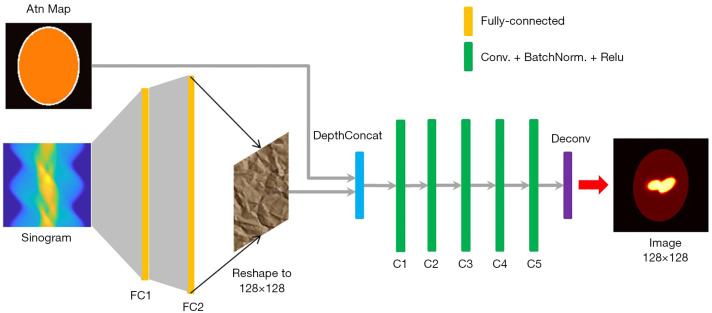
Using AI to perform an end-to-end SPECT image reconstruction is also of interest (39-42). An ANN consisting of two fully connected layers followed by five convolutional layers was designed to accept SPECT projection data and attentuation map at its input, and to output a 2-D quantitative activity image showing the distribution of the 123I- Ioflupane in the brain. The network architecure is shown in Figure 3. The input is composed of two channels to accept the sinogram and the attenuation map respectively (their matrix size can be different). The sinogram will pass througth two fully-connected layers to reconstruct a raw image which will then concatenate with the attenuation map for compensation and noise reduction in the convolutional layers. Regarding the NN training, different from all previous examples using merely a few tens of patient data to conduct the training, 20,000 synthetic phantom images were employed in NN training and patient data was used for validation. The large training data pool significantly reduced the probability of overfitting, which is a serious issue in AI systems development. The reconstruction image by the developed NN was found to outperform the one reconstructed by the traditional OSEM algorithm validated using both the Zubal phantom data (43) and patient data.
Figure 3.
The architecture of the ANN presented in (39) and the reconstructed brain image showing the biomarker in the striatum using patient data.
Another interesting example was by Huertas-Fernández et al. who developed two machine learning models in 2015 for differentiating vascular parkinsonism and PD (44), still using the data from 123I FP-CIT DAT SPECT. The study included 80 patients with vascular parkinsonism and 164 patinets with PD who underewent 123I FP-CIT DAT SPECT studies. Signicant differences were found in the region of interest (ROI) analysis (counterpart of the whole-brain analysis method) between vascular parkinsonism and PD in 123I FP-CIT uptake in the more affected side of the putamen and the ipsilateral caudate. The diagnostic accuracy of the logistic regression model using ROI data was 90.3% and of the SVM model using statistical parametric mapping (SPM) data was 90.4%. The conclusion was that the models built with ROI data and SPM data from 123I FP-CIT SPECT provide great discrimination accuracy between vascular parkinsonism and PD.
Cardiac imaging
SPECT is widely used in cardiac imaging to investigate myocardial perfusion, metabolism, viability, and innervation. Therefore, AI cardiac SPECT has attracted a great deal of attention in the last few years. In 2013, Ciecholewski presented a high-efficient SVM method to diagnose ischemic heart disease (45). Heart images acquired by SPECT were classified by the SVM system with a higher accuracy than using principal component analysis (PCA) and NNs. But the NN investigated in the article had only one hidden layer, raising some questions as to the robustness of the conclusions.
Other than diagnosis using ML approaches based on SPECT images, another topic that has been recently intensively researched is prediction of cardiac death in heart failure. In 2019, Haro Alonso et al. (46) developed several ML models to estimate a patient’s risk of cardiac death based on adenosine or exercise stressed myocardial perfusion SPECT (MPS). Accuracy of the models was measured by area under the receiver operating characteristic (ROC) curve (AUC). The performance of all ML models outperformed the conventional baseline logistic regression (LR) method for prediction of cardiac death in patients undergoing MPS, and SVM method was found to yield the best AUC among all ML models. In contrast, Nakajima et al. proposed another ML system incorporating 123I-metaiodobenzylguanidine (MIBG) myocardial innervation SPECT imaging to differentially predict risk of life-threatening arrhythmic events (ArE) and heart failure death (HFD) (47,48). The ANN was found diagnostically accurate and comparable to nuclear cardiology expert interpretation, and better than conventional semi-quantitative defect scoring. The details of their ANN architecture, however, were not presented in the article.
Another contribution of AI is to improve the scan efficiency (49,50) through benefitting from the powerful image-analysis capability of convolutional NN (CNN) technique. The acquisition time in SPECT can be either reduced by a decrease of acquisition time per projection, or a reduction of the number of angular projections. The obtained sinogram is then processed by a fully-trained CNN to recover the full sinogram from a normal SPECT scan (Figure 4). Shiri et al. (50) found that the predicted full-time sinogram had better quality than the predicted full-angle sinogram, when implementing a residual CNN to perform the transformation from the half-time sinogram and half-angle sinogram, respectively. The clinical benefit of such an application can be significant, including reduced radiation dose, improved patient comfort, and increased clinical efficiency.
Figure 4.
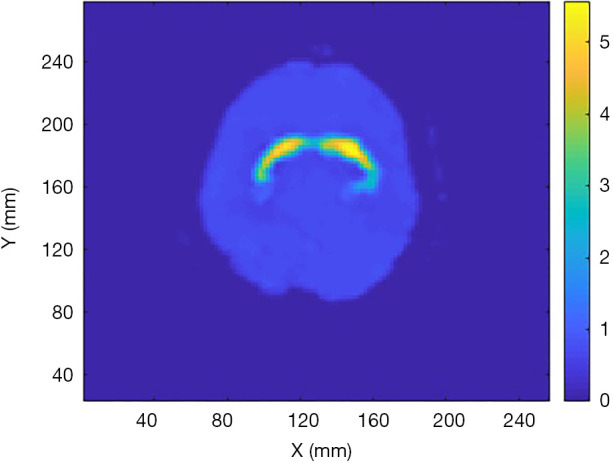
Reconstructed image by the developed ANN in (39) using patient data.
In addition, attenuation map estimation is also an interesting application of AI. Attenuation correction using CT or transmission scan generated attenuation maps is often used in SPECT imaging to increase accuracy and to enable quantitative analysis. However, the use of CT scans increases radiation dose to patients, and significant artifacts may occur due to misregistration between SPECT and CT scans as a result of patient motion. Moreover, there are many existing SPECT systems that do not have a CT scanner or transmission scan capability. Recently, ML methods have been proposed to estimate the attenuation map directly from SPECT emission data. In Yale University, Shi et al. (51) developed a deep NN to perform such estimation. Both photopeak window and scatter window SPECT images were used as the input of the network. The CT-based attenuation maps were used as labels and 65 cardiac SPECT/CT images were employed for the NN training and testing. Two training strategies, standard training and adversarial-mode training were applied in the NN development. In the adversarial mode, the generator was a U-net like network accepting primary SPECT image patch and the scatter SPECT image patch and generating attenuation map at its output. The generated attenuation map as well as the measured (real) attenuation map were fed to the discriminator to make a judgement, labelled 0 for generated or 1 for real. The generator and the discriminator were trained together until a Nash equilibrium was reached. The result was that highly reliable attenuation maps can be acquired by the CNN method to facilitate attenuation correction for SPECT-only scanners. The team also addressed that when only primary window SPECT data were available with the absence of scatter data, it was still feasible to generate accurate attenuation maps using the adversarial training mode, but impossible with the standard training mode.
Prostate cancer
A Sweden-US collaboration team (52) has recently developed a deep learning algorithm which is called by the team prostate-specific membrane antigen with AI (PSMA-AI), to identify men with low risk of prostate cancer who are potential surveillance candidates, based on a PSMA-targeted small molecule SPECT/CT imaging agent (99mTc-MIP-1404). SPECT/CT images were retrospectively analysed by the PSMA-AI system which calculates the uptake of 99mTc-MIP-1404 against the background reference. Upon adding PSMA-AI to their previously developed prediction model (based on a multivariable logistic regression method), the positive predictive value was increased from 70% to 77%, and the negative predictive value from57% to 74%. In a second experiment, the system was used to read SPECT/CT images from 464 patients with very low, low, or intermediate risk of prostate cancer and make an assessment (53). The result was compared with manual assessment and higher AUCs were achieved by the PSMA-AI system.
Thyroid disease
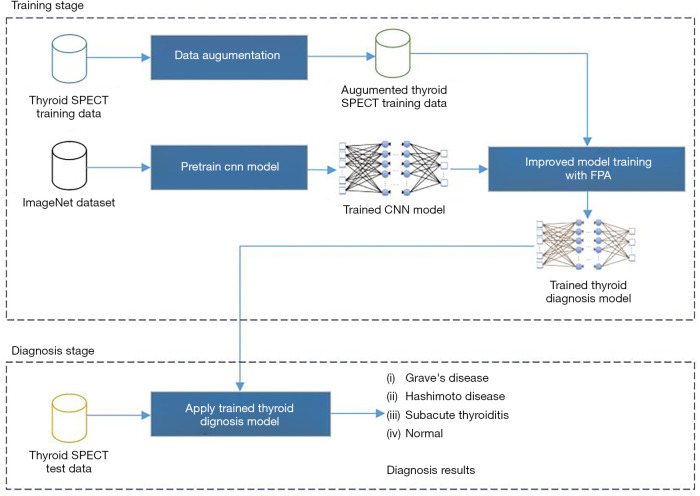
Thyroid disease is the second largest disease in the endocrine field and SPECT has been a vital diagnosis tool in recognizing thyroid diseases. In order to apply a computer-aided system to thyroid disease diagnosis, researchers from China developed a NN to read SPECT images and then classify images into specific diseases according to characteristics in the SPECT images learned by the NN (54,55). A transfer-learning strategy was employed: a CNN that has been pretrained with ImageNet database was retrained by using SPECT patient images to fine-tune the network, including 780 samples of Grave’s disease, 438 samples of Hashimoto thyroiditis, 810 samples of subacute thyroiditis, and 860 normal cases, with label pre-signed to each SPECT image. Detailed training scheme is shown in Figure 5. The CNN was then expected to identify such four diseases given SPECT images after the training procedures. Their experimental results showed that the developed CNN was efficient for the diagnosis of thyroid disease with SPECT images and could provide superior performance relative to other CNN methods.
Figure 5.
The training strategy of the thyroid SPECT diagnosis AI system developed in (55).
Bone lesions
99mTc-MDP SPECT has been well-established in the diagnosis of lumbar stress injury. However, the low quality of 99mTc-MDP SPECT images hampers lesion detectability and leads to a high interobserver variability. A deep learning-based diagnostic system was thus developed by Petibon et al. (56) to improve lesion detection and reduce variability in interpretation. FBP reconstruction images were fed to a CNN to make a classification. The performance of the developed CNN was compared to nuclear medicine physicians using ROC and localization ROC analyses based on 65 adolescent athletes (mean age 14.9 years) SPECT images. Five physicians reviewed the test images to provide a confidence rating of a lesion presence or absence. The developed CNN offered similar performance to that of physicians in detecting positive lumber lesions. The mean area under the ROC curve was 0.893 for physicians and 0.905 for the CNN. For LROC, the mean area under the curve was 0.811 for the CNN and 0.787 for physicians.
Role in projection simulation
MC simulation of SPECT imaging is a useful method to precisely model how particles propagate and the interactions inside the medium, e.g., a patient’s body, and the SPECT detector system, i.e., photon interactions in the collimator and the scintillator (crystal). It has been widely used in SPECT imaging research. However, such a method is extremely computationally expensive because one must track many photon histories to generate low-noise estimates.
In 2018, Sarrut et al. proposed an NN algorithm for accelerating the MC simulation (57). Usually, a MC simulation can be thought to comprise of two steps: (I) tracking the particles inside the patient, and (II) tracking the particles in the SPECT detector including the digitization chain of the readout electronic components. As addressed by the team, the first step may be accelerated by employing a parallel algorithm running on GPUs, so the team’s focus was on developing an NN to accelerate the second step. Specifically, since photon interactions in the detector can be approximated by the angular response functions (ARF) (58,59), the purpose of the work was to essentially develop an NN to learn the ARF of a collimator-detector system. The input of the NN is the photon energy and direction (incident angles), and the output was the photon detection probability within an energy window. The NN was composed of three fully-connected layers with each consisting of 400 neurons. The training dataset was generated from a MC simulation, and the training of the NN took 35–45 min on a Nvidia Titan XP graphics processing unit (GPU) (Nvidia, Santa Clara, CA, USA). As a result, the overall speed-up of a complete SPECT simulation was between 80 and 400 times compared to a non-NN involved conventional simulation, depending on the energy window.
Another example where AI played a role in SPECT was by Xiang et al., who developed a deep CNN (DCNN) for fast scatter estimation (60). In general MC simulation-based scatter estimation is accurate but time consuming. Commonly used energy-window-based scatter estimation is simple to implement but suffers from inaccuracy. The team aimed to develop an NN to estimate the scatter projection given the SPECT emission projection (90Y bremsstrahlung) and the projection of the attenuation map. The training dataset was acquired from a high-count MC simulation (1 billion per projection), which modeled the Siemens Intevo SPECT/CT system (5/8’’ crystal, 105–195 KeV acquisition window, 128 views, 128×80 matrix, 4.8×4.8 mm pixel size). Scaling of the input to the network is required because the total counts in the emission projections may vary enormously between MC simulations and real clinic measurements. The estimated scatter projection was then provided to an OSEM reconstruction to compensate for the scatter. The network architecture developed by the team was a 13-layer NN consisting of separate paths for accepting the emission and attenuation projection respectively, and then concatenated before the final convolution steps. The method was evaluated by a simulated sphere phantom with a lung insert, measurements of a liver phantom, and patient data after 90Y radioembolization. The developed DCNN was found to be able to estimate the scatter in 90Y bremsstrahlung SPECT/CT with accuracy similar to MC-based estimation, but at a fraction of the time. The DCNN could generate patient scatter projection for 128 views within 1 min using a single processor on a desktop computer which is about 3 orders of magnitude faster than estimating bremsstrahlung scatter by MC.
Outlook
Although the birth of AI applications for SPECT imaging emerged approximately 30 years ago, there has been a marked acceleration in recent years. Nearly 80% of publications have occurred in or after the year of 2018, benefiting from the significant progress of the ML theory in the 2010s and the development of GPU computing. Unlike in the last century when applications were restricted to image reconstruction and data restoration, many new applications are emerging, including diagnosis, projection-data augmentation for quick SPECT examination, future risk forecasting, attenuation map generation, and MC SPECT simulation acceleration, as illustrated in the above sections.
Thus, one trend of the use of AI in SPECT medical imaging which we can confirm at present is the diversity. It can be estimated that many new applications that have not been investigated or imagined by people in the past, are emerging and the speed of emergence is accelerating. In addition, the rapid growth of ML theory will benefit AI SPECT research in term of new training algorithms to develop more accurate and reliable systems. As more and more clinic/patient data are used in AI system development, the risk of overfitting will be reduced. The growth of CPU and GPU computational capability will further increase the efficiency of AI systems. It can be expected that more efficient and more accurate SPECT AI techniques are emerging to benefit scientific research, clinical patient care, and society as a whole sooner than imagined.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This work is supported by NIH under grant R01NS094227 and U01CA140204. We also thank Nvidia Corporation for the Higher Education and Research Grants program to support this work.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Footnotes
Provenance and Peer Review: This article was commissioned by the editorial office, Annals of Translational Medicine for the series “Artificial Intelligence in Molecular Imaging”. The article has undergone external peer review.
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-5988
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-5988). The series “Artificial Intelligence in Molecular Imaging” was commissioned by the editorial office without any funding or sponsorship. SPR served as the unpaid Guest Editor of the series and serves as an unpaid editorial board member of Annals of Translational Medicine from Mar 2020 to Feb 2022. Dr. Shao reports grants and non-financial support from Nvidia Corporation, during the conduct of the study. The authors have no other conflicts of interest to declare.
References
- 1.Gullberg GT, Christian PE, Zeng GSL. Cone beam tomography of the heart using sinle-photon smission-computed tomography. Invest Radiol 1991;26:681-8. 10.1097/00004424-199107000-00014 [DOI] [PubMed] [Google Scholar]
- 2.Lalush DS, Tsui BMW. Block-iterative techniques for fast 4D reconstruction using a priori motion models in gated cardiac SPECT. Phys Med Biol 1998;43:875-86. 10.1088/0031-9155/43/4/015 [DOI] [PubMed] [Google Scholar]
- 3.Chen J, Garcia EV, Folks RD, et al. Onset of left ventricular mechanical contraction as determined by phase analysis of ECG-gated myocardial perfusion SPECT imaging: development of a diagnostic tool for assessment of cardiac mechanical dyssynchrony. J Nucl Cardiol 2005;12:687-95. 10.1016/j.nuclcard.2005.06.088 [DOI] [PubMed] [Google Scholar]
- 4.Garcia EV, Faber TL, Esteves FP. Cardiac dedicated ultrafast SPECT cameras: new designs and clinical aimplications. J Nucl Med 2011;52:210-7. 10.2967/jnumed.110.081323 [DOI] [PubMed] [Google Scholar]
- 5.Nicolas JM, Catafau AM, Estruch R, et al. Regional cerebral blood flow-SPECT in chronic-alcoholism – relation to neuropsychological testing. J Nucl Med 1993;34:1452-9. [PubMed] [Google Scholar]
- 6.Sanna G, Piga M, Terryberry JW, et al. Central nervous system involvement in systemic lupus erythematosus: cerebral imaging and serological profile in patients with and without overt neuropsychiatric manifestations. LUPUS 2000;9:573-83. 10.1191/096120300678828695 [DOI] [PubMed] [Google Scholar]
- 7.Perri M, Erba P, Volterrani D, et al. Octreo-SPECT/CT imaging for accurate detection and localization of suspected neuroendocrine tumors. Q J Nucl Med Mol Imaging 2008;52:323-33. [PubMed] [Google Scholar]
- 8.Keidar Z, Israel O, Krausz Y. SPECT/CT in tumor imaging: technical aspects and clinical applications. Semin Nucl Med 2003;33:205-18. 10.1053/snuc.2003.127310 [DOI] [PubMed] [Google Scholar]
- 9.Christian JA, Partridge M, Mioutskkou E, et al. The incoporation of SPECT functional lung imagign into inverse radiotherapy planning for non-small lung cancer. Radiother Oncol 2005;77:271-7. 10.1016/j.radonc.2005.08.008 [DOI] [PubMed] [Google Scholar]
- 10.Lerman H, Metser U, Lievshitz G, et al. Lymphoscintigraphic sentinel node identification in patients with breast cancer: the role of SPECT-CT. Eur J Nucl Med Mol Imaging 2006;33:329-37. 10.1007/s00259-005-1927-4 [DOI] [PubMed] [Google Scholar]
- 11.Bhushan KR, Misra P, Liu F, et al. Detection of breast cacer microcalcifications using a dual-modality SPECT/NIR fluoresent probe. J Am Chem Soc 2008;130:17648-9. 10.1021/ja807099s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fukuyama H, Ouchi Y, Matsuzaki S, et al. Brain functional activity during gait in normal subjects: A SPECT study. Neurosci Lett 1997;228:183-6. 10.1016/S0304-3940(97)00381-9 [DOI] [PubMed] [Google Scholar]
- 13.Dietze MMA, Branderhorst W, Kunnen B, et al. Accelerated SPECT image reconstruction with FBP and an image enhancement convolutional neural network. EJNMMI Phys 2019;6:14. 10.1186/s40658-019-0252-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Floyd CR. An artificial neural network for SPECT image reconstruction. IEEE Trans Med Imaging 1991;10:485-7. 10.1109/42.97600 [DOI] [PubMed] [Google Scholar]
- 15.Hoffman EJ, Ricci AR, van der Stee M, et al. ECAT III – basic design condierations. IEEE Trans Nucl Sci 1983;15:729-33. 10.1109/TNS.1983.4332366 [DOI] [Google Scholar]
- 16.Pelc NJ, Chesler DA. Utilization of cross-plane rays for three-dimensional reconstruction by filtered back-projection. J Comput Assist Tomogr 1979;3:385-95. 10.1097/00004728-197906000-00015 [DOI] [PubMed] [Google Scholar]
- 17.Gopal SS, Hebert TJ. Pre-reconstruction restoration of SPECT projection images by a neural network. IEEE Trans. Nucl Sci 1994;41:1620-5. 10.1109/23.322958 [DOI] [Google Scholar]
- 18.El Fakhri G, Moore SC, Maksud P, et al. Absolute active quantitation in simultaneous 123I/99mTc brain SPECT. J Nucl Med 2001;42:300-8. [PubMed] [Google Scholar]
- 19.Kogure D, Matsuda H, Ohnishi T, et al. Longitudinal evaluation of early Alzheimer’s disease using brain perfusion SPECT. J Nucl Med 2000;41:1155-62. [PubMed] [Google Scholar]
- 20.Illan A, Gorriz JM, Ramirez J, et al. Projecting independent components of SPECT images for computer aided diagnosis of Alzheimer’s disease. Pattern Recognition Lett 2010;31:1342-7. 10.1016/j.patrec.2010.03.004 [DOI] [Google Scholar]
- 21.Rondina JM, Ferreira LK, de Souza Duran FL, et al. Selecting the most relevant brain regions to discriminate Alzheimer’s disease patients from healthy controls using multiple kernel learning: A compariosn across functional and structural imaging modalities and atlases. Neuroimage Clin 2017;17:628-41. 10.1016/j.nicl.2017.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Höller Y, Bathke AC, Uhl A, et al. Combining SPECT and Quatitative EEG analysis for the automated differential diagosis of diorders with amnestic symptoms. Front Aging Neurosci 2017;9:290. 10.3389/fnagi.2017.00290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Świetlik D, Bialowas J. Application of artificial neural networks to identify Alzheimer’s disease using cerebral perfusion SPECT data. Int J Environ Res Public Health 2019;16:1303. 10.3390/ijerph16071303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Illan IA, Gorriz JM, Ramirez J, et al. Automatic assistance to Parkinson’s disease diagnosis in DaTScan SPECT imaging. Med Phys 2012;39:5971-80. 10.1118/1.4742055 [DOI] [PubMed] [Google Scholar]
- 25.Anita S, Priya PA. Three dimensional analysis of SPECT images for diagnosing early Parkinson's disease using radial basis function kernel – extreme learning machine. Curr Med Imaging Rev 2019;15:461-70. 10.2174/1573405614666171219154154 [DOI] [PubMed] [Google Scholar]
- 26.Oliveira FPM, Faria DB, Costa DC, et al. Extraction, selection and comparison of features for an effective automated computer-aided diagnosis of Parkinson's disease based on [I-123]FP-CIT SPECT images. Eur J Nucl Med Mol Imaging 2018;45:1052-62. 10.1007/s00259-017-3918-7 [DOI] [PubMed] [Google Scholar]
- 27.Mabrouk R, Chikhaoui B, Bentabet L. Machine learning based classification using clinical and DaTSCAN SPECT imaging features: a study on Parkinson's disease and SWEDD. IEEE Trans Radiat Plasma Med Sci 2019;3:170-7. 10.1109/TRPMS.2018.2877754 [DOI] [Google Scholar]
- 28.Choi H, Ha S, Im H, et al. Refining diagnosis of Parkinson's disease with deep learning-based interpretation of dopamine transporter imaging. NeuroImage Clin 2017;16:586-94. 10.1016/j.nicl.2017.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim DH, Wit H, Thurston M. Artificial intelligence in the diagnosis of Parkinson's disease from ioflupane-123 single-photon emission computed tomography dopamine transporter scans using transfer learning. Nucl Med Commun 2018;39:887-93. 10.1097/MNM.0000000000000890 [DOI] [PubMed] [Google Scholar]
- 30.Segovia F, Gorriz JM, Ramirez J. Using deep neural networks along with dimensionality reduction techniques to assist the diagnosis of neurodegenerative disorders. Log J IGPL 2018;26:618-28. 10.1093/jigpal/jzy026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor JC, Romanowski C, Lorenz E, et al. Computer-aided diagnosis for (I-123) FP-CIT imaging: impact on clinical reporting. EJNMMI Res 2018;8:36. 10.1186/s13550-018-0393-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ortiz A, Munilla J, Martinez M, et al. Parkinson’s disease detection using isosurfaces-based features and convolutional neural networks. Front Neuroinform 2019;13:48. 10.3389/fninf.2019.00048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pahuja G, Nagabhushan TN, Prasad B. Early detection of Parkinson’s disease by using SPECT imaging and biomarkers. J Intell Syst 2020;29:1329-44. 10.1515/jisys-2018-0261 [DOI] [Google Scholar]
- 34.Tang J, Yang B, Adams MP, et al. Artificial neural network-based prediction of outcome in Parkinson's disease patients using DaTscan SPECT imaging features. Mol Imaging Biol 2019;21:1165-73. 10.1007/s11307-019-01334-5 [DOI] [PubMed] [Google Scholar]
- 35.Hudson HM, Larkin RS. Accelerated image reconstruction using ordered subsets of projection data. IEEE Trans Med Imaging 1994;13:601-9. 10.1109/42.363108 [DOI] [PubMed] [Google Scholar]
- 36.Hudson HM, Hutton BF, Larkin RS. Accelerated EM reconstruction using ordered subsets. J Nucl Med 1992;33:960. [Google Scholar]
- 37.Leung KH, Salmanpour MR, Saberi A, et al. Using deep learning to predict outcome of patients with Parkinson’s disease. IEEE NSS/MIC Proc. 2018: 1-4. [Google Scholar]
- 38.Leung K, Shao W, Solnes Lilja, et al. A deep learning-based approach for disease detection in the projection space of DAT-SPECT images of patients with Parkinson’s disease. J Nucl Med 2020;61:509. [Google Scholar]
- 39.Shao W, Pomper MG, Du Y. A learned reconstruction network for SPECT imaging. IEEE Trans Radiat Plasma Med Sci 2021;5:26-34. 10.1109/TRPMS.2020.2994041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shao W, Leung K, Du Y. SPECT image reconstruction a learnt neural network. J Nucl Med 2020;61:1478. [Google Scholar]
- 41.Shao W, Du Y. SPECT image reconstruction by deep learning using a two-step training method. J Nucl Med 2019;60:1353. [Google Scholar]
- 42.Shao W, Rowe S, Du Y. SPECTnet: a deep leaning neural network for SPECT image reconstraction. Ann Transl Med 2020. [Epub ahead of print]. doi: . 10.21037/atm-20-3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zubal IG, Harrell CR, Smith EO, et al. Computerized three-dimensional segmented human anatomy. Med Phys 1994;21:299-302. 10.1118/1.597290 [DOI] [PubMed] [Google Scholar]
- 44.Huertas-Fernández I, García-Gómez FJ, García-Solís D, et al. Machine learning models for the differential diagnosis of vascular parkinsonism and Parkinson's disease using [(123)I]FP-CIT SPECT. Eur J Nucl Med Mol Imaging 2015;42:112-9. 10.1007/s00259-014-2882-8 [DOI] [PubMed] [Google Scholar]
- 45.Ciecholewski M. Ischemic heart disease detection using selected machine learning methods. Inter J Computer Math 2013;90:1734-59. 10.1080/00207160.2012.742189 [DOI] [Google Scholar]
- 46.Haro Alonso D, Wernick MN, Yang Y, et al. Prediction of cardiac death after ademosine myocardial perfusion SPECT based on machine learning. J Nucl Cardiol 2019;26:1746-54. 10.1007/s12350-018-1250-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakajima K, Nakata T, Doi T, et al. Machine learning-based risk model using I-123-metaiodobenzlguanidine to differentially predict modes of cardiac death in hear failure. J Nucl Cardiol 2020. [Epub ahead of print]. doi: . 10.1007/s12350-020-02173-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakajima K, Kudo T, Nakata T, et al. Diagnostic accuracy of an artificial neural network compared with statistical quantitation of myocardial perfusion images: A Japanese multicenter study. Eur J Nucl Med Mol Imaging 2017;44:2280-9. 10.1007/s00259-017-3834-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garcia EV. Deep learning, another important tool for improving acqusition efficiency in SPECT MPI imaging. J Nucl Cardiol 2020. [Epub ahead of print]. doi: . 10.1007/s12350-020-02188-z [DOI] [PubMed] [Google Scholar]
- 50.Shiri I, Sabet KA, Arabi H, et al. Standard SPECT myocardial perfusion estimation from half-time acquisitions using deep convolutional residual neural networks. J Nucl Cardiol 2020. [Epub ahead of print]. doi: . 10.1007/s12350-020-02119-y [DOI] [PubMed] [Google Scholar]
- 51.Shi L, Onofrey JA, Liu H, et al. Deep learning-based attenuation map generation for myocardial perfusion SPECT. Eur J Nucl Med Mol Imaging 2020;47:2383-95. 10.1007/s00259-020-04746-6 [DOI] [PubMed] [Google Scholar]
- 52.Meyer A, Stambler N, Sjostrand K, et al. Deep learning algorithm improves identification of men with low risk prostate cancer using PSMA-targeted (TC)-T-99M-MIP-1404 SPECT/CT. J Urol 2020;203:1195-6. 10.1097/JU.0000000000000967.06 [DOI] [Google Scholar]
- 53.Pouliot F, Sjostrand K, Stambler N, et al. Prospective evaluation of a novel deep learning algorithm (PSMA-AI) in the assessment of 99mTc-MIP-1404 SPECT/CT in patients with low or intermediate risk prostate cancer. Eur J Nucl Med Mol Imaging 2020;46:756. [Google Scholar]
- 54.Ma L, Ma C, Liu Y, et al. Diagnosis of thyroid diseases using SPECT images based on convolutional neural network. J Med Imag Health Info 2018;8:1684-9. 10.1166/jmihi.2018.2493 [DOI] [Google Scholar]
- 55.Ma L, Ma C, Liu Y, et al. Thyroid diagnosis from SPECT images using convolutional neural network with optimization. Comput Intell Neurosci 2019;2019:6212759. 10.1155/2019/6212759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Petibon Y, Fahey F, Cao X, et al. Deep learning-based detection of bone lesions in 99mTc-MDP SPECT: comparison with human observers. J Nucl Med 2020;61:510. [Google Scholar]
- 57.Sarrut D, Krah N, Badel JN, et al. Learning SPECT detector angular response function with neural network for accelerating Monte-Carlo simulations. Phys Med Biol 2018;63:205013. 10.1088/1361-6560/aae331 [DOI] [PubMed] [Google Scholar]
- 58.Song X, Segars WP, Du Y, et al. Fast modelling of the collimator-detector response in Monte Carlo simulation of SPECT imaging using the angular response function. Phys Med Biol 2005;50:1791-804. 10.1088/0031-9155/50/8/011 [DOI] [PubMed] [Google Scholar]
- 59.Rydén T, Heydorn Lagerlöf J, Hemmingsson J, et al. Fast GPU-based Monte Carlo code for SPECT/CT reconstructions generates improved 177Lu images. EJNMMI Phys 2018;5:1. 10.1186/s40658-017-0201-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xiang H, Lim H, Fessler JA, et al. A deep learning neural network for fast and accurate scatter estimation in quantitative SPECT/CT under challenging scatter conditions. Eur J Nucl Med Mol Imaging 2020;47:2956-67. 10.1007/s00259-020-04840-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as