Abstract
Background
Fast progression (FP), hyperprogressive disease (HPD), and early death (ED) are the newly reported cancer progression patterns in response to immune checkpoint inhibitor (ICI) treatment. This study aimed to investigate the clinical and genomic characteristics of FP, HPD, and ED following the ICI treatment of advanced non-small cell lung cancer (NSCLC).
Methods
We retrospectively reviewed 117 patients with advanced NSCLC who were treated with ICIs from March 2017 to October 2019. FP was defined as (I) time to treatment failure (TTF) <1.5 months; and (II) ≥50% increase in the sum of the longest diameter (SLD) of target lesions. HPD was defined as (I) TTF <2 months; and (II) ≥50% change in tumor growth rate compared with before ICI initiation. ED was defined as overall survival (OS) <3 months. Tissue samples from 18 FP/HPD/ED patients and 5 partial response (PR) patients were subjected to genomic profiling. Genomic data from 693 tumor mutational burden- and histology-matched lung cancer samples were retrieved from an internal database as a control.
Results
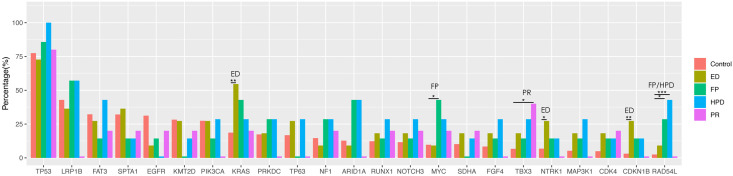
FP, HPD, and ED occurred in 7.21%, 9.38%, and 11.97% patients, respectively. The progression-free survival was comparable among the 3 groups. The median overall survival for FP, HPD, and ED were 3.19, 11.2, and 1.84 months, respectively. The genomic landscape revealed 1 EGFR amplification, 1 ALK fusion, 6 KRAS mutations, 1 ERBB2 amplification, 1 MET amplification, and 1 RET fusion among the 18 patients with FP/HPD/ED. Compared with the Control group, ED patients showed higher mutation frequencies for KRAS (P<0.01), CDKN1B (P<0.01), and NTRK1 (P=0.04). Mutations in RAD54L (P=0.018) and MYC (P=0.04) were more common in FP patients; HPD patients showed more frequent RAD54L mutations (P<0.001).
Conclusions
We demonstrated different genomic characteristics across different progression patterns following ICI treatment, which might assist clinicians in the prediction of a patient’s response, identifying candidates for more effective ICI therapy.
Keywords: Hyperprogressive disease (HPD), fast progression (FP), early death (ED), immune checkpoint inhibitor (ICI), non-small cell lung cancer (NSCLC)
Introduction
Immune checkpoint inhibitors (ICIs) have demonstrated promising anticancer efficacy and superior survival outcomes compared with standard chemotherapy in patients with advanced non-small cell lung cancer (NSCLC) (1-3). However, the overall response rate to ICIs is only approximately 20% in unselected NSCLC patients. Even after screening patients for biomarkers, such as programmed death-ligand 1 (PD-L1) expression or tumor mutational burden (TMB), approximately 50% of patients fail to respond to ICI treatment (1,2,4,5). Dynamic and complicated interactions that occur between the immune system and cancer cells can result in heterogeneous responses to ICIs among patients (6). A subset of patients who respond poorly to ICI therapy due to inherent resistance has even presented with accelerated tumor progression (7,8). Few effective therapeutic options have been identified for these patients. Recently, some case reports have suggested that NSCLC patients with rapid tumor progression in response to ICI therapy may benefit from corticosteroid or salvage chemotherapy (9,10). However, the efficacies of these treatments merit further investigation among larger cohorts.
Recently, several novel progression patterns in response to ICIs have been reported, including fast progression (FP), hyperprogressive disease (HPD), and early death (ED) (11-13). FP is classified as a greater than 50% increase in the sum of the longest diameter (SLD) of the target lesions on computed tomography (CT) images within 6 weeks after ICI therapy initiation (13). Inoue et al. (12) defined ED as any death within 12 weeks after ICI treatment initiation. HPD is a new progression pattern, characterized by an accelerated tumor growth rate (TGR) after immunotherapy, defined as progression at the first evaluation and a >2-fold increase in the TGR compared with before ICI treatment (11,14). Although defined differentially, these progression patterns can display overlap. FP has been reported to occur in 11% of HPD patients, correlated with more aggressive features and worse survival. ED within 3 months was observed in 46% of HPD patients (14).
Studies have identified several genomic alternations (KRAS and STK11 concurrent mutations, MDM2 family amplification, and EGFR mutations) that have been associated with poor prognosis or accelerated progression in lung cancer patients treated with ICIs (15,16). However, the molecular characteristics associated with different progression patterns have not been well-elucidated or compared, which might serve as potential biomarkers to predict the response to immunotherapy.
In the present study, we examined the incidence of HPD/FP/ED in 117 NSCLC patients treated with ICI and explored differences in the clinical and genomic characteristics associated with the different progression patterns.
We present the following article in accordance with the STROBE reporting checklist (available at http://dx.doi.org/10.21037/atm-20-6910).
Methods
Patient information and study design
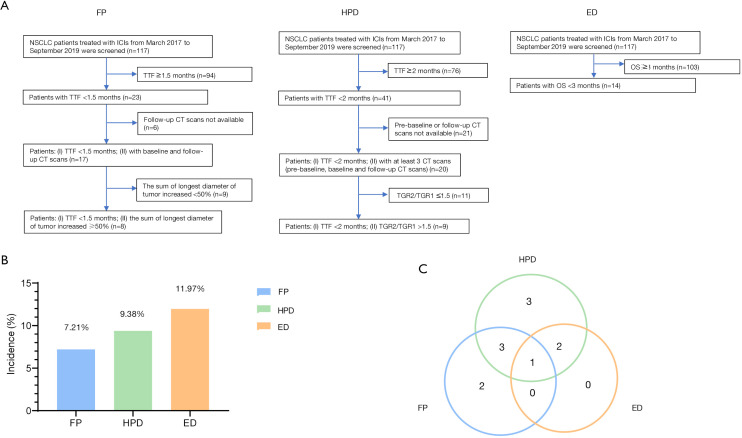
This study retrospectively evaluated 117 patients with advanced NSCLC who received a programmed death 1 (PD-1)/PD-L1 inhibitor, either as a single agent or combined with cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) from March 2017 to October 2019 at Shanghai Chest Hospital. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Shanghai Chest Hospital institutional review board (No. IS2118) and all patients provided written informed consent before the performance of invasive procedures and immunotherapy. Each patient’s progression pattern was assessed according to the following inclusion criteria. FP: (I) time to treatment failure (TTF) <1.5 months; (II) at least two CT scans (one at baseline and one after ICI start); (III) ≥50% increase in the SLD. HPD: (1) TTF <2 months; (II) at least three CT scans (one prior to ICI treatment, one at baseline, and one after treatment); (III) a change in the tumor growth rate (TGR) >50% compared with that prior to ICI initiation (TGR2/TGR1 >1.5). ED: overall survival (OS) <3 months. As illustrated in Figure 1A, 8 patients were defined as FP, 14 as ED, and 9 as HPD.
Figure 1.
Definitions and incidences of different progression patterns. (A) The definitions of FP, HPD, and ED. (B) The incidences of FP, HPD, and ED in NSCLC patients treated with ICIs. (C) A Venn diagram showing patients with FP, HPD, and ED. Only patients with three consecutive CT images (prior-treatment, baseline, post-treatment), who qualified for TGR calculations, were included in the Venn diagram. FP, fast progression; HPD, hyperprogressive disease; ED, early death; NSCLC, non-small cell lung cancer; TGR, tumor growth rate.
Epidemiological and clinical characteristics were collected from all eligible HPD/FP/ED patients and were compared among groups according to progression pattern, including age, gender, smoking history, plasma lactate dehydrogenase (LDH) level, number of metastatic sites, pathology, TNM stage, metastasis, PD-L1 expression, drug target, and line of treatment.
Additionally, we selected 5 patients with partial response (PR) from the 117 ICI-treated patients for genomic profiling. The genomic data from an additional 693 TMB- and histology-matched lung cancer samples were retrieved from our internal database for use as the Control group for mutational comparisons (Figure S1).
Radiological evaluation
To assess tumor progression, CT scans were performed every 4–6 weeks after immunotherapy, and efficacy was evaluated according to the Response Evaluation Criteria in Solid Tumors version 1.1 (RECIST 1.1). Progression-free survival (PFS) was defined as the time from ICI treatment to disease progression, and OS was calculated from the start of ICI treatment to the date of death or the last follow-up.
All patients with TTF less than two months after immunotherapy were selected for calculating TGR. TGR is calculated according to the definition by Champiat et al. (11) and any lesion that could be measured was included in the analysis (no more than two new lesions per organ) (6). TGR1 was assessed before ICI therapy initiation. TGR2 was assessed within 2 months after ICI initiation.
Next-generation sequencing (NGS)
Tissue samples from 36 patients (18 HPD/FP/ED, 5 PR, and 13 non-HPD/FP/ED) were subjected to genomic profiling by NGS. Genomic DNA was extracted from formalin-fixed paraffin-embedded (FFPE) specimens using the QIAamp DNA FFPE tissue kit (Qiagen, Hilden, Germany), according to the manufacturer’s protocol. NGS libraries were prepared using a commercially available, capture-based, targeted sequencing panel (Burning Rock Biotech, Guangzhou, China), which targeted 520 cancer-related genes. Indexed DNA libraries were sequenced using NextSeq500 (Illumina, Inc., San Diego, CA, USA) with paired-end reads and an average sequencing depth >1,000×.
Data analysis
Sequencing data were mapped to the reference human genome (hg19) using the Burrows-Wheeler Aligner, v.0.7.10 (17). Local alignment optimization, duplication marking, and variant calling were performed using the Genome Analysis Tool Kit, v.3.2 (18), and VarScan, v.2.4.3 (19). Variants with a depth <100 were filtered out. Variants with a population frequency over 0.1% in the ExAC, 1,000 Genomes, dbSNP, or ESP6500SI-V2 databases were grouped as single-nucleotide polymorphisms and excluded from further analysis. Remaining variants were annotated with ANNOVAR (2016-02-01 release) (20) and SnpEff, v.3.6 (21). DNA translocation analysis was performed using Factera, v.1.4.3 (22). The copy number variation (CNV) was called with an in-house algorithm based on sequencing depth. The TMB per patient was computed as the ratio between the total number of detected nonsynonymous mutations and the total coding region size of the panel.
Statistical analysis
Statistical analysis was performed using R, version 3.3.3, software. Differences between groups were analyzed using Fisher’s exact test, paired, two-tailed, Student’s t-test, or analysis of variance, as appropriate. Kaplan-Meier analysis was used to estimate survival functions, and a log-rank test was used to determine differences in the survival curves between groups. A P value of <0.05 was considered significant.
Results
The frequency and clinical characteristics of patients with HPD, ED, and FP
Among 117 patients treated with ICIs, 11.97% (14/117) died within 3 months after treatment and were classified as ED (Figure 1B). Patients defined as HPD and FP accounted for 9.38% (9/96) and 7.21% (8/111) of the cohort, respectively. Among all patients showing progression pattern(s), only 11 provided 3 consecutive CT scans and were eligible for all FP, ED, and HPD assessments (Figure 1C). Three and two of these eleven patients met the criteria for both FP and HPD and for both HPD and ED, respectively. One patient qualified for all 3 definitions. The clinical characteristics of patients exhibited no significant differences among the HPD, ED, and FP groups, including age (P=0.771), LDH levels (P=0.612), pathology (P=0.560), gender (P=0.729), and smoking status (P=0.396, Table 1).
Table 1. Clinical characteristics of patients with different progression patterns.
| Characteristics | HPD (n=9) | ED (n=14) | FP (n=8) | P |
|---|---|---|---|---|
| Median age (range), years | 63 (44 to 80) | 69 (40 to 79) | 62 (58 to 80) | 0.771# |
| Median LDH (range) | 227 (183 to 346) | 209 (140 to 774) | 265 (209 to 483) | 0.612# |
| Number of metastatic sites before ICIs, n (%) | ||||
| ≤2 | 7 (77.8) | 7 (50.0) | 5 (62.5) | 0.368* |
| >2 | 2 (22.2) | 7 (50.0) | 3 (37.5) | |
| Pathology, n (%) | 0.560* | |||
| Adenocarcinoma | 5 (55.6) | 6 (42.9) | 6 (75.0) | |
| Squamous | 4 (44.4) | 6 (42.9) | 2 (25.0) | |
| NSCLC | 0 (0.0) | 2 (14.3) | 0 (0.0) | |
| Gender, n (%) | 0.729* | |||
| Female | 0 (0.0) | 1 (7.1) | 1 (12.5) | |
| Male | 9 (100.0) | 13 (92.9) | 7 (87.5) | |
| Smoking status, n (%) | ||||
| Never-smoker | 0 (0.0) | 3 (21.4) | 2 (25.0) | 0.396* |
| Smoker | 9 (100.0) | 11 (78.6) | 6 (75.0) | |
| TNM stage, n (%) | ||||
| IV | 9 (100.0) | 14 (100.0) | 8 (100.0) | – |
| Brain metastases, n (%) | ||||
| No | 9 (100.0) | 12 (85.7) | 6 (75.0) | 0.251* |
| Yes | 0 (100.0) | 2 (14.3) | 2 (25.0) | |
| Bone metastasis, n (%) | ||||
| No | 5 (55.6) | 7 (50.0) | 5 (62.5) | 0.900* |
| Yes | 4 (44.4) | 7 (50.0) | 3 (37.5) | |
| PD-L1 expression, n (%) | ||||
| Positive | 9 (100.0) | 14 (100.0) | 8 (100.0) | – |
| Drug targets, n (%) | 0.932* | |||
| PD-1 | 4 (44.4) | 5 (35.7) | 4 (50.0) | |
| PD-L1 | 5 (55.6) | 6 (42.9) | 4 (50.0) | |
| PD-1 + CTLA-4 | 0 (0.0) | 1 (7.1) | 0 (0.0) | |
| PD-L1 + CTLA-4 | 0 (0.0) | 2 (14.3) | 0 (0.0) | |
| Treatment line, n (%) | 0.697* | |||
| First line | 4 (44.4) | 9 (64.3) | 4 (50.0) | |
| Second line | 5 (55.6) | 4 (28.6) | 4 (50.0) | |
| Third line | 0 (0.0) | 1 (7.1) | 0 (0.0) |
*, P value was calculated by Fisher’ exact test. #, P value was calculated by ANOVA. FP, fast progression; HPD, hyperprogressive disease; ED, early death; LDH, lactate dehydrogenase; NSCLC, non-small cell lung cancer; ICI, immune checkpoint inhibitor; PD-1, programmed cell death-1; PD-L1, programmed cell death-1 ligand; CTLA-4, cytotoxic T-lymphocyte associated protein 4.
Dynamic changes in tumor lesions among HPD, ED, and FP patients
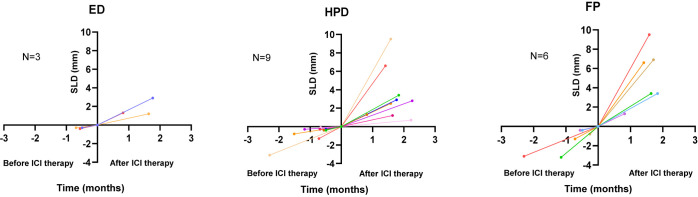
We plotted the SLD of target lesions before and after ICI treatment among the 11 patients with 3 consecutive CT images available (prior-treatment, baseline, post-treatment). Among ED and HPD patients (Figure 2), the increase in SLD per unit of time was greater after ICI therapy compared with the rate of increase before treatment. Interestingly, one patient with FP (green line) showed a decline in the SLD increase rate after ICI treatment compared with before ICI treatment (Figure 2).
Figure 2.
Variations in the sum of the SLD of target lesions among ED, HPD, and FP patients before and after ICI treatment. Only patients with three consecutive CT images (prior-treatment, baseline, post-treatment) were included. FP, fast progression; HPD, hyperprogressive disease; ED, early death; SLD, sum of the longest diameter.
Survival analysis among patients with HPD, ED, and FP
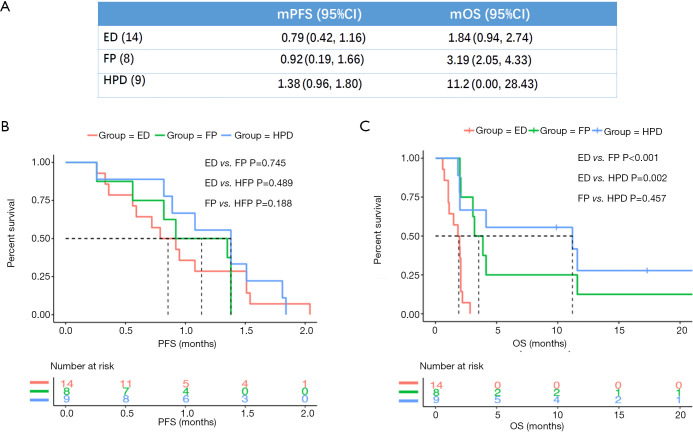
The PFS and OS of patients with different progression patterns were assessed and compared (Figure 3). HPD, ED, and FP patients showed comparable median PFS (mPFS) (Figure 3A,B). ED patients had a shorter median OS (mOS) [1.84 months; 95% CI: 0.94–2.74 months, Figure 3A,C) compared with HPD patients (11.2 months; 95% CI: 0.00–28.43 months; P=0.002) and FP patients (3.19 months; 95% CI: 2.05–4.33 months; P<0.001). No significant difference in OS was found between FP and HPD patients (P=0.457).
Figure 3.
PFS and OS for patients with ED, FP, and HPD. (A) Median PFS and OS; (B) PFS curve; (C) OS curve. PFS, progression-free survival; OS, overall survival; FP, fast progression; HPD, hyperprogressive disease; ED, early death.
Genomic landscapes of HPD, ED, and FP patients
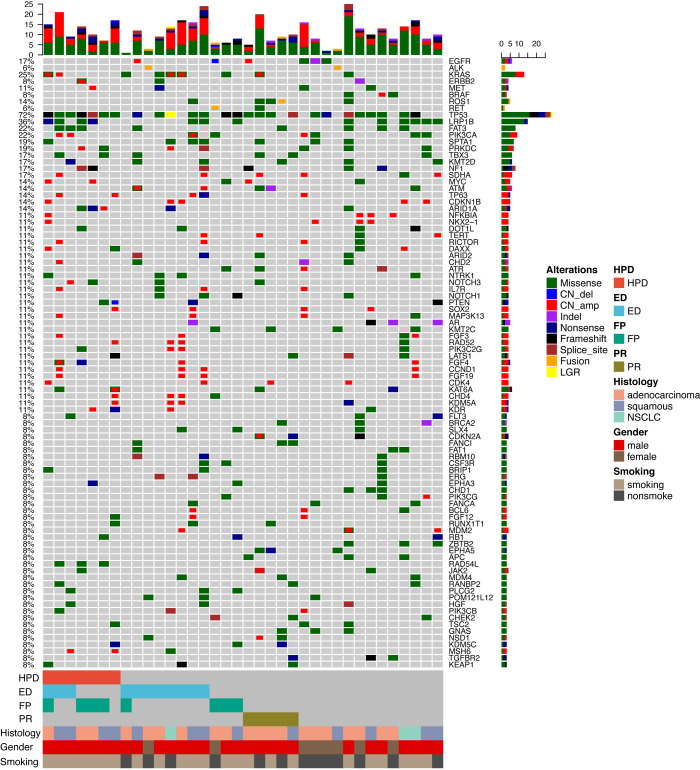
To investigate the genomic landscapes of patients with different progression patterns, we performed NGS on tumor tissue samples from 36 patients, including 18 with HPD/ED/FP, 5 PR patients, and 13 non-HPD/ED/FP/PR patients. The landscape of driver genes in 18 FP/ED/HPD patients revealed 1 EGFR amplification, 1 ALK fusion, 6 KRAS mutations, 1 ERBB2 amplification, 1 MET amplification, and 1 RET fusion. Two ED patients harbored concomitant STK11 and KRAS mutations. MDM2 amplification and MDM4 mutation co-occurred in 1 ED patient. One patient with both ED and HPD harbored an MDM4 mutation. In the PR patients, we also identified a ROS1 fusion, a KRAS mutation, and an EGFR amplification, in one patient each (Figure 4).
Figure 4.
Genomic alterations in patients with HPD, ED, FP, PR, and non-HPD/ED/FP/PR. The histology, gender, and smoking characteristics are summarized at the bottom of the panel. The frequencies on the left of the panel include all alterations for each gene (missense, deletion, amplification, indel, nonsense, frameshift, splice site, fusion, large genomic rearrangement). HPD/ED/FP, n=18; PR, n=5; non-HPD/ED/FP/PR, n=13. FP, fast progression; HPD, hyperprogressive disease; ED, early death; PR, partial response.
Mutational comparison among patients with HPD, ED, and FP
We compared the mutational profiles of 18 HPD/ED/FP patients with 693 TMB- and histology-matched lung cancer samples that were retrieved from our internal database (Control group). ED patients had a higher mutation frequency in KRAS (P<0.01), CDKN1B (P<0.01), and NTRK1 (P=0.04) compared with the Control group (Figure 5). Mutations in RAD54L (P=0.018) and MYC (P=0.04) occurred more commonly in FP patients than in the Control group. HPD patients also showed a higher rate of RAD54L mutations than the Control group (P<0.001). Patients with PR harbored more frequent TBX3 mutations than the Control group (P=0.04). We also compared the mutation profile between each progression group and the remaining genomically profiled ICI-treated patients in our cohort (Figure S2). Mutations in ARID1A occurred more commonly in HPD patients than in non-HPD patients (P=0.04, Figure S2A). Compared with non-ED patients, ED patients displayed a higher KRAS mutation frequency (P=0.012, Figure S2B).
Figure 5.
Comparison of the genomic alterations associated with different progression patterns. For comparison, 5 patients with PR and 693 TMB- and histology-matched lung cancer samples (Control) from an internal database were selected as controls. The Y-axis represents the mutation frequency of each gene in each group. ***, P<0.001; **, P<0.01; *, P<0.05. FP, fast progression; HPD, hyperprogressive disease; ED, early death; PR, partial response.
Discussion
To our knowledge, these new progression patterns (FP/HPD/ED) following ICI treatment have rarely been reported. We retrospectively analyzed the incidence of HPD/FP/ED in 117 NSCLC patients treated with ICI and identified genomic characteristics associated with the different progression patterns.
In the present study, we discovered an incidence of 11.97% and 7.21% for ED and FP, respectively, among NSCLC patients treated with ICI (Figure 1B), which were comparable with previously reported rates of 18.6% (38/201, P=0.122) and 10.4% (44/425, P=0.313) (12,13). The incidence of HPD was reported to range from 9% to 29% (8,11,23,24), compared with 9.38% in our study. The different tumor types and inconsistent definitions of HPD used for different studies might explain these diverse results.
Ferrara et al. (14) reported that 6/6 FP patients and 21/46 ED patients were also classified as HPD based on TGR analysis. In our study, 3/6 FP patients and 2/3 ED patients were also classified as HPD, and 1 patient qualified for all 3 definitions (Figure 1C). Although only 3 of the 14 ED patients were eligible for HPD assessment in this study, all 3 experienced HPD after receiving ICI therapy, indicating an overlap between ED patients and HPD. However, whether the remaining 11 ED patients also experienced HPD remains unknown. The lack of post-treatment CT images for the 6/11 remaining ED patients could partially be attributed to the extremely rapid disease progression among these patients. Therefore, the HPD rate in our study might be underestimated.
We observed that ED patients showed a significantly inferior mOS (1.84 months) compared with that for FP patients (3.19 months) and HPD patients (11.2 months) (Figure 3A). Ferrara et al. (8) reported that patients who developed HPD within the first 6 weeks after treatment with a PD-1/PD-L1 inhibitor had an mOS of 3.4 months (n=23; 95% CI: 2.8–7.5 months). The mOS of FP patients was significantly shorter than that of HPD patients (0.7 vs. 1.6 months, P=0.02) (14). The mOS in our study appeared to be longer than those reported in previous studies, which may be due to our small sample size and the fact that a higher proportion of patients in our study received immunotherapy as a first- or second-line treatment. This finding may also suggest that these three patterns of immunotherapy-related outcomes may differ in the prognosis, which may require further analysis of more clinical data in the future.
The genomic landscape revealed several driver gene alterations (including EGFR deletion and amplification, ALK fusion, MET amplification, and RET fusion) that could be detected in our HPD/FP/ED patients (Figure 4). It has been reported that constitutive oncogenic signaling induces PD-L1 expression in driver gene -mutant NSCLCs (25,26). Previous studies (27,28) have revealed an association between NSCLC patients harboring EGFR mutations or ALK-fusions and an inferior response to ICI treatment, which suggested that lack of concurrent PD-L1 expression and CD8+ tumor-infiltrating T-lymphocytes (TILs) within the microenvironments of these driver gene-mutated tumors may underlie these observations. However, Yoshimura et al. (29) described favorable responses to ICI in two patients with MET-amplified lung cancer and demonstrated an increase in CD8+ TILs in MET-amplified tumors. Besides, the low tumor mutational burden in driver gene-mutant tumors might diminish the efficacy of ICI (30). Collectively, the interplay and crosstalk between oncogenic signaling and PD-1/PD-L1 axis, as well as tumor microenvironment and tumor mutation burden confer the complex regulatory mechanisms and explain the heterogeneous impacts of oncogenic mutation on ICI response. The underlying biology remains elusive and requires further investigation.
The mutational comparison showed that RAD54L mutations were more common in patients with HPD (P<0.001) and FP (P=0.018) than in the TMB- and histology-matched Control group (Figure 5). RAD54L, located on chromosome 1p32, is a member of the DEAD-like helicase superfamily and plays an important role in repairing damaged DNA (31,32). Li et al. (33) reported that the RAD54L C157T polymorphism was an adverse genotype with a significant effect on the OS and clinical prognosis of pancreatic cancer patients. However, little is known about the association between the RAD54L gene and immunotherapy in lung cancer. Further studies examining large samples remain necessary to verify the correlation between RAD54L and immunotherapy-induced HPD/FP in lung cancer.
The tumor microenvironment plays an important role in immunotherapy. MYC activation promotes the rapid release of interleukin-23 (IL-23) and chemokine (C-C motif) ligand 9 (CCL9), driving the rapid loss of T, B, and NK cells, the recruitment of PD-L1+ macrophages, and angiogenesis (34). Kortlever et al. (34) indicated that the PD-L1 blockade could reverse the loss of T and B cells but did not significantly affect the MYC-induced recruitment of macrophages, angiogenesis, or the loss of NK cells in the tumor environment. In our study, MYC mutations were more commonly identified in FP patients whose tumors increased within a short timeframe (Figure 5). The resistance to the anti-PD-1/PD-L1 antibody may be associated with the activation of MYC. However, the predictive value of MYC mutations in patients treated with ICIs requires further investigation.
A previous study reported that the high expression level of the CDKN1B gene, encoding the p27Kip1 protein, served as a good prognostic marker in breast cancer (35,36). However, we observed a higher frequency of CDKN1B amplification in lung cancer patients with ED (Figure 5). We also found that the mutation frequencies for KRAS and NTRK1 were higher in patients with ED than in other patients (Figure 5, Figure S2B). KRAS mutations have been identified in 10–30% of lung cancer patients and have been demonstrated to be associated with poor prognosis (37). However, Jeanson et al. (38) reported comparable OS between KRAS-mutant and KRAS-wild-type NSCLC patients treated with ICIs. The inconsistent prognostic values of KRAS mutations may be due to heterogeneous immunological features, such as PD-L1 expression, in KRAS-mutant NSCLCs. The diversity of mutation co-occurrence may also affect the differences in ICI efficacy (39). KRAS-mutant lung adenocarcinomas with concomitant STK11/LKB1 often have high rates of KEAP1 mutational inactivation and expressed lower levels of immune markers, including PD-L1. KRAS-mutant tumors with TP53 mutation demonstrate higher mutational burden, inflammatory markers, immune checkpoint effector molecules, and improved survival. A recent study also demonstrated that the co-occurrence of KRAS and STK11/LKB1 mutations were a major driver of primary resistance to a PD-1 monoclonal antibody, associated with a shorter OS (15). Concordantly, the co-occurrence of KRAS and STK11 mutations was identified in 2 patients with ED in our study. NTRK1 promotes the invasion, metastasis, and growth of tumor cells (40). Previous studies have shown that NTRK1 overexpression can reduce the infiltration of T cells in the tumor microenvironment and promote the exhaustion of CD8+ T cells, partially explaining the reduced response to anti-PD-1/PD-L1 inhibitors (40). Moreover, Konen et al. (40) indicated that the transcript level and protein activity of NTRK1 were increased in tumors treated with the anti-PD-1 antibody. These observations support our finding that NTRK1 mutations may be associated with poor prognosis in response to immunotherapy.
Our study was also limited in the following aspects. First, due to the retrospective nature of this study, HPD and FP could not be assessed in patients who lacked consecutive CT scans, which may have resulted in the underestimation of the frequency of HPD and FP. Second, the mutational comparison analysis lacked an effective control group who also received ICI but responded favorably to the treatment because only a few non-HPD/FP/ED patients in the cohort were genomically profiled. Third, our study was a single-center study with a small sample size, which might introduce selection bias. Therefore, larger, prospective, multi-center studies remain necessary to further explore the clinicopathological and genetic characteristics of patients with different progression patterns. Finally, the differential mutational profiles demonstrated in our study do not necessarily drive the different progression patterns. Therefore, the impacts and predictive roles of these genomic mutations require further investigations using functional experiments and prospective studies in the future.
Conclusions
In conclusion, our study revealed that the FP, HPD, and ED occurred in 7.21%, 9.38%, and 11.97% of NSCLC patients treated with ICIs, respectively. The patients with ED had shorter OS compared with HPD and FP patients. We also demonstrated distinctive genomic characteristics among patients with different progression patterns, which might further assist clinicians in the prediction of a patient’s response to ICIs, identifying candidates for effective ICI therapy.
Supplementary
The article’s supplementary files as
Acknowledgments
We want to appreciate all patients and their families for their contributions to the study.
Funding: This work was supported by the Shanghai Committee of Science and Technology, China (grants: 19411970900) and Shanghai Chest Hospital Collaborative Innovation Project (grants: YJXT20190204).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Shanghai Chest Hospital institutional review board (No. IS2118) and all patients provided written informed consent before the performance of invasive procedures and immunotherapy.
Footnotes
Reporting Checklist: The authors have completed the STROBE Statement reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-6910
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-6910
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-6910). The authors have no conflicts of interest to declare.
References
- 1.Borghaei H, Paz-Ares L, Horn L, et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. N Engl J Med 2015;373:1627-39. 10.1056/NEJMoa1507643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brahmer J, Reckamp KL, Baas P, et al. Nivolumab versus Docetaxel in Advanced Squamous-Cell Non-Small-Cell Lung Cancer. N Engl J Med 2015;373:123-35. 10.1056/NEJMoa1504627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mok TSK, Wu YL, Kudaba I, et al. Pembrolizumab versus chemotherapy for previously untreated, PD-L1-expressing, locally advanced or metastatic non-small-cell lung cancer (KEYNOTE-042): a randomised, open-label, controlled, phase 3 trial. Lancet 2019;393:1819-30. 10.1016/S0140-6736(18)32409-7 [DOI] [PubMed] [Google Scholar]
- 4.Wu YL, Lu S, Cheng Y, et al. Nivolumab Versus Docetaxel in a Predominantly Chinese Patient Population With Previously Treated Advanced NSCLC: CheckMate 078 Randomized Phase III Clinical Trial. J Thorac Oncol 2019;14:867-75. 10.1016/j.jtho.2019.01.006 [DOI] [PubMed] [Google Scholar]
- 5.Reck M, Rodriguez-Abreu D, Robinson AG, et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N Engl J Med 2016;375:1823-33. 10.1056/NEJMoa1606774 [DOI] [PubMed] [Google Scholar]
- 6.Kim Y, Kim CH, Lee HY, et al. Comprehensive Clinical and Genetic Characterization of Hyperprogression Based on Volumetry in Advanced Non-Small Cell Lung Cancer Treated With Immune Checkpoint Inhibitor. J Thorac Oncol 2019;14:1608-18. 10.1016/j.jtho.2019.05.033 [DOI] [PubMed] [Google Scholar]
- 7.Syn NL, Teng MWL, Mok TSK, et al. De-novo and acquired resistance to immune checkpoint targeting. Lancet Oncol 2017;18:e731-41. 10.1016/S1470-2045(17)30607-1 [DOI] [PubMed] [Google Scholar]
- 8.Ferrara R, Mezquita L, Texier M, et al. Hyperprogressive Disease in Patients With Advanced Non-Small Cell Lung Cancer Treated With PD-1/PD-L1 Inhibitors or With Single-Agent Chemotherapy. JAMA Oncol 2018;4:1543-52. 10.1001/jamaoncol.2018.3676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanazu M, Edahiro R, Krebe H, et al. Hyperprogressive disease in patients with non-small cell lung cancer treated with nivolumab: A case series. Thorac Cancer 2018;9:1782-7. 10.1111/1759-7714.12894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ogawara D, Soda H, Iwasaki K, et al. Remarkable response of nivolumab-refractory lung cancer to salvage chemotherapy. Thorac Cancer 2018;9:175-80. 10.1111/1759-7714.12543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Champiat S, Dercle L, Ammari S, et al. Hyperprogressive Disease Is a New Pattern of Progression in Cancer Patients Treated by Anti-PD-1/PD-L1. Clin Cancer Res 2017;23:1920-8. 10.1158/1078-0432.CCR-16-1741 [DOI] [PubMed] [Google Scholar]
- 12.Inoue T, Tamiya M, Tamiya A, et al. Analysis of Early Death in Japanese Patients With Advanced Non-small-cell Lung Cancer Treated With Nivolumab. Clin Lung Cancer 2018;19:e171-6. 10.1016/j.cllc.2017.09.002 [DOI] [PubMed] [Google Scholar]
- 13.Gandara DR, Reck M, Morris S, et al. Fast progression in patients treated with a checkpoint inhibitor (cpi) vs chemotherapy in OAK, a phase III trial of atezolizumab (atezo) vs docetaxel (doc) in 2L1 NSCLC. Ann Oncol 2018;29:x7. 10.1093/annonc/mdy511 [DOI] [Google Scholar]
- 14.Ferrara R, Mezquita L, Texier M, et al. Fast-progression (FP), hyper-progression (HPD) and early deaths (ED) in advanced non-small cell lung cancer (NSCLC) patients (pts) upon PD-(L)-1 blockade (IO). Journal of Clinical Oncology 2019;37. 10.1200/JCO.2019.37.15_suppl.9107 [DOI] [Google Scholar]
- 15.Skoulidis F, Goldberg ME, Greenawalt DM, et al. STK11/LKB1 Mutations and PD-1 Inhibitor Resistance in KRAS-Mutant Lung Adenocarcinoma. Cancer Discov 2018;8:822-35. 10.1158/2159-8290.CD-18-0099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kato S, Goodman A, Walavalkar V, et al. Hyperprogressors after Immunotherapy: Analysis of Genomic Alterations Associated with Accelerated Growth Rate. Clin Cancer Res 2017;23:4242-50. 10.1158/1078-0432.CCR-16-3133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754-60. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297-303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koboldt DC, Zhang Q, Larson DE, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 2012;22:568-76. 10.1101/gr.129684.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164. 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cingolani P, Platts A, Wang le L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80-92. 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newman AM, Bratman SV, Stehr H, et al. FACTERA: a practical method for the discovery of genomic rearrangements at breakpoint resolution. Bioinformatics 2014;30:3390-3. 10.1093/bioinformatics/btu549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saada-Bouzid E, Defaucheux C, Karabajakian A, et al. Hyperprogression during anti-PD-1/PD-L1 therapy in patients with recurrent and/or metastatic head and neck squamous cell carcinoma. Ann Oncol 2017;28:1605-11. 10.1093/annonc/mdx178 [DOI] [PubMed] [Google Scholar]
- 24.Kim CG, Kim KH, Pyo KH, et al. Hyperprogressive disease during PD-1/PD-L1 blockade in patients with non-small-cell lung cancer. Ann Oncol 2019;30:1104-13. 10.1093/annonc/mdz123 [DOI] [PubMed] [Google Scholar]
- 25.Ota K, Azuma K, Kawahara A, et al. Induction of PD-L1 Expression by the EML4-ALK Oncoprotein and Downstream Signaling Pathways in Non-Small Cell Lung Cancer. Clin Cancer Res 2015;21:4014-21. 10.1158/1078-0432.CCR-15-0016 [DOI] [PubMed] [Google Scholar]
- 26.Akbay EA, Koyama S, Carretero J, et al. Activation of the PD-1 pathway contributes to immune escape in EGFR-driven lung tumors. Cancer Discov 2013;3:1355-63. 10.1158/2159-8290.CD-13-0310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rizvi H, Sanchez-Vega F, La K, et al. Molecular Determinants of Response to Anti-Programmed Cell Death (PD)-1 and Anti-Programmed Death-Ligand 1 (PD-L1) Blockade in Patients With Non-Small-Cell Lung Cancer Profiled With Targeted Next-Generation Sequencing. J Clin Oncol 2018;36:633-41. 10.1200/JCO.2017.75.3384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gainor JF, Shaw AT, Sequist LV, et al. EGFR Mutations and ALK Rearrangements Are Associated with Low Response Rates to PD-1 Pathway Blockade in Non-Small Cell Lung Cancer: A Retrospective Analysis. Clin Cancer Res 2016;22:4585-93. 10.1158/1078-0432.CCR-15-3101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoshimura K, Inoue Y, Tsuchiya K, et al. Elucidation of the relationships of MET protein expression and gene copy number status with PD-L1 expression and the immune microenvironment in non-small cell lung cancer. Lung Cancer 2020;141:21-31. 10.1016/j.lungcan.2020.01.005 [DOI] [PubMed] [Google Scholar]
- 30.Dong ZY, Zhang JT, Liu SY, et al. EGFR mutation correlates with uninflamed phenotype and weak immunogenicity, causing impaired response to PD-1 blockade in non-small cell lung cancer. Oncoimmunology 2017;6:e1356145. 10.1080/2162402X.2017.1356145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rasio D, Murakumo Y, Robbins D, et al. Characterization of the human homologue of RAD54: a gene located on chromosome 1p32 at a region of high loss of heterozygosity in breast tumors. Cancer Res 1997;57:2378-83. [PubMed] [Google Scholar]
- 32.Bugreev DV, Hanaoka F, Mazin AV. Rad54 dissociates homologous recombination intermediates by branch migration. Nat Struct Mol Biol 2007;14:746-53. 10.1038/nsmb1268 [DOI] [PubMed] [Google Scholar]
- 33.Li D, Frazier M, Evans DB, et al. Single nucleotide polymorphisms of RecQ1, RAD54L, and ATM genes are associated with reduced survival of pancreatic cancer. J Clin Oncol 2006;24:1720-8. 10.1200/JCO.2005.04.4206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kortlever RM, Sodir NM, Wilson CH, et al. Myc Cooperates with Ras by Programming Inflammation and Immune Suppression. Cell 2017;171:1301-15.e14. 10.1016/j.cell.2017.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cusan M, Mungo G, De Marco Zompit M, et al. Landscape of CDKN1B Mutations in Luminal Breast Cancer and Other Hormone-Driven Human Tumors. Front Endocrinol (Lausanne) 2018;9:393. 10.3389/fendo.2018.00393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guan X, Wang Y, Xie R, et al. p27(Kip1) as a prognostic factor in breast cancer: a systematic review and meta-analysis. J Cell Mol Med 2010;14:944-53. 10.1111/j.1582-4934.2009.00730.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jancik S, Drabek J, Radzioch D, et al. Clinical relevance of KRAS in human cancers. J Biomed Biotechnol 2010;2010:150960. 10.1155/2010/150960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jeanson A, Tomasini P, Souquet-Bressand M, et al. Efficacy of Immune Checkpoint Inhibitors in KRAS-Mutant Non-Small Cell Lung Cancer (NSCLC). J Thorac Oncol 2019;14:1095-101. 10.1016/j.jtho.2019.01.011 [DOI] [PubMed] [Google Scholar]
- 39.Skoulidis F, Byers LA, Diao L, et al. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov 2015;5:860-77. 10.1158/2159-8290.CD-14-1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Konen JM, Rodriguez BL, Fradette JJ, et al. Ntrk1 Promotes Resistance to PD-1 Checkpoint Blockade in Mesenchymal Kras/p53 Mutant Lung Cancer. Cancers (Basel) 2019;11:462. 10.3390/cancers11040462 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as