Abstract
Background
This study aimed to investigate the specific vaginal microbiome in the differential diagnosis of endometriosis/adenomyosis (EM/AM)-associated chronic pelvic pain (CPP) from other types of CPP, and to explore the role of the vaginal microbiome in the mechanism of EM/AM-associated CPP.
Methods
We recruited 37 women with EM/AM-associated CPP, 25 women with chronic pelvic pain syndrome (CPPS) without EM/AM, and 66 women without CPPS into our study. All of the participants were free from human papillomavirus (HPV) infection. Sequencing of barcoded 16S rRNA gene fragments (V4) was used to determine the vaginal microbiome composition on the Illumina HiSeq2500 System. Taxonomic and functional bioinformatics analyses were performed using t-test, linear discriminant analysis effect size (LEfSe), MetaStat, and PICRUSt algorithms.
Results
At the species level, EM/AM-associated CPP was found to be associated with a predominance of Clostridium butyricum, Clostridium disporicum, Alloscardovia omnicolens, and Veillonella montpellierensis, and a concomitant paucity of Lactobacillus jensenii, Lactobacillus reuteri, and Lactobacillus iners. When the relative abundance of Clostridium disporicum was over 0.001105% and that of Lactobacillus reuteri was under 0.1911349%, the differential diagnostic sensitivity and specificity were 81.08% and 52.0%, respectively. When serum CA125 was combined, the sensitivity increased to 89.19%, but the specificity remained at 52.0%. The PICRUSt results identified 7 differentially regulated pathways within the 3 groups that may be of relevance.
Conclusions
Compared to that of CPPS patients without EM/AM and women without CPPS, the vaginal microbiome of patients with EM/AM-associated CPP shows significantly higher alpha (phylogenetic) diversity, as well as higher counts of Clostridium butyricum, Clostridium disporicum, Alloscardovia omnicolens, and Veillonella montpellierensis. These differences in the vaginal microbiome may interfere with local functional pathways, which could provide a direction for innovative metabolite-specific targeted treatment. The combination of vaginal biomarkers and serum CA125 may provide an original method to differentiate EM/AM-associated CPP.
Keywords: Chronic pelvic pain (CPP), endometriosis/adenomyosis (EM/AM), 16S rRNA, function prediction, vaginal microbiome
Introduction
Chronic pelvic pain syndrome (CPPS) is one of the most common pain problems experienced by women and is associated with several gynecological conditions (1). CPPS, which is defined as intermittent or continuous pain in the lower abdomen or pelvis lasting at least 6 months, can have negative cognitive, behavioral, sexual, and emotional consequences. CPP disorders are debilitating conditions which affect 38 in every 1,000 women (2), and account for 2–10% of all gynecological outpatient appointments, of which about 20% of cases undergo diagnostic laparoscopy (3), and 10–15% qualify for a hysterectomy (4). However, CPPS is still poorly understood and difficult to diagnose.
As a multifactorial syndrome, CPPS is often associated with systemic symptoms and it has several etiological and potentiating contributing factors including infection, inflammation, and some other pivotal factors. According to the Royal College of Obstetricians and Gynecologists (RCOG), pain can be etiologically categorized as either gynecological or non-gynecological (5). Gynecological pain includes endometriosis/adenomyosis (EM/AM), pelvic congestion syndrome, leiomyoma, malignant disease, pelvic inflammatory disease (PID), and adhesions. Both EM and AM are defined as the growth of ectopic endometrial tissue outside the uterine cavity.
For several gynecological diseases, there are currently no available or effective biomarkers (6). Although some diseases, such as AM and EM, can be diagnosed by routine noninvasive methods, patients are usually at a late stage at the time of diagnosis when such methods are used. Complaints of CPPS are frequent in gynecological clinics (7); however, there are currently no validated biomarkers to distinguish EM/AM-associated chronic pelvic pain (CPP) from CPPS induced by other disorders. Ambiguous diagnosis offers a poor reference for targeted and effective non-surgical treatment. Therefore, an accurate noninvasive biomarker is sought for clinicians to make correct diagnoses and choose appropriate therapeutic regimens.
Along with the development of microbiology and the deep understanding of the role of microorganisms in diseases, researchers began to realize the value of microorganisms as potential diagnostic biomarkers. Some studies have indicated that vaginal or cervical microbiota might be reference factors in the detection of upper reproductive tract diseases (8). However, microorganism research regarding patients with CPP is scarce. In light of this, the present study aimed to explore the relationship between the vaginal microbiome and CPP, specifically EM/AM-associated CPP. We also aimed to uncover potential vaginal microbiome biomarkers which could help in the diagnosis and differentiation of EM/AM-associated CPP. The trial was registered in ClinicalTrials.gov, NCT03786588. We present the following article in accordance with the Materials and Design Analysis Reporting (MDAR) and the STrengthening the Reporting of OBservational studies in Epidemiology (STROBE) reporting checklist (available at http://dx.doi.org/10.21037/atm-20-4586).
Methods
Ethics
This study was conducted according to the Declaration of Helsinki (as revised in 2013). It was approved by the Ethics Committee of Peking Union Medical College Hospital (JS-1844), and informed consent was obtained from all participants.
Study population
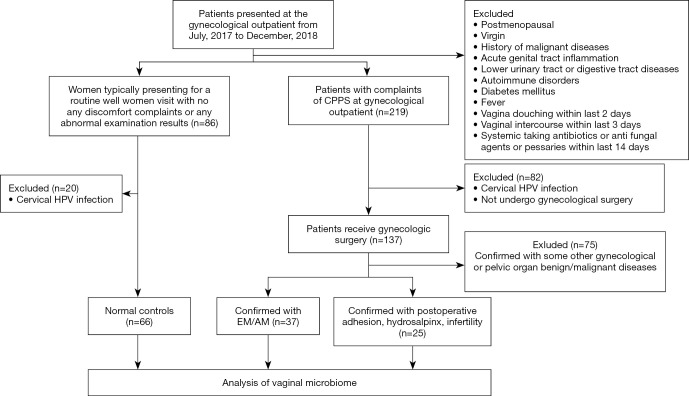
Patients who visited the clinics of the Department of Obstetrics and Gynecology of Peking Union Medical College Hospital between July 2017 and December 2018 with complaints of CPPS and received gynecological surgery were recruited to this study. The inclusion criteria were as follows: premenopausal women who were not menstruating, pregnant, or in puerperium were included. Thirty-seven women with CPPS confirmed with EM/AM by exploratory laparoscopy or surgical pathology were defined as group A; 25 women with CPPS confirmed with adhesions, hydrosalpinx, or infertility were defined as group B; and 66 women without CPPS (most of whom had visited for routine examinations or health counselling) were enrolled as normal controls, defined as group C. Cervical human papillomavirus (HPV) tests were shown to be negative for all participants. The exclusion criteria were as follows: (I) women who had never had sexual intercourse; (II) postmenopausal women; (III) women with cervical HPV infection, a fever, or a history of malignant diseases; (IV) women who were diagnosed with acute lower genital tract inflammation, or with diseases of the lower urinary or digestive tract; and (V) women with autoimmune disorders or diabetes mellitus. Furthermore, all participants should also meet the following requirements: no systemic antibiotics or antifungal agents or pessaries within the 14 days prior to sampling; no vaginal intercourse within the 3 days prior to sampling; and no vaginal douching within the 2 days prior to sampling. The Consolidated Standards for Reporting Trials (CONSORT) flow diagram is shown in Figure 1.
Figure 1.
CONSORT flow diagram of the study. Lower urinary tract or digestive tract disease refers to participants without relevant medical history or systematic clinical manifestations. Female with fever refers to participants whose body temperature exceeded 37.5 °C. Gynecologic benign diseases include deformity of the reproductive tract, uterine myoma, benign ovarian cyst, and mesangial cyst of the fallopian tube. Gynecologic malignant diseases refer to ovarian cancer, endometrial cancer, cervical cancer, or fallopian tube carcinoma. Pelvic organ benign/malignant diseases refer to those affecting the urinary or digestive system, or retroperitoneal tumor. CONSORT, Consolidated Standards for Reporting Trials; AM, adenomyosis; CPPS, chronic pelvic pain syndrome; EM, endometriosis; HPV, human papillomavirus.
Specimen collection
Specimens were collected from participants on their first visit to the clinics. The vagina and cervix were fully exposed by a sterile, disposable speculum without lubricant, and samples were obtained from the posterior vaginal fornix. The sterile swab samples were stored immediately at −80 °C for subsequent DNA extraction. At the same time, ThinPrep® Pap testing (Hologic, Inc., Marlborough, MA, USA) and the Cobas® 4800 System HPV Genotyping Test (Roche Molecular Diagnostics, Pleasanton, CA, USA) were also performed.
Extraction and amplification
The procedure of DNA extraction and the amplification of bacterial 16S rRNA V4 gene regions are described in detail in the supplementary materials (Appendix 1), together with the Illumina sequencing and bioinformatics data analysis.
Statistical analysis
Clinical data analysis was performed in SPSS 23.0 software (SPSS Inc., Chicago, IL, USA). T tests were used to analyze continuous variables, while the chi-square test was adopted for categorical variables. Statistical significance was considered to exist when P<0.05. To identify the functional pathways that differed between the groups, Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) was used (9) to predict the gene families comprising a metagenome from 16S rRNA sequencing data. PICRUSt was used to infer the gene function spectrum of their common ancestors based on Greengenes phylogeny and the genetic information on the operational taxonomic unit (OTU). Finally, the metabolic function of the microbiome was predicted by mapping the sequenced microbial composition into the database.
Results
Sociodemographic and clinical baseline characteristics
The sample size was in accordance with those of previous studies. The mean age was 39.89±6.24 years in group A, 37.56±5.48 in group B, and 38.23±7.80 in group C; no significant age difference existed between the 3 groups (P=0.368). Furthermore, no significant differences were found regarding parity and gravidity, menstrual phase, or contraceptive method between the groups (P=0.077, P=0.191, P=0.270, and P=0.216, respectively). Table 1 shows the clinical baseline characteristics of the women from the 3 groups. The flow diagram is shown in Figure 1.
Table 1. Participant characteristics.
| Characteristics | Group A (n=37) | Group B (n=25) | Group C (n=66) | Total (n=128) | P value |
|---|---|---|---|---|---|
| Age, years (mean ± SD) | 39.89±6.24 | 37.56±5.48 | 38.23±7.80 | 38.58±6.97 | |
| Parity, n/N (%) | 0.076 | ||||
| Nulliparous | 8/37 (21.6) | 12/25 (48.0) | 25/66 (37.9) | 45/128 (35.2) | |
| Parous | 29/37 (78.4) | 13/ 25 (52.0) | 41/66 (62.1) | 83/128 (64.8) | |
| Situation of fertility | |||||
| Parity | 1.03±0.70 | 0.65±0.71 | 0.74±0.69 | 0.81±0.71 | 0.077 |
| Gravidity | 1.78±1.23 | 1.21±1.14 | 1.69±1.50 | 1.63±1.37 | 0.191 |
| Phase of menstrual cycle, n/N (%) | 0.270 | ||||
| Follicular | 12/37 (32.4) | 5/25 (20.0) | 22/66 (33.3) | 39/128 (30.5) | |
| Luteal | 15/37 (40.5) | 16/25 (64.0) | 35/66 (53.0) | 66/128 (51.6) | |
| NA | 10/37 (27.0) | 4/25 (16.0) | 9/66 (13.6) | 23/128 (18.0) | |
| Contraception, n/N (%) | 0.216 | ||||
| Nil | 26/37 (70.3) | 16/25 (64.0) | 33/66 (50.0) | ||
| Condoms | 10/37 (27.0) | 7/25 (28.0) | 25/66 (37.9) | ||
| IUD | 1/37 (2.7) | 2/25 (8.0) | 8/66 (12.1) | ||
IUD, Intrauterine device; SD, standard deviation. P-value is calculated by the Pearson Chi-Square and t-test.
Microbiome community diversity
Identification of vaginal microbiota
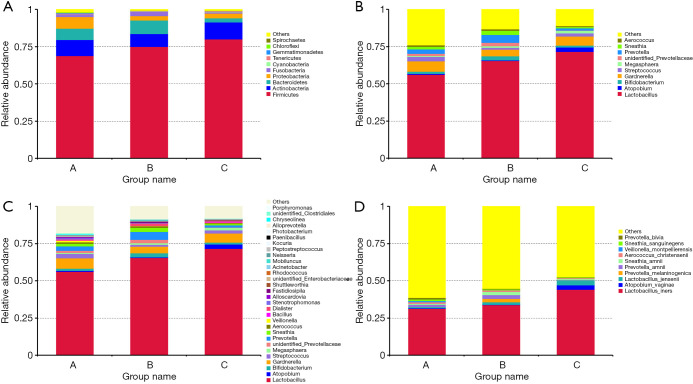
This study detected 57 phyla, 1,017 genera, and 919 species.The distribution of the vaginal bacteria at each of these levels is shown in Figure 2.
Figure 2.
Bar chart showing vaginal microbiome composition in participants from the 3 groups. (A) The relative abundance of the top 10 phyla. (B) The relative abundance of the top 10 genera. (C) The relative abundance of the top 30 genera. (D) The relative abundance of the top 10 species.
The structure of the vaginal microbiome in the 3 groups
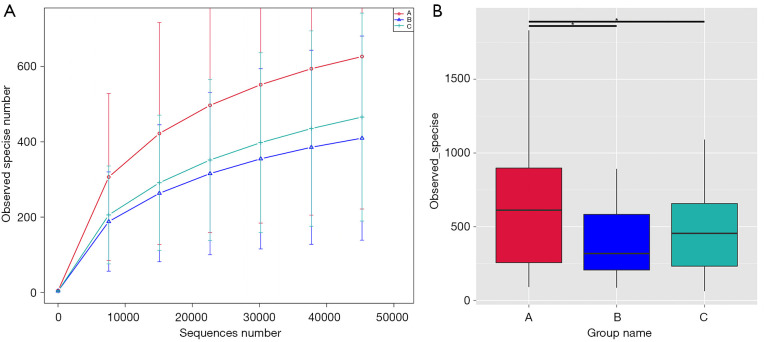
The alpha (phylogenetic)-diversity of microbial OTUs in the 3 groups was determined from samples. As shown in Figure 3A, group A had the highest microbiome diversity, followed by group C and group B. Based on PD_whole_tree (Figure 3B), which was calculated by QIIME software (Version 1.7.0, University of Colorado, Boulder, CO, USA), the vaginal microbiota diversity in group A was significantly richer than those in groups B and C, but there was no significant difference between group B and C (A vs. B, P=0.0278; A vs. C, P=0.0430; B vs. C, P=0.7396).
Figure 3.
Alpha diversity analysis of vaginal microbiome composition in participants from the 3 groups. (A) Rarefaction curve of vaginal microbiome diversity in the 3 groups; error bars represent standard deviation. Total number of observed species: group A: 626 species; group B: 409 species; and group C: 465 species. (B) Bar chart showing microbiota diversity in the 3 groups.
Identification of the vaginal microbiome composition in the 3 groups
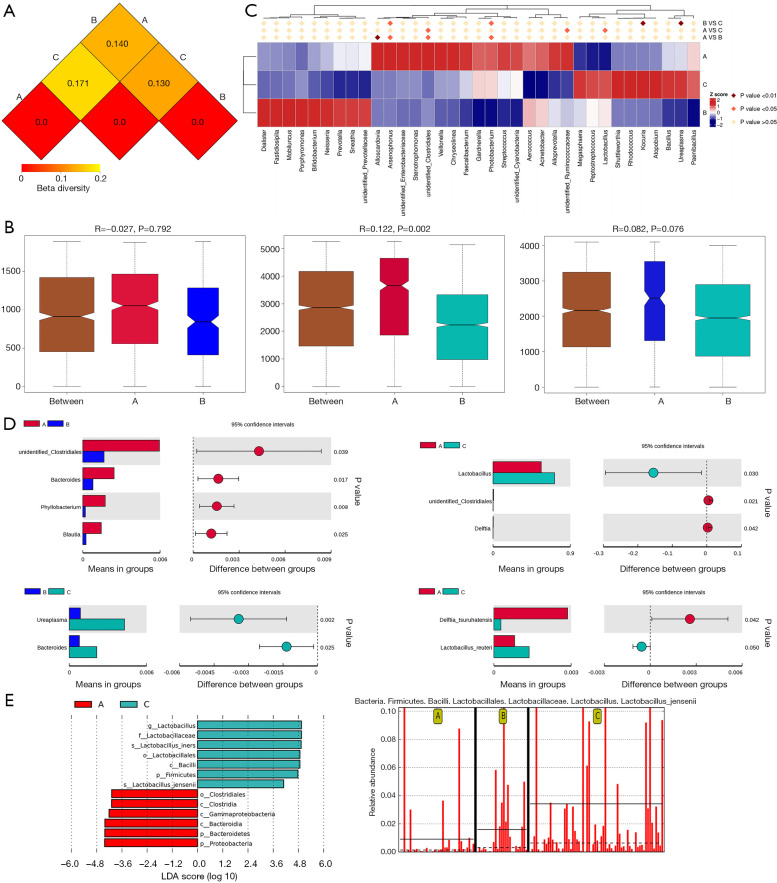
A distance matrix heatmap, which was based on the weighted UniFrac distance, revealed that the difference in the vaginal microbiome was greatest between group A and group C, while it was least different between group B and group C (Figure 4A). An analysis of similarity (ANOSIM) test, which was performed using QIIME software (version 1.7.0), revealed there to be a significant difference in composition between groups A and C (R=0.122, P=0.002). However, there was no significant difference between groups A and B (P=0.792), or between groups B and C (R=0.122, P=0.076) (Figure 4B). When the vaginal microbiome compositions of the 3 groups were compared simultaneously using the method of analysis of molecular variance (AMOVA), a significant difference was detected (P=0.027).
Figure 4.
Vaginal microbiomes composition within the 3 groups. (A) Distance matrix heatmap. The number in the graph is the difference coefficient between the 2 samples. The difference coefficient is positively correlated with the difference in microbiome diversity. (B) Bar chart showing the intergroup difference analysis of vaginal microbiome by ANOSIM analysis. (C) Significant differences in the abundance of genera within groups by MetaStat analysis. (D) T test analysis of vaginal microbiome composition within the groups at the genera and species levels. (E) LEfSe analysis of vaginal microbiomes within groups. A significant effect size was considered when the clades had statistical significance (P<0.05) and an LDA score >±4. Prefixes represent abbreviations for the taxonomic rank of each taxa: phylum (p_), class (c_), order (o_), family (f_), genus (g_), and species (s_). ANOSIM, analysis of similarity; LDA, linear discriminant analysis.
Several analytic methods were used to identify the specific clades of the microbiome in the 3 groups, including the linear discriminant analysis effect size (LEfSe) algorithm, the t test, and MetaStat analysis. With respect to the top 30 genera, group A showed the lowest relative abundance of Megasphaera, followed by group B. The relative abundances of Lactobacillus and Shuttleworthia were significantly lower in group A than in group C. Also, the relative abundances of unidentified Clostridiales and Alloscardovia were significantly higher in group A than in group B or C. Group A also had higher relative abundances of Veillonella and Stenotrophomonas than group C (Table S1, Figure 4C,D,E).
Regarding the top 30 genera, 26 species showed a significant difference in relative abundance between group A and group B, including 3 which had a relative abundance above 0.1%. The relative abundances of 36 species were found to differ significantly between groups A and C, and 6 had a relative abundance above 0.1%. A total of 22 species showed significant differences between group B and group C, but none had relative abundance above 0.1% (Table 2, Figure 4C,D,E). When the 3 groups were analyzed together, group A was found to have the lowest relative abundance of Lactobacillus jensenii, while Clostridium butyricum was significantly over-represented in group A (Table 3).
Table 2. Statistically different genera and species of the top 30 genera between the 3 groups.
| Taxa | Relative abundance | Value | Method of analysis | ||
|---|---|---|---|---|---|
| Group A | Group B | Group C | |||
| g__Alloscardovia; | 0.005482 | 0.000078 | 0.0494129526387591* | MetaStat analysis | |
| s__Alloscardovia_omnicolens | |||||
| g_unidentified_Clostridiales | 0.005946 | 0.001649 | 0.0385635457112232# | t-test analysis | |
| g__Alloscardovia | 0.005495 | 0.000078 | 0.0313232534451& | LEfSe analysis | |
| g__Lactobacillus. | 0.009102 | 0.015988 | 0.0466384014478& | ||
| s__Lactobacillus_jensenii | |||||
| g__unidentified_Clostridiales. | 0.003145 | 0.000857 | 0.0458592442122& | ||
| s__Clostridium_disporicum | |||||
| g__Lactobacillus | 0.561734 | 0.716614 | 0.0295794229242957# | t-test analysis | |
| g__unidentified_Clostridiales | 0.005946 | 0.001314 | 0.020573151880915# | ||
| g__Lactobacillus; | 0.000805 | 0.001369 | 0.0496299579455244# | ||
| s__Lactobacillus_reuteri | |||||
| g__Megasphaera | 0.006245 | 0.016048 | 0.0175645262151& | LEfSe analysis | |
| g__Veillonella | 0.009851 | 0.002642 | 0.0420310009006& | ||
| g__Stenotrophomonas | 0.008724 | 0.003283 | 0.0000568575884661& | ||
| g__Alloscardovia | 0.005495 | 0.000171 | 0.00547979034357& | ||
|
g__Lactobacillus.
s__Lactobacillus_iners |
0.311857 | 0.441428 | 0.0190787524907& | ||
| g__Lactobacillus. | 0.009102 | 0.034176 | 0.0000291877469495& | ||
| s__Lactobacillus_jensenii | |||||
| g__Sneathia. | 0.005893 | 0.00667 | 0.043367592503& | ||
| s__Sneathia_amnii | |||||
| g__Veillonella. | 0.008356 | 0.001802 | 0.0381148044568& | ||
| s__Veillonella_montpellierensis | |||||
| g__Alloscardovia. | 0.005482 | 0.000171 | 0.00547979034357& | ||
| s__Alloscardovia_omnicolens | |||||
| g__Megasphaera | 0.009965 | 0.016048 | 0.0415655213735& | LEfSe analysis | |
| g__Shuttleworthia | 0.000604 | 0.00221 | 0.0072300237267& | ||
*, Q value; #, P value; &, Kruskal-Wallis test P value.
Table 3. Statistically different genera and species of the top 30 genera between the 3 groups.
| Relative abundance | Kruskal-Wallis test P value | LEfSe analysis | |||
|---|---|---|---|---|---|
| Group A | Group B | Group C | |||
| Taxa (genus) | |||||
| g__Megasphaera | 0.006245 | 0.009965 | 0.016048 | 0.0223508462253 | |
| g__Stenotrophomonas | 0.008724 | 0.002758 | 0.003283 | 0.000312479146166 | |
| g__Shuttleworthia | 0.000621 | 0.000604 | 0.00221 | 0.0121196322448 | |
| Taxa (species) | |||||
| g__Lactobacillus | 0.009102 | 0.015988 | 0.034176 | 0.000198940154373; LDA=4.1586381629 | |
| s__Lactobacillus_jensenii | |||||
| g__unidentified_Clostridiales | 0.000564 | 0.000169 | 0.000167 | 0.0288725220546 | |
| s__Clostridium_butyricum | |||||
All of the significant different genera and species have relative abundance more than 0.001.
The diagnostic efficacy of the vaginal microbiome
Receiver operating characteristic (ROC) curves were drawn to determine the cut-off for relative abundance of potential microbiome biomarkers for distinguishing EM/AM-associated CPP from other types of CPPS. When the relative abundance of Clostridium disporicum was over 0.001105% and that of Lactobacillus reuteri was under 0.1911349%, the differential diagnostic sensitivity and specificity for EM/AM-associated CPP were 81.08% and 52.00%, respectively. Moreover, the sensitivity increased to 89.19% when the 2 potential microbiome biomarkers and serum CA125 were combined (Table S1). In the prospective validation trial, the sensitivity and specificity for differentiating EM/AM-associated CPP from other types of CPPS were 70.00% and 53.62%, respectively, when the relative abundance of Clostridium disporicum was over 0.001105% and that of Lactobacillus reuteri was under 0.1911349%. The diagnostic sensitivity and specificity of clinical serum CA125 were 46% and 100.00%, respectively, and the sensitivity increased to 86.00% when serum CA125 and vaginal biomarkers were combined (Table S1).
Predictive functional profiling of vaginal microbiome
Functional profiling of the vaginal microbiomes of the 3 groups was performed using the PICRUSt algorithm. Figure S1 shows the dysregulation of several functional pathways revealed by post-hoc analysis using Storey correction. On level 1, cellular processes were overexpressed in group A compared with group B (P=0.011), while genetic information processing was overexpressed in group B compared with group A (P=0.003). Differentially regulated pathways of potential relevance between groups A and C included metabolism, cellular processes, and organismal systems, which were overexpressed in group A compared to group C, while genetic information processing and environmental information processing were overexpressed in group C compared with group A (P<0.001 to P=0.046). On level 2, in terms of the relative abundance of predicted gene functions of the top 10, compared with group A, group B showed upregulation in pathways including replication and repair, translation, and nucleotide metabolism (P=0.002 to P=0.007). Compared with group C, group A showed upregulation in pathways including amino acid metabolism, energy metabolism, metabolism of cofactors and vitamins, and cellular processes and signaling, whereas membrane transport, carbohydrate metabolism, replication and repair, translation, and nucleotide metabolism were overexpressed in group C compared with group A (P<0.001 to P=0.007). Cellular processes and signaling were overexpressed in group B compared to group C, while membrane transport was overexpressed in group C compared to group B (P=0.038 and P=0.047, respectively) (Figures S2-S4).
Discussion
Worldwide, 6–27% of women experience CPP (10). It is estimated that chronic pain in the minor pelvic region affects 3.8% of women of any age and 12% of women of reproductive age (11). Despite the etiology being unknown for a considerable proportion of patients (12), EM/AM remains 1 of the most common causes of CPP. At present, many effective conservative treatments are available for EM/AM, such as compound oral contraceptive (COC), gonadotropin-releasing hormone (GnRHa), and the levonorgestrel intrauterine system. Distinguishing EM/AM-associated CPP through noninvasive methods is the greatest challenge to accurate and timely diagnosis. Therefore, a unique, consistent biomarker is needed to aid in the identification of these patients.
The interplay and symbiotic relationship between human bodies and microorganisms is becoming clearer. This relationship has been widely investigated and preliminary results have been obtained. Gut microbiome dysbiosis has been discovered to play a role in a variety of diseases and conditions (13-15). Urine microbiota (16) or expressed prostatic secretion microbiome (17) is associated with chronic prostatitis (CP)/CPPS. Furthermore, the relationship between genital tract microbiome and some gynecologic diseases has also been investigated. Overgrowth of anaerobic and facultative bacteria in the vagina, or bacterial vaginosis, and an increase in proinflammatory cytokines are implicated as risk factors for the development of infectious and obstetric complications (18,19). The presence of Atopobium vaginae and Porphyromonas sp. in the uterine cavity with a simultaneous high vaginal pH has been reported to be correlated with endometrial cancer (20). The presence of pathogenic bacteria in the pelvic cavity of patients with pelvic inflammatory diseases (PID) can be assumed to promote the development of endometriosis by causing excessive endometrial inflammation (21). However, the relationship between the vaginal microbiome and CPPS has yet to be studied. The vaginal ecosystem in human beings involves a dynamic in which the microbes and the host mutually affect each other (22). Our study focused on the potential specific biomarkers for distinguishing EM/AM-associated CPP from other phenotypes of CPPS. The PICRUSt algorithm was also used to predict the functional bioinformation.
The participants in the 3 groups were age-matched, and there were no significant differences regarding their parity, gravidity, menstrual phase, and method of contraception. The striking finding of this pilot study was the significantly higher alpha diversity in patients with EM/AM-associated CPP compared to the other 2 groups (A vs. B, P=0.0278; A vs. C, P=0.0430). Currently, it is unclear why the vaginal microbiome of women in the EM/AM group showed greater alpha diversity. The participants in our study had not received any antibiotics within the 2 weeks prior to sampling, and patients with autoimmune disorders and diabetes mellitus were excluded. The alpha diversity analysis was far from able to explain the differences in bacteria between the 3 groups, so we performed beta diversity analysis to identify specific bacteria biomarkers. We found that several individual bacterial taxa were over or under-represented in the vaginal microbiome of females with EM/AM-associated CPP compared to the other 2 groups. Clostridium butyricum, Clostridium disporicum, Alloscardovia omnicolens, and Veillonella montpellierensis were isolated in high numbers from vaginal excretions, while Lactobacillus jensenii, Lactobacillus reuteri, and Lactobacillus iners were isolated in low numbers. These characteristic changes in the microbiome may contribute to the etiology of CPPS and could serve as potential biomarkers for the diagnosis of EM/AM-associated CPP and as therapeutic targets.
Although the cause and effect cannot be clarified from the association alone, our results indicate that females with EM/AM-associated CPP can be distinguished from those without EM/AM based on the structure of their vaginal microbiome. The differential diagnostic sensitivity and specificity of serum CA125 were 40.54% and 100.0%, respectively. Thus, serum CA125 held satisfying specificity while its sensitivity was by no means poor. When the vaginal microorganism biomarkers were combined, the diagnostic sensitivity increased to 89.19%, indicating that it is easier to diagnose EM/AM-associated CPP in patients with normal levels of serum CA125. In the prospective validation trial, it was validated that serum CA125 holds low sensitivity but satisfying specificity, and equivalent diagnostic sensitivity and specificity were achieved when serum CA125 was combined with vaginal microbiome biomarkers. It is widely accepted that a lactobacilli-dominated vaginal microbiome can protect the host from adverse reproductive health outcomes. Although not all lactobacilli are necessarily stable or healthy, anti-inflammatory bacteria such as the lactobacillus species may be important for well-being and could play a role in the development and severity of CPPS. Reuterin can be produced by Lactobacillus reuteri (23), through which L. reuteri plays a modulating role in the growth of all enteric microflora (24). L. reuteri is of great interest as a bacteriocin production probiotic and is used in animal nutrition as well as in yoghurt products and pharmaceutic preparations. Clostridium disporicum, a saccharolytic species, is known to be an ursodeoxycholic acid producer; however, the functional reports on this opportunistic bacterium are limited. C. disporicum is a gram-positive obligately anaerobic bacillus, and can contain 2 subterminal spores. There is a severe lack of reports on the isolation of C. disporicum from human specimens. It was isolated from a 75-year-old diabetic female patient following a ring pessary insertion for uterine prolapse by Woo et al. in 2001 (25), and the first case of intra-abdominal infection caused by C. disporicum was reported in 2013 (26).
Although the role of the microbiome has been revealed in some diseases, studies on vaginal microbiota in women with CPPS have been inadequate. Therefore, we also explored the etiology of EM/AM-associated CPP using the PICRUSt algorithm. The etiology may be related to the metabolic actions of the changed bacterial populations which disrupt normal physiology. The vagino-uterine microbiota in patients with AM has been reported to be especially enriched in pathways involved in flagella assembly and biosynthesis of aromatic amino acids (8). The results of our study are in line with this report that the vaginal microbiota of women with EM/AM-associated CPP are statistically enriched in pathways involved in amino acid metabolism. Bacteria accumulating in cervical mucus and their consequent entry into the uterine cavity may lead to the occurrence of endometriosis via LPS/TLR4, lipopeptide/TLR2, or TLR6 engagement of the innate immune system (27). Differentially regulated pathways between the 3 groups that may be of relevance included cellular processes and signaling, which were enriched in the EM/AM group compared to the other 2 groups; in comparison, replication and repair, membrane transport, nucleotide metabolism, and translation pathways were down-regulated in the EM/AM group compared to the other 2 groups. Furthermore, amino acid metabolism, energy metabolism, and metabolism of cofactors and vitamins were enriched in the EM/AM group compared to the other 2 groups. Given the current understanding of the role of the microbiome and the mechanism by which it affects systemic inflammation and modulates pain response, as well as its putative role in psychosocial stress, it is not impossible that the vaginal microbiome plays a role in the etiology of EM/AM-associated CPP and the occurrence of pelvic pain. The potential microbiome biomarkers may be used to confirm suspected cases of EM/AM-associated CPP.
There are some limitations to this pilot study. Although we tried to collect information on the most relevant comorbidities, other unmeasured factors may have existed and impacted the results. As our study was cross-sectional, it was impossible to determine the causes and effects of changes in the vaginal microbiome in the occurrence of CPPS. Using PICRUSt allowed us to perform functional profiling of our data, but it is admittedly inferior to the more expensive direct metabolomic techniques and the macrogenome, which can identify specific bacteria leading to differences in metabolism. Our study was also limited by its small sample size, despite the long timespan and the equivalent result in the validation trial. However, the participants were strictly screened and samples were collected under consistent conditions. Even with a small sample size, we ultimately managed to identify significant microbiome differences between the 3 groups as well as differential abundant taxa of potential vaginal microbiome biomarkers in the differential diagnosis of CPPS. A large sample size is needed to further verify our findings and to increase their statistical power. Since the microbiome of adjacent organs might also influence pelvic pain through organ visceral sensory pathways (28,29), the microbiome compositions of the gut and uterine tract should also be analyzed to explore their relationships with CPPS.
Conclusions
In summary, a predominance of Clostridium butyricum, Clostridium disporicum, Alloscardovia omnicolens, and Veillonella montpellierensis with a concomitant paucity of Lactobacillus jensenii, Lactobacillus reuteri, and Lactobacillus iners may be related to EM/AM-associated CPP, and these vaginal microorganism may be considered as potential contributing pathogens and prospective biomarkers. The combination of Clostridium disporicum with Lactobacillus reuteri and serum CA125 had a satisfactory differential diagnostic effect, which was also validated in this study. Besides, our findings provide important insights into the etiology and manifestation of EM/AM-associated CPP, and give solid validation for biomarker development for the early detection of EM/AM with pelvic pain and the phenotype of females with CPPS.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This study was funded by National Key R&D Program of China (2017YFC1001200), CAMS Innovation Fund for Medical Sciences (CIFMS) (2016-I2M-1-002), and Chinese Academy of Medical Sciences Initiative for Innovative Medicine (CAMS-2017-I2M-1-002).
Ethical Statement: The authors are accountable for all aspects of this work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethics Committee of Peking Union Medical College Hospital (JS-1844) and informed consent was provided by all participants.
Footnotes
Reporting Checklist: The authors have completed the MDAR and STROBE reporting checklists (available at http://dx.doi.org/10.21037/atm-20-4586).
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-4586
Peer Review File: Available at http://dx.doi.org/10.21037/atm-20-4586
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-4586). The authors have no conflicts of interest to declare.
References
- 1.Chao MT, Abercrombie PD, Nakagawa S, et al. Prevalence and use of complementary health approaches among women with chronic pelvic pain in a prospective cohort study. Pain Med 2015;16:328-40. 10.1111/pme.12585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheong Y, Saran M, Hounslow JW, et al. Are pelvic adhesions associated with pain, physical, emotional and functional characteristics of women presenting with chronic pelvic pain? A cluster analysis. BMC Womens Health 2018;18:11. 10.1186/s12905-017-0509-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perry CP. Current concepts of pelvic congestion and chronic pelvic pain. JSLS 2001;5:105-10. [PMC free article] [PubMed] [Google Scholar]
- 4.Gelbaya TA, El-Halwagy HE. Focus on primary care: chronic pelvic pain in women. Obstet Gynecol Surv 2001;56:757-64. 10.1097/00006254-200112000-00002 [DOI] [PubMed] [Google Scholar]
- 5.No G G. The Initial Management of Chronic Pelvic Pain. Royal College of Obstetricians and Gynaecologists 2012;41:1-16. [Google Scholar]
- 6.Govorov I, Sitkin S, Pervunina T, et al. Metabolomic Biomarkers in Gynecology: A Treasure Path or a False Path? Curr Med Chem 2020;27:3611-22. 10.2174/0929867326666190104124245 [DOI] [PubMed] [Google Scholar]
- 7.Allaire C, Williams C, Bodmer-Roy S, et al. Chronic pelvic pain in an interdisciplinary setting: 1-year prospective cohort. Am J Obstet Gynecol 2018;218:114.e1-12. 10.1016/j.ajog.2017.10.002 [DOI] [PubMed] [Google Scholar]
- 8.Chen C, Song X, Wei W, et al. The microbiota continuum along the female reproductive tract and its relation to uterine-related diseases. Nat Commun 2017;8:875. 10.1038/s41467-017-00901-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Langille MG, Zaneveld J, Caporaso JG, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 2013;31:814-21. 10.1038/nbt.2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahangari A. Prevalence of chronic pelvic pain among women: an updated review. Pain physician 2014;17:E141-7. 10.36076/ppj.2014/17/E141 [DOI] [PubMed] [Google Scholar]
- 11.Triolo O, Lagana AS, Sturlese E. Chronic pelvic pain in endometriosis: an overview. J Clin Med Res 2013;5:153-63. 10.4021/jocmr1288w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mathias SD, Kuppermann M, Liberman RF, et al. Chronic pelvic pain: prevalence, health-related quality of life, and economic correlates. Obstet Gynecol 1996;87:321-7. 10.1016/0029-7844(95)00458-0 [DOI] [PubMed] [Google Scholar]
- 13.Arora HC, Eng C, Shoskes DA. Gut microbiome and chronic prostatitis/chronic pelvic pain syndrome. Ann Transl Med 2017;5:30. 10.21037/atm.2016.12.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schulberg J, De Cruz P. Characterisation and therapeutic manipulation of the gut microbiome in inflammatory bowel disease. Intern Med J 2016;46:266-73. 10.1111/imj.13003 [DOI] [PubMed] [Google Scholar]
- 15.Geurts L, Neyrinck AM, Delzenne NM, et al. Gut microbiota controls adipose tissue expansion, gut barrier and glucose metabolism: novel insights into molecular targets and interventions using prebiotics. Benef Microbes 2014;5:3-17. 10.3920/BM2012.0065 [DOI] [PubMed] [Google Scholar]
- 16.Shoskes DA, Altemus J, Polackwich AS, et al. The Urinary Microbiome Differs Significantly Between Patients With Chronic Prostatitis/Chronic Pelvic Pain Syndrome and Controls as Well as Between Patients With Different Clinical Phenotypes. Urology 2016;92:26-32. 10.1016/j.urology.2016.02.043 [DOI] [PubMed] [Google Scholar]
- 17.Xiao J, Ren L, Lv H, et al. Atypical microorganisms in expressed prostatic secretion from patients with chronic prostatitis/chronic pelvic pain syndrome: microbiological results from a case-control study. Urol Int 2013;91:410-6. 10.1159/000350934 [DOI] [PubMed] [Google Scholar]
- 18.Giakoumelou S, Wheelhouse N, Cuschieri K, et al. The role of infection in miscarriage. Hum Reprod Update 2016;22:116-33. 10.1093/humupd/dmv041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gillet E, Meys JF, Verstraelen H, et al. Bacterial vaginosis is associated with uterine cervical human papillomavirus infection: a meta-analysis. BMC Infect Dis 2011;11:10. 10.1186/1471-2334-11-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walther-António MR, Chen J, Multinu F, et al. Potential contribution of the uterine microbiome in the development of endometrial cancer. Genome Med 2016;8:122. 10.1186/s13073-016-0368-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tai FW, Chang CY, Chiang JH, et al. Association of Pelvic Inflammatory Disease with Risk of Endometriosis: A Nationwide Cohort Study Involving 141,460 Individuals. J Clin Med 2018;7:379. 10.3390/jcm7110379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith SB, Ravel J: The vaginal microbiota, host defence and reproductive physiology. J Physiol 2017;595:451-63. 10.1113/JP271694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Talarico TL, Dobrogosz WJ. Chemical characterization of an antimicrobial substance produced by Lactobacillus reuteri. Antimicrob Agents Chemother 1989;33:674-9. 10.1128/AAC.33.5.674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen CY, Chen SW, Wang HT. Effect of supplementation of yeast with bacteriocin and Lactobacillus culture on growth performance, cecal fermentation, microbiota composition, and blood characteristics in broiler chickens. Asian-Australas J Anim Sci 2017;30:211-20. 10.5713/ajas.16.0203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woo PC, Fung AM, Lau SK, et al. Group G beta-hemolytic streptococcal bacteremia characterized by 16S ribosomal RNA gene sequencing. J Clin Microbiol 2001;39:3147-55. 10.1128/JCM.39.9.3147-3155.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Plassart C, Mauvais F, Heurte J, et al. First case of intra-abdominal infection with Clostridium disporicum. Anaerobe 2013;19:77-8. 10.1016/j.anaerobe.2012.12.002 [DOI] [PubMed] [Google Scholar]
- 27.Akiyama K, Nishioka K, Khan KN, et al. Molecular detection of microbial colonization in cervical mucus of women with and without endometriosis. Am J Reprod Immunol 2019;82:e13147. 10.1111/aji.13147 [DOI] [PubMed] [Google Scholar]
- 28.Rudick CN, Jiang M, Yaggie RE, et al. O-antigen modulates infection-induced pain states. PLoS One 2012;7:e41273. 10.1371/journal.pone.0041273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malykhina AP. Neural mechanisms of pelvic organ cross-sensitization. Neuroscience 2007;149:660-72. 10.1016/j.neuroscience.2007.07.053 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as