Abstract
Background
Inflammatory bowel disease (IBD) is a chronic nonspecific intestinal disease. Our previous work showed that long non-coding RNA (LncRNA) nuclear enriched abundant transcript 1 (NEAT1) plays an important role in IBD. In the current study, we aimed to explore the underlying mechanism by which NEAT1 participates in the development of the disease.
Methods
Real-time quantitative polymerase chain reaction (RT-qPCR) and western blotting were used to detect the expression of NEAT1 and tumor necrosis factor superfamily member 1B (TNFRSF1B) in clinical specimens and dextran sulfate sodium (DSS) colitis mice. Inflammatory cell models were established by stimulating human normal intestinal epithelial cell line NCM460 and human colon cancer cell line HT-29 with tumor necrosis factor alpha (TNF-α). Expressions of inflammatory cytokines such as interleukin-8 (IL-8) and monocyte chemoattractant protein-1 (MCP-1) were detected by enzyme-linked immunosorbent assay (ELISA) or RT-qPCR, TNFRSF1B, NF-κB p65 and p-NF-κB p65 followed by the knockdown or overexpression of NEAT1 and TNFRSF1B were analyzed by western blotting, and the regulatory effects of NEAT1 on TNFRSF1B were detected by RNA pull-down experiments and RNA-decay assay. The translocation of NF-κB p65 to the nucleus was detected by immunofluorescence.
Results
In patients’ specimens and DSS colitis mouse models, NEAT1 and TNFRSF1B expression were up-regulated compared with the control group. TNF-α stimulation increased NEAT1 and TNFRSF1B expression and activated NF-κB signaling pathway by increasing the translocation of NF-κB p65 to the nucleus. In the presence of TNF-α stimulation, NEAT1 knockdown reduces the expression of TNFRSF1B and the translocation of NF-κB p65, thereby relieves cell inflammation. These effects can be reversed by the overexpression of TNFRSF1B.In addition, NEAT1 is involved in inflammatory response by up-regulating the mRNA levels of TNFRSF1B, and knocking down NEAT1 can alleviate inflammation by down-regulating TNFRSF1B. Moreover, NEAT1 co-precipitates TNFRSF1B mRNA in RNA-pulldown assay, and the presence of NEAT1 stabilizes the mRNA of TNFRSF1B.
Conclusions
Our results showed that LncRNA NEAT1 promotes NF-κB p65 translocation and mediates intestinal inflammation by regulating TNFRSF1B.
Keywords: Nuclear enriched abundant transcript 1 (NEAT1), tumor necrosis factor superfamily member 1B (TNFRSF1B), NF-κB, ulcerative colitis
Introduction
Inflammatory bowel disease (IBD), which is classified as ulcerative colitis (UC) and Crohn’s disease (CD), is a chronic nonspecific intestinal inflammatory disease. The incidence of UC varies in different race and gender. Studies have shown that in western countries, the incidence of UC in women before age 45 is not significantly different from that in men, thereafter, the incidence of UC in men was significantly higher than in women. In the Asia-Pacific region, men are more likely to develop UC from adolescence to middle and late middle age (1,2). The pathogenesis of UC mainly attributed to environmental and genetic factors, and the homeostasis between the innate and the adaptive immune systems of the gastrointestinal tract and the intestinal microbes (3).
Long noncoding RNA are noncoding RNAs with a length of more than 200 bp, and contribute to the etiology of various diseases such as respiratory diseases, inflammatory diseases and tumors (4-7). NEAT1, as a lncRNA, is involved in many pathophysiological processes such as immune response, tumorigenesis and infection (8), but its effect on inflammation is debatable. Studies have confirmed that NEAT1 may be involved in the development of colitis by acting on miR-204-5p (9). On the contrary, in other inflammatory diseases, such as acute-on-chronic liver failure, traumatic brain injury, NEAT1 is downregulated (10,11). Our previous study has demonstrated that NEAT1 expression is upregulated in the mouse model of inflammatory bowel disease and modulates intestinal inflammation by regulating exosome-mediated macrophage polarization, knocking down NEAT1 can relieve intestinal inflammation in mice (12), but the mechanism by which NEAT1 participates in intestinal inflammation is still elusive.
TNFRSF1B is a member of the tumor necrosis factor receptor superfamily (TNFRSF), also known as TNFR2. TNFRSF contain several cysteine-rich structural domains in the extracellular membrane, and functions mostly as homologous trimers and binds to cytokines include TNF-α, leukotriene (LT), factor associated suicide ligand (FasL), CD40L, nerve growth factor (NGF), etc. TNFRSF1B, which is one of the major TNF-α receptors, is involved in the regulation of tissue regeneration (13,14). However, the relationship between TNFRSF1B and intestinal inflammation has not yet been definitively concluded. Holtmann et al. reported that tumor necrosis factor mediated colitis in mice mainly through binding to TNFRSF1B, and promoted the loss of mucosal barrier and the progression of colitis (15). In TNFRSF1B knockout mice, there was significant protection against colitis related-barrier loss (16), however, some contrary views suggest that TNFR signaling plays a key role in promoting mucosal healing of chronic colitis. TNFRSF1B can alleviate colitis by inhibiting the proliferation of CD8+T cells in the colon (17,18). Mice injected with iTreg cells which knocked out TNFRSF1B showed more severe colonic inflammation (19).
In this study, the upregulation of NEAT1 and TNFRSF1B in the inflammatory and control groups were verified in vivo and in vitro. We investigated whether NEAT1 participates in intestinal inflammation by regulating TNFRSF1B expression and activating the NF-κB pathway. The results of this study may reveal a novel role for NEAT1 in regulating IBD, especially of UC.
We present the following article in accordance with the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/atm-21-34).
Methods
Mice
Female C57BL/6 mice (7 weeks, 17–18 g) were purchased from the laboratory animals department of Central South University, and kept in specific pathogen-free (SPF) environment at 22 to 26 °C and with a 12:12 light/dark cycle, 40–70% humidity, free drinking and eating. All animal experiments were approved by the Ethics Committee of the Third Xiangya Hospital of Central South University (No: 2019-S000), and performed according to the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Cells and culture media
Human normal colonic epithelial cell line NCM460 were acquired from the Cancer Research Institute of Central South University and human colon cancer cell line HT-29 were purchased from American Type Culture Collection.NCM460 was cultured in DMEM medium (cat. no. SH30022.01; HyClone) with 10% fetal bovine serum (cat. no. 04-001-1A/B; Biological Industries), and HT-29 was cultured in RPMI1640 medium (cat. no. SH30809.01; HyClone) with 10% fetal bovine serum. Both were cultured at 37 °C and was supplied with 5% CO2. Inflammation cell models were induced by human recombinant TNF-α (cat. no. 300-01A; PeproTech, Inc.) with the concentration gradient of set as follows: 0, 5, 10, 20 and 50 ng/mL for 24 h, according to previous studies (20).
Human tissue samples
The colon biopsy tissues at the site of inflammation and para-inflammatory tissues of the 3 patients were collected from the Center of Gastroenterology and Endoscopy, The Third Xiangya Hospital of Central South University. The demography is shown in Table 1. The tissue specimens were immediately washed with sterile PBS and placed at -80°C for subsequent testing. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Third Xiangya Hospital, Central South University (No.: 2019-S000) and informed consent was taken from all the patients.
Table 1. Demographic data of three UC patients.
| Patient’s number | Sex | Age | BMI (kg/m2) | Type | UCEIS† | Anti-TNF therapies |
|---|---|---|---|---|---|---|
| Patient 1 | Male | 38 | 18.92 | Chronic recurrent type | 3 | No |
| Patient 2 | Male | 45 | 18.82 | Chronic recurrent type | 3 | No |
| Patient 3 | Female | 61 | 17.71 | Chronic recurrent type | 2 | No |
†UCEIS means Ulcerative Colitis Endoscopic Index of Severity.
DSS induced colitis in mice
Eight female mice were randomly divided into two groups (n=4 per group): control group and DSS group. All mice in the DSS group received 3% DSS (cat. no. 160110; MP Biomedicals), while the control group mice received purified water. The mice were weighed at a fixed time every day to observe their weight changes and were sacrificed on the 7th day. Colon tissues of mice were collected, washed with sterile PBS, and put into −80 °C for subsequent detection.
RT-qPCR
The expressions of NEAT1, TNFRSF1B, IL-8 and MCP-1 were detected by real-time quantitative PCR (RT-qPCR). Briefly, TRIzol reagent (cat. no. 15596026; Invitrogen) was used to extract total RNA from NCM460 and HT-29 cells or mice intestinal tissues according to the manufacturer’s instructions. Reverse transcription kit (Thermo Fisher Scientific, Inc.) was used to get cDNA, the reaction procedure was: 42 °C for 60 minutes, 10 °C for 5 minutes, qPCR was performed using the SYBR Green qPCR Master mix (Vazyme Biotech Co., Ltd.). The procedure was: 95 °C for 30 seconds, and then 95 °C for 5 seconds and 60 °C for 30 seconds to 39 cycles, take β-actin as internal reference, used 2-ΔΔCq method to calculate the relative expression of various molecular (21) (Primer sequences are shown in Table S1).
Western blotting
RIPA lysis buffer (cat. no. SL1020-100ML; Coolaber) and protease inhibitor cocktail (cat. no. B14001;Bimake.cn)were mixed at the ratio of 100:1 and then added to intestinal tissue of patients, mouse intestinal tissue or NCM460 and HT-29 cells, cracked on the ice for 40 min, supernatant protein was taken after 12,000 rpm 20 min at 4 °C. The protein concentration of each sample was determined by BCA method (22). The volume of each protein sample was determined by the final 50 µg protein. Add to 10% SDS-PAGE gel for electrophoresis, 80 V for 30 min, 120 V for 1 h, transferred for 90 min onto polyvinylidene difluoride membranes (Merck KGaA), uses 5% BSA (Wuhan Goodbio Technology Co., Ltd.) to block membranes for 1 h and incubated them overnight at 4 °C. The detailed list of antibodies are as follows: TNFRSF1B Rabbit pAb (1:1,000; cat. no. 19272-1-AP; Proteintech Group, Inc.), NF-κB p65 Rabbit pAb (1:1,000; Cat. No. 10745-1-AP; Proteintech Group, Inc.), p-NF-κB p65 Rabbit pAb (1:1,000; cat. no. ab28856; Abcam, Inc.), β-actin Mouse mAb (1:2,000; cat. no. 66009-1-Ig; Proteintech Group, Inc.), IL1R1 Rabbit pAb (1:1,000; cat. no. bs-20697R; Bioss, Inc.), Caspase-4 Rabbit pAb (1:2,000; cat. no. BA2212; Boster, Inc.) followed with corresponding HRP-conjugated Affinipure Goat-Rabbit/Mouse IgG Secondary Antibody (1:4,000; cat. no. SA00001-2/SA00001-2; ProteinTech Group, Inc.) incubated at 37 °C for 1 h, and finally luminescence solution was added. Protein bands were displayed with Merck KGaA kit, and β-actin protein was used as internal reference. Image J (version 1.52 V; National Institutes of Health) was used to analyze the gray value of the protein stripe to quantitatively calculate the relative expression level of the protein.
Enzyme-linked immunosorbent assay (ELISA)
Inflammatory factors IL-8 and MCP-1 in the cell supernatant were determined by human IL-8 and MCP-1 ELISA kits (Amoy LunChangShuoBiotech, co., Ltd). Standard curves were drawn according to the instructions, and OD value of the samples was substituted into the calculated concentration value. The experiment was repeated for three times.
RNA pull-down assay
RNA pull-down assay was conducted by using Pierce™ Magnetic RNA-Protein Pull-Down Kit (cat. no. 20164, Thermo Fisher Scientific) according to the instructions. NEAT1 and negative control RNA chain was synthesized in vitro and then labeled with biotin. The RNA sequence is as follows: 5'-GGAGGGTTTCACAACCCTAATGTCCACACTCGGTGACACGGTT-3' (Biotin-NEAT1 probe) and 5'-UUCUCCGAACGUGUCACGUUUCUCCGGCTCACCTCGUTT-3' (Biotin-NC probe). Biotin-labeled RNA was mixed with cell extract and incubated with streptavidin agarose beads. The retrieved RNA from RNA-binding protein complexes were collected to RT-qPCR analysis.
Cell transfection
TNFRSF1B overexpressed plasmid, NEAT1 and TNFRSF1B small interfering RNA and negative control (NC) were synthesized by Shanghai GenePharma, Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) was used as transfection reagent, TNFRSF1B overexpressed plasmid, TNFRSF1B small interference RNA and negative control NC were controlled at the final concentration of 50 nM in the experiment, and samples were collected 48 hours after transfection for subsequent detection.
Immunofluorescence (IF)
Cells were washed with 1× PBS and fixed with 4% paraformaldehyde. After washing with PBS, the cells were blocked with 1% BSA at room temperature for 20 min. Then the cells were incubated in primary antibody anti-NF-κB p65 (1:100; cat. no. 10745-1-AP; Proteintech Group, Inc.), settle at 4 °C overnight, washed with PBS three times, 5 min each, then the cells were incubated with DyLight 488-antibody conjugates Goat Anti-Rabbit IgG secondary antibodies (1:500; cat. no. A23220; Abbkine, Inc.) for 1 h at room temperature. Further washed with PBS, the cells were finally incubated with DAPI and observed with laser confocal scanning microscope (Olympus Corporation).
RNA-decay assay
The cells were treated with corresponding knockdown or overexpression treatment in the presence of 5 µg/mL actinomycin D. Cells were collected at 2, 4, 6, 8 and 12 h, respectively. RNA was finally extracted according to the instructions of RNA extraction methods for RT-qPCR detection, 18sRNA as the internal reference, calculate according to the previous research methods and plot the RNA stability curve (23).
H&E and Histology and immunohistochemistry (IHC)
H&E: the colonic tissues of mice were fixed with formaldehyde for 24 hours and embedded in paraffin, cut into sections about 3–5 µm thick, stained with hematoxylin for 30 sec and stained with eosin for 2 min, and observed the pathological tissues and inflammation. Immunohistochemistry: Paraffin-embedded colonic tissues of 8 mice were placed in citric acid repair solution (0.01M, pH6.0). Put it into a pressure cooker at 100 °C for 10–15 min and cool it to room temperature. Primary anti-p-NF-κB p65 (1:100; cat. no. ab 28856; Abcam, Inc.) and TNFRS1B (1:500; cat. no. 19272-1-AP; Proteintech, Inc.) were incubated at 4 °C overnight, and then incubated with secondary biotin-conjugated Abs (cat. no. KIT-9710, Mai New Biotechnology Development Company) at 37 °C for 1 h. The immunohistochemical reaction was observed with 3,3,0-diaminobenzidine (DAB) for 3–5 min. The positive color was brown or yellow, captured one picture under 200× and 400× field, respectively.
Prediction of NEAT1 target gene in UC
We downloaded GSE87466 dataset from the Gene Expression Omnibus database (GEO) (https://www.ncbi.nlm.nih.gov/geo) which has 21 healthy mucosal biopsies and 87 ulcerative colitis mucosal biopsies and collected 5,988 up-regulated genes in UC, after data standardization by the limma package in R language (version 3.4.1) and averaging of different probe data of genes, the correlation between these up-regulated genes and NEAT1 were analyzed by R software, we concluded that the correlation coefficient of NEAT1 and TNFRSF1B ranked top 10 (data not shown). Finally, we used GraphPad Prism (Version 8.0.2; GraphPad Software Inc.) to draw a scatter plot and the result showed that NEAT1 and TNFRSF1B are positively correlated (r=0.6237, P<0.0001) (Figure S1).
Statistical analysis
Use GraphPad Prism (Version 8.0.2; GraphPad Software Inc.) to analyze data, and the results were presented as mean ± standard deviation. The differences between each group were calculated and evaluated by Student’s t test or one-way ANOVA. P<0.05 and below were considered statistically significant, and differences between two or three groups could be considered.
Results
NEAT1 and TNFRSF1B expression were up-regulated by inflammation
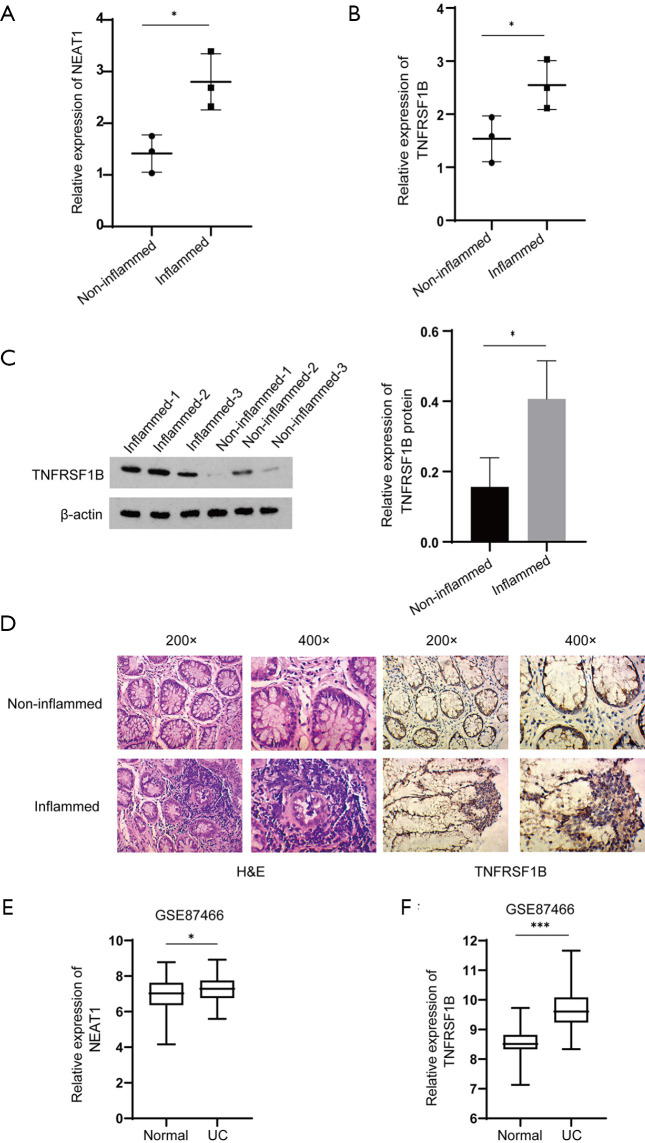
In order to verify the expression of NEAT1 and TNFRSF1B in UC patients, we collected tissues from the lesion and the adjacent site from three patients diagnosed with ulcerative colitis. The RT-qPCR results indicate that in the lesion tissues, the expression of NEAT1 was upregulated compared with the adjacent site (2.80±0.31 vs. 1.41±0.21), as well as TNFRSF1B (2.55±0.27 vs. 1.54±0.25) (Figure 1A,B), The western blot results showed that the expression of TNFRSF1B protein was upregulated compared with the adjacent site (0.41±0.06 vs. 0.16±0.05) (Figure 1C). These results are also consistent with immunohistochemistry staining (Figure 1D).
Figure 1.
Expression of NEAT1, TNFRSF1B in the lesion and adjacent site of UC patients. (A, B) Relative mRNA expression of NEAT1 and TNFRSF1B in intestinal inflammatory tissues detected by RT-qPCR. (C) The protein expression of TNFRSF1B in intestinal inflammatory tissues and para-inflammatory tissues detected by WB. (D) Tissue samples from UC patients and their corresponding para-inflammatory tissues, from left to right are H&E, TNFRSF1B. (E,F) Relative expression of NEAT1 and TNFRSF1B in GSE87466. *P<0.05. ***P<0.001. UC, ulcerative colitis; NEAT1, nuclear enriched abundant transcript 1; TNFRSF1B, tumor necrosis factor superfamily member 1B; RT-qPCR, real-time quantitative polymerase chain reaction; WB, western blot; H&E, hematoxylin-eosin staining; GSE, Gene Expression Omnibus Series.
We also explored the expression of NEAT1 and TNFRSF1B in GEO database, we used R language to standardize the data and average the different probe data of genes from GEO database and found that TNFRSF1B is among the top 10 genes that co-expressed with NEAT1 in UC, the rest are PTGS2, C3, CXCL1, CXCL2, TIMP1, IRAK2, TLR1, FPR1 and CXCL3. Then we used GraphPad Prism 8 to analyze the value of the expression of NEAT1 and TNFRSF1B and the results showed that NEAT1 and TNFRSF1B are positively correlated (r=0.6237, P<0.0001) and the expression of NEAT1 and TNFRSF1B were significantly up-regulated in tissues from UC patients (7.25±0.07 vs. 6.86±0.25 and 9.65±0.07 vs. 8.53±0.11) (Figure 1E,F).
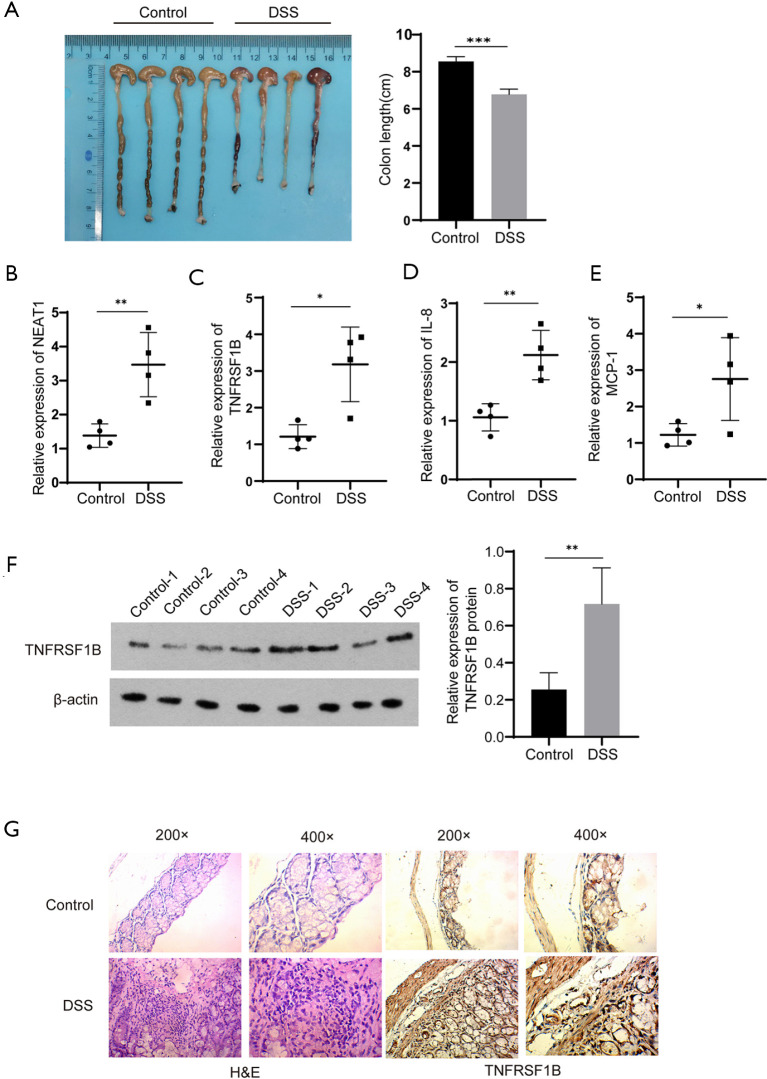
We further explore the expression of NEAT1 and TNFRSF1B in DSS-induced mouse model of acute colitis. Compared with the control group, all mice in the DSS group present significant hyperemic and shortened colon (6.78±0.14 cm vs. 8.55±0.13 cm) (Figure 2A). Meanwhile, the RNA levels of NEAT1 (3.47±0.47 vs. 1.38±0.17), TNFRSF1B (3.18±0.51 vs. 1.21±0.16), IL-8 (2.12±0.21 vs. 1.06±0.12) and MCP-1 (2.76±0.57 vs. 1.22±0.15) quantified by RT-qPCR was significantly increased (Figure 2B,C,D,E). Similar results were observed in western blot detection of TNFRSF1B protein expression (0.72±0.11 vs. 0.26±0.05) and immunohistochemical of TNFRSF1B (Figure 2F,G).
Figure 2.
Expression of NEAT1, TNFRSF1B, IL-8 and MCP-1 in mice. (A) Colon length in DSS model mice and control mice. (B, C) Relative mRNA expression of NEAT1 and TNFRSF1B. (D, E) Relative mRNA expression of IL-8 and MCP-1 in intestinal tissue of DSS model mice and control mice. (F) TNFRSF1B in DSS model mice and control mice tested by western blot. (G) Tissue samples from DSS model mice and control mice, from left to right are H&E, TNFRSF1B. *P<0.05, **P<0.01, ***P<0.001. NEAT1, nuclear enriched abundant transcript 1; TNFRSF1B, tumor necrosis factor superfamily member 1B; RT-qPCR, real-time quantitative polymerase chain reaction; DSS, dextran sulfate sodium; IL-8, interleukin-8; MCP-1, monocyte chemoattractant protein-1, H&E, hematoxylin-eosin staining.
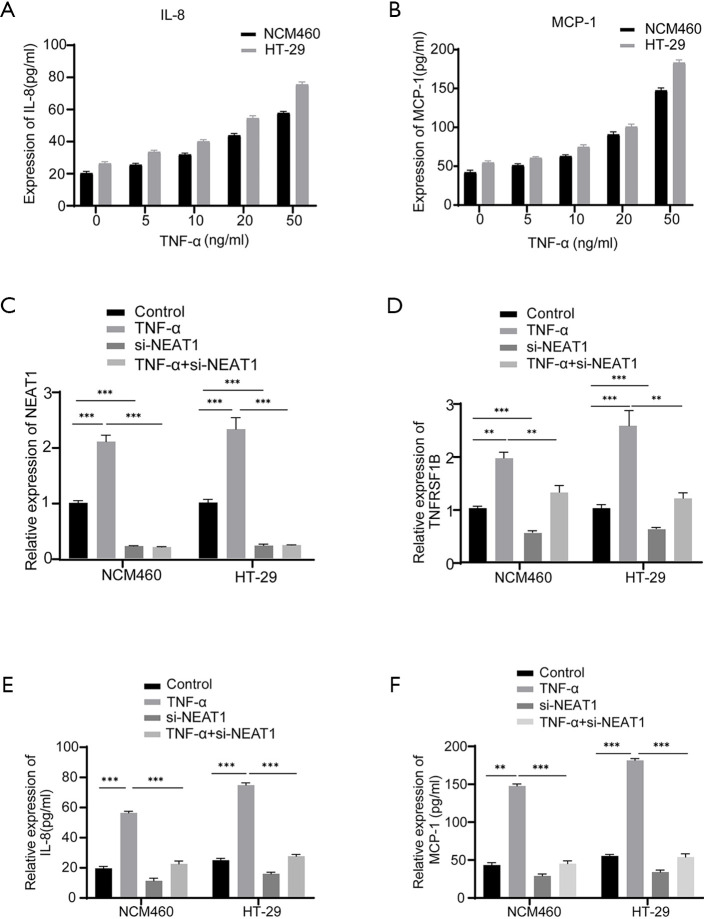
NEAT1 regulates TNFRSF1B expression
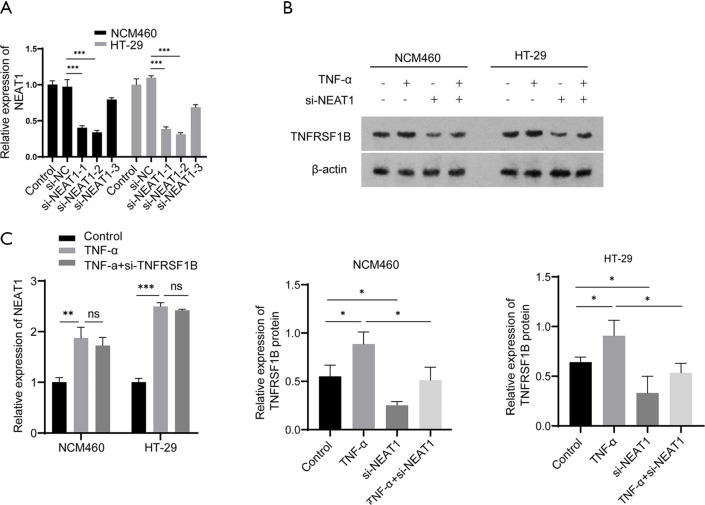
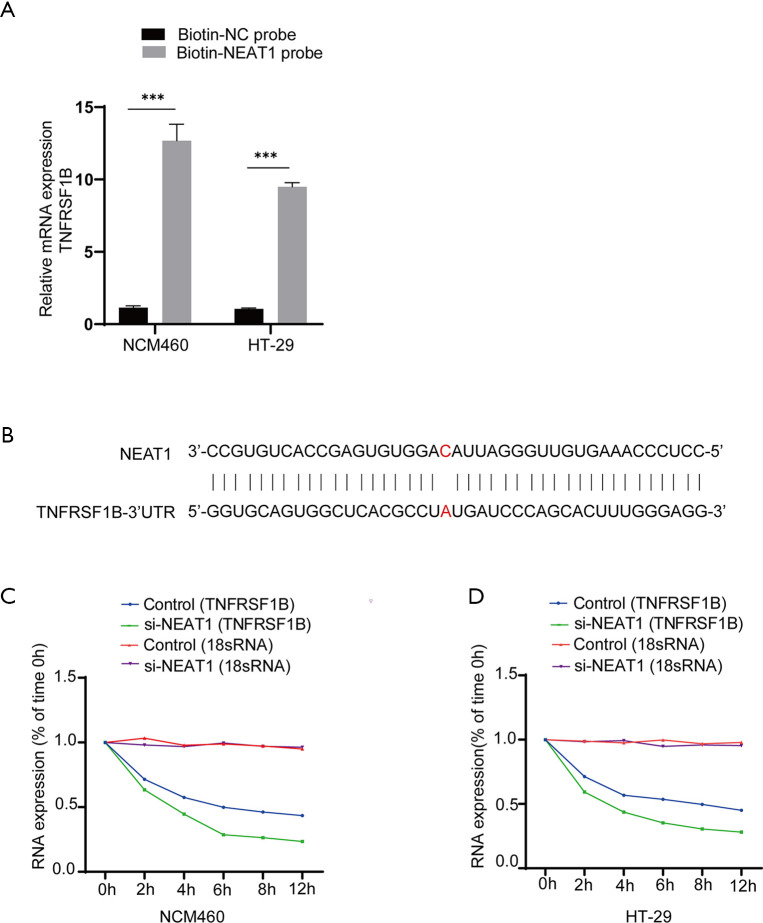
Next, we verified the relationship between NEAT1 and TNFRSF1B by manipulating the level of NEAT1 and TNFRSF1B. The knockdown efficiency of three candidates small interfering RNAs si-NEAT1-1, si-NEAT1-2 and si-NEAT1-3 was detected by RT-qPCR, the results showed that NEAT1 expression was decreased to 0.40±0.02 and 0.34±0.02 in NCM460, 0.39±0.02 and 0.31±0.01 in HT-29, respectively with si-NEAT1-1 and si-NEAT1-2, which was significantly decreased compared with the negative control group, so the mixture of si-NEAT1-1 and si-NEAT-2 was used in the following experiments to ensure maximum knockdown efficiency (Figure 3A). The results show that knocking down NEAT1 decreased the level of TNFRSF1B (0.25±0.02 vs. 0.55±0.07 in NCM460 and 0.33±0.10 vs. 0.64±0.03 in HT-29) (Figure 3B), while silencing TNFRSF1B did not alter the expression of NEAT1 (1.73±0.09 vs. 1.87±0.02 in NCM460 and 2.42±0.01 vs. 2.50±0.04 in HT-29) (Figure 3C), indicating that NEAT1 regulates the level of TNFRSF1B. We further analyzed how NEAT1 regulates the expression level of TNFRSF1B. First, we conducted RNA pull-down experiments and followed by RT-qPCR, the results showed that TNFRSF1B contents precipitated by Biotin labelled-NEAT1 probe (Biotin-NEAT1 probe) were significantly more abundant than their counterparts precipitated by the control probe that does not contain NEAT1(Biotin-NC probe) (12.68±0.66 vs. 1.14±0.07 in NCM460 and 9.50±0.16 vs. 1.06±0.03 in HT-29) (Figure 4A). Then we predicted the sequence of NEAT1 binding to TNFRSF1B mRNA on Freiburg RNA Tools IntaRNA-RNA-RNA interaction and found that NEAT1 is likely to combine with the reverse complementary of the 3'UTR end of the TNFRSF1B mRNA (Figure 4B). To further verify whether NEAT1 alters the pre-transcriptional or post-transcriptional level of TNFRSF1B, we conducted RNA-decay experiments, and the results showed that after the introduction of actinomycin D, the mRNA stability of TNFRSF1B in the NEAT1 knockdown group was significantly decreased compared with the control group (Figure 4C,D), which indicates that NEAT1 stabilizes the mRNA of TNFRSF1B. Taken together, these results indicated that NEAT1 could regulate the expression of TNFRSF1B.
Figure 3.
NEAT1 can regulate the expression of TNFRSF1B. (A) The knockdown efficiency of three siRNAs targeting NEAT1 detected by RT-qPCR. (B) The relative expression of TNFRSF1B protein in different groups. (C) The expression of NEAT1 in different groups detected by RT-qPCR. *P<0.05, **P<0.01, ***P<0.001, ns represents no significance. NEAT1, nuclear enriched abundant transcript 1; TNFRSF1B, tumor necrosis factor superfamily member 1B; RT-qPCR, real-time quantitative polymerase chain reaction; TNF-α, tumor necrosis factor-alpha. NC, negative control.
Figure 4.
NEAT1 can stabilize the mRNA of TNFRSF1B. (A) The retrieved mRNA of TNFRSF1B from RNA-binding protein complexes obtained by RNA pull-down assay and analyzed by RT-qPCR. Biotin-NC probe is the negative control probe labeled with biotin. Biotin-NEAT1 probe is a biotin-labeled probe targeting NEAT1. (B) The predicted site that NEAT1 binds to the 3’UTR end of TNFRSF1B. (C,D) The mRNA expression of TNFRSF1B in NCM460 cells and HT-29 cells with time after the addition of actinomycin D, 18sRNA as internal reference because it is stable in the cell and is not affected by nuclease. ***P<0.001. NEAT1, nuclear enriched abundant transcript 1; TNFRSF1B, tumor necrosis factor superfamily member 1B; RT-qPCR, real-time quantitative polymerase chain reaction; TNF-α, tumor necrosis factor-alpha. NC, negative control.
NEAT1 participates in inflammatory response via regulating TNFRSF1B expression
Since TNFRSF1B is receptor of TNF-α, we next explored whether NEAT1 is involved in TNF-α mediated inflammation by regulating TNFRSF1B. The results show that in the case of TNF-α induced inflammation indicated by the upregulation of inflammatory cytokines IL-8 and MCP-1 (Figure 5A,B), the expression of both NEAT1 and TNFRSF1B increased significantly, while NEAT1 knockdown blocked the elevation of TNFRSF1B (1.30±0.07 vs. 2.01±0.06 in NCM460 and 1.19±0.06 vs. 2.55±0.17 in HT-29), IL-8 (22.64±1.09 vs. 56.47±0.64 pg/mL in NCM460 and 27.69±0.66 vs. 74.87±0.88 pg/mL in HT-29) and MCP-1 (45.25±2.16 vs. 147.8±1.54 pg/mL in NCM460 and 53.94±2.52 vs. 181.5±1.37 pg/mL in HT-29) even in the presence of TNF-α induction (Figure 5C,D,E,F). These results showed that NEAT1 knockdown could down-regulate the expression level of TNFRSF1B and relieve the degree of cell inflammation.
Figure 5.
NEAT1 is involved in inflammation by regulating TNFRSF1B. (A, B) ELISA was used to analyze the expression of IL-8 and MCP-1 in different groups. (C,D) The mRNA expression of NEAT1 and TNFRSF1B in different groups detected by RT-qPCR. (E,F) The expression of IL-8 and MCP-1 in different groups detected by ELISA. **P<0.01, ***P<0.001. NEAT1, nuclear enriched abundant transcript 1; TNFRSF1B, tumor necrosis factor superfamily member 1B; IL-8, interleukin-8; MCP-1, monocyte chemoattractant protein-1; TNF-α, tumor necrosis factor-alpha; RT-qPCR, real-time quantitative polymerase chain reaction; ELISA, enzyme-linked immunosorbent assay.
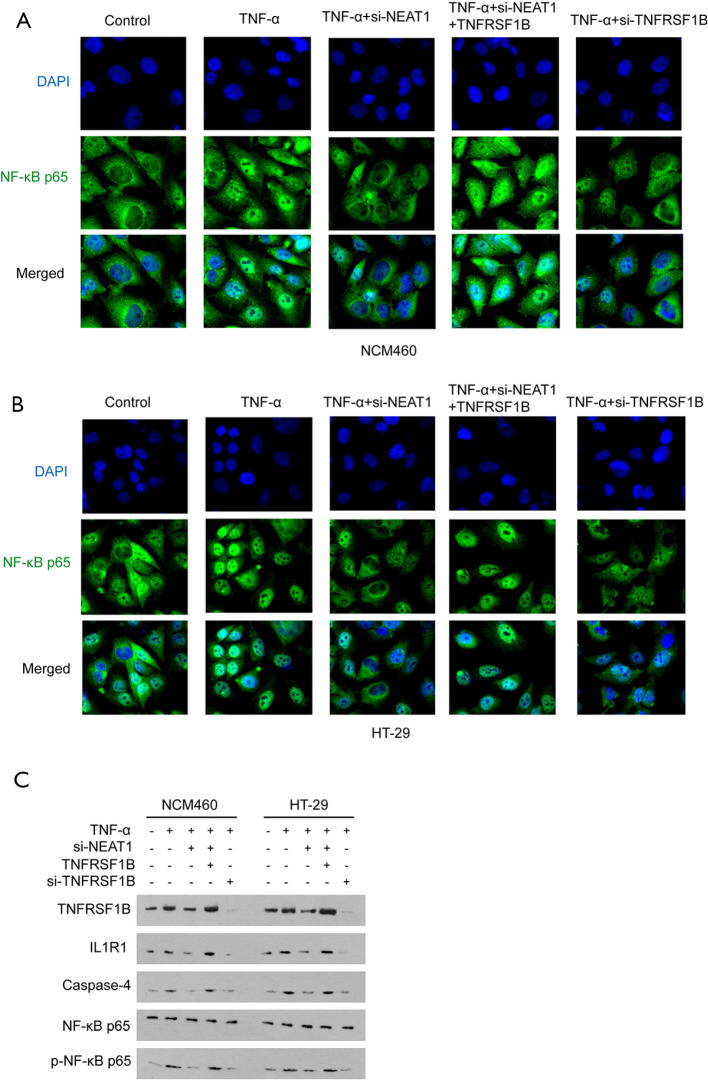
NEAT1 promotes the translocation of NF-κB p65 by regulating TNFRSF1B and participates in TNF-α mediated inflammation
NF-κB p65 is a known down-stream molecule of TNFRSF1B. After activation by TNFRSF1B, NF-κB p65 translocated from cytoplasm to the cell nucleus, and induces the transcription of varies genes, including caspase-4, which mediates cell pyroptosis (24). NF-κB p65 also initiates the inflammatory loop by recruiting IL1R1, a pro-inflammatory effector in IBD (25,26). It was observed that TNF-α increased the nucleus entry of NF-κB p65, indicated by immunofluorescence, and this effect is enhanced by overexpressing TNFRSF1B, even in the presence of NEAT1 knocking down (Figure 6A,B). As expected, TNF-α stimulation increased IL1R1 and Caspase-4 in NCM460 and HT-29 cells, as well as the expression of p-NF-κB p65 protein. However, NF-κB p65 did not differ significantly among the groups, because only when NF-κB p65 is modified by phosphorylation and entry into the nucleus can it participate in the inflammatory response. Meanwhile, knocking down of NEAT1 alleviates the upregulation of IL1R1, Caspase-4 and p-NF-κB p65. However, overexpression of TNFRSF1B could reverse the effect caused by NEAT1 knockdown, increasing the inflammation level (Figure 6C).
Figure 6.
NEAT1 promotes the translocation of NF-κB p65 by regulating TNFRSF1B and participates in TNF-α mediated inflammation. (A) Translocation of NF-κB p65 to the nucleus in NCM460, NF-κB p65 (green) and DAPI stained nuclei (blue) under 200× field. (B) Translocation of NF-κB p65 to the nucleus in HT-29 under 200× field. (C) The protein expression levels of TNFRSF1B, NF-κB p65 and p-NF-κB p65, as well as proliferation and inflammation-related factors IL1R1 and Caspase-4 in different groups. NEAT1, nuclear enriched abundant transcript 1; IL1R1, interleukin 1 receptor type I; TNFRSF1B, tumor necrosis factor superfamily member 1B; TNF-α, tumor necrosis factor-alpha; NF-κB, nuclear factor-kappa B.
Discussion
LncRNAs are considered to be an important regulatory molecule in IBD (27). It has been reported that lncRNAs regulates inflammatory cytokines release and plays an important role on intestinal mucosal integrity (28). Our previous studies have shown that NEAT1 expression is up-regulated in IBD, and NEAT1 is involved in IBD by regulating intestinal epithelial barrier and exosome-mediated macrophage polarization (12). Another study showed that NEAT1 improved the therapeutic effect of PDT in IBD by NEAT1-miRNA-204-5p-PI3K-Akt axis (9). In this study, we found that NEAT1 mediated intestinal inflammation by regulating TNFRSF1B and the distribution of NF-κB p65.
LncRNAs modulate the targets’ transcription by several different mechanisms. In this study, we observed that NEAT1 could regulate the expression level of TNFRSF1B by stabilizing its mRNA. LncRNAs could work as ceRNAs to compete the shared mRNA transcripts with miRNA and therefore prevents the target mRNA from degeneration. Therefore, we analyzed the potential miRNAs by searching the LncRNA-mRNA target prediction database DIANA-LncBase v2 (29) (updated in 2016), a total of 1,386 candidate miRNAs that may be regulated by NEAT1. Furthermore, we obtained the list of TNFRSF1B upstream regulatory miRNA in TarBase v8 database (30) (updated in 2018) of DIANA Tools and obtained 18 candidate miRNAs. The downstream candidate miRNAs of NEAT1 and the upstream regulatory miRNA of TNFRSF1B showed no overlaps (Figure S2) (See https://cdn.amegroups.cn/static/public/atm-21-34-1.xlsx for the specific list of miRNAs), suggests a possibility that NEAT1 may directly bind to the mRNA of TNFRSF1B and participate in the inflammatory response. At the same time, NEAT1 is a key components of paraspeckle, a stress-responsive complex that retains the target mRNA in the nuclear to arrest its translation (31). Previous studies have shown that NEAT1 stabilizes ELF3 mRNA to help reduce its degradation but does not affect its transcription (32). Therefore, our study might also explain NEAT1 involved in intestinal inflammation by regulating TNFRSF1B mRNA level. Also, we’ll discuss in detail whether NEAT1 acts directly on the mRNA of TNFRSF1B in future study.
The role of TNFRSF1B in the inflammation process is controversial. Previous studies have shown that TNFRSF1B expression is up-regulated in the mouse model or patients with IBD, suggesting that TNFRSF1B plays a pro-inflammatory role in colitis (33), a similar observation has had been documented that in the absence of TNFRSF1B, DSS-induced colitis in mice showed relieved inflammation (34), this may be explained by the cytotoxic effects of TNFRSF1B through enhanced TNFRSF1A and programmed necrosis (35). TNFRSF1B is also reported to increase the binding of TNFRSF1A with TNF and exacerbating the inflammatory process (36). However, contrary opinions also hold that TNFRSF1B plays an anti-inflammatory role in promoting intestinal epithelial barrier recovery and mucosal healing in colitis (17-19,37). These contradictory results suggest the effect of TNFRSF1B may be context-dependent. In the current study, we believe that TNFRSF1B plays a proinflammatory role in intestinal inflammation.
In summary, we found that NEAT1 promotes NF-κB p65 entry into the nucleus by regulating TNFRSF1B and thus plays a pro-inflammatory role. This study may provide novel insights for understanding the etiology of IBD.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This work was supported by the National Nature Science Foundation of China (grant Nos. 81670504 and 81970494) and Science Foundation of Hunan Province (grant No. 2020JJ4842).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Third Xiangya Hospital, Central South University (No: 2019-S000) and informed consent was taken from all the patients. All animal experiments were approved by the Ethics Committee of the Third Xiangya Hospital of Central South University (No: 2019-S000), and performed according to the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Footnotes
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/atm-21-34
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-21-34
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-21-34). The authors have no conflicts of interest to declare.
References
- 1.Shah SC, Khalili H, Gower-Rousseau C, et al. Sex-Based Differences in Incidence of Inflammatory Bowel Diseases-Pooled Analysis of Population-Based Studies From Western Countries. Gastroenterology 2018;155:1079-89.e3. 10.1053/j.gastro.2018.06.043 [DOI] [PubMed] [Google Scholar]
- 2.Shah SC, Khalili H, Chen CY, et al. Sex-based differences in the incidence of inflammatory bowel diseases-pooled analysis of population-based studies from the Asia-Pacific region. Aliment Pharmacol Ther 2019;49:904-11. 10.1111/apt.15178 [DOI] [PubMed] [Google Scholar]
- 3.Kobayashi T, Siegmund B, Le Berre C, et al. Ulcerative colitis. Nat Rev Dis Primers 2020;6:74. 10.1038/s41572-020-0205-x [DOI] [PubMed] [Google Scholar]
- 4.Geng H, Bu H-F, Liu F, et al. In Inflamed Intestinal Tissues and Epithelial Cells, Interleukin 22 Signaling Increases Expression of H19 Long Noncoding RNA, Which Promotes Mucosal Regeneration. Gastroenterology 2018;155:144-55. 10.1053/j.gastro.2018.03.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma D, Cao Y, Wang Z, et al. CCAT1 lncRNA Promotes Inflammatory Bowel Disease Malignancy by Destroying Intestinal Barrier via Downregulating miR-185-3p. Inflamm Bowel Dis 2019;25:862-74. 10.1093/ibd/izy381 [DOI] [PubMed] [Google Scholar]
- 6.Mineo M, Lyons SM, Zdioruk M, et al. Tumor Interferon Signaling Is Regulated by a lncRNA INCR1 Transcribed from the PD-L1 Locus. Mol Cell 2020;78:1207-23.e8. 10.1016/j.molcel.2020.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qiao C, Yang L, Wan J, et al. Long noncoding RNA ANRIL contributes to the development of ulcerative colitis by miR-323b-5p/TLR4/MyD88/NF-kappaB pathway. Biochem Biophys Res Commun 2019;508:217-24. 10.1016/j.bbrc.2018.11.100 [DOI] [PubMed] [Google Scholar]
- 8.Wang Z, Li K, Huang W. Long non-coding RNA NEAT1-centric gene regulation. Cell Mol Life Sci 2020;77:3769-79. 10.1007/s00018-020-03503-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang K, Zhang Z, Liu K, et al. Neat1-miRNA204-5p-PI3K-AKT axis as a potential mechanism for photodynamic therapy treated colitis in mice. Photodiagnosis Photodyn Ther 2018;24:349-57. 10.1016/j.pdpdt.2018.10.020 [DOI] [PubMed] [Google Scholar]
- 10.Xu Y, Cao Z, Ding Y, et al. Long Non-coding RNA NEAT1 Alleviates Acute-on-Chronic Liver Failure Through Blocking TRAF6 Mediated Inflammatory Response. Front Physiol 2019;10:1503. 10.3389/fphys.2019.01503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhong J, Jiang L, Huang Z, et al. The long non-coding RNA Neat1 is an important mediator of the therapeutic effect of bexarotene on traumatic brain injury in mice. Brain Behav Immun 2017;65:183-94. 10.1016/j.bbi.2017.05.001 [DOI] [PubMed] [Google Scholar]
- 12.Liu R, Tang A, Wang X, et al. Inhibition of lncRNA NEAT1 suppresses the inflammatory response in IBD by modulating the intestinal epithelial barrier and by exosome-mediated polarization of macrophages. Int J Mol Med 2018;42:2903-13. 10.3892/ijmm.2018.3829 [DOI] [PubMed] [Google Scholar]
- 13.Fischer R, Kontermann R, Maier O. Targeting sTNF/TNFR1 Signaling as a New Therapeutic Strategy. Antibodies 2015;4:48-70. 10.3390/antib4010048 [DOI] [Google Scholar]
- 14.Fischer R, Kontermann RE, Pfizenmaier K. Selective Targeting of TNF Receptors as a Novel Therapeutic Approach. Front Cell Dev Biol 2020;8:401. 10.3389/fcell.2020.00401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holtmann MH, Douni E, Schütz M, et al. Tumor necrosis factor-receptor 2 is up-regulatedon lamina propria T cells in Crohn’s disease andpromotes experimental colitis in vivo. Eur J Immunol 2002;32:3142-51. [DOI] [PubMed] [Google Scholar]
- 16.Su L, Nalle SC, Shen L, et al. TNFR2 activates MLCK-dependent tight junction dysregulation to cause apoptosis-mediated barrier loss and experimental colitis. Gastroenterology 2013;145:407-15. 10.1053/j.gastro.2013.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bradford EM, Ryu SH, Singh AP, et al. Epithelial TNF Receptor Signaling Promotes Mucosal Repair in Inflammatory Bowel Disease. J Immunol 2017;199:1886-97. 10.4049/jimmunol.1601066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Punit S, Dubé PE, Liu CY, et al. Tumor Necrosis Factor Receptor 2 Restricts the Pathogenicity of CD8+ T Cells in Mice With Colitis. Gastroenterology 2015;149:993-1005.e2. 10.1053/j.gastro.2015.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang S, Xie C, Chen Y, et al. Differential roles of TNFα-TNFR1 and TNFα-TNFR2 in the differentiation and function of CD4+Foxp3+ induced Treg cells in vitro and in vivo periphery in autoimmune diseases. Cell Death Dis 2019;10:27. 10.1038/s41419-018-1266-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar A, Chatterjee I, Gujral T, et al. Activation of Nuclear Factor-kappaB by Tumor Necrosis Factor in Intestinal Epithelial Cells and Mouse Intestinal Epithelia Reduces Expression of the Chloride Transporter SLC26A3. Gastroenterology 2017;153:1338-50.e3. 10.1053/j.gastro.2017.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods 2001;25:402-8. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 22.Noble JE, Bailey MJA. Chapter 8 Quantitation of Protein. Guide to Protein Purification, 2nd Edition. Methods in Enzymology, 2009. p. 73-95. [DOI] [PubMed] [Google Scholar]
- 23.Huang H, Weng H, Sun W, et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol 2018;20:285-95. 10.1038/s41556-018-0045-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hersoug LG, Moller P, Loft S. Role of microbiota-derived lipopolysaccharide in adipose tissue inflammation, adipocyte size and pyroptosis during obesity. Nutr Res Rev 2018;31:153-63. 10.1017/S0954422417000269 [DOI] [PubMed] [Google Scholar]
- 25.Chassaing B, Van de Wiele T, De Bodt J, et al. Dietary emulsifiers directly alter human microbiota composition and gene expression ex vivo potentiating intestinal inflammation. Gut 2017;66:1414-27. 10.1136/gutjnl-2016-313099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X, Tan Y, Zhang Y, et al. The novel glycyrrhetinic acid-tetramethylpyrazine conjugate TOGA induces anti-hepatocarcinogenesis by inhibiting the effects of tumor-associated macrophages on tumor cells. Pharmacol Res 2020;161:105233. 10.1016/j.phrs.2020.105233 [DOI] [PubMed] [Google Scholar]
- 27.Zacharopoulou E, Gazouli M, Tzouvala M, et al. The contribution of long non-coding RNAs in Inflammatory Bowel Diseases. Dig Liver Dis 2017;49:1067-72. 10.1016/j.dld.2017.08.003 [DOI] [PubMed] [Google Scholar]
- 28.Chen SW, Wang PY, Liu YC, et al. Effect of Long Noncoding RNA H19 Overexpression on Intestinal Barrier Function and Its Potential Role in the Pathogenesis of Ulcerative Colitis. Inflamm Bowel Dis 2016;22:2582-92. 10.1097/MIB.0000000000000932 [DOI] [PubMed] [Google Scholar]
- 29.Paraskevopoulou MD, Vlachos IS, Karagkouni D, et al. DIANA-LncBase v2: indexing microRNA targets on non-coding transcripts. Nucleic Acids Res 2016;44:D231-8. 10.1093/nar/gkv1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karagkouni D, Paraskevopoulou MD, Chatzopoulos S, et al. DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res 2018;46:D239-45. 10.1093/nar/gkx1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin Y, Schmidt BF, Bruchez MP, et al. Structural analyses of NEAT1 lncRNAs suggest long-range RNA interactions that may contribute to paraspeckle architecture. Nucleic Acids Res 2018;46:3742-52. 10.1093/nar/gky046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feng Y, Gao L, Cui G, et al. LncRNA NEAT1 facilitates pancreatic cancer growth and metastasis through stabilizing ELF3 mRNA. Am J Cancer Res 2020;10:237-48. [PMC free article] [PubMed] [Google Scholar]
- 33.Mizoguchi E, Mizoguchi A, Takedatsu H, et al. Role of tumor necrosis factor receptor 2 (TNFR2) in colonic epithelial hyperplasia and chronic intestinal inflammation in mice. Gastroenterology 2002;122:134-44. 10.1053/gast.2002.30347 [DOI] [PubMed] [Google Scholar]
- 34.Wang K, Han G, Dou Y, et al. Opposite role of tumor necrosis factor receptors in dextran sulfate sodium-induced colitis in mice. PLoS One 2012;7:e52924. 10.1371/journal.pone.0052924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chan FK, Shisler J, Bixby JG, et al. A role for tumor necrosis factor receptor-2 and receptor-interacting protein in programmed necrosis and antiviral responses. J Biol Chem 2003;278:51613-21. 10.1074/jbc.M305633200 [DOI] [PubMed] [Google Scholar]
- 36.Louis A., Tartaglia DP, David V. Goeddel. Ligand passing: the 75-kDa tumor necrosis factor (TNF) receptor recruits TNF for signaling by the 55-kDa TNF receptor. J Biol Chem 1993;265:18542-8. [PubMed] [Google Scholar]
- 37.Dayer Schneider J, Seibold I, Saxer-Sekulic N, et al. Lack of TNFR2 expression by CD4+ T cells exacerbates experimental colitis. Eur J Immunol 2009;39:1743-53. 10.1002/eji.200839132 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as