Abstract
Due to their capability to transport chemicals or proteins into target cells, cell‐penetrating peptides (CPPs) are being developed as therapy delivery tools. However, and despite their interesting properties, arginine‐rich CPPs often show toxicity for reasons that remain poorly understood. Using a (PR)n dipeptide repeat that has been linked to amyotrophic lateral sclerosis (ALS) as a model of an arginine‐rich CPP, we here show that the presence of (PR)n leads to a generalized displacement of RNA‐ and DNA‐binding proteins from chromatin and mRNA. Accordingly, any reaction involving nucleic acids, such as RNA transcription, translation, splicing and degradation, or DNA replication and repair, is impaired by the presence of the CPPs. Interestingly, the effects of (PR)n are fully mimicked by protamine, a small arginine‐rich protein that displaces histones from chromatin during spermatogenesis. We propose that widespread coating of nucleic acids and consequent displacement of RNA‐ and DNA‐binding factors from chromatin and mRNA accounts for the toxicity of arginine‐rich CPPs, including those that have been recently associated with the onset of ALS.
Keywords: ALS, arginine‐rich peptides, chromatin, mRNA, protamine
Subject Categories: RNA Biology, Protein Biosynthesis & Quality Control
The toxic side effects of basic peptides used as pharmaceutic delivery tools, originates from their protamine‐like coating of nucleic acids.
Introduction
Efforts to develop methods to introduce nucleic acids or proteins into cells started in the mid‐XXth century, with the realization that alkaline conditions favored the infectivity of poliovirus RNA in HeLa cells (Sprunt et al, 1959). Soon thereafter, it was found that the addition of basic proteins such as histones or protamine also enhanced the uptake of RNA or ALBUMIN by tumor cells in culture (Smull & Ludwig, 1962; Ryser & Hancock, 1965). These early studies already noted that arginine‐rich histone fractions were more efficient than lysine‐rich ones in stimulating the cellular uptake of proteins. In fact, several factors with arginine‐rich domains such as fibroblast growth factor (bFGF), pancreatic Ribonuclease A (RNase A), or the cytokine midkine (MK) are known to freely enter into cells (reviewed in (Fuchs & Raines, 2006)). The field of protein transfection regained attention in 1988 when 2 groups independently discovered that the HIV‐1 trans‐activator protein TAT could enter cells, accumulate at nuclei and nucleoli and be functional in trans‐activating HIV‐1 RNA transcription (Frankel & Pabo, 1988; Green & Loewenstein, 1988). The arginine‐rich dodecapeptide GRKKRRQRRRPQ was later defined as the minimum functional sequence from TAT, which when fused to any given protein enables its cellular uptake (Park et al, 2002). Today, cell‐penetrating peptides (CPPs) are intensively being developed due to their capacity to facilitate the uptake of nucleic acids, small molecules, proteins, or even viruses (reviewed in (Borrelli et al, 2018)).
Despite the interesting properties of arginine‐rich CPPs, their development has been in part limited by their toxicity. For instance, and despite its clinical use as an antidote of the anticoagulant heparin, protamine has important side effects which are driving the search for alternative heparin antidotes (Sokolowska et al, 2016). In addition, HIV‐1 TAT is neurotoxic in vitro, an observation that has been proposed as a potential explanation for AIDS‐associated neurodegeneration (King et al, 2006). Interestingly, studies done in the last decade have revealed that the pathogenicity of the most frequent mutation found in patients of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD), which occurs at a gene named C9ORF72, might also be related to the production of arginine‐rich CPPs. C9ORF72 mutations involve the expansion of a GGGGCC hexanucleotide within the first intron of the gene, which is amplified to hundreds or even thousands of copies in ALS/FTD patients (DeJesus‐Hernandez et al, 2011; Renton et al, 2011). Through repeat‐associated non‐AUG (RAN) translation sense and antisense GGGGCC expansions are translated into several dipeptide repeats (DPR), including (PR)n and (GR)n (Ash et al, 2013; Mori et al, 2013; Zu et al, 2013). Significantly, synthetic (PR)n and (GR)n peptides added to culture media enter cells accumulate at nucleoli and kill cells, thus behaving as toxic CPPs (Kwon et al, 2014). Accordingly, the expression of these DPRs is toxic in cells and animal models (Kwon et al, 2014; Mizielinska et al, 2014; Wen et al, 2014; Chew et al, 2015; Stopford et al, 2017; Swaminathan et al, 2018; Zhang et al, 2018). While the toxicity of arginine‐rich CPPs seems to underlie several pathologies, the mechanisms by which this occurs remain unknown. We here reveal that the exposure to arginine‐rich CPPs leads to a generalized displacement of RNA‐ and DNA‐binding factors from chromatin and RNA, providing a mechanism that explains their toxicity and widespread effects on reactions involving nucleic acids.
Results
(PR)20 peptides impair the assembly of 80S ribosomal particles on mRNA
Proteomic and genetic studies have revealed that arginine‐rich DPRs arising from C9ORF72 mutations have a particular impact on mRNA translation (Kanekura et al, 2016; Lopez‐Gonzalez et al, 2016; Chai & Gitler, 2018; Zhang et al, 2018). To further investigate how these DPRs affect translation, we performed a proteomic characterization of ribosomes purified from HeLa cells exposed to synthetic (PR)20 peptides. Ribosome purification was done by immunoprecipitation of a streptavidin‐binding peptide (SBP)‐tagged RPS9. The most prominent observation in ribosomes isolated from (PR)20‐treated cells was a generalized decrease in the abundance of L ribosomal proteins (RPLs) from the 60S subunit (Fig 1A, Appendix Table S1), which was not due to a reduction in the total levels of these factors (Fig 1B). Since ribosome purification was performed by pulling down RPS9, a component of the 40S ribosomal subunit, the observed reduction in 60S factors suggested that the peptide could be impairing the assembly of 80S ribosomes from 40S and 60S subunits. Accordingly, ribosome profiling from HeLa cells treated with (PR)20 peptides revealed an accumulation of polysome halfmers (Fig 1C), which is indicative of an incomplete assembly of 80S ribosomes at sites of translation initiation. Interestingly however, (PR)20 did not impair the in vitro assembly of 80S particles from purified 40S and 60S subunits in the absence of mRNA, as evaluated by electron microscopy, arguing that the peptide did not directly affect the components of the 40S or 60S subunits (Fig 1D and E).
Figure 1. Translation inhibition by (PR)20 .
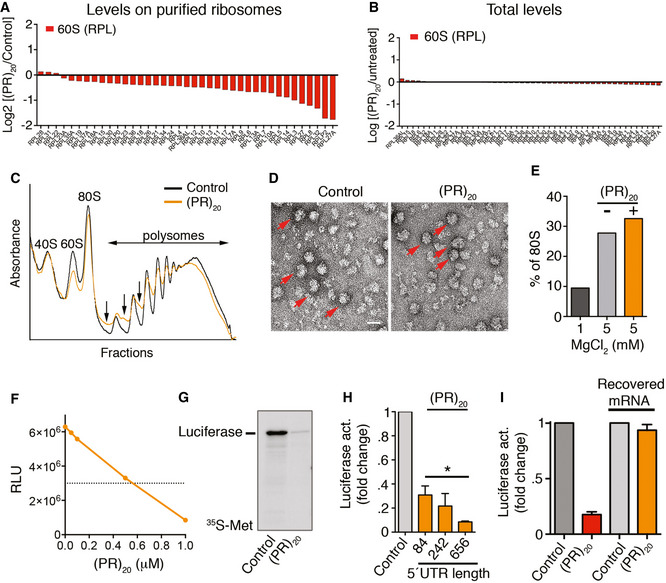
- Protein levels of RPL factors in ribosomes purified from Hela RPS9SBP cells exposed to 10 μM of (PR)20 for 16 h, as identified by LC‐MS/MS.
- Protein levels of RPL factors in the input extracts used for ribosome purification from Hela RPS9SBP cells exposed to 10 μM of (PR)20 for 16 h, as identified by LC‐ MS/MS.
- Representative polysome profiles obtained from HeLa cells untreated or treated with 10 μM of (PR)20 for 16 h. The presence of halfmers is indicated (arrows).
- Electron microscopy images from purified 40S and 60S ribosomal complexes (1 pmol each) assembled in vitro in the presence of MgCl2, and in the presence or absence of 5 pmol of (PR)20. Assembled 80S particles are indicated (red arrows). Scale bar (white) represents 10 nm.
- Quantification of 80S particles identified in (D) (n = 1,000) in non‐assembly (1 mM MgCl2) or assembly (5 mM MgCl2) conditions.
- In vitro translation of 100 ng of luciferase mRNA (quantified by Relative Luciferase Units [RLU]) in the presence of increasing doses of (PR)20.
- In vitro translation of 100 ng of luciferase mRNA in the presence or absence of 0.5 μM (PR)20. Translation products were labeled with [35S]‐Met/Cys and analyzed by SDS–PAGE and autoradiography.
- In vitro translation of 100 ng of luciferase mRNA with different 5′ UTR lengths in the presence (orange columns) or absence (grey column) of 0.5 μM (PR)20 (n = 3). Data represent mean values ± SD (*P < 0.05; t‐test).
- In vitro translation of 100 ng of luciferase mRNA in the presence or absence of 0.5 μM (PR)20. In the right two columns, the mRNA was extracted from a translation reaction done in the presence of the DPR, and subsequently used in a new translation reaction performed in the absence of (PR)20 (n = 3). Data represent mean values ± SD.
To further investigate how (PR)20 impairs translation, we performed in vitro translation reactions in rabbit reticulocyte lysates. Consistent with previous work (Kanekura et al, 2016), in vitro translation of a luciferase mRNA was impaired by (PR)20 in a dose‐dependent manner (Fig 1F and G). The presence of polysome halfmers and the lower abundance of RPLs on ribosomes purified from (PR)20‐treated cells led us hypothesize that the peptide could be preventing the assembly 80S ribosomes by binding to mRNA at translation initiation sites, as this is the step where 40S and 60S subunits assemble into 80S ribosomes in cells. In fact, arginine‐rich peptides have very high biochemical affinity for RNA (Tan & Frankel, 1995) and ALS‐associated arginine‐rich DPRs specifically have been shown to bind to and form aggregates in the presence of RNA (Kanekura et al, 2016; Boeynaems et al, 2017; White et al, 2019). In support of this model, extending the length of the non‐coding 5′UTR of the luciferase mRNA led to a further decrease in translation by (PR)20 (Fig 1H), which we reasoned could be due to an increased probability of DPR binding to the 5′UTR impairing initiation. Of note, the binding to (PR)20 does not irreversibly alter RNA molecules, as mRNA purified from (PR)20‐treated reticulocyte extracts was efficiently translated in a subsequent in vitro translation reaction performed in the absence of the DPR (Fig. 1I). Collectively, these experiments indicate that the mechanism by which (PR)n peptides impair translation is due to interactions of the peptide with mRNA that prevent the assembly of 40S and 60S subunits into 80S ribosomes at active polysomes.
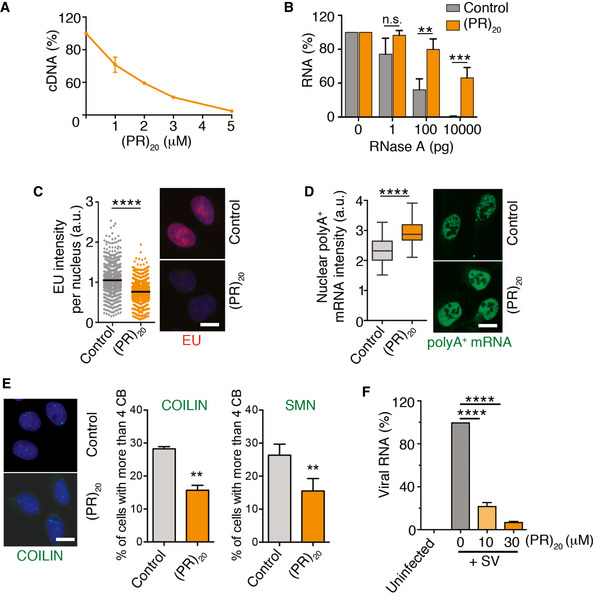
A general effect of (PR)20 peptides in reactions using DNA or RNA substrates
Besides translation, a number of cellular processes using RNA intermediates have been reported to be altered by C9ORF72‐associated arginine‐rich peptides including splicing (Kwon et al, 2014; Yin et al, 2017), rRNA biosynthesis (Kwon et al, 2014), and mRNA export (Rossi et al, 2015). While each of these effects has been proposed to be due to the binding of the DPRs to specific factors such as the splicing regulator U2 snRNP (Yin et al, 2017), we reasoned that if the effects of arginine‐rich peptides on translation were due to RNA coating, any reaction using an RNA template should be affected. Supporting this view, in vitro reactions using RNA substrates such as reverse transcription or RNase A‐mediated RNA degradation were impaired by (PR)20 or (GR)20 peptides in a dose‐dependent manner, but not by (GA)20 or BSA (Fig 2A and B, and Appendix Fig S1A and B). Regarding the cellular effects of (PR)20 related to RNA reactions, we could confirm an overall decrease in RNA biosynthesis (Kwon et al, 2014) (Fig 2C) together with an accumulation of polyadenylated mRNA in the nucleus that is indicative of deficient mRNA export (Rossi et al, 2015) (Fig 2D) in U2OS cells exposed to (PR)20. In addition, the presence of the peptide decreased the number of Cajal Bodies identified by COILIN or SMN, which are membrane‐less nuclear organelles made up of RNA and proteins that are involved in various aspects of RNA metabolism including snRNA modifications, histone mRNA assembly, or telomeric RNA processing (Fig 2E) (Nizami et al, 2010). Noteworthy, the effects of (PR)20 are not restricted to cellular RNAs as the intracellular replication of Sindibis virus, which contains a single‐stranded RNA genome, in Baby Hamster Kidney cells (BHK‐21) was also impaired by the presence of (PR)20 (Fig 2F), further supporting the notion that (PR) DPRs have a general effect on cellular reactions using RNA substrates.
Figure 2. (PR)20 peptides impair reactions using RNA substrates in vitro and in vivo .
-
AReverse transcription of RNA (500 ng) with oligo dT in the presence of increasing doses of (PR)20. Data represent the fluorometric quantification of the resultant cDNA (n = 3), mean values ± SD.
-
BPercentage of RNA (1 μg) remaining after a 15′ digestion with increasing doses of RNase A in the presence or absence of (PR)20 (5 μM; n = 3). Data represent mean values ± SEM.
-
C, DHigh‐throughput microscopy (HTM)‐mediated analysis of 5‐ethynyl‐uridine (EU) (C) and polyA+ mRNA (D) levels per nucleus found in U2OS cells exposed to 10 μM (PR)20 for 16 h. EU was added 30′ prior to fixation (n = 3). Black lines indicate median values in both panels. In (D), boxes define upper and lower quartiles and whiskers mark the highest and lowest observations.
-
EEffect of (PR)20 (10 μM, 16 h) in the number of Cajal Bodies (CB) identified by immunofluorescence with anti‐COILIN and anti‐SMN antibodies (n = 3). Data represent mean values ± SD.
-
FAccumulation of Sindibis virus (SV) genomic RNA in BHK‐21 cells in the absence or presence of increasing doses of (PR)20. The amount of viral and cellular RNA was quantified from total RNA isolated 4 h after infection by quantitative real‐time PCR (qRT–PCR) with specific primers (n = 3). Data represent mean values ± SEM.
Data information: **P < 0.01; ***P < 0.001; ****P < 0.0001; t‐test. Representative images from these analyses (C–E) are provided in each case. Scale bar (white) indicates 2–5 μm. See also Appendix Fig S1.
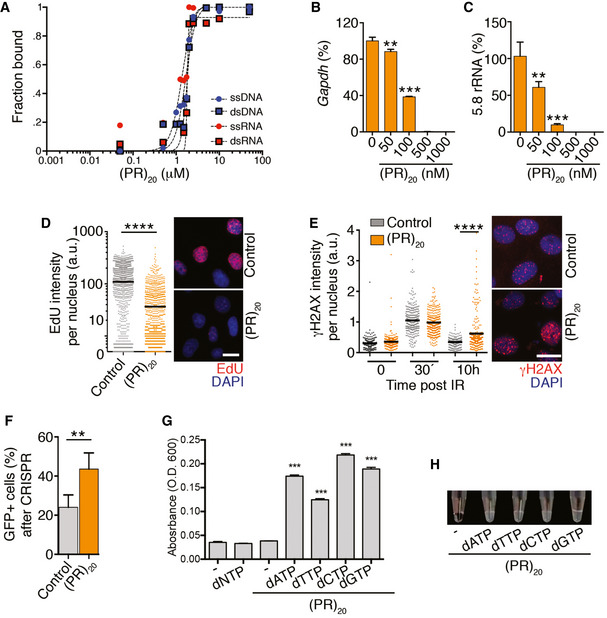
In addition to RNA, arginine‐rich peptides are also known to bind avidly to DNA and promote its compaction (Tan & Frankel, 1995; Mascotti & Lohman, 1997; DeRouchey et al, 2013). Consistently, electrophoretic mobility assays (EMSA) revealed a similar affinity of (PR)20 for DNA and RNA in single‐ or double‐stranded forms (Fig 3A). We thus explored the impact of (PR)20 in reactions using DNA substrates. In vitro, (PR)20 and (GR)20, but not BSA or (GA)20, inhibited DNA polymerase chain reactions in a dose‐dependent manner (Fig 3B and C and Appendix Fig S1C). When added to U2OS, the presence of (PR)20 reduced DNA replication rates (Fig 3D) and impaired the repair of DNA breaks generated by ionizing radiation as measured with antibodies detecting the phosphorylated form of histone H2AX (γH2AX; Fig 3E). The efficiency of gene deletion of an EGFP cDNA by CRISPR/Cas9 using a sgRNA against EGFP was also significantly affected by (PR)20 (Fig 3F), although this observation could reflect effects of the peptide on DNA and/or RNA. Interestingly, (PR)20 also bound to dNTPs in vitro, which could certainly contribute to the effects of the peptide on nucleic acid reactions (Fig 3G and H). In contrast to the widespread effects of arginine‐rich peptides on RNA and DNA metabolism, the presence of (PR)20 did not affect the efficiency of biochemical reactions not using nucleic acids such as an in vitro phosphatase assay (Appendix Fig S2) or the previously mentioned in vitro assembly of 80S ribosomal particles from purified 40S and 60S subunits (Appendix Fig S1E and F). Collectively, the experiments presented above revealed that arginine‐rich DPRs have a general effect in limiting cellular reactions that use nucleic acid substrates.
Figure 3. Effects of (PR)20 on DNA‐based reactions.
-
AQuantification of EMSA assays evaluating the binding of (PR)20 to 19 nt‐long ssDNA, ssRNA, dsDNA and dsRNA molecules. Each probe (0.2 μM) was incubated with increasing concentrations of (PR)20 for 10′. Curve‐fitting was performed using non‐linear regression with the Hill equation.
-
B, CPercentage of GAPDH or 5.8 rRNA levels quantified by qPCR in reactions containing increasing doses of (PR)20 (n = 3).Data represent mean values ± SEM.
-
DHTM‐mediated analysis of 5‐ethynyl‐deoxyuridine (EdU) levels per nucleus found in U2OS cells exposed to 10 μM (PR)20 for 4 h. EdU was added 30′ prior to fixation. Representative images from these analyses are provided (right). Black lines indicate median values. Scale bar (white) indicates 5 μm.
-
EHTM‐mediated analysis of γH2AX levels per nucleus in U2OS cells exposed to 3 Gy of IR in the presence or absence of 10 μM (PR)20 for the indicated times. Note the accumulation of cells with persistent DNA damage (γH2AX) 10 h after IR in the presence of the DPR. Representative images from these analyses are provided (right). Black lines indicate median values. Scale bar (white) indicates 5 μm.
-
FEfficiency of CRISPR‐mediated gene deletion quantified in mouse embryonic stem cells stably expressing EGFP, a doxycycline‐inducible Cas9 and an EGFP‐targeting sgRNA. The percentage of EGFP‐positive cells was quantified by flow cytometry 72 h after inducing Cas9 expression with doxycycline in the presence or absence of 10 μM (PR)20 (n = 3). Data represent mean values ± SEM.
-
G, HTurbidity of solutions containing (PR)20 mixed with the different dNTPs measured by the optical density at 600 nm (O.D. 600) (G) (n = 3). Data represent mean values ± SEM. A representative image of these assays is provided in (H).
Data information: **P < 0.01; ***P < 0.001; ****P < 0.0001; t‐test. See also Appendix Fig S2.
Protamine recapitulates the cellular and biochemical effects of (PR)20 and (GR)20
We next explored if the effects observed with (PR)20 or (GR)20 peptides would be similar to those triggered by other arginine‐rich CPPs. To this end, we evaluated the effects of protamine, a sperm‐specific polypeptide that has the highest percentage of arginine content from the animal proteome (24 or its 51 aminoacids are arginines) (Balhorn, 2007). Like (PR)20 or (GR)20, protamine is also a CPP (Scheicher et al, 2015) and is known to be toxic for poorly understood reasons (Sokolowska et al, 2016). Interestingly, while protamine is known for its DNA‐binding properties during spermatogenesis, EMSA assays revealed a similar affinity of protamine for DNA and RNA (Fig 4A). In addition, and like (PR)20 or (GR)20, protamine had an inhibitory effect on DNA‐ or RNA‐based reactions such as PCR and RNA degradation (Fig 4B and C). In what regards to the cellular effects of protamine, these were remarkably similar to those associated with (PR)20 and (GR)20 peptides. Accordingly, exogenously added protamine entered into U2OS cells, accumulated at nucleoli as evidenced by its colocalization with UBF1 (Fig 4D) and reduced transcription and translation rates (Fig 4E and F). Exposure to protamine also triggered abnormal splicing (Fig 4G), an effect that was previously observed with (PR)20 (Kwon et al, 2014), and that we could also detect with (PR)20 and (GR)20 but not (GA)20 or (PK)20 peptides (Fig 4H). In summary, the widespread cellular effects on nucleic acid metabolism that have been associated with ALS‐associated (PR) or (GR) DPRs are a general property of arginine‐rich CPPs.
Figure 4. Protamine inhibits reactions using nucleic acid templates.
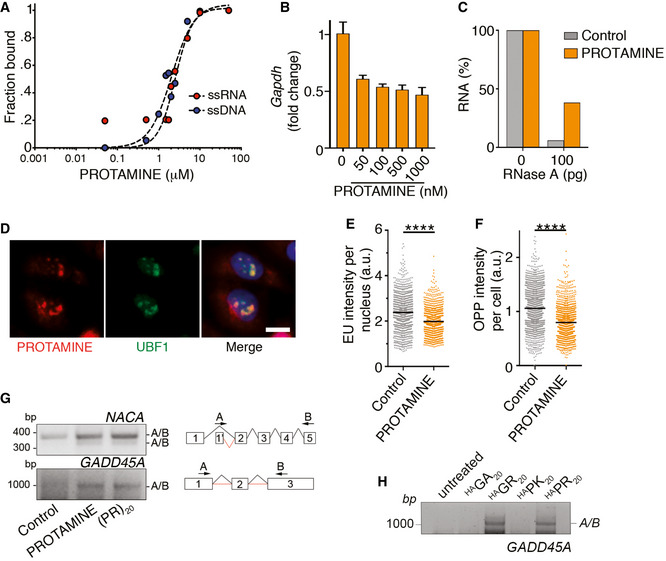
-
AQuantification of EMSA assays evaluating the binding of protamine to 19 nt‐long ssDNA and ssRNA molecules. Each probe (0.2 μM) was incubated with increasing concentrations of (PR)20 for 10′. Curve fitting was performed using non‐linear regression with the Hill equation.
-
BPercentage of GAPDH levels quantified by qPCR in reactions containing increasing doses of protamine (n = 3). Data represent mean values ± SEM.
-
CPercentage of RNA (1 μg) remaining after a 15′ digestion with increasing doses of RNase A in the presence or absence of protamine (5 μM). Data derive from one representative experiment that was repeated twice.
-
DImmunofluorescence of Cy3‐labelled salmon protamine (red) and the nucleolar factor UBF1 (green) in U2OS cells treated with 30 μM Cy3‐protamine for 4 h. Scale bar (white) indicates 2.5 μm.
-
E, FHTM‐mediated quantification of the EU levels per nucleus (E) and O‐propargyl‐puromycin (OPP) levels per cell (F) in U2OS cells exposed to 30 μM protamine for 16 (E) or 24 (F) h. EU and OPP were added 30′ prior to fixation. Black lines indicate median values (****P < 0.0001; t‐test).
-
GRT–PCR of NACA and GADD45A mRNAs in U2OS cells treated with protamine (30 μM) or (PR)20 (20 μM) using primers in non‐consecutive exons to monitor alternative splicing events. A scheme of the primers used is provided on the right side of the panel.
-
HRT–PCR of GADD45A mRNAs in U2OS cells treated with HA‐tagged (PR)20, (GR)20, (PK)20 and (GA)20 peptides (20 μM) using the pipeline defined in (G).
Exposure to arginine‐rich CPPs leads to a generalized displacement of RNA‐ and DNA‐binding factors
The biological function of protamine is to bind all genomic DNA, without any particular sequence bias, where it displaces histones and High Mobility Group (HMG) proteins from chromatin allowing a higher compaction of the sperm genome. Given the similar phenotypes triggered by protamine and arginine‐rich CPPs in somatic cells, we reasoned that these could be due to a widespread effect of the DPRs in displacing DNA‐binding factors from chromatin. Consistent with this hypothesis, proteomic analyses showed a highly significant overlap in the changes that are induced by (PR)20 or protamine on chromatin‐bound factors (Fig 5A and B). Moreover, linker histone H1 variants or High Mobility Group (HMG) proteins were among the proteins that showed a highest reduction in their chromatin‐bound levels upon (PR)20 or protamine exposure, further supporting that (PR)20 peptides phenocopy the cellular effects of protamine (Appendix Table S2). Next, and given that protamine and (PR)20 also bind to RNA (Figs 2A and 4A), we evaluated whether these peptides had a similar effect on displacing RNA‐binding factors from cellular RNAs. Indeed, and through the use of a proteomic pipeline that allows an analysis of the RNA‐bound proteome (Castello et al, 2013), we observed that both protamine and (PR)20 exposure led to a generalized reduction in the amount of RNA‐binding factors that were bound to RNA (Fig 5C). In addition, and similar to what happens on chromatin, there was a highly significant correlation between the factors that were displaced from RNA upon treatment with protamine or (PR)20 (Fig 5D and Appendix Table S3). Interestingly, the list of proteins that are displaced from RNA by both peptides included well‐known ALS‐associated RNA‐binding proteins such as TARDBP (TDP‐43) or FUS. In summary, the presence of arginine‐rich peptides such as protamine or (PR)20 leads to a generalized displacement of RNA‐ and DNA‐binding factors from cellular RNA and chromatin, providing a unifying mechanism that explains the widespread defects in nucleic acid metabolism that are observed upon exposure to these peptides.
Figure 5. Arginine‐rich peptides trigger a widespread displacement of proteins from chromatin and RNA.
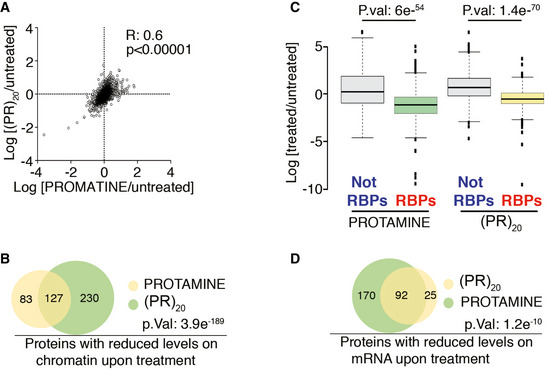
- Distribution of the chromatin‐bound levels of all proteins identified by proteomics in U2OS cells treated with protamine (30 μM) or (PR)20 (20 μM) for 90′. Numbers indicate the Pearson's correlation coefficient (R) and the P‐value, which was obtained by a t‐test to evaluate the Pearson correlation between both samples.
- Venn diagram illustrating the overlap among the proteins that show reduced levels on chromatin upon treatment of U2OS cells with protamine (30 μM) or (PR)20 (20 μM) for 90′. The P‐value was obtained by a hypergeometric test. See also Table S2.
- Distribution of the levels of all proteins identified after isolation of the RNA‐bound proteome in U2OS cells treated with protamine (30 μM) or (PR)20 (20 μM) for 180′. RNA‐binding proteins (RBPs) were identified as those significantly enriched after the isolation of mRNA‐binding factors. Note that treatment with either peptide leads to a selective reduction in the levels of RBPs bound to mRNA that is not observed for the rest of the factors (Not RBPs) (n = 2). P values were calculated with two‐sided two‐sample t‐tests assuming equal variances. Center lines indicate median values; box limits indicate the 25th and 75th percentiles; whiskers extend 1.5 times the interquantile range from the 25th and 75th percentiles and outliers are represented by dots.
- Venn diagram illustrating the overlap among the proteins that show reduced levels on mRNA upon treatment of U2OS cells with protamine (30 μM) or (PR)20 (20 μM) for 90′. The P‐value was obtained by a hypergeometric test. See also Appendix Table S2.
Non‐coding oligonucleotides or heparin rescue of the toxic effects of arginine‐rich CPPs
Based on the model of generalized DNA and RNA binding, we investigated whether the effects of arginine‐rich peptides could be alleviated by the presence of non‐coding oligonucleotides that could scavenge the DPR and thus limit the amount of free peptide that is able to interfere with nucleic acid metabolism. In support of this hypothesis, addition of a 646 nucleotide (nt) RNA‐lacking ATG rescued the effect of (PR)20 on in vitro translation reactions in a dose‐dependent manner (Fig 6A). Similarly, a non‐coding ssDNA oligonucleotide rescued the effects of (PR)20 peptides in translation (Fig 6B) and reduced the toxicity of the DPR in U2OS cells (Fig 6C). Importantly, oligonucleotides did not prevent the entry of (PR)20 into the nucleus or its accumulation at nucleoli (Fig 6D and Appendix Fig S3A). On the contrary, the presence of the DPR enabled the entry of ssDNA and, to a lesser extent, ssRNA, into cells, and their accumulation at nucleoli (Fig 6E and Appendix Fig S3B). Similar observations were made with (GR)20 but not with (GA)20 or (PK)20 peptides (Appendix Fig S3C–F). The data from (PK)20 peptides are particularly interesting as even if this peptide behaves as a CPP and enters into cells, it does not accumulate at nucleoli nor has the capacity to transfect oligonucleotides into the cell, highlighting the unique affinity of arginines for nucleic acids. Of note, arginine‐rich histone fractions and protamine were the first transfection reagent ever discovered (Smull & Ludwig, 1962), and arginine‐rich CPPs have in fact been developed as non‐lipidic transfection reagents for decades (Emi et al, 1997; Schwarze et al, 1999). Interestingly, the capacity to transfect nucleic acids correlates with toxicity as protamine, (PR)20 and (GR)20 but not (GA)20 or (PK)20 displayed toxicity in clonogenic assays in U2OS cells (Appendix Fig S3G).
Figure 6. Non‐coding oligonucleotides rescue the effects of arginine‐rich CPPs.
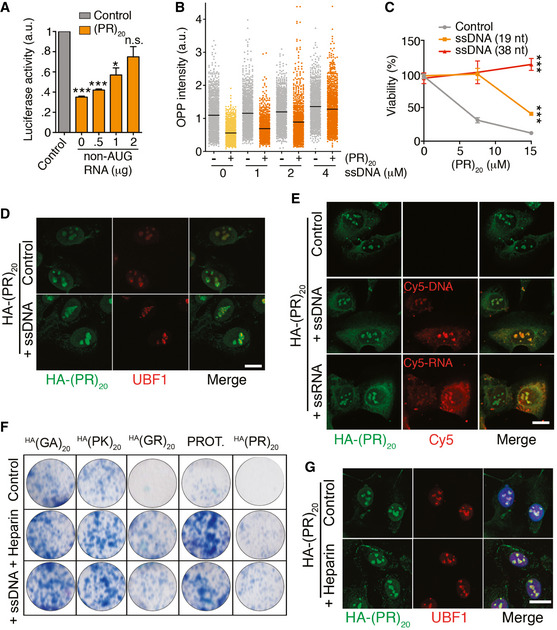
- In vitro translation of a luciferase mRNA with or without of 0.5 μM (PR)20 and in the presence of increasing amounts of a 646 nt non‐coding RNA (n = 3). Data represent mean values ± SEM.
- HTM‐mediated quantification of OPP levels per cell (PR)20 (10 μM, 16 h) alone or together with a 38 nt ssDNA oligonucleotide at the indicated doses. Black lines indicate median values.
- Percentage of viable cells as evaluated with a CellTiter‐Glo luminescent assay in U2OS cells treated with increasing doses of (PR)20 alone or together with 2 μM of ssDNA oligonucleotides (19 or 38 nt; n = 3). Data represent mean values ± SEM.
- Immunofluorescence of HA‐(PR)20 (green) and the nucleolar factor UBF1 (red) in U2OS cells treated with 7.5 μM HA‐(PR)20 alone or together with 2 μM of a 19 nt ssDNA oligonucleotide for 8 h.
- Immunofluorescence of HA‐(PR)20 (green) and Cy5 (red) in U2OS cells treated with 7.5 μM HA‐(PR)20 alone or together with 4 μM of Cy5‐labeled 19 nt ssDNA or ssRNA oligonucleotides for 8 h.
- Clonogenic survival assay of U2OS cells exposed to 7.5 μM with the indicated peptides or protamine (PROT.) alone or together with 2 μM of a 38 nt ssDNA oligonucleotide or 0.5 μM Heparin.
- Immunofluorescence of HA‐(PR)20 (green) and the nucleolar factor UBF1 (red) in U2OS cells treated with 7.5 μM HA‐(PR)20 alone or together with heparin (0.5 μM).
Data information: See also Appendix Fig S3. Scale bar (white) in (D), (E) and (G) indicates 2.5 μm. *P < 0.1; ***P < 0.001; t‐test. See Appendix Fig S3 for an HTM‐mediated quantification of the image analyses.
The properties of protamine gave us an additional idea as to how to rescue the toxicity of arginine‐rich CPPs. Since protamine is clinically used as antidote of the anticoagulant heparin, we tested whether heparin could revert the toxic effects of protamine and other arginine‐rich CPPs. In fact, heparin rescued the viability of U2OS cells exposed to protamine, (PR)20, and (GR)20 in clonogenic assays (Fig 6F). However, and in contrast to the effects of oligonucleotides, the use of heparin partly reduced the amount of the DPR that entered into cells (Appendix Fig S3H). We believe that the effects of heparin could reflect the fact that cell entry of CPPs is thought to be initiated by binding of the peptides to heparanated receptors on the cell surface (Borrelli et al, 2018). Consistently, in the presence of heparin some of the peptide accumulated at the cell membrane (Fig 6G).
Our model of generalized DNA and RNA binding by arginine‐rich CPPs implies that the mechanism of their toxicity should be similar regardless of the cell type. Nevertheless, since (PR) and (GR) DPR toxicity have been associated with ALS, we finally tested the effects of heparine and ssDNA oligonucleotides in two independent models of motoneurons: motoneurons differentiated from mouse embryonic stem cells (mMN) and differentiated NSC34 cells, which derive from a fusion of mouse spinal cord motoneurons with mouse neuroblastoma (NSC34DIFF). Exposure to protamine or (PR)20 was toxic on both models, whereas (GA)20 or (PK)20 peptides were not (Appendix Fig S4). Moreover, and as in U2OS, heparin or a 38 nt‐long ssDNA oligonucleotide significantly rescued the toxicity of (PR)20 peptides in mMN and NSC34DIFF cells (Fig 7A–F). Both strategies also rescued neurite lengths motoneurons, which were severely reduced by the CPP (Fig 7A and D). Collectively, these experiments illustrate that the cellular toxicity of arginine‐rich CPPs can be alleviated by heparin or non‐coding oligonucleotides in several cell types, including motoneurons.
Figure 7. Rescue of the toxicity of (PR)20 in mouse motor neurons.
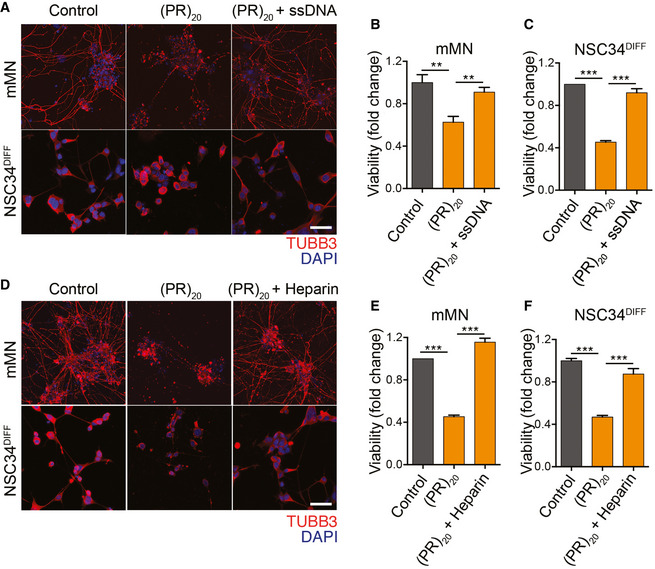
-
AImmunofluorescence of the neuronal marker TUBB3 (red) in mouse motor neurons differentiated from mESC (mMN) or from NSC34 cells (NSC34DIFF) exposed to 2.5 μM (PR)20 for 24 and 48 h, respectively, with or without 2 μM of a non‐coding 38 nt oligonucleotide. Nuclei were stained with DAPI (blue). Scale bar (white), 10 μm. Note that besides its impact on overall survival, the presence of the oligonucleotide also had a marked effect in rescuing neurite lengths that had been shortened by the peptide.
-
B, CCell viability quantified CellTiter‐Glo luminescence assays mMN (B) and NSC34DIFF (C) treated as in (A) (n = 3). Data represent mean values ± SEM.
-
DImmunofluorescence of the neuronal marker TUBB3 (red) in mouse motor neurons differentiated from mESC (mMN) or from NSC34 cells (NSC34DIFF) exposed to 2.5 μM (PR)20 for 24 and 48 h, respectively, with or without heparin (0.5 μM). Nuclei were stained with DAPI (blue). Scale bar (white), 10 μm.
-
E, FCell viability quantified CellTiter‐Glo luminescence assays mMN (E) and NSC34DIFF (F) treated as in (D) (n = 3). Data represent mean values ± SEM.
Data information: **P < 0.01; ***P < 0.001. See also Appendix Fig S4.
Discussion
Facilitating the delivery of nucleic acids or proteins into cells is an intense area of biomedical research, particularly in these days where the use of biological agents is gaining momentum for the treatment of human disease (Bruce & McNaughton, 2017). In this context, while arginine‐rich oligopeptides are among the most efficient CPPs, their clinical applications have been limited by their toxicity as first noticed during the medical use of protamine (Sokolowska et al, 2016). Interestingly, the toxic properties of arginine‐rich CPPs have also been exploited in the animal kingdom. For instance, PR‐39 is an antibacterial arginine‐rich peptide that is present in the small intestine of pigs, the toxicity of which was early on associated to its effects on DNA synthesis and translation (Boman et al, 1993). The discovery of toxic arginine‐rich CPPs in a subset of ALS/FTD patients has now made the study of this phenomenon even more relevant. Still, identifying the fundamental mechanism by which arginine‐rich CPPs kill cells has remained elusive.
The work presented here provides a unifying mechanism to explain the toxic effects of arginine‐rich CPPs. We propose that the high affinity of arginine‐rich peptides for RNA or DNA leads to a widespread coating of nucleic acids in cells, with a consequent generalized displacement of RNA‐ or DNA‐binding factors from chromatin and RNA. This model implies that any reaction using nucleic acid substrates such as RNA transcription, splicing, and translation or DNA replication and repair will be affected by arginine‐rich CPPs, and in fact alterations on all of these reactions have been linked to ALS/FTD (Hardiman et al, 2017). Based on the remarkable similarities that we found on the effects of protamine and (PR)20 peptides in displacing factors from chromatin and RNA, we propose that the cellular effects driven by (PR)n and (GR)n peptides in cells from ALS/FTD patients would be equivalent to what could be expected if protamine were to be accidentally expressed in adult motoneurons. Of note, the fact that arginine‐rich CPPs also bind to nucleotides can certainly contribute to the effects of these peptides in nucleic acid metabolism and should be taken into consideration when analyzing their cellular effects.
Our model helps to further understand a number of observations that have been linked to the biology of arginine‐rich DPRs. For instance, proteomic studies have consistently found an enrichment of RNA‐binding factors among (PR)n interactors (Kanekura et al, 2016; Lin et al, 2016; Yin et al, 2017). However, the fact that (PR)20 and (GR)20 peptides protect RNA from degradation might affect the interpretation of these studies, since a control step during the purification of protein–protein interactions is to degrade RNA in order to exclude indirect interactions that mediated by binding to the same RNA. We thus believe that RNA molecules—and not proteins—are the primary target of arginine‐rich CPPs, which in addition explains why the peptides accumulate at nucleoli as this is where the highest concentration of RNA exists in cells. The very high affinity of arginine‐rich peptides for RNA should also contribute to the reported property of these CPPs to undergo phase separation (Boeynaems et al, 2017; Jafarinia et al, 2020), as these reactions are seeded by RNA. In addition to their own phase separation, C9ORF72‐associated argine‐rich DPRs perturb the integrity of membrane‐less organelles such as the nucleolus or stress granules, which we reason can be driven by displacing the RNA‐binding proteins that mediate the phase separation of these structures (Lee et al, 2016; Boeynaems et al, 2017; White et al, 2019). Besides RNA interactions, the very high affinity or arginine‐rich CPPs for DNA can also contribute to their impact on chromatin. For example, a GFP‐(PR)50 peptide was found to accumulate at heterochromatin leading to the loss of HP1α (Zhang et al, 2019), which was proposed to be due to a specific interaction of (PR)50 peptides with HP1α. We believe however that this observation reflects a more general effect of arginine‐rich peptides on displacing chromatin‐bound factors, which is consistent with our proteomic analysis indicating that histones and HMG proteins are among the top factors that are displaced by protamine or (PR)20 peptides.
Finally, in support of our model of nucleic acid coating by arginine‐rich CPPs, we here show that their toxicity can be alleviated by non‐coding oligonucleotides that can scavenge the peptides and limit their effective concentration available to interact with cellular nucleic acids. While heparin also limits the toxicity of arginine‐rich peptides, it is important to note that it also affects their cellular uptake. This observation could nevertheless be relevant in the context of the paracrine toxic effects that have been reported for C9ORF72‐derived DPRs, although the relevance of this phenomenon to the disease remains to be proven (Westergard et al, 2016; Zhou et al, 2017). Paracrine toxicity of arginine‐rich peptides has also been proposed to contribute to AIDS‐related neurodegeneration driven by the arginine‐rich TAT protein from the HIV‐1 virus (King et al, 2006). In any case, we want to end our discussion with a clear statement in that by no means we want our results to be interpreted as a suggestion for therapy in ALS/FTD or any other disease. We simply used these experiments to provide further support to the model that the toxicity of arginine‐rich CPPs is due to widespread DNA and RNA binding. Collectively, the experiments presented here provide a unifying mechanism that explains the widespread effects of arginine‐rich CPPs in nucleic acid metabolism and provides some initial proof‐of‐principle ideas as to how this knowledge could be used to limit their toxicity in mammalian cells.
Materials and Methods
Cell culture
U2OS, BHK‐21, RPE, HeLa‐SBP, HeLa‐RPS9SBP, and NSC34 cells stably expressing RPS9‐Flag‐TEV‐SBP were cultivated in standard DMEM medium supplemented with 10% FBS, 2 mM l‐glutamine, and 1% penicillin/streptomycin. U2OS, BHK‐21, RPE, and NSC34 cells were acquired from ATCC as the parental HeLa from which stable transfectants were made. mES and the previously described mouse ESCas9 cells (Ruiz et al, 2016) were grown on gelatin in DMEM (high glucose) supplemented with 15% knockout serum replacement (Invitrogen), LIF (1,000 U/ml), 0.1 mM non‐essential amino acids, 1% glutamax, and 55 mM β‐mercaptoethanol. All cell lines are regularly tested for mycoplasma contamination.
Motor neuron differentiation
For NSC34 differentiation, cells were seeded onto plates containing DMEM supplemented with 10% FBS, 2 mM l‐glutamine, and 1% penicillin/streptomycin. On the next day, the media was exchanged to Neurobasal medium (Thermo Fisher scientific, 21103049) containing a B‐27 supplement (Thermo Fisher scientific, 17504044) for 48h. For the differentiation of spinal motor neurons from mESCs, we used the previously described protocol (Wichterle et al, 2002). Briefly, embryo bodies (EBs) were generated by culturing mESCs in suspension in non‐adherent plates under constant shaking over 2 days. Differentiation was promoted by exposing EBs to the smoothened agonist SAG (500 nM, PeproTech #9128694) and retinoic acid (100 nM, Sigma # R2625‐100MG) for 5 days, at the end of which they were dissociated and plated in 96‐well plates at 200,000 cells/cm2. The plates were previously coated with poly‐l‐ornithine (Sigma‐Merck #A‐004‐C), followed by laminin (Sigma # L2020‐1MG). mMN cultures were maintained in medium supplemented with GDNF and BDNF (10 ng/ml, PeproTech, #450‐02, #450‐10).
Peptides and oligonucleotides
(PR)20, (GR)20, (PK)20, and (GA)20 dipeptide repeats with a C‐terminal HA epitope tag were synthesized at Genscript. Protamine isolated from salmon sperm was obtained from Sigma (P4005). Fluorophore‐labelled oligonucleotides were synthesized by Sigma, with the following sequences: Cy3‐5′‐DNA (CCACTGCACCGCTGCTAGG); Cy5‐5′‐DNA (CCTAGCAGCGGTTGCAGTGG); Cy3‐5′‐RNA (CCACUGCACCGCUGCUAGG); and Cy5‐RNA (CCUAGCAGCGGUUGCAGUGG).
Immunofluorescence and high throughput microscopy
For immunofluorescence using HA (Roche; #11867423001), COILIN (kind gift of Angus Lamond), TUBB3 (Biolegend; #801202), SMN (Santa Cruz; #sc‐32313), and UBF (Santa Cruz; #sc‐13125) antibodies, cells were fixed with 4% PFA prepared in PHEM buffer (60 mM Pipes, 25 mM Hepes, 10 mM EGTA, 2 mM MgCl2 pH 6.9) containing 0.2% of Triton X‐100, and permeabilized with 0.5% Triton X‐100 after fixation. For HTM, cells were grown on μCLEAR bottom 96‐well plates (Greiner Bio‐One) and immunofluorescence of γH2AX (Millipore; #05‐636) was performed using standard procedures. Analysis of DNA replication by EdU, transcription by EU, and translation by OPP incorporation were done using Click‐It kits (Invitrogen) following manufacturer's instructions. All secondary antibodies were from Life Technologies (Anti‐Mouse IgG‐488; #A11029, Anti‐Mouse IgG‐555; #A21422, Anti‐Rabbit IgG‐488; #A11008, Anti‐Rabbit IgG‐555; #A31572). In all cases, images were automatically acquired from each well using an Opera High‐Content Screening System (Perkin Elmer). A 20× or 40× magnification lens was used, and images were taken at non‐saturating conditions. Images were segmented using DAPI signals to generate masks matching cell nuclei from which the mean signals for the rest of the stainings were calculated.
Cell viability
4,000 cells were seeded per well in a 96‐well tissue culture plate and treated with the indicated concentrations of (PR)20 alone or together with 4 μM of 19 or 38 nt‐long Cy5‐RNA or Cy5‐DNA oligonucleotides. 36 h after the treatment, cell viability was measured using a luminescent system (CellTiter‐Glo, Promega), according to the manufacturer's protocol. Viability is plotted as percentages compared with untreated controls.
Clonogenic assays
2,000 U2OS cells were plated in six‐well tissue culture plates in culture medium. The following day, cells were incubated with 10 μM (PR)20 alone or in combination with 2 μM of 19 (CCACTGCACCGCTGCTAGG) or 38 (CCACTGCACCGCTGCTAGGATCGCCTGAAATCGTTGGC) nucleotide‐length ssDNA molecules. After 2 days, the medium was changed and cells were then grown for 8 additional days in untreated medium. At the end of the experiments, cells were fixed and stained with 0.4% methylene blue in methanol for 30 min.
Peptide labeling
The Cy®3 Mono 5‐pack kit (Sigma) was used for the labeling of protamine. Briefly, 0.150 mM salmon protamine was incubated with 0.850 mM of Cy3 NHS Ester in 20 mM Hepes and 150 mM NaCl buffer overnight at 4°C, followed by the addition of 1 M Tris pH 7.5 in order to quench the reaction. Cy3‐protamine was purified using 3K Amicon Ultra 0.5 ml centrifugal filters (Sigma).
Immunoprecipitation
10 × 106 Hela‐SBP and Hela RPS9SBP cells were lysed in cold RNA‐IP lysis buffer (50 mM Tris pH 8.0, 150 mM NaCl, 1 mM MgCl2, 2% NP‐40, 10% glycerol, and freshly added Complete protease inhibitors) and incubated with 30 μl of Strep‐Tactin beads (IBA) for 1 h at 4°C with constant shaking. Beads were washed 6 times in NET2 buffer (50 mM Tris pH 7.5, 150 mM NaCl, 0.5% Triton X‐100).
Chromatin fractionation
After treatment with PR20 or protamine, cells were washed twice with ice‐cold phosphate‐buffered saline (PBS), resuspended in 180 μl of ice‐cold hypotonic lysis buffer (10 mM HEPES, pH 7.9, 10 mM KCl, 0.1 mM EDTA containing protease and phosphatase inhibitors), and incubated on ice for 10 min, followed by addition of 20 μl of Nonidet P‐40; after 3 min at room temperature, cells were vortexed and the cytosolic fraction was obtained by centrifugation for 5 min at 2,500 g. The nuclear pellet was resuspended in high‐salt‐concentration extraction buffer (20 mM HEPES, pH 7.9, 0.4 M NaCl, 1 mM EDTA containing protease and phosphatase inhibitors) and incubated with shaking at 4°C for 1 h. For chromatin fraction, the pellet was then extracted in 50 mM Tris, pH 7.5, 8 M urea, and 1% CHAPS.
Isolation of RNA‐binding proteins
The systems‐wide purification of RNA‐binding proteins was developed as performed reported (Castello et al, 2013). Briefly, 5 × 150 cm2 dishes of 60% confluent U2OS cells were treated for 4 h with 20 μM of PR20 or Protamine. RBPs and polyadenylated RNAs were crosslinked by irradiating the cells with 0.15 J/cm2 (~ 1 min) at 254 nm UV, and non‐irradiated cells were used as a control. Cells were lysated and proteins covalently to mRNA were captured with oligo(dT) magnetic beads. After stringent washes, the mRNA interactome was determined by quantitative mass spectrometry (MS).
Mass spectrometry
IP samples were eluted in urea, FASP‐digested with Lys‐C/trypsin, and analyzed by LC‐MS/MS. Label‐free quantification was performed using MaxQuant. Chromatin and whole‐cell extract samples were trypsin‐digested using S‐traps, isobaric‐labeled with iTRAQ 8‐plex (chromatin) or TMT 11‐plex (whole cell) reagents and pre‐fractionated with RP‐HPLC at high pH. Fractions were then analyzed by LC‐MS/MS, and raw data were processed with MaxQuant. Statistical analyses were performed with Perseus and ProStar.
For RNA‐bound proteomics, proteins were digested by means of standard FASP protocol. Briefly, proteins were reduced (15 mM TCEP, 30 min, RT), alkylated (50 mM CAA, 20 min in the dark, RT), and sequentially digested with Lys‐C (Wako; o/n at RT) and trypsin (Promega; 6 h at 37°C). Resulting peptides were desalted using Sep‐Pak C18 cartridges (Waters). LC‐MS/MS was done by coupling an UltiMate 3000 HPLC system to a Q Exactive Plus mass spectrometer (Thermo Fisher Scientific). Peptides were loaded into a trap column Acclaim™ PepMap™ 100 C18 LC Columns 5 μm, 20 mm length) for 3 min at a flow rate of 10 μl/min in 0.1% FA. Then, peptides were transferred to an analytical column (PepMap RSLC C18 2 μm, 75 μm × 50 cm) and separated using a 89 min effective curved gradient (buffer A: 0.1% FA; buffer B: 100% ACN, 0.1% FA) at a flow rate of 250 nl/min from 2 to 42.5% of buffer B. The mass spectrometer was operated in a data‐dependent mode, with an automatic switch between MS (350–1,400 m/z) and MS/MS scans using a top 15 method (intensity threshold signal ≥ 3.8e4, z ≥ 2). An active exclusion of 26.3 s was used. Peptides were isolated using a 2 Th window and fragmented using higher‐energy collisional dissociation (HCD) with a normalized collision energy of 27. Protein interaction networks arising from proteomic experiments were analyses with STRING (Szklarczyk et al, 2015).
Polysome analyses
Hela RPS9SBP cells were treated with either water as control or 10 μM of (PR)20 during 16 h. Ribosomes were stalled by addition of 100 μg/ml cycloheximide (CHX) for 5 min, and cells were lysed in polysome lysis buffer (15 mM Tris–HCl pH 7.4, 15 mM MgCl2, 300 mM NaCl, 1% Triton X‐100, 0.1% β‐mercaptoethanol, 200 U/ml RNAsin [Promega], 1 complete Mini Protease Inhibitor Tablet [Roche] per 10 ml). Nuclei were removed by centrifugation (9,300 g, 4°C, 10 min), and the cytoplasmic lysate was loaded onto a sucrose density gradient (17.5–50% in 15 mM Tris–HCl pH 7.4, 15 mM MgCl2, 300 mM NaCl and, for fractionation from BMDM, 200 U/ml Recombinant RNAsin Ribonuclease Inhibitor, Promega). After ultracentrifugation (2.5 h, 126,000 g at 4°C in a SW60Ti rotor), gradients were eluted with a Teledyne Isco Foxy Jr. system into 16 fractions of similar volume.
In vitro ribosome assembly
Ribosomal subunits were purified from BHK‐21 cells using previously described procedures (Pisarev et al, 2007). Briefly, the ribosomal fraction (P100) was treated with 1 mM puromycin for 15 min on ice and then for 15 min at 37ºC. The suspension was adjusted to 0.5 M KCl and loaded onto 10 to 30% sucrose density gradients which were centrifuged at 61,255 g for 16 h at 4ºC in a SW40 rotor. The peaks corresponding to 40S and 60S subunits were identified by optical density at 260 nm. Finally, fractions were concentrated using YM‐100 centricons. 1 pmol of 40S and 60S were incubated in assembly buffer (25 mM of Tris–HCl pH7.5, 100 mM KCl, 5 mM MgCl2, and 3mM of DTT) for 45 min at 30ºC. To control for non‐assembly, the concentration of MgCl2 in the buffer was reduced to 1 mM.
Electron microscopy
For negative staining, 3 μl of purified 40S + 60S ribosome fractions or alternatively 3 μl of 40S + 60S ribosome fractions in the presence of (PR)20 (1:10 molar ratio) were deposited onto freshly glow‐discharged carbon‐coated 400 mesh copper electron microscopy (EM) grids (Electron Microscopy Sciences) and retained on the grid for 1 min. Afterward, grids were washed with three distinct 50 μl drops of MilliQ water. The excess of water was removed with filter paper, and the grids were placed on the top of three different 40 μl drops of 2% uranyl acetate and stained on the last drop for 1 min. The grids were then gently stripped for 4 s and air dried. Finally, grids were visualized on a Tecnai 12 transmission electron microscope (Thermo Fisher Scientific Inc.) with a lanthanum hexaboride cathode operated at 120 keV. Images were recorded at 61,320 nominal magnification on a 4kx4k TemCam‐F416 CMOS camera (TVIPS).
In vitro translation
In vitro translation was performed in nuclease‐treated rabbit reticulocyte lysates (RRL; Promega cat #L4960) using a luciferase mRNAs in the presence of the indicated concentrations of (PR)20. Reactions were incubated for 60 min at 30ºC, and a 5 μl alicuot was used to measure luciferase activity in 100 μl of reaction buffer (15 mM K2HPO4, 15 mM MgSO4, 4 mM EGTA pH 8, 4 mM DTT, 2 mM ATP, and 0.1 mM luciferin) in a Berthold Luminometer. For radioactive measurements, in vitro translation was performed in nuclease‐treated RRL (Promega) using 150 ng of luciferase mRNA in the presence of 15 μCi of [35S]‐Met for 60 min at 30°C. Samples were denatured in sample buffer and analyzed by SDS–PAGE and autoradiography.
In situ hybridization
In situ hybridization was carried out as previously described (Palanca et al, 2014). Briefly, U2OS cells were fixed with 4% PFA prepared in PHEM buffer. An oligo dT(50)‐mer, 5′‐end‐labeled with biotin (MWG‐Biotech, Germany) was used as a probe for in situ hybridization to poly(A) RNA. After hybridization, cells were washed and the hybridization signal was detected with FITC‐avidin for 30 min. All samples were mounted with the ProLong anti‐fading medium (Invitrogen).
RNA degradation assay
RNA was extracted from mouse embryonic stem cells using the Absolutely RNA microprep kit (Agilent). 1 μg of RNA was pre‐incubated with 4.5 μg of (PR)20 or water for 10 min at room temperature, and subsequently treated with serially diluted RNase A (Qiagen) at 37°C for 15 min, at the end of which the reaction was halted by RNasin ribonuclaease inhibitor (Promega) and 1% SDS. The resulting mixture was column‐purified using the RNA Clean & Concentrator‐5 kit (Zymo Research), according to manufacturer's instructions. The recovered RNA was analyzed by agarose gel electrophoresis and quantified by image analysis (ImageJ, NIH, USA).
Reverse transcription
500 ng of RNA isolated from mESC was pre‐incubated on ice for 5 min with increasing concentrations of (PR)20. The mixture was then used as template for cDNA synthesis using the SuperScript III First‐Strand Synthesis System (Thermo Fisher Scientific) according to manufacturer's instructions. The reaction mixture was subsequently treated with RNase A (15 min at 37°C), solubilized with SDS 1%, and finally purified using PCR cleanup columns (Qiagen). The yield of cDNA was visualized by agarose gel electrophoresis and quantified by fluorometry using the Qubit kit (Thermo Fisher Scientific), according to manufacturer's instructions.
qRT–PCR
cDNA generated from mESC was pre‐incubated on ice for 5 min with increasing concentrations of (PR)20. Out of this mixture, 50 ng of cDNA per reaction was used. The real‐time PCR was carried out using the 2X SYBR Select Master Mix (Life Technologies) in MicroAmp© Optical 384‐Well plates (Applied Biosystems) in the QuantStudio™ 6 Flex Real‐Time PCR System (Thermo Fisher) using standard protocols. The sequences of the primers used are as follows: Gapdh F TTCACCACCATGGAGAAGGC, Gapdh R CCCTTTTGGCTCCACCCT, 5.8S F GTCGATGAAGAACGCAGCTA, 5.8S R AACCGACGCTCAGACAGG, 18S F CTGGATACCGCAGCTAGGAA, 18S R GAATTTCACCTCTAGCGGCG, Eif1 F TGGTACTGTAATTGAGCATCCAG, Eif1 R CCTTAGCCAGCCCAATCTCT. Each value was normalized against that of the untreated reaction of the corresponding gene.
Viral infection
BHK‐21 cells were infected with Sindbis virus (Alphavirus, ssRNA genome with positive polarity) at a MOI of 20 viral PFU/cell in the absence or presence of (PR)20. Cells were collected 4 h after infection and total RNA was purified using RNeasy Mini Kit (Qiagen). Virus‐specific primers (F:GGTACTGGAGACGGATATCGC and R:CGATCAAGTCGAGTAGTGGTTG) were used to detect viral RNA by qRT–PCR, which were normalized against cellular GAPDH.
Splicing
U2OS cells were plated at a density of 2.4 × 104 cells/ml on six‐well plates and treated on the following day with either 20 μM of (PR)20 or 30 μM protamine. RT–PCR was performed as described above using previously described primers to detect alternative splicing events at GADD45A and NACA mRNAs (Kwon et al, 2014).
EMSA
To analyze the binding of (PR)20 or protamine to ssDNA and ssRNA, 0.2 μM of Cy3‐RNA (CCACUGCACCGCUGCUAGG) or Cy3‐DNA (CCACTGCACCGCTGCTAGG) oligonucleotides were incubated with increasing amounts of (PR)20 for 10 min at room temperature. To evaluate the binding of (PR)20 or protamine to dsDNA and dsRNA, Cy3‐RNA and Cy3‐DNA oligonucleotides were annealed with the complementary RNA (CCUAGCAGCGGUUGCAGUGG) or DNA (CCTAGCAGCGGTTGCAGTGG) molecule prior to incubation with (PR)20. Reactions were supplemented with 4 μl of 6× loading buffer (30% glycerol, bromphenol blue, xylene cyanol) and were resolved by polyacrylamide gel electrophoresis in 4–12% TBE Gels (Invitrogen) in 1%TBE buffer at 100 V for 45 min at 4°C. The gels were scanned, and Cy3 intensity was measured using Typhoon Trio (GE Health Care). Band quantification was performed using ImageJ (Schneider et al, 2012) and data fitting was performed using GraphPad Prism 7.
CRISPR/Cas9 efficiency
The previously described ESCas9 mESC line (Ruiz et al, 2016) which carries a Doxycycline‐inducible Cas9 was co‐infected with lentiviral vectors expressing EGFP (pLVTHM, Addgene, 12247) and an EGFP‐targeting sgRNA cloned in the pKLV‐U6gRNA‐PGKpuro2ABFP backbone (Addgene, 50946). Two independent clones with stable expression of all components were seeded on gelatin. After 6 h, Doxycycline (1 μg/ml) and/or (PR)20 (10 μM) were added to the medium. 72 h later, cells were recovered and the percentage of GFP‐positive cells was quantified by flow cytometry using the FlowJo software (BD).
Phosphatase assay
Briefly, RPE cells were lysate with RIPA (150 mM NaCl, 25 mM Tris–HCl, 1 mM EDTA, 0.5 mM EGTA, 1% Triton X‐100, 0.1% SDS, and protease inhibitors) and 100 mg of protein was used for each condition. PP2A Immunoprecipitation Phosphatase Assay kit was used for determining phosphatase activity following manufactured instructions. The dephosphorylation of a KRpTIRR peptide activity was measured by absorbance of Malakita green.
dNTP binding
The binding of peptides to dNTPs was done by measuring absorbance of peptide‐dNTP solutions at 600 nm using NanoDrop 2000 (Thermo Scientific, Waltham, MA, USA) according to the manufacturer's instructions.
Author contributions
Most experiments, data analyses, and preparation of the figures: VL and OS; In vitro translation and viral infection experiments: ID‐L and IV; Proteomic analyses: EZ and JM; Polysome analyses: MH and GS; RPS9SBP‐expressing HeLa cells: BJ; Electron microscopy experiments: JB; Phosphatase assays: AG; DNA‐ and RNA‐binding experiments: RF‐L; Study supervision and manuscript writing: OF‐C.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Review Process File
Acknowledgements
We would want to thank Drs. André Nussenzweig an Jordi Carreras‐Puigvert for insightful comments on the manuscript. Research was funded by Fundación Botín, by Banco Santander through its Santander Universities Global Division and by grants from the Spanish Ministry of Science, Innovation and Universities (RTI2018‐102204‐B‐I00, co‐financed with European FEDER funds) and the European Research Council (ERC‐617840) to OF; DKFZ NCT3.0 Integrative Project in Cancer Research grant (NCT3.0_2015.54 DysregPT) and SFB 1036/TP07 from the Deutsche Forschungsgemeinschaft to G.S.
The EMBO Journal (2021) 40: e103311.
Data availability
Correspondence and request of materials should be addressed to O.F. Mass spectrometry proteomics datasets associated with this work are available in the following databases:
Impact of oligoarginine peptides in ribosome composition and chromatin proteomes: PRIDE PXD010555 (http://www.ebi.ac.uk/pride/archive/projects/PXD010555).
Impact of oligoarginine peptides in RNA‐binding proteomes: PRIDE PXD014085 (http://www.ebi.ac.uk/pride/archive/projects/PXD014085).
References
- Ash P, Bieniek K, Gendron T, Caulfield T, Lin W‐L, DeJesus‐Hernandez M, van Blitterswijk M, Jansen‐West K, Paul J, Rademakers R et al (2013) Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron 77: 639–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balhorn R (2007) The protamine family of sperm nuclear proteins. Genome Biol 8: 227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeynaems S, Bogaert E, Kovacs D, Konijnenberg A, Timmerman E, Volkov A, Guharoy M, De Decker M, Jaspers T, Ryan VH et al (2017) Phase separation of C9orf72 dipeptide repeats perturbs stress granule dynamics. Mol Cell 65: 1044–1055.e1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boman HG, Agerberth B, Boman A (1993) Mechanisms of action on Escherichia coli of cecropin P1 and PR‐39, two antibacterial peptides from pig intestine. Infect Immun 61: 2978–2984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrelli A, Tornesello AL, Tornesello ML, Buonaguro FM (2018) Cell penetrating peptides as molecular carriers for anti‐cancer agents. Molecules 23: 295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce VJ, McNaughton BR (2017) Inside job: methods for delivering proteins to the interior of mammalian cells. Cell Chem Biol 24: 924–934 [DOI] [PubMed] [Google Scholar]
- Castello A, Horos R, Strein C, Fischer B, Eichelbaum K, Steinmetz LM, Krijgsveld J, Hentze MW (2013) System‐wide identification of RNA‐binding proteins by interactome capture. Nat Protoc 8: 491–500 [DOI] [PubMed] [Google Scholar]
- Chai N, Gitler AD (2018) Yeast screen for modifiers of C9orf72 poly(glycine‐arginine) dipeptide repeat toxicity. FEMS Yeast Res 18: foy024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew J, Gendron TF, Prudencio M, Sasaguri H, Zhang Y‐J, Castanedes‐Casey M, Lee Cw, Jansen‐West K, Kurti A, Murray Me et al (2015) Neurodegeneration. C9ORF72 repeat expansions in mice cause TDP‐43 pathology, neuronal loss, and behavioral deficits. Science 348: 1151–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus‐Hernandez M, Mackenzie I, Boeve B, Boxer A, Baker M, Rutherford N, Nicholson A, Finch NiCole A, Flynn H, Adamson J et al (2011) Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p‐linked FTD and ALS. Neuron 72: 245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRouchey J, Hoover B, Rau DC (2013) A comparison of DNA compaction by arginine and lysine peptides: a physical basis for arginine rich protamines. Biochemistry 52: 3000–3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emi N, Kidoaki S, Yoshikawa K, Saito H (1997) Gene transfer mediated by polyarginine requires a formation of big carrier‐complex of DNA aggregate. Biochem Biophys Res Commun 231: 421–424 [DOI] [PubMed] [Google Scholar]
- Frankel AD, Pabo CO (1988) Cellular uptake of the tat protein from human immunodeficiency virus. Cell 55: 1189–1193 [DOI] [PubMed] [Google Scholar]
- Fuchs SM, Raines RT (2006) Internalization of cationic peptides: the road less (or more?) traveled. Cell Mol Life Sci 63: 1819–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M, Loewenstein PM (1988) Autonomous functional domains of chemically synthesized human immunodeficiency virus tat trans‐activator protein. Cell 55: 1179–1188 [DOI] [PubMed] [Google Scholar]
- Hardiman O, Al‐Chalabi A, Chio A, Corr EM, Logroscino G, Robberecht W, Shaw PJ, Simmons Z, van den Berg LH (2017) Amyotrophic lateral sclerosis. Nat Rev Dis Primers 3: 17085 [DOI] [PubMed] [Google Scholar]
- Jafarinia H, van der Giessen E, Onck PR (2020) Phase separation of toxic dipeptide repeat proteins related to C9orf72 ALS/FTD. Biophys J 119: 843–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanekura K, Yagi T, Cammack AJ, Mahadevan J, Kuroda M, Harms MB, Miller TM, Urano F (2016) Poly‐dipeptides encoded by the C9ORF72 repeats block global protein translation. Hum Mol Genet 25: 1803–1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King JE, Eugenin EA, Buckner CM, Berman JW (2006) HIV tat and neurotoxicity. Microbes Infect 8: 1347–1357 [DOI] [PubMed] [Google Scholar]
- Kwon I, Xiang S, Kato M, Wu L, Theodoropoulos P, Wang T, Kim J, Yun J, Xie Y, McKnight SL (2014) Poly‐dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science 345: 1139–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K‐H, Zhang P, Kim HJ, Mitrea DM, Sarkar M, Freibaum BD, Cika J, Coughlin M, Messing J, Molliex A et al (2016) C9orf72 dipeptide repeats impair the assembly, dynamics, and function of membrane‐less organelles. Cell 167: 774–788.e717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Mori E, Kato M, Xiang S, Wu L, Kwon I, McKnight SL (2016) Toxic PR poly‐dipeptides encoded by the C9orf72 repeat expansion target LC domain polymers. Cell 167: 789–802.e712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez‐Gonzalez R, Lu Y, Gendron TF, Karydas A, Tran H, Yang D, Petrucelli L, Miller BL, Almeida S, Gao FB (2016) Poly(GR) in C9ORF72‐related ALS/FTD compromises mitochondrial function and increases oxidative stress and DNA damage in iPSC‐derived motor neurons. Neuron 92: 383–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascotti DP, Lohman TM (1997) Thermodynamics of oligoarginines binding to RNA and DNA. Biochemistry 36: 7272–7279 [DOI] [PubMed] [Google Scholar]
- Mizielinska S, Gronke S, Niccoli T, Ridler Ce, Clayton El, Devoy A, Moens T, Norona Fe, Woollacott I, Pietrzyk J et al (2014) C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine‐rich proteins. Science 345: 1192–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Weng S‐M, Arzberger T, May S, Rentzsch K, Kremmer E, Schmid B, Kretzschmar HA, Cruts M, Van Broeckhoven C et al (2013) The C9orf72 GGGGCC repeat is translated into aggregating dipeptide‐repeat proteins in FTLD/ALS. Science 339: 1335–1338 [DOI] [PubMed] [Google Scholar]
- Nizami Z, Deryusheva S, Gall JG (2010) The Cajal body and histone locus body. Cold Spring Harb Perspect Biol 2: a000653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanca A, Casafont I, Berciano MT, Lafarga M (2014) Reactive nucleolar and Cajal body responses to proteasome inhibition in sensory ganglion neurons. Biochim Biophys Acta 1842: 848–859 [DOI] [PubMed] [Google Scholar]
- Park J, Ryu J, Kim KA, Lee HJ, Bahn JH, Han K, Choi EY, Lee KS, Kwon HY, Choi SY (2002) Mutational analysis of a human immunodeficiency virus type 1 Tat protein transduction domain which is required for delivery of an exogenous protein into mammalian cells. J Gen Virol 83: 1173–1181 [DOI] [PubMed] [Google Scholar]
- Pisarev AV, Unbehaun A, Hellen CU, Pestova TV (2007) Assembly and analysis of eukaryotic translation initiation complexes. Methods Enzymol 430: 147–177 [DOI] [PubMed] [Google Scholar]
- Renton A, Majounie E, Waite A, Simón‐Sánchez J, Rollinson S, Gibbs J, Schymick J, Laaksovirta H, van Swieten J, Myllykangas L et al (2011) A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21‐linked ALS‐FTD. Neuron 72: 257–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi S, Serrano A, Gerbino V, Giorgi A, Di Francesco L, Nencini M, Bozzo F, Schinina ME, Bagni C, Cestra G et al (2015) Nuclear accumulation of mRNAs underlies G4C2‐repeat‐induced translational repression in a cellular model of C9orf72 ALS. J Cell Sci 128: 1787–1799 [DOI] [PubMed] [Google Scholar]
- Ruiz S, Mayor‐Ruiz C, Lafarga V, Murga M, Vega‐Sendino M, Ortega S, Fernandez‐Capetillo O (2016) A genome‐wide CRISPR screen identifies CDC25A as a determinant of sensitivity to ATR inhibitors. Mol Cell 62: 307–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryser HJ, Hancock R (1965) Histones and basic polyamino acids stimulate the uptake of albumin by tumor cells in culture. Science 150: 501–503 [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheicher B, Schachner‐Nedherer AL, Zimmer A (2015) Protamine‐oligonucleotide‐nanoparticles: recent advances in drug delivery and drug targeting. Eur J Pharm Sci 75: 54–59 [DOI] [PubMed] [Google Scholar]
- Schwarze SR, Ho A, Vocero‐Akbani A, Dowdy SF (1999) In vivo protein transduction: delivery of a biologically active protein into the mouse. Science 285: 1569–1572 [DOI] [PubMed] [Google Scholar]
- Smull CE, Ludwig EH (1962) Enhancement of the plaque‐forming capacity of poliovirus ribonucleic acid with basic proteins. J Bacteriol 84: 1035–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolowska E, Kalaska B, Miklosz J, Mogielnicki A (2016) The toxicology of heparin reversal with protamine: past, present and future. Expert Opin Drug Metab Toxicol 12: 897–909 [DOI] [PubMed] [Google Scholar]
- Sprunt K, Redman WM, Alexander HE (1959) Infectious ribonucleic acid derived from enteroviruses. Proc Soc Exp Biol Med 101: 604–608 [DOI] [PubMed] [Google Scholar]
- Stopford MJ, Higginbottom A, Hautbergue GM, Cooper‐Knock J, Mulcahy PJ, De Vos KJ, Renton AE, Pliner H, Calvo A, Chio A et al (2017) C9ORF72 hexanucleotide repeat exerts toxicity in a stable, inducible motor neuronal cell model, which is rescued by partial depletion of Pten. Hum Mol Genet 26: 1133–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan A, Bouffard M, Liao M, Ryan S, Callister JB, Pickering‐Brown SM, Armstrong GAB, Drapeau P (2018) Expression of C9orf72‐related dipeptides impairs motor function in a vertebrate model. Hum Mol Genet 27: 1754–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta‐Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP et al (2015) STRING v10: protein‐protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43: D447–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan R, Frankel AD (1995) Structural variety of arginine‐rich RNA‐binding peptides. Proc Natl Acad Sci USA 92: 5282–5286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X, Tan W, Westergard T, Krishnamurthy K, Markandaiah SS, Shi Y, Lin S, Shneider NA, Monaghan J, Pandey UB et al (2014) Antisense proline‐arginine RAN dipeptides linked to C9ORF72‐ALS/FTD form toxic nuclear aggregates that initiate in vitro and in vivo neuronal death. Neuron 84: 1213–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westergard T, Jensen BK, Wen X, Cai J, Kropf E, Iacovitti L, Pasinelli P, Trotti D (2016) Cell‐to‐cell transmission of dipeptide repeat proteins linked to C9orf72‐ALS/FTD. Cell Rep 17: 645–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MR, Mitrea DM, Zhang P, Stanley CB, Cassidy DE, Nourse A, Phillips AH, Tolbert M, Taylor JP, Kriwacki RW (2019) C9orf72 Poly(PR) dipeptide repeats disturb biomolecular phase separation and disrupt nucleolar function. Mol Cell 74: 713–728.e716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichterle H, Lieberam I, Porter JA, Jessell TM (2002) Directed differentiation of embryonic stem cells into motor neurons. Cell 110: 385–397 [DOI] [PubMed] [Google Scholar]
- Yin S, Lopez‐Gonzalez R, Kunz RC, Gangopadhyay J, Borufka C, Gygi SP, Gao FB, Reed R (2017) Evidence that C9ORF72 dipeptide repeat proteins associate with U2 snRNP to Cause Mis‐splicing in ALS/FTD patients. Cell Rep 19: 2244–2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y‐J, Gendron TF, Ebbert MTW, O’Raw AD, Yue M, Jansen‐West K, Zhang Xu, Prudencio M, Chew J, Cook CN et al (2018) Poly(GR) impairs protein translation and stress granule dynamics in C9orf72‐associated frontotemporal dementia and amyotrophic lateral sclerosis. Nat Med 24: 1136–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Guo L, Gonzales PK, Gendron TF, Wu Y, Jansen‐West K, O'Raw AD, Pickles SR, Prudencio M, Carlomagno Y et al (2019) Heterochromatin anomalies and double‐stranded RNA accumulation underlie C9orf72 poly(PR) toxicity. Science 363: eaav2606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Lehmer C, Michaelsen M, Mori K, Alterauge D, Baumjohann D, Schludi MH, Greiling J, Farny D, Flatley A et al (2017) Antibodies inhibit transmission and aggregation of C9orf72 poly‐GA dipeptide repeat proteins. EMBO Mol Med 9: 687–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, Liu Y, Banez‐Coronel M, Reid T, Pletnikova O, Lewis J, Miller TM, Harms MB, Falchook AE, Subramony SH et al (2013) RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci USA 110: E4968–E4977 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Review Process File
Data Availability Statement
Correspondence and request of materials should be addressed to O.F. Mass spectrometry proteomics datasets associated with this work are available in the following databases:
Impact of oligoarginine peptides in ribosome composition and chromatin proteomes: PRIDE PXD010555 (http://www.ebi.ac.uk/pride/archive/projects/PXD010555).
Impact of oligoarginine peptides in RNA‐binding proteomes: PRIDE PXD014085 (http://www.ebi.ac.uk/pride/archive/projects/PXD014085).


