Abstract
Epigenetics is an emerging field, due to its relevance in the regulation of a wide range of biological processes. The Su(Var)3–9, Enhancer-of-zeste and Trithorax (SET) and Myeloid, Nervy, and DEAF-1 (MYND) domain-containing (SMYD) proteins, named SMYD1, SMYD2, SMYD3, SMYD4 and SMYD5, are enzymes that catalyse methylation of histone and non-histone substrates, thereby playing a key role in gene expression regulation in many biological contexts, such as muscle development and physiology, haematopoiesis and many types of cancer. This review focuses on a relatively unexplored aspect of SMYD family members - their relation with immunology. Here, immunology is defined in the broadest sense of the word, including basic research on macrophage function or host immunity against pathogen infection, as well as clinical studies, most of which are centred on blood cancers.
Keywords: SMYD, Methyltransferases, Epigenetics, Immunology, Cancer, Muscle
SMYD, methyltransferases, epigenetics, immunology, cancer, muscle.
1. Introduction
The Su(Var)3–9, Enhancer-of-zeste and Trithorax (SET) and Myeloid, Nervy, and DEAF-1 (MYND) domain-containing (SMYD) histone methyltransferases are a family of proteins that is composed by five members in mice and humans: SMYD1, SMYD2, SMYD3, SMYD4 and SMYD5. The SET domain has lysine-specific methyltransferase activity, while the MYND domain contains a zinc-finger motif able to bind proline-rich regions and mediate protein-protein interactions, as well as DNA binding. Thus, these enzymes act on histone and non-histone targets to regulate many biological processes, including muscle development and cancer (Spellmon et al., 2015). Indeed, SMYD2 (Yi et al., 2019) and SMYD3 (Bottino et al., 2020) are considered to have oncogenic properties in a myriad of cancer types, although they are dispensable for normal mouse development and adult life, probably due to its redundancy (Bagislar et al., 2016). In addition, SMYD proteins are very important for muscle physiopathology (Du et al., 2014). Here, we will focus on their function in immunology, in the broadest sense of the word, including basic mechanisms and clinical conditions related to the immune system.
2. Innate immune cells: macrophages
SMYD2, SMYD3 and SMYD5 seem to play a role in macrophages. Xu et al. identified SMYD2 as a negative regulator of macrophage activation and M1 polarization. Smyd2 upregulation abrogated macrophage production of interleukin-6 (IL-6) and tumour necrosis factor-alpha (TNFα), among other proinflammatory cytokines, by catalysing H3K36 dimethylation on Tnf and Il6 promoters and suppression of nuclear factor kappa B subunit 1 (NFκB) and extracellular signal-regulated kinase (ERK) signalling (Xu et al., 2015). Li et al. studied macrophages in pathological conditions, such as exposure to the plastics industry widely-used chemicals bisphenol A and phthalate. These chemicals affect peritoneal macrophages by hampering their capacity to produce proinflammatory cytokines, express scavenger receptors and phagocytise. SMYD2 seemed to be involved in this process since SMYD2 knockdown or inhibition of its methyltransferase activity led to an expected decrease in H3K36 dimethylation and activation of IL-6 and TNFα expression, although the phagocytic capacity of the macrophages was unexpectedly reduced (Li et al., 2018).
In regard to SMYD3, Yıldırım-Buharalıoglu et al. observed that 18 h after interferon gamma (IFNγ) treatment of colony-stimulating factor 1 (CSF1)-differentiated primary human macrophages, SMYD3 mRNA levels decreased, in parallel with a reduction in cell proliferation (Yildirim-Buharalioğlu et al., 2017). Furthermore, under hyperglycaemic conditions, SMYD3, together with SET7/9, activated S100 calcium binding protein A12 (S100A12) expression by methylation of the S100A12 promoter in classically activated M1 macrophages (Mossel et al., 2020).
With respect to SMYD5, Stender et al. demonstrated that SMYD5 is associated with the nuclear receptor corepressor (NCoR) complex and recruited to Toll-like receptor (TLR)4-responsive genes in macrophages, where it actively participates in the basal repression of TLR4-responsive promoters by catalysing histone 4 lysine 20 trimethylation (H4K20me3). When TLR4 is activated, PHD finger protein 2 (PHF2) demethylates H4K20me3 to induce TLR4-dependent gene expression. According to their results, SMYD5 may be a negative regulator of macrophage inflammatory responses (Stender et al., 2012).
3. Host-pathogen interactions
In addition to macrophages, SMYD2 and SMYD3 are also implicated in host-pathogen interactions. Infection with Leishmania donovani, the intracellular parasite responsible for leishmaniasis, activated the expression of Smyd2 via c-Myc in murine cell lines and primary macrophages. This parasitic infection also caused histone 3 lysine 36 (H3K36) dimethylation at the TNFα promoter, probably by the enzymatic action of SMYD2. Pharmacological inhibition of SMYD2 using AZ505 enhanced the protective inflammatory response in infected macrophage cell lines and decreased parasite multiplication in infected mice. Thus, SMYD2, along with other methyltransferases, aids Leishmania donovani in the process of infecting the host (Parmar et al., 2020). Moreover, using human CD4+ cells lines and T cells from human immunodeficiency virus type 1 (HIV-1)-infected donors, Boehm et al. demonstrated that SMYD2 mediates H4K20 monomethylation in the HIV-1 promoter located in the 5′ long terminal repeat. H4K20 monomethylation promotes association of the reader protein L3MBTL histone methyl-lysine binding protein 1 (L3MBTL1) with the 5’ long terminal repeat. The presence of SMYD2 is required for this association and necessary for HIV latency, since SYMD2 knockdown or inhibition reactivates the virus. Thus, SMYD2 positively regulates HIV-1 latency (Boehm et al., 2017).
SMYD3 plays a role in host immunity in relation to the human T-cell lymphotropic virus type 1 (HTLV-1, also known as human T-cell leukaemia type 1). This virus is linked to leukemogenesis, among other pathological processes. Yamamoto et al. revealed that there is endogenous SMYD3 expression in T cell lines and primary T cells, in which it directly interacts with HTLV-1 Tax and supports its cytoplasmic localisation. By controlling Tax subcellular localization, SMYD3 permits or hampers its interaction with cytoplasmic or nuclear proteins (Yamamoto et al., 2011). Additionally, Nagata et al. claimed that SMYD3 is involved in the epigenetic regulation of inducible regulatory T (iTreg) cells. SMYD3 catalysed histone 3 lysine 4 (H3K4) trimethylation in the promoter region and conserved the noncoding DNA sequence of the foxp3 gene and regulated its expression in a transforming growth factor-beta1/mothers against decapentaplegic homolog 3 (TGFβ1/Smad3)-dependent manner. Hence, inhibition of SMYD3 by siRNA impaired iTreg cell formation in vitro. Accordingly, SMYD3 KO mice infected with the respiratory syncytial virus displayed exaggerated inflammatory responses and aggravation of the disease, due to the key role of iTreg in this viral infection (Nagata et al., 2015).
4. Blood cell cancers
Regarding immune cell-related cancer, recurrent mutations in SMYD1 were found in a group of patients with splenic marginal zone lymphoma, which is a type of B-cell non-Hodgkin lymphoma (Peveling-Oberhag et al., 2015). The mechanism by which this mutations affect SMYD1 function remains to be clarified and, therefore, it is unknown if SMYD1 behaves as an oncogene or as a tumour suppressor in lymphoma.
With reference to malignant haematopoiesis, Velinder et al. showed that SMYD2 methylates growth factor independent 1 transcription repressor (GFI1), which is a master regulator of normal and malignant haematopoiesis. The proposed mechanism was that this specific methylation in GFI1 protein provokes recruitment of lysine demethylase 1A (LSD1) and repression of the transcription of GFI1-targeted genes, leading to normal and/or malignant haematopoiesis (Velinder et al., 2016).
High expression levels of SMYD2 were described in many types of leukaemia by Brown et al. In B-cell acute lymphoblastic leukaemia there was a positive correlation between high SMYD2 and low overall patient survival. Haematopoietic stem cell-specific SMYD2 KO mice displayed aberrant haematopoiesis caused by apoptosis induction and changes in gene expression. Indeed, SMYD2 controls gene expression programmes related to wingless-related integration site (WNT) regulation of haematopoietic stem cells proliferation, in part by its association with Frizzled Receptor 2 (FZD2) (Brown et al., 2020). Another study confirmed the previous results (Brown et al., 2020); SMYD2 was overexpressed in acute lymphoblastic leukaemia compared to non-malignant bone marrow and high SMYD2 expression negatively correlated with overall survival in acute lymphoblastic leukaemia. A decrease in SMYD2 expression was observed 29 days after chemotherapy in most of the patients who responded to the treatment (leukaemia remission was determined by the presence of less than 5% of leukaemic cells in bone marrow) (Sakamoto et al., 2014). Similar results were obtained by Zhang et al. who observed overexpression of SMYD2 in the bone marrow of children with B lineage acute lymphoblastic leukaemia, which was associated with bad prognosis (including increased white blood cell count and less overall survival and tumour-free survival) and a reduction of SMYD2 expression levels after remission (Zhang et al., 2020). Animal studies demonstrated that, although germ-line SMYD2 KO mice are viable, healthy and display normal haematopoiesis, SMYD2 deficiency delayed and reduced penetrance of death due to MLL-AF9/NRasG12D-induced acute myeloid leukaemia. After disease establishment, both wild-type (WT) and SMYD2 KO cells gave rise to indistinguishable acute myeloid leukaemia phenotypes, but SMYD2 deletion caused competitive disadvantages over their WT counterparts in vitro and in vivo. The authors also described direct binding of Myc to Smyd2 promoter as a possible molecular mechanism for this phenotype (Bagislar et al., 2016). Moreover, Oliveira-Santos et al. found that both SMYD2 and SMYD3 were upregulated in malignant B cells from the same patients with chronic lymphocytic leukaemia, indicating that these two SMYD proteins may be controlled by the same molecular mechanism in this biological context. Interestingly, patients with very low expression of SMYD2 and SMYD3 displayed elevated white blood cell counts and a complex karyotype (Oliveira-Santos et al., 2016). Furthermore, Hay et al. noticed that premalignant thymus from RUNX family transcription factor 2 (RUNX2)/myelocytomatosis oncogene (MYC) transgenic mice overexpress Smyd2 and, by extrapolating data from murine primary fibroblasts, they speculated that SMYD2 (but not SMYD5) may inhibit the senescence-like growth arrest caused by RUNX upregulation in RUNX2/MYC lymphoma, thereby acting as an oncogene (Hay et al., 2019). Indeed, altogether, these data indicate that SMYD2 has an oncogenic role in leukaemia.
On the contrary, Zipin-Roitman et al. observed that low expression of SMYD2 in acute myeloid leukaemia patients negatively correlated with treatment response and the probability of tumour-free survival. In addition, human acute myeloid leukaemia cells devoid of SMYD2 were more resistant to DNA damage caused by irradiation compared to control cells, and the mechanism leading to this resistance seemed to be the induction of transient quiescence. The decrease in SMYD2 levels led to a compensatory increase of SET domain-containing protein (SET)7/9 methyltransferase levels, suggesting an interplay between the two enzymes that induces quiescence of leukaemia cells, which then become more resistant to chemotherapy (DNA damage) (Zipin-Roitman et al., 2017).
SMYD3 plays a role in all three principal types of blood cancers: leukaemia, lymphoma and myeloma. Liu et al. described a mechanism by which SMYD3 participates in the development of Hodgkin lymphoma via H3K4 methylation at the promoter of 15-Lipoxygenase-1 (15-LOX-1). Deregulation of this protein is observed in many cancer and pathological immunological conditions, including lymphoma (Liu et al., 2012). SMYD3 is upregulated by signal transducer and activator of transcription 3 (STAT3) in chronic lymphocytic leukaemia. The STAT3-SMYD3 axis promotes carcinogenesis, since high SMYD3 levels show a negative correlation with suppression of cell proliferation and invasion capacity. Downregulation of the phosphorylation of STAT3 by means of the STAT3 inhibitor WP1066 suppressed STAT3 binding to the SMYD3 promoter, consequently inhibiting SMYD3 gene expression (Ma et al., 2019; Lin et al., 2019). Ectopic overexpression of SMYD3 in a human leukaemia cell line induced mRNA expression of the c-Met oncogen, thereby providing a rationale to test SMYD3 inhibitors for the treatment of leukaemia (Zou et al., 2009).
Moreover, knockdown of SMYD3 caused mRNA and protein levels upregulation of C-X-C motif chemokine ligand 9 (CXCL9), C-X-C motif chemokine ligand 10 (CXCL10), C-X-C motif chemokine ligand 11 (CXCL11) and transporter 1, ATP binding cassette subfamily B member (TAP1) in human papilloma virus-negative squamous cell carcinoma of the head and neck cell lines, suggesting a role of SMYD3 in cytokine release (Vougiouklakis et al., 2017).
Natural compounds can affect SMYD3 expression in cancer. Zhao et al. attempted to elucidate the anti-tumoural mechanism of action of triptolide (the main active component of extracts from the Chinese herb Tripterygium wilfordii Hook.F) in a human multiple myeloma cell line and found that triptolide downregulates SMYD3 (Zhao et al., 2010). Furthermore, the isoquinoline plant alkaloid berberine also downregulated SMYD3 gene expression in the same cell line (Wang et al., 2016).
5. Immune responses in rheumatoid arthritis
Immune cells are directly involved in inflammatory disorders such as rheumatoid arthritis. The TNFα inhibitor etanercept (commonly used for the treatment of rheumatoid arthritis) diminished the protein levels of SMYD2 and also H3K36 trimethylation in the C–C motif chemokine ligand 2 (CCL2) promoter region in a lipopolysaccharide (LPS)-stimulated human monocytic cell line. These and other data presented by the authors indicate that the mechanism of action of this drug involves suppression of LPS-induced CCL2 expression by inhibiting SMYD2 and other methyltransferases (Lin et al., 2017).
Nothing is known about the function of SMYD4 in relation to the immune system, either in health or in disease. There is only one article that mentions SMYD4 as a methyltransferase upregulated in TNFα-stimulated synovial fibroblasts from patients with rheumatoid arthritis compared to equally stimulated synovial fibroblasts from patients with osteoarthritis. No further insights about the biological meaning of this alteration in SMYD4 expression are offered by the authors (Araki et al., 2018).
6. SMYD1 and immune responses in other biological contexts
Finally, not-so-obvious relations of Smyd1 with the immune system have been described. Due to its key role in cardiac and skeletal muscle development and regeneration (Du et al., 2014), muscle-specific Smyd1 KO mice exhibited multiple defects of skeletal muscle, that finally led to perinatal death. Interestingly, trunk muscles were substituted with brown adipose tissue (BAT), and downregulation of key adaptive and innate immune transcription factors for BAT development and functionality, such as Il-6, TNFα, chemokine (C–C motif) ligand 6 (Ccl6), chemokine (C–C motif) ligand 7 (Ccl7), chemokine (C–C motif) ligand 9 (Ccl9), forkhead box P3 (Foxp3), BAF chromatin remodelling complex subunit BCL11A (Bcl11A), RUNX family transcription factor 1 (RunX1) and core binding factor β (Cbfβ), was observed (Rasmussen and Tucker 2018). Furthermore, whole-genome resequencing of yak populations (Bos grunniens, ruminant animals that are evolutionarily adapted to high altitudes and, therefore, hypoxia) identified copy number variations in SMYD1, which may be involved in immune responses, as well as hypoxia adaptation (Wang et al., 2019). A cluster of patients with AnWj-negative blood group antigen, which is associated with lymphoid malignancies, immunologic disorders and autoimmune haemolytic anaemia, presented a R320Q substitution in SMYD1. This gene variant seems to be the base of an inherited form of the AnWJ-negative blood group phenotype, although the molecular mechanism is unknown (Yahalom et al., 2018). It has also been suggested that SMYD1 is involved in the pathogenesis of velocardiofacial syndrome, a disease with heterogeneous genotypes and phenotypes, which could include immune deficiency (Wu et al., 2019).
7. Conclusions and further directions
Most of the data about SMYD protein function in immunology are related to blood cancers, since the involvement of these methyltransferases in cancer is unquestionable. Nevertheless, basic research regarding their function in immune cells is scarce, with few publications studying macrophages and T cells.
Since few studies explored the role of SMYD4 and SMYD5 in immunology, here we have mainly focused on SMYD1, SMYD2 and SMYD3. The murine Smyd1 gene, also known as Bop, was described in the ‘90s (Hwang and Gottlieb 1995). The Smyd1 gene was linked to T lymphocytes, and the authors demonstrated that this gene produces a presumably non-coding cDNA and two protein-coding variants; one protein variant expressed in cytotoxic T lymphocytes that derives from alternative splicing, as well as a second protein variant expressed in cardiac and skeletal muscle, the transcription of which is activated by a different promoter (Hwang and Gottlieb 1997). Although the role of the cytotoxic T lymphocytes variant has not been further explored, mutations on SMYD1 have been linked to a wide range of diseases where the immune system plays a key function, suggesting that SMYD1 coordinates immune responses by mechanisms yet to be discovered.
Most of the data concerning the relationship between SMYD2 and the immune system refer to its methyltransferase activity on H3K36, which was initially described to restrain cell proliferation (Brown et al., 2006). Nonetheless, SMYD2 enzymatic activity is not restricted to H3K36, and this enzyme can also act on histone 4 lysine 20 (H4K20), among other targets (Boehm et al., 2017; Yi et al., 2019). Through its methylatse activity, SMYD2 helps Leishmania donovani to infect the host organism (Parmar et al., 2020) and activates HIV-1 latency (Boehm et al., 2017). Furthermore, the majority of the data indicates that SMYD2 acts as an oncogene in blood malignancies, although there is some controversy. Despite that, selective SMYD2 inhibitors, such as compound LLY-507 (Nguyen et al., 2015), which have been successfully used to inhibit cell growth in other cancer types (Bayo et al., 2019; Kojima et al., 2020), could be tested in cell cultures and animals models of blood malignancies.
In addition, SMYD3 activity is linked to the immune system in many ways, including macrophage function, T cells and blood cancers. Its role as an oncogene seems to be relevant to immune system-related malignancies and the use of selective SMYD3 inhibitors will confirm that it is a cancer driver. Furthermore, SMYD3 function in iTreg is worth studying, since it might provide insights into host-pathogen interactions, but also a better understanding of tumorigenesis, since iTregs are recruited y many tumours (Deng 2018).
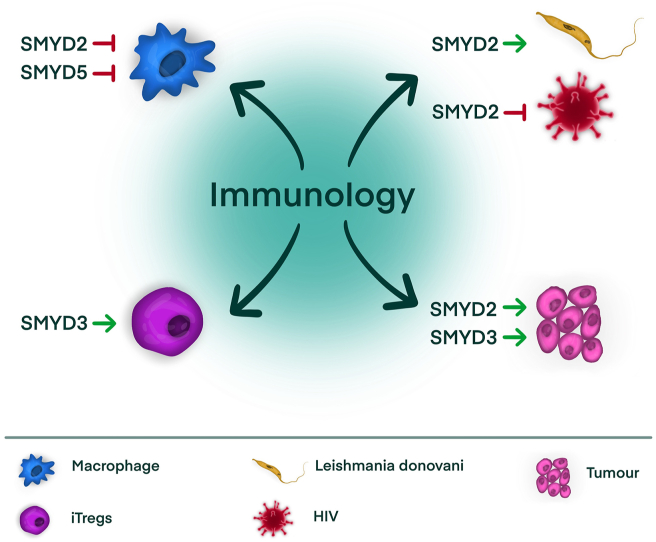
In conclusion, all the data presented in this review (summarised in Figure 1 and Table 1) indicate that SMYD family members do have relevant functions in immunology, in health and disease, but they need to be further explored. Future research may also assess whether they can be targeted in pathologies in which the immune system is crucial.
Figure 1.
Schematic representation of the main findings regarding SMYD proteins in immunology. Abbreviations: iTreg; inducible regulatory T cells, HIV; human immunodeficiency virus.
Table 1.
Studies on SMYD proteins and the main findings related to immunology.
| SMYD | Model | Main findings | References |
|---|---|---|---|
| SMYD1 | pGL-2 and pGL-IO cell lines | SMYD1 expression in cytotoxic T lymphocytes | (Hwang and Gottlieb 1997) |
| SMYD1 | Muscle-specific Smyd1 KO mice, N = 3 | Downregulation of Il-6, TNFα, Ccl6, Ccl7, Ccl9, Foxp3, Bcl11A, RunX1 and Cbfβ in brown adipose tissue | (Rasmussen and Tucker 2018) |
| SMYD1 | Yak, N = 48 | Copy number variations in SMYD1 | (Wang et al., 2019) |
| SMYD1 | AnWj- negative humans, N = 6 | R320Q substitution in SMYD1 | (Yahalom et al., 2018) |
| SMYD1 | Patients with velocardiofacial syndrome, N = 88 | Identification of SMYD1 as a putative pathogenic gene | (Wu et al., 2019) |
| SMYD1 | Patients with splenic marginal zone lymphoma, N = 26 | Recurrent mutations in SMYD1 | (Peveling-Oberhag et al., 2015) |
| SMYD2 | Monkey Cos7L cell line | SMYD2 methylation of GFI1 provokes recruitment of LSD1 and repression of the transcription of GFI1 target genes, leading to normal and/or malignant haematopoiesis | (Velinder et al., 2016) |
| SMYD2 | C57BL/6 mice, N = 3 | Upregulation of Smyd2 abrogates macrophage production of IL-6 and TNFα by catalysing H3K36 dimethylation of Tnf and Il6 promoters and suppression of NFκB and ERK signalling | (Xu et al., 2015) |
| SMYD2 | CD-1 female mice, N = 6-10 | SMYD2 catalyses H3K36 dimethylation and inhibits IL-6 and TNFα expression after bisphenol A and phthalate exposure of peritoneal macrophages | (Li et al., 2018) |
| SMYD2 | Human monocyte THP-1 cell line and human primary monocytes | The TNFα inhibitor etanercept acts by suppressing LPS-induced CCL2 expression by decreasing protein levels of SMYD2 and H3K36me3 in the CCL2 promoter region | (Lin et al., 2017) |
| SMYD2 | Patients with leukaemia, N = 50 Haematopoietic stem cell-specific SMYD2 KO mice, N = 3-8 |
SMYD2 overexpression in many types of leukaemia Positive correlation between high SMYD2 and low overall patient survival in B-cell acute lymphoblastic leukaemia Aberrant haematopoiesis in haematopoietic stem cell-specific SMYD2 KO mice |
(Brown et al., 2020) |
| SMYD2 | Patients with leukaemia, N = 83 |
SMYD2 overexpression in acute lymphoblastic leukaemia Positive correlation between high SMYD2 and low overall patient survival in acute lymphoblastic leukaemia Decrease in SMYD2 expression 29 days after chemotherapy related to treatment response |
(Sakamoto et al., 2014) |
| SMYD2 | Patients with leukaemia, N = 134 |
SMYD2 overexpression in B lineage acute lymphoblastic leukaemia, associated with bad prognosis Reduction of SMYD2 expression levels after remission |
(Zhang et al., 2020) |
| SMYD2 | SMYD2 KO mice, N = 12 | Normal haematopoiesis in SMYD2 KO mice Reduced penetrance of death due to MLL-AF9/NRasG12D-induced acute myeloid leukaemia in SMYD2 KO mice Competitive disadvantages of SMYD2 KO acute myeloid leukaemia cells |
(Bagislar et al., 2016) |
| SMYD2 | Patients with leukaemia, N = 59 |
SMYD2 and SMYD3 upregulation chronic lymphocytic leukaemia Residual expression of SMYD2 and SMYD3 correlated with elevated white blood cells count and a complex karyotype |
(Oliveira-Santos et al., 2016). |
| SMYD2 | RUNX2/MYC transgenic mice, N = 3 | Smyd2 overexpression in premalignant thymus from RUNX2/MYC transgenic mice | (Hay et al., 2019) |
| SMYD2 | Patients with leukaemia and hematopoietic TEX cell line | Low expression of SMYD2 in acute myeloid leukaemia negatively correlates with treatment response and probability of tumour-free survival SMYD2 KO human acute myeloid leukaemia cells are more resistant to DNA damage caused by irradiation due to transient quiescence |
(Zipin-Roitman et al., 2017) |
| SMYD2 | Female BALB/c mice, N = 3 Murine J774 macrophage cell line |
Smyd2 upregulation and H3K36 dimethylation at TNFα promoter after infection with Leishmania donovani SMYD2 inhibition protects against Leishmania donovani infection |
(Parmar et al. 2020) |
| SMYD2 | CD4+ T cells from patients infected HIV-1, HEK293T and J-Lat cell lines | SMYD2 mediates H4K20 monomethylation in the HIV-1 promoter located in the 5’ long terminal repeat. H4K20me1 promotes association of L3MBTL1 with the 5′ long terminal repeat SYMD2 inhibition reactivated HIV-1 |
(Boehm et al., 2017) |
| SMYD3 | Human primary monocytes | Decreased SMYD3 mRNA and cell proliferation 18 h after IFNγ treatment of CSF1 differentiated primary human macrophages | (Yildirim-Buharalioğlu et al., 2017) |
| SMYD3 | Patients with diabetes (T1DM and T2DM), N = 19-21 | Under hyperglycaemic conditions, SMYD3 activates S100A12 expression by methylation of S100A12 promoter in M1 macrophages | (Mossel et al., 2020) |
| SMYD3 | Many cell lines | SMYD3 in T cell lines and primary T cells directly interacts with HTLV-1 Tax and supports Tax cytoplasmic localisation | (Yamamoto et al., 2011) |
| SMYD3 | Female C57BL/6 mice, N = 3 | SMYD3 catalyses H3K4me3 in the promoter region of the foxp3 gene and regulates its expression in iTreg cells Inhibition of SMYD3 impairs iTreg cell formation. SMYD3 KO mice infected with respiratory syncytial virus display exaggerated inflammatory responses and aggravation of the disease |
(Nagata et al., 2015) |
| SMYD3 | Human Hodgkin lymphoma L1236 and L428 cell lines | SMYD3 participates in Hodgkin lymphoma development via H3K4 methylation at the promoter of 15-LOX-1 | (Liu et al., 2012) |
| SMYD3 | Human chronic lymphocytic leukaemia MEC1 cell line Patients with leukaemia, N = 20 Many human chronic lymphocytic leukaemia cell lines |
SMYD3 upregulation by STAT3 in chronic lymphocytic leukaemia The STAT3-SMYD3 axis promotes carcinogenesis, since high SMYD3 levels showed a negative correlation with suppression of cell proliferation and invasion capacity Downregulation of the phosphorylation of STAT3 suppresses STAT3 binding to the SMYD3 promoter and SMYD3 expression |
(Ma et al., 2019; Lin et al., 2019) |
| SMYD3 | Human leukaemia K562 and HL- 60 cell lines | Overexpression of SMYD3 in a leukaemia cell line induces c-Met expression | (Zou et al., 2009) |
| SMYD3 | Patients with human papilloma virus-negative squamous cell carcinoma of the head and neck, N = 364, and derived cell lines | Inhibition of SMYD3 causes CXCL9, CXCL10, CXCL11 and TAP1 upregulation in human papilloma virus -negative squamous cell carcinoma of the head and neck cell lines | (Vougiouklakis et al., 2017) |
| SMYD3 | Human multiple myeloma U266 cell line | Triptolide downregulates SMYD3 in a human multiple myeloma cell line | (Zhao et al., 2010) |
| SMYD3 | Human multiple myeloma U266 cell line | Berberine downregulates SMYD3 in a human multiple myeloma cell line | (Wang et al., 2016) |
| SMYD4 | Patients with rheumatoid arthritis | SMYD4 overexpression in TNFα-stimulated synovial fibroblasts from patients with rheumatoid arthritis | (Araki et al., 2018) |
| SMYD5 | Primary murine macrophages and many cell lines | SMYD5 basal repression of TLR4-responsive promoters in macrophages by catalysing H4K20me3 | (Stender et al., 2012) |
N indicates the number of subjects per experimental group. Abbreviations: KO; knockout, Il-6; interleukin-6, TNFα; tumour necrosis factor-alpha, Ccl6; chemokine (C–C motif) ligand 6, Ccl7; chemokine (C–C motif) ligand 7, Ccl9; chemokine (C–C motif) ligand 9, Foxp3; forkhead box P3, Bcl11A; BAF chromatin remodelling complex subunit BCL11A, RunX1; RUNX family transcription factor 1, Cbfβ; core binding factor β, GFI1; growth factor independent 1 transcription repressor, LSD1; lysine demethylase 1A, H3K36me2; histone 3 lysine 36 dimethylation, NFκB; nuclear factor kappa B subunit 1, ERK; extracellular signal-regulated kinase, LPS; lipopolysaccharide, CCL2; C–C motif chemokine ligand 2, H3K36me3; histone 3 lysine 36 trimethylation, RunX2; RUNX family transcription factor 2, Myc; myelocytomatosis oncogene, HIV-1; immunodeficiency virus type 1, H4K20me1; histone 4 lysine 20 monomethylation, L3MBTL1; L3MBTL histone methyl-lysine binding protein 1, IFNγ; interferon gamma, CSF1; colony-stimulating factor 1, T1DM; type 1 diabetes, T2DM; type 2 diabetes, S100A12; S100 calcium binding protein A12, HTLV-1; human T-cell lymphotropic virus type 1/human T-cell leukaemia type 1, H3K4me3; histone 3 lysine 4 trimethylation, iTreg; inducible regulatory T cells, H3K4me; histone 3 lysine 4 methylation, 15-LOX-1; 15-Lipoxygenase-1, STAT3; signal transducer and activator of transcription 3, Cxcl9; C-X-C motif chemokine ligand 9, Cxcl10; C-X-C motif chemokine ligand 10, Cxcl11; C-X-C motif chemokine ligand 11, Tap1; transporter 1, ATP binding cassette subfamily B member, TLR4; Toll-like receptor 4, H4K20me3; histone 4 lysine 20 trimethylation.
Declarations
Author contribution statement
All authors listed have significantly contributed to the development and the writing of this article.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Data availability statement
No data was used for the research described in the article.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
References
- Araki Yasuto, Yoshimi Aizaki, Sato Kojiro, Oda Hiromi, Kurokawa Riki, Mimura Toshihide. Altered gene expression profiles of histone lysine methyltransferases and demethylases in rheumatoid arthritis synovial fibroblasts. Clin. Exp. Rheumatol. 2018;36(2):314–316. [PubMed] [Google Scholar]
- Bagislar Sevgi, Sabò Arianna, Kress Theresia R., Doni Mirko, Nicoli Paola, Campaner Stefano, Bruno Amati. Smyd2 is a myc-regulated gene critical for MLL-AF9 induced leukemogenesis. Oncotarget. 2016;7(41):66398–66415. doi: 10.18632/oncotarget.12012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayo Juan, Fiore Esteban J., Dominguez Luciana M., Alejandrina Real, Malvicini Mariana, Rizzo Manglio, Atorrasagasti Catalina. A comprehensive study of epigenetic alterations in hepatocellular carcinoma identifies potential therapeutic targets. J. Hepatol. 2019;71(1):78–90. doi: 10.1016/j.jhep.2019.03.007. [DOI] [PubMed] [Google Scholar]
- Boehm Daniela, Jeng Mark, Camus Gregory, Gramatica Andrea, Schwarzer Roland, Johnson Jeffrey R., Hull Philip A. SMYD2-Mediated histone methylation contributes to HIV-1 latency. Cell Host Microbe. 2017;21(5):569–579. doi: 10.1016/j.chom.2017.04.011. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottino Cinzia, Peserico Alessia, Cristiano Simone, Caretti Giuseppina. SMYD3: an oncogenic driver targeting epigenetic regulation and signaling pathways. Cancers. 2020;12(1):142. doi: 10.3390/cancers12010142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown Mark A., Edwards Melissa A., Alshiraihi Ilham, Geng Huimin, Dekker Joseph D., Tucker Haley O. The lysine methyltransferase SMYD2 is required for normal lymphocyte development and survival of hematopoietic leukemias. Gene Immun. 2020;21(2):119–130. doi: 10.1038/s41435-020-0094-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown Mark A., Sims Robert J., Gottlieb Paul D., Tucker Philip W. Identification and characterization of Smyd2: a split SET/MYND domain-containing histone H3 lysine 36-specific methyltransferase that interacts with the Sin3 histone deacetylase complex. Mol. Canc. 2006;5(1):26. doi: 10.1186/1476-4598-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Guoping. Tumor-infiltrating regulatory T cells: origins and features. Am. J. Clin. Experim. Immunol. 2018;7(5):81–87. http://www.ncbi.nlm.nih.gov/pubmed/30498624 [PMC free article] [PubMed] [Google Scholar]
- Du Shao Jun, Tan Xungang, Zhang Jianshe. SMYD proteins: key regulators in skeletal and cardiac muscle development and function. Anat. Rec. 2014;297(9):1650–1662. doi: 10.1002/ar.22972. [DOI] [PubMed] [Google Scholar]
- Hay Jodie, Gilroy Kathryn, Huser Camille, Kilbey Anna, Mcdonald Alma, Amanda MacCallum, Holroyd Ailsa, Cameron Ewan, Neil James C. Collaboration of MYC and RUNX2 in lymphoma simulates T-cell receptor signaling and attenuates P53 pathway activity. J. Cell. Biochem. 2019;120(10):18332–18345. doi: 10.1002/jcb.29143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang Inkyu, Gottlieb Paul D. Bop: a new T-cell-restricted gene located upstream of and opposite to mouse CD8b. Immunogenetics. 1995;42(5):353–361. doi: 10.1007/BF00179396. [DOI] [PubMed] [Google Scholar]
- Hwang Inkyu, Gottlieb Paul D. The Bop gene adjacent to the mouse CD8b gene encodes distinct zinc-finger proteins expressed in CTLs and in muscle. J. Immunol. 1997;158(3):1165–11674. (Baltimore, Md.: 1950) [PubMed] [Google Scholar]
- Kojima Machiko, Sone Kenbun, Oda Katsutoshi, Hamamoto Ryuji, Kaneko Syuzo, Oki Shinya, Kukita Asako. The histone methyltransferase SMYD2 is a novel therapeutic target for the induction of apoptosis in ovarian clear cell carcinoma cells. Oncology Letters. 2020;20(5):1–8. doi: 10.3892/ol.2020.12014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Quanxi, Lawrence Catherine R., Nowak Romana A., Flaws Jodi A., Bagchi Milan K., Bagchi Indrani C. Bisphenol A and phthalates modulate peritoneal macrophage function in female mice involving SYMD2-H3K36 dimethylation. Endocrinology. 2018;159(5):2216–2228. doi: 10.1210/en.2017-03000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Fujia, Wu Danjuan, Fang Dan, Chen Yao, Zhou Haitao, Ou Caiwen. STAT3-Induced SMYD3 transcription enhances chronic lymphocytic leukemia cell growth in vitro and in vivo. Inflamm. Res. 2019;68(9):739–749. doi: 10.1007/s00011-019-01257-5. [DOI] [PubMed] [Google Scholar]
- Lin Yi Ching, Yu Chih Lin, Ming Yii Huang, Po Lin Kuo, Cheng Chin Wu, Min Sheng Lee, Chong Chao Hsieh. Tumor necrosis factor-alpha inhibitors suppress CCL2 chemokine in monocytes via epigenetic modification. Mol. Immunol. 2017;83:82–91. doi: 10.1016/j.molimm.2017.01.009. [DOI] [PubMed] [Google Scholar]
- Liu Cheng, Xu Dawei, Han Hongya, Fan Yidong, Schain Frida, Xu Zhonghua, Claesson Hans Erik, Björkholm Magnus, Jan Sjöberg. Transcriptional regulation of 15-lipoxygenase expression by histone H3 lysine 4 methylation/demethylation. PloS One. 2012;7(12):1–9. doi: 10.1371/journal.pone.0052703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Wei, Zhang Yingying, Qi Yu, Guo Shidong. STAT3 promotes chronic lymphocytic leukemia progression through upregulating SMYD3 expression. Arch. Med. Sci. 2019;15(5):1163–1175. doi: 10.5114/aoms.2018.77733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossel Dieuwertje M., Moganti Kondaiah, Riabov Vladimir, Weiss Christel, Kopf Stefan, Cordero Julio, Dobreva Gergana. Epigenetic regulation of S100A9 and S100A12 expression in monocyte-macrophage system in hyperglycemic conditions. Front. Immunol. 2020;11(June):1–19. doi: 10.3389/fimmu.2020.01071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata D. E.De Almeida, Ting H.A., Cavassani K.A., Schaller M.A., Mukherjee S., Ptaschinski C., Kunkel S.L., Lukacs N.W. Epigenetic control of foxp3 by SMYD3 H3K4 histone methyltransferase controls ITreg development and regulates pathogenic T-cell responses during pulmonary viral infection. Mucosal Immunol. 2015;8(5):1131–1143. doi: 10.1038/mi.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen Hannah, Allali-Hassani Abdellah, Stephen Antonysamy, Chang Shawn, Chen Lisa Hong, Curtis Carmen, Spencer Emtage. LLY-507, a cell-active, potent, and selective inhibitor of protein-lysine methyltransferase SMYD2. J. Biol. Chem. 2015;290(22):13641–13653. doi: 10.1074/jbc.M114.626861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira-Santos Wilson, Rabello Doralina Amaral, Antônio Roberto Lucena-Araujo, Morato de Oliveira Fábio, Magalhaes Rego Eduardo, Pittella Silva Fábio, Saldanha-Araujo Felipe. Residual expression of SMYD2 and SMYD3 is associated with the acquisition of complex karyotype in chronic lymphocytic leukemia. Tumor Biol. 2016;37(7):9473–9481. doi: 10.1007/s13277-016-4846-z. [DOI] [PubMed] [Google Scholar]
- Parmar Naveen, Chandrakar Pragya, Kar Susanta. Leishmania donovani subverts host immune response by epigenetic reprogramming of macrophage M(lipopolysaccharides + IFN-γ)/M(IL-10) polarization. J. Immunol. 2020;204(10):2762–2778. doi: 10.4049/jimmunol.1900251. [DOI] [PubMed] [Google Scholar]
- Peveling-Oberhag, Jan Franziska Wolters, Döring Claudia, Walter Dirk, Sellmann Ludger, Scholtysik René, Lucioni Marco. Whole exome sequencing of microdissected splenic marginal zone lymphoma: a study to discover novel tumor-specific mutations. BMC Canc. 2015;15(1):1–10. doi: 10.1186/s12885-015-1766-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen Tara, Tucker Haley. Loss of SMYD1 results in perinatal lethality via selective defects within myotonic muscle descendants. Diseases. 2018;7(1):1. doi: 10.3390/diseases7010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto Luis Henrique Toshihiro, Vieira de Andrade Rosangela, Felipe Maria Sueli Soares, Motoyama Andrea Barretto, Pittella Silva Fabio. SMYD2 is highly expressed in pediatric acute lymphoblastic leukemia and constitutes a bad prognostic factor. Leuk. Res. 2014;38(4):496–502. doi: 10.1016/j.leukres.2014.01.013. [DOI] [PubMed] [Google Scholar]
- Spellmon Nicholas, Holcomb Joshua, Trescott Laura, Sirinupong Nualpun, Yang Zhe, Spellmon Nicholas, Holcomb Joshua, Trescott Laura, Sirinupong Nualpun, Yang Zhe. Structure and function of SET and MYND domain-containing proteins. Int. J. Mol. Sci. 2015;16(1):1406–1428. doi: 10.3390/ijms16011406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stender Joshua D., Pascual Gabriel, Wen Liu, Kaikkonen Minna U., Do Kevin, Spann Nathanael J., Boutros Michael, Perrimon Norbert, Rosenfeld Michael G., Glass Christopher K. Control of proinflammatory gene programs by regulated trimethylation and demethylation of histone H4K20. Mol. Cell. 2012;48(1):28–38. doi: 10.1016/j.molcel.2012.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velinder Matthew, Singer Jason, Bareyan Diana, Meznarich Jessica, Tracy Christopher M., Fulcher James M., McClellan David. GFI1 functions in transcriptional control and cell fate determination require SNAG domain methylation to recruit LSD1. Biochem. J. 2016;473(19):3355–3369. doi: 10.1042/BCJ20160558. [DOI] [PubMed] [Google Scholar]
- Vougiouklakis Theodore, Bao Riyue, Nakamura Yusuke, Saloura Vassiliki. Protein methyltransferases and demethylases dictate CD8+ T-cell exclusion in squamous cell carcinoma of the head and neck. Oncotarget. 2017;8(68):112797–112808. doi: 10.18632/oncotarget.22627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Hui, Chai Zhixin, Hu Dan, Ji Qiumei, Xin Jinwei, Zhang Chengfu, Zhong Jincheng. A global analysis of CNVs in diverse yak populations using whole-genome resequencing. BMC Genom. 2019;20(1):1–12. doi: 10.1186/s12864-019-5451-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Zhixiang, Liu Yuan, Xue Yong, Hu Haiyan, Ye Jieyu, Li Xiaodong, Lu Zhigang, Meng Fanyi, Liang Shuang. Berberine acts as a putative epigenetic modulator by affecting the histone code. Toxicol. Vitro. 2016;36:10–17. doi: 10.1016/j.tiv.2016.06.004. [DOI] [PubMed] [Google Scholar]
- Wu Dandan, Chen Yang, Chen Qiming, Wang Guoming, Xu Xiaofeng, Peng A., Jin Hao, He Jinguang, Huang Li, Dai Jiewen. Clinical presentation and genetic profiles of Chinese patients with velocardiofacial syndrome in a large referral centre. J. Genet. 2019;98(2) [PubMed] [Google Scholar]
- Xu Guiliang, Liu Guilin, Xiong Sidong, Liu Haiyan, Chen Xi, Zheng Biao. The histone methyltransferase Smyd2 is a negative regulator of macrophage activation by suppressing interleukin 6 (IL-6) and tumor necrosis factor α (Tnf-α) J. Biol. Chem. 2015;290(9):5414–5423. doi: 10.1074/jbc.M114.610345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahalom Vered, Pillar Nir, Zhao Yingying, Modan Shirley, Fang Mingyan, Yosephi Lydia, Asher Orna. SMYD1 is the underlying gene for the AnWj-negative blood group phenotype. Eur. J. Haematol. 2018;101(4):496–501. doi: 10.1111/ejh.13133. [DOI] [PubMed] [Google Scholar]
- Yamamoto Keiyu, Ishida Takaomi, Nakano Kazumi, Yamagishi Makoto, Yamochi Tadanori, Tanaka Yuetsu, Furukawa Yoichi, Nakamura Yusuke, Watanabe Toshiki. SMYD3 interacts with HTLV-1 Tax and regulates subcellular localization of Tax. Canc. Sci. 2011;102(1):260–266. doi: 10.1111/j.1349-7006.2010.01752.x. [DOI] [PubMed] [Google Scholar]
- Yi Xin, Xue Jun Jiang, Fang Ze Min. Histone methyltransferase SMYD2: ubiquitous regulator of disease. Clin. Epigenet. 2019;11(1):112. doi: 10.1186/s13148-019-0711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim-Buharalioğlu, Gökçe, Mark Bond, Sala-Newby Graciela B., Charles C.T.Hindmarch, Newby Andrew C. Regulation of epigenetic modifiers, including KDM6B, by interferon-γ and interleukin-4 in human macrophages. Front. Immunol. 2017;8(FEB):1–18. doi: 10.3389/fimmu.2017.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Ping, Jin Fei Ruan, Weng Wenwen, Tang Yongmin. Overexpression of SET and MYND domain-containing protein 2 (SMYD2) is associated with poor prognosis in pediatric B lineage acute lymphoblastic leukemia. Leuk. Lymphoma. 2020;61(2):437–444. doi: 10.1080/10428194.2019.1675875. [DOI] [PubMed] [Google Scholar]
- Zhao Fei, Chen Yan, Zeng Linglan, Li Rui, Zeng Rong, Lu Wen, Liu Yuan, Zhang Chun. Role of triptolide in cell proliferation, cell cycle arrest, apoptosis and histone methylation in multiple myeloma U266 cells. Eur. J. Pharmacol. 2010;646(1–3):1–11. doi: 10.1016/j.ejphar.2010.05.034. [DOI] [PubMed] [Google Scholar]
- Zipin-Roitman Adi, Aqaqe Nasma, Yassin Muhammad, Biechonski Shahar, Amar Mariam, Mark F., van Delft, Gan Olga I. SMYD2 lysine methyltransferase regulates leukemia cell growth and regeneration after genotoxic stress. Oncotarget. 2017;8(10):16712–16727. doi: 10.18632/oncotarget.15147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Jia Ning, Shu Zhen Wang, Jia Sen Yang, Xue Gang Luo, Jing Hang Xie, Tao Xi. Knockdown of SMYD3 by RNA interference down-regulates c-met expression and inhibits cells migration and invasion induced by HGF. Canc. Lett. 2009;280(1):78–85. doi: 10.1016/j.canlet.2009.02.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.