Abstract
Background:
Physiologic aging has been associated with gut dysbiosis. Although short exercise interventions have been linked to beneficial changes in gut microbiota in younger adults, limited data are available from older populations. We hypothesized that exercise would produce beneficial shifts in microbiota and short-chain fatty acid (SCFA) levels in older persons.
Methods:
Stool samples were collected before and at completion of a supervised 24-week cardiovascular and resistance exercise intervention among 50–75-year-old participants. SCFA levels were analyzed by gas chromatography and microbiome by 16S rRNA gene sequencing. Negative binomial regression models compared pre- and post-differences using false discovery rates for multiple comparison.
Results:
A total of 22 participants provided pre-intervention samples; 15 provided samples at study completion. At baseline, the majority of participants were men (95%), mean age 58.0 (8.8) years, mean body mass index 27.4 (6.4) kg/m2. After 24 weeks of exercise, at the genus level, exercise was associated with significant increases in Bifidobacterium (and other unidentified genera within Bifidobacteriaceae), Oscillospira, Anaerostipes, and decreased Prevotella and Oribacterium (p < 0.001). Stool butyrate increased with exercise [5.44 (95% confidence interval 1.54, 9.24) mmol/g, p = 0.02], though no significant differences in acetate or propionate (p ⩾ 0.09) were seen.
Conclusion:
Our pilot study suggested that an exercise intervention is associated with changes in the microbiome of older adults and a key bacterial metabolite, butyrate. Although some of these changes could potentially reverse age-related dysbiosis, future studies are required to determine the contribution of changes to the microbiome in the beneficial effect of exercise on overall health of older adults. Clinical Trials NCT02404792
Keywords: aging, exercise, microbiome, physical function, stool metabolites
Background
The last decade has experienced a surge of interest in the role that the gut microbiome plays in health and disease. The microbiome and measures of gut health and intestinal permeability have been linked to multiple different disease processes and comorbidities. With aging, additional shifts are seen in the microbiome, including decreased abundance of beneficial bacteria [e.g. Bifidobacterium spp. and short-chain fatty acid (SCFA) producers] and increased abundance of bacteria with inflammatory potential (pathobionts, e.g. Proteobacteria spp.).1,2 While many studies link health outcomes to the gut microbiome, few studies focus on alterations specific to older adults, and even fewer identify effective interventions that alter the microbiome in older adults and improve overall health.
Exercise has numerous benefits on overall health and physical function among persons of all ages, including improvements in physical function, and decrease in cardiovascular disease and diabetes. Exercise also has known beneficial effects on the gut, including decreased transit time and lower rates of colon cancer. 3 As recently reviewed, 3 numerous animal studies have consistently shown that exercise training also alters the composition and function of the gut microbiome, with an increase in the relative abundance (RA) of butyrate-producing organisms and increased production of butyrate, a short chain fatty acid (SCFA) that promotes gut barrier integrity and intestinal homeostasis. 3 Studies in humans are primarily limited to cross-sectional studies exploring associations between self-reported physical activity or fitness and the microbiome. Few human studies have demonstrated whether initiating exercise has an impact on the microbiome among previously sedentary adults. A 6-week aerobic exercise intervention in 17 sedentary women (mean age 37 years) demonstrated alterations in the microbiome including an increase in RA of Akkermansia and a decrease in Proteobacteria. 4 A separate 6-week aerobic exercise intervention found differing responses among lean (n = 18) or obese participants (n = 14), though both groups had increases in stool butyrate and butyrate-producing taxa. 5 An 8-week intervention study of young individuals found no significant differences in the microbiota when comparing aerobic exercise, whey protein, or both (n = 30/group). 6
Only two small studies, to the best of our knowledge, have explored the impact of exercise on gut microbiota in older adults. A total of 33 Japanese men (aged 62–76 years) assigned to 5 weeks of supervised cycling on a stationary bike had a significant decrease in Clostridium difficile and a significant increase in Oscillospira. 7 Sedentary older women (⩾65 years) assigned to 12 weeks of brisk walking (n = 18) or core exercises (n = 14), experienced an increase in Bacteroides, in the brisk walking group only. 8 Notably, all of the prior interventional studies were 12 weeks or less, limiting data on the durability of changes in the microbiome, and no published studies tested the combination of aerobic and resistance exercise, as is recommended for older adults. 9 As the type, intensity, and duration of exercise, and underlying dietary differences between populations may impact microbiome changes in response to exercise, changes in these prior studies may not be generalizable to other populations.
We have previously demonstrated significant improvements in physical function following a 24-week supervised exercise intervention combining both aerobic and resistance exercise in older adults (ages 50–75 years). 10 In this pilot study, we hypothesized that our 24-week combination intervention in older adults would have findings unique to other studies, with a beneficial shift in microbiota and increased stool SCFA.
Materials and methods
The Exercise for Healthy Aging Study enrolled older adults with and without HIV infection from April 2014 to May 2017 (Clinical Trials NCT02404792). 10 All participants were aged 50–75 years, sedentary (<60 min of physical activity each week for the preceding 6 months by self-report), had a body mass index (BMI) between 20 kg/m2 and 40 kg/m2, and had no contraindications to initiating an exercise regimen (e.g. severe mobility limitation, unstable angina, supplemental oxygen requirement, uncontrolled hypertension). Participants with diabetes had hemoglobin A1c of 7.5% or less; sex hormone supplementation was restricted to stable, physiologic doses for ⩾3 months prior to study entry, and intramuscular testosterone was excluded. In a microbiome pilot study, participants could volunteer to complete a dietary log and provide stool samples at two time points. As we saw significant differences in the microbiome at baseline by HIV serostatus, for this analysis we focused only on those without HIV. The study procedures were approved by the Colorado Multiple Institutional Review Board (CO 14-2207). Written informed consent was obtained from all participants.
As previously published, each participant attended supervised exercise sessions 3 times/week for 24 weeks at the University of Colorado Anschutz Medical Campus Exercise Research Laboratory. 10 Participants began with a 2-week supervised, low-intensity exercise acclimation for machine familiarization consisting of 20–30 min of treadmill walking at 30–40% of maximal aerobic capacity (VO2 max) and three sets of eight repetitions of four weight-assisted machine (Cybex) exercises (e.g. bench press, leg press, lateral pulldown, and a rotating fourth exercise) at low intensity [40–50% of the one repetition maximum (1-RM), the maximum weight that can be lifted one time with proper range of motion]. After 2 weeks, participants increased cardiovascular endurance exercise intensity to 40–50% VO2 max and time by 5 min every week to achieve a goal of 50 min/session by the end of 12 weeks. Resistance exercise was increased to 60–70% of 1-RM; 1-RM was reassessed every 3 weeks and target weight loads adjusted as needed. At week 12, VO2 max measurements were repeated and participants were randomized to either continue moderate-intensity exercise or advance to high-intensity exercise (60–70% of week 13 VO2 max and >80% 1-RM) for the remaining 12 weeks. Randomization was created in REDCap and used a random allocation sequence created by the study statistician. The investigators enrolling participants were blinded to the allocation sequence, in blocks by sex, age, and HIV serostatus. Investigators collecting stool samples and completing microbiome profiling and SCFA extraction/analysis were blinded to the randomization.
Microbiome profiling and SCFA extraction/analysis
A self-collected stool sample was obtained prior to the start of intervention and immediately stored in the participant’s home freezer until the next visit (typically within a few days), when samples were transferred to a −80°C freezer. All samples were processed within 6 months of storage. Stool bacterial profiles were generated by broad-range amplification and sequence analysis of bacterial 16S rRNA genes as previously published. 11 DNA was extracted using the QIAamp PowerFecal Pro DNA Isolation Kit (QIAGEN, Venlo, The Netherlands). Broad-range polymerase chain reaction (PCR) amplicons were generated using barcoded primers targeting the V3V4 variable region of the 16S rRNA gene: primers 338F (5′ ACTCCTACGGGAGGCAGCAG) and 806R (5′ GGACTACHVGGGTWTCTAAT). Illumina paired-end sequencing was performed on the MiSeq platform using a 600-cycle (v.3) reagent kit. Paired-end reads were quality-filtered, demultiplexed, merged, and classified using SINA (1.3.0-r23838) as previously described. 11 Operational taxonomic units were produced by clustering sequences with identical taxonomic assignments. Sequences that could not be classified to the genus level were assigned to the lowest taxonomic level to which they could be classified (e.g. phylum/order/family). These higher level bins were appended with the term ‘other’ (e.g. ‘Bifidobacteriaceae other’) to clarify that they represented a subset of unclassified genera within a family/order/phylum, rather than an entire family/order/phylum. Samples were sequenced to the depth of 131085 sequences/sample (interquartile range [IQR]: 115717, 167681); all samples had Goods coverage scores greater than 99.976%, indicating excellent depth of sequence coverage. All 16S rRNA gene sequences and associated metadata were submitted to the National Center Biotechnology Information (NCBI) sequence read archive under project accession number PRJNA657420. Stool SCFA was measured using gas chromatography as previously described:11,12 7 g of stool taken from disparate sections of each sample were dissolved in purified water (1:5, weight to volume) and homogenized by vortexing.
Dietary analysis
Participants completed a 3-day dietary survey designed to quantify macronutrient intake during the period of stool collection. Raw dietary data were converted to caloric intake by the Nutrition Core at the Colorado Clinical and Translational Sciences Institute. 11 Dietary measurements were averaged over 3 days.
Statistical analysis
This was a pilot study to test the feasibility of stool collection in the setting of an exercise intervention, and to collect data on the effect size of exercise on the microbiome and SCFAs with exercise and by exercise intensity to power larger studies focused on the microbiome. As sample collection began after the parent study was underway, samples were only available on a limited number of participants and sample size calculation was not carried out. Due to the small sample size, primary outcome was change in the microbiota and SCFAs with the intervention (moderate/high intensity combined); the effect of randomized exercise intensity was considered secondary. RA was calculated as the number of sequences for a specific taxon standardized to the total number of sequences in that sample; samples with counts above 0 in less than 2% of the population were collapsed into a single ‘rare’ category. Diversity measures (alpha diversity by Sobs, Shannon diversity, and evenness), dietary measures, and SCFA levels were modeled in linear mixed models with a random intercept for each subject with time points (pre- versus post-intervention) as predictors. Alpha diversity indices were calculated using rarefaction and bootstrapping with 100 replicate resamples using the tidyMicro package in R. 13 Differences between beta diversity pre- versus post-intervention were tested using a permutation-based multiple analysis of variance (PERMANOVA) with 1000 permutations constrained to be within individual to account for the repeated measures study design. The adonis function in the vegan R package was used to implement this test with the Bray–Curtis distance metric. 14 Relationships between individual taxa before and after exercise were evaluated using generalized linear mixed models assuming a negative binomial distribution and log link function and a random intercept for each subject, with a false discovery rate correction applied at each taxonomic level to control for multiple comparisons. Effect sizes less than 1 indicate a decrease and greater than 1 indicate an increase in the outcome. All models were adjusted for age, to account for age-related differences we have previously observed in this cohort, and then for exercise intensity, as a secondary outcome. 11
Results
Stool samples were provided by 22 participants at baseline and 15 participants (7 high-intensity and 8 moderate-intensity exercise) at study completion. As shown in Table 1, the majority of participants were men, white, and non-Hispanic, and, on average, had an overweight BMI. Participants without study completion data were similar to those with data, with the exception of a younger age (55 years 10 versus 62 years; 9 p = 0.09). No participants had inflammatory bowel disease, irritable bowel syndrome, or used probiotics.
Table 1.
Baseline characteristics of participants.
| Age, years | 58.0 (55.0, 63.8) |
| Male sex | 21 (95%) |
| Body mass index, kg/m2 | 27.4 (24.6, 31.0) |
| Race (%) | |
| White | 17 (77) |
| Other or more than one race | 4 (18) |
| Hispanic or Latino ethnicity | 3 (14) |
| Sexual preference (%) | |
| Heterosexual | 16 (73) |
| Men who have sex with men | 6 (27) |
| Smoking status (%) | |
| Current/prior smoker | 8 (36) |
| Never smoker | 14 (64) |
| Food consumed per day, g | 2395 (1561, 2880) |
| Fiber consumed per day, g | 19.7 (14.8, 25.2) |
Data reported as number (frequency) or median (interquartile range).
Exercise-associated changes in stool bacterial communities
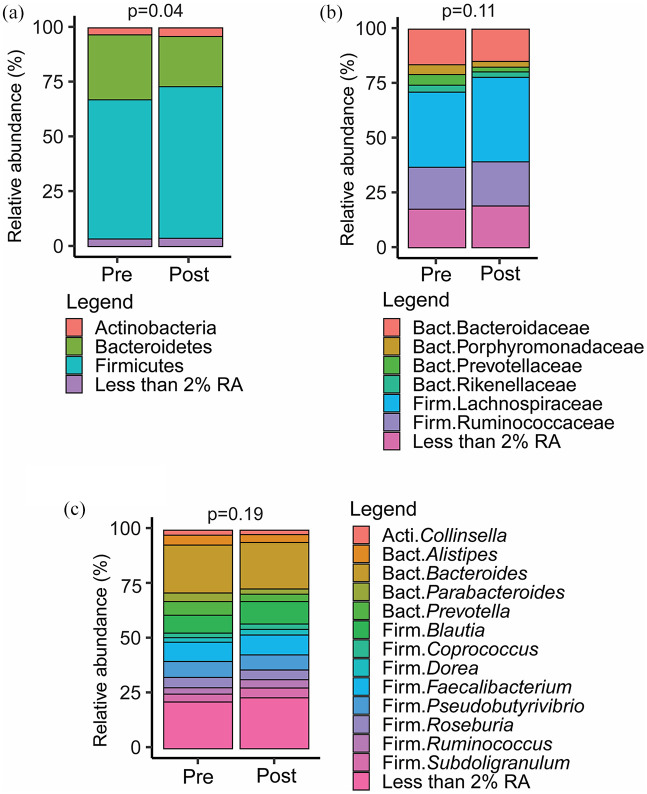
Measures of microbial alpha diversity, including stool microbiome richness (Sobs) and overall diversity (Shannon diversity and evenness) were minimally changed between baseline and week 24 of supervised exercise after adjustment for age [Sobs: estimate 0.92 (95% confidence interval [CI] −2.35, 4.20), p = 0.59; Shannon evenness (H/Hmax): 0.01 (−0.02, 0.04), p = 0.50; Shannon’s H: −0.07 (−0.11, 0.24), p = 0.47]. In contrast, significant changes in beta-diversity (i.e. differences in the overall community composition) pre- and post-exercise intervention (restricted to the n = 15 with samples at both time points) were observed across phyla (p = 0.04) by PERMANOVA; no differences were noted at the family (p = 0.11) or genus (p = 0.19) levels (Figure 1).
Figure 1.
Stacked bar charts representing mean RA at the (a) phylum, (b) family, and (c) genus levels in individuals with 16sRNA sequencing data pre- and post-exercise intervention (N = 15). Taxa with RA <2% were collapsed into a single category.
PERMANOVA tests stratified by individual were conducted to test whether beta-diversity changed significantly from pre- to post-intervention.
Acti, Actinobacteria; Bact, Bacteroidetes; Firm, Firmicutes; RA, relative abundance.
The exercise effects on the RA of specific taxa were determined with negative binomial linear mixed models. The robustness of the models was further explored with adjustment separately by age and exercise intensity. In unadjusted models, we found no significant changes in individual phyla. Notably, Firmicutes tended to increase [estimate 1.12 (95% CI 1.01, 1.25)] and Bacteroidetes tended to decrease [0.71 (0.49, 1.04)], although neither reached statistical significance (p ⩾ 0.11). Similarly, no significant changes were observed at the phylum level when adjusting for age or for exercise intensity (data not shown).
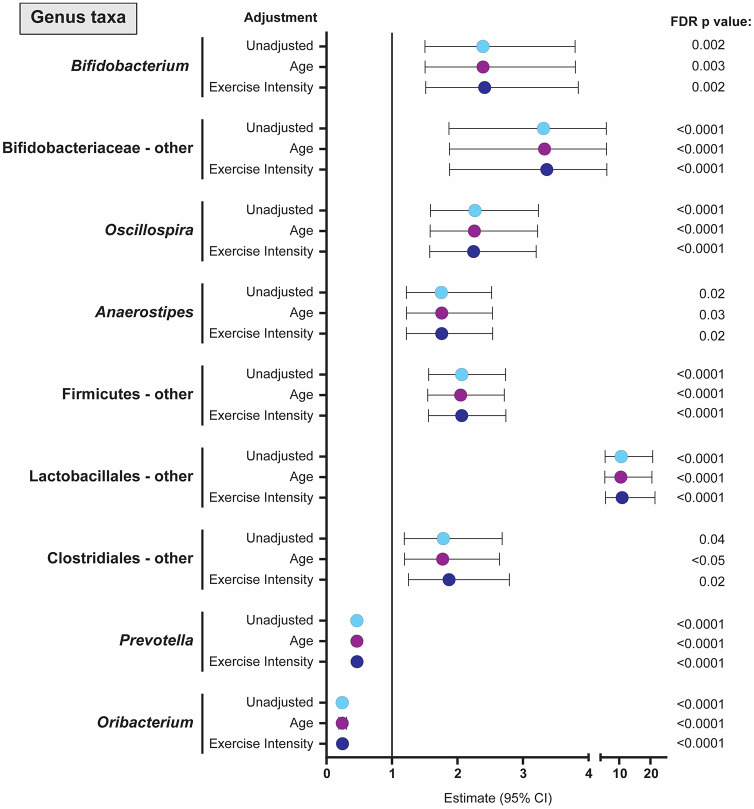
At the genus level, exercise was associated with significant increases in Bifidobacterium (and other unclassified genera within Bifidobacteriaceae), Oscillospira, Anaerostipes, and other genera within Firmicutes, Lactobacillales and Clostridiales taxa in unadjusted models, as well as those adjusted for age and exercise intensity (Figure 2). Exercise was associated with significant decreases in Prevotella and Oribacterium in both unadjusted and adjusted models. Although Solobacterium increased with the exercise intervention [3.72 (3.67, 3.78), p < 0.0001], significance was lost in adjusted models. Increasing age (but not the intervention) was associated with increased abundance of genera belonging to the RC9 gut group [6.80 (6.77, 6.84), p < 0.0001]. Higher intensity exercise (but not the intervention alone) was associated with lower Succinivibrio [0.33 (0.26, 0.42), p < 0.0001].
Figure 2.
Forest plot showing the estimate and 95% CIs for all genus level taxa where the exercise intervention had a significant effect on RA. Values are shown for unadjusted analyses (light blue dot) as well as estimates (95% CI) when adjusted for age (green dot) or exercise intensity (purple dot). Estimates above 1 suggest higher RA of genus taxa following exercise intervention with higher levels at older ages when adjusting for age and higher levels in the high-intensity group when adjusting for exercise intensity. Estimates below 1 indicate lower RA with lower levels at higher ages or in the high-intensity exercise group. ‘Other’ refers to all sequences that were classified to the taxa level detailed, but could not be further classified to specific genus. CI, confidence interval; FDR, false discovery rate; RA, relative abundance.
With the exercise intervention, increases were seen with stool butyrate, propionate, and acetate concentrations, although only butyrate reached statistical significance (Table 2). As changes in diet can impact changes in stool bacterial communities and SCFAs, we also examined changes in self-reported dietary intake between baseline and week 24. No significant differences were seen in the total energy consumed, % protein, % carbohydrates, % fat, fiber (soluble and insoluble), or fat to fiber ratio from baseline to week 24 (all p ⩾ 0.26) (Table 3).
Table 2.
Effect of exercise intervention on stool short-chain fatty acid levels.
| Short-chain fatty acid (mmol/g) |
Mean (SD) pre-exercise | Mean (SD) post-exercise | Estimate of exercise effect | 95% CI | p value |
|---|---|---|---|---|---|
| Acetate | 37.7 (14.1) | 42.7 (13.5) | 5.40 | −3.25, 14.04 | 0.24 |
| Butyrate | 7.7 (6.1) | 15.2 (5.9) | 5.44 | 1.54, 9.34 | 0.02 |
| Propionate | 10.2 (4.9) | 12.5 (4.5) | 2.44 | −0.22, 5.09 | 0.09 |
CI, confidence interval; SD; standard deviation.
Table 3.
Change in self-reported dietary intake with the exercise intervention.
| Dietary measure | Estimate of exercise intervention effect (95% CI, p value) | Estimate of age effect* (95% CI, p value) | Estimate of exercise and age interaction effect* (95% CI, p value) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Total energy (Kcal) | −174.21 | −488.84, 140.42 | 0.30 | 18.22 | −12.53, 48.97 | 0.26 | |||
| % Carbohydrates | 0.33 | −3.31, 3.98 | 0.86 | 0.22 | −0.46, 0.90 | 0.54 | |||
| % Protein | 0.72 | −2.3, 3.74 | 0.65 | −0.16 | −0.41, 0.09 | 0.22 | |||
| Fat/fiber ratio | −0.15 | −1.03, 0.72 | 0.74 | 0.02 | −0.11, 0.14 | 0.81 | |||
| % Fat | 22.13 | −8.65, 52.91 | 0.18 | 0.03 | −0.52, 0.59 | 0.91 | −0.36 | −0.86, 0.14 | 0.18 |
| Fiber (g) | 32.08 | −6.44, 70.6 | 0.13 | 0.17 | −0.29, 0.62 | 0.48 | −0.56 | −1.19, 0.07 | 0.11 |
| Soluble fiber (g) | 7.74 | −4.20, 19.68 | 0.23 | 0.05 | −0.09, 0.19 | 0.48 | −0.15 | −0.34, 0.05 | 0.17 |
| Insoluble fiber (g) | 25.54 | −4.69, 55.77 | 0.12 | 0.19 | −0.18, 0.55 | 0.33 | −0.43 | −0.93, 0.06 | 0.11 |
All models were also tested for an interaction between the exercise intervention and age; when there was an interaction detected (p < 0.20), these values are also reported.
CI, confidence interval.
Discussion
In a pilot study among older sedentary adults, after adjusting for age effects, we demonstrated that 24 weeks of a supervised, combined aerobic and resistance exercise intervention was associated with significant increases in abundances of bacteria with reported gut health benefits (including Bifidobacterium, Oscillospira, Anaerostipes) and decreases in commensal bacterial species with reported inflammatory properties (including Prevotella and Succinivibrio). We acknowledge that our study sample was small, primarily male, dietary intake was ad libitum throughout the intervention and, due to the small sample size, we were unable to adjust for many key covariates that potentially could have impacted the gut microbiome. However, several provocative findings merit further discussion and investigation in larger studies of sedentary older adults.
First, readers may ask what the clinical relevance is of the specific microbiota that decreased with exercise in our cohort. A greater abundance of Prevotella spp. in the microbiome (not limited to gut) has been associated with a plant-rich, high-carbohydrate and high-fiber diet, and lower frailty in some studies,15–18 but has also been linked to multiple inflammatory diseases. 19 Similar to Prevotella, Succinivibrio (decreased in models adjusted for exercise intensity) has also been associated with a plant-rich, high-fiber diet.20,21
Among the bacteria that increased with the exercise intervention, Bifidobacterium spp. are generally considered to be beneficial, gut-protective bacteria associated with decreased intestinal inflammation. 22 Bifidobacteria promote by cross-feeding greater production of anti-inflammatory metabolites (such as butyrate) in the presence of adequate fiber,23,24 and are involved in the metabolism of folate and riboflavin, which may exert beneficial effects on muscle. 15 Of note, physiological aging has been associated with decreased abundance of various Bifidobacterium spp.1,2 suggesting that exercise may improve some features of age-associated dysbiosis. Greater abundances of Oscillospira spp. have been associated with both beneficial (leanness, 25 lower prevalence of inflammatory bowel disease26,27) and harmful (Parkinson’s disease, 28 lower dietary fiber content 29 ) effects. The genus Anaerostipes also includes butyrate-producing bacteria, though clinical associations are limited.30,31 Thus, based on prior associations in the literature, our supervised exercise intervention in older adults was associated with increases in some bacteria with beneficial effects on gut health and a decrease in some potential pathobionts.
The majority of evidence for exercise-related changes in the gut microbiome comes from pre-clinical models.3,32 Studies in humans have been primarily cross-sectional, linking the gut microbiome to greater self-reported physical activity levels or to exercise endurance. In these studies, the gut microbiome varied by the study population: for example, compared with healthy controls, professional rugby players had lower abundances of the phylum Bacteroidetes and genus Bacteroides, and higher Akkermansia in addition to a higher abundance of bacteria that decreased with exercise in our study (Prevotella and Succinivibrio), perhaps due to age, differences in dietary composition, muscle mass, or geography. 33 Young adults with greater maximal aerobic capacity had a higher Firmicutes:Bacteroidetes ratio 34 and a lower RA of Bacteroides spp. 35 Although Firmicutes tended to increase and Bacteroidetes tended to decrease in the present study, neither were significantly different with exercise. Higher levels of physical activity were associated with greater Bifidobacterium in young women, similar to the increase we observed with our intervention. 36
Few studies have reported microbiome changes within the context of initiating exercise, particularly among older adults. A 5-week cycling intervention among elderly Japanese men was associated with significant increases in Oscillospira, 7 a genus that also increased in our study, and a decrease in Clostridium difficile. The increase in Bifidobacteria that we observed with exercise was similarly seen in a 6-week exercise intervention among younger women. 4 In contrast, we found minimal change in Bacteroides, while two other studies found increases in Bacteroides in some participants: Bacteroides increased with a brisk walking intervention (but not core muscle strengthening) among older women 8 and increased among individuals with obesity (but decreased in lean individuals) in a 6-week aerobic exercise intervention. 5 Whereas some microbiome changes were similar to those seen in our population, differences may be due to sex, age, body fat distribution, intensity of exercise, regional differences in diet, or the impact of co-medications more commonly prescribed among older adults.
Perhaps more important than the changes in bacterial communities are the functional changes in the microbiota and resulting metabolites (i.e. SCFAs). While the composition of the gut microbiome is a window into gut health, the functions of certain organisms are dependent upon dietary content, age, the inflammatory environment within the gut, stool transit time, and alterations in the gut immune system. The SCFA butyrate is clearly linked to exercise-related outcomes: butyrate is anti-inflammatory, promotes the activation of several pathways linked to adenosine triphosphate (ATP) production, and protects against skeletal muscle catabolism.15,37 Interestingly, although we did not see exercise-induced changes in genera with the most abundant butyrate-producing species (e.g. Faecalibacterium, Roseburia, Eubacterium), we did detect significant changes in the SCFA butyrate and a modest, non-significant increase in propionate with exercise in our small study of older persons. Studies in young adults have similarly found that greater physical activity levels 38 or exercise interventions4,5 were associated with higher levels or an increase in butyrate-producing species, SCFAs, or increased expression of butyryl-CoA:acetate CoA-transferase and methylmalonyl-CoA decarboxylase gene rRNA. The mechanisms by which exercise leads to increased stool butyrate are primarily speculative, but may include altered gut immunity, improved colonic transit time, or alterations in the microbiome induced by increased exercise-induced lactate.3,39 Thus the increase in butyrate with the exercise intervention may provide additional evidence and mechanisms underlying the numerous health benefits of exercise, particularly among older adults.
We also note that the existing literature linking physical activity or exercise to changes in the gut microbiome is limited nearly exclusively to younger populations, but changes that occur in the intestinal barrier, the microbiome, and the effect of interventions on the microbiome can differ considerably by age. Weakening of the intestinal barrier with age is suggested by lower levels of tight junction proteins and bacterial products such as lipopolysaccharide (LPS) and LPS receptors among older compared with younger adults. 40 A prior small study similarly found a reduction of gene expression involved in SCFA production among centenarians. 41
Limitations of this pilot study should be noted. Our small sample size limited adjustment for potential confounders, thus our results should be considered exploratory and confirmed in larger cohorts. Our cohort was nearly all male and did not include a non-exercise control group. Diet was ad libitum, dietary reports did not precisely overlap with the time of stool collection, and the time from collection of stool and transfer from home freezer to −80°C freezer varied and may have impacted the SCFA results. As the study was not specifically developed for microbiome assessments, no additional data on bowel habits were collected.
Conclusion
In summary, multiple studies have demonstrated shifts in either gut microbiota or microbial metabolites in association with increased physical activity. Results from our pilot study suggest that exercise resulted in changes in gut microbial composition and the metabolite butyrate in older adults, thus may reverse some reported features of age-related dysbiosis. Taken as a whole, our study provides yet another potential mechanism by which exercise exerts health benefits.
Acknowledgments
We thank the participants for the time contributed to this study.
Footnotes
Author contributions: KME and CMJ developed and implemented the parent study; KME, JL, RJ, MK, DNF, and CCW contributed to the design of the pilot study. KME and JL coordinated the collection of participant data; CER, DNF, YT, JH, and BH performed the microbiome, SCFA, or nutritional analyses; KME, JL, RJ, MK, DNF, SD and CCW contributed to the initial statistical analysis and interpretation. KME prepared the initial draft and all authors contributed to editing and review of the final manuscript.
Availability of data and materials: All 16S rRNA gene sequences and associated metadata were submitted to the NCBI sequence read archive under project accession number PRJNA657420. Primary endpoints regarding the exercise intervention were posted on ClinicalTrials.gov.
Conflict of interest statement: KME has received research funding (to the University of Colorado) from Gilead Sciences, and consulting payments from Theratechnologies, Gilead Sciences, and ViiV Pharmaceuticals.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the Gilead Sciences Research Scholars Program in HIV https://researchscholars.gilead.com/en/hiv_portal/program-overview (to KME), the National Institute of Aging of the National Institutes of Health https://www.nia.nih.gov/research
(K23AG050260) to KME, T32 AG279-15 to JL, the University of Colorado GI and Liver Innate Immune Program https://www.ucdenver.edu/academics/colleges/medicalschool/programs/Innate-Immune-Program/Pages/Pilot-Program-Funding.aspx
(KME, DNF, CER, and CCW), and the National Center for Advancing Translational Sciences, Colorado CTSA https://ncats.nih.gov/ctsa Grant Number UL1TR002535. The funding sources had no a role in data collection, analysis, or interpretation, trial design, or patient recruitment. No payments were made in the writing of this manuscript. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
ORCID iD: Kristine M. Erlandson  https://orcid.org/0000-0003-0808-6729
https://orcid.org/0000-0003-0808-6729
Contributor Information
Kristine M. Erlandson, Department of Medicine, Division of Infectious Diseases, University of Colorado Anschutz Medical Campus, 12700 E. 19th Avenue, Mail Stop B168, Aurora, CO 80045, USA.
Jay Liu, Department of Medicine, University of Colorado Anschutz Medical Campus, Aurora, CO, USA.
Rachel Johnson, Department of Biostatistics and Informatics, Colorado School of Public Health, Aurora, CO, USA.
Stephanie Dillon, Department of Medicine, University of Colorado Anschutz Medical Campus, Aurora, CO, USA.
Catherine M. Jankowski, College of Nursing, University of Colorado Anschutz Medical Campus, Aurora, CO, USA
Miranda Kroehl, Department of Biostatistics and Informatics, Colorado School of Public Health, Aurora, CO, USA.
Charles E. Robertson, Department of Medicine, University of Colorado Anschutz Medical Campus, Aurora, CO, USA
Daniel N. Frank, Department of Medicine, University of Colorado Anschutz Medical Campus, Aurora, CO, USA
Yunus Tuncil, Food Engineering Department, Ordu University, Ordu, Turkey.
Janine Higgins, Department of Pediatrics, University of Colorado Anschutz Medical Campus, Aurora, CO, USA.
Bruce Hamaker, Department of Food Science, Purdue University, West Lafayette, IN, USA.
Cara C. Wilson, Department of Medicine, University of Colorado Anschutz Medical Campus, Aurora, CO, USA
References
- 1. Dillon SM, Wilson CC. What is the collective effect of aging and HIV on the gut microbiome? Curr Opin HIV AIDS 2020; 15: 94–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aleman FDD, Valenzano DR. Microbiome evolution during host aging. PLoS Pathog 2019; 15: e1007727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mailing LJ, Allen JM, Buford TW, et al. Exercise and the gut microbiome: a review of the evidence, potential mechanisms, and implications for human health. Exerc Sport Sci Rev 2019; 47: 75–85. [DOI] [PubMed] [Google Scholar]
- 4. Munukka E, Ahtiainen JP, Puigbo P, et al. Six-week endurance exercise alters gut metagenome that is not reflected in systemic metabolism in over-weight women. Front Microbiol 2018; 9: 2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Allen JM, Mailing LJ, Niemiro GM, et al. Exercise alters gut microbiota composition and function in lean and obese humans. Med Sci Sports Exerc 2018; 50: 747–757. [DOI] [PubMed] [Google Scholar]
- 6. Cronin O, Barton W, Skuse P, et al. A prospective metagenomic and metabolomic analysis of the impact of exercise and/or whey protein supplementation on the gut microbiome of sedentary adults. mSystems 2018; 3: e00044-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Taniguchi H, Tanisawa K, Sun X, et al. Effects of short-term endurance exercise on gut microbiota in elderly men. Physiol Rep 2018; 6: e13935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Morita E, Yokoyama H, Imai D, et al. Aerobic exercise training with brisk walking increases intestinal bacteroides in healthy elderly women. Nutrients 2019; 11: 868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Department of Health and Human Services. Physical activity guidelines for Americans. 2nd ed. Washington, DC: US Department of Health and Human Services, https://health.gov/paguidelines/second-edition/pdf/Physical_Activity_Guidelines_2nd_edition.pdf (2018, accessed 14 January 2020). [Google Scholar]
- 10. Erlandson KM, MaWhinney S, Wilson M, et al. Physical function improvements with moderate or high-intensity exercise among older adults with or without HIV infection. Aids 2018; 32: 2317–2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu J, Johnson R, Dillon S, et al. Among older adults, age-related changes in the stool microbiome differ by HIV-1 serostatus. EBioMedicine 2019; 40: 583–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bishehsari F, Engen PA, Preite NZ, et al. Dietary fiber treatment corrects the composition of gut microbiota, promotes SCFA production, and suppresses colon carcinogenesis. Genes (Basel) 2018; 9: 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Carpenter C, Frank DN, Williamson K. tidyMicro: a pipeline for microbiome analysis and visualization. https://cran.r-project.org/package=tidyMicro (2020, accessed 4 April 2020). [DOI] [PMC free article] [PubMed]
- 14. Oksanen J, Guillaume Blanchet F, Friendly M, et al. vegan: community ecology package. R package version 2.5-7. https://cran.r-project.org/package=vegan (2020, accessed 4 April 2020).
- 15. Ticinesi A, Lauretani F, Milani C, et al. Aging gut microbiota at the cross-road between nutrition, physical frailty, and sarcopenia: is there a gut-muscle axis? Nutrients 2017; 9: 1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wu GD, Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011; 334: 105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ley RE. Gut microbiota in 2015: prevotella in the gut: choose carefully. Nat Rev Gastroenterol Hepatol 2016; 13: 69–70. [DOI] [PubMed] [Google Scholar]
- 18. Gemikonakli G, Mach J, Hilmer SN. Interactions between the aging gut microbiome and common geriatric giants: polypharmacy, frailty and dementia. J Gerontol A Biol Sci Med Sci 2021; 76: 1019–1028. [DOI] [PubMed] [Google Scholar]
- 19. Larsen JM. The immune response to Prevotella bacteria in chronic inflammatory disease. Immunology 2017; 151: 363–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tandon D, Haque MM, Saravanan R, et al. A snapshot of gut microbiota of an adult urban population from Western region of India. PLoS One 2018; 13: e0195643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. De Filippo C, Di Paola M, Ramazzotti M, et al. Diet, environments, and gut microbiota. A preliminary investigation in children living in rural and urban Burkina Faso and Italy. Front Microbiol 2017; 8: 1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Villanueva-Millan MJ, Perez-Matute P, Oteo JA. Gut microbiota: a key player in health and disease. A review focused on obesity. J Physiol Biochem 2015; 71: 509–525. [DOI] [PubMed] [Google Scholar]
- 23. Kuo SM. Does modification of the large intestinal microbiome contribute to the anti-inflammatory activity of fermentable fiber? Curr Dev Nutr 2018; 2: nzx004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. De Vuyst L, Leroy F. Cross-feeding between bifidobacteria and butyrate-producing colon bacteria explains bifdobacterial competitiveness, butyrate production, and gas production. Int J Food Microbiol 2011; 149: 73–80. [DOI] [PubMed] [Google Scholar]
- 25. Garcia-Mantrana I, Selma-Royo M, Alcantara C, et al. Shifts on gut microbiota associated to mediterranean diet adherence and specific dietary intakes on general adult population. Front Microbiol 2018; 9: 890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Walters WA, Xu Z, Knight R. Meta-analyses of human gut microbes associated with obesity and IBD. FEBS Lett 2014; 588: 4223–4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Konikoff T, Gophna U. Oscillospira: a central, enigmatic component of the human gut microbiota. Trends Microbiol 2016; 24: 523–524. [DOI] [PubMed] [Google Scholar]
- 28. Petrov VA, Saltykova IV, Zhukova IA, et al. Analysis of gut microbiota in patients with Parkinson’s disease. Bull Exp Biol Med 2017; 162: 734–737. [DOI] [PubMed] [Google Scholar]
- 29. Lin D, Peters BA, Friedlander C, et al. Association of dietary fibre intake and gut microbiota in adults. Br J Nutr 2018; 120: 1014–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Doumatey AP, Adeyemo A, Zhou J, et al. Gut microbiome profiles are associated with type 2 diabetes in urban Africans. Front Cell Infect Microbiol 2020; 10: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yang Q, Lin SL, Kwok MK, et al. The roles of 27 genera of human gut microbiota in ischemic heart disease, type 2 diabetes mellitus, and their risk factors: a Mendelian randomization study. Am J Epidemiol 2018; 187: 1916–1922. [DOI] [PubMed] [Google Scholar]
- 32. Mitchell CM, Davy BM, Hulver MW, et al. Does exercise alter gut microbial composition? A systematic review. Med Sci Sports Exerc 2019; 51: 160–167. [DOI] [PubMed] [Google Scholar]
- 33. Clarke TB. Microbial programming of systemic innate immunity and resistance to infection. PLoS Pathogens 2014; 10: e1004506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Durk RP, Castillo E, Marquez-Magana L, et al. Gut microbiota composition is related to cardiorespiratory fitness in healthy young adults. Int J Sport Nutr Exerc Metab 2019; 29: 249–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yang Y, Shi Y, Wiklund P, et al. The association between cardiorespiratory fitness and gut microbiota composition in premenopausal women. Nutrients 2017; 9: 792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bressa C, Bailen-Andrino M, Perez-Santiago J, et al. Differences in gut microbiota profile between women with active lifestyle and sedentary women. PLoS One 2017; 12: e0171352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Leonel AJ, Alvarez-Leite JI. Butyrate: implications for intestinal function. Curr Opin Clin Nutr Metab Care 2012; 15: 474–479. [DOI] [PubMed] [Google Scholar]
- 38. Barton W, Penney NC, Cronin O, et al. The microbiome of professional athletes differs from that of more sedentary subjects in composition and particularly at the functional metabolic level. Gut 2018; 67: 625–633. [DOI] [PubMed] [Google Scholar]
- 39. Okamoto T, Morino K, Ugi S, et al. Microbiome potentiates endurance exercise through intestinal acetate production. Am J Physiol Endocrinol Metab 2019; 316: E956–E966. [DOI] [PubMed] [Google Scholar]
- 40. Lustgarten MS. Classifying aging as a disease: the role of microbes. Front Genet 2016; 7: 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rampelli S, Candela M, Turroni S, et al. Functional metagenomic profiling of intestinal microbiome in extreme ageing. Aging (Albany NY) 2013; 5: 902–912. [DOI] [PMC free article] [PubMed] [Google Scholar]