Summary
Skeletal muscle insulin resistance is a central defect in the pathogenesis of type 2 diabetes (T2D). Here, we analyzed skeletal muscle proteome in 148 vastus lateralis muscle biopsies obtained from men covering all glucose tolerance phenotypes: normal, impaired fasting glucose (IFG), impaired glucose tolerance (IGT) and T2D. Skeletal muscle proteome was analyzed by a sequential window acquisition of all theoretical mass spectra (SWATH-MS) proteomics technique. Our data indicate a downregulation in several proteins involved in mitochondrial electron transport or respiratory chain complex assembly already in IFG and IGT muscles, with most profound decreases observed in T2D. Additional phosphoproteomic analysis reveals altered phosphorylation in several signaling pathways in IFG, IGT, and T2D muscles, including those regulating glucose metabolic processes, and the structure of muscle cells. These data reveal several alterations present in skeletal muscle already in prediabetes and highlight impaired mitochondrial energy metabolism in the trajectory from prediabetes into T2D.
Subject areas: Molecular biology, Diabetology, Proteomics
Graphical abstract
Highlights
-
•
Skeletal muscle proteome from men with all stages of glucose tolerance was analyzed
-
•
Phosphoproteomics reveal altered phosphorylation in IFG, IGT, and T2D muscles
-
•
OXPHOS proteins are decreased in prediabetic muscles, with most decrease in T2D
Molecular biology; Diabetology; Proteomics
Introduction
The prevalence of type 2 diabetes (T2D) has reached epidemic proportions worldwide. As a chronic metabolic disease, T2D contributes to increased morbidity and mortality, leading to increased economic strain on health care as well as human suffering (Zheng et al., 2017; Unnikrishnan et al., 2017). Despite intensive efforts, currently available methods to prevent and treat T2D are far from optimal, and a significant proportion of patients doesnot meet treatment goals (Unnikrishnan et al., 2017, American Diabetes Association, 2018). Thus, more research is urgently needed to understand the pathogenesis of T2D in more detail.
Beyond its obvious role in locomotion, skeletal muscle is the main site of glucose uptake and accounts for ∼80% of total insulin-stimulated glucose disposal (Defronzo et al., 1981; Defronzo et al., 1985). Skeletal muscle insulin resistance is a central and early defect in the pathogenesis of T2D (Eriksson et al., 1989, 1992; Cline et al., 1999; Defronzo et al., 1985). Therefore, detailed knowledge about the factors regulating insulin action in muscle is likely to improve the treatment of people with insulin resistance, and help prevent the development of T2D. To get further insight into the role of the skeletal muscle in the pathogenesis of T2D, we have previously obtained vastus lateralis skeletal muscle biopsies from 271 Finnish individuals spanning all glucose tolerance phenotypes (normal glucose tolerance [NGT], impaired fasting glucose [IFG], impaired glucose tolerance [IGT], and T2D subjects) (Scott et al., 2016). We have profiled their muscle transcriptome with high-depth strand-specific mRNA sequencing and performed dense array-based genotyping. We found that the expression of several different gene groups, such as genes involved in ER protein localization or cellular respiration, was decreased in T2D muscles (Scott et al., 2016). The observed decrease in genes involved in cellular respiration is in good agreement with earlier studies on muscle transcriptome that demonstrated decreased expression of genes regulating oxidative metabolism and mitochondrial function in insulin resistance and T2D (Mootha et al., 2003; Patti et al., 2003). Taken together, studies on carefully characterized humans exhibiting normal, prediabetic as well as diabetic glucose metabolism, are powerful approaches to reveal important and novel mechanisms contributing to insulin resistance and T2D.
Genes function as building instructions to proteins which ultimately regulate and carry out cellular functions. As alterations in mRNA expression may not be accurately reflective of the respective protein concentration (Edfors et al., 2016), it is crucial to analyze potential differences in protein expression between the different glucose tolerance phenotypes to get insight into the potential causes leading to metabolic alterations. To this end, several relatively small studies, with 6–11 individuals per different phenotype (e.g., lean, obese, and type 2 diabetic), have used proteomics techniques. These studies have revealed important differences in skeletal muscle of insulin resistant individuals, such as an increase in expression of proteins involved in glycolysis, a decrease in mitochondrial proteins, and alterations in muscle structural proteins (Højlund et al., 2003; Hwang et al., 2010; Giebelstein et al., 2012; Chae et al., 2018). However, a shortcoming in the previous studies on skeletal muscle proteome may have been limited proteome coverage, as for example the most expressed proteins, such as contractile proteins, have been quantified, while the low-abundance regulatory proteins have remained undetected (Deshmukh, 2016). The recent development of quantitative proteomic techniques using liquid chromatography-tandem mass spectrometry combined with enhanced bioinformatic analyses has enabled deeper coverage of the proteome (Deshmukh, 2016; Loukovaara et al., 2015). This advance in the field is crucial for understanding the complex molecular interactions at tissue level that predispose to insulin resistance.
Most proteomic studies have been analyzed using data-dependent acquisition (DDA) method where a selected number of peptides are sequentially selected for MS2 analysis. However, the stochastic precursor selection of DDA can lead to inconsistent detection of peptides and the corresponding proteins in the sample cohort, resulting in many missing values in the data (Gillet et al., 2012; Krasny and Huang, 2021). In the present study, we have analyzed 148 skeletal muscle proteomes from men who cover all glucose tolerance phenotypes: NGT, IFG, IGT, as well as T2D, using a data-independent acquisition method, a sequential window acquisition of all theoretical mass spectra (SWATH-MS) proteomics technique (Gillet et al., 2012; Krasny and Huang, 2021). SWATH-MS is an emerging mass spectrometric method that offers a high degree of quantitative accuracy, proteomic coverage, reproducibility of proteome coverage, and sample throughput, as well as a low number of missing values. Our data indicate a strong downregulation in several proteins involved in mitochondrial electron transport or respiratory chain complex assembly already in IFG and IGT muscles, with most profound decreases observed in T2D. These data highlight impaired mitochondrial energy metabolism in the trajectory from prediabetes into T2D.
Results
SWATH-MS-based quantitative analysis of human muscle samples
We obtained vastus lateralis skeletal muscle biopsies from 148 men whose glucose tolerance covered all phenotypes from normal to T2D (Table 1), and we performed extensive proteome profiling of the muscle samples using label-free quantitative SWATH-MS proteomic analysis. The workflow used is shown in Figure 1A. Type 2 diabetic men had good glycemic control and they were slightly older than men with normal glucose tolerance. Men with IGT or T2D had higher body weight and had a significantly greater BMI compared to normal glucose tolerant men (Table 1).
Table 1.
Male volunteers participating in the study
| NGT (n = 85) | IFG (n = 25) | IGT (n = 14) | T2D (n = 24) | |
|---|---|---|---|---|
| Age (y) | 51 ± 1 | 53 ± 2 | 55 ± 2 | 57 ± 2a |
| Weight (kg) | 83.1 ± 1.6 | 86.5 ± 3.1 | 97.6 ± 2.8b | 95.4 ± 3.1b |
| Height (cm) | 180 ± 1 | 178 ± 1 | 180 ± 2 | 179 ± 1 |
| BMI (kg/m2) | 25.7 ± 0.4 | 27.3 ± 0.8 | 30.3 ± 1.1b | 29.8 ± 1.0c |
| Waist (cm) | 95 ± 1 | 100 ± 2 | 108 ± 3c | 106 ± 3c |
| Hip (cm) | 99 ± 1 | 100 ± 1 | 106 ± 2b | 103 ± 1 |
| WHR | 0.95 ± 0.01 | 0.99 ± 0.01 | 1.02 ± 0.02b | 1.03 ± 0.02c |
| fB-glucose (mmol/L) (n) | 5.3 ± 0.08 (24) | 5.4 ± 0.2 (10) | 5.0 ± 0.3 (5) | 6.7 ± 0.3c, d, e (11) |
| fP-glucose (mmol/L) (n) | 5.5 ± 0.05 (61) | 6.3 ± 0.2 b (15) | 6.3 ± 0.2 a (9) | 7.6 ± 0.4 c, d, e (13) |
| HbA1c (%) | 5.3 ± 0.03 | 5.4 ± 0.07 | 5.6 ± 0.09 | 6.3 ± 0.12 c, d, e |
| HbA1c (mmol/mol) | 35 ± 0.3 | 35 ± 0.8 | 37 ± 1.0 | 45 ± 1.3c, d, e |
| Fasting serum insulin (mU/l) | 8 ± 1 | 13 ± 2 a | 18 ± 2 c | 18 ± 3 c |
| Fasting serum cholesterol (mmol/L) | 4.4 ± 0.1 | 4.4 ± 0.1 | 4.3 ± 0.2 | 4.1 ± 0.2 |
| Fasting serum triglycerides (mmol/L) | 1.2 ± 0.1 | 1.3 ± 0.1 | 1.8 ± 0.2 b | 1.5 ± 0.1 a |
| Fasting serum HDL cholesterol (mmol/L) | 1.4 ± 0.03 | 1.3 ± 0.05 | 1.1 ± 0.05 c | 1.2 ± 0.05 |
| Fasting serum LDL cholesterol (mmol/L) | 1.7 ± 0.05 | 1.7 ± 0.06 | 1.7 ± 0.14 | 1.5 ± 0.09 |
| HOMA-IR | 1.4 ± 0.1 | 2.6 ± 0.4 a | 3.4 ± 0.5 c | 4.2 ± 0.6 c, f |
Data are given as mean ± SEM, one-way ANOVA followed by Sidak's post hoc test for multiple comparisons.
p < 0.05 vs NGT.
p < 0.01 vs NGT.
p < 0.001 vs NGT.
p < 0.001 vs IFG.
p < 0.001 vs IGT.
p < 0.01 vs IFG.
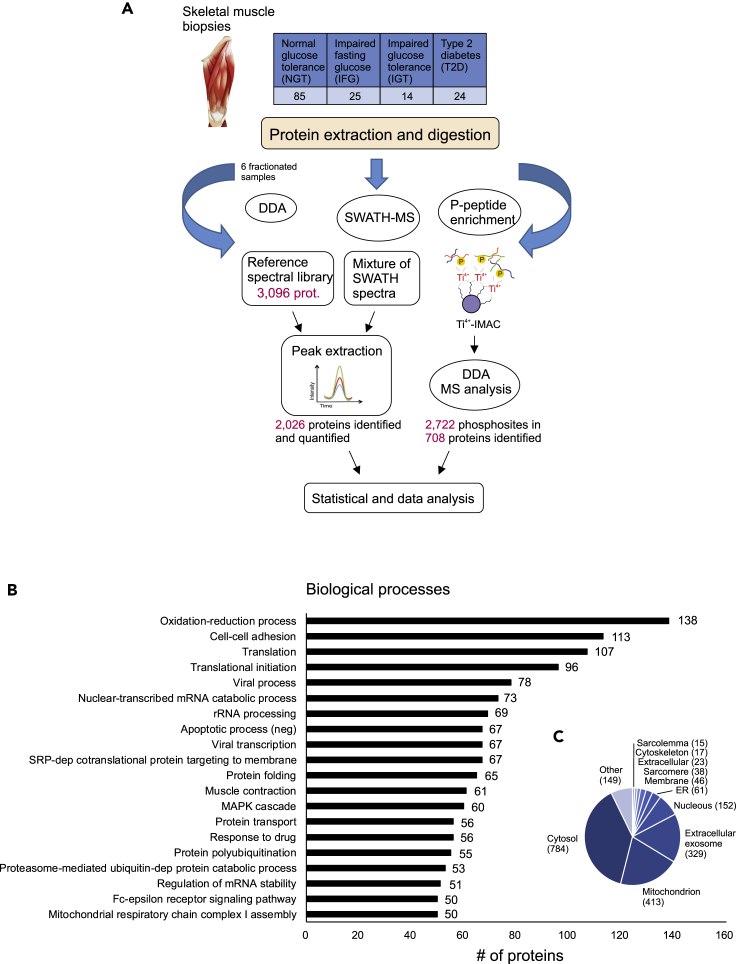
Figure 1.
Quantitative SWATH-MS analysis of human muscle samples
(A) Muscle biopsies were collected from 148 men whose glucose tolerance covered all phenotypes from normal to type 2 diabetes. Muscle proteins were extracted and digested with trypsin. For spectral library generation, the resulting peptides were fractionated and analyzed using data-dependent acquisition mode (DDA). The spectral library built was then used to extract the peptide and the quantification information of the SWATH runs. In addition, six samples from each glucose tolerance group were subjected to phosphopeptide analysis. Statistical analysis and bioinformatics approaches were used to understand the biological relevance of the differentially expressed proteins.
(B and C) (B) The DAVID bioinformatics tool was used to classify all 2,026 identified proteins as per their biological processes and to predict cellular location (C).
The SWATH-MS data analysis depends on a reference assay library, which must be sample-specific and be of sufficient compositional depth to enable extensive peptide identification. Therefore, a muscle-sample-specific reference spectral library was first generated by DDA analysis of the six fractionated muscle samples that covered all glucose tolerance phenotypes: NGT (n = 2), a combination of IFG and IGT (n = 1), and T2D (n = 3). This resulted in a spectral library consisting of 3,096 human proteins. In quantitative SWATH-MS experiments, a total of 2,026 human proteins were identified and quantified across all samples (see Table S1). The most abundant proteins in samples are typical sarcomere proteins, such as actins, myosins, tropomyosins, and titin. The identified proteins were associated with typical muscle functions, such as oxidation-reduction, cell-cell adhesion, translation, muscle contraction, and mitochondrial electron transport (Figure 1B). In addition, the subcellular localization of the identified proteins was a characteristic of muscle tissue (Figure 1C), including mitochondria, cytosol, sarcomere, and sarcolemma. This corresponds well with the fact that skeletal muscle proteins are compartmentalized in different compartments including microsome, sarcolemma, cytosol, contractile apparatus, and mitochondria (Ohlendieck, 2011).
Comparative proteomics contrasting all glucose tolerance phenotypes
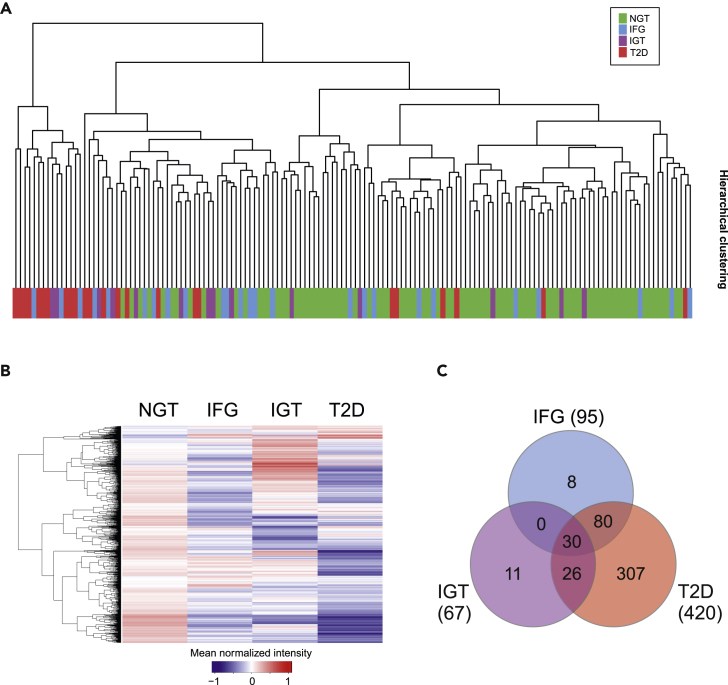
To get an overall view of protein profiles of the skeletal muscle samples in different glucose tolerance phenotypes, we performed hierarchical clustering of all quantified proteins based on the abundance of the proteome profile (Figure 2A). Most prediabetes or T2D samples accumulated on the other side of the dendrogram, in separate from samples with normal glucose tolerance. A similar result was seen in principal component analysis that showed some grouping of samples. However, no clear difference between sample types could be observed (see Figure S1). In spite of a high similarity in the protein profiles at the individual level, differences were detected between sample groups. The unique protein profile of each glucose tolerance category was visualized by a heatmap of the average of protein abundance in different sample groups (Figure 2B).
Figure 2.
Changes in skeletal muscle proteome in different glucose tolerance phenotypes
(A) Hierarchical clustering of all quantified proteins based on the abundance of the proteome profile.
(B) In the Z-score heatmap the expression levels of all 2,026 quantified proteins are shown as the mean of the SWATH peak area for each sample group.
(C) Venn diagrams showing the overlap between significantly different proteins in IFG, IGT, and T2D proteomes compared with NGT proteome. The number in brackets indicates how many proteins were significantly altered (q < 0.05) in each sample group.
Because we detected mean differences in prediabetes and T2D proteomes compared with NGT proteomes, we next performed one-way ANOVA analysis to find the proteins that are present in significantly altered concentrations in the different glucose tolerance sample groups. In comparison with NGT samples, the relative abundance of 95 and 67 proteins was significantly altered (q < 0.05) in IFG and IGT samples, respectively (Figure 2C and Table S2). However, more profound changes in the proteome were detectable in T2D samples, where 420 proteins were significantly changed when compared with NGT. The changes in protein abundance are mainly moderate and decreasing relative to NGT, as shown in heatmap in Figure 2B.
Biological processes behind differentially expressed proteins
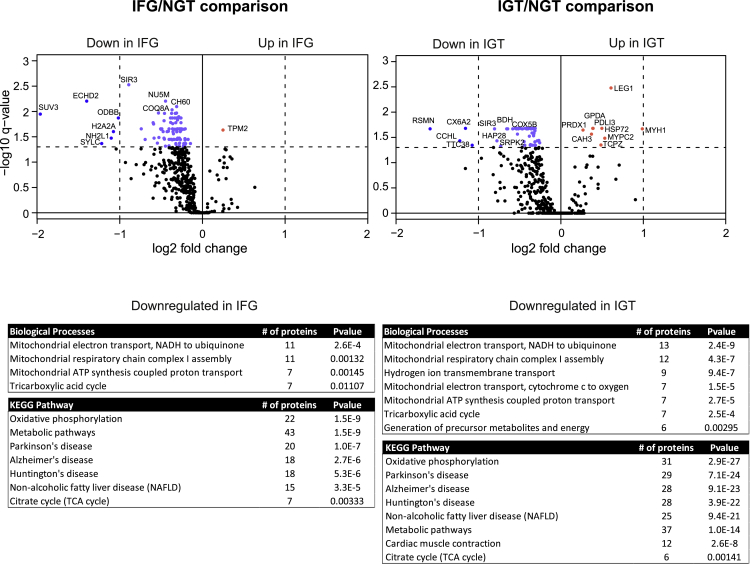
When the proteomes of prediabetic IFG and IGT were compared with NGT, the majority of the altered proteins were found to be reduced (Figure 3). Only one protein in IFG comparison and nine proteins in IGT comparison were upregulated in prediabetic condition. To understand the functional association of these altered proteins, the proteins were classified as per their involvement in different biological processes and KEGG pathways using DAVID bioinformatics resources (Figure 3). The muscle library was used here as a background database. Our data indicate a strong downregulation in mitochondrial energy metabolism in IFG and IGT samples because several proteins involved in mitochondrial electron transport (11 proteins in IFG and 13 in IGT) or mitochondrial respiratory chain complex I assembly (11 proteins in IFG and 12 in IGT) were decreased when compared with NGT. In addition, KEGG pathway analysis revealed a downregulation of oxidative phosphorylation (22 proteins in IFG and 31 in IGT) and other pathways related to energy metabolism.
Figure 3.
Comparison between prediabetes and NGT proteomes
The volcano plots of differentially expressed muscle proteins between IFG and NGT (left panel) or IGT and NGT (right panel). p-value was set to match q-value < 0.05. The light blue dots indicate significantly down-regulated proteins with FC < 2, and the blue dots with FC > 2. The red dots indicate significantly up-regulated proteins. Down-regulated proteins in the prediabetes samples were categorized according to their biological processes and KEGG pathways using DAVID bioinformatics tool. The most significant events are listed with Benjamini corrected p-value.
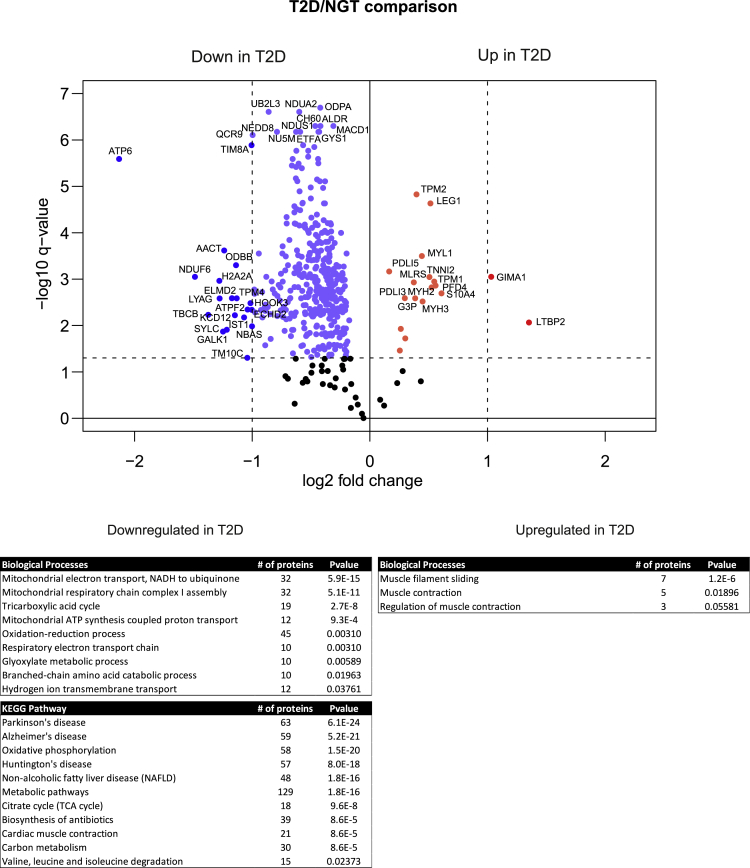
More differences in proteome level were detected when T2D samples were compared with NGT (Figure 4). Similarly, the expression levels of the most proteins were reduced relative to NGT, but we could also detect a group of proteins with higher abundance. The downregulated proteins were strongly associated with energy metabolism as seen in GO enrichment and KEGG pathway analysis. We could detect a decrease in several proteins involved in aerobic respiration, such as tricarboxylic acid cycle (the citric acid cycle, 19 proteins) and the electron transport system (32 proteins), as well as proteins linked to branched-chain amino acid catabolism (10 proteins). Interestingly, many proteins with oxidoreductase activity involved in oxidation-reduction (redox) processes (45 proteins) were also found to be decreased in T2D. Redox equilibrium is essential for cellular homeostasis, being a key modulator of skeletal muscle plasticity/dysfunction in response to exercise or metabolic diseases (Espinosa et al., 2016).
Figure 4.
Comparison between type 2 diabetes and NGT proteomes
The volcano plots of differentially expressed muscle proteins between T2D and NGT. p-value was set to match q-value < 0.05. The light blue dots indicate significantly down-regulated proteins with FC < 2 and the blue dots with FC > 2. The red dots indicate significantly upregulated proteins. Downregulated and upregulated proteins in the T2D samples were categorized as per their biological processes and KEGG pathways using DAVID bioinformatics tool. The most significant events are listed with Benjamini corrected p-value.
A total 18 upregulated proteins were also detected in T2D samples. These proteins were involved in muscle filament sliding (7 proteins) and muscle contraction (5 proteins). These muscle function proteins, including myosin (MYH2, MYH3, MYL1), tropomyosin (TPM1, TPM2), and troponin 1 (TNNI2), were all slightly upregulated in T2D samples, indicating that type 2 diabetic metabolism affects muscle structure and function (Figure 4).
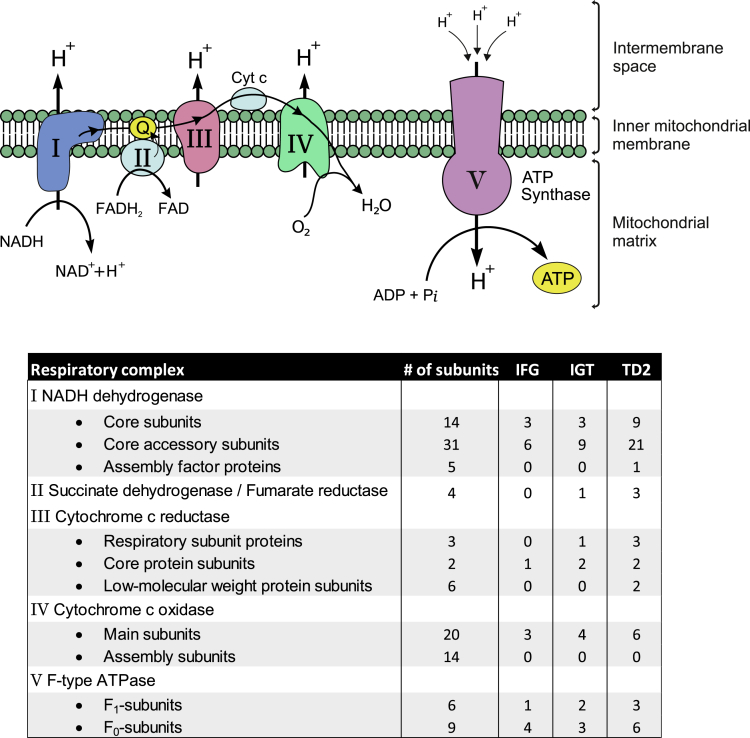
Oxidative phosphorylation
Proteomics analysis of skeletal muscle samples from different glucose tolerance phenotypes revealed changes in energy metabolic processes, especially in oxidative phosphorylation (OXPHOS, Figure 5). Oxidative phosphorylation is a highly efficient metabolic pathway to produce large amounts of ATP. In this process, electrons from NADH or FADH2 are transferred to O2 simultaneously pumping protons out from the mitochondrial matrix by a set of enzymes which is called the electron transport chain (complexes I to IV). In the final step, ATP is synthesized when protons flow back to the mitochondrial matrix through ATP synthase (complex V). All complexes of the respiratory chain consist of several subunits, and we observed a decrease in the several different subunits (Figure 5). Some changes were apparent already in prediabetic conditions, but the most obvious changes were seen in T2D samples. Especially, 31 subunits of 50 in complex I (NADH dehydrogenase) and 7 subunits of 11 in complex III (cytochrome c reductase) were downregulated in T2D samples. In addition, a number of subunits of both F1 and F0 domains of ATP synthase were reduced in T2D samples compared with the NGT group. These data are consistent with previous observations regarding obese and type 2 diabetic skeletal muscle (Højlund et al., 2003; Hwang et al., 2010; Giebelstein et al., 2012; Chae et al., 2018). Reduced OXPHOS gene expression has been observed also in skeletal muscle in people with IGT (Osler et al., 2015; Mootha et al., 2003).
Figure 5.
Oxidative phosphorylation is downregulated in skeletal muscle in prediabetes and type 2 diabetes
The oxidative phosphorylation system (OXPHOS) of the mitochondrial inner membrane is composed of five enzymes (respiratory complexes I–V). All respiratory enzymes consist of several subunits (the total number of subunits shown in the table), and we observed a decrease in most subunits already in prediabetes (IFG, IGT). Number of proteins with significant differences compared with NGT for each sample group is shown in the table.
Phosphorylation in skeletal muscle proteome
To complement the quantitative results, we characterized the general phosphorylation events in a subset of skeletal muscle samples representing all different glucose tolerance phenotypes (6 men in each glucose tolerance group NGT, IFG, IGT, and T2D) using phosphoproteomic analysis. We identified a total of 2722 distinct phosphorylated amino acids in 708 phosphoproteins (see Table S3), indicating that several proteins were phosphorylated at multiple sites. A large proportion of the identified phosphopeptides originated from highly abundant myofibrillar proteins. This is in agreement with previous publications that demonstrate that sarcomeric proteins are overrepresented in phosphoanalysis (Højlund et al., 2009; Potts et al., 2017). For example, in our study, the protein with the most phosphorylation sites was titin, in which we identified 384 distinct phosphorylation sites. This result is not unexpected because titin is a giant protein that contributes to approximately 16% of the total protein mass in skeletal muscle (Deshmukh et al., 2015). Moreover, titin is highly phosphorylated: more than 960 phosphosites in human titin are reported in the PhosphoSitePlus database (www.phosphosite.org). Other sarcomere proteins with multiple phosphorylated sites, such as myosins (225 distinct phosphosites), actins (19 phosphosites), and tropomyosins (34 phosphosites), were also detected.
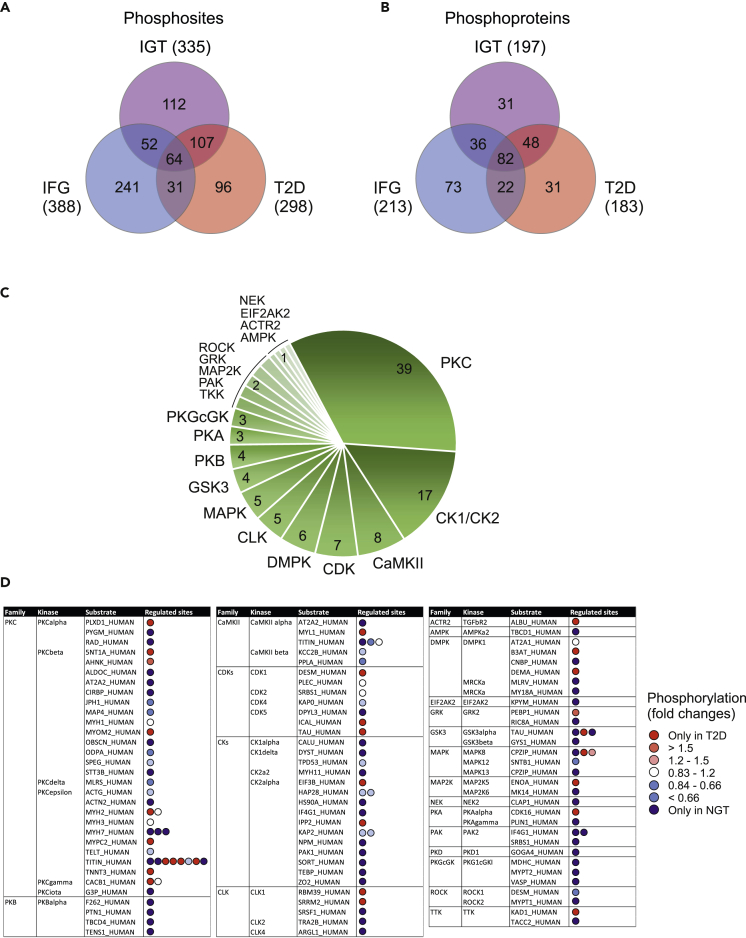
Next, we compared these identified phosphosites in samples from men with prediabetic IFG and IGT, or T2D with samples from men with normal glucose tolerance to identify the skeletal muscle proteins where phosphorylation is altered during progression to diabetes. We identified 388, 335, and 298 phosphosites that exhibited a major alteration in abundance (p-value < 0.05, or observed only in another condition) in IFG, IGT, and T2D sample groups, respectively (Figure 6A). These phosphosites were linked to 213, 197, and 183 different phosphoproteins, respectively (Figure 6B). KEGG pathway overrepresentation analysis of these proteins with altered phosphorylation revealed an enrichment of signaling pathways regulating glucose metabolic processes, muscle contraction, and the structure of muscle cells, correlating well with SWATH-quantification results (see Table S4). Alterations were identified in the glycolysis/gluconeogenesis and insulin signaling pathway events. Interestingly, a clear change in phosphorylation was already evident in IFG, indicating that changes in skeletal muscle are already evident at an early stage in dysregulated glucose metabolism.
Figure 6.
Phosphorylation in skeletal muscle proteome
(A and B) Venn diagram representing the number of phosphosites (A) or phosphoproteins (B) with a significant change in abundance (p-value < 0.05, or observed only in another condition) in the IFG, IGT and T2D sample groups compared with NGT.
(C) NetworKIN software was used to predict kinases that are dysregulated in type 2 diabetes. 298 phosphorylated sites that exhibited an alteration in abundance in T2D samples were used as input. A total of 20 distinct kinase families associated with 115 phosphorylated sites was predicted. The numbers indicate the total number of phosphorylation sites linked to the specific kinases.
(D) The more specific details about NetworKIN predicted kinase substrates. All the substrates listed here have significant differences between T2D and NGT samples. Red spot means that the substrate is phosphorylated more in T2D samples, indicating more active kinases in T2D. Blue spot indicates more active kinase in NGT samples (more phosphorylation in NGT samples). Some substrate proteins have multiple regulated phosphorylation sites (in sequence order in the figure). Phosphosites are indicated in Table S3.
To investigate which kinases controlled the phosphorylation of proteins in T2D samples, we used NetworKIN software, which combines protein-associated network and preferred recognition motif information to predict the likelihood of kinase-substrate relationships (Horn et al., 2014). This analysis predicted a kinase in 115 of 298 detected phosphosites in T2D samples (Figure 6C). The analysis revealed a very high prediction for protein kinase C (PKC) kinases (PKCα, PKCβ, PKCδ, PKCε, PKCγ), regulating a total of 39 substrate phosphosites. Also casein kinases (CK) 1 and 2 were heavily enriched, having 17 substrate sites. In addition, the NetworKIN algorithm predicted Ca2+/calmodulin-dependent protein kinase II, (CaMKII), cyclin-dependent kinases (CDKs), myotonin-protein kinase (DMPK), CLK, and MAPKs as likely candidates involved in the diabetes process. The more detailed analysis of the phosphorylation of these kinase substrates indicates that many kinases were less active in diabetes samples compared with NGT samples (Figure 6D). For example, with a few exceptions, phosphorylation of PKCβ and CKs substrates was markedly reduced or in some cases not detectable at all in T2D samples. Similarly, many substrates of PKB and CaMKII were less phosphorylated in T2D samples. On the other hand, based on NetworKIN analysis, more active kinases, such as PKCε, were also detected. PKCε phosphorylates sarcomere proteins, such as myosins and titin, indicating a regulatory role in contractile function and the structure of muscles. We also observed a slight increase in the activity of CDK kinases in T2D samples.
Changes in proteome with aging
We had the opportunity to analyze skeletal muscle proteome in paired muscle biopsies obtained several years apart from two men: one man with NGT (7-year interval) and one man with T2D (9-year interval). Expression of 20 proteins was upregulated in both men as they were elderly (see Table S5). The Jupiter microtubule-associated homolog 1 (JPT1) protein abundance increased 185-fold in T2D muscle during the 9-year follow-up.
Unique differentially expressed proteins
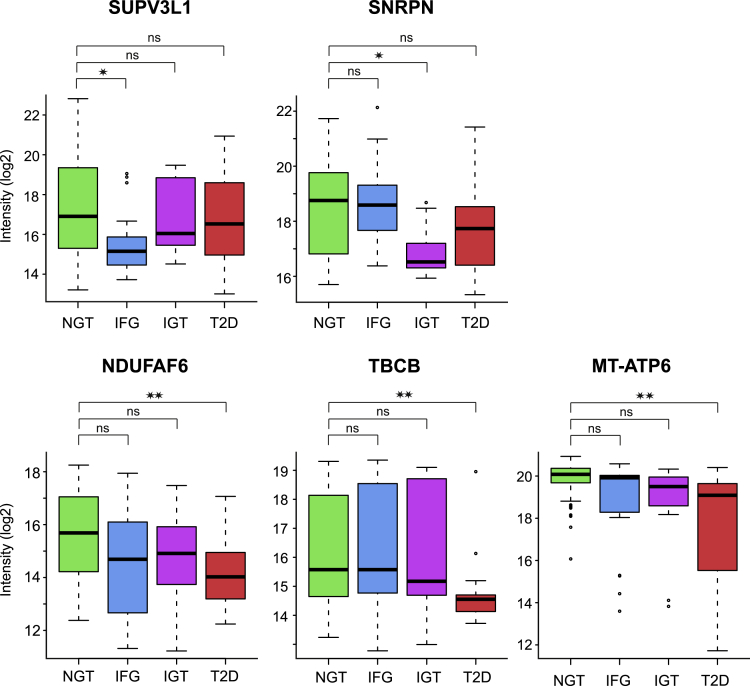
We identified interesting individual proteins that displayed a significant decrease (q < 0.05) when comparing the prediabetic or T2D samples with the NGT samples (Figure 7). ATP-dependent RNA helicase SUPV3L1, mitochondrial (SUPV3L1) regulates mitochondrial RNA metabolism and it also unwinds dsDNA (Minczuk et al., 2002; Szczesny et al., 2010). SUPV3L1 protein expression was uniquely reduced in IFG muscles (3.9 times less compared with NGT). Small nuclear ribonucleoprotein-associated protein N (SNRPN) is a part of the spliceosome, and it regulates pre-mRNA processing and may be involved in tissue-specific alternative splicing (Jing et al., 2015). SNRPN expression was uniquely downregulated in IGT muscles (3 times less compared with NGT). ATP synthase subunit a (MT-ATP6) is a part of the mitochondrial membrane ATP synthase (complex V) (Ganetzky et al., 2019). NDUFAF6 is crucial in the assembly of mitochondrial NADH:ubiquinone oxidoreductase complex (complex I) (Pagliarini et al., 2008, Mckenzie et al., 2011), and tubulin-folding cofactor B regulates tubulin synthesis (Tian et al., 1997). The expression of MT-ATP6 (4.3 times less compared with NGT), NDUFAF6 (2.8 times less compared with NGT), and tubulin-folding cofactor B (2.6 times less compared with NGT) was uniquely downregulated in T2D muscles.
Figure 7.
Unique differentially expressed proteins
Box-and-whisker plots of differentially expressed individual proteins that displayed a significantly different abundance (q < 0.05) in the prediabetic or T2D samples compared with the NGT samples. Box-and-whisker plots indicate median and interquartile range; the whiskers extend to the most extreme data point which is no more than 1.5 times the interquartile range from the box. ATP-dependent RNA helicase SUPV3L1 protein expression was uniquely reduced in IFG muscles. Small nuclear ribonucleoprotein-associated protein N (SNRPN) expression was uniquely downregulated in IGT muscles. ATP synthase subunit a (MT-ATP6), NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 (NDUFAF6) and tubulin-folding cofactor B (TBCB) were significantly downregulated only in T2D muscles. ∗ = q < 0.05; ∗∗ = q < 0.01, ns = nonsignificant differences, one-way ANOVA followed by Dunnett's post hoc test with the NGT group as the reference group.
Multiple linear regression analysis
Multiple linear regression was used to estimate the effect of the subjects' glucose tolerance status, age, HOMA-IR (log2), and physical activity level on protein intensities (see Table S6). The regression model reached overall significance (F-test q < 0.05) for 524 proteins, with the highest number of proteins affected by glucose tolerance status and physical activity level (200 and 143 proteins, respectively; q < 0.05), while age and HOMA-IR contributed to only a few of the proteins (3 and 5 proteins, respectively; q < 0.05). The proteins affected by glucose tolerance status and physical activity level were mostly different, with an overlap of 29 proteins (see Figure S2). Galectin-1 (LGALS1) was increased with increasing HOMA-IR and decreased by increasing physical activity level (q < 0.0001 for both). Glyceraldehyde-3-phosphate dehydrogenase (q < 0.01), glycogen debranching enzyme (AGL, q < 0.01), and L-lactate dehydrogenase A chain (q < 0.05) were increased with increasing HOMA-IR, whereas D-beta-hydroxybutyrate dehydrogenase, mitochondrial (q < 0.01) was decreased with increasing HOMA-IR, independent of other variables in the model.
Glycogen synthase levels were decreased by worsening of glucose tolerance status from NGT, IFG, IGT to T2D, independent of other variables in the model (q < 0.01). Moreover, concentrations of many proteins involved in mitochondrial energy metabolism such as isocitrate dehydrogenase [NAD] subunit beta, mitochondrial (IDH3B) (q < 0.01), NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 10, mitochondrial (NDUFA10) (q < 0.01), and ATP synthase subunit a (MT-ATP6) (q < 0.01) were decreased by worsening of glucose tolerance status, with no statistically significant contribution by other variables in the model.
There were also mitochondrial proteins that were impacted by both glucose tolerance, as well as physical activity level. NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 3 (NDUFB3) and NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 2 (NDUFA2) were downregulated by worsening glucose tolerance (q < 0.05 and q < 0.01, respectively) and increased by level of physical activity (q < 0.0001 for both), independent of age and HOMA-IR. Citrate synthase, mitochondrial (CS) was also decreased by worsening glucose tolerance (q < 0.05) and increased by physical activity level (q < 0.05) with no statistically significant contribution by other variables in the model (see Table S6).
Discussion
Skeletal muscle insulin resistance is a central and early defect in the pathogenesis of T2D (Eriksson et al., 1989, 1992; Cline et al., 1999). Therefore, detailed knowledge about regulation of insulin action in muscle is likely to improve treatment of people with insulin resistance and help prevent the development of T2D. Here, we report skeletal muscle proteome in 148 vastus lateralis muscle biopsies obtained from men covering all glucose tolerance phenotypes: from normal glucose tolerance, IFG, IGT, and T2D. To the best of our knowledge, this is to date the largest proteomic analysis of human skeletal muscle.
Our results indicate a strong downregulation in proteins involved in mitochondrial energy metabolism and oxidative phosphorylation in prediabetic (IFG and IGT) as well as T2D samples. We observed a decrease in the several different subunits in the complexes of the respiratory chain (Figure 5), with changes apparent also in prediabetic conditions. However, the most obvious changes were seen in T2D muscle samples. Multiple linear regression analysis revealed an independent contribution of worsening of glucose tolerance status to the decrease in intensities of many proteins involved in mitochondrial energy metabolism. There were also mitochondrial proteins where both worsening of glucose tolerance (a decrease) and increase in physical activity level (an increase) contributed to protein intensities. HOMA-IR contributed significantly to only a few protein intensities in skeletal muscle samples, which may be unsurprising because HOMA-IR is rather a surrogate marker for hepatic insulin resistance (Matthews et al., 1985; Singh and Saxena, 2010). Overall, our data are in good agreement with previous studies which have shown a downregulation of mitochondrial proteins in skeletal muscles from obese and T2D people (Højlund et al., 2003; Hwang et al., 2010; Giebelstein et al., 2012; Chae et al., 2018). Reduced gene expression of mitochondrial complex components has been observed also in skeletal muscle in people with IGT (Osler et al., 2015; Mootha et al., 2003). Our study expands these previous observations, as we observed a reduction in mitochondrial proteins already in skeletal muscles from men with IFG. These data suggest that the downregulation of mitochondrial proteins precedes the development of hyperglycemia, in agreement with reduced expression of OXPHOS genes in skeletal muscle from men with IGT (Mootha et al., 2003).
Analysis of skeletal muscle phosphoproteome complements our findings and reveals an enrichment of altered proteins in signaling pathways regulating glucose metabolic processes, muscle contraction, and the structure of muscle cells (see Tables S3 and S4 and Figure 6). As examples, basal phosphorylation of TBC1D1 (at Ser 237) and TBC1D4 (Akt Susbtrate 160) (at Ser 341 and at Thr 568), signaling effectors involved in insulin and AMPK signaling to GLUT4 translocation and glucose transport (Hatakeyama et al., 2019), was increased in NGT as compared with T2D muscles, as these phosphorylations were not detected in T2D muscles (see Table S3 and Figure 6). On the other hand, phosphorylation of myosin light chain 1/3 (at Thr 65) or troponin T (at Ser 159) was increased in T2D as compared with NGT muscles, as these phosphorylations were not detected in NGT muscles. Dysregulation in basal phosphorylation of proteins in T2D muscle is in agreement with the global analysis of phosphoproteome in iPS-cell-derived-induced myoblasts from T2D donors which has revealed that there are several alterations in basal phosphorylation of proteins regulating various cellular functions such as RNA metabolism, vesicle-mediated transport, and cell cycle in T2D (Batista et al., 2020). In our study, alterations in phosphorylation were also evident in muscles from men with IFG or IGT, indicating that changes in skeletal muscle proteome are initiated at an early stage during the development of dysregulation in glucose metabolism. Collectively, our data suggest that reduced mitochondrial electron transport and oxidative phosphorylation in skeletal muscle may contribute to insulin resistance and may predispose to deterioration of glucose tolerance from normal toward the diabetic spectrum. These observations highlight the importance of physical activity as an important nonpharmaceutical method to improve metabolic health and reduce risk for insulin resistance and T2D, as aerobic endurance training is associated with many beneficial physiological changes, such as mitochondrial biogenesis, changes in skeletal muscle proteins involved in energy metabolism, reduced cardiovascular risk profile, and improved insulin sensitivity (Egan and Zierath, 2013; Hussey et al., 2013; Robinson et al., 2017).
We had the opportunity to analyze skeletal muscle proteome in biopsies obtained from two men several years apart. This is a number too low to draw any firm conclusions. However, we observed upregulation of 20 proteins that were common in both men, likely to be related to aging. Interestingly, JPT1 protein expression was increased 185-fold in skeletal muscle biopsy from the man with T2D with advancing age. JPT1 expression has originally been identified in murine hematopoietic and brain cells (Tang et al., 1997), and JPT1 has been reported to inhibit AKT-GSK3β signaling in cancer cells (Varisli et al., 2011). AKT-GSK3β signaling is centrally involved in the regulation of insulin action on glycogen synthesis. In our study, an increase in JPT1 protein expression in type 2 diabetic skeletal muscle over a time period of 9 years was associated with the deterioration of metabolic control of this individual, as his overall glucose control had attenuated (HbA1c 5.9% vs 6.8%) despite intensification of his antihyperglycemic treatment. Thus, increased JPT1 protein expression may have contributed to worsening of glycemic control via impaired nonoxidative glucose disposal (glycogen synthesis), owing to inhibition of AKT-GSK3β signaling. Clearly, the potential role of JPT1 in the inhibition of glycogen synthesis and insulin sensitivity in skeletal muscle needs to be explored in future studies.
Our data set also revealed interesting individual proteins that were uniquely altered in skeletal muscle in different glucose tolerance phenotypes. For example, ATP-dependent RNA helicase SUPV3L1, mitochondrial (SUPV3L1) was uniquely downregulated in skeletal muscle biopsies from men with IFG. SUPV3L1 regulates mitochondrial RNA metabolism and it also unwinds dsDNA (Minczuk et al., 2002; Szczesny et al., 2010). SUPV3L1 protein expression has been reported to be increased in skeletal muscle from nondiabetic obese (mean BMI 34.2 vs 25.1 kg/m2) people (Formentini et al., 2017). In our study, men with NGT (mean BMI 25.7 kg/m2) or IFG (mean BMI 27.3 kg/m2) were of similar BMI. Therefore, our data suggest that the presence of glucose dysregulation, IFG, results in decreased SUPV3L1 expression. This may impact mitochondrial RNA metabolism in IFG. SNRPN is a part of the spliceosome. It regulates pre-mRNA processing and may be involved in tissue-specific alternative splicing (Jing et al., 2015). SNRPN expression was uniquely downregulated in IGT muscle, suggesting impairment in mRNA metabolism in this condition. Interestingly, analysis of phosphoproteome in iPSC-derived myoblasts reveals a dysregulated basal phosphorylation of proteins involved in mRNA metabolism and mRNA splicing in T2D (Batista et al., 2020). Our results suggest that abnormalities in mRNA metabolism are present also in prediabetes.
ATP synthase subunit a (MT-ATP6), NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 (NDUFAF6), and tubulin-folding cofactor B were uniquely downregulated in T2D muscle. ATP synthase subunit a (MT-ATP6) is a part of the mitochondrial membrane ATP synthase (complex V). This subunit of complex V contains the pore that releases the proton gradient in the mitochondrial inner membrane. Mutations in MT-ATP6 gene are the most common cause of complex V deficiency and have been described to lead to several biochemical alterations such as decreased basal respiration, increased ROS, and reduced ATP synthesis (Ganetzky et al., 2019). NDUFAF6 is crucial in the assembly of mitochondrial NADH:ubiquinone oxidoreductase complex (complex I). Knockdown of NDUFAF6 gene, also known as C8orf38, leads to a profound reduction in complex I abundance and activity, comparable with the knockdown of the known complex I assembly factor NDUFAF1. Mutation in the NDUFAF6/C8orf38 gene causes an inherited mitochondrial complex I deficiency in humans (Leigh syndrome) (Pagliarini et al., 2008, Mckenzie et al., 2011). Thus, decreased expression of MT-ATP6 and NDUFAF6 in T2D muscle is in agreement with the generally observed OXPHOS defect in T2D (Højlund et al., 2003; Hwang et al., 2010; Giebelstein et al., 2012; Chae et al., 2018; Mootha et al., 2003; Patti et al., 2003). Microtubules are a part of the cytoskeletal networks in the eukaryotic cell. Tubulin-folding cofactor B regulates tubulin synthesis (Tian et al., 1997). Our observation of the reduced expression of tubulin-folding cofactor B in T2D muscle raises a possibility that the generation of microtubules is compromised in T2D muscle. Intact microtubule cytoskeleton is required for example for proper GLUT4 trafficking into plasma membrane and insulin-stimulated glucose uptake (Fletcher et al., 2000). Thus, reduced tubulin-folding cofactor B may lead to impaired microtubule formation in skeletal muscle and thus contribute to T2D pathogenesis.
Limitations of the study
For the comparison of the proteome in paired muscle biopsies over time, the number of subjects was very low, allowing only for discovery of tentative associations with aging or worsening of glucose metabolic control. Another limitation is that this is a cross-sectional study, and therefore, it remains uncertain whether the observed progressive reduction in mitochondrial proteins is the cause or the consequence of deterioration of glucose tolerance from normal, IFG, IGT, to T2D. While there is a consensus on skeletal muscle mitochondrial dysfunction in insulin resistance and T2D, it remains much debated if the decrease in mitochondrial oxidative metabolism is causal to insulin resistance and diabetic metabolism (Goodpaster, 2013; Holloszy, 2013; Patti and Corvera, 2010; Pinti et al., 2019). For example, a study of subjects with severe insulin resistance owing to mutations in the insulin receptor suggests that defective insulin signaling may lead to impaired mitochondrial function in skeletal muscle (Sleigh et al., 2011). On the other hand, excessive exercise training led to impairment in mitochondrial function in skeletal muscle with concomitant decrease in glucose tolerance (Flockhart et al., 2021). Owing to the descriptive nature of our study, the findings can be considered exploratory, and there is a need for further mechanistic studies regarding the novel targets with altered expression between the different glucose tolerance groups. The overall strength of the study is the large cohort of clinically characterized men. There was a clear decrease in mitochondrial proteins in muscles from men with prediabetes or T2D, which is in agreement with previous studies (Højlund et al., 2003; Hwang et al., 2010; Giebelstein et al., 2012; Chae et al., 2018). As myoblasts cultured from induced pluripotent stem cells from donors with T2D retain the insulin-resistant phenotype and demonstrate key defects of impaired insulin signaling, glucose uptake, and mitochondrial respiration (Batista et al., 2020), the available data collectively suggest a tight association between insulin resistance and impaired mitochondrial function.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Human skeletal muscle biopsies | This paper | N/A |
| Chemicals, peptides and recombinant proteins | ||
| UREA | Sigma Aldrich | Cat#U5378 |
| Ammonium bicarbonate, NH4HCO3 | Sigma Aldrich | Cat#A6141 |
| Tris(2-carboxyethyl)phosphine | Sigma Aldrich | Cat#C4706 |
| Iodoacetamide | Fluka BioChemica | Cat#57670 |
| Sequencing Grade Modified Trypsin | Promega | Cat#V5113 |
| Formic acid | Fluka | Cat#56302-10X1ML |
| Acetonitrile | Merck | Cat#1000292500 |
| Trifluoroacetic acid | Fluka | Cat#40967-10x1ml |
| 10% ammonia | Fluka | Cat#17837 |
| Critical commercial assays | ||
| BCA protein assay kit | Pierce, Thermo Scientific | Cat#23225 |
| iRT-peptide kit | Biognosys | Ki-3002-2 |
| Software and algorithms | ||
| SPSS Statistics | IBM | version 25 |
| RStudio | RStudio Team https://www.rstudio.com/ | RStudio version 1.4.1106 |
| R | R Core Team https://www.r-project.org/ | R version 4.0.54 |
| ProteinPilot software | AB SCIEX | Paragon algorithm (v4.5) |
| PeakView | AB SCIEX | version 2.1 with SWATH quantitation plug-in |
| MaxQuant | Cox and Mann. 2008 | version 1.6.0.16 |
| Human Uniprot Database | www.uniprot.org | release 11_2018 with 20,347 entries |
| Human Uniprot Database | www.uniprot.org | release 01_2020 with 20,303 entries |
| DAVID bioinformatics resources | https://david.ncifcrf.gov/ | version 6.8 |
| VENNY 2.1 | https://bioinfogp.cnb.csic.es/tools/venny/ | version 2.1 |
| PhosphoSitePlus® | www.phosphosite.org | version 6.5.9.3 |
| NetworKIN software | Horn et al., 2014 | version 2.1 |
| Other | ||
| FastPrep-24 5G bead homogenizer | MP Biomedicals | 116005500 |
| Agilent 3100 OFFGEL fractionator | Agilent Technologies | RRID:SCR_019388 |
| TripleTOF® 6600 Quadrupole Time-Of-Flight instrument | Sciex | TripleTOF 6600 |
| Eksigent nanoLC with a Turbo V Source | Sciex | Turbo V Source |
| Q Exactive ESI-quadrupole-orbitrap mass spectrometer | Thermo Fisher Scientific | RRID:SCR_020571 |
| EASY-nLC 1000 nanoflow LC | Thermo Fisher Scientific | RRID:SCR_014993 |
| zirconia beads | BioSpec Products | 1 mm beads |
| IPG strips | GE Healthcare | 24 cm pH 3–10 |
| C18 microspin columns | Nest Group | HUM S18V |
| YMC-Triart C18 column | YMC | 12nm, 3μm, 150x0.3mm |
| Acclaim PepMap™100 | Thermo Fisher Scientific | 100 μm×2cm, 3μm, 100Å |
| Acclaim PepMap™100 | Thermo Fisher Scientific | 75 μm×15cm, 2μm, 100Å |
Resource availability
Lead contact
Further information and requests for resources should be directed to the lead contact, Heikki Koistinen (heikki.koistinen@helsinki.fi), University of Helsinki and Helsinki University Hospital, Helsinki, Finland.
Materials availability
This study did not generate new unique reagents.
Data and code availability
Raw mass spectrometry data reported in this study cannot be deposited in a public repository because of the EU-GDPR regulations. The summary statistics describing these data have been published as supplemental tables (Tables S1, S2, and S3) with this paper and are publicly available as of the date of publication.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and subject details
Subjects
The study protocol was reviewed and approved by the Ethics Committee of the Department of Medicine, Helsinki University Hospital, and written informed consent was obtained from all subjects before participation. The reported investigations have been carried out in accordance with the principles of the Declaration of Helsinki. We obtained muscle biopsies from 148 men (aged 20–70 years): 24 men with T2D, 14 men with impaired glucose tolerance (either isolated IGT, or a combination of IFG and IGT), 25 men with impaired fasting glucose (IFG), and 85 men with normal glucose tolerance (NGT) (Table 1). To characterize glucose tolerance (WHO criteria), a standard oral glucose tolerance test (OGTT) with 75 g glucose was performed on a separate study day (Table 2). Seventeen men had previously been diagnosed with T2D and did not participate in OGTT. Seven men were found to have a previously undiagnosed T2D, had not yet received antihyperglycemic treatment, and are considered here as diet-treated. Overall, the duration of diabetes was 4 ± 1 years. T2D was managed with a diet (n=14), metformin (n=3), a combination of metformin and DPP4 inhibitor (n=3), a combination of metformin and insulin (n=1), a combination of metformin, sulphonylurea and insulin (n=1), or a combination of metformin, GLP-1 receptor agonist and insulin (n=1). One man with T2D was studied twice 9 years apart. At the time of his first examination his weight was 76 kg, BMI 25.2 kg/m2, his T2D was managed with metformin only, and his HbA1c was 5.9%. At the time of the second study, his weight was 71.5 kg, BMI 24.0 kg/m2, his diabetes was managed with a combination of metformin, sulphonylurea, and DPP4 inhibitor, and his HbA1c was 6.8%. One man with normal glucose tolerance was studied twice 7 years apart. His body weight (76 vs 75.5 kg), BMI (24.4 vs 24.8 kg/m2), and HbA1c (5 % vs 5.5%) were stable during this period (0 vs 7 years, respectively). Proteomic data for one time point only are reported in the main analysis (Tables S1 and S2); proteomic data on paired muscle biopsies are shown in Table S5. One man with IFG and one man with T2D were found to have previously untreated primary hypothyroidism, and one man with NGT was found to have untreated vitamin B12 deficiency owing to atrophic gastritis at the time of the biopsy. Analysis of the data set without these 3 samples revealed that changed proteins and associated biological pathways were essentially the same as in the analysis of the whole cohort. Therefore, data from these 3 samples are included in this report.
Table 2.
Results of oral glucose tolerance tests
| NGT | IFG | IGT | T2D | |
|---|---|---|---|---|
| fB-glucose (mmol/L) (n) | 5.1 ± 0.06 (24) | 5.9 ± 0.07 a (10) | 5.6 ± 0.3 b (4) | ND |
| 2h-B-glucose (mmol/L) (n) | 4.8 ± 0.2 (24) | 5.4 ± 0.1 (10) | 7.3 ± 0.3a, c (4) | ND |
| fP-glucose (mmol/L) (n) | 5.5 ± 0.03 (61) | 6.3 ± 0.04 a (15) | 6.2 ± 0.1 a (10) | 7.5 ± 0.2 a, d, e (7) |
| 2hP-glucose (mmol/L) (n) | 5.4 ± 0.1 (61) | 6.2 ± 0.2 (15) | 8.8 ± 0.3 a, d (10) | 11.2 ± 1.5 a, d, f (7) |
Oral glucose tolerance tests were performed on a day separate from muscle biopsy. Data are given as mean ± SEM, one-way ANOVA followed by Sidak's post hoc test for multiple comparisons.
p < 0.001 vs NGT.
p < 0.05 vs NGT.
p < 0.01 vs IFG.
p < 0.001 vs IFG.
p < 0.001 vs IGT.
p < 0.01 vs IGT.
Method details
Muscle biopsy
All men were instructed to avoid strenuous exercise for 72 h before the muscle biopsy. The participants reported to the clinical research unit after an overnight fast, and a venous blood sample was taken from the antecubital vein. Serum samples were stored at -70 °C for subsequent analysis. Muscle biopsies were obtained from vastus lateralis muscle under local anesthesia (5–10 mg/ml lidocaine hydrochloride) (Kuoppamaa et al., 2008; Scott et al., 2016). A portion of the biopsy was directly frozen in liquid nitrogen and stored at -140 °C for proteomic analyses.
Sample preparation
20–80 mg muscle tissue pieces were homogenized in 8.0 M UREA/50 mM NH4HCO3 using FastPrep-24 5G bead homogenizer (MP Biomedicals) with 1mm zirconia beads (BioSpec Products). Total protein content of the homogenates was measured using a BCA protein assay kit (Pierce, Thermo Scientific), and 100 μg of total protein per sample was taken for SWATH analysis and an additional 500 μg from six selected samples, covering all sample types, for the ion library building. The proteins were reduced with Tris(2-carboxyethyl)phosphine (TCEP; Sigma Aldrich), alkylated with iodoacetamide, trypsin-digested with Sequencing Grade Modified Trypsin (Promega) using a 1:100 enzyme:protein ratio at 37°C o/n, and then, desalted with C18 microspin columns (Nest Group).
The ion library generation
For the muscle library building, tryptic peptides from six selected muscle samples were fractionated with an Agilent 3100 OFFGEL fractionator (Agilent Technologies), using 24 cm pH 3–10 IPG strips (GE Healthcare). The strips were focused at a maximum of 8000 V, 50 μA, 200 mW, until 50 kVh was reached. The runs took approximately 24–36 h. A total of eight fractions per sample were collected, desalted with C18 columns, and analyzed separately with a TripleTOF® 6600 Quadrupole Time-Of-Flight instrument (Sciex) coupled to an Eksigent nanoLC with a Turbo V Source, working in microspray mode. Reference peptides from the iRT-kit (Biognosys) were spiked into each fraction. YMC-Triart C18 column (12 nm, 3 μm, 150 x 0.3 mm) was used for peptide separation. The MS analysis was performed in data-dependent acquisition (DDA) in positive ion mode, using a linear 60-min gradient from 5 to 35 % buffer B (0.1 % formic acid in acetonitrile). Survey scans were acquired in 250 ms and the top 30 ions above the intensity threshold of 150 counts were selected for subsequent MS/MS scans (100–1500 m/z, 100 ms accumulation time per MS/MS).
The raw MS data were processed with ProteinPilot software using Paragon algorithm (v4.5, AB SCIEX [Shilov et al., 2007]). Searches were performed against the reviewed human UniProt database (release 11/2018) supplemented with porcine trypsin and iRT-peptide sequences (total of 20,347 entries). Carbamidomethylation (+57.021 Da) of cysteine residues was used as static modification and oxidation (+15.994 Da) of methionine was used as dynamic modification. Precursor mass tolerance and fragment mass tolerance were set to less than 20 ppm and 0.1 Da, respectively. For ProteinPilot search, the “Thorough ID” mode was selected, which automatically adjusts the mass tolerance to fit the high-resolution MS and MS/MS data. A maximum of two missed cleavages was allowed and the results were filtered to a maximum false discovery rate (FDR) of 1%. The resulting human muscle specific spectral library contains 1,140,750 spectra from 80,041 distinct peptides useful to quantify 3,096 proteins, including iRT reference peptide mix and trypsin used for digestion.
SWATH analysis and peak extraction
SWATH-MS analysis, a data-independent acquisition method, was performed using the same instrument used for muscle library building in DDA mode. Precursor ion selection was done in the 400–1250 m/z range, with a variable window width strategy (from 6 to 50 Da). Peptide activation was performed using collision induced dissociation, using nitrogen as inert gas, with rolling collision energy and 5 eV of energy spread. The accumulation time was set to 250 ms for MS1 and 100 ms for MS2 scan. The entire duty cycle was approximately 3.1 s.
Peak extraction of the SWATH data was performed using PeakView (version 2.1) with SWATH quantitation plug-in (SCIEX). Reference peptides from the iRT-kit (Biognosys) spiked into each sample were used to calibrate the retention time of extracted peptide peaks. The settings used were as follows: the maximum number of peptides per protein, 25; the number of transitions or fragment ions per peptide, 6; a peptide confidence threshold, 90; FDR 1 %; XIC (Extracted Ion Chromatogram) retention time window, 10; m/z tolerance, 75 ppm; shared and modified peptides were excluded. After SWATH peak extraction, the transition ion peak areas, peptide peak areas, and protein peak areas were exported in Excel format for further statistical analysis.
Other determinations
The circumference of the waist was measured to the nearest 0.5 cm midway between the lower rib margin and the iliac crest, and the circumference of the hip was measured at the level of the trochanters with a soft measuring tape (Koistinen et al., 2001; Kuoppamaa et al., 2008). Blood glucose concentration (participants 1–50) was analyzed by glucose dehydrogenase method (Precision-G Blood Glucose Testing System, Medisense, Abbott, IL, USA), and plasma glucose concentration (participants 51-148) was analyzed by hexokinase method (Roche Modular, F. Hoffman-La Roche Ltd, Rotkreuz, Switzerland). HbA1c was determined by an immunological method or by high-performance liquid chromatography. Fasting serum insulin concentration was determined by chemiluminescence immunoassay (Liaison XL, DiaSorin, Saluggia, Italy). Fasting serum glucose; total, HDL, and LDL cholesterol; and triglyceride concentrations were determined by proton NMR metabolomics (Nightingale Health Ltd, Helsinki, Finland) (Locke et al., 2019). Homeostatic model assessment of insulin resistance (HOMA-IR) was calculated from fasting serum glucose and insulin concentrations by the formula: HOMA-IR (Matthews et al., 1985; Singh and Saxena, 2010).
Mass spectrometry analysis of phosphorylation sites
For phosphoproteomics analysis, 300 μg of total protein per sample was digested, as described previously. Phosphopeptide enrichment was performed using immobilized metal ion affinity chromatography with titanium (IV) ion (Ti4+-IMAC). The IMAC material was prepared and used essentially as described (Zhou et al., 2013). Briefly, Ti4+-IMAC beads were loaded onto GELoader tips (Thermo Fisher Scientific). The protein digests were dissolved in a loading buffer (80% ACN, 6% trifluoroacetic acid) and added into the spin tips. The columns were washed and the bound phosphopeptides were eluted with 10% ammonia, followed by purification with C18 microspin columns before LC-MS/MS analysis.
The LC-MS/MS analysis of phosphorylated peptides was performed on a Q Exactive ESI-quadrupole-orbitrap mass spectrometer coupled to an EASY-nLC 1000 nanoflow LC (Thermo Fisher Scientific), using Xcalibur version 3.1.66.10 (Thermo Fisher Scientific). The C18-packed precolumn (Acclaim PepMap™100, 100 μm×2 cm, 3 μm, 100 Å; Thermo Fisher Scientific) and the C18-packed analytical column (Acclaim PepMap™100, 75 μm×15 cm, 2 μm, 100 Å) were used. The phosphopeptides were separated by a 120-min linear gradient. The total measurement time was 140 min per sample. The mass spectrometry analysis was performed as DDA in positive-ion mode. MS spectra were acquired from m/z 300 to m/z 2000 with a resolution of 70,000, with full AGC target value of 3,000,000 ions and a maximal injection time of 120 ms in profile mode. The ten most abundant ions with charge states from 2+ to 7+ were selected for subsequent fragmentation (HCD), and MS/MS spectra were acquired with a resolution of 17,500, with an AGC target value of 5000, a maximal injection time of 120 ms and the lowest mass fixed at m/z 120 in centroid mode. Dynamic exclusion duration was 30 s. Six fully independent replicates were performed for each glucose tolerance category.
Raw data were processed with MaxQuant version 1.6.0.16 (Cox and Mann, 2008) against the human component of the Uniprot Database (release 01_2020 with 20,303 entries) using the Andromeda search engine (Cox et al., 2011). Carbamidomethylation (+57.021 Da) of cysteine residues was used as static modification. Phosphorylation of serine/threonine/tyrosine (+79.966 Da) and oxidation (+15.994 Da) of methionine were used as dynamic modification. Precursor mass tolerance and fragment mass tolerance were set to less than 20 ppm and 0.1 Da, respectively. A maximum of two missed cleavages was allowed. The results were filtered to a maximum FDR of 0.05 and further based on the phosphorylation localization probability at the cutoff value of 0.75 with exception of two adjacent sites where the cutoff value was 0.5. Student’s t-test was used to detect statistically significant changes between sample groups, and differences with p-value < 0.05 were considered significant. In the comparison analysis, detection of two phosphosites of six replicates was demanded.
Functional annotations
Gene Ontology annotations were obtained from DAVID bioinformatics resources (Huang et al., 2009; Huang et al., 2008). Pathway analysis was performed by mapping the proteins with significantly altered expression levels with pathways in the Kyoto Encyclopedia of Genes and Genomes database. Comparisons between datasets were calculated using VENNY 2.1 online tool (https://bioinfogp.cnb.csic.es/tools/venny/). Identified phosphorylation sites and substrates were compared to kinase-substrate data extracted from PhosphoSitePlus® (v6.5.9.3; www.phosphosite.org). Kinase prediction was made using NetworKIN software (Horn et al., 2014).
Quantification and statistical analysis
The clinical data are given as mean±SEM (Tables 1 and 2). Potential differences in clinical variables between the study groups were analyzed by one-way ANOVA followed by Sidak’s post hoc test for multiple comparisons (IBM SPSS Statistics, version 25).
Proteomics data were analyzed with RStudio version 1.4.1106 (RStudio Team, 2020) running with R version 4.0.54 (R Core Team, 2014). The data were separated to two data sets: one containing only the data from men that were sampled twice (Table S5) and one for the main comparison between the different glucose tolerance groups (main data set, Tables S1 and S2). The earlier muscle samples of the twice-sampled men were removed from the main dataset. The following steps were performed separately on these two data sets.
The protein peak areas were normalized by total area normalization: the sums of protein peak areas for each sample were calculated, and the normalization coefficient for each sample was the maximum of these summed protein peak areas divided by the total peak area of the corresponding sample. The missing values, 30 in total in the main data set and none in the other, were imputed with k nearest neighbors imputation (kNN, k = 10). Finally, the data were log2 transformed.
One-way ANOVA was used for protein abundance comparison across the different glucose tolerance groups (Results, figure legends 2-4, and 7; and Tables S1 and S2). The ANOVA p-values were multiple hypothesis corrected with FDR (Benjamini-Hochberg) correction, and the proteins for which q-value < 0.05 were subjected to Dunnett’s post hoc test with the NGT group as the reference group. The p-values from the post hoc test were FDR corrected, and proteins with q-value < 0.05 were considered significantly differentially expressed. Fold changes were calculated using arithmetic means of the log2 transformed protein intensities.
For hierarchical clustering (Figure 2) and principal component analysis (PCA, Figure S1), the log2-transformed data was scaled and centered to mean 0 and variance 1. Hierarchical clustering was performed with Euclidean distance metric and complete linkage.
When analyzing the effect of glucose tolerance status on phosphoproteome (Figure 6 and Table S3), 2-sided t-tests with Welch approximation for unequal variances were used and differences with a p-value < 0.05 were considered significant.
Fold-changes for the twice-sampled men’s proteomes were calculated using arithmetic means of the log2 transformed protein intensities. For the pairwise comparison of protein abundance between two individual samples, statistical testing could not be performed, and therefore, only fold change > 5 was used as a tentative criterion of significance (see Table S5).
Multiple linear regression was used to estimate the effect of the following clinical parameters on protein intensities: subjects glucose tolerance status, age, log2-HOMA-IR and physical activity level. In order to estimate the dose-response effect of the level of physical activity on protein intensities, the subjects were graded as sedentary (0= no reported physical activity, or 1= walking) or physically active (more strenuous physical activity than walking: 2= 30 min–4 h per week, and 3= more than 4 h per week) as per their self-reported physical activity. Glucose tolerance status was dummy coded as follows: 0=NGT; 1=IFG; 2=IGT; 3=T2D. Normalized, log2 transformed protein intensities were used. FDR multiple hypothesis correction was used separately for each clinical variable, and q-values lower than 0.05 were considered significant. Detailed results of the regression analysis are presented in Table S6.
Acknowledgments
The authors thank all the volunteers who participated in the study. For the graphical abstract, images were obtained from Reactome icon library (Cristoffer Sevilla, CC BY 4.0, https://creativecommons.org/licenses/by/4.0/; reprinted with modifications) and from Sciex (reprinted with permission). This research has been supported by grants from Academy of Finland (grant no:s 127093 and 258753 to HAK, and grant no:s 288475 and 294173 to MV), grants from Diabetes Wellness Sverige (grant no 598-174) (to HAK), Finnish Research Foundation for Cardiovascular Disease (to HAK), Finnish Cultural Foundation (to HAK), Finnish Diabetes Research Foundation (to HAK), Finska Läkaresällskapet (to HAK), Finnish Medical Foundation (to HAK), Helsinki University Hospital (funding from the hospital administration and VATR grants TYH7104, TYH2009126, TYH2014219, TYH2017129, TYH2018110, TYH2019223, TYH2021317) (to HAK), Jalmari and Rauha Ahokas Foundation (to HAK), Laboratoriolääketieteen edistämissäätiö sr (to HAK), Liv och Hälsa Foundation (to HAK), Maud Kuistila Foundation (to HAK), Novo Nordisk Foundation (to HAK), Paulo Foundation (to HAK), Sigrid Jusélius Foundation (to HAK), University of Helsinki, Research Council Support Action (to HAK); and grants from the Finnish Cancer Foundation (to MV), the University of Helsinki Three-year Research Grant (to MV), Biocentrum Finland (to MV), Magnus Ehrnrooth Foundation (to MV), and Instrumentarium Research Foundation (to MV).
Author contributions
Conceptualization: HAK; Sample collection: HAK; Methodology: TÖ, JT and MV; Data analysis: TÖ, JT and MV; Interpretation of results: TÖ, JT, ND, SM, MV and HAK; Writing (original draft preparation): TÖ, and HAK; Writing (review and editing): JT, ND, SM and MV. Supervision: MV and HAK. Funding acquisition: MV and HAK.
Declaration of interests
The authors declare no competing interests.
Published: July 23, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102712.
Supplemental information
Document S1. Figures S1 and S2
References
- American Diabetes Association Improving care and promoting health in populations: standards of medical care in diabetes—2018. Diabetes Care. 2018;41:S7–S12. doi: 10.2337/dc18-S001. [DOI] [PubMed] [Google Scholar]
- Batista T.M., Jayavelu A.K., Wewer Albrechtsen N.J., Iovino S., Lebastchi J., Pan H., Dreyfuss J.M., Krook A., Zierath J.R., Mann M., Kahn C.R. A cell-autonomous signature of dysregulated protein phosphorylation underlies muscle insulin resistance in type 2 diabetes. Cell Metab. 2020;32:844–859.e5. doi: 10.1016/j.cmet.2020.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae S., Kim S.-J., Do Koo Y., Lee J.H., Kim H., Ahn B.Y., Ha Y.-C., Kim Y.-H., Jang M.G., Koo K.-H. A mitochondrial proteome profile indicative of type 2 diabetes mellitus in skeletal muscles. Exp. Mol. Med. 2018;50:129. doi: 10.1038/s12276-018-0154-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline G.W., Petersen K.F., Krssak M., Shen J., Hundal R.S., Trajanoski Z., Inzucchi S., Dresner A., Rothman D.L., Shulman G.I. Impaired glucose transport as a cause of decreased insulin-stimulated muscle glycogen synthesis in type 2 diabetes. N. Engl. J. Med. 1999;341:240–246. doi: 10.1056/NEJM199907223410404. [DOI] [PubMed] [Google Scholar]
- Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- Defronzo R.A., Gunnarsson R., Bjorkman O., Olsson M., Wahren J. Effects of insulin on peripheral and splanchnic glucose metabolism in noninsulin-dependent (type II) diabetes mellitus. J. Clin. Invest. 1985;76:149–155. doi: 10.1172/JCI111938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defronzo R.A., Jacot E., Jequier E., Maeder E., Wahren J., Felber J.P. The effect of insulin on the disposal of intravenous glucose. Results from indirect calorimetry and hepatic and femoral venous catheterization. Diabetes. 1981;30:1000–1007. doi: 10.2337/diab.30.12.1000. [DOI] [PubMed] [Google Scholar]
- Deshmukh A. Proteomics of skeletal muscle: focus on insulin resistance and exercise biology. Proteomes. 2016;4:6. doi: 10.3390/proteomes4010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmukh A.S., Murgia M., Nagaraj N., Treebak J.T., Cox J., Mann M. Deep proteomics of mouse skeletal muscle enables quantitation of protein isoforms, metabolic pathways, and transcription factors. Mol. Cell Proteomics. 2015;14:841–853. doi: 10.1074/mcp.M114.044222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edfors F., Danielsson F., Hallström B.M., Käll L., Lundberg E., Pontén F., Forsström B., Uhlén M. Gene-specific correlation of RNA and protein levels in human cells and tissues. Mol. Syst. Biol. 2016;12:883. doi: 10.15252/msb.20167144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan B., Zierath J.R. Exercise metabolism and the molecular regulation of skeletal muscle adaptation. Cell Metab. 2013;17:162–184. doi: 10.1016/j.cmet.2012.12.012. [DOI] [PubMed] [Google Scholar]
- Eriksson J., Franssila-Kallunki A., Ekstrand A., Saloranta C., Widen E., Schalin C., Groop L. Early metabolic defects in persons at increased risk for non-insulin-dependent diabetes mellitus. N. Engl. J. Med. 1989;321:337–343. doi: 10.1056/NEJM198908103210601. [DOI] [PubMed] [Google Scholar]
- Eriksson J., Koranyi L., Bourey R., Schalin-Jantti C., Widen E., Mueckler M., Permutt A.M., Groop L.C. Insulin resistance in type 2 (non-insulin-dependent) diabetic patients and their relatives is not associated with a defect in the expression of the insulin-responsive glucose transporter (GLUT-4) gene in human skeletal muscle. Diabetologia. 1992;35:143–147. doi: 10.1007/BF00402546. [DOI] [PubMed] [Google Scholar]
- Espinosa A., Henríquez-Olguín C., Jaimovich E. Reactive oxygen species and calcium signals in skeletal muscle: a crosstalk involved in both normal signaling and disease. Cell Calcium. 2016;60:172–179. doi: 10.1016/j.ceca.2016.02.010. [DOI] [PubMed] [Google Scholar]
- Fletcher L.M., Welsh G.I., Oatey P.B., Tavaré J.M. Role for the microtubule cytoskeleton in GLUT4 vesicle trafficking and in the regulation of insulin-stimulated glucose uptake. Biochem. J. 2000;352 Pt 2:267–276. [PMC free article] [PubMed] [Google Scholar]
- Flockhart M., Nilsson L.C., Tais S., Ekblom B., Apró W., Larsen F.J. Excessive exercise training causes mitochondrial functional impairment and decreases glucose tolerance in healthy volunteers. Cell Metab. 2021;33:957–970.e6. doi: 10.1016/j.cmet.2021.02.017. [DOI] [PubMed] [Google Scholar]
- Formentini L., Ryan A.J., Gálvez-Santisteban M., Carter L., Taub P., Lapek J.D., Jr., Gonzalez D.J., Villarreal F., Ciaraldi T.P., Cuezva J.M., Henry R.R. Mitochondrial H(+)-ATP synthase in human skeletal muscle: contribution to dyslipidaemia and insulin resistance. Diabetologia. 2017;60:2052–2065. doi: 10.1007/s00125-017-4379-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganetzky R.D., Stendel C., Mccormick E.M., Zolkipli-Cunningham Z., Goldstein A.C., Klopstock T., Falk M.J. MT-ATP6 mitochondrial disease variants: phenotypic and biochemical features analysis in 218 published cases and cohort of 14 new cases. Hum. Mutat. 2019;40:499–515. doi: 10.1002/humu.23723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giebelstein J., Poschmann G., Højlund K., Schechinger W., Dietrich J.W., Levin K., Beck-Nielsen H., Podwojski K., Stühler K., Meyer H.E., Klein H.H. The proteomic signature of insulin-resistant human skeletal muscle reveals increased glycolytic and decreased mitochondrial enzymes. Diabetologia. 2012;55:1114–1127. doi: 10.1007/s00125-012-2456-x. [DOI] [PubMed] [Google Scholar]
- Gillet L.C., Navarro P., Tate S., Röst H., Selevsek N., Reiter L., Bonner R., Aebersold R. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol. Cell Proteomics. 2012;11 doi: 10.1074/mcp.O111.016717. O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodpaster B.H. Mitochondrial deficiency is associated with insulin resistance. Diabetes. 2013;62:1032–1035. doi: 10.2337/db12-1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama H., Morino T., Ishii T., Kanzaki M. Cooperative actions of Tbc1d1 and AS160/Tbc1d4 in GLUT4-trafficking activities. J. Biol. Chem. 2019;294:1161–1172. doi: 10.1074/jbc.RA118.004614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloszy J.O. Deficiency" of mitochondria in muscle does not cause insulin resistance. Diabetes. 2013;62:1036–1040. doi: 10.2337/db12-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn H., Schoof E.M., Kim J., Robin X., Miller M.L., Diella F., Palma A., Cesareni G., Jensen L.J., Linding R. KinomeXplorer: an integrated platform for kinome biology studies. Nat. Methods. 2014;11:603–604. doi: 10.1038/nmeth.2968. [DOI] [PubMed] [Google Scholar]
- Huang D.W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2008;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hussey S.E., Sharoff C.G., Garnham A., Yi Z., Bowen B.P., Mandarino L.J., Hargreaves M. Effect of exercise on the skeletal muscle proteome in patients with type 2 diabetes. Med. Sci. Sports Exerc. 2013;45:1069–1076. doi: 10.1249/MSS.0b013e3182814917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang H., Bowen B.P., Lefort N., Flynn C.R., de Filippis E.A., Roberts C., Smoke C.C., Meyer C., Højlund K., Yi Z., Mandarino L.J. Proteomics analysis of human skeletal muscle reveals novel abnormalities in obesity and type 2 diabetes. Diabetes. 2010;59:33–42. doi: 10.2337/db09-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Højlund K., Bowen B.P., Hwang H., Flynn C.R., Madireddy L., Geetha T., Langlais P., Meyer C., Mandarino L.J., Yi Z. In vivo phosphoproteome of human skeletal muscle revealed by phosphopeptide enrichment and HPLC−ESI−MS/MS. J. Proteome Res. 2009;8:4954–4965. doi: 10.1021/pr9007267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Højlund K., Wrzesinski K., Larsen P.M., Fey S.J., Roepstorff P., Handberg A., Dela F., Vinten J., Mccormack J.G., Reynet C., Beck-Nielsen H. Proteome analysis reveals phosphorylation of ATP synthase β-subunit in human skeletal muscle and proteins with potential roles in type 2 diabetes. J. Biol. Chem. 2003;278:10436–10442. doi: 10.1074/jbc.M212881200. [DOI] [PubMed] [Google Scholar]
- Jing J., Zhao Y., Wang C., Zhao Q., Liang Q., Wang S., Ma J. Effect of small nuclear ribonucleoprotein-associated polypeptide N on the proliferation of medulloblastoma cells. Mol. Med. Rep. 2015;11:3337–3343. doi: 10.3892/mmr.2015.3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koistinen H.A., Remitz A., Gylling H., Miettinen T.A., Koivisto V.A., Ebeling P. Dyslipidemia and a reversible decrease in insulin sensitivity induced by therapy with 13-cis-retinoic acid. Diabetes Metab. Res. Rev. 2001;17:391–395. doi: 10.1002/dmrr.222. [DOI] [PubMed] [Google Scholar]
- Krasny L., Huang P.H. Data-independent acquisition mass spectrometry (DIA-MS) for proteomic applications in oncology. Mol. Omics. 2021;17:29–42. doi: 10.1039/d0mo00072h. [DOI] [PubMed] [Google Scholar]
- Kuoppamaa H., Skrobuk P., Sihvo M., Hiukka A., Chibalin A.V., Zierath J.R., Koistinen H.A. Globular adiponectin stimulates glucose transport in type 2 diabetic muscle. Diabetes Metab. Res. Rev. 2008;24:554–562. doi: 10.1002/dmrr.883. [DOI] [PubMed] [Google Scholar]
- Locke A.E., Steinberg K.M., Chiang C.W.K., Service S.K., Havulinna A.S., Stell L., Pirinen M., Abel H.J., Chiang C.C., Fulton R.S. Exome sequencing of Finnish isolates enhances rare-variant association power. Nature. 2019;572:323–328. doi: 10.1038/s41586-019-1457-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loukovaara S., Nurkkala H., Tamene F., Gucciardo E., Liu X., Repo P., Lehti K., Varjosalo M. Quantitative proteomics analysis of vitreous humor from diabetic retinopathy patients. J. Proteome Res. 2015;14:5131–5143. doi: 10.1021/acs.jproteome.5b00900. [DOI] [PubMed] [Google Scholar]
- Matthews D.R., Hosker J.P., Rudenski A.S., Naylor B.A., Treacher D.F., Turner R.C. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28:412–419. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- Mckenzie M., Tucker E.J., Compton A.G., Lazarou M., George C., Thorburn D.R., Ryan M.T. Mutations in the gene encoding C8orf38 block complex I assembly by inhibiting production of the mitochondria-encoded subunit ND1. J. Mol. Biol. 2011;414:413–426. doi: 10.1016/j.jmb.2011.10.012. [DOI] [PubMed] [Google Scholar]
- Minczuk M., Piwowarski J., Papworth M.A., Awiszus K., Schalinski S., Dziembowski A., Dmochowska A., Bartnik E., Tokatlidis K., Stepien P.P., Borowski P. Localisation of the human hSuv3p helicase in the mitochondrial matrix and its preferential unwinding of dsDNA. Nucleic Acids Res. 2002;30:5074–5086. doi: 10.1093/nar/gkf647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha V.K., Lindgren C.M., Eriksson K.F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstrale M., Laurila E. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Ohlendieck K. Skeletal muscle proteomics: current approaches, technical challenges and emerging techniques. Skeletal Muscle. 2011;1:6. doi: 10.1186/2044-5040-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osler M.E., Fritz T., Caidahl K., Krook A., Zierath J.R., Wallberg-Henriksson H. Changes in gene expression in responders and nonresponders to a low-intensity walking intervention. Diabetes Care. 2015;38:1154–1160. doi: 10.2337/dc14-2606. [DOI] [PubMed] [Google Scholar]
- Pagliarini D.J., Calvo S.E., Chang B., Sheth S.A., Vafai S.B., Ong S.-E., Walford G.A., Sugiana C., Boneh A., Chen W.K. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patti M., Butte A., Crunkhorn S., Cusi K., Berria R., Kashyap S., Miyazaki Y., Kohane I., Costello M., Saccone R. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: potential role of PGC1 and NRF1. Proc. Natl. Acad. Sci. U S A. 2003;100:8466–8471. doi: 10.1073/pnas.1032913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patti M.E., Corvera S. The role of mitochondria in the pathogenesis of type 2 diabetes. Endocr. Rev. 2010;31:364–395. doi: 10.1210/er.2009-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinti M.V., Fink G.K., Hathaway Q.A., Durr A.J., Kunovac A., Hollander J.M. Mitochondrial dysfunction in type 2 diabetes mellitus: an organ-based analysis. Am. J. Physiol. Endocrinol. Metab. 2019;316:E268–E285. doi: 10.1152/ajpendo.00314.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.M., Dasari S., Konopka A.R., Johnson M.L., Manjunatha S., Esponda R.R., Carter R.E., Lanza I.R., Nair K.S. Enhanced protein translation underlies improved metabolic and physical adaptations to different exercise training modes in young and old humans. Cell Metab. 2017;25:581–592. doi: 10.1016/j.cmet.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts G.K., McNally R.M., Blanco R., You J.-S., Hebert A.S., Westphall M.S., Coon J.J., Hornberger T.A. A map of the phosphoproteomic alterations that occur after a bout of maximal-intensity contractions. J. Physiol. 2017;595:5209–5226. doi: 10.1113/JP273904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . R Foundation for Statistical Computing; 2014. R: A Language and Environment for Statistical Computing. [Google Scholar]
- RStudio Team . Integrated Development for R. RStudio, PBC; Boston, MA: 2020. RStudio. [Google Scholar]
- Scott L.J., Erdos M.R., Huyghe J.R., Welch R.P., Beck A.T., Wolford B.N., Chines P.S., Didion J.P., Narisu N., Stringham H.M. The genetic regulatory signature of type 2 diabetes in human skeletal muscle. Nat. Commun. 2016;7:11764. doi: 10.1038/ncomms11764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilov I.V., Seymour S.L., Patel A.A., Loboda A., Tang W.H., Keating S.P., Hunter C.L., Nuwaysir L.M., Schaeffer D.A. The paragon algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol. Cell Proteomics. 2007;6:1638–1655. doi: 10.1074/mcp.T600050-MCP200. [DOI] [PubMed] [Google Scholar]
- Singh B., Saxena A. Surrogate markers of insulin resistance: a review. World J. Diabetes. 2010;1(2):36–47. doi: 10.4239/wjd.v1.i2.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleigh A., Raymond-Barker P., Thackray K., Porter D., Hatunic M., Vottero A., Burren C., Mitchell C., McIntyre M., Brage S. Mitochondrial dysfunction in patients with primary congenital insulin resistance. J. Clin. Invest. 2011;121:2457–2461. doi: 10.1172/JCI46405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczesny R.J., Borowski L.S., Brzezniak L.K., Dmochowska A., Gewartowski K., Bartnik E., Stepien P.P. Human mitochondrial RNA turnover caught in flagranti: involvement of hSuv3p helicase in RNA surveillance. Nucleic Acids Res. 2010;38:279–298. doi: 10.1093/nar/gkp903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Lai Y.H., Han X.D., Wong P.M., Peters L.L., Chui D.H. Murine Hn1 on chromosome 11 is expressed in hemopoietic and brain tissues. Mamm. Genome. 1997;8:695–696. doi: 10.1007/s003359900540. [DOI] [PubMed] [Google Scholar]
- Tian G., Lewis S.A., Feierbach B., Stearns T., Rommelaere H., Ampe C., Cowan N.J. Tubulin subunits exist in an activated conformational state generated and maintained by protein cofactors. J. Cell Biol. 1997;138:821–832. doi: 10.1083/jcb.138.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unnikrishnan R., Pradeepa R., Joshi S.R., Mohan V. Type 2 diabetes: demystifying the global epidemic. Diabetes. 2017;66:1432–1442. doi: 10.2337/db16-0766. [DOI] [PubMed] [Google Scholar]
- Varisli L., Gonen-Korkmaz C., Debelec-Butuner B., Erbaykent-Tepedelen B., Muhammed H.S., Bogurcu N., Saatcioglu F., Korkmaz K.S. Ubiquitously expressed hematological and neurological expressed 1 downregulates Akt-mediated GSK3β signaling, and its knockdown results in deregulated G2/M transition in prostate cells. DNA Cell Biol. 2011;30:419–429. doi: 10.1089/dna.2010.1128. [DOI] [PubMed] [Google Scholar]
- Zheng Y., Ley S.H., Hu F.B. Global aetiology and epidemiology of type 2 diabetes mellitus and its complications. Nat. Rev. Endocrinol. 2017;14:88–98. doi: 10.1038/nrendo.2017.151. [DOI] [PubMed] [Google Scholar]
- Zhou H., Ye M., Dong J., Corradini E., Cristobal A., Heck A.J., Zou H., Mohammed S. Robust phosphoproteome enrichment using monodisperse microsphere-based immobilized titanium (IV) ion affinity chromatography. Nat. Protoc. 2013;8:461–480. doi: 10.1038/nprot.2013.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Document S1. Figures S1 and S2
Data Availability Statement
Raw mass spectrometry data reported in this study cannot be deposited in a public repository because of the EU-GDPR regulations. The summary statistics describing these data have been published as supplemental tables (Tables S1, S2, and S3) with this paper and are publicly available as of the date of publication.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.