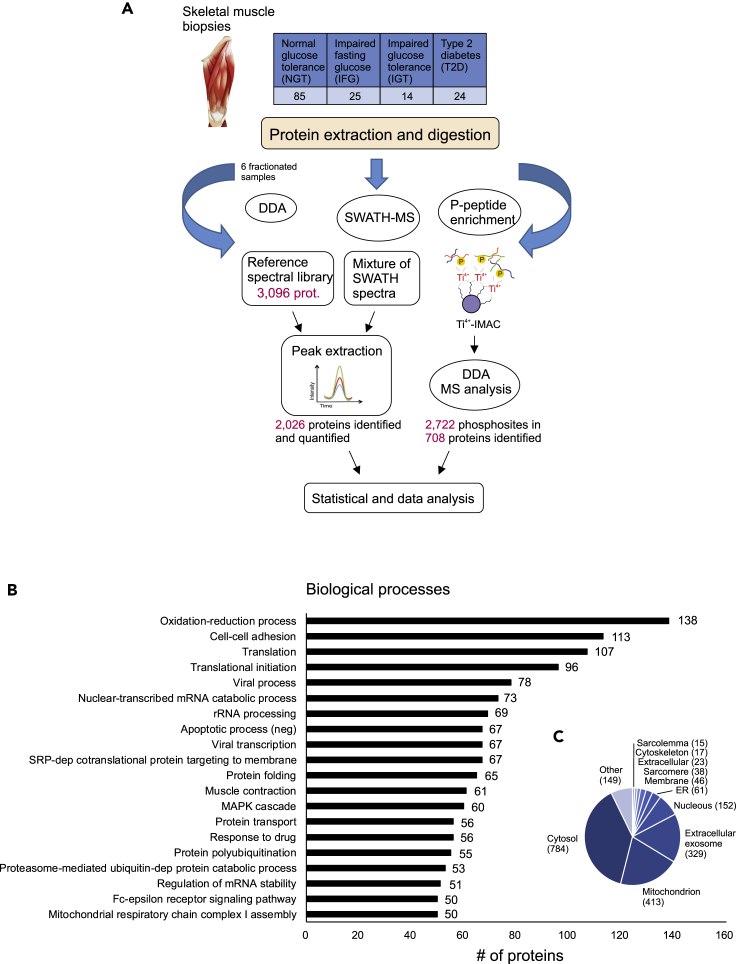
Figure 1.
Quantitative SWATH-MS analysis of human muscle samples
(A) Muscle biopsies were collected from 148 men whose glucose tolerance covered all phenotypes from normal to type 2 diabetes. Muscle proteins were extracted and digested with trypsin. For spectral library generation, the resulting peptides were fractionated and analyzed using data-dependent acquisition mode (DDA). The spectral library built was then used to extract the peptide and the quantification information of the SWATH runs. In addition, six samples from each glucose tolerance group were subjected to phosphopeptide analysis. Statistical analysis and bioinformatics approaches were used to understand the biological relevance of the differentially expressed proteins.
(B and C) (B) The DAVID bioinformatics tool was used to classify all 2,026 identified proteins as per their biological processes and to predict cellular location (C).