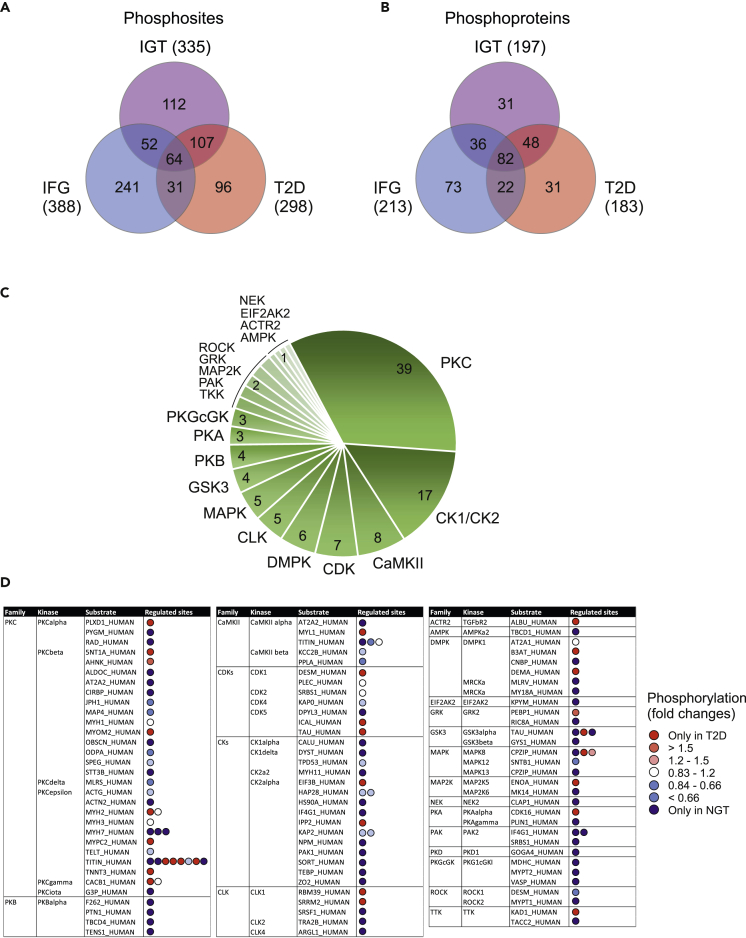
Figure 6.
Phosphorylation in skeletal muscle proteome
(A and B) Venn diagram representing the number of phosphosites (A) or phosphoproteins (B) with a significant change in abundance (p-value < 0.05, or observed only in another condition) in the IFG, IGT and T2D sample groups compared with NGT.
(C) NetworKIN software was used to predict kinases that are dysregulated in type 2 diabetes. 298 phosphorylated sites that exhibited an alteration in abundance in T2D samples were used as input. A total of 20 distinct kinase families associated with 115 phosphorylated sites was predicted. The numbers indicate the total number of phosphorylation sites linked to the specific kinases.
(D) The more specific details about NetworKIN predicted kinase substrates. All the substrates listed here have significant differences between T2D and NGT samples. Red spot means that the substrate is phosphorylated more in T2D samples, indicating more active kinases in T2D. Blue spot indicates more active kinase in NGT samples (more phosphorylation in NGT samples). Some substrate proteins have multiple regulated phosphorylation sites (in sequence order in the figure). Phosphosites are indicated in Table S3.