Summary
The neuromuscular junction is a synapse critical for muscle strength and coordinated motor function. Unlike CNS injuries, motor neurons mount robust regenerative responses after peripheral nerve injuries. Conversely, motor neurons selectively degenerate in diseases such as amyotrophic lateral sclerosis (ALS). To assess how these insults affect motor neurons in vivo, we performed ribosomal profiling of mouse motor neurons. Motor neuron-specific transcripts were isolated from spinal cords following sciatic nerve crush, a model of acute injury and regeneration, and in the SOD1G93A ALS model. Of the 267 transcripts upregulated after nerve crush, 38% were also upregulated in SOD1G93A motor neurons. However, most upregulated genes in injured and ALS motor neurons were context specific. Some of the most significantly upregulated transcripts in both paradigms were chemokines such as Ccl2 and Ccl7, suggesting an important role for neuroimmune modulation. Collectively these data will aid in defining pro-regenerative and pro-degenerative mechanisms in motor neurons.
Subject area: Biological sciences, Neuroscience, Sensory neuroscience, Techniques in neuroscience
Graphical abstract

Highlights
-
•
A translatomic method evaluated motor neuron gene expression after injury and in ALS
-
•
This approach highlighted shared and divergent pathways during injury and degeneration
-
•
Motor neurons upregulate chemokines during axon regeneration and ALS degeneration
-
•
Regeneration upregulates neuropeptides, whereas ALS triggers developmental pathways
Biological sciences; Neuroscience; Sensory neuroscience; Techniques in neuroscience;
Introduction
Motor neuron cell bodies reside in the central nervous system but extend their axons to innervate skeletal muscle in the periphery. Many peripheral axons, including motor axons, display a great deal of regenerative capacity. In response to injury, motor neurons can initiate axon regeneration to allow for axon growth and reinnervation of neuromuscular junctions (NMJs). However, regeneration following severe acute trauma or chronic denervation from underlying neurodegenerative disease is often insufficient and can profoundly impact neuromuscular activity and function (Grinsell and Keating, 2014; Zarei et al., 2015). Amyotrophic lateral sclerosis (ALS), also called Lou Gerhrig's disease, is typically a rapidly progressing disease that is invariably fatal. ALS rather selectively affects upper and lower motor neurons, leading to a loss of motor control and muscle wasting. Approximately 10% of ALS cases are familial, while the remaining 90% of cases are sporadic indicating there is no clear inheritance pattern (Zarei et al., 2015, 2020). The first gene found to be associated with ALS was superoxide dismutase 1 (SOD1), and mice engineered to overexpress the G93A-SOD1 (SOD1G93A) mutation develop motor neuron disease similar to human ALS (Gurney et al., 1994; Rosen et al., 1993; Ticozzi et al., 2011; Tu et al., 1996; Turner and Talbot, 2008). More than 20 genes have since been identified with ALS-associated mutations, many of which are de novo mutations present in patients with sporadic ALS (Ferraiuolo et al., 2011). From the analysis of these genetic pathways, several molecular mechanisms have been suggested to contribute to the pathogenesis of ALS, such as axonal transport defects, toxic protein aggregation, impaired nucleocytoplasmic transport, and inflammation (Blokhuis et al., 2013; Boulis et al., 2017; Ferraiuolo et al., 2011; Philips and Robberecht, 2011; Sun et al., 1975; Takahashi et al., 1972; Warita et al., 1999; Williamson and Cleveland, 1999; Wosiski-Kuhn et al., 2019; Zhang et al., 2015). More recently, genes involved in RNA transport, splicing and translation have been linked to ALS, suggesting that deficiencies in RNA metabolism contribute to motor neuron degeneration (Gitcho et al., 2008; Kwiatkowski et al., 2009; Neumann et al., 2006; Sreedharan et al., 2008; Vance et al., 2009). Therefore, understanding motor neuron-specific translational responses to ALS neurodegeneration may help identify new therapeutic targets.
One of the challenges to understanding the etiology of ALS is that there are significant cell non-autonomous components to disease progression (Ilieva et al., 2009). This has been well demonstrated for SOD1 mutations in which the expression of mutant SOD1 in astrocytes and microglia does not affect the onset of disease but significantly contributes to disease progression (Boillee et al., 2006; Wang et al., 2009; Yamanaka et al., 2008). This underscores the need to evaluate motor neuron degeneration in the context of their complex environment. Traditional methods commonly used to investigate cell-type-specific changes in gene expression from tissues include in situ hybridization, laser capture microdissection (LCM), and enzymatic dissociation followed by fluorescence-activated cell sorting (FACS). Although these methodologies have provided important insights into gene regulation, there are limitations associated with their usefulness in assessing gene expression in complex tissues (Chen et al., 2019a). While in situ hybridization allows for the precise localization of RNA, it is not generally amenable to high-throughput profiling to identify novel transcripts, and it is less quantitative than other methods. Conversely, LCM and FACS sorting can be used on a broader scale for gene discovery, but each of these techniques come with their own challenges. Contamination by the proximity of adjacent cells can occur with LCM and the enzymatic dissociation process required for FACS purification can alter gene expression profiles. An added challenge posed by FACS purification of adult neuronal populations is that axotomy combined with the stress of cell sorting often greatly reduces cell survival. Finally, in addition to the challenges posed by these methods, gene transcript number is not necessarily correlated with protein expression (Vogel and Marcotte, 2012). Together, these limitations create a ceiling for understanding the impacts of gene expression in specific populations of cells in vivo.
To overcome these challenges, we used RiboTag mice to explore motor neuron-specific gene expression in vivo. In this model, ribosomal protein L22 (Rpl22) is genetically modified to express Rpl22-HA (Rpl22HA) protein in a Cre-dependent manner (Sanz et al., 2009). As a member of the 60S ribosomal subunit, Rpl22HA is incorporated into ribosomal complexes and can be immunoprecipitated with an HA antibody. Isolation and purification of ribosomal-bound mRNA transcripts allows for the assessment of gene expression in a cell-type specific manner (De Gendt et al., 2014; Itoh et al., 2018; Sanz et al., 2009; Shigeoka et al., 2016; Srinivasan et al., 2016; Zhu et al., 2017). Although this approach does not permit analysis of the entire transcriptome, it reveals a subset of ribosome-associated genes that are presumably undergoing translation. Importantly, a recent study using the RiboTag model estimated that roughly 85% of captured mRNAs from adult retinal axons were undergoing active translation (Shigeoka et al., 2016). To drive Rpl22HA expression in spinal motor neurons, the RiboTag mouse line was crossed with mice that express Cre under the Choline acetyltransferase (Chat) protomer (Rossi et al., 2011). In characterizing this mouse model, we were able to demonstrate that Rpl22HA;ChATCre mice exhibit strong HA immunoreactivity in spinal motor neurons. Furthermore, we could isolate high-quality, motor neuron-specific mRNA transcripts from HA-tagged ribosomes immunoprecipitated from spinal cord tissue. After demonstrating the specificity of this model, we then used it to examine how gene expression in motor neurons is altered in response to a denervating injury and during neurodegeneration in the hSOD1G93A model of ALS.
Results
Rpl22HA is faithfully expressed in cholinergic neurons of the mouse spinal cord
The NMJ serves as the interface between the motor neuron and skeletal muscle. Motor neuron cell bodies located in the ventral horn of the spinal cord integrate incoming sensory and descending higher-order information, and upon activation result in skeletal muscle contraction. To examine gene expression in motor neurons, we crossed RiboTag homozygous mice into the ChATCre mouse line. Choline acetyltransferase (ChAT) is an enzyme required for the synthesis of acetylcholine (ACh), the principle neurotransmitter utilized by motor neurons. Therefore, we expected this strategy to direct Rpl22HA expression in all cholinergic neurons, including motor neurons.
Rpl22HA expression in the lumbar spinal cords of adult RiboHET;ChATCre and RiboHET;ChATWT mice was examined (Figures 1 and S1). Staining transverse spinal cord sections with HA (red), the neuronal marker NeuN (Green), and ChAT (blue) allowed us to identify four major populations of Rpl22HA positive cells (Figure S1A). In addition to ventrally located NeuN+ChAT+ somatic motor neurons that project axons to skeletal muscle (Figure S1E), HA immunofluorescence was observed in two other cholinergic populations. HA+ pre-motor interneurons, previously described to play a neuromodulatory role during locomotive behavior (Zagoraiou et al., 2009), were found around the central canal (Figure S1B), while HA+ preganglionic visceral motor neurons that project axons to sympathetic ganglia of the autonomic nervous system (Stifani, 2014) were found in the intermediolateral column in the lateral horn (Figure S1C). We also observed a rare and unexpected population of HA+NeuN+ChAT− cells in the ventral horn of the spinal cord (Figures S1D and S1F). Although the origin and function of these cells is unclear, they likely represent a population of interneurons that were cholinergic at some point during development but have since stopped expressing ChAT.
Figure 1.
Rpl22HA expression in motor neurons is Cre-dependent
(A and B) Lumbar spinal cords from RiboHET;ChATWT (n = 2, (A)) and RiboHET;ChATCre (n = 3, (B)) mice were serially sectioned and immunolabeled with antibodies to NeuN (green), ChAT (blue), and HA (red). ChAT+ motor neurons in the ventral horn were HA+ in RiboHET;ChATCre mice, but not RiboHET;ChATWT littermates.
(B′) Higher power image of motor neurons from the inset indicated in (B) Motor neurons were categorized into αMNs (NeuN+ChAT+, asterisk) and γMNs (NeuN−ChAT+, arrowheads).
(C) Nearly all MNs were HA+ in RiboHET;ChATCre mice, while no HA+ expression was observed in RiboHET;ChATWT mice. Scale bar represents 100 μm (A and B), 25 μm (B′). Error bars represent the mean ± s.e.
To more thoroughly investigate the extent of Rpl22HA expression in somatic motor neurons, we examined Rpl22HA expression in serial sections throughout the lumbar spinal cord. ChAT immunostaining was observed in the ventral horn of both RiboHET;ChATCre and RiboHET;ChATWT mice. Rpl22HA expression, however, was only observed in RiboHET; ChATCre mice (Figures 1A and 1B). Somatic motor neurons can be classified into two main populations. Alpha motor neurons (αMNs) innervate extrafusal muscle fibers at the NMJ to produce muscle contractions (Kanning et al., 2010), while gamma motor neurons (γMNs) innervate intrafusal muscle fibers at the muscle spindle to provide proprioceptive feedback as the muscle is stretched during contraction (Hulliger, 1984; Kanning et al., 2010). Staining for NeuN allowed us to distinguish between alpha and gamma motor neurons, both of which were robustly labeled with Rpl22HA (αMNs, NeuN+ChAT+ Figure 1B′, asterisk; γMNs, NeuN−ChAT+ Figure 1B′, arrowheads) (Friese et al., 2009). Further quantification of these populations revealed that ChAT+ neurons in the ventral horn of RiboHET;ChATCre and RiboHET;ChATWT mice contained a similar percentage of αMNs (76.9% ± 1.21% vs. 82.5% ± 2.71%) and γMNs (23.1% ± 1.21% vs. 17.5% ± 0.722). Importantly, while almost every motor neuron in RiboHET;ChATCre mice were identified as HA+, very few cells (αMNs: 2.55% ± 0.72%, γMNs: 0.59% ± 0.30%) were HA−. Additionally, no HA expression was observed in RiboHET;ChATWT motor neurons (Figure 1C).
To further examine the expression of Rpl22HA in motor neurons, we co-immunolabeled spinal cord sections for HA (red) and either glial fibrillary acidic protein (GFAP, blue, Figure S1G) or microtubule-associated protein (MAP2, green, Figure S1H). The absence of Rpl22HA can be appreciated in GFAP+ astrocytes, while positive Rpl22HA staining was observed extending into MAP2+ motor neuron dendrites (see arrowheads). Overall, RiboHET;ChATCre mice display Rpl22HA immunolabeling selectively in cholinergic neurons in the spinal cord and not in other cell types, such as astrocytes.
Immunoprecipitation and isolation of motor neuron specific mRNA transcripts
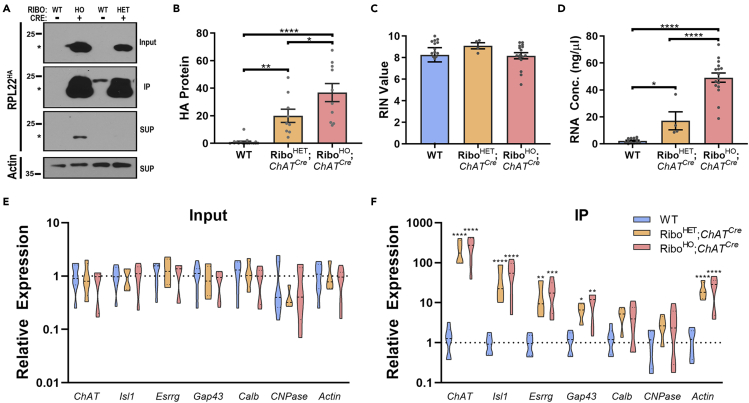
After immunohistochemical characterization of Rpl22HA localization to motor neurons, we evaluated whether Rpl22HA protein could be immunoprecipitated from spinal cord tissue lysates. Whole spinal cords were collected from RiboHO;ChATCre, RiboHET;ChATCre, and wild-type (WT) mice of various genotypes (see STAR Methods). Since all control groups behaved similarly, the results were averaged and displayed as one WT group. Enrichment of Rpl22HA protein after HA immunoprecipitation (IP) was determined by Western blotting. Prior to HA IP, Rpl22HA protein was detected in the input lysate (Figure 2A). HA IP increased the amount of Rpl22HA protein detected in the immunoprecipitate, while simultaneously depleting supernatant (SUP) levels of Rpl22HA (Figure 2A). Over a series of experiments, the amount of HA detected in the IP was quantified and normalized to the amount of actin in the SUP. A significant, gene dose-dependent increase in Rpl22HA protein was observed among the various genotypes (WT, 1.00 ± 0.594; RiboHET;ChATCre, 19.97 ± 4.826; RiboHO;ChATCre, 36.85 ± 6.559, Figure 2B). To further demonstrate the specificity of Rpl22HA IP and confirm that it resulted in the isolation of intact ribosomal complexes from spinal cord lysates, we performed co-immunoprecipitation experiments. Cytosolic proteins known not to be associated with ribosomes, and that were abundant in the input and supernatants following Rpl22HA IP, were not detected in the immunoprecipitated product (Figures S2A, S2D, and S2E). Furthermore, both 40S (Rps2, Rps17) and 60S (Rpl36) ribosomal components were readily detected in the co-IP of spinal cord lysates isolated in a Cre-dependent manner (Figures S2B–S2D).
Figure 2.
Rpl22HA protein and mRNA can be isolated from motor neurons in RiboTag;ChATCre mice
(A) Rpl22HA immunoprecipitation (IP) was performed on whole spinal cords from RiboHO;ChATCre, RiboHET;ChATCre, and wild-type (WT) littermate mice. HA protein could be detected in the input lysates (top panel), was increased after IP (second panel), and depleted from the supernatant (SUP, third panel). The level of actin in the SUP was used as a loading control (bottom panel).
(B) The amount of Rpl22HA in the IP was normalized to actin, which revealed a gene dose-dependent increase in the amount of isolated HA protein from RiboHO;ChATCre (n = 10) and RiboHET;ChATCre (n = 9) mice compared to WT littermates (n = 17).
(C and D) Bioanalyzer analysis demonstrated that high quality (C) and a gene dose-dependent amount (D) of mRNA could be isolated after Rpl22HA IP from WT (n = 14), RiboHET;ChATCre (n = 5), and RiboHO;ChATCre (n = 16) mice.
(E and F) RT-qPCR was performed on RNA isolated in input lysates (E) or after Rpl22HA IP (F) from WT (n = 7), RiboHET;ChATCre (n = 4) and RiboHO;ChATCre (n = 6) mice. Genes expressed in motor neurons (ChAT, Isl1, Esrrg, and Gap43) are enriched in RiboHET;ChATCre and RiboHO;ChATCre mice compared to WT IPs, and none of these genes were enriched in input lysates. Genes expressed in interneurons (Calb) and oligodendrocytes (CNPase) were not enriched in the IPs.
Error bars represent the mean ± s.e. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001.
We next examined whether RNA could be obtained from immunoprecipitated ribosomal complexes. Spinal cords from WT, RiboHETChATCre, and RiboHO;ChATCre mice were lysed, immunoprecipitated with an HA antibody, and associated RNAs were purified. Isolated RNA was then analyzed by Bioanalyzer to determine RNA quality (Figure 2C) and concentration (Figure 2D). High quality and intact RNA were routinely isolated from all mice analyzed (average RIN values for WT, 8.26 ± 0.661; RiboHET;ChATCre, 9.10 ± 0.280; RiboHO;ChATCre, 8.18 ± 0.282). Furthermore, like Rpl22HA protein, we observed that a gene dose-dependent amount of RNA was isolated from immunoprecipitated ribosomes (WT, 2.24 ± 0.443; RiboHET;ChATCre, 17.09 ± 6.657; RiboHO;ChATCre, 49.15 ± 3.398, Figure 2D).
To validate that immunoprecipitated RNA contained transcripts specific to motor neurons we used reverse transcription quantitative polymerase chain reaction (RT-qPCR) to assess the expression of several positive and negative control genes (Figures 2E and 2F). All genes examined were expressed at similar levels in the input lysate regardless of genotype (Figure 2E). However, after Rpl22HA IP, RiboHET;ChATCre and RiboHO;ChATCre mice exhibited a significant enrichment for the motor neuron specific transcripts ChAT and Islet1 (Is1, Figure 2F). The γMN marker estrogen related receptor gamma (Esrrg) (Friese et al., 2009) and injury-responsive gene growth-associated protein 43 (Gap43) (Chong et al., 1992; Skene and Willard, 1981) were also detected in motor neurons, albeit to a lower extent (Figure 2F). Importantly, the interneuron marker Calbindin (Calb) (Floyd et al., 2018; Merkulyeva et al., 2016) and the oligodendrocyte marker 2′3′-cyclic-nucleotide 3′phosphodiesterase (CNPase) (Lappe-Siefke et al., 2003) were not enriched (Figure 2F). Taken together, Rpl22HA protein and associated mRNA transcripts can be purified from RiboHET;ChATCre and RiboHO;ChATCre spinal cords in a gene-dose dependent manner and this methodology allows for the isolation of highly enriched motor neuron-specific transcripts in vivo.
RiboTag;ChATCre mouse models can be employed to investigate the motor neuron translatome in response to injury and disease
To induce an acute injury response in motor neurons, a unilateral sciatic nerve crush was performed on 3-month-old RiboHO;ChATCre mice (Figure 3A). Injured (INJ) and uninjured (UNJ) tissues were isolated at different time points after injury (Figure 3B). We then used RT-qPCR analysis of total RNA isolated from the tibialis anterior (TA) muscle to monitor the extent and consistency of muscle denervation and reinnervation (Figures 3C–3E). In agreement with previous studies (Bowen et al., 1998; Valenzuela et al., 1995), we found that the postsynaptic marker muscle-associated receptor tyrosine kinase (MuSK) was highly upregulated in response to muscle denervation, with peak expression observed at day 3 after injury (D3, Figure 3C). Conversely, a major structural component of the myelin sheath, myelin protein zero (MPZ) (Gupta et al., 1988), was significantly decreased from D1-D7 but rebounded by D21 (Figure 3D). Gap43 is known to be expressed in regenerating motor axons and Schwann cells after injury (Chong et al., 1992; Curtis et al., 1992), and accordingly we found that Gap43 expression peaked at D7 after injury (Figure 3E). In addition to the nerve crush paradigm, we also employed a nerve transection protocol where a large segment of the nerve was removed to preclude axon regeneration. As expected, in the absence of muscle reinnervation both Gap43 and MuSK remained significantly upregulated at 21 days after nerve transection (D21-t), while MPZ remained downregulated (Figures 3C and 3D).
Figure 3.
The sciatic nerve crush model elicits reproducible changes in gene expression
(A) The sciatic nerve was unilaterally crushed at the mid-thigh, resulting in denervation of motor neurons in lumbar sections L3-L6.
(B) Injured (INJ) and uninjured (UNJ) tibialis anterior muscles were collected at several time points after nerve crush.
(C–E) RNA isolated on different days after nerve crush (D1-D50) or nerve transection (D21-t) was subjected to RT-qPCR for muscle-associated receptor tyrosine kinase (MuSK, (C)), myelin protein zero (MPZ, (D)), and growth-associated protein 43 (Gap43, (E)).
Error bars represent the mean ± s.e. ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001.
To study degenerative motor neuron disease, we crossed the hSOD1G93A ALS mouse model with RiboTag;ChATCre mice. This generated RiboHO;ChATCre;hSOD1WT (RiboWT) and RiboHO;ChATCre;hSOD1G93A (RiboG93A) littermate mice (Figure 4A). The overall disease progression in RiboG93A mice was similar to previous reports (Gurney et al., 1994; Tu et al., 1996), with the early symptomatic phase beginning around 3 months of age and a total lifespan of approximately 5 months (Figure 4B). To examine the degree of muscle denervation in RiboG93A mice, NMJs of the extensor digitorum longus (EDL) muscle were imaged at various ages. Regardless of age, a similar number of NMJs were identified by α-bungarotoxin (BTX, Figure 4C). Co-staining endplates with Tuj1 (Figure 4D) and Synapsin (Figure 4E) allowed us to score the extent of innervation as fully innervated (Innerv), partially innervated (Partial), or denervated (Den). Given that innervation in RiboWT mice was similar at all time points, the results were averaged into one WT group. NMJs from RiboG93A and WT mice had similar innervation patterns at the 1-month time point. Denervation became apparent at 3 months of age and was the most severe at the 5-month time point. In addition to characterizing NMJ denervation, we also examined microgliosis in RiboG93A mice (Figures 4F and 4G). Longitudinal lumbar spinal cords were stained with HA (green) to visualize motor neurons and the microglial markers CD68 (red, Figure 4F) and CD11b (red, Figure 4G). Low levels of CD68 and CD11b were present in WT and 1-month old RiboG93A mice. However, similar to the time course of NMJ denervation, there was marked induction of microgliosis at 3 months of age that worsened by the 5-month time point (Figures 4F and 4G).
Figure 4.
The hSOD1G93A mouse model of ALS exhibits stereotyped neurodegeneration
(A) The hSOD1G93A model of ALS was crossed into the RiboTag; ChATCre strain to produce RiboHO;ChATCre;hSOD1WT (RiboWT) and RiboHO;ChATCre;hSOD1G93A (RiboG93A) littermates.
(B) Tissues were collected from 1-, 3-, 4-, and 5-month-old RiboG93A and RiboWT mice, representing presymptomatic, early symptomatic, symptomatic, and end-stage time points.
(C–E) Neuromuscular junctions (NMJs) were analyzed in extensor digitorum longus (EDL) muscles. The total number of analyzed NMJs was not altered at any time point (C). Tuj1 (D) and Synapsin (E) staining were used to quantify NMJ innervation as fully innervated (Innerv), partially innervated (Partial), or denervated (Den). Denervation started at 3 months of age and became progressively worse thereafter.
(F and G) Microgliosis in the ventral horn of the spinal cord was observed at 3 months of age in RiboG93A mice by immunolabeling longitudinal sections with HA to visualize motor neurons (green) and the microglial markers CD68 (red, (F) or CD11b (red, (G). Scale bar represents 100 μm. Error bars represent the mean ± s.e.
With these nerve crush and ALS models characterized, we used RNA-seq following Rpl22HA IP to assess how injury and disease alter the motor neuron translatome. Motor neuron gene expression was examined at D7 after sciatic nerve crush (corresponding to maximal regeneration, Figure 3) and in tissues from 1-month (presymptomatic), 3-month (early symptomatic), and 4-month (symptomatic) RiboG93A mice or RiboWT littermates (Figure 4). To generate samples for sequencing, spinal cords from D7 injured RiboHO;ChATCre mice, and RiboG93A and RiboWT mice were isolated. To restrict analysis to motor neurons affected by nerve crush, lumbar spinal cord corresponding to levels 3–6 (L3-L6) was separated into INJ and UNJ hemispheres and processed separately, while the entire lumbar spinal cord (L1-L6) was processed from RiboG93A and RiboWT mice. For all samples, mRNA from input lysates and Rpl22HA IPs was isolated, converted to cDNA libraries, and sequenced. An average of 83.6% of the total reads obtained could be uniquely mapped to the mouse genome (mm10) generating an average of 64 million high-quality, mapped reads per sample (Figure S3A). Hierarchical clustering and heatmaps of significantly changed genes demonstrate a high correlation among biological replicates from D7 INJ (Figures S3B and S3F) and 4-month-old RiboG93A IP samples (Figures S3E and S3G), while 1- and 3-month-old RiboG93A IP samples exhibited less correlation (Figures S3C, S3D, and S3G). After Rpl22HA IP, ChAT expression was consistent across all samples and, as expected, Sod1 was expressed at much higher levels in RiboG93A mice (Figure S3H).
To broadly examine the enrichment of motor neuron-specific genes after Rpl22HA IP in our sequencing data, we performed DEG analysis on the UNJ IP and UNJ input samples. This identified 4,097 genes enriched in motor neurons and 4,195 genes enriched in the input. Functionally grouped gene ontology (GO) analysis of the top 500 differentially expressed genes expressed in the IP were compared to the top 500 differentially expressed genes in the input using ClueGO (Bindea et al., 2009). In agreement with the known metabolic demand on motor neurons, we found an enrichment of GO terms relating to ATP metabolic processes, proton transmembrane transport, and translation (Figures S4A and S4B). Conversely, the input was enriched with GO terms not expected in motor neurons, including glial cell differentiation, myelin maintenance, and GABAergic synaptic transmission (Figures S4A and S4C). We next selected various genes that were known to be expressed by either motor neurons or glial cells. Among all control comparisons (UNJ IP:Input, and 1-month, 3-month, and 4-month-old RiboWT IP:Input) motor neuron genes [ChAT, Motor neuron and pancreas homeobox 1 (Mnx1), Islet1 (Is1), Islet2 (Is2), Calcitonin-related polypeptide alpha (Calca), Chondrolectin (Chodl), Matrix metallopeptidase 9 (MMP9), Sonic hedgehog signaling molecule (Shh), Neurofilament heavy (Nefh), Neuregulin1 (Nrg1)] were consistently enriched, while glial genes [Glial fibrillary acidic protein (Gfap), Aldehyde dehydrogenase 1 family member L1 (Aldh1l1), Myelin associated glycoprotein (Mag), SRY-box transcription factor 10 (Sox10), Cd83, Integrin subunit alpha M (Itgam or Cd11b), Adhesion G protein-coupled receptor E1 (Adgre1), C-X3-C motif chemokine receptor 1 (Cx3cr1), Cd68, and Transmembrane protein 119 (Tmem119)] were de-enriched (Figures S4D–G).
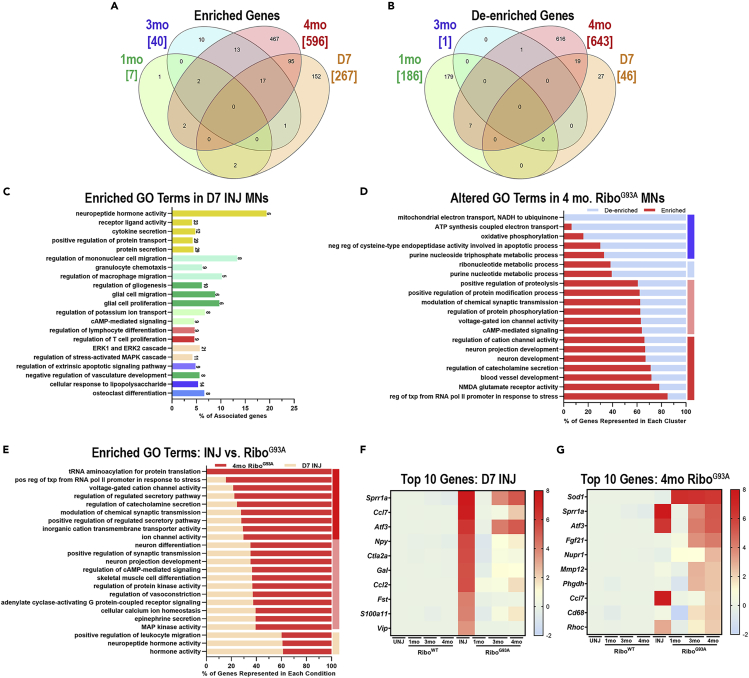
To determine how injury and neurodegeneration impact motor neuron gene expression, we examined the genes enriched by Rpl22HA IP in INJ and RiboG93A mice. For all comparisons, DEG analysis was conducted by comparing the experimental condition relative to its matched negative control (INJ IP vs. UNJ IP, 1-month RiboG93A vs. 1-month RiboWT, etc.). The number of enriched (Figure 5A) and de-enriched (Figure 5B) genes in all comparisons is illustrated in Venn Diagrams. Gene expression in 1- and 3-month-old RiboG93A motor neurons was largely similar to their age-matched controls. However, by 4 months of age, the number of DEGs in symptomatic motor neurons was much greater. After nerve injury most DEGs (85%) observed were found to be enriched in INJ motor neurons, compared to DEGs that were de-enriched. Interestingly, only a fraction (38%) of the 267 genes enriched by sciatic nerve crush was also enriched in 4-month-old degenerating RiboG93A motor neurons. In contrast, most DEGs were context-specific and only expressed in injured (58%) or 4-month RiboG93A (78%) motor neurons.
Figure 5.
Comparison of injured and diseased motor neurons
(A and B) Differential gene expression (DEG) analysis was performed on Rpl22HA IP datasets from INJ and 1-, 3-, and 4-month-old RiboG93A mice using their respective UNJ or age-matched RiboWT IP as a control. The number of enriched (A) and de-enriched (B) DEGs are displayed in Venn diagrams. The total number of DEGs in each condition is displayed in brackets, and the number of shared DEGs is indicated within the overlapping regions.
(C) GO analysis illustrates biological processes upregulated in injured motor neurons. The x axis represents the percent of DEGs within a GO term category, while the number to the right of each horizontal bar is the total number of observed DEGs.
(D) ClueGO analysis was used to compare genes enriched- and de-enriched in 4-month-old RiboG93A motor neurons. For each category, the percent of de-enriched genes are displayed in blue, while the percent of enriched genes are in red. The vertical bars on the right-hand-side illustrate categories that are highly de-enriched (dark blue), moderately de-enriched (light blue), moderately enriched (light pink), and highly enriched (red).
(E) Enriched genes identified in 4-month-old diseased (red) and injured (orange) motor neurons were compared using ClueGO analysis. Each category on the y axis is a GO term and the percentage of genes observed in diseased compared to injured motor neurons is displayed.
(F and G) Heat maps display the average gene expression across all samples analyzed for the top 10 most enriched genes in injured (F) and diseased (G) motor neurons.
Given the limited number of DEGs in 1- and 3-month-old RiboG93A mice, we choose to focus our attention on D7 INJ (injured) and 4-month RiboG93A (degenerating) motor neurons. GO analysis of injured motor neurons revealed that significantly changed GO terms included neuropeptide hormone activity, protein transport and secretion, cytokine secretion, immune cell migration, glial cell proliferation and migration, and MAPK signaling (Figure 5C). Genes enriched in degenerating motor neurons, on the other hand, were associated with regulation of transcription, excitability and neurotransmission (catecholamine secretion, NMDA glutamate receptor activity, voltage-gated ion channel activity), neural development and development of neural projections (red bars, Figure 5D). Conversely, genes associated with metabolic processes and oxidative phosphorylation tended to be de-enriched in degenerating motor neurons (blue bars, Figure 5D). We also directly compared the genes enriched in injured and degenerating motor neurons (Figure 5E). ClueGO analysis was used to determine how many genes in injured vs. degenerating motor neurons were assigned to specific GO terms. While most GO terms were generally associated with degenerating motor neurons, leukocyte migration and neuropeptide hormone activity GO terms were more associated with injured motor neurons.
Finally, we examined the top 10 genes enriched after injury (Figure 5F) and in diseased motor neurons (Figure 5G). In all conditions, low baseline gene expression was observed in all control tissues (INJ and RiboWT motor neurons). From this analysis, we could identify different patterns of gene expression. While some genes were enriched in both conditions [Small proline rich protein 1A (Sprr1a), Activating transcription factor 3 (Atf3), C-C motif chemokine ligand 7 (Ccl7), C-C motif chemokine ligand 2 (Ccl2), Neuropeptide Y (Npy), S100 calcium binding protein A11 (S100a11), and Ras homolog family member C (Rhoc)], other genes were specific to injury [Cytotoxic T lymphocyte-associated protein 2 alpha (Ctla2a), Galanin (Gal), Follistatin (Fst), and Vasoactive intestinal peptide (Vip)] or ALS degeneration [Fibroblast growth factor 21 (Fgf21), Nuclear protein 1, transcriptional regulator (Nupr1), Matrix Metallopeptidase 12 (Mmp12), Phosphoglycerate dehydrogenase (Phgdh), and Cd68]. Of note, 70% of the genes that were most enriched in 4-month-old RiboG93A mice were already enriched at the 3-month time point (Figure 5G), indicating that despite the higher variability in gene expression, some DEGs begin to emerge by 3 months of age.
Chemokines are dynamically upregulated in motor neurons following sciatic nerve crush
To validate genes enriched in injured motor neurons 7 days after sciatic nerve crush, we used a combination of RT-qPCR and fluorescence in situ hybridization (FISH). INJ and UNJ lumbar hemispheres were isolated at several time points after injury from RiboHO;ChATCre mice and Rpl22HA IPs were performed. Atf3 and Gap43 were selected to confirm our RNA-seq analysis of the injury response in motor neurons because they are well known to be upregulated in peripheral neurons after axonal injury (Chong et al., 1992; Curtis et al., 1992), and we found them to be enriched in motor neurons (Atf3, 134-fold; Gap43, 5.67-fold) after sciatic nerve crush. In agreement with the RNA-seq expression, RT-qPCR analysis demonstrated that from D1-D7 after injury there was a dramatic increase in Atf3 (>200-fold) in RNA isolated from the Rpl22HA IP (Figure 6A), while the increase in Gap43 (D1, 1.96 ± 0.183; D3, 5.82 ± 1.578; D7, 9.93 ± 1.610) was smaller and more gradual (Figure 6B). Both Atf3 and Gap43 expression levels returned to baseline by the D21 time point when NMJs were largely reinnervated, whereas failure of reinnervation after a nerve transection led to persistent elevation of both genes (Atf3, 115.47 ± 23.889; Gap43, 15.36 ± 2.22, Figures 6A and 6B). The expression of Atf3 and Gap43 was also examined in the input lysates that were collected prior to Rpl22HA IP. Upregulation of Atf3 could be detected in the input, albeit to a lesser extent than in the IP (<40-fold vs. >200-fold), whereas the upregulation of Gap43 in the input was not detected (Figures 6A and 6B). These data can be explained by the fact that motor neurons only represent a small fraction of the total cells in spinal cord. While large-scale changes in the IP (i.e. Atf3) can be detected at reduced expression levels in the input without Rpl22HA enrichment, genes that still exhibit robust expression after injury (i.e. the 10-fold induction of Gap43 in IP samples) are not sufficiently induced such that changes can be detected in the input. Together, this highlights the importance of the RiboTag strategy to identify changes in motor neuron gene expression (or regulation) from complex tissues.
Figure 6.
Chemokines are upregulated in injured motor neurons
(A and B) INJ and UNJ lumbar hemispheres were isolated from RiboHO;ChATCre mice D1-D50 after nerve crush or 21 days after nerve transection (D21-t). After Rpl22HA IP, RT-qPCR was used to examine Atf3 (A) and Gap43 (B) gene expression in both the IP and the input relative to the UNJ control. For simplicity, asterisks indicate statistical significance as compared to the D50 when reinnervation is largely complete.
(C) Quantification of the percentage of Gap43+ motor neurons that co-expressed Ccl4, Ccl2, or Ccl7 at D3 and D7 after injury based on the data in D and E (n = 5).
(D) Fluorescent in situ hybridization (FISH) was performed on WT lumbar spinal cord sections at D3 (n = 3) and D7 (n = 2) after sciatic nerve crush. Ccl7 (red) or Ccl12 (blue) were not expressed in the UNJ contralateral spinal cord, were upregulated by injury at both D3 and D7 time points, and only Ccl7 was observed in Gap43+ (green) injured motor neurons.
(E) Similar analysis as in D demonstrated that both Ccl2 (blue) and Ccl4 (red) were observed in D3 and D7 injured Gap43+ (green) motor neurons. Scale bar represents 50 μm.
Error bars represent the mean ± s.e. ∗∗∗∗p ≤ 0.0001, ∗∗p ≤ 0.01.
We next used FISH to validate that DEGs identified by RNA-seq are localized to motor neurons in 3-month-old C57BL/6 wild-type mice after injury. Because our RNA-seq analysis identified several GO terms related to immune system function, we choose to look at the expression of several chemokines at D3 and D7 after nerve crush. Ccl7, Ccl2, and Ccl12 have been previously reported to go up in the dorsal root ganglia (DRG) after injury (Zhao et al., 2020; Zigmond and Echevarria, 2019). In our analysis, Ccl7 and Ccl2 were among the most enriched genes in injured motor neurons (Ccl7, 403-fold; Ccl2, 90-fold; Figure 5F), Ccl4 was modestly enriched (2.5-fold) and Ccl12 expression was not significantly changed after injury. Spinal cord sections were labeled with probes to these cytokines in conjunction with a probe to ChAT to demonstrate that they were indeed upregulated in INJ motor neurons (Figures S5B and S5C). We then quantified the extent of cytokine upregulation after injury. Gap43 was used as a marker for injured motor neurons because, together with Sprr1a, it defines all regenerating motor neurons following sciatic nerve crush (Figure S5A). Consistent with our previous results, both Ccl7 and Ccl2 were upregulated in a majority of Gap43+ motor neurons at both D3 and D7 time points (Ccl7, 78.09 ± 1.158%; Ccl2, 78.59 ± 2.343%, Figures 6C–6E), while Ccl4 was only upregulated in a small population of motor neurons (12.64 ± 2.090%, Figures 6C and 6E). Interestingly, although we confirmed that Ccl12 was not induced in motor neurons after injury, we did find it increased in cells within close proximity to motor neurons (Figures 6D, and S5B).
RiboG93A motor neurons upregulate stress and chemokine markers
Similar to the analysis of injured motor neurons, we validated changes identified by RNA-Seq in RiboG93A motor neurons at different ages. Lumbar spinal cords were isolated from 1-, 3-, and 5-month-old RiboG93A and RiboWT littermate controls and Rpl22HA enrichment was performed. Markers of cholinergic neurons were used to examine gene expression in the IP and input samples (Figures 7A–7C). ChAT is a general marker of all cholinergic neurons and, given that cell death in ALS predominantly affects αMNs, it is perhaps not surprising that we did not observe a significant decrease in ChAT mRNA in the IP at any age in RiboG93A mice, although there was a slight decrease in overall ChAT mRNA in the input at 5 months of age (Figure 7A). We next examined NeuN and Esrrg expression as differential markers of motor neuron survival because immunostaining of cholinergic populations in the spinal cord demonstrated that NeuN is more robustly expressed in αMNs compared to other cholinergic populations (Figure S1A), and γMNs that express Esrrg are known to be resistant to degeneration in ALS (Lalancette-Hebert et al., 2016). Consistent with these observations, we detected a significant decrease in NeuN mRNA in IPs from late stage 5-month-old RiboG93A motor neurons (RiboWT, 1.00 ± 0.028, RiboG93A 5-mo, 0.452 ± 0.033, Figure 7B) indicative of αMN death, but no change in Esrrg expression (Figure 7C). We also examined changes in Atf3 and Fgf21, which were both identified in our RNA-seq screen as two of the most enriched genes in degenerating motor neurons (Figure 5G) and are implicated in ALS neurodegeneration (Logsiger et al., 2007; Vlug et al., 2005) and neuronal stress responses (Restelli et al., 2018). Accordingly, both genes were significantly increased at 3 and 5 months of age in RiboG93A mice (Atf3, RiboWT, 1.00 ± 0.064, 1-month, 0.95 ± 0.196, 3-mo, 33.79 ± 5.923, 5-mo, 89.08 ± 7.994; Fgf21, RiboWT, 1.00 ± 0.052, 1-month, 1.65 ± 0.572, 3-mo, 28.77 ± 2.851, 5-mo, 48.85 ± 2.971, Figures 7D and 7E).
Figure 7.
Stress and chemokine response in ALS degenerating motor neurons
(A–E) RT-qPCR was used to examine motor neuron gene expression after Rpl22HA IP from 1-, 3-, and 5-month-old RiboG93A mice relative to RiboWT littermate controls. Neither ChAT (A) nor the γMN marker Esrrg (C) was altered at any time point; however, a decrease in NeuN was observed at the 5-month time point (B) and an increase in Atf3 (D) and Fgf21 (E) were observed at both 3 and 5 months of age in RiboG93A motor neurons. Asterisks above the different RiboALS time points in A-E indicate statistical significance as compared to the average of RiboWT littermate controls from all time points.
(F) Increased Fgf21 was confirmed by FISH. Fgf21 transcript (blue) colocalized with Chat+ (red), degenerating motor neurons.
(G) Fgf21 transcript levels were scored as negative (Fgf21-), low (Fgf21lo), or high (Fgf21hi) and the percentage of Chat+ motor neurons was quantified in RiboWT (each time point, n = 3), RiboALS mice (1-, 4-mo, n = 3; 3-mo, n = 2; 5-mo, n = 4).
(H) Ccl7 and Ccl12 expression were similarly examined by FISH. At 4- and 5-month time points, rare Chat+ (red) motor neurons in RiboG93A mice exhibited Ccl7 (green) expression. Ccl12 (blue) was generally increased, however expression was not observed in motor neurons.
(I) The percentage of Ccl7 positive neurons were scored as in (G) Scale bar represents 50 μm.
Error bars represent the mean ± s.e. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001.
Fgf21 is produced by multiple tissues, including liver, pancreas, muscle, adipose tissue, and thymus (BonDurant and Potthoff, 2018), and has been reported to be produced by glial cells in vitro (Leng et al., 2016; Makela et al., 2014). To confirm that Fgf21 mRNA is localized in motor neurons during ALS neurodegeneration we performed FISH on longitudinal spinal cord sections from RiboWT and RiboG93A tissues isolated at various ages. The majority of RiboWT and 1-month-old RiboG93A motor neurons did not express Fgf21 (Fgf21-), while a small fraction exhibited low levels of Fgf21 transcript expression (Fgf21lo). A robust upregulation of Fgf21 expression (Fgf21hi) was only observed in 3-, 4-, and 5-month-old RiboG93A ChAT+ motor neurons (Figures 7F and 7G). We also used FISH to examine the chemokines Ccl7 and Ccl12. Like injured motor neurons, Ccl7 was identified as one of the top 10 most upregulated genes in 4-month-old RiboG93A motor neurons, although the extent of upregulation was less than that observed after sciatic nerve crush (4-mo RiboG93A, 16-fold vs. D7 INJ, 403-fold, Figures 5F and 5G). Consistent with our RNA-seq findings, we did not detect Ccl7 expression in RiboWT or 1- and 3-month-old RiboG93A motor neurons, and readily detected sparse, but strong labeling in ChAT+ motor neurons at 4- and 5-month-old time points (Figures 7H and 7I). Immunolabelling of ChAT+ motor neurons with antibodies to CCL7 confirmed that a subset of degenerating neurons expressed CCL7 in 5-month-old RiboG93A motor neurons and not RiboWT neurons (0.00% for RiboWT mice and 1.28% ± 0.29 for 5-month-old RiboG93A motor neurons, Figures S6B and S6C). Ccl12, which was not identified as upregulated in the RNA-seq data, was modestly upregulated in cells nearby motor neurons in RiboG93A spinal cord at 3 months and was increasingly upregulated with age (Figure 7H), suggesting that Ccl12 is upregulated by cells neighboring motor neurons in response to stress. In addition to expression in motor neurons, we also observed rare Ccl7 expression in cells adjacent to motor neurons. Interestingly, these cells were only observed at late stages in 5-month-old RiboG93A mice, and Ccl7 expression did not overlap with cells that express Ccl12 (Figure S6A).
Finally, we also examined transcript expression in two lines of TDP-43 mutant mice by FISH. A moderately progressing line (TDP-43Q331K, Figure S6D) (Arnold et al., 2013) and a more rapidly progressing line (TDP-43A315T, Figure S6E) (Wegorzewska et al., 2009) were analyzed at early symptomatic ages. Fgf21+ChAT+ motor neurons were observed in both the moderately and rapidly progressing mouse models of ALS (Figures S6F and S6H). We also observed Ccl7+ChAT+ motor neurons in TDP-43A315T spinal cords to a greater extent than in TDP-43Q331K spinal cords (Figures S6G and S6I). Importantly, these results extend our findings to different models of ALS neurodegeneration.
Discussion
Motor neurons play a requisite function in orchestrating movement as they represent the final common pathway that conveys neural information to skeletal muscle. Although motor neurons possess significant regenerative capacity in response to peripheral injuries, they remain susceptible to degeneration in the face of severe trauma or neurodegenerative disease, such as ALS. Here, we define a strategy using ribosomal profiling to directly assess motor neuron gene expression. By applying RNA-seq following sciatic nerve crush and in an ALS mouse model we also provide a direct comparison of the regenerating and degenerating motor neuron translatome.
Interestingly, other methods of ribosomal profiling in motor neurons have been recently reported using the bacTRAP system (Sun et al., 2015). One advantage of the RiboTag mouse model is that the tagged ribosomes are expressed from an endogenous allele. This both avoids competition of the exogenous allele for ribosome occupancy (Shigeoka et al., 2016) and allows for the enrichment of Rpl22HA protein and associated RNA in a gene dose-dependent manner (Figures 2A–2D). Moreover, by using homozygous Rpl22HA expression, significantly more RNA was isolated from small tissue samples while not interfering with gene expression (Figure 2F), altering the time course of regeneration after sciatic nerve crush (Figure 3) (Bowen et al., 1998; Curtis et al., 1992; Gupta et al., 1988; Valenzuela et al., 1995), or neurodegeneration in the SOD1G93A ALS mouse model (Figure 4) (Gurney et al., 1994).
Regenerating axons projecting within the periphery and central nervous system exhibit dramatically different properties. The expression of regeneration-associated genes (RAGs) in peripheral axons enable axon growth and reinnervation of target tissues, while regeneration of central axons is blunted due to a combination of inhibitory intrinsic and extrinsic factors (He and Jin, 2016; Plunet et al., 2002). As such, understanding the mechanisms that govern peripheral nerve regeneration may lead to improvements in regeneration after central nervous system injury or in neurodegenerative disease. Historically, high-throughput analysis of gene expression in spinal motor neurons after injury has been hampered by the fact that they represent a relatively rare cell population in the spinal cord. The RiboTag;ChATCre mouse model allowed us to examine gene expression by RNA-seq in regenerating motor neurons 7 days after nerve crush. DEG analysis found that 267 genes are enriched following sciatic nerve crush, while only 46 genes were de-enriched (Figure 5A). Closer examination of the injury-induced (enriched) genes revealed that many corresponded to known RAGs, including: Atf3, Sprr1a, Gap43, Basp1 (CAP-23), Cebpd (C/EBP-Delta), Sox11, and Socs3 (Figures 5F, 6A, 6B, and Table S1) (Ma and Willis, 2015). Subsequent pathway analysis of all injury-induced genes further demonstrated a prominent role for neuropeptide hormone activity, cytokine secretion, immune cell function, and glial cell proliferation, and migration in regenerating spinal motor neurons (Figure 5C). Neuropeptide production in response to axotomy has been well documented in many peripheral neurons. Following injury, neurons of the superior cervical ganglion (SCG) and DRG upregulate Gal, Vip, Adcyap1 (PACAP), Tac1 (substance P), Calca (CGRP), and Cck (cholecystokinin). Upregulation of Npy has been reported in the DRG, but its expression is downregulated in the SCG following injury (Zigmond, 2012). Neuropeptide expression in spinal motor neurons has also been previously reported and our analysis confirms that Gal, Vip, Adcyap1, and Calca are significantly induced in lumbar motor neurons following sciatic nerve crush (Figure 5F and Table S1) (Sala et al., 1995; Zigmond, 2012). We also identified the upregulation of Npy (Figure 5F), Grp (gastrin releasing peptide), Nms (neuromedin s), and Nts (neurotensin) in injured spinal motor neurons (Table S1). Although Cck has been described to be downregulated in lumbar motor neurons we did not observe any change in Cck expression in our analysis (Table S1).
In addition to neuropeptides, there was a striking increase of injury-responsive chemokines. Ccl7 (MCP-3) and Ccl2 (MCP-1) were among the top 10 most enriched genes after injury (Figure 5F). Ccl2 expression by Schwann cells has long been known to recruit monocytes to the site of axonal injury (Zigmond and Echevarria, 2019), while injury-induced neuronal expression of Ccl2 leads to monocyte infiltration of peripheral ganglia (Lindborg et al., 2018; Schreiber et al., 2001; Wang et al., 2018). Here, we demonstrate that Ccl2 and Ccl7 mRNA are upregulated in the majority (78%) of regenerating Gap43+ spinal motor neurons (Figures 6C–6E). Although the precise role of chemokine expression in motor neurons remains to be determined, it is possible that the secretion of Ccl2/7 from motor axons contributes to recruitment of monocytes to the injury site (Zigmond and Echevarria, 2019). Alternatively, it may activate microglia and/or other cells surrounding motor neurons to promote regeneration independent of peripheral axonal debris clearance (Niemi et al., 2013). Interestingly, C-C chemokine receptor type 2 (CCR2), the predominant receptor for both Ccl2 and Ccl7, is expressed by microglia (El Khoury et al., 2007), and infiltrating myeloid cells after nerve transection (Rotterman et al., 2019). DEG analysis of input lysate samples from injured versus uninjured spinal cords revealed an induction of markers of microglial activation (Cd68, Cd11b, F4/80), but Ccr2 expression was not detected at baseline or in response to injury (Table S1). Although this does not preclude a role for CCR2 in CCL2/7 signaling, it raises the possibility that signaling may occur through atypical chemokine receptors (Gschwandtner et al., 2019). RNAseq analysis also identified a modest upregulation in the chemokine Ccl4 after injury. Examination of Ccl4 expression by FISH demonstrated that a small subpopulation (12%) of motor neurons upregulate Ccl4 after sciatic nerve crush. While the function of Ccl4 signaling after injury is unknown, it highlights the sensitivity of the RiboTag;ChATCre model.
In contrast to acute injury models, neurodegenerative diseases such as ALS exhibit chronic motor neuron dysfunction. Notably, the first sign of pathology is motor axon withdrawal from NMJs, which begins prior to the onset of clinical symptoms (Fischer et al., 2004). To examine the motor neuron translatome during different phases of neurodegeneration, we crossed the RiboTag;ChATCre mouse model to SOD1G93A mice. At presymptomatic (1 month) and early symptomatic ages (3 months), we found relatively few changes in RiboG93A motor neuron gene expression (Figures 5A and 5B). This was surprising given that previous studies detected numerous changes in motor neuron gene expression using methods such as LCM (Nardo et al., 2013). Although it is unclear what underlies this difference, it may reflect the greater variability in gene expression at early ages (Figures S3C, S3D, and S3G) and/or a more general difference in changes to the transcriptome versus ribosome-associated transcript expression. Nevertheless, extensive changes in gene expression were found in symptomatic, 4-month-old RiboG93A motor neurons (596 enriched and 643 de-enriched genes, Figures 5A and 5B). In accordance with the high energetic demand placed on motor neurons, we observed that de-enriched genes belonged to pathways related to ATP synthesis, oxidative phosphorylation, and metabolic processes (Figure 5D). Conversely, enriched genes were associated with protein misfolding and aggregation (regulation of transcription from RNA pol II promoter in response to stress) and excitotoxicity (NMDA glutamate receptor activity), both processes that are highly associated with neurodegeneration in ALS. As mentioned above, ribosomal profiling of motor neurons in the more slowly progressing SOD1G37R mouse model of ALS was recently reported (Sun et al., 2015). Of the genes identified in 8-month-old SOD1G37R mice, which corresponds to disease onset, only 29% were also enriched in our ribosomal profiling of 4-month-old RiboG93A mice. Although it is difficult to discern if the low degree of overlap is due to the ALS models themselves or the age that was analyzed, many of the most upregulated genes (Fgf21, Atf3, Nupr1, Phgdh) were altered in both models (Figure 5D) (Sun et al., 2015).
Originally Fgf21 was identified in liver and thymus but has recently been reported to be expressed by neurons in response to ER stress and in mouse models of tauopathy and prion disease (Restelli et al., 2018). In our study, Fgf21 was detected at disease onset and increased with disease severity (Figure 6E). Using in situ hybridization we demonstrated that Fgf21 induction was confined to ChAT+ motor neurons in both SOD1 and TDP43 mouse models of ALS (Figures 6F, S7C, and S7D). Most FGF family members are considered paracrine factors because they contain a heparin binding domain that results in their retention in the extracellular matrix. FGF21, in contrast, belongs to a subfamily characterized by endocrine signaling because absence of the heparin binding domain allows it to readily diffuse through tissues and into the peripheral circulation (BonDurant and Potthoff, 2018). Indeed, during stress conditions such as fasting, FGF21 is released by the liver into the circulation, and stimulates gluconeogenesis and fatty acid oxidation in order to maintain glucose homeostasis (Martinez-Garza et al., 2019). Overexpression of FGF21 in transgenic mice, however, leads to osteopenia, increased sympathetic drive and weight loss and can cause muscle atrophy and weakness (Bookout et al., 2013; Inagaki et al., 2007; Oost et al., 2019). What role Fgf21 has in the progression of ALS will be important to elucidate in future studies, especially considering recent findings that FGF21 can protect neurons from tau hyperphosphorylation, oxidative stress and apoptosis in Alzheimer's disease models (Chen et al., 2019b).
Two other genes that were among the most upregulated in RiboG93A motor neurons were Mmp12 and Ccl7 (Figure 5G). Matrix metalloproteinases (MMPs) are a large family of zinc-dependent proteases that degrade basement membrane and extracellular matrix components and function in immune processes by cleaving growth factors, chemokines, and cytokines (Brkic et al., 2015). While MMPs are known to play important roles in tissue remodeling and regeneration, dysregulation of MMP activity has been described in many neurodegenerative diseases (Brkic et al., 2015). Although Mmp12 is expressed by microglia in SOD1G93A mice (Chiu et al., 2013), it is also expressed by neurons, including motor neurons in the rat facial nucleus after axotomy (Sajjan et al., 2014), and abnormal or excessive Mmp12 expression could destabilize synaptic structures by acting on the basal lamina. Ccl7 (and to a lesser extent Ccl2, Table S1) were also upregulated in 4-month-old RiboG93A motor neurons. As discussed above, these cytokines were also observed among the most upregulated genes after sciatic nerve crush. Closer examination of Ccl7 by in situ hybridization and immunolabelling demonstrated that it was indeed expressed by ChAT+ motor neurons in symptomatic 4- and 5-month old RiboG93A mice and in the more aggressive TDP-43A315T mouse model (Figures 7H, 7IS6B, S6G, and S6I). Despite being among the most upregulated genes in RiboG93A motor neurons, it was surprising that Ccl7 was only expressed in a small percentage of motor neurons (Figures 7H and 7I). This finding may reflect asynchronous waves of degeneration that occur among motor neurons over time or perhaps differential responses that occur within specific motor neuron subpopulations. Finally, recent studies suggest that Ccl12 (MCP-5) is expressed in DRG neurons after injury (Zhao et al., 2020) and in SOD1G93A motor neurons (Nardo et al., 2013). In our study, Ccl12 was not differentially expressed in D7-injured or RiboG93A motor neurons following Rpl22HA IP, but it was upregulated in cells surrounding motor neurons (Figures 6C, 7H, and S7I). Although Ccl12 expression has been reported in SOD1G93A microglia (Chiu et al., 2013), what function it plays in the non-cell autonomous response to injury and neurodegeneration is not known. Together, these data raise important questions about chemokine signaling in motor neurons. Future investigation is needed to determine if motor neuron chemokine expression is necessary for axon regeneration. In the context of neurodegeneration, it is possible that chemokine expression is detrimental, given it is restricted to fully symptomatic ALS mice. However, if chemokine signaling is beneficial for axonal regeneration, chemokine expression may act to slow the progression of degeneration in ALS by recruiting macrophages and microglia that promote tissue repair.
The emergence of single-cell sequencing technology enables us to dissect complex gene expression patterns among heterogeneous cell populations. This has revealed much about gene expression dynamics and aided in the identification of rare cell populations. Despite these advancements, many scRNA-seq approaches have technical challenges such as high dropout rates that increase noise and mask weakly expressed genes (Chen et al., 2019a). Given that protein abundance is predominately controlled at the level of translation, and there is a low correlation between mRNA and protein for many genes (Schwanhausser et al., 2011; Zhao et al., 2019), translatomic approaches offer unique insights into the regulation of gene expression. Therefore, the analysis presented here represents an important resource that will help inform future research. By focusing of how gene expression is altered in regenerating and degenerating neurons, our analysis will help to distinguish between genes that represent adaptive, protective responses to neurodegeneration as compared to gene expression changes that may be maladaptive. Importantly, this strategy can be easily applied to different ALS mouse models in future research to further assist in the identification of shared and divergent neurodegenerative mechanisms that underlie heterogeneity in ALS and neurodegenerative disease.
Limitations of the study
Some genes we identified as enriched in response to ALS degeneration included markers for microglial activation, such as Cd68 and Tmem119. While the Rpl22HA IPs allow for a significant enrichment of motor neuron transcripts, there is a small amount of high-quality, non-specific transcript that associates with the IP (Figure 2C). Therefore, it is possible that the microglial markers we identified were enriched due to their increased abundance during neurodegeneration. Analysis of Cd68 induction by RT-qPCR and FISH was preformed (data not shown), but the close proximity of microglia and motor neurons during degeneration made it difficult to conclusively determine if all signal detected was from microglial contamination or if motor neurons also contribute to the increased expression levels.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| HA (for IPs) | BioLegend | Cat#MMS-101P |
| Actin | Santa Cruz | Cat#SC-1616-G |
| HA (for immunoblotting) | Cell Signaling | Cat#3724 |
| Fasn | Cell Signaling | Cat#3189 |
| Acot1 | Abcam | Cat#ab100915 |
| Rps2 | Bethyl Labs | Cat#A303-794 |
| Rpl36 | Abcam | Cat#ab241584 |
| Rps17 | Abcam | Cat#ab128671 |
| ChAT | Millipore | Cat#AB144P |
| NeuN | Millipore | Cat#MAB377 |
| CD11b | BioLegend | Cat#MCA711 |
| CD68 | BioLegend | Cat#MCA1957 |
| Synapsin | Cell Signaling | Cat#5297S |
| Tuj1 | Sigma-Aldrich | Cat#T8578 |
| MCP3 (CCL7) | Abcam | Cat#ab228979 |
| Chemicals, peptides, and recombinant proteins | ||
| Protein G Magnetic beads | Fisher | Cat#S1430S |
| Donkey Serum | Jackson Immunoresearch | Cat#9624464 |
| M.O.M. Blocking agent | Vector laboratories (Fisher) | Cat# MKB22131 |
| Critical commercial assays | ||
| RNAScope Multiplex Fluorescent Reagent Kit v2 | ACD | Cat#323100 |
| SMART-Seq v4 Ultra Low Input RNA kit | Takara | Cat#R400752 |
| SMARTer ThruPLEX DNA-Seq kit | Takara | Cat#R400675 |
| Superscript III First-Strand Synthesis SuperMix | Invitrogen | Cat#18080400 |
| Deposited data | ||
| Raw and Analyzed sequence data | This paper | GEO: GSE162028 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J | Jackson Laboratories | Stock#000664 |
| Mouse: RiboTag: B6N.129-Rpl122tm1.1Psam/J | Jackson Laboratories | Stock#011029 |
| Mouse: ChAT-IRES-Cre: B6;129S6-ChATtm(Cre)/Lowl/J | Jackson Laboratories | Stock#006410 |
| Mouse: B6.SOD1-G93A: B6.Cg-Tg(SOD1∗G93A)1Gur/J | Jackson Laboratories | Stock#004435 |
| Mouse: B6N.Cg-Tg(Prnp-TARDBP∗Q331K)103Dwc/J | Jackson Laboratories | Stock#017933 |
| Mouse: B6.Cg-Tg(Prnp-TARDPBP∗A315T)95Balo/J | Jackson Laboratories | Stock#010700 |
| Oligonucleotides | ||
| Primers for Real-time RT-PCR, see Table S1 | This Paper | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Brian Pierchala (brpierch@iu.edu).
Materials availability
Individual mouse lines are available from Jackson Labs, and combinations of these will be provided upon request.
Data and code availability
The sequencing data generated in this study has been deposited into the Gene Expression Omnibus (GEO) of NCBI and is available as accession number GSE162028. Bioinformatics comparisons and lists of differentially expressed genes are included in the Table S1, contained in the Supplemental information.
Experimental model and subject details
Animals
All procedures performed on mice were approved by the Institutional Animal Care and Use Committees (IACUCs) of the University of Michigan and Indiana University School of Medicine. Mice were maintained in a specific pathogen-free (SPF) facility that included environmental enrichment and were provided food and water ad libitum. Wild-type C57BL/6J (000664), RiboTag (B6N.129-Rpl122tm1.1Psam/J, 011029), ChAT-IRES-Cre (B6;129S6-ChATtm(Cre)/Lowl/J, 006410), B6.SOD1-G93A (B6.Cg-Tg(SOD1∗G93A)1Gur/J, 004435), B6N.Cg-Tg(Prnp-TARDBP∗Q331K)103Dwc/J (TDP-43Q331K, 017933), and B6.Cg-Tg(Prnp-TARDPBP∗A315T)95Balo/J (TDP-43A315T, 010700) mice were obtained from Jackson Laboratories (Bar Harbor, ME). RiboTag conditional mice were crossed with ChAT-Cre mice to generate Rpl22HA/+;ChATCre/+ (RiboHET;ChATCre) and Rpl22HA/HA;ChATCre/+ (RiboHO;ChATCre) experimental mice. Additionally, C57BL/6J mice and Rpl22+/+;ChATCre (RiboWT;ChATCre), Rpl22HA/+;ChAT+/+ (RiboHET;ChATWT), and Rpl22HA/HA;ChATWT mice were used as wild-type (WT) controls. For experiments to study neurodegeneration in motor neurons, B6.SOD1-G93A mice were crossed into Rpl22HA/HA;ChATCre line to create a triple transgenic line (hSOD1G93A;RiboHO;ChATCre, simplified as RiboG93A and RiboWT). All mice were genotyped according to publicly available protocols and both male and female mice were analyzed in similar numbers for all experiments. Adult mice (2-3 months of age) were analyzed for all method establishment experiments and nerve crush experiments (Figures 1, 2, 3, 6, S1, S2, and S5). For experiments examining ALS model mice (Figures 4, 5, 7, S3, S4, and S6), mice were 1-, 3-, 4- or 5-months-old, as described in the Figure Legends.
Method details
Nerve crush
Mice were deeply anesthetized with 2-3% isoflurane mixed with oxygen and the surgical site was shaved and disinfected. A small incision at the level of the mid-thigh was made through the skin and overlying musculature to expose the sciatic nerve. The exposed nerve was then crushed by applying pressure with Moria Ultra Fine Forceps (#11399-80, Fine Science Tools). Following the crush procedure, the muscle was closed with a suture and the skin was closed with staples. In a small number of experiments, a nerve transection was performed instead of a nerve crush. In these instances, the same procedure was performed except that a 3-5mm piece of nerve was excised as opposed to being crushed with forceps.
Tissue preparation
Mice were transcardially perfused with phosphate-buffered saline (PBS). The entire spinal cord was rapidly isolated by hydraulic extrusion (Kennedy et al., 2013) and then the lumbar segment was further dissected for subsequent analysis. For RNA and protein extraction, spinal cord and muscle tissues were snap frozen in liquid nitrogen. For immunohistological experiments, spinal cord was post-fixed in 4% paraformaldehyde (PFA, Electron Microscopy Sciences) at 4°C overnight. Tissues were washed with PBS, soaked for at least one day in 30% sucrose at 4°C, and then frozen for cryosectioning.
HA immunoprecipitation and RNA isolation
Homogenization and HA IP were largely performed as previously described (Sanz et al., 2009). Briefly, tissues were placed into IP buffer (50 mM Tris, pH 7.4, 100 mM KCl, 12 mM MgCl2, 1% Nonidet P-40 [NP-40] supplemented with 200 U/mL Promega RNasin, 1 mg/mL heparin, 100μg/mL cycloheximide, and protease inhibitor mixture [Sigma-Aldrich]) and subjected to Dounce homogenization. Samples were then rotated at 4°C for 15-20 minutes followed by centrifugation at 10,000 x g for 10 min at 4°C to generate a postmitochondrial supernatant. A 50μl aliquot of the input was saved and processed separately for protein and/or RNA analysis. The remainder of the sample was precleared with Protein G magnetic beads (25 μl of beads per 200 μl of supernatant, New England Biolabs) for 1 hour under constant rotation at 4°C. After preclearing, samples were separated from the magnetic beads and incubated with an HA antibody (1:150 dilution, #MMS-101P, BioLegend) by toppling at 4°C for 4 hours. Protein G magnetic beads were then added and allowed to incubate under rotation at 4°C overnight. The following day, samples were placed on a magnetic rack and supernatants were collected. The IP product associated with the magnetic beads was then washed 3 times in high salt buffer (50 mM Tris, pH 7.4, 300 mM KCl, 12 mM MgCl2, 1% NP-40 supplemented with 100 μg/mL cycloheximide). RNA associated with the HA IP was then isolated using the RNeasy Plus Micro Kit (Qiagen) per manufacturer instructions.
Bioanalyzer analysis, cDNA preparation, and RT-qPCR
The concentration of isolated RNA was determined by running samples on a 2100 Bioanalyzer Instrument (Agilent Genomics). cDNA was then synthesized with the remainder of the RNA sample (∼12 μl) using Superscript III First-Strand Synthesis SuperMix (Invitrogen). Subsequent RT-qPCR was performed using a 7900HT Fast Real-Time PCR System (Applied Biosystems) with the appropriate primer set (Table S1) and FastStart Universal SYBR Green Master Mix (Roche). All samples were run in triplicate and the resulting CTs were averaged. Data were normalized to a loading control (B2m, Figure 3 and Actin, 6 and 7), while normalization to a loading control was not performed in Figure 2. Following normalization data were log2 transformed and then relative expression was calculated for each gene set examined (Figure 2: average WT; Figure 3 average of all UNJ; Figure 7: average RiboHO). For Figure 6, the graph reflects the ddCT in injured/uninjured motor neurons.
HA immunoprecipitation for western blotting
Spinal cord tissues were placed into immunoprecipitation buffer with glycerol (10% glycerol, C0mplete protease inhibitors [Roche], and sodium vanadate in Tris buffered saline [TBS], pH 6.8). Tissues were homogenized with the Tissue Lyser II (Qiagen) set at 30 Hz for 5min. Samples were then detergent extracted by the addition of NP-40 (to a 1% final concentration) with constant rotation for 30 min at 4°C. Insoluble material was removed by centrifugation for 5 min at maximum speed in a microfuge. The samples were then precleared with Protein G agarose beads (Roche) for 1 hour. HA antibody (1:150 dilution, #MMS-101P, BioLegend) and Protein G agarose beads were then added overnight under constant rotation at 4°C. The next day samples were lightly centrifuged (5000 rpm, 5 min) and the supernatant was collected and denatured in 2x sodium dodecyl sulfate (SDS) sample buffer (20% glycerol, 4% SDS, 1% β-mercaptoethanol, and bromophenol blue in TBS, pH 6.8) by heating for 10 min at 100°C. The immunoprecipitated lysates were washed 3 times with the glycerol-based immunoprecipitation buffer followed by denaturation in SDS sample buffer as described for the supernatants.
Protein isolation after RNA lysis and processing
In some instances, protein was co-purified with RNA isolated using the RNeasy Plus Micro Kit (Qiagen). Once RNA binds to the RNA-binding cup, protein in the flow-through was precipitated with ice-cold acetone for 30 min on ice. Samples were centrifuged for 10 min at max speed to pellet the protein precipitate. Protein pellets were then washed with 100% ice-cold ethanol, centrifuged again, and resuspended in water. Finally, the isolated protein was denatured by adding an equal volume of 2x SDS sample buffer and heating for 10 min at 100°C.
Immunoblotting
All samples were resolved on 12 or 14% (SDS)-polyacrylamide gel electrophoresis (SDS-PAGE) gels and transferred to polyvinylidene fluoride (PVDF) membranes. Membranes were blocked in 4% Milk in Tris buffered saline, pH 7.4 containing 0.1% Tween-20 (TBS-T) for 1 hour at room temperature. Primary antibodies [Actin (1:1,000, #SC-1616-G, Santa Cruz), HA (1:1,000, #3724, Cell Signaling), Fasn (1:1000, #3189, Cell Signaling), Acot1 (1:1000, #ab100915, Abcam), Rps2 (1:1000, #A303-794, Bethyl Labs), Rpl36 (1:1000, #ab241584, Abcam), Rps17 (1:1000, #ab128671, Abcam)] were diluted in 3% bovine serum albumin (BSA) and incubated overnight at 4°C. The following day, membranes were washed and incubated with the appropriate horseradish peroxidase (HRP)-linked secondary antibodies (1:10,000, Jackson ImmunoResearch) in 3% BSA followed by visualization with a chemiluminescent substrate (Thermo Scientific). Western blot quantification was performed by densitometric analysis using ImageJ (NIH). The signal intensity in arbitrary units was determined for HA protein in the IP and the amount of actin in the supernatant. After normalization to actin, HA protein expression in RiboHET;ChATCre and RiboHO;ChATCre mice was displayed relative to wild-type controls.
Immunostaining
30 μm transverse (immunofluorescent) or 20 μm longitudinal (in situ) cryosections were cut using a CM1950 cryostat (Leica Biosystems). For ChAT and NeuN immunostaining antigen retrieval was performed prior to permeabilization and blocking by boiling sections in 10 mM sodium citrate buffer, pH 6.0 for 3 min. Sections were permeabilized and blocked in PBS containing 0.2% Triton-X, 1% BSA, 10% donkey serum (Jackson ImmunoResearch), and M.O.M blocking reagent (Vector Laboratories) for 1 hour at room temperature. Slides were then incubated with primary antibodies [(HA (1:100, #3724, Cell Signaling), ChAT (1:100, #AB144P, Millipore), NeuN (1:100, #MAB377, Millipore), CD11b (1:100, #MCA711, Biolegend), CD68 (1:100, #MCA1957, Biolegend), Synapsin (1:250, #5297S, Cell Signaling), Tuj1 (1:500, #T8578, Sigma-Aldrich)], MCP3 (CCL7, 1:100, #ab228979, Abcam) in PBS containing 0.2% Triton-X, 1% BSA overnight at 4°C (for most antibodies) or at room temperature (for ChAT and NeuN staining). The following day sections were washed in PBS and stained with the fluorescently conjugated α-bungarotoxin (for NMJ analysis) and/or appropriate secondary antibodies (Biotium) in PBS containing 0.2% Triton-X for 1 hour at room temperature. After PBS washes, slides were cover slipped with DAPI mounting medium (Southern Biotech) and imaged on a confocal microscope (Leica SP5).
In situ hybridization
Sections were soaked in PBS for 5 min and then baked at 60°C for 45 min. The sections were fixed to the slides with 4% PFA for 1 hour at room temperature. Alcohol dehydration was then performed in 50%, 70%, and 2x 100% ethanol for 5 min each. Slides were air-dried and incubated for 10 min with RNAScope® hydrogen peroxide. Tissue sections were then antigen retrieved at 95°C in RNAScope® Target Retrieval solution (Advanced Cell Diagnostics, ACD). Slides were baked a final time at 60°C for 45 min. In situ hybridization was subsequently performed with the RNAScope® Multiplex Fluorescent Reagent Kit v2 according to the manufacturer guidelines. Probes used for all experiments include: Gap43 (318621-C2), Ccl7 (446821), Ccl12 (437521-C3), Ccl2 (311791-C3), Ccl4 (421071), Sprr1a (426871-C2), Chat (408731-C2), Fgf21 (460931-C3), Cd68 (316611), and Tmem119 (472901-C3).
Image analysis
Spinal cord or muscle sections were imaged at 20x magnification with high resolution (2048x2048) and a z-step size of 1.5 μm (NMJ) or 2.0 μm (motor neuron) and maximum projection (LAS Software, Leica Biosystems) images were generated. NMJ analysis was performed on all in-plane bungarotoxin+ endplates from serial sections (5-6 sections per slide) and junctions were scored as fully, partially, or not innervated. Injured motor neuron analysis after nerve crush was performed on serial sections of longitudinal spinal cord spaced ∼60 μm apart. All sections containing Gap43+ motor neurons were imaged and every Gap43+ motor neuron was scored for the presence or absence of cytokine transcript expression (Figure 6C, Ccl7; Figure 6D, Ccl2 and Ccl4). Fgf21 expression in RiboWT and RiboALS motor neurons was performed on two longitudinal spinal cord sections spaced ∼80 μm apart. All Chat+ motor neurons were imaged and scored for absence, low, or high levels of Fgf21 transcript expression. For each given analysis, identical confocal and threshold settings were applied to samples and images, respectively. Imaging and scoring were performed by a blinded observer that was unaware of genotype or time point being analyzed.
cDNA library construction and RNA-seq
The purity and quality of isolated RNA was confirmed by Bioanalyzer or TapeStation (Agilent Genomics). Samples were then submitted to the University of Michigan Sequencing Core for cDNA library construction and sequencing. Briefly, the SMART-Seq v4 Ultra Low input RNA kit (Takara) was used to synthesize cDNA from 4ng of RNA using 11 cycles of cDNA amplification, according to manufacturer’s protocols. The cDNA was sheared by Covaris and quantitated by Qbit dsDNA HS Assay kit (ThermoFisher). Libraries were prepared from 5 ng cDNA using SMARTer ThruPLEX DNA-Seq kit (Takara) with 8 cycles of PCR according to manufacturer’s protocols. Final libraries were checked for quality and quantity by the Bioanalyzer or TapeStation (Agilent Genomics) and qPCR using Kapa’s library quantification kit for Illumina Sequencing platforms (#KK4835, Kapa Biosystems). Libraries were sequenced on an Illumina HiSeq 4000 sequencer with single-end, 51 cycle reads. Quality of data was assessed with FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Reads were mapped to the mouse genome (mm10) with STAR (Dobin et al., 2013) and bam files were generated with SAMtools (Li and Durbin, 2009). Statistical analysis was done using cufflinks (Trapnell et al., 2012). Differentially expressed genes were defined by FDR-corrected p-values less than 0.05. Data were visualized and extracted using cummeRbund (Trapnell et al., 2012). Heatmaps were generated using ComplexHeatmap (Gu et al., 2016), Venn diagrams were generated using InteractiVenn (interactivenn.net) and gene ontology was analyzed using ClueGO (Bindea et al., 2009, 2013).
Quantification and statistical analysis
All statistical analyses were performed using Prism 8 software (GraphPad). One- or two-way ANOVA was used for all analyses. Significant differences among pairwise comparisons were identified by Tukey’s post hoc tests. All graphs and error bars represent the mean ± standard error (se). The details on sample sizes (n) are included in the Figure Legends, and extent of significance are ∗p ≤ 0.05, ∗∗ p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗ p ≤ 0.0001, which is also included in the Figure Legends.
Acknowledgments
Financial support was provided to J.L.S. by the National Institute of Dental and Craniofacial Research grant T32 DE007057 and to E.A.F. by a University of Michigan Rackham Graduate School Regents Fellowship and a National Research Service Award from the National Institute of Neurological Disorders and Stroke (NINDS, NS103378). These experiments were supported by an NINDS grant R01 NS089585 (B.A.P.)
Author contributions
J.L.S. designed the study and conducted most of the experiments with the help of W.M.S., A.M.M. and R.E.I. E.A.F., J.L.S., and A.A. carried out bioinformatic analyses of RNAseq data. B.A.P. supervised the project and experiments. J.L.S., W.M.S. and B.A.P. analyzed the data and wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: July 23, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102700.
Supplemental information
This table contains all of the differentially expressed genes identified in the RNAseq analysis, displayed in separate pages as the experimental condition (injury or ALS model) in comparison to control samples.
References
- Arnold E.S., Ling S.C., Huelga S.C., Lagier-Tourenne C., Polymenidou M., Ditsworth D., Kordasiewicz H.B., McAlonis-Downes M., Platoshyn O., Parone P.A. ALS-linked TDP-43 mutations produce aberrant RNA splicing and adult-onset motor neuron disease without aggregation or loss of nuclear TDP-43. Proc. Natl. Acad. Sci. U S A. 2013;110:E736–E745. doi: 10.1073/pnas.1222809110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindea G., Galon J., Mlecnik B. CluePedia Cytoscape plugin: pathway insights using integrated experimental and in silico data. Bioinformatics. 2013;29:661–663. doi: 10.1093/bioinformatics/btt019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindea G., Mlecnik B., Hackl H., Charoentong P., Tosolini M., Kirilovsky A., Fridman W.H., Pages F., Trajanoski Z., Galon J. ClueGO: a cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokhuis A.M., Groen E.J., Koppers M., van den Berg L.H., Pasterkamp R.J. Protein aggregation in amyotrophic lateral sclerosis. Acta Neuropathol. 2013;125:777–794. doi: 10.1007/s00401-013-1125-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boillee S., Yamanaka K., Lobsiger C.S., Copeland N.G., Jenkins N.A., Kassiotis G., Kollias G., Cleveland D.W. Onset and progression in inherited ALS determined by motor neurons and microglia. Science. 2006;312:1389–1392. doi: 10.1126/science.1123511. [DOI] [PubMed] [Google Scholar]
- BonDurant L.D., Potthoff M.J. Fibroblast growth factor 21: a versatile regulator of metabolic homeostasis. Annu. Rev. Nutr. 2018;38:173–196. doi: 10.1146/annurev-nutr-071816-064800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bookout A.L., de Groot M.H.M., Owen B.M., Lee S., Gautron L., Lawrence H.L., Ding X., Elmquist J.K., Takahashi J.S., Mangelsdorf D.J. FGF21 regulates metabolism and circadian behavior by acting on the nervous system. Nat. Med. 2013;19:1147–1153. doi: 10.1038/nm.3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulis N., O’Connor D., Donsante A., editors. Molecular and Cellular Therapies for Motor Neuron Diseases. Academic Press; 2017. pp. 61–99. [Google Scholar]
- Bowen D.C., Park J.S., Bodine S., Stark J.L., Valenzuela D.M., Stitt T.N., Yancopoulos G.D., Lindsay R.M., Glass D.J., Distefano P.S. Localization and regulation of MuSK at the neuromuscular junction. Dev. Biol. 1998;199:309–319. doi: 10.1006/dbio.1998.8936. [DOI] [PubMed] [Google Scholar]
- Brkic M., Balusu S., Libert C., Vandenbroucke R.E. Friends or Foes: matrix metalloproteinases and their multifaceted roles in neurodegenerative diseases. Med. Inflam. 2015;2015:620581. doi: 10.1155/2015/620581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Ning B., Shi T. Single-cell RNA-seq technologies and related computational data analysis. Front. Genet. 2019;10:317. doi: 10.3389/fgene.2019.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Chen S.-T., Sun Y., Xu Z., Wang Y., Yao S.-Y., Yao W.-B., Gao X.-D. Fibroblast growth factor 21 ameliorates neurodegeneration in rat and cellular models of Alzheimer’s disease. Redox Biol. 2019;22:101133. doi: 10.1016/j.redox.2019.101133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu I.M., Morimoto E.T.A., Goodarzi H., Liao J.T., O’Keeffe S., Phatnani H.P., Muratet M., Carroll M.C., Levy S., Tavazoie S. A neurodegeneration-specific gene expression signature and immune profile of acutely isolated microglia from an ALS mouse model. Cell Rep. 2013;4:385–401. doi: 10.1016/j.celrep.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong M.S., Fitzgerald M., Winter J., Hu-Tsai M., Emson P.C., Wiese U., Woolf C.J. GAP-43 mRNA in rat spinal cord and dorsal root ganglia neurons: developmental changes and re-expression following peripheral nerve injury. Eur. J. Neurosci. 1992;4:883–895. doi: 10.1111/j.1460-9568.1992.tb00115.x. [DOI] [PubMed] [Google Scholar]
- Curtis R., Stewart H.J., Hall S.M., Wilkin G.P., Mirsky R., Jessen K.R. GAP-43 is expressed by nonmyelin-forming Schwann cells of the peripheral nervous system. J. Cell Biol. 1992;116:1455–1464. doi: 10.1083/jcb.116.6.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gendt K., Verhoeven G., Amieux P.S., Wilkinson M.F. Genome-wide identification of AR-regulated genes translated in Sertoli cells in vivo using the RiboTag approach. Mol. Endocrinol. 2014;28:575–591. doi: 10.1210/me.2013-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNAseq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Khoury J., Toft M., Hickman S.E., Means T.K., Terada K., Geula C., Luster A.D. Ccr2 deficiency impairs microglial accumulation and accelerates progression of Alzheimer-like disease. Nat. Med. 2007;13:432–438. doi: 10.1038/nm1555. [DOI] [PubMed] [Google Scholar]
- Ferraiuolo L., Kirby J., Grierson A.J., Sendtner M., Shaw P.J. Molecular pathways of motor neuron injury in amyothrophic lateral sclerosis. Nat. Rev. Neurol. 2011;7:616–630. doi: 10.1038/nrneurol.2011.152. [DOI] [PubMed] [Google Scholar]
- Fischer L.R., Culver D.G., Tennant P., Davis A.A., Wang M., Castellano-Sanchez A., Khan J., Polak M.A., Glass J.D. Amyotrophic lateral sclerosis is a distal axonopathy: evidence in mice and man. Exp. Neurol. 2004;185:232–240. doi: 10.1016/j.expneurol.2003.10.004. [DOI] [PubMed] [Google Scholar]
- Floyd T.L., Dai Y., Ladle D.R. Characterization of calbindin D28k expressing interneurons in the ventral horn of the mouse spinal cord. Dev. Dyn. 2018;247:185–193. doi: 10.1002/dvdy.24601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friese A., Kaltschmidt J.A., Ladle D.R., Sigrist M., Jessell T.M., Arber S. Gamma and alpha motor neurons distinguished by expression of transcription factor Err3. Proc. Natl. Acad. Sci. U S A. 2009;106:13588–13593. doi: 10.1073/pnas.0906809106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitcho M.A., Baloh R.H., Chakraverty S., Mayo K., Norton J.B., Levitch D., Hatanpaa K.J., White C.L., 3rd, Bigio E.H., Caselli R. TDP-43 A315T mutation in familial motor neuron disease. Ann. Neurol. 2008;63:535–538. doi: 10.1002/ana.21344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinsell D., Keating C.P. Peripheral nerve reconstruction after injury: a review of clinical and experimental therapies. Biomed. Res. Int. 2014;2014:698256. doi: 10.1155/2014/698256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gschwandtner M., Derler R., Midwood K.S. More than just attractive: how CCL2 influences myeloid cell behavior beyond chemotaxis. Front. Immunol. 2019;10:2759. doi: 10.3389/fimmu.2019.02759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z., Eils R., Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- Gupta S.K., Poduslo J.F., Mezei C. Temporal changes in PO and MBP gene expression after crush-injury of the adult peripheral nerve. Brain Res. 1988;464:133–141. doi: 10.1016/0169-328x(88)90005-8. [DOI] [PubMed] [Google Scholar]
- Gurney M.E., Pu H., Chiu A.Y., Dal Canto M.C., Polchow C.Y., Alexander D.D., Caliendo J., Hentati A., Kwon Y.W., Deng H.X. Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science. 1994;264:1772–1775. doi: 10.1126/science.8209258. [DOI] [PubMed] [Google Scholar]
- He Z., Jin Y. Intrinsic control of axon regeneration. Neuron. 2016;90:437–450. doi: 10.1016/j.neuron.2016.04.022. [DOI] [PubMed] [Google Scholar]
- Hulliger M. The mammalian muscle spindle and its central control. Rev. Physiol. Biochem. Pharmacol. 1984;101:1–110. doi: 10.1007/BFb0027694. [DOI] [PubMed] [Google Scholar]
- Ilieva H., Polymenidou M., Cleveland D.W. Non-cell autonomous toxicity in neurodegenerative disorders: ALS and beyond. J. Cell Biol. 2009;187:761–772. doi: 10.1083/jcb.200908164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki T., Dutchak P., Zhao G., Ding X., Gautron L., Parameswara V., Li Y., Goetz R., Mohammadi M., Esser V. Endocrine regulation of the fasting response by PPARalpha-mediated induction of fibroblast growth factor 21. Cell Metab. 2007;5:415–425. doi: 10.1016/j.cmet.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Itoh N., Itoh Y., Tassoni A., Ren E., Kaito M., Ohno A., Ao Y., Farkhondeh V., Johnsonbaugh H., Burda J. Cell-specific and region-specific transcriptomics in the multiple sclerosis model: focus on astrocytes. Proc. Natl. Acad. Sci. U S A. 2018;115:E302–E309. doi: 10.1073/pnas.1716032115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanning K.C., Kaplan A., Henderson C.E. Motor neuron diversity in development and disease. Annu. Rev. Neurosci. 2010;33:409–440. doi: 10.1146/annurev.neuro.051508.135722. [DOI] [PubMed] [Google Scholar]
- Kennedy H.S., Jones C.r., Caplazi P. Comparison of standard laminectomy with an optimized ejection method for the removal of spinal cords from rats and mice. J. Histotechnol. 2013;36:86–91. doi: 10.1179/014788813X13756994210382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski T.J., Jr., Bosco D.A., Leclerc A.L., Tamrazian E., Vanderburg C.R., Russ C., Davis A., Gilchrist J., Kasarskis E.J., Munsat T. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- Lalancette-Hebert M., Sharma A., Lyashchenko A.K., Shneider N.A. Gamma motor neurons survive and exacerbate alpha motor neuron degeneration in ALS. Proc. Natl. Acad. Sci. U S A. 2016;113:E8316–E8325. doi: 10.1073/pnas.1605210113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappe-Siefke C., Goebbels S., Gravel M., Nicksch E., Lee J., Braun P.E., Griffiths I.R., Nave K.A. Disruption of Cnp1 uncouples ologodendroglial functions in axonal support and myelination. Nat. Genet. 2003;33:366–374. doi: 10.1038/ng1095. [DOI] [PubMed] [Google Scholar]
- Leng Y., Wang J., Wang Z., Liao H.M., Wei M., Leeds P., Chuang D.M. Valproic acid and other HDAC inhibitors upregulate FGF21 gene expression and promote process elongation in glia by inhibiting HDAC2 and 3. Int. J. Neuropsychopharmacol. 2016;19:035. doi: 10.1093/ijnp/pyw035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindborg J.A., Niemi J.P., Howarth M.A., Liu K.W., Moore C.Z., Mahajan D., Zigmond R.E. Molecular and cellular identification of the immune response in peripheral ganglia following nerve injury. J. Neuroinflammat. 2018;15:192. doi: 10.1186/s12974-018-1222-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logsiger C.S., Boillee S., Cleveland D.W. Toxicity from different SOD1 mutants dysregulates the complement system and the neuronal regenerative response in ALS motor neurons. Proc. Natl. Acad. Sci. US A. 2007;104:7319–7326. doi: 10.1073/pnas.0702230104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T.C., Willis D.E. What makes a RAG regeneration associated? Front. Mol. Neurosci. 2015;8:43. doi: 10.3389/fnmol.2015.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makela J., Tselykh T.V., Maiorana F., Eriksson O., Do H.T., Mudo G., Korhonen L.T., Belluardo N., Lindholm D. Fibroblast growth factor-21 enhances mitochondrial functions and increases the activity of PGC-1alpha in human dopaminergic neurons via Sirtuin-1. SpringerPlus. 2014;3:2. doi: 10.1186/2193-1801-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Garza U., Torres-Oteros D., yarritu-Gallego A., Marrero P.F., Haro D., Relat J. Fibroblast growth factor 21 and the adaptive response to nutritional challenges. Int. J. Mol. Sci. 2019;20:1–21. doi: 10.3390/ijms20194692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkulyeva N., Veshchitskii A., Makarov F., Gerasimenko Y., Musienko P. Distribution of 28 kDa calbindin-immunopositive neurons in the cat spinal cord. Front. Neuroanat. 2016;9:166. doi: 10.3389/fnana.2015.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardo G., Iennaco R., Fusi N., Heath P.R., Marino M., Trolese M.C., Ferraiuolo L., Lawrence N., Shaw P.J., Bendotti C. Transcriptomic indicies of fast and slow disease progression in two mouse models of amyotrophic lateral sclerosis. Brain. 2013;136:3305–3332. doi: 10.1093/brain/awt250. [DOI] [PubMed] [Google Scholar]
- Neumann M., Sampathu D.M., Kwong L.K., Truax A.C., Micsenyi M.C., Chou T.T., Bruce J., Schuck T., Grossman M., Clark C.M. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- Niemi J.P., DeFrancesco-Lisowitz A., Roldan-Hernandez L., Lindborg J.A., Mandell D., Zigmond R.E. A critical role for macrophages near axotomized neuronal cell bodies in stimulating nerve regeneration. J. Neurosci. 2013;33:16236–16248. doi: 10.1523/JNEUROSCI.3319-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oost L.J., Kustermann M., Armani A., Blaauw B., Romanello V. Fibroblast growth factor 21 controls mitophagy and muscle mass. J. Cach Sarco Mus. 2019;10:630–642. doi: 10.1002/jcsm.12409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philips T., Robberecht W. Neuroinflammation in amyotrophic lateral sclerosis: role of glial activation in motor neuron disease. Lancet Neurol. 2011;10:253–263. doi: 10.1016/S1474-4422(11)70015-1. [DOI] [PubMed] [Google Scholar]
- Plunet W., Kwon B.K., Tetzlaff W. Promoting axonal regeneration in the central nervous system by enhancing the cell body response to axotomy. J. Neurosci. Res. 2002;68:1–6. doi: 10.1002/jnr.10176. [DOI] [PubMed] [Google Scholar]
- Restelli L.M., Oettinghaus B., Halliday M., Agca C., Licci M., Sironi L., Savoia C., Hench J., Tolnay M., Neutzner A. Neuronal mitochondrial dysfunction activates the integrated stress response to induce fibroblast growth factor 21. Cell Rep. 2018;24:1407–1414. doi: 10.1016/j.celrep.2018.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen D.R., Siddique T., Patterson D., Figlewicz D.A., Sapp P., Hentati A., Donaldson D., Goto J., O’Regan J.P., Deng H.-X. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- Rossi J., Balthasar N., Olson D., Scott M., Berglund E., Lee C.E., Choi M.J., Lauzon D., Lowell B.B., Elmquist J.K. Melanocortin-4 receptors expressed by cholinergic neurons regulate energy balance and glucose homeostasis. Cell Metab. 2011;13:195–204. doi: 10.1016/j.cmet.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotterman T.M., Akhter E.T., Lane A.R., MacPherson K.P., Garcia V.V., Tansey M.G., Alvarez F.J. Spinal motor circuit synaptic plasticity after peripheral nerve injury depends on microglia activation and a CCR2 mechanism. J. Neurosci. 2019;39:3412–3433. doi: 10.1523/JNEUROSCI.2945-17.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajjan S., Holsinger R.M., Fok S., Ebrahimkhani S., Rollo J.L., Banati R.B., Graeber M.B. Up-regulation of matrix metallopeptidase 12 in motor neurons undergoing synaptic stripping. Neuroscience. 2014;274:331–340. doi: 10.1016/j.neuroscience.2014.05.052. [DOI] [PubMed] [Google Scholar]
- Sala C., Andreose J.S., Fumagalli G., Lomo T. Calcitonin gene-related peptide: possible role in formation and maintenance of neuromuscular junctions. J. Neurosci. 1995:520–528. doi: 10.1523/JNEUROSCI.15-01-00520.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz E., Yang L., Su T., Morris D.R., McKnight G.S., Amieux P.S. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc. Natl. Acad. Sci. U S A. 2009;106:13939–13944. doi: 10.1073/pnas.0907143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber R.C., Krivacic K., Kirby B., Vaccariello S.A., Wei T., Ransohoff R.M., Zigmond R.E. Monocyte chemoattractant protein (MCP)-1 is rapidly expressed by sympathetic ganglion neurons following axonal injury. Neuroreport. 2001;12:601–606. doi: 10.1097/00001756-200103050-00034. [DOI] [PubMed] [Google Scholar]
- Schwanhausser B., Busse D., Li N., Dittmar G., Schuchardt J., Wolf J., Chen W., Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Shigeoka T., Jung H., Jung J., Turner-Bridger B., Ohk J., Qiaojin Lin J., Amieux P.S., Holt C.E. Dynamic axonal translation in developing and mature visual circuits. Cell. 2016;166:1–12. doi: 10.1016/j.cell.2016.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene J.H., Willard M. Axonally transported proteins associated with axon growth in rabbit central and peripheral nervous systems. J. Cell Biol. 1981;89:96–103. doi: 10.1083/jcb.89.1.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreedharan J., Blair I.P., Tripathi V.B., Hu X., Vance C., Rogelj B., Ackerley S., Durnall J.C., Williams K.L., Buratti E. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan R., Lu T.Y., Chai H., Xu J., Huang B.S., Golshani P., Coppola G., Khakh B.S. New transgenic mouse lines for selectively targeting astrocytes and studying calcium signals in astrocyte processes in situ and in vivo. Neuron. 2016;92:1181–1195. doi: 10.1016/j.neuron.2016.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stifani N. Motor neurons and the generation of spinal motor neuron diversity. Front. Cell Neurosci. 2014;8:293. doi: 10.3389/fncel.2014.00293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C.N., Araoz C., Lucas G., Morgan P.N., White H.J. Amyotrophic lateral sclerosis. Inclusion bodies in a case of the classic sporadic form. Ann. Clin. Lab Sci. 1975;5:38–44. [PubMed] [Google Scholar]
- Sun S., Ling S.-C., Ferraiuolo L., McAlonis-Downes M., Zou Y., Drenner K., Wang Y., Ditsworth D., Tokunaga S., Kopelevich A. Translational profiling identifies a cascade of damage initiated in motor neurons and spreading to glia in mutant SOD1-mediated ALS. Proc. Natl. Acad. Sci. U S A. 2015;112:E6993–E7002. doi: 10.1073/pnas.1520639112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Nakamura H., Okada E. Hereditary amyotrophic lateral sclerosis. Histochemical and electron microscopic study of hyaline inclusions in motor neurons. Arch. Neurol. 1972;27:292–299. doi: 10.1001/archneur.1972.00490160020003. [DOI] [PubMed] [Google Scholar]
- Ticozzi N., Tiloca C., Morelli C., Colombrita C., Poletti B., Doretti A., Maderna L., Messina S., Ratti A., Silani V. Genetics of familial amyotrophic lateral sclerosis. Arch. Ital. Biol. 2011;149:65–82. doi: 10.4449/aib.v149i1.1262. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu P.H., Raju P., Robinson K.A., Gurney M.E., Trojanowski J.Q., Lee V.M. Transgenic mice carrying a human mutant superoxide dismutase transgene develop neuronal cytoskeletal pathology resembling human amyotrophic lateral sclerosis lesions. Proc. Natl. Acad. Sci. U S A. 1996;93:3155–3160. doi: 10.1073/pnas.93.7.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner B.J., Talbot K. Transgenics, toxicity and therapeutics in rodent models of mutant SOD1-mediated familial ALS. Prog. Neurobiol. 2008;85:94–134. doi: 10.1016/j.pneurobio.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Valenzuela D.M., Stitt T.N., DiStefano P.S., Rojas E., Mattsson K., Compton D.L., Nunez L., Park J.S., Stark J.L., Gies D.R. Receptor tyrosine kinase specific for the skeletal muscle lineage: expression in embryonic muscle, at the neuromuscular junction, and after injury. Neuron. 1995;15:573–584. doi: 10.1016/0896-6273(95)90146-9. [DOI] [PubMed] [Google Scholar]
- Vance C., Rogelj B., Hortobagyi T., De Vos K.J., Nishimura A.L., Sreedharan J., Hu X., Smith B., Ruddy D., Wright P. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlug A.S., Teuling E., Haasdijk E.D., French P., Hoogenraad C.C., Jaarsma D. ATF3 expression precedes death of spinal motoneurons in amyotrophic lateral sclerosis-SOD1 transgenic mice and correlates with c-Jun phosphorylation, CHOP expression, somato-dendritic ubiquitination and Golgi fragmentation. Eur. J. Neurosci. 2005;22:1881–1894. doi: 10.1111/j.1460-9568.2005.04389.x. [DOI] [PubMed] [Google Scholar]
- Vogel C., Marcotte E.M. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Sharma K., Grisotti G., Roos R.P. The effect of mutant SOD1 dismutase activity on non-cell autonomous degeneration in familial amyotrophic lateral sclerosis. Neurobiol. Dis. 2009;35:234–240. doi: 10.1016/j.nbd.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhang S., Liu T., Wang H., Liu K., Wang Q., Zeng W. Sarm1/Myd88-5 regulates neuronal intrinsic immune response to traumatic axonal injuries. Cell Rep. 2018;23:716–724. doi: 10.1016/j.celrep.2018.03.071. [DOI] [PubMed] [Google Scholar]
- Warita H., Itoyama Y., Abe K. Selective impairment of fast anterograde axonal transport in the peripheral nerves of asymptomatic transgenic mice with a G93A mutant SOD1 gene. Brain Res. 1999;819:120–131. doi: 10.1016/s0006-8993(98)01351-1. [DOI] [PubMed] [Google Scholar]
- Wegorzewska I., Bell S., Cairns N.J., Miller T.M., Baloh R.H. TDP-43 mutant transgenic mice develop features of ALS and frontotemporal lobar degeneration. Proc. Natl. Acad. Sci. U S A. 2009;106:18809–18814. doi: 10.1073/pnas.0908767106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson T.L., Cleveland D.W. Slowing of axonal transport is a very early event in the toxicity of ALS-linked SOD1 mutants to motor neurons. Nat. Neurosci. 1999;2:50–56. doi: 10.1038/4553. [DOI] [PubMed] [Google Scholar]
- Wosiski-Kuhn M., Lyon M.S., Caress J., Milligan C. Inflammation, immunity, and amyotrophic lateral sclerosis: II. immune-modulating therapies. Muscle Nerve. 2019;59:23–33. doi: 10.1002/mus.26288. [DOI] [PubMed] [Google Scholar]
- Yamanaka K., Chun S.J., Boillee S., Fujimori-Tonou N., Yamashita H., Gutmann D.H., Takahashi R., Misawa H., Cleveland D.W. Astrocytes as determinants of disease progression in inherited amyotrophic lateral sclerosis. Nat. Neurosci. 2008;11:251–253. doi: 10.1038/nn2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagoraiou L., Akay T., Martin J.F., Brownstone R.M., Jessell T.M., Miles G.B. A cluster of cholinergic premotor interneurons modulates mouse locomotor activity. Neuron. 2009;64:645–662. doi: 10.1016/j.neuron.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarei M., Pizarro-Delgado J., Barroso E., Palomer X., Vazquez-Carrera M. Targeting FGF21 for the treatment of nonalcoholic steatohepatitis. Trends Pharmacol. Sci. 2020;41:199–208. doi: 10.1016/j.tips.2019.12.005. [DOI] [PubMed] [Google Scholar]
- Zarei S., Carr K., Reiley L., Diaz K., Guerra O., Altamirano P.F., Pagani W., Lodin D., Orozco G., Chinea A. A comprehensive review of amyotrophic lateral sclerosis. Surg. Neurol. Int. 2015;6:171. doi: 10.4103/2152-7806.169561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Donnelly C.J., Haeusler A.R., Grima J.C., Machamer J.B., Steinwald P., Daley E.L., Miller S.J., Cunningham K.M., Vidensky S. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature. 2015;525:56–61. doi: 10.1038/nature14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Duan L.J., Sun Q.L., Gao Y.S., Yang Y.D., Tang X.S., Zhao D.Y., Xiong Y., Hu Z.G., Li C.H. Identification of key pathways and genes in L4 dorsal root ganglia after sciatic nerve injury via microarray analysis. J. Invest. Surg. 2020;33:172–180. doi: 10.1080/08941939.2018.1452996. [DOI] [PubMed] [Google Scholar]
- Zhao J., Qin B., Nikolay R., Spahn C.M.T., Zhang G. Translatomics: the global view of translation. Int. J. Mol. Sci. 2019;20:212. doi: 10.3390/ijms20010212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Lyapichev K., Lee D.H., Motti D., Ferraro N.M., Zhang Y., Yahn S., Soderblom C., Zha J., Bethea J.R. Macrophage transcriptional profile identifies lipid catabolic pathways that can Be therapeutically targeted after spinal cord injury. J. Neurosci. 2017;37:2362–2376. doi: 10.1523/JNEUROSCI.2751-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond R.E. gp130 cytokines are positive signals triggering changes in gene expression and axon outgrowth in peripheral neurons following injury. Front. Mol. Neurosci. 2012;4:62. doi: 10.3389/fnmol.2011.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond R.E., Echevarria F.D. Macrophage biology in the peripheral neurvous system after injury. Prog. Neurobiol. 2019;173:102–121. doi: 10.1016/j.pneurobio.2018.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This table contains all of the differentially expressed genes identified in the RNAseq analysis, displayed in separate pages as the experimental condition (injury or ALS model) in comparison to control samples.
Data Availability Statement
The sequencing data generated in this study has been deposited into the Gene Expression Omnibus (GEO) of NCBI and is available as accession number GSE162028. Bioinformatics comparisons and lists of differentially expressed genes are included in the Table S1, contained in the Supplemental information.