Highlights
-
•
The higher OPN mRNA and protein expression in patients with primary bone cancer.
-
•
The serum OPN differentiate the patients from the controls with 100% sensitivity.
-
•
The OPN local and circulating level was correlated with bone tumor severity.
Abbreviations: OPN, Osteopontin; SOX9, SRY-Box Transcription Factor 9; cDNA, Complementary DNA; PBMC, Peripheral blood mononuclear cells; ANOVA, One-way analysis of variance; ELISA, Enzyme-linked immunosorbent assay; qRT-PCR, Quantitative Real-time transcription-polymerase chain reaction; ROC, receiver operating characteristic; AUC, area under the curve; CI, confidence interval; MMP9, Matrix metallopeptidase 9; S100A8, S100 calcium-binding protein A8; PBS, phosphate-buffered saline; HRP, horseradish peroxidase; OCT, Optimal Cutting Temperature; DAPI, 4′,6-Diamidine-2′-phenylindole dihydrochloride; EMT, epithelial-mesenchymal transition; HIF-1α, hypoxia-inducible factor-1 alpha
Keywords: Osteopontin, Bone tumors, Osteosarcoma, Ewing Sarcoma, Chondrosarcoma
Abstract
Purpose
The development of novel and efficient biomarkers for primary bone cancers is of grave importance.
Methods
The expression pattern of osteopontin (OPN) was investigated in the 153 patients with benign (n = 72) and malignant (n = 81) primary bone cancers. Both local and circulating OPN mRNA expression levels and their protein concentration in serum and tumor site were assessed using real-time qRT-PCR, ELISA, and immunohistochemistry techniques, respectively. As a control, 29 healthy individuals were considered. The number of 153 tumor tissue specimens and the 153 paired margins were taken on surgical resection from the patients. 153 blood samples were also drained from all participants, then peripheral blood mononuclear cells (PBMC) and sera were separated.
Results
The mean mRNA expression was significantly higher in all of the cancerous tissues than the paired margins and the PBMC of the patients than the controls. Consistently, the protein concentrations of OPN in serum and tumor tissues were significantly higher in the patients. Furthermore, the malignant cases had significantly elevated the mRNA levels and the protein compared to the benign cases. OPN could potentially differentiate the patients from the controls with 100% sensitivity and specificity in serum. Moreover, OPN could predict some of the malignant cases' clinicopathological features, including metastasis, recurrence, grade, and response to chemotherapy.
Conclusions
In conclusion, OPN might be involved in the pathogenesis of primary bone tumors and can be considered as a potential biomarker to bone cancer diagnosis.
1. Introduction
Primary bone and joint malignancies are the third leading cause of death in cancer patients under 20 years of age [1]. These tumors were responsible for 3,500 new cancer cases and 1,660 deaths in the United States in 2019 [2]. Primary skeletal tumors are mainly diagnosed through imaging and biopsy; therefore, less invasive diagnostic techniques are urgently demanded. Moreover, the handling of bone tumors is considered a big challenge for oncologists and surgeons due to their diverse histological natures and different clinical manifestations. Additionally, because of the limited ability to remove or manage the tumors, the mortality rate is relatively high in these patients. Although tremendous progress has been made within the past decade, there are currently no specific markers that can reliably be employed to diagnose bone tumors. Such biomarkers would significantly decrease mortality and increase limb salvage strategies. Furthermore, early detection of either recurrent or metastatic disease can contribute to timely decisions and action to treat the tumor, improving patient prognosis [3], [4].
Osteopontin (OPN) is a secreted non-collagenous, sialic acid-rich, chemokine-like, matricellular phosphoglycoprotein that facilitates cell-matrix interactions and promotes tumor progression [5]. It is expressed by many normal cells, particularly those originating from bone and immune systems, including fibroblasts, osteoblasts, osteocytes, hypertrophic chondrocytes, dendritic cells, macrophages, smooth muscle cells, and activated T cells [6]. OPN is a multifunctional cytokine that plays a significant role in cell proliferation, survival, drug resistance, invasion, and stem-like behavior[7]. Considering its vital role in regulating cell fate, its abnormal expression and/or splicing is involved in undesirable alterations in disease pathologies, specifically cancer[8]. In this regard, OPN has been identified in various types of human carcinomas, where its overexpression was associated with a colon[9], bladder [10], breast[11], gastric , and liver[12] cancer progression. . Furthermore, using the MICROMAX complementary DNA (cDNA) microarray system, Wong et al. (2001) identified 30 putative genes, including OPN, that are differentially overexpressed in ovarian cancer cell lines[13]. It has also been demonstrated that OPN increased expression induced tumor cell invasion and migration through induction of PI3K signaling and Matrix metallopeptidase 9 (MMP9) activity in colon cancer[9]. Moreover, the simultaneous over-expression of OPN and up-regulation of MMP9 and S100 calcium-binding protein A8 (S100A8) was associated with tumor grade and shorter survival time of patients with bladder cancer[10].
Despite its crucial role in the pathophysiology of primary bone tumors, the diagnostic and prognostic values of OPN have not yet been adequately addressed in the patients with various primary skeletal cancers, and the available studies are mainly limited to a single type of tumor, with osteosarcoma dominating the literature landscape [14]. More importantly, the efficacy of OPN in predicting outcomes of the disease remains unclear. Therefore, we aimed at these shortcomings and designed this comparative study to 1) determine the gene and protein expression pattern of OPN in different malignant and benign primary bone cancers 2) determine the the relevance ofOPN expression to the clinicopathological features of the malignant cases, including metastasis, recurrence, grade, and response to chemotherapy. To get valuable insight into the efficacy of OPN as a biomarker in primary bone cancer , its level was measured in three different sites, including the tissue specimens (tumor and paired margin), peripheral blood mononuclear cells (PBMC), and serum.
2. Materials & Methods
2.1. Patients and sample collection
A total of 153 patients suffering from malignant (n = 81) and benign (n = 72) primary bone cancers were enrolled in the study. The demographic data and history of the participants can be found in table 1. Moreover, 29 healthy age and sex-matched individuals were considered as the control group. . Following surgical resection, fresh pair of the tumor and marginal tissue were taken from all the 153 patients who were subjected to surgery at Shafa Orthopedic Hospital. A part of the tumor and margin tissue of each patient was transferred to the pathology department for further histological evaluations and immunohistochemistry evaluations and the rest were immediately transferred to the lab in a cool situation and kept at −80 for gene expression evaluation. Furthermore, to evaluate the serum level of OPN, the 6 ml of peripheral blood were drained from all the patients and controls and transferred to the laboratory immediately , and subjected to serum separation by centrifuge for 10 min at approximately 1000g [15]. The serum samples were stored at −80 °C until evaluation with appropriate indications and labeling. Moreover, the total amount of 6 ml peripheral blood was taken from the patients and healthy controls and applied for the peripheral blood mononuclear cell (PBMC) separation. The clinic- pathological features of the patients with malignant/benign bone tumors are summarized in table 1 and the biochemical profile of the patients is presented in table 2.
Table 1.
The demographic data of the patients with different bone cancers.
| Variable | Age | Gender | Tumor grade | Metastasis | Chemotherapy | Recurrence | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient | Group | ≤40 | ≥40 | Male | Female | Low | High | Positive | Negative | Positive | Negative | Positive | Negative |
| Osteosarcoma (n = 27) | 12 | 15 | 13 | 14 | 9 | 18 | 10 | 17 | 18 | 9 | 7 | 20 | |
| Chondrosarcoma(n = 27) | 0 | 27 | 16 | 11 | 15 | 12 | 8 | 19 | 0 | 27 | 0 | 0 | |
| Ewing sarcoma (n = 27) | 15 | 12 | 17 | 10 | 8 | 19 | 10 | 17 | 15 | 12 | 7 | 20 | |
| Exostosis (n = 24) | 13 | 11 | 15 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Giant Cell Tumor (n = 24) | 8 | 16 | 16 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Osteochondroma (n = 24) | 6 | 18 | 15 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
Table 2.
The biochemical profile of the enrolled patients (n = 153).
| Standard deviation ± Mean | No. of patients | Normal range | Parameters |
|---|---|---|---|
| 150.36 ± 3.24 17.58 ± 2.37 268.25 ± 3.31 |
124 7 22 |
Normal range: 24–170 Low: 24 ≤ High: 170≥ |
Creatine phosphokinase (U/L) |
| 196.72 ± 4.59 0 756.32 ± 21.26 504.82 ± 11.74 |
118 0 19 16 |
Normal: Male 80–306 Female 64–306 High: Male 306≥ Female 306≥ |
Alkaline phosphatase (U/L) |
| 301.94 ± 11.37 1257 ± 22.38 |
127 26 |
Normal: 480 < High: 480≥ |
Lactate dehydrogenase (U/L) |
| 9.65 ± 0.39 7.86 ± 0.53 11.63 ± 0.78 |
125 11 17 |
Normal: 8.6–10.3 Low: 8.6 ≤ High: 10.3> |
Calcium (Serum) (mg/dL) |
| 141.35 ± 1.99 133.62 ± 1.79 |
137 16 |
Normal: 136–145 Low: 145≤ |
Sodium (Serum) (mmol/L) |
| 4.41 ± 0.93 2.41 ± 0.18 5.06 ± 0.34 |
88 20 45 |
Normal: 2.7–4.5 Low: 2.7≤ High: 4.5≥ |
Inorganic Phosphorus (mg/dL) |
| 4.33 ± 0.35 1.62 ± 0.62 7.56 ± 0.92 |
140 7 6 |
Normal: 3.5–5.5 Low: 3.5≤ High: 5.5≥ |
Potassium (mmol/L) |
2.2. Separation of PBMC
Ficoll-Hypaque density gradient centrifugation method (Sigma Chemical Co, St Louis, MO, USA, Cat No. # 10771) was used for PBMC separation from the blood samples[16]. Based on this method, freshly drawn 6 ml of peripheral blood samples of each patient and control were collected and diluted 1:1 in phosphate-buffered saline (PBS). Then the diluted blood was gently added to the tube containing Ficoll (3 ml) and centrifuged at 2700 g for 40 min in a refrigerated centrifuge. After centrifugation, the layer containing peripheral blood mononuclear cells were removed and poured into a separate tube. The cells were washed with (PBS) and finally centrifuged at 3200 g for 5 min and counted by hemocytometer. The same number of cells were used for RNA extraction.
2.3. Assessment of biochemical profile and serum OPN level
Creatine phosphokinase (Cat No. # 1050016), alkaline phosphatase (Cat No. # 5018) , lactate dehydrogenase (Cat No. # 1400022), calcium (Cat No. # 1100008), inorganic phosphorus (Cat No. # 140020) were measured using the available standard assay kits (PARS AZMUN, Tehran, Iran) according to the manufacturer's recommendations. sodium and potassium. . Blood electrolytes including sodium and potassium were measured photometrically. Briefly, an ion Selective Electrode is placed in the sample and due to the passage of the desired ions through a membrane (due to the concentration gradient), a potential difference occurs on both sides of the membrane. The voltage of this current is proportional to the logarithm of the desired electrolyte concentration. Additionally, serum OPN level was measured using a Human Osteopontin Assay Kit (IBL, Japan) (Cat No. # 27158) with an analytical sensitivity of 5 ng/mL (Interassay CV: 7.8% & Intraassay CV: 4.7%). The basis of this kit is a sandwich method using a monoclonal antibody. Anti-OPN antibodies were coated directly on the solid surface of the plates and following adding a serum and the provided standards to the plate, the secondary antibodies conjugated to HRP (horseradish peroxidase) were applied. The HRP antigen–antibody complex produces color by acting on the substrate. Tetramethyl benzidine was used as a staining agent (chromogen). The color intensity was proportional to the amount of OPN in the serum. The optical absorption of standards and samples was read at a wavelength of 450 nm via a microplate reader.
2.4. RNA extraction, cDNA synthesis, and Real-Time PCR
Total RNA was extracted from the tissue samples (tumors and margins) and PBMC via Trizol (Invitrogen, Grand Island, USA Cat No. # 15596026) according to our previously described procedure for measuring the SOX9 gene expression [17]. Total RNA (1 µg of each sample) was used to synthesize cDNA with PrimeScript First Strand cDNA Synthesis Kit (Takara, Japan, Cat No. # 6110A) according to the manufacturer’s instructions. OPN expression levels were quantitated with SYBR Premix Ex Taq II (Takara, Japan, Cat No. # RR820A), which was performed in Applied Biosystems Step One Plus, Real-time system (Applied Biosystems, USA). The sequences of the designed primers are shown in table 3. The reactions were set up as follows: 95 °C for 5 min following 40 cycles at 95 °C for 5 s, 55 °C for 20 s, and 60 °C for 35 s. Beta-Actin was used as a housekeeping gene to normalize the expression levels of the OPN gene. The formula RQ = 2-ΔCT was used to analyze the relative gene expression levels after normalization with the endogenous controls and the OPN and beta-actin melt curves were considered as primer specificity.
Table 3.
The primer sets used in this study for qRT-PCR assay.
| Gene | Primers | Primer sequence | Tm |
|---|---|---|---|
| OPN | Forward Reverse |
5′– CTCGAACGACTCTGATGATGT-3′ 5′– TGTCAGGTCTGCGAAACTTCT −3′ |
58 |
| Beta-Actin | Forward Reverse |
5-GAT CTC CTT CTG CAT CCT GT-3′ 5′-TGG GCA TCC ACG AAA CTA C- 3′ |
57 |
2.5. Immunohistochemistry
The local protein expression level of OPN in tumor and marginal tissues were assessed using immunohistochemistry. Briefly, the frozen tissue blocks were prepared using Optimal Cutting Temperature (OCT) embedding medium and fixed in 4% paraformaldehyde, and incubated in 20% sucrose. The cryotome was used to prepare 10 nm tissue sections and the slides were washed with PBS and exposed to Triton 3% for 30 min. Triton is used to induce cell membrane permeable to antibodies. In the next step, 10% goat serum was added to the cells for half an hour to block the non-specific antigenic sites. The dilution of 1:100 of a monoclonal antibody of OPN (Santa Cruz Biotechnology, USA, Cat No. # SC-21742) was used for staining in a proper incubation time and the goat anti-mouse IgG-FITC secondary antibody (Santa Cruz Biotechnology, USA, Cat No. # SC-2010) was applied with a delusion of 1:200 in dark at 37 °C. Following adequate washing with PBS (3 times), the 4′,6-Diamidine-2′-phenylindole dihydrochloride (DAPI) (Sigma, Germany,
Cat No. # 28718–90-3) was used to stain the nuclei and then evaluated with a fluorescent microscope. The staining intensity of OPN was quantified using ImageJ and reported as the percentage of positive reactivity[18].
2.6. Statistical analysis
Statistical analysis was performed using commercially available software (GraphPad Prism version 8.0.2, GraphPad Software, San Diego, California). All data were examined for normal distribution using an examination of the residuals and a D'agostino-Pearson omnibus test. Considering the non-Gaussian distribution of the data, multiple comparisons were carried out using the nonparametric Kruskal-Wallis test by controlling the false discovery rate and calculating exact P-values (q-values). Side-to-side comparisons were conducted using the parametric unpaired t-test and nonparametric Mann–Whitney U test. The receiver operating characteristic (ROC) curves and calculation of area under the curve (AUC) were applied to calculate the diagnostic value of OPN gene expression in tumor and normal tissue and protein level in serum. Accordingly, the sensitivity and specificity at various cut- off points of OPN level (gene and protein) were calculated and the optimal cut-off value was determined based on the Youden index. In the Youden index, the maximum vertical distance from the point (x, y) on the diagonal line of the ROC curve is used [19], [20]. The selection of cutoff points for OPN at each site was undertaken using ROC-curve analysis. A cut-off value is a value that has the maximum sensitivity and specificity of the tested marker that discriminate between two groups (patient and healthy subjects) and is a separator scale. To better evaluate the diagnostic value of OPN, ROC analysis was performed twice, including the control group to evaluate the diagnostic accuracy of the parameter to differentiate the patients from the healthy individuals and excluding the control to assess the ability to discriminates between different stages of diseases (benign Vs. malignant). P-values < 0.05 were considered statistically significant.
3. Results
3.1. Evaluation of demographic characteristics and biochemical profiles of patients with various types of bone cancer
The clinic- pathological features of the patients with different types of malignant and benign bone tumors are summarized in table 1. As it is obvious, the equal number of most prevalent types of malignant bone tumors including osteosarcoma, Ewing's Sarcoma, and chondrosarcoma also benign bone tumors including osteochondroma, Giant cell Tumor, and exostosis were enrolled in the study. There was no significant difference in the age distribution of patients between the groups and in each type of bone cancer, there were patients aged 40 and under. Only in the chondrosarcoma group, all participating patients were over 40 years old and in the osteochondroma and Giant cell Tumor groups, more patients were over 40 years old. In this study, both males and females participated in each group of patients ,and notably, most of the patients with benign tumors were male. Patients' tumor grade was also evaluated as a measure of the severity of their disease and 18, 12 and 19 out of 27 patients had high-grade tumors in osteosarcoma, chondrosarcoma and Ewing's Sarcoma group, subsequently. Also, 17, 19 , and 17 out of 27 patients had a metastatic tumor in osteosarcoma, chondrosarcoma, and Ewing's Sarcoma group, subsequently. In addition, the standard combination of doxorubicin, cisplatin, and methotrexate received as a chemotherapy regimen by patients with osteosarcoma while the chemotherapy regimen of Ewing's Sarcoma patients was the combination of vincristine, cyclophosphamide, and doxorubicin and patients with osteochondroma received no chemotherapy regimen before the surgery. Accordingly, 18 and 15 patients out of 27 patients in osteosarcoma and Ewing's Sarcoma group received chemotherapy regimens. Local tumor recurrence is a critical complication in malignant bone tumors [21] and in our survey, 7 patients out of 27 patients in each group of osteosarcoma and Ewing's Sarcoma had recurrent tumors. The biochemical profile of the patients is presented in table 2. The creatine phosphokinase as an indicator of muscle damage and heart attack showed to be normal in 124 patients (150.36 ± 3.24) while 22 patients were in a high range (268.25 ± 3.31) of this marker. Alkaline phosphatase as an indicator of liver disease and bone disorders was also measured in patients and showed to be normal in 118 patients with bone tumors (196.72 ± 4.59). The level of Lactate dehydrogenase as an indicator of injuries and heart failure was in the normal range in 127 patients (301.94 ± 11.37), while in the high range in 26 patients (1257 ± 22.38). The level of electrolytes such as calcium, sodium, and potassium was normal in the majority of patients: 125 patients had normal calcium levels (9.65 ± 0.39), 137 patients had normal sodium levels (141.35 ± 1.99) and 140 patients had normal potassium level (4.33 ± 0.35). As indicated, most patients (77–91%) are well within the normal range for the measured biochemical parameters, except for inorganic phosphorus, in which only ≈ 57% of the patients had normal values.
3.2. The OPN expression level in different types of primary bone tumors
As shown in Table 4, the OPN mRNA expression levels were significantly higher in the cancerous tissues than the paired margins for all the six subtypes of patients (P < 0.0001). The OPN expression was also significantly higher in the three malignant tumors (mean = 3.60, 95% CI = 3.27–3.93) than the benign ones (mean = 1.10, 95% CI = 0.99–1.21). Moreover, the osteosarcoma patients showed significantly elevated levels of the mRNA compared to the chondrosarcoma patients. However, no significant differences were observed among the benign subtypes. Comparison of the margins with each other revealed that the mRNA expression is non-significantly lower in the benign patients than the malignant counterparts (P > 0.05). As indicated in table 5, both the malignant and benign patients had significantly higher OPN expression levels than healthy individuals. Furthermore, the mRNA expression was found to be significantly higher in the malignant patients (mean = 0.76, 95% CI = 0.68–0.84) than the benign ones (mean = 0.31, 95% CI = 0.27–0.35). Although the osteosarcoma patients tended to have higher mRNA levels, no significant differences were observed among the different malignant or benign subtypes. Table 5 also represents that parallel to OPN expression levels in PBMC; its protein concentrations were significantly higher in the patient cases than the controls. Consistently, its concentration was also assessed to be significantly higher in the malignant patients (mean = 178.12, 95% CI = 155.20–201.05) than the benign ones (mean = 68.02, 95% CI = 62.11–73.93). Interestingly, similar to OPN mRNA expression in tumors, inter-subtype differences were noticed among the malignant patients, as OPN concentrations were remarkably higher in sera of the osteosarcoma patients than the other patients (p < 0.05). Additionally, the exostosis patients had significantly lower levels of protein than the others. The cutoff point, sensitivity, specificity, and AUC are shown in table 6. The optimal cutoff point was set by the ROC method to > 1.875 for the OPN mRNA expression in tumor tissues to discriminate between the malignant and benign patients with 92% and 94% sensitivity and specificity, respectively. The mRNA expression in PBMC and the corresponding protein levels in serum were also able to differentiate between the benign and malignant patients, with acceptable sensitivity and specificity (Fig. 1). Of significant importance, the protein showed the highest possible accuracy to discriminate the patients from the healthy individuals.
Table 4.
The assessed OPN mRNA expression levels in the tumors and paired margins of patients with primary bone cancers. Mean (95% CI).
| Malignancy | Type | Marginal tissue | Cancerous tissue |
|---|---|---|---|
| Malignant | Osteosarcoma | 0.11 (0.04–0.17) | 4.40 (3.71–5.09)a |
| Chondrosarcoma | 0.22 (0.08–0.36) | 2.99 (2.56–3.42)b | |
| Ewing sarcoma | 0.28 (0.09–0.46) | 3.41 (2.92–3.91)ab | |
| Benign | Exostosis | 0.08 (0.01–0.15) | 0.93 (0.80–1.06)c |
| Giant Cell Tumor | 0.07 (0.03–0.11) | 1.34 (1.11 – 1.57)c | |
| Osteochondroma | 0.06 (0.02–0.11) | 1.09 (0.92–1.26)c |
Means within a column with different superscript letters (a-c) denote significant differences (p < 0.05).
Margins Vs. tumors are significantly different (P < 0.0001) within each row.
Table 5.
The assessed OPN mRNA expression levels in the PBMC and protein concentrations in the sera of the healthy individuals and patients with primary bone cancers. Mean (95% CI).
| Malignancy | Type | PBMC | Serum |
|---|---|---|---|
| Healthy | Control | 0.05 (0.03–0.06)a | 11.71 (10.06–13.35)a |
| Malignant | Osteosarcoma | 0.88 (0.71–1.02)b | 263.68 (218.92–308.41)b |
| Chondrosarcoma | 0.72 (0.58–0.86)b | 126.43 (106.83–145.92)c | |
| Ewing sarcoma | 0.69 (0.57–0.81)b | 144.41(113.14–175.83)c | |
| Benign | Exostosis | 0.29 (0.22–0.37)c | 55.99 (46.48–65.50)d |
| Giant Cell Tumor | 0.34 (0.25–0.44)c | 74.87 (61.92–87.82)e | |
| Osteochondroma | 0.30 (0.23–0.36)c | 73.19 (66.15 – 80.24)e |
Means within a column with different superscript letters (a-c) denote significant differences (p < 0.05).
Table 6.
The ROC curve analysis of OPN for determination of diagnostic sensitivity and specificity in the patients with primary bone cancers.
| Cutoff point | Sensitivity (%) | Specificity (%) | AUC | P-value | ||
|---|---|---|---|---|---|---|
| Bone | Benign Vs. malignant | > 1.875 | 92 | 94 | 0.98 | <0.0001 |
| PBMC | Control Vs. benign | > 0.09 | 95 | 97 | 0.98 | <0.0001 |
| Control Vs. malignant | > 0.10 | 100 | 97 | 0.99 | <0.0001 | |
| Benign Vs. malignant | > 0.43 | 86 | 75 | 0.88 | <0.0001 | |
| Serum | Control Vs. benign | > 26.54 | 100 | 100 | 1 | <0.0001 |
| Control Vs. malignant | > 31.08 | 100 | 100 | 1 | <0.0001 | |
| Benign Vs. malignant | > 90.74 | 81 | 85 | 0.90 | <0.0001 |
Fig. 1.
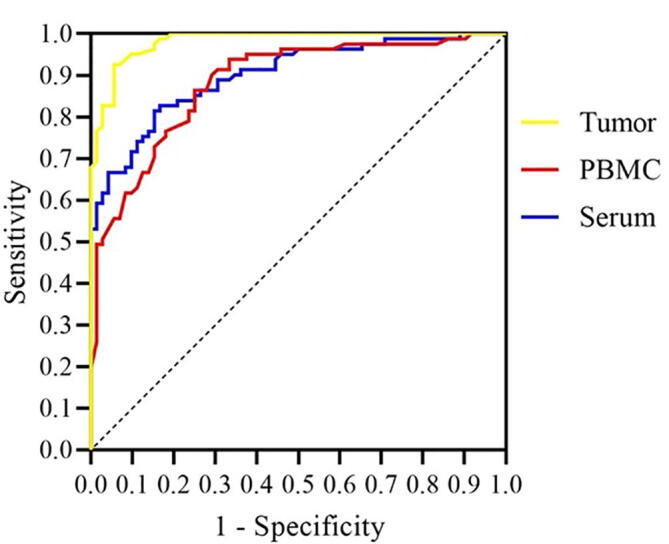
Comparison of the diagnostic potential of OPN to differentiate between malignant and benign patients.
3.3. The OPN differential expression in bone cancer patients with different demographic features
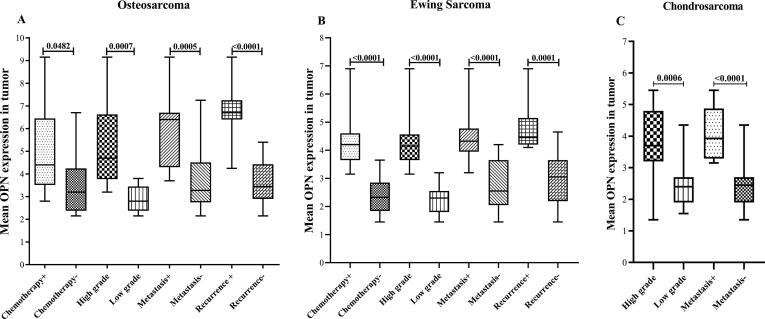
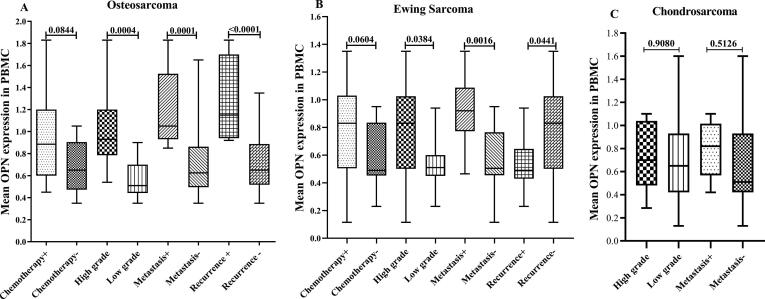
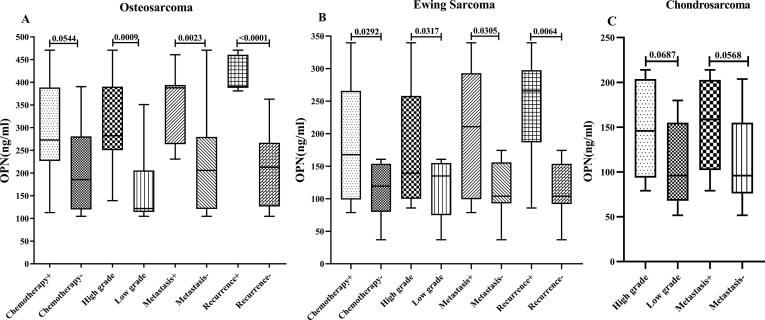
As shown in Fig. 2, the OPN mRNA expression levels in tumor tissue were significantly different for all of the assessed clinicopathological features in the malignant cases. To be specific, the mRNA expression in the patients with metastatic, high grade, and recurrent tumor were notably elevated. However, the expression in PBMC had no significant difference between the patients undergoing chemotherapy compared to the untreated cases (Fig. 3). Moreover, chondrosarcoma patients suffering from metastatic and high-grade tumors had statistically similar OPN mRNA levels to the patients with non-metastatic and low-grade tumors in their PBMC (Fig. 3). Consistently, the OPN protein concentrations in serum had no significant difference between the patients mentioned above (Fig. 4). Interestingly, OPN in PBMC and serum differentiated metastatic, high-grade, and recurrent osteosarcoma and Ewing sarcoma from the matched counterparts. Like OPN expression in PBMC, the serum protein concentration had no significant difference between the osteosarcoma patients undergoing chemotherapy and the untreated cases (P = 0.0544). However, the protein level was significantly lower in the treated Ewing sarcoma patients than the matched untreated patients (P = 0.0292).
Fig. 2.
The assessed OPN mRNA expression levels in tumor for the different clinicopathological variables in the patients with malignant bone cancers The OPN mRNA expression level was evaluated in different types of malignant bone tumors including osteosarcoma (a), Ewing sarcoma(b), and chondrosarcoma (c) for the different clinicopathological variables (chemotherapy status, tumor grade, tumor metastasis, and recurrent) in the patients. The Statistical differences between groups are shown by P values.
Fig. 3.
The assessed OPN mRNA expression levels in PBMC for the different clinicopathological variables in the patients with malignant bone cancers. The OPN mRNA expression level was evaluated in PBMC of patients in different types of malignant bone tumors including osteosarcoma (a), Ewing sarcoma(b) and chondrosarcoma (c) for the different clinicopathological variables (chemotherapy status, tumor grade, tumor metastasis and recurrent) in the patients. The Statistical differences between groups are shown by P values.
Fig. 4.
The assessed OPN protein expression levels in serum for the different clinicopathological variables in the patients with malignant bone cancers The OPN protein level in serum was evaluated in patients with different types of malignant bone tumors including osteosarcoma (a), Ewing sarcoma(b) and chondrosarcoma (c) for the different clinicopathological variables (chemotherapy status, tumor grade, tumor metastasis and recurrent) in the patients. The Statistical differences between groups are shown by P values.
3.4. The protein level of OPN in bone tumor tissues
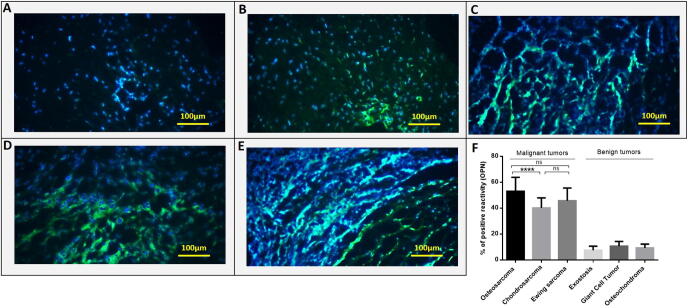
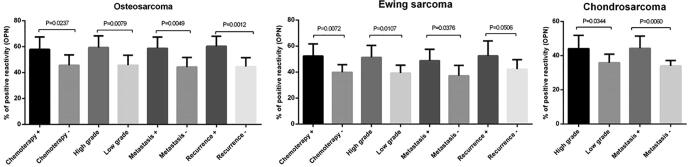
To determine whether the protein level of OPN obeys the same expression pattern as the OPN gene in tumor and marginal bone tissues, the OPN protein level was assessed using immunohistochemistry in different types of primary bone tumors. The representative images of OPN immunohistochemistry staining in primary bone tumor tissues are illustrated in Fig. 5. Based on our data, OPN protein expressed more in tumor tissue in each group compared to the matched marginal tissue (P < 0.0001). Also, in malignant bone tumors, the protein level of OPN over-expressed compared to the benign bone tumors and the difference was statistically significant (P < 0.0001). Osteosarcoma tumors expressed the highest level of OPN protein compared to Ewing's Sarcoma (P = 0.06) and chondrosarcoma (P = 0.0008) tumors (Fig. 5F). Our data showed that in tumor tissues of patients who received chemotherapy, the level of OPN protein was higher compared to the tumor tissues of patients without the history of chemotherapy in osteosarcoma (P = 0.0237) ,and Ewing's Sarcoma (P = 0.0072). OPN protein overexpressed in high-grade osteosarcoma (P = 0.0079), Ewing's Sarcoma (P = 0.0107) ,and chondrosarcoma (P = 0.0344) tumors compared to the low-grade tumors in each group. Also, the metastatic tumors expressed a higher level of OPN protein compared to the non-metastatic tumors in osteosarcoma (P = 0.0049), Ewing's Sarcoma (P = 0.0376), and chondrosarcoma (P = 0.0060) group. Our data showed up-regulation of OPN protein in recurrent osteosarcoma (P = 0.0012) and Ewing's Sarcoma (P = 0.0506) bone tumors compared to the non-recurrent tumors (Fig. 6).
Fig. 5.
The OPN protein level in primary bone tumors. The OPN differential protein expression was assessed in different bone tumor and margin tissues and the OPN expression level was indicated as the percentage of positive reactivity. Representative images showing the expression of OPN in normal bone tissue; with < 10% immunoreactivity, ×100 (a), tumor tissue; with 32% immunoreactivity, ×100.,(b), tumor tissue; with 44% immunoreactivity, ×100.(c), tumor tissue with 53.6% immunoreactivity, ×100.(d) and tumor tissue with 57.1% immunoreactivity, ×100.(e). The OPN protein expression pattern in malignant, benign and margin tissues is shown (f). The Statistical differences between groups are shown by P values.
Fig. 6.
The OPN protein expression in patients with different malignant bone tumors. The protein level of OPN in tumor tissues of patients with different types of malignant bone tumors was assessed. The over-expression of OPN was detected in chemotherapy positive, high- grade, metastatic and recurrent tumors in osteosarcoma (a), Ewing sarcoma(b) and chondrosarcoma (c). The Statistical differences between groups are shown by P values.
4. Discussion
Although promising progress has been made in diagnostic techniques, particularly medical imaging, the morbidity and mortality rates are still high in the patients with primary bone tumors due to lack of specific biomarkers, nonspecific symptoms, and late patient referring which lead to failure of timely detection and any therapeutic strategy [4]. There are currently several biomarkers for the diagnosis and prognosis of primary bone tumors, which can be divided into three main classes, including serologic, genetic, and histologic. The serological biomarkers are established on reflecting the osteoblastic and osteoclastic activity of the bone. These markers come with major limitations, including nonspecificity (false positive and false negative results), time-consuming procedure, and unaffordable costs[22]; therefore, cost-effective, highly sensitive, and specific biomarkers should urgently be investigated. Tackling the aforementioned shortages, our team has recently investigated the efficacy of a few novel biomarkers. Accordingly, we observed that both local and circulating SOX9 were able to differentiate between the malignant and benign tumors with a strong and positive correlation with tumor severity, malignancy, size, and chemotherapy suggesting its potential diagnostic and prognostic values [17]. Recent studies point to the role of OPN as an effective mediator in increasing the growth of cancer cells[23]. Although OPN is a non-collagenous bone matrix protein, as versatile protein associated with multiple intracellular signaling pathways, it plays an important role in controlling the growth of cancer cells[24]. The role of OPN in the pathogenesis of cancer is important from various aspects including its role in the innate and adaptive immune system and cytokine production[25], cell differentiation[26], macrophage recruitment[27], cell adhesion, and cell migration[28]. Evidence from previous studies suggests that OPN is involved in multiple stages of cancer progression from tumor cell proliferation through binding to CD44 and inducing anti-apoptotic proteins to stimulate angiogenesis by inducing VEGF and facilitate cell detachment by inducing epithelial-mesenchymal transition (EMT) phenotype possibly through inducing MMPs[29], [30], [31]. The results of the expression pattern of OPN in different cancers show that OPN can play an effective role in the pathogenesis of cancer. In accordance, significantly elevated OPN protein levels were observed in tumors originating from various human body organs, including breast, ovary, endometrium, esophagus, stomach, pancreas, bile duct, liver, colon, kidney, bladder, prostate, head and neck, and lung [32]. Consistently, we observed remarkably higher expression levels of the OPN mRNA and protein in the patients, highlighting the pivotal role of OPN in the pathophysiology of primary bone cancers. According to our experiments, OPN was significantly different between the malignant and the benign patients suggesting a diagnostic potential confirmed by the ROC method. More importantly, we noticed that the OPN protein could differentiate between the cases and controls with 100 sensitivity and specificity, inspiring it to be employed as an accurate and non-invasive diagnostic tool. In agreement with our findings, Kim et al. (2002) measured OPN expression in both ovarian cancerous cell lines and tissue specimens as well as its serum concentration and demonstrated that the mRNA expression was significantly higher in the cancerous cells/tissues compared to those of normal/controls. Its protein concentration was also considerably higher in cancer patients than the healthy individuals, patients with benign diseases, or other gynecological cancers[33]. In support of our data, it is postulated that attachment of OPN to the integrin and CD44 receptors mediate activation of PI3K/Akt signaling pathway that results in tumor cell survival in favor of tumor progression[34]. Osteosarcoma is the most frequent malignant bone neoplasm accounting for 30–80% of primary skeletal sarcomas, followed by chondrosarcoma and Ewing sarcoma[22]. Interestingly, we observed that osteosarcoma patients had the highest levels of OPN in the tissue, PBMC, and serum samples. Both increased and decreased OPN expression have been reported in osteosarcoma cell lines; each one has different interpretations. The lower expression indicates that the majority of osteosarcoma cells fail to undergo terminal osteogenic differentiation, thereby promoting osteosarcoma growth, and on the other hand, the elevated expression can be indicative of the enhanced metastatic ability of osteosarcoma cells [35], [36]. Benign tumors of the bone are not cancerous and would not metastasize to other regions of the body. However, they can occur in any part of the skeleton and can still be dangerous as they may grow and compress healthy bone tissue [37]. In the current study, the lowest OPN levels were measured in the samples taken from patients with exostosis. Generally, among the different primary bone cancers, osteosarcoma tends to be more aggressive and invasive than the other five cancers. Conversely, exostosis tends to be more benign and gentle than the others [38]. Our preliminary research shows that the OPN level is proportional to the degree of aggressiveness of primary bone tumors, but further investigations should be conducted on a larger number of samples to confirm this hypothesis. Based on previous reports, it is postulated that OPN may involve in shaping the tumor microenvironment and can mediate inducing hypoxia-inducible factor-1 alpha (HIF-1α) pathway and EMT phenotype that results in enhanced metastasis in breast and ovarian cancer [39], [40]. The same mechanism may be assumed for primary bone cancer, and our evidence has shown that OPN is highly expressed in patients with metastasis and disease severity, but further studies are needed to substantiate this theory.The treatment of osteosarcoma includes preoperative chemotherapy, followed by surgical removal and postoperative chemotherapy. Currently, the degree of necrosis in response to preoperative chemotherapy is used as a reliable prognostic tool to manage postoperative chemotherapy. The patients are considered to be good responders to the treatment (good prognosis) when the necrosis rate is higher than 90%. Modifying postoperative chemotherapy can promote the outcomes of poor responders, and despite the massive efforts, the achievements are not satisfactory. Therefore, more efficient prognostic tools are required to timely predict response to the chemotherapy and guide the treatment on the right track. To overcome this pitfall, researchers are vastly investigating various molecular markers [41]. Our experiments revealed that the OPN mRNA expression in the tissue samples from osteosarcoma patients receiving chemotherapy was around 1.5 folds lower than the untreated counterparts. Moreover, serum OPN was measured to be around two folds lower in the osteosarcoma patients without tumor recurrence than the recurrent counterparts. In accordance, the association between OPN and chemoresistance is investigated, recently. It was demonstrated that OPN can activate the autophagy cell death pathway through activation of NF- κB pathway in pancreatic cancer cells leading to chemoresistance to gemcitabine[42]. Moreover, it seems that the anti- apoptotic effect of OPN may play role in the response of cancer cells to chemotherapy. For example, it was shown that down-regulation of OPN enhanced the sensitivity of breast cancer cells to the doxorubicin that might be occurred by activating the p38 MAPK pathway[34], [43].
According to our experiments, OPN mRNA expression in PBMC and its protein level in serum was around 1.7 folds higher in osteosarcoma patients with metastatic disease than those without metastatic disease. In support of our findings, Wong et al. (2000) measured significantly elevated OPN mRNA expression in osteosarcoma patients compared to healthy individuals. More importantly, they noticed that 6 of the patients with peripheral blood OPN mRNA expression exceeding the highest level found in the healthy subjects developed clinical metastasis within 12 months after the diagnosis. It appears that the elevated mRNA expression may stem from an increased number of circulating osteosarcoma cells, and these observations can potentially be employed as a biomarker for the diagnosis of osteosarcoma micrometastases and evaluation of prognosis [44].
In the current study, OPN mRNA expression was assessed in tumor tissue specimens (local) and PBMC (circulating). The lower mRNA expression in the circulation than the local one is attributable to the lower populations of undifferentiated cells and cancer stem cells in blood than tumors [45]. According to the available evidence, only a small fraction of the cancer cells may be released into the bloodstream, making it difficult for detection and evaluation[46]. Considering that mRNA expression might not always end up in the protein translation due to several genetic and epigenetic mediators, therefore, in the current study, OPN was assessed at both mRNA and protein levels to have better insight into the diagnostic and prognostic efficacy of OPN. Multiple lines of shreds of evidence, suggest that OPN can potentially be employed to manage and monitor the disease in a non-invasive and straightforward way. In agreement, the results of a meta-analysis by Sun et al. (2018) demonstrate that OPN can serve as a prognostic biomarker and a potential therapeutic target for hepatocellular carcinoma as its level was significantly associated with poor overall survival and disease-free survival [47]. Another study shows that OPN levels measured in serum and ascites of women with epithelial ovarian cancer can reflect the disease outcomes and significantly contribute to surgical planning [48]. However, our preliminary study was conducted using a relatively small sample size (153 patients); however, further studies with larger sample sizes are still required to confirm OPN as a biomarker for primary skeletal tumors. We also could not perform survival analysis, which is one of the critical features of an efficient prognostic biomarker. This limitation should be addressed in future studies.
5. Conclusion
In conclusion, OPN showed a different expression pattern in bone tumors with different degrees of deterioration and severity and a significant increase in its expression in malignant tumors with high grade and metastasis could indicate its effective role in bone tumor development. our experiments demonstrated that OPN may have diagnostic potentials in bone cancer that should be validated by further studies. Particularly, the OPN protein could differentiate the patients from the healthy individuals with the highest possible accuracy and OPN can be employed to manage and monitor patients with malignant primary bone tumors.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgement
This work was financially supported by the Iran University of Medical Sciences (Grant Number: 96–01-30–30638). We deeply appreciate all the patients who made this study possible by generously providing us with their tissue/blood samples.
Ethics approval and consent to participate
The project was ethically approved by the ethics committee of the Vice president of research of Iran University of Medical Sciences with ethics committee code: IR.IUMS.REC.1396.30638. All participants were informed before surgery and following the informed consent, they were included in the study. The signed consent form for each patient is available.
Author contributions
AH and BS carried out the experiments and collected the samples, AN and SAF contributed to data analysis and drafting the manuscript, AM provided the samples and contributed to clinical interpretation, KHJ and MB performed surgery, HK contributed to statistical analysis, VS contributed to data analysis and clinical interpretation, MTY designed and supervised the study and contributed to data analysis and drafting the manuscript.
References
- 1.Niu X. Primary Bone Tumors: Epidemiologic Comparison of 9200 Patients Treated at Beijing Ji Shui Tan Hospital, Beijing, China, With 10 165 Patients at Mayo Clinic, Rochester. Minnesota. Arch Pathol Lab Med. 2015;139(9):1149–1155. doi: 10.5858/arpa.2014-0432-OA. [DOI] [PubMed] [Google Scholar]
- 2.Siegel, R.L., K.D. Miller, and A. Jemal, Cancer statistics, 2019. CA: A Cancer Journal for Clinicians, 2019. 69(1): p. 7-34. [DOI] [PubMed]
- 3.Wan-Ibrahim W.I. Biomarkers for Bone Tumors: Discovery from Genomics and Proteomics Studies and Their Challenges. Molecular medicine (Cambridge. Mass.) 2016;21(1):861–872. doi: 10.2119/molmed.2015.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown H.K., Schiavone K., Gouin F., Heymann M.-F., Heymann D. Biology of Bone Sarcomas and New Therapeutic Developments. Calcified tissue international. 2018;102(2):174–195. doi: 10.1007/s00223-017-0372-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lamort A.-S., Giopanou I., Psallidas I., Stathopoulos G.T. Osteopontin as a Link between Inflammation and Cancer: The Thorax in the Spotlight. Cells. 2019;8(8):815. doi: 10.3390/cells8080815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Icer M.A., Gezmen-Karadag M. The multiple functions and mechanisms of osteopontin. Clin Biochem. 2018;59:17–24. doi: 10.1016/j.clinbiochem.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 7.Caputo S., Bellone M. Osteopontin and the immune system: another brick in the wall. Cell Mol Immunol. 2018;15(4):405–407. doi: 10.1038/cmi.2017.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shevde L.A., Samant R.S. Role of osteopontin in the pathophysiology of cancer. Matrix Biology. 2014;37:131–141. doi: 10.1016/j.matbio.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei R., Wong J.P.C., Lyu P., Xi X., Tong O., Zhang S.-D., Yuen H.F., Shirasawa S., Kwok H.F. In vitro and clinical data analysis of Osteopontin as a prognostic indicator in colorectal cancer. J Cell Mol Med. 2018;22(9):4097–4105. doi: 10.1111/jcmm.13686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wong J.P.C., Wei R., Lyu P., Tong O.L.H., Zhang S.D., Wen Q., Yuen H.F., El-Tanani M., Kwok H.F. Clinical and in vitro analysis of Osteopontin as a prognostic indicator and unveil its potential downstream targets in bladder cancer. Int J Biol Sci. 2017;13(11):1373–1386. doi: 10.7150/ijbs.21457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao H., Chen Q., Alam A., Cui J., Suen K.C., Soo A.P., Eguchi S., Gu J., Ma D. The role of osteopontin in the progression of solid organ tumour. Cell Death Dis. 2018;9(3) doi: 10.1038/s41419-018-0391-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao D.X. Osteopontin as potential biomarker and therapeutic target in gastric and liver cancers. World J Gastroenterol. 2012;18(30):3923–3930. doi: 10.3748/wjg.v18.i30.3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong K.-K., Cheng R.S., Mok S.C. Identification of differentially expressed genes from ovarian cancer cells by Micromax™ cDNA microarray system. Biotechniques. 2001;30(3):670–675. doi: 10.2144/01303dd05. [DOI] [PubMed] [Google Scholar]
- 14.Li Y.-S., Deng Z.-H., Zeng C., Lei G.-H. Role of osteopontin in osteosarcoma. Med Oncol. 2015;32(1) doi: 10.1007/s12032-014-0449-y. [DOI] [PubMed] [Google Scholar]
- 15.Tuck M.K., Chan D.W., Chia D., Godwin A.K., Grizzle W.E., Krueger K.E., Rom W., Sanda M., Sorbara L., Stass S., Wang W., Brenner D.E. Standard operating procedures for serum and plasma collection: early detection research network consensus statement standard operating procedure integration working group. J Proteome Res. 2009;8(1):113–117. doi: 10.1021/pr800545q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuss I.J., Kanof M.E., Smith P.D., Zola H. Isolation of whole mononuclear cells from peripheral blood and cord blood. Curr Protoc Immunol. 2009;85(1) doi: 10.1002/0471142735.2009.85.issue-110.1002/0471142735.im0701s85. [DOI] [PubMed] [Google Scholar]
- 17.Hosseini A. The local and circulating SOX9 as a potential biomarker for the diagnosis of primary bone cancer. Journal of Bone Oncology. 2020;23 doi: 10.1016/j.jbo.2020.100300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crowe A., Yue W. Semi-quantitative Determination of Protein Expression using Immunohistochemistry Staining and Analysis: An Integrated Protocol. Bio Protoc. 2019;9(24) doi: 10.21769/BioProtoc.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Obuchowski N.A., Bullen J.A. Receiver operating characteristic (ROC) curves: review of methods with applications in diagnostic medicine. Phys Med Biol. 2018;63(7):p. 07tr01. doi: 10.1088/1361-6560/aab4b1. [DOI] [PubMed] [Google Scholar]
- 20.Hajian-Tilaki K. Receiver Operating Characteristic (ROC) Curve Analysis for Medical Diagnostic Test Evaluation. Caspian J Intern Med. 2013;4(2):627–635. [PMC free article] [PubMed] [Google Scholar]
- 21.Hasan O., Ali M., Mustafa M., Ali A., Umer M. Treatment and recurrence of giant cell tumors of bone - A retrospective cohort from a developing country. Ann Med Surg (Lond) 2019;48:29–34. doi: 10.1016/j.amsu.2019.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Evola, F.R., et al., Biomarkers of Osteosarcoma, Chondrosarcoma, and Ewing Sarcoma. Frontiers in pharmacology, 2017. 8: p. 150-150 [DOI] [PMC free article] [PubMed]
- 23.Rangaswami H., Bulbule A., Kundu G.C. Osteopontin: role in cell signaling and cancer progression. Trends Cell Biol. 2006;16(2):79–87. doi: 10.1016/j.tcb.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 24.ELTANANI M., CAMPBELL F., KURISETTY V., JIN D., MCCANN M., RUDLAND P. The regulation and role of osteopontin in malignant transformation and cancer. Cytokine Growth Factor Rev. 2006;17(6):463–474. doi: 10.1016/j.cytogfr.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 25.Higashi A., Dohi Y., Uraoka N., Sentani K., Uga S., Kinoshita H., Sada Y., Kitagawa T., Hidaka T., Kurisu S., Yamamoto H., Yasui W., Kihara Y. The Potential Role of Inflammation Associated with Interaction between Osteopontin and CD44 in a Case of Pulmonary Tumor Thrombotic Microangiopathy Caused by Breast Cancer. Intern Med. 2015;54(22):2877–2880. doi: 10.2169/internalmedicine.54.4749. [DOI] [PubMed] [Google Scholar]
- 26.Chen Q. Fate decision of mesenchymal stem cells: adipocytes or osteoblasts? Cell Death Differ. 2016;23(7):1128–1139. doi: 10.1038/cdd.2015.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Remmerie A., Martens L., Thoné T., Castoldi A., Seurinck R., Pavie B., Roels J., Vanneste B., De Prijck S., Vanhockerhout M., Binte Abdul Latib M., Devisscher L., Hoorens A., Bonnardel J., Vandamme N., Kremer A., Borghgraef P., Van Vlierberghe H., Lippens S., Pearce E., Saeys Y., Scott C.L. Osteopontin Expression Identifies a Subset of Recruited Macrophages Distinct from Kupffer Cells in the Fatty Liver. Immunity. 2020;53(3):641–657.e14. doi: 10.1016/j.immuni.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wai P.Y., Kuo P.C. The role of Osteopontin in tumor metastasis. J Surg Res. 2004;121(2):228–241. doi: 10.1016/j.jss.2004.03.028. [DOI] [PubMed] [Google Scholar]
- 29.Zhou Y., Dai D.L., Martinka M., Su M., Zhang Y.i., Campos E.I., Dorocicz I., Tang L., Huntsman D., Nelson C., Ho V., Li G. Osteopontin expression correlates with melanoma invasion. J Invest Dermatol. 2005;124(5):1044–1052. doi: 10.1111/j.0022-202X.2005.23680.x. [DOI] [PubMed] [Google Scholar]
- 30.Leali D., Dell’Era P., Stabile H., Sennino B., Chambers A.F., Naldini A., Sozzani S., Nico B., Ribatti D., Presta M. Osteopontin (Eta-1) and fibroblast growth factor-2 cross-talk in angiogenesis. J Immunol. 2003;171(2):1085–1093. doi: 10.4049/jimmunol.171.2.1085. [DOI] [PubMed] [Google Scholar]
- 31.Ng L., Wan T.-H., Lam C.-C., Chow A.-M., Wong S.-M., Man J.-W., Li H.-S., Cheng N.-M., Pak R.-H., Cheung A.-K., Yau T.-C., Lo O.-H., Foo D.-C., Poon J.-C., Poon R.-P., Pang R.-C., Law W.-L., Samant R. Post-operative plasma osteopontin predicts distant metastasis in human colorectal cancer. PLoS One. 2015;10(5):e0126219. doi: 10.1371/journal.pone.0126219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coppola D., Szabo M., Boulware D., Muraca P., Alsarraj M., Chambers A.F., Yeatman T.J. Correlation of osteopontin protein expression and pathological stage across a wide variety of tumor histologies. Clin Cancer Res. 2004;10(1):184–190. doi: 10.1158/1078-0432.ccr-1405-2. [DOI] [PubMed] [Google Scholar]
- 33.Kim J.-H. Osteopontin as a Potential Diagnostic Biomarker for Ovarian Cancer. JAMA. 2002;287(13):1671–1679. doi: 10.1001/jama.287.13.1671. [DOI] [PubMed] [Google Scholar]
- 34.Yang L. Down-regulation of osteopontin expression by RNA interference affects cell proliferation and chemotherapy sensitivity of breast cancer MDA-MB-231 cells. Mol Med Rep. 2012;5(2):373–376. doi: 10.3892/mmr.2011.679. [DOI] [PubMed] [Google Scholar]
- 35.Luo X., Chen J., Song W.-X., Tang N.i., Luo J., Deng Z.-L., Sharff K.A., He G., Bi Y., He B.-C., Bennett E., Huang J., Kang Q., Jiang W., Su Y., Zhu G.-H., Yin H., He Y., Wang Y.i., Souris J.S., Chen L., Zuo G.-W., Montag A.G., Reid R.R., Haydon R.C., Luu H.H., He T.-C. Osteogenic BMPs promote tumor growth of human osteosarcomas that harbor differentiation defects. Laboratory investigation. 2008;88(12):1264–1277. doi: 10.1038/labinvest.2008.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Velupillai P., Sung C.K., Tian Y.u., Dahl J., Carroll J., Bronson R., Benjamin T., Sugden B. Polyoma virus-induced osteosarcomas in inbred strains of mice: host determinants of metastasis. PLoS Pathogens. 2010;6(1):e1000733. doi: 10.1371/journal.ppat.1000733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hakim D.N., Pelly T., Kulendran M., Caris J.A. Benign tumours of the bone: A review. Journal of bone oncology. 2015;4(2):37–41. doi: 10.1016/j.jbo.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bukhari, M.H., S. Qamar, and F. Batool, Differential Diagnosis of Osteogenic Tumors in the Context of Osteosarcoma, in Osteosarcoma–Diagnosis, Mechanisms, and Translational Developments. 2019, IntechOpen.
- 39.Song G. Osteopontin promotes ovarian cancer progression and cell survival and increases HIF-1alpha expression through the PI3-K/Akt pathway. Cancer Sci. 2008;99(10):1901–1907. doi: 10.1111/j.1349-7006.2008.00911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raja R., Kale S., Thorat D., Soundararajan G., Lohite K., Mane A., Karnik S., Kundu G.C. Hypoxia-driven osteopontin contributes to breast tumor growth through modulation of HIF1α-mediated VEGF-dependent angiogenesis. Oncogene. 2014;33(16):2053–2064. doi: 10.1038/onc.2013.171. [DOI] [PubMed] [Google Scholar]
- 41.Man T.-K., Chintagumpala M., Visvanathan J., Shen J., Perlaky L., Hicks J., Johnson M., Davino N., Murray J., Helman L., Meyer W., Triche T., Wong K.-K., Lau C.C. Expression profiles of osteosarcoma that can predict response to chemotherapy. Cancer research. 2005;65(18):8142–8150. doi: 10.1158/0008-5472.CAN-05-0985. [DOI] [PubMed] [Google Scholar]
- 42.Yang M.C. Blockade of autophagy reduces pancreatic cancer stem cell activity and potentiates the tumoricidal effect of gemcitabine. Mol Cancer. 2015;14:179. doi: 10.1186/s12943-015-0449-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pang H., Cai L.i., Yang Y., Chen X., Sui G., Zhao C. Knockdown of osteopontin chemosensitizes MDA-MB-231 cells to cyclophosphamide by enhancing apoptosis through activating p38 MAPK pathway. Cancer Biother Radiopharm. 2011;26(2):165–173. doi: 10.1089/cbr.2010.0838. [DOI] [PubMed] [Google Scholar]
- 44.Wong I.H., Chan A.T., Johnson P.J. Quantitative analysis of circulating tumor cells in peripheral blood of osteosarcoma patients using osteoblast-specific messenger RNA markers: a pilot study. Clin Cancer Res. 2000;6(6):2183–2188. [PubMed] [Google Scholar]
- 45.Dylla S.J., Beviglia L., Park I.-K., Chartier C., Raval J., Ngan L., Pickell K., Aguilar J., Lazetic S., Smith-Berdan S., Clarke M.F., Hoey T., Lewicki J., Gurney A.L., Gilliland D.G. Colorectal Cancer Stem Cells Are Enriched in Xenogeneic Tumors Following Chemotherapy. PLOS ONE. 2008;3(6):e2428. doi: 10.1371/journal.pone.0002428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tayoun, Faugeroux, Oulhen, Aberlenc, Pawlikowska, Farace CTC-derived models: a window into the seeding capacity of circulating tumor cells (CTCs) Cells. 2019;8(10):1145. doi: 10.3390/cells8101145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun, T., et al., Prognostic value of osteopontin in patients with hepatocellular carcinoma: A systematic review and meta-analysis. Medicine, 2018. 97(43): p. e12954-e12954 [DOI] [PMC free article] [PubMed]
- 48.Cerne, K., et al., Potential of osteopontin in the management of epithelial ovarian cancer. Radiology and oncology, 2019. 53(1): p. 105-115 [DOI] [PMC free article] [PubMed]