Abstract
We report on the physicochemical processes and the products of DNA damage involved in Ne-22 ion-beam radiation of hydrated (12 ± 3 H2O/nucleotide) salmon testes DNA at 77 K. Free radicals trapped at 77 K were identified using electron spin resonance (ESR) spectroscopy. The measurement of DNA damage using two different techniques of mass spectrometry revealed the formation of numerous DNA products. Results obtained by ESR spectroscopy showed that as the linear energy transfer (LET) of the ion-beam radiation increases along the beam track, the production of DNA radicals correspondingly increases until just before the Bragg peak is reached. Yields of DNA products along the ion-beam track were in excellent agreement with the radical production. This work is the first to use the combination of ESR spectroscopy and mass spectrometric techniques enabling a better understanding of mechanisms of radiation damage to DNA by heavy ion beams detailing the formation of DNA free radicals and their subsequent products.
Introduction
The ever-increasing use of ion-beam irradiation, especially with protons and relatively heavy ions for cancer therapy, and a renewed interest in extended space missions have resulted in increased interest in the biological effects of ion beams.1 The biological effects of ionizing radiation, including low linear energy transfer (LET) γ- and X-rays and high-LET energetic ion beams, are largely initiated by damage to the cellular DNA.2 Radiation damage to DNA by ionizing radiation can be broadly classified as arising from two effects, direct effects and indirect effects.2−6 Damage by direct effects has two components, the first in which damage is caused by direct ionization of the DNA itself, resulting in the formation of cation radicals and anion radicals on the DNA structure.2−4 A second component of direct effects (called the quasi-direct effect) results from ionization of the first few water molecules of solvation (ca. 10–12 H2O/nucleotide).3,5−7 In this case, the holes and electrons formed in the solvation shell are efficiently transferred to DNA, thereby forming additional cation radicals and anion radicals on the DNA.
Indirect damage is mediated by reactive free radicals formed by the ionization of the surrounding bulk water, principally •OH, H•, and eaq–.3,4 These may react with DNA constituents leading to DNA base and 2′-deoxyribose products, strand breaks, unaltered base release, tandem lesions, 8,5′-cyclopurine-2′-deoxynucleosides, and interstrand cross-links.4,8−12 In this current work, the DNA was hydrated (designated by Γ) to Γ = 12 ± 3 H2O/nucleotide. Thus, no bulk water is present, and the indirect effect is largely suppressed. As a result, the direct effects predominate.3,5−7 Studies indicate that ca. 50% of the DNA damage in cells is due to direct-type (direct and quasi-direct) effects.3
To fully understand the radiation effects of ion-beam irradiation on DNA and the spatial arrangement of the damage, the highly heterogeneous deposition of energy that occurs with energetic ions must be taken into account.2,3,13−17 An oft-used model for the energy deposition invokes a track structure in which the energetic ion, through relatively short-range interactions with electrons in the target material, ionizes atoms of the target forming a dense, cylindrical track of ionized atoms called the track core.18 The core is the high-LET region of the track. The electrons that result from these initial ionizations undergo multiple collisions with atoms in the target, resulting in further ionizations. Although many electrons stay within the high energy density core (Figure 1) and largely recombine with holes, the most energetic of them escape from the core as δ-rays and cause further ionizations in a low-LET region of the track called the penumbra.18 The energy deposition in the penumbra is very much like that that occurs with low-LET radiation such as γ-rays and X-rays.2,3,13−18 The radius of the core and penumbra both increase with energy, and the penumbra radius is very much larger than that of the core.18
Figure 1.
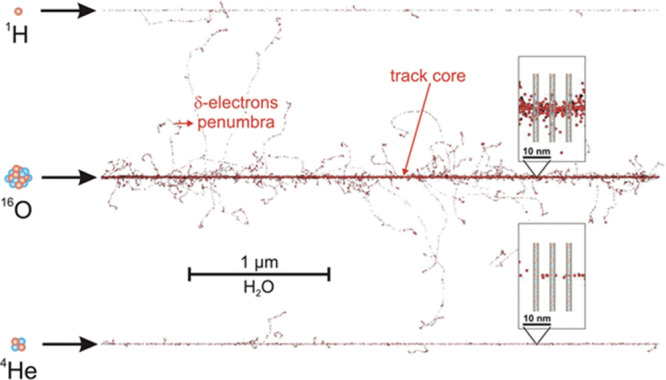
PARTRAC simulated track structure for three different ions (H, O-16, and He-4), all with energy 6.25 MeV/u. The insets show a high-density ionization segment of the track (top inset) and a lower density ionization area (bottom inset), superimposed on schematic DNA segments. Reproduced from ref (18) with the permission of the Danish Royal Academy of Science and Letters.
In this work, we report on the investigation of physicochemical processes including the formation of DNA radicals and the resulting products in DNA upon Ne-22 ion-beam radiation damage at 77 K to hydrated highly polymerized salmon testes DNA. We used a combination of electron spin resonance (ESR) spectroscopy to elucidate the DNA radicals formed (Figure 2) and gas chromatography-tandem mass spectrometry (GC–MS/MS) and liquid-chromatography tandem mass spectrometry (LC–MS/MS) with isotope dilution to identify and quantify a plethora of DNA-base damage products including 8,5′-cyclopurine-2′-deoxynucleosides.
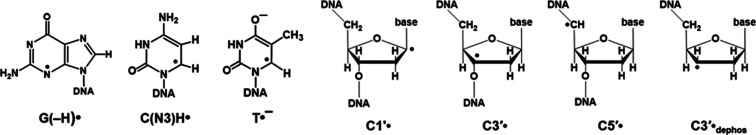
Figure 2.
Radicals considered in this work.
Results
Measurement of DNA Radicals by ESR Spectroscopy
The aim of this work was to investigate the mechanisms of direct-type DNA damage by heavy-ion radiation by means of the formation of DNA free radicals and products using ESR spectroscopy, GC–MS/MS, and LC–MS/MS. The combined use of these techniques for this purpose has not been reported previously. Highly polymerized salmon testes DNA hydrated to Γ= 12 ± 3 H2O/nucleotide was exposed to Ne-22 ion irradiation at 77 K. DNA radicals trapped in DNA at 77 K were elucidated using ESR spectroscopy.
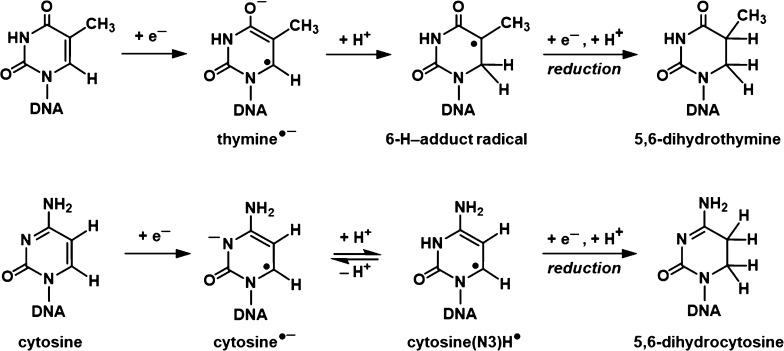
Figure 3 shows the ESR spectra from Ne-22 ion-irradiated H2O-hydrated DNA. In earlier work, we have thoroughly characterized spectra such as these from both γ-irradiated and ion-beam-irradiated DNA.2,3,5,6,14−17,20,21 Previous studies indicated that the spectra are composites that result from at least seven radicals (Figure 2). These are three base radicals, G(−H)•/G•+, C(N3)H•/C•–, and T•–, which originate from ion-radical precursors. The radiation causes loss of an electron from guanine leading to the guanine radical cation (G•+), which undergoes reversible intrabase pair proton transfer from N1 to the N3 of the base-paired cytosine, forming the neutral free radical G(−H)• and diamagnetic cation C(N3)H+.2,3,13,38,39 The addition of an electron to cytosine results in the formation of the free radical C•– which undergoes reversible protonation at N3, to form the radical C(N3)H•. T•– results from capture of an electron by thymine.
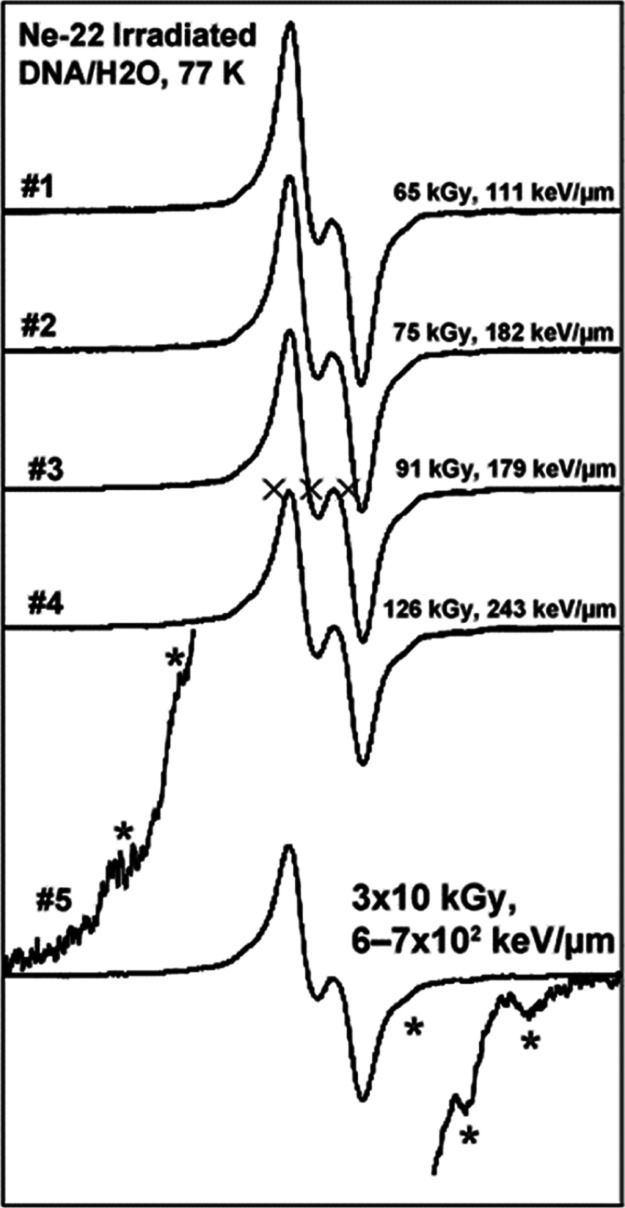
Figure 3.
ESR spectrum, at 77 K, of hydrated [Γ = (12 ± 3) H2O/nucleotide] salmon testes DNA, using Ne-22 ion irradiation at 77 K. The Ne-22 ion energy at the entrance to the sample packet was 1.14 GeV. The sample number indicates the position in the sample packet, #1 at the entrance and #5 at the Bragg peak. Doses and LETs for each spectrum are indicated. Each spectrum is a composite arising from at least seven radicals (Figure 2). The wings of the spectrum for the sample given the highest dose have been expanded to show line components (indicated by *) assigned to radicals on the 2′-deoxyribose sugar (Figure 2). Three X’s indicate Fremy Salt resonances, with the central marker at g = 2.0056 and AN = 13.09 G.
The three trapped base radicals form predominantly in the ion-track penumbra. This is because in the densely ionized core, base anion and cation radicals form near one another and Coulombic attractions drive their fast recombination.2,3,13 Consequently, DNA-base radicals are not observed in significant amounts at 77 K in the core.2,3,13−17 On the other hand, in the sparsely ionized penumbra, about 30% of the base radicals escape recombination and are trapped at 77 K.2,3,13−17 The three 2′-deoxyribose radicals (C1′•, C3′•, and C5′•) are formed principally via an ionization-deprotonation pathway, i.e., fast deprotonation of 2′-deoxyribose cation radicals before recombination can occur, as part of an oxidative damage mechanism.2,3,13−17,20 On the other hand, C3′•dephos is formed by dissociative electron attachment via the reaction of both radiation-produced quasi-free electrons and low energy electrons with DNA, possibly through the agency of excited states, in a reductive damage mechanism.2,3,13−17,40−44 Most of the 2′-deoxyribose radicals are formed in the track core, but a small fraction is also formed in the track penumbra.2,3,13−17 These four 2′-deoxyribose radicals (C1′•, C3′•, C5′•, and C3′•dephos) are uncharged and not as susceptible to recombination as are the base radicals.2,3,13−17,20
The relative amount of the radicals (base vs 2′-deoxyribose) trapped depends on the LET of the radiation. At higher LETs, more energy is generally deposited in the core and less in the penumbra, and the fraction of base radicals found in the cohort of radicals decreases.2,3,13−17,20 Analysis of the 2′-deoxyribose radical cohort using a specific benchmark spectrum for C5′• (Figure S3) indicates that C5′• constitutes ca. 25% of the trapped 2′-deoxyribose radicals in ion-beam-irradiated hydrated DNA.14 This is highly pertinent to the formation of the 8,5′-cyclopurine-2′-deoxynucleosides (see sections entitled “Measurement of DNA Products” and “Discussion”). Figure 4 shows the measured yields (μmol/kg) of trapped radicals as the ion beam penetrates into the sample packet. The seven blue bars in the graph represent the seven ESR samples in the sample packet. Product yield results, which were determined for thinner samples along the path than the ESR samples, indicate that the Bragg peak occurs at about 4.8–5.2 mm in the packet. To construct the two red bars, 98% of the total yield found in the fifth sample in the packet is imputed to the first ca. 0.4 mm of the sample, in recognition of the fact that the last ca. 0.9 mm of the sample is unirradiated. The 2% of the yield allocated to the last 0.9 mm of the fifth sample gives the background yield for this packet. It should be noted here that the total yield of trapped radicals at the Bragg peak is lower than that found in the (fourth) pre-Bragg peak sample. In addition, the small yields beyond the Bragg Peak are expected from beam fragmentation.
Figure 4.
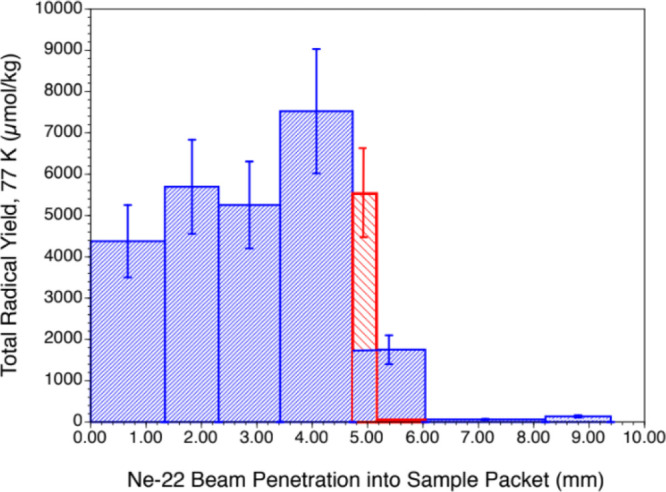
Yields of trapped radicals (77 K) in Ne-22-irradiated, hydrated [Γ = 12 ± 3 H2O/nucleotide] salmon testes DNA. Yields in the seven samples used. Only the first five samples in the packet were irradiated, the fifth only partially. Thus, the Bragg peak for the Ne-22 ion is located in the fifth sample at ca. 5.2 mm penetration depth. The yield in the fifth sample is recalculated in recognition of the fact that the beam penetrated only to ca. 5.2 mm into the sample packet, leaving most of the fifth sample unirradiated. The uncertainties depicted represent the typical ca. 20% variation seen in measuring radical yields.
Figure 5 shows the dose response of trapped radicals at 77 K for Ne-22-irradiated DNA. The three curves shown represent the yields of (a) the four 2′-deoxyribose radicals treated as a group (ΣdR•), (b) the three base radicals treated as a group [G(−H)•/G•+ + C(N3)H• + T•–], and (c) the sum of all seven radicals, i.e., all of the radicals observed near g = 2.00 (Figure 3). Because of unanticipated handling problems with H2O-hydrated samples, this curve is constructed from the ESR spectra from pre-Bragg peak samples for the Ne-22-irradiated DNA samples hydrated with D2O rather than H2O. We have found that there is little observable difference between the behavior of D2O- versus H2O-hydrated samples insofar as radical yields are concerned. Those yield curves which tend to or reach a plateau are fit to eq S2. For the total radical yield, G = 0.13 μmol/J with k = 5.5 × 10–6 Gy–1. The benchmark for the sum of the 2′-deoxyribose radicals yields G = 0.052 μmol/J; since the fit for the 2′-deoxyribose radicals is linear, k is zero. The curve for the sum of the base radical yields is drawn for the eye only. The G-value for the total trapped radicals at 77 K in similarly prepared and handled γ-irradiated samples is 0.25 μmol/J.20 Ne-22 beam-irradiated samples have a lower G-value than γ-irradiated samples due to rapid recombination of base radicals in the ion-beam core, which leads to a diminution in trapped base radicals.
Figure 5.
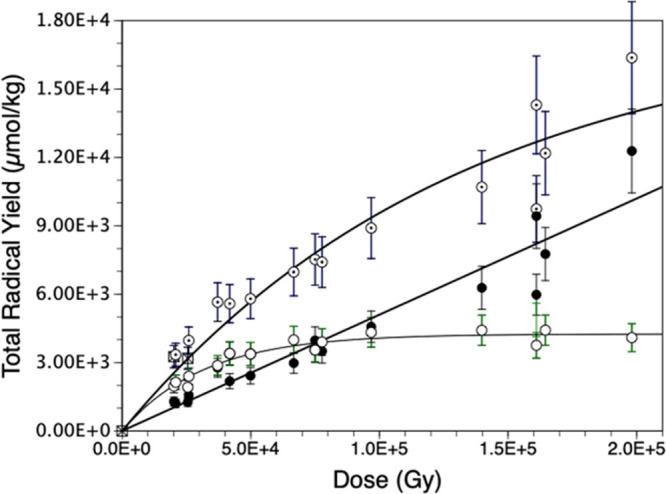
Dose response of trapped radicals in Ne-22-irradiated D2O-hydrated [Γ = 12 ± 3 H2O/nucleotide] salmon testes DNA (77 K). Here, the label ⊙ represents the total radical yield, “●” represents the sum of sugar radicals, Σ(C1′• + C3′• + C5′• + C3′•dephos), and the label “○” represents the sum of base radicals Σ[G(−H)•/G•+ + C(N3H)• + T•–]. Uncertainties are estimates based on typical spin-counting variability.
Measurement of DNA-Base Damage Products
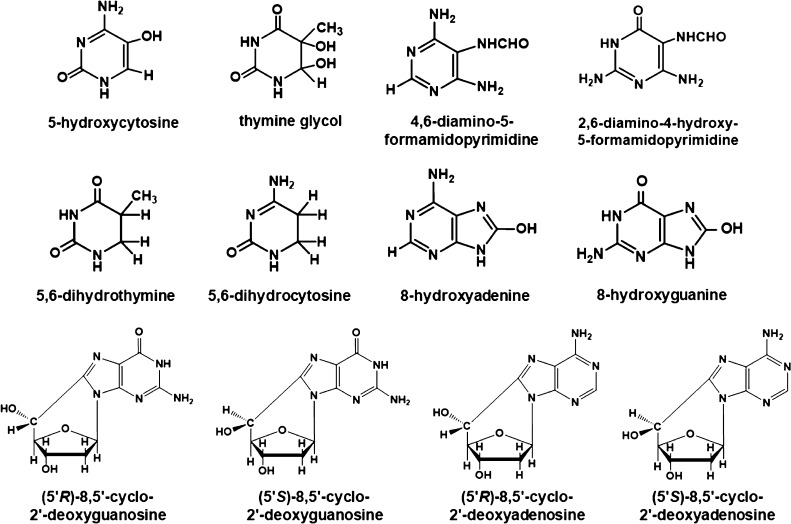
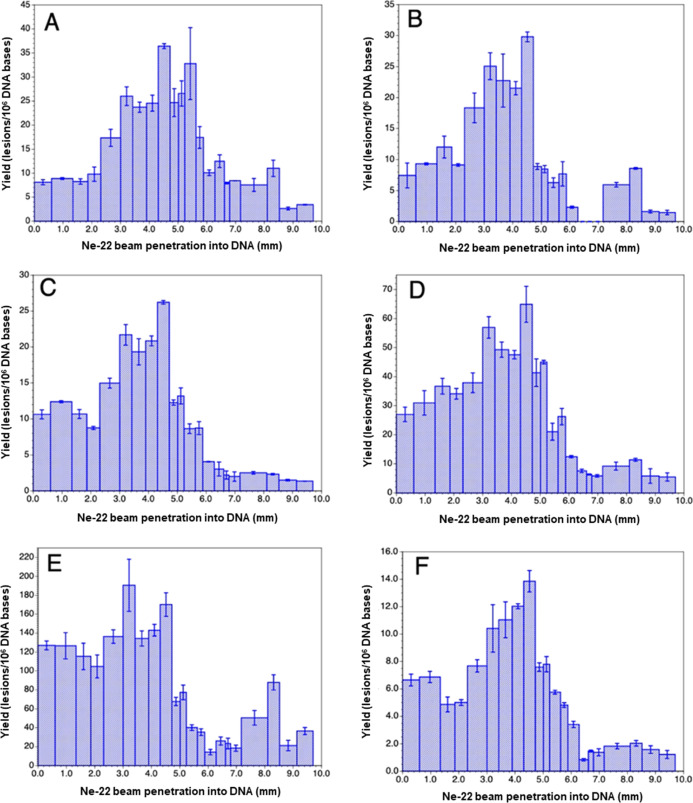
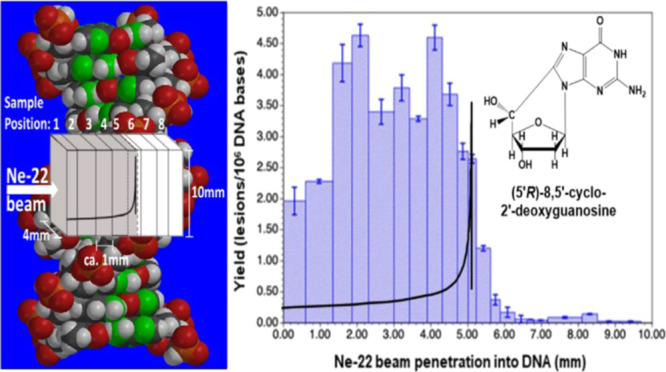
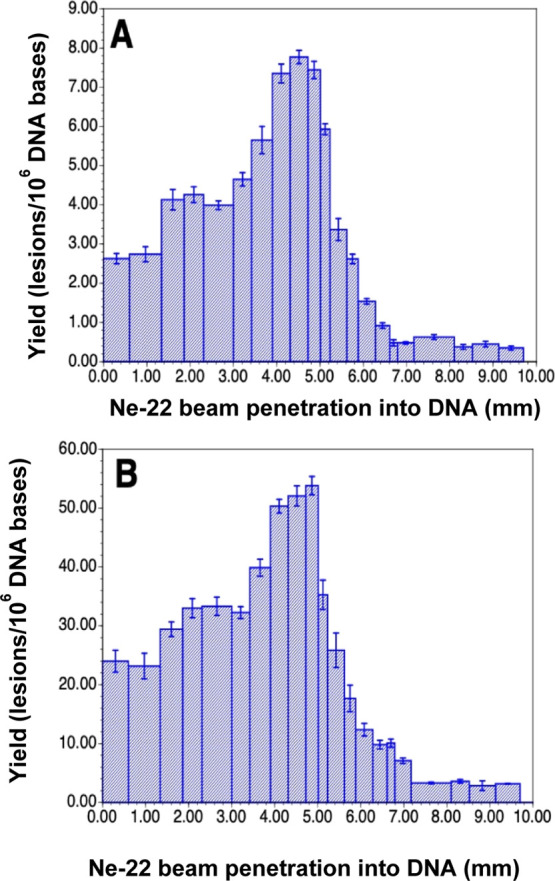
DNA-base damage products (Figure 6) in the DNA samples were identified and quantified using two different mass spectrometric techniques. LC–MS/MS with isotope dilution was used for the measurement of the 8,5′-cyclo-2′-deoxynucleosides, i.e., (5′R)-8,5′-cyclo-2′-deoxyadenosine (R-cdA), (5′S)-8,5′-cyclo-2′-deoxyadenosine (S-cdA), (5′R)-8,5′-cyclo-2′-deoxyguanosine (R-cdG), and (5′S)-8,5′-cyclo-2′-deoxyguanosine (S-cdG), and the purine-derived lesions 8-hydroxyadenine (8-OH-Ade) and 8-hydroxyguanine (8-OH-Gua) (also called 8-oxo-Gua) as their 2′-deoxynucleosides, i.e., 8-hydroxy-2′-deoxyadenosine (8-OH-dA) and 8-hydroxy-2′-deoxyguanosine (8-OH-dG), respectively. The measurement of the pyrimidine-derived lesions, 5-hydroxycytosine (5-OH-Cyt), thymine glycol (ThyGly), 5,6-dihydrothymine (5,6-diHThy), 5,6-dihydrocytosine (5,6-diHCyt), and 5,6-dihydrouracil (5,6-diHUra), and purine-derived lesions 4,6-diamino-5-formamidopyrimidine (FapyAde) and 2,6-diamino-4-hydroxy-5-formamidopyrimidine (FapyGua) was achieved using GC–MS/MS with isotope dilution. The reason for the use of GC–MS/MS for the measurement of these products is that this technique is the best suitable one among available analytical techniques for this purpose (see the Supporting Information and the cited literature). It should be noted that 5,6-diHUra is produced to some extent by deamination of 5,6-diHCyt under acidic conditions (see Experimental section in the Supporting Information). Figure 6 illustrates the structures of these compounds. The yields of 8,5′-cyclopurine-2′-deoxynucleosides with Ne-22 beam irradiation along the ion-beam track are shown in Figure 7. Doses for these samples within the beam range are shown in Table S1. A steady increase in the yields was observed up to 4 mm along the radiation path followed by some decrease up to 5 mm. Afterward, the yields sharply decreased to slightly above the levels in unirradiated DNA samples (less than 0.05 lesions/106 DNA bases). The Bragg peak was likely to be at ca. 5.2 mm of the radiation path (Figure 7). The low damage levels observed beyond the Bragg peak are likely due to inhomogeneity in the samples and/or fragmentation of the ion beam. Table S1 in the Supporting Information shows the yields of 8,5′-cyclopurine-2′-deoxynucleosides in Ne-22-irradiated DNA. In all cases, the yields of the R-diastereomers were observed to be greater than those of the S-diastereomers of both cdA and cdG. The R/S-ratios practically remained constant for all the samples within acceptable uncertainties (see the average values of the R/S-ratio with the uncertainties in Table S1).
Figure 6.
Structures of DNA-base lesions found in Ne-22-irradiated hydrated DNA.
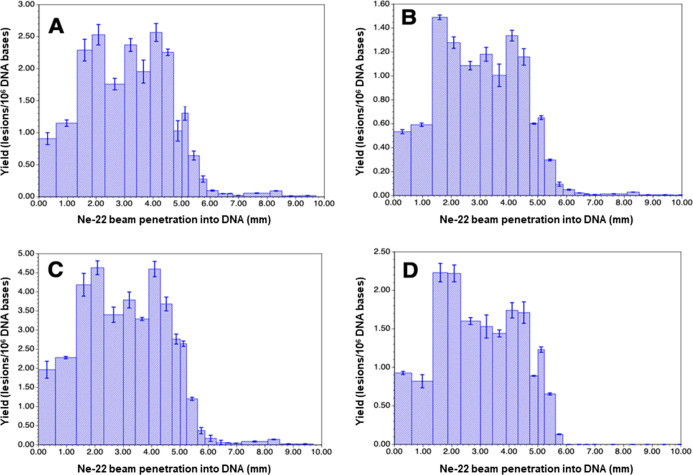
Figure 7.
Yields of 8,5′-cyclopurine-2′-deoxynucleosides in Ne-22 beam-irradiated hydrated (Γ = 12 ± 3 H2O/nucleotide) salmon testes DNA (measured by LC–MS/MS with isotope dilution). The Bragg peak location is judged to be in the 5.0–5.2 mm region. For each of the samples, the yield is calculated using the full sample mass. However, it should be noted that the sample containing the Bragg peak may not be uniformly irradiated throughout the sample depth. Uncertainties are standard deviations. Doses for those samples within the beam range are shown in Table S1. (A) R-cdA, (B) S-cdA, (C) R-cdG, and (D) S-cdG.
For comparison, we also measured the yields of these compounds in calf thymus DNA γ-irradiated at 10 Gy in N2O-saturated aqueous solution and in hydrated salmon testes DNA in the solid state γ-irradiated at room temperature at 20 kGy. Table S2 in the Supporting Information shows the yields in these samples. The yields in DNA samples γ-irradiated at 10 Gy in N2O-saturated aqueous solution were much greater than those in hydrated DNA γ-irradiated with 20 kGy at room temperature. At the highest level of damage, the yields of cdA and cdG in DNA samples irradiated with Ne-22 ions at 77 K were similar to those in calf thymus DNA samples γ-irradiated at 10 Gy in aqueous solutions (compare Tables S1 and S2 in the Supporting Information). In making these comparisons, it should be pointed out that the radiation chemistry in aqueous solutions is quite different from that in hydrated DNA in the solid state. One important distinction is that irradiation of an aqueous solution at room temperature produces copious amounts of •OH that in the absence of radical scavengers, migrate to and react with the DNA.2−4,13,42 It should be emphasized that the striking difference between the DNA samples γ-irradiated in aqueous solutions and those γ-irradiated at room temperature in the solid state or Ne-22 beam-irradiated at 77 K was in the ratios of the R-diastereomers to the S-diastereomers of both cdA and cdG. The yields of the S-diastereomers were greater than those of the R-diastereomers in DNA γ-irradiated in aqueous solutions, with the R/S ratios being 0.90 for cdA and 0.73 for cdG (Table S2 in the Supporting Information). In contrast, the R-diastereomers dominated over the S-diastereomers in hydrated DNA samples γ-irradiated with 20 kGy at room temperature or Ne-22 beam-irradiated at 77 K with average R/S ratios being greater than 2 for both cdA and cdG (compare Tables S2 and S4 in the Supporting Information). In the case of the other DNA-base lesions including 5,6-dihydropyrimidines, a similar trend of the yields was observed for Ne-22 beam-radiation doses at 77 K as was observed for the 8,5′-cyclo-2′-deoxynucleosides. Figures 8 and 9 and Table S3 in the Supporting Information show the yields of these lesions in DNA samples. The yield of 5,6-diHCyt represents the total levels of 5,6-diHCyt and 5,6-diHUra because the latter is formed by deamination of the former by acidic treatment prior to GC–MS/MS analysis (see Experimental section in the Supporting Information). Table S4 in the Supporting Information shows the yields of DNA-base lesions in calf thymus DNA γ-irradiated at 10 Gy in N2O-saturated aqueous solution and in hydrated salmon testes DNA γ-irradiated at room temperature to 20 kGy. In the case of the purine-derived lesions, the yields were somewhat similar between the two types of DNA samples. In contrast, the yields of the pyrimidine-derived lesions 5-OH-Cyt and ThyGly were significantly lower in hydrated DNA than those in DNA γ-irradiated in aqueous solutions.
Figure 8.
Yields of DNA-base products in Ne-22 beam-irradiated hydrated (Γ = 12 ± 3 H2O/nucleotide) salmon testes DNA. For each of the samples, the yield is calculated using the full sample mass. However, it should be noted that the sample containing the Bragg peak may not be uniformly irradiated throughout the sample depth. Doses for those samples within the beam range are shown in Table S3 in the Supporting Information. (A) 5-OH-Cyt, (B) ThyGly, (C) FapyAde, (D) FapyGua (these four products were measured by GC–MS/MS with isotope dilution), (E) 8-OH-dG, and (F) 8-OH-dA (these two products were measured by LC–MS/MS with isotope dilution). Uncertainties are standard deviations.
Figure 9.
Yields of (A) 5,6-diHThy and (B) 5,6-diHCyt in Ne-22 beam-irradiated hydrated (Γ = 12 ± 3 H2O/nucleotide) salmon testes DNA (measured by GC–MS/MS with isotope dilution). Uncertainties are standard deviations.
Discussion
In this work, hydrated DNA samples (Γ = 12 ± 3 H2O/nucleotide) were irradiated at 77 K by an initial 1.14 GeV Ne-22 beam. The ion energy degrades along its path through the stacked samples and drops to zero at the path end just after the Bragg peak. Smaller yields found in samples beyond the Bragg peak are in part a result of fragmentation of the parent ion and the dose delivered by the fragment ions. ESR spectroscopy was used to determine the yield of DNA radicals trapped in hydrated DNA samples at 77 K. Subsequently, upon warming of the same samples, GC–MS/MS and LC–MS/MS analyses were carried out to identify and quantify resulting DNA products. By analyses of ESR spectra recorded at 77 K, we determined the yields of two separate cohorts of radicals that are responsible for the experimental spectra: (a) the combined yield of the individual base radicals as the DNA-base ion radicals (and their conjugate species in prototropic equilibrium) and (b) the combined yield of the known four radicals of the 2′-deoxyribose moiety, C1′•, C3′•, C5′•, and C3′•dephos.2,3,13−17 C1′•, C3′•, and C5′• are produced by (a) ionization of 2′-deoxyribose, followed by deprotonation,2,3,13−17,42 (b) charge and spin transfer from the excited base cation radicals to 2′-deoxyribose-producing cation radicals of 2′-deoxyribose, followed by deprotonation,2,3,13,14,42 and (c) phosphate-to-2′-deoxyribose hole transfer, followed by deprotonation.42,43 On the other hand, the formation of C3′•dephos is associated with a radiation-produced low-energy electron-mediated dissociative electron attachment process.2,3,13−17,41
Using the trapped radical yields in Ne-22 beam-irradiated hydrated DNA samples that were obtained by ESR at 77 K and the physical track structure model described above, we, in earlier work, developed a DNA Radiation Chemistry Track Structure Model that delineates the spatial position, within the ion-beam track, of the early free radicals responsible for the formation of clustered lesions, including double-strand breaks, and the often mutagenic and/or cytotoxic DNA products found at room temperature.2,3,13−17,41 This model posits that the DNA-base radicals and/or their charged precursors undergo rapid recombination in the track core and, therefore, the trapped surviving DNA-base radicals observed at 77 K are formed largely in the track penumbra.2,3,13−17,41 In contrast, most of the 2′-deoxyribose radicals that are formed in the core survive at 77 K.2 In addition, a small fraction of the trapped 2′-deoxyribose radicals is formed in the penumbra.2,3,13−17,41 Therefore, our DNA Radiation Chemistry Track Structure Model posits that after ion-beam irradiation, most of the trapped DNA-neutral radicals observed at 77 K form in the core and nearly all the charged base ion radicals form in the penumbra.2,3,13−17,41
In this work, the initial free radicals that are trapped at 77 K in hydrated, anoxic DNA after Ne-22 ion-beam irradiation are investigated and described, as a variety of potentially mutagenic and/or lethal products formed when the irradiated samples are warmed to room temperature and the initial radicals react to form products. Radical yields and product yields are reported. The likely reactions that lead from the initial radicals to the products are provided, and the spatial arrangements, in the ion-beam track, of the initial radicals and the products formed are described. Finally, the pertinence of these results to various base lesions including 8,5′-cyclopurine-2′-deoxynucleosides and 5,6-dihydropyrimidines are presented for the first time. The combination of ESR spectroscopy and product analyses of Ne-22 ion-beam-irradiated hydrated DNA has led to the following salient points:
The location of maximum damage is found to be just before the Bragg peak: ESR measurements presented in this work have shown that as the LET of the ion-beam radiation increases, reaching a peak at the Bragg peak, at which point the ions come to rest, the production of cation, anion, and neutral radicals of DNA increases along the ion-beam track, reaching a maximum in the sample just before that containing the Bragg peak. With regard to the formation of C3′•dephos (see Figure 2 for its structure), excitations increase significantly just before and at the Bragg peak relative to the beam plateau region.40
This would tend to increase the probability for the formation of immediate strand breaks from low-energy electrons (LEEs), which can involve transient anion excited states.41−44 It has been shown that C3′•dephos and the phosphoryl radical (DNA–OP(•)O2–), which are the direct products from a strand break, result from LEEs through the formation of an excited-state transient negative ion (TNI)13−17,40−44 and that the concentrations of these radicals are significantly higher in ion-beam-irradiated samples than in γ-irradiated ones.13−17 In fact, both are observed in our Ne-22-irradiated DNA/D2O samples. This suggests that increased strand breaks may occur near the Bragg peak in ion-beam-irradiated DNA. The yields of DNA-base damage products along the ion-beam track were in excellent agreement with the radical production. Because the probability of recombination of DNA radicals in the core increases due to the rise in concentration of proximate ion radicals, the location of the highest energy deposition, at the Bragg peak, should show different damage from the plateau region of the beam. Furthermore, the yields of radicals and products found at the Bragg Peak are lower than those found in the sample(s) just before the Bragg Peak. This leads us to conclude that the location of the maximum damage, insofar as the radicals and products described here are concerned, is not at the Bragg Peak but just before the peak in energy deposition and, thereby, the peak in dose. This, we think, is an important finding of our work. The lower yield at the Bragg peak is a result of the fact that as the ion slows to a stop at the Bragg peak, the track “pencils down” to a point, increasing the concentration of ionizations. This, in turn, increases ion recombination, thereby significantly lowering the damage yield at the Bragg Peak.
We note that the product yields are calculated using the mass of the whole sample. As noted earlier, the sample containing the Bragg peak may not have been irradiated for all of its depth and that the product yield for the irradiated section of the sample will be higher than that reported for the whole sample. See Figure 5 for a similar effect in ESR radical yields. Based on the observed yields, this does not affect the conclusion that pre-Bragg peak samples typically have higher yields of trapped radicals (77 K) and product yields than the sample containing the Bragg peak.
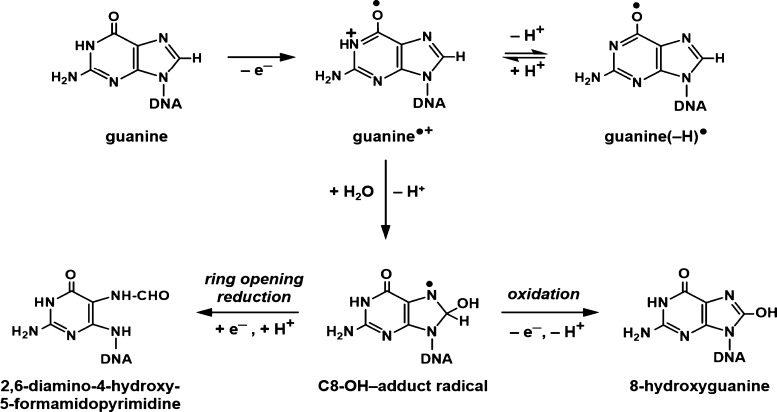
Yields of purine-derived DNA-base lesions, i.e., FapyGua, 8-OH-Gua, FapyAde, and 8-OH-Ade: the mechanism of the formation of the guanine-derived products is shown in Figure 10. In the first step of this mechanism, G•+ is formed by one-electron oxidation and is in prototropic equilibrium with G(−H)•, which is dominant at 77 K.2,3,13,38,39 Although we could not precisely determine the yields of trapped G(−H)•/G•+ in Ne-22 ion-beam-irradiated hydrated DNA, computer deconvolution of the ESR spectra of the first four samples depicted in Figure 4 indicated that G(−H)•/G•+ comprises ca. 25% of the trapped radicals at 77 K. On the other hand, the yield of products FapyGua and 8-OH-Gua that originate from G•+ and G(−H)• (Figure 10) adds to 13 ± 2% of the yield of trapped radicals at 77 K. This indicates that roughly half of the trapped G•+ is converted to FapyGua and 8-OH-Gua when samples are warmed. Since G•+ and G(−H)• radicals are in prototropic equilibrium, the complete conversion of G(−H)• to FapyGua and 8-OH-Gua is possible. Figure 10 also shows the mechanism of the formation of these products by OH– addition (addition of H2O followed by deprotonation) to G•+ giving rise to the C8–OH-adduct radical of Gua, which then undergoes one-electron oxidation to yield 8-OH-Gua or one-electron reduction to yield FapyGua, depending on experimental conditions.3,4,13,21 The formation of the adenine-derived lesions, 8-OH-Ade and FapyAde, occurs via analogous pathways from the adenine cation (A•+) radical. Extensive ESR spectral analyses of both γ-irradiated and ion-beam-irradiated (at 77 K) hydrated DNA samples have not led to any observable trapped A•+ radicals at 77 K. Nevertheless, the observed formation of 8-OH-Ade and FapyAde suggests that A•+ is formed upon Ne-22 ion-beam irradiation at 77 K at about 15% of G•+ levels based on the product yields. These levels are not in sufficient intensity to be resolved in the ESR spectra.
Figure 10.
Formation of FapyGua and 8-OH-Gua from the guanine cation radical, G•+.
Moreover, Gua has the lowest one-electron reduction potential (E7 = midpoint potential = 1.29 V4) and the lowest ionization energy among the DNA bases; as a consequence, the hole localizes on Gua.2−8,13,38−40,42 These properties of Gua also explain the yields of FapyGua and 8-OH-Gua being much greater than those of FapyAde and 8-OH-Ade in DNA samples that were Ne-22 ion-beam-irradiated at 77 K (Tables S3 and S4 in the Supporting Information). However, runs of Ade (i.e., sequences such as AAA) in DNA would likely have trapped A•+ which would account for the modest levels of products found for Ade.
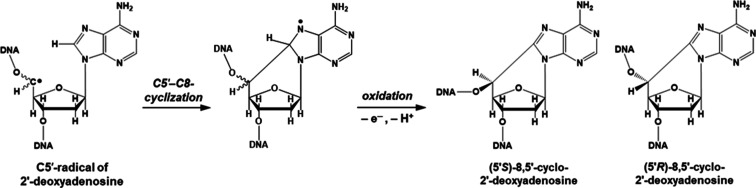
Formation of R-cdA, S-cdA, R-cdG, and S-cdG in Ne-22 beam-irradiated hydrated DNA: the 2′-deoxyribose radical, C5′• (Figure 2), has been detected previously in hydrated DNA exposed to γ- and ion-beam radiations.13,14 Using a benchmark ESR spectrum for C5′• (Figure S3 in the Supporting Information), it was estimated that C5′• makes up ca. 25% (ion-beam-irradiated) to ca. 40% (γ-irradiated) of the 2′-deoxyribose radicals (ΣdR•) trapped at 77 K.14 Since, for each ESR sample, the yield of 2′-deoxyribose radicals, as determined by spectral deconvolution using the ΣdR benchmark spectrum can be estimated (Figure 6), the yield of C5′• could also be estimated. Once formed, C5′• can undergo several reactions: (a) unaltered base release along with strand break formation,3,4,45 (b) 8,5′-intramolecular cyclization followed by one-electron oxidation, leading to the formation of R-cdA, S-cdA, R-cdG, and S-cdG,8−10 and (c) cross-link production.4,8−13 When compared with the measured yields of 8,5′-cyclopurine-2′-deoxynucleosides [Σ(R-cdA + S-cdA + R-cdG + S-cdG)], it is found that approximately (4.1 ± 1.1)% of the trapped C5′• reacts to form these products. The reactions leading to the formation of R-cdA and S-cdA from C5′• radicals in DNA are shown in Figure 11. The formation of R-cdG and S-cdG occurs via analogous reactions. It should be pointed out that R-cdA, S-cdA, R-cdG, and S-cdG were identified in ion-beam-irradiated DNA at 77 K for the first time in this work.
Figure 11.
Reactions leading from the C5′• of 2′-deoxyadenosine to R-cdA and S-cdA.
Our results show that for R-cdA, S-cdA, R-cdG, and S-cdG measured in DNA samples γ-irradiated in aqueous solutions and in irradiated cells at ambient temperature,9 the yields of the S-diastereomers were found to be greater than those of the R-diastereomers. This was confirmed in the present work for the yields of cdG and cdA in DNA γ-irradiated in aqueous solutions (Table S2 in the Supporting Information). However, hydrated DNA samples of Ne-22 ion-irradiated at 77 K or γ-irradiated at room temperature exhibited greater yields of the R-diastereomers than those of the S-diastereomers for both cdA and cdG (Tables S1 and S4 in the Supporting Information). This is quite an interesting phenomenon which is due to a stereoisomeric effect on the formation of the covalent bond between C5′• of 2′-deoxyribose and C8 of Ade or Gua in the same nucleoside in the hydrated DNA vs that of DNA in solution. Our ESR studies established that the conformations of C5′• in γ-irradiated hydrated DNA and in ion-beam-irradiated DNA are quite similar.13,14 This finding explains our observation of greater yields of R-diastereomers than those of the S-diastereomers for both cdA and cdG in hydrated DNA samples either γ-irradiated at room temperature or Ne-22 beam-irradiated at 77 K.
Based on these results, the R/S ratio of 8,5′-cyclopurine-2′-deoxynucleosides may be diagnostic for the direct or indirect effect of ionizing radiations. In addition, recent theoretical calculations46 predict that the R/S ratio of the radicals (Figure 11) leading to the formation of the R and S forms of cdG is ca. 2, and the corresponding R/S ratio of the diamagnetic product cdG is ca. 0.3. These results predict the trend observed in Table S2 and suggest that the R/S ratio of the radicals is maintained in the hydrated DNA in forming the diamagnetic cdG.
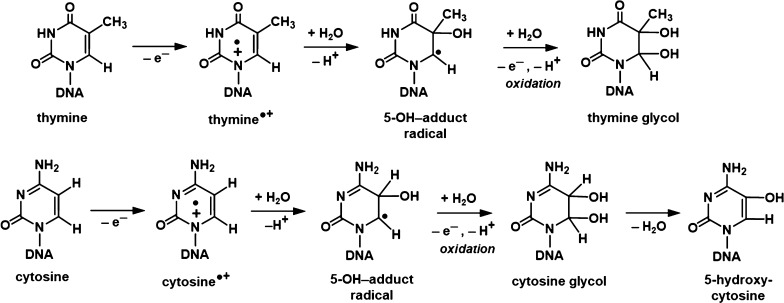
Formation of pyrimidine-derived lesions, ThyGly and 5-OH-Cyt: the hydration (OH– addition, i.e., H2O addition followed by deprotonation) of pyrimidine cation radicals or their deprotonated forms, i.e., T•+/T(−H)• and C•+/C(−H)•, gives rise to 5-OH-adduct radicals,47 the oxidation of which results in the formation of ThyGly and CytGly, respectively, as shown in Figure 12.4,8,48 The dehydration of CytGly leads to 5-OH-Cyt.4,8,48 Extensive analyses of ESR spectra of both γ-irradiated and ion-beam-irradiated hydrated DNA in the literature and in our results in Figure 4 do not indicate the existence of any observable line components due to any trapped pyrimidine cation radicals at 77 K.2−4,13,38−40,42 In fact, there is compelling evidence that any pyrimidine cation radicals that are initially formed in irradiated DNA quickly undergo hole transfer to guanine to form G•+.2−4,13,38−40,42 This indicates that the initial step shown in Figure 12, the addition of water to the pyrimidine cation radical, occurs on a relatively fast time scale, before completion of the hole transfer away from the cation radical can occur (ca. 10 ps).41 Water residents in the major and minor grooves of the DNA, with a residence time of ca. 1 ns,49 are likely candidates for this initial step. The identification of ThyGly and 5-OH-Cyt attests to the formation of T•+ and C•+ in Ne-22 beam-irradiated hydrated DNA (Figure 12). The fact that the yields of these products in Ne-22 beam-irradiated hydrated DNA are much lower than those of Gua-derived lesions (Table S3) may point to the fast hole transfer from T•+ and C•+ to Gua.2−4,13,38−40,42
Figure 12.
Formation of ThyGly from the thymine cation radical, T•+, and formation of 5-OH-Cyt from the cytosine cation radical, C•+.
Formation of pyrimidine-derived lesions 5,6-diHThy and 5,6-diHCyt: the mechanisms of the formation of 5,6-diHThy and 5,6-diHCyt are shown in Figure 13. These products originate with the pyrimidine anion radicals, T•– and C•–, which are formed by one-electron addition to Thy and Cyt, respectively, and which are part of the trapped radical cohort observed at 77 K (Figures 3, 4, and 5). It should be mentioned that 5,6-diHThy and 5,6-diHCyt have been identified previously in γ-irradiated hydrated DNA.50 They were identified in ion-beam-irradiated DNA at 77 K for the first time in this work. The yields of 5,6-diHCyt are 6–9 times those of 5,6-diHThy in the irradiated samples. Analysis of the percentages of C(N3)H•/C•– and T•– in Ne-22-irradiated hydrated DNA, at 77 K, indicated that the yields of C(N3)H•/C•–were higher than T•– yields, consistent with the finding of higher yields of 5,6-diHCyt relative to 5,6-diHThy.
Figure 13.
Formation of 5,6-diHThy from the thymine anion radical, T•–, and formation of 5,6-diHCyt from the cytosine anion radical, C•–.
Spatial positioning of 8,5′-cyclopurine-2′-deoxynucleosides and DNA-base lesions in the ion-beam track: based on the ESR spectroscopy results, it is possible to suggest the spatial positioning, in the ion track, of the products discussed in this work. R-cdA, S-cdA, R-cdG, and S-cdG are all thought to originate from C5′• (Figure 11), which is largely formed in the ion-beam core, which indicates that the cyclized products are largely located in the core. These helix-distorting tandem lesions are repaired by nucleotide excision repair and, if not repaired, can lead to adverse biological effects such as reducing transcription, blocking replication. They are also highly mutagenic, leading to G → A and A → G transitions and G → T and A → T transversions.10,34,51 The formation of C5′• radicals in the small volume of the ion-beam core is likely to lead to the formation of damage clusters and an increased difficulty in repair of the resulting 8,5′-cyclopurine-2′-deoxynucleosides, especially in clustered damage sites, which, in turn, suggests an increase in the cytotoxicity of these lesions in ion-beam-irradiated DNA, relative to low-LET radiation. Furthermore, owing to the large amount of energy deposited in the low volume core, clustered damage along the track core is an expected result.52 Unlike the 8,5′-cyclopurine-2′-deoxynucleosides, the DNA-base lesions originate from DNA-base radicals, which are formed almost entirely in the beam penumbra. Through both intra- and intertrack interactions, these products may also contribute to the formation of damage clusters. It is often conjectured that the dose in the penumbra decreases as 1/r2 from the center of the track, thus significant amounts of energy are deposited in the penumbra close to the core. DNA-base lesions formed near or in the core can, in a single track, also contribute to damage cluster formation. The DNA-base lesions identified in Ne-22 beam-irradiated DNA possess highly mutagenic and cytotoxic effects.53−55 All these DNA-base damage products may contribute to the mutagenic and cytotoxic effects of high-LET ion-beam irradiations in vivo.56
In summary, this work is the first to use the combination of ESR spectroscopy, LC–MS/MS and GC–MS/MS, and to report the formation of DNA products that had not been reported previously in DNA ion-beam-irradiated at 77 K. Our results show that both DNA radicals and DNA-base damage products formed via both oxidative and reductive pathways (e.g., Figure 10) increase along the track until just before the Bragg peak is reached; in addition, the DNA radical and product yields are reduced at the Bragg Peak. This work thus enables a better understanding of the mechanisms of radiation damage to DNA along the ion-beam track in terms of the formation of DNA radicals, the stable products that are formed from the radicals, and the location of products in the ion track.
Experimental Section
The experimental part is divided in sections, viz., (a) the materials used, (b) hydrated DNA sample preparation,14−17,19−21 (c) Ne-22 ion-beam and γ-irradiation (Figures S1 and S2, eqs S1 and S2),20−24 (d) electron spin resonance (ESR) spectroscopy and computer analyses,14−17,19−21 and (e) measurements of DNA lesions.25−37 All these are described in the Supporting Information.
Acknowledgments
Certain commercial reagents and instrumentation are identified throughout to adequately describe the experimental procedures. In no case does such an identification imply an endorsement by NIST, nor does NIST suggest that the materials or equipment so identified are necessarily the best available for the purposes described herein.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.1c01954.
Experimental section, depiction of a sample packet containing eight DNA samples, photograph of the plexiglass sample holder, benchmark ESR spectrum, eqs S1 and S2, yields of 8,5′-cyclopurine-2′-deoxynucleosides in Ne-22-irradiated DNA, yields of 8,5′-cyclopurine-2′-deoxynucleosides in DNA γ-irradiated in aqueous solutions or in the hydrated solid state, yields of DNA-base lesions in Ne-22-irradiated DNA, and yields of DNA-base lesions in γ-irradiated in aqueous solutions or in the hydrated solid state (PDF)
Author Contributions
All authors contributed to this work equally and the final version has been approved by all authors.
A.A., D.B., and M.D.S. thank the National Cancer Institute of the National Institutes of Health (grant RO1CA045424) for support. A.A. and M.D.S. thank REF, CBR at OU for support. D.B., M.D.S., and A.A. acknowledge the National Superconducting Cyclotron Laboratory (NSCL) at Michigan State University for its help and support. A.A. also acknowledges support by the National Science Foundation under grant no. CHE-1920110. The operation of the National Superconducting Cyclotron Laboratory at Michigan State University is supported by the NSF under grant PHY-1565546.
The authors declare no competing financial interest.
Supplementary Material
References
- Ebner D. K.; Kamada T. The Emerging Role of Carbon-Ion Radiotherapy. Front. Oncol. 2016, 6, 140. 10.3389/fonc.2016.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevilla M. D.; Becker D.; Kumar A.; Adhikary A. Gamma and ion-beam irradiation of DNA: Free radical mechanisms, electron effects, and radiation chemical track structure. Radiat. Phys. Chem. 2016, 128, 60–74. 10.1016/j.radphyschem.2016.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikary A.; Becker D.; Sevilla M. D.. Electron Spin Resonance of Radicals in Irradiated DNA. Applications of EPR in Radiation Research; Lind A., Shiotani M., Eds.; Springer-Verlag: Berlin, 2014; pp 299–352. [Google Scholar]
- von Sonntag C.Free-Radical-Induced DNA Damage and Its Repair; Springer: Heidelberg, 2006. [Google Scholar]
- Becker D.; Vere T. L.; Sevilla M. D. ESR detection at 77 K of the hydroxyl radical in the hydration layer of gamma-irradiated DNA. Radiat. Res. 1994, 140, 123–129. 10.2307/3578577. [DOI] [PubMed] [Google Scholar]
- Vere T. L.; Becker D.; Sevilla M. D. Yields of • OH in Gamma-Irradiated DNA as a Function of DNA Hydration: Hole Transfer in Competition with • OH Formation. Radiat. Res. 1996, 145, 673–680. 10.2307/3579357. [DOI] [PubMed] [Google Scholar]
- Sevilla M. D.; Becker D.; Razskazovskii Y. Electron and hole transfer within DNA and its hydration layer. Nukleonika 1997, 42, 283. [Google Scholar]
- Dizdaroglu M.; Jaruga P. Mechanisms of free radical-induced damage to DNA. Free Radic. Res. 2012, 46, 382–419. 10.3109/10715762.2011.653969. [DOI] [PubMed] [Google Scholar]
- Dizdaroglu M.; Coskun E.; Jaruga P. Measurement of oxidatively induced DNA damage and its repair, by mass spectrometric techniques. Free Rad. Res. 2015, 49, 525–548. 10.3109/10715762.2015.1014814. [DOI] [PubMed] [Google Scholar]
- Chatgilialoglu C.; Ferreri C.; Geacintov N. E.; Krokidis M. G.; Liu Y.; Masi A.; Shafirovich V.; Terzidis M. A.; Tsegay P. S. 5′,8-Cyclopurine Lesions in DNA Damage: Chemical, Analytical, Biological, and Diagnostic Significance. Cells 2019, 8, 513. 10.3390/cells8060513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romieu A.; Gasparutto D.; Cadet J. Synthesis and characterization of oligodeoxynucleotides containing the two 5R and 5S diastereomers of (5′S,6S )-5′,6-cyclo-5,6-dihydrothymidine; radiation-induced tandem lesions of thymidine. J. Chem. Soc., Perkin Trans. 1 1999, 1257–1264. 10.1039/a901812c. [DOI] [Google Scholar]
- Jaruga P.; Xiao Y.; Nelson B. C.; Dizdaroglu M. Measurement of (5′R)- and (5′S)-8,5′-cyclo-2′-deoxyadenosines in DNA in vivo by liquid chromatography/isotope-dilution tandem mass spectrometry. Biochem. Biophys. Res. Commun. 2009, 386, 656–660. 10.1016/j.bbrc.2009.06.107. [DOI] [PubMed] [Google Scholar]
- Becker D.; Kumar A.; Adhikary A.; Sevilla M. D.. Gamma- and Ion-beam DNA Radiation Damage: Theory and Experiment. DNA Damage, DNA Repair and Disease; Dizdaroglu M., Lloyd R. S., Eds.; Royal Society of Chemistry: London, 2020; Vol. 2, pp 426–457. [Google Scholar]
- Adhikary A.; Becker D.; Palmer B. J.; Heizer A. N.; Sevilla M. D. Direct Formation of the C5′-Radical in the Sugar-Phosphate Backbone of DNA by High-Energy Radiation. J. Phys. Chem. B 2012, 116, 5900–5906. 10.1021/jp3023919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker D.; Razskazovskii Y.; Callaghan M. U.; Sevilla M. D. Electron Spin Resonance of DNA Irradiated with a Heavy-Ion Beam ( 16 O 8+ ): Evidence for Damage to the Deoxyribose Phosphate Backbone. Radiat. Res. 1996, 146, 361–368. 10.2307/3579297. [DOI] [PubMed] [Google Scholar]
- Becker D.; Bryant-Friedrich A.; Trzasko C.; Sevilla M. D. Electron spin resonance study of DNA irradiated with an argon-ion beam: evidence for formation of sugar phosphate backbone radicals. Radiat. Res. 2003, 160, 174–185. 10.1667/rr3037. [DOI] [PubMed] [Google Scholar]
- Becker D.; Adhikary A.; Tetteh S. T.; Bull A. W.; Sevilla M. D. Kr-86 ion-beam irradiation of hydrated DNA: free radical and unaltered base yields. Radiat. Res. 2012, 178, 524–537. 10.1667/rr3066.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauptner A.; et al. Spatial Distribution of DNA Double-Strand Breaks from Ion Tracks, in Invited lectures presented at a symposium arranged by the Royal Danish Academy of Sciences and Letters, Copenhagen, 1– 5 May, Peter S., Ed.; The Royal Danish Academy of Sciences and Letters, Copenhagen, 2006. Matematisk-fysiske Meddelelser 52, Det Kongelige Danske Videnskabernes Selskab. [Google Scholar]
- Swarts S. G.; Sevilla M. D.; Becker D.; Tokar C. J.; Wheeler K. T. Radiation-Induced DNA Damage as a Function of Hydration: I. Release of Unaltered Bases. Radiat. Res. 1992, 129, 333–344. 10.2307/3578034. [DOI] [PubMed] [Google Scholar]
- Shukla L. I.; Pazdro R.; Becker D.; Sevilla M. D. Sugar Radicals in DNA: Isolation of Neutral Radicals in Gamma-Irradiated DNA by Hole and Electron Scavenging. Radiat. Res. 2005, 163, 591–602. 10.1667/rr3347. [DOI] [PubMed] [Google Scholar]
- Shukla L. I.; Adhikary A.; Pazdro R.; Becker D.; Sevilla M. D. Formation of 8-oxo-7,8-dihydroguanine-radicals in γ-irradiated DNA by multiple one-electron oxidations. Nucleic Acids Res. 2004, 32, 6565–6574. 10.1093/nar/gkh989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarasov O. B.; Bazin D. LISE++: Radioactive beam production with in-flight separators. Nucl. Instrum. Methods Phys. Res., Sect. B 2008, 266, 4657–4664. 10.1016/j.nimb.2008.05.110. [DOI] [Google Scholar]
- Ziegler J. F.; Ziegler M. D.; Biersack J. P. SRIM - The stopping and range of ions in matter (2010). Nucl. Instrum. Methods Phys. Res., Sect. B 2010, 268, 1818–1823. 10.1016/j.nimb.2010.02.091. [DOI] [Google Scholar]; http://www.srim.org
- Sümmerer K. Improved empirical parametrization of fragmentation cross sections. Phys. Rev. C 2012, 86, 014601. 10.1103/physrevc.86.014601. [DOI] [Google Scholar]; Erratum: Improved empirical parametrization of fragmentation cross sections [Phys. Rev. C86, 014601 (2012)]. Phys. Rev. C 2013, 87, 039903. 10.1103/physrevc.87.039903
- http://www.nist.gov/srm/index.cfm and https://www-s.nist.gov/srmors/view_detail.cfm?srm=2396.
- Dizdaroglu M.; Coskun E.; Jaruga P. Repair of oxidatively induced DNA damage by DNA glycosylases: Mechanisms of action, substrate specificities and excision kinetics. Mutat. Res., Rev. Mutat. Res. 2017, 771, 99–127. 10.1016/j.mrrev.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy P. T.; Jaruga P.; Kirkali G.; Tuna G.; Nelson B. C.; Dizdaroglu M. Identification and quantification of human DNA repair protein NEIL1 by liquid chromatography/isotope-dilution tandem mass spectrometry. J. Proteome Res. 2013, 12, 1049–1061. 10.1021/pr301037t. [DOI] [PubMed] [Google Scholar]
- Jaruga P.; Coskun E.; Kimbrough K.; Jacob A.; Johnson W. E.; Dizdaroglu M. Biomarkers of oxidatively induced DNA damage in dreissenid mussels: A genotoxicity assessment tool for the Laurentian Great Lakes. Environ. Toxicol. 2017, 32, 2144–2153. 10.1002/tox.22427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dizdaroglu M. The use of capillary gas chromatography-mass spectrometry for identification of radiation-induced DNA base damage and DNA base-amino acid cross-links. J. Chromatogr. 1984, 295, 103–121. 10.1016/s0021-9673(01)87602-0. [DOI] [PubMed] [Google Scholar]
- Dizdaroglu M. Application of capillary gas chromatography-mass spectrometry to chemical characterization of radiation-induced base damage of DNA: implications for assessing DNA repair processes. Anal. Biochem. 1985, 144, 593–603. 10.1016/0003-2697(85)90158-7. [DOI] [PubMed] [Google Scholar]
- Jaruga P.; Birincioglu M.; Rodriguez H.; Dizdaroglu M. Mass Spectrometric Assays for the Tandem Lesion 8,5′-Cyclo-2′-deoxyguanosine in Mammalian DNA. Biochemistry 2002, 41, 3703–3711. 10.1021/bi016004d. [DOI] [PubMed] [Google Scholar]
- Birincioglu M.; Jaruga P.; Chowdhury G.; Rodriguez H.; Dizdaroglu M.; Gates K. S. DNA base damage by the antitumor agent 3-amino-1,2,4-benzotriazine 1,4-dioxide (tirapazamine). J. Am. Chem. Soc. 2003, 125, 11607–11615. 10.1021/ja0352146. [DOI] [PubMed] [Google Scholar]
- Dizdaroglu M.; Jaruga P.; Rodriguez H. Identification and quantification of 8,5′-cyclo-2′-deoxy-adenosine in DNA by liquid chromatography/ mass spectrometry. Free Radical Biol. Med. 2001, 30, 774–784. 10.1016/s0891-5849(01)00464-6. [DOI] [PubMed] [Google Scholar]
- Jaruga P.; Dizdaroglu M. 8,5′-Cyclopurine-2′-deoxynucleosides in DNA: Mechanisms of formation, measurement, repair and biological effects. DNA Repair 2008, 7, 1413–1425. 10.1016/j.dnarep.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Nelson V. C. Synthesis of isotopically labelled DNA degradation products for use in mass spectrometric studies of cellular DNA damage. J. Labelled Compd. Radiopharm. 1996, 38, 713–723. . [DOI] [Google Scholar]
- White E.; Krueger P. M.; McCloskey J. A. Mass spectra of trimethylsilyl derivatives of pyrimidine and purine bases. J. Org. Chem. 1972, 37, 430–438. 10.1021/jo00968a023. [DOI] [PubMed] [Google Scholar]
- Reddy P. T.; Jaruga P.; Nelson B. C.; Lowenthal M.; Dizdaroglu M. Stable isotope-labeling of DNA repair proteins, and their purification and characterization. Protein Expression Purif. 2011, 78, 94–101. 10.1016/j.pep.2011.02.011. [DOI] [PubMed] [Google Scholar]
- Adhikary A.; Khanduri D.; Sevilla M. D. Direct Observation of the Hole Protonation State and Hole Localization Site in DNA-Oligomers. J. Am. Chem. Soc. 2009, 131, 8614–8619. 10.1021/ja9014869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikary A.; Sevilla M. D. Comment on “Theoretical Study of Polaron Formation in Poly(G)-Poly(C) Cations”. J. Phys. Chem. B 2011, 115, 8947–8948. 10.1021/jp202664j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z.; Sevilla M. D.. Studies of excess electron and hole transfers. Long Range Charge Transfer in DNA II; Schuster G. B., Ed.; Springer, 2004; Vol. 237, pp 103–127. [Google Scholar]
- Kumar A.; Becker D.; Adhikary A.; Sevilla M. D. Reaction of Electrons with DNA: Radiation Damage to Radiosensitization. Int. J. Mol. Sci. 2019, 20, 3998. 10.3390/ijms20163998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J.; Denisov S. A.; Adhikary A.; Mostafavi M. Ultrafast Processes Occurring in Radiolysis of Highly Concentrated Solutions of Nucleosides/Tides. Int. J. Mol. Sci. 2019, 20, 4963. 10.3390/ijms20194963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J.; Denisov S. A.; Marignier J.-L.; Pernot P.; Adhikary A.; Seki S.; Mostafavi M. Ultrafast Electron Attachment and Hole Transfer Following Ionizing Radiation of Aqueous Uridine Monophosphate. J. Phys. Chem. Lett. 2018, 9, 5105–5109. 10.1021/acs.jpclett.8b02170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J.; Kumar A.; Muroya Y.; Yamashita S.; Sakurai T.; Denisov S. A.; Sevilla M. D.; Adhikary A.; Seki S.; Mostafavi M. Quasi-free electron attachment to nucleoside in solution: excited anion radical formation and its dissociation. Nat. Commun. 2019, 10, 102. 10.1038/s41467-018-08005-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudgal M.; Rishi S.; Lumpuy D. A.; Curran K. A.; Verley K. L.; Sobczak A. J.; Dang T. P.; Sulimoff N.; Kumar A.; Sevilla M. D.; Wnuk S. F.; Adhikary A. Prehydrated One-Electron Attachment to Azido-Modified Pentofuranoses: Aminyl Radical Formation, Rapid H-Atom Transfer, and Subsequent Ring Opening. J. Phys. Chem. B 2017, 121, 4968–4980. 10.1021/acs.jpcb.7b01838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell M.; Kumar A.; Sevilla M. D. Electron-Induced Repair of 2′-Deoxyribose Sugar Radicals in DNA: A Density Functional Theory (DFT) Study. Int. J. Mol. Sci. 2021, 22, 1736. 10.3390/ijms22041736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikary A.; Kumar A.; Heizer A. N.; Palmer B. J.; Pottiboyina V.; Liang Y.; Wnuk S. F.; Sevilla M. D. Hydroxyl ion addition to one-electron oxidized thymine: Unimolecular interconversion of C5 to C6 OH-adducts. J. Am. Chem. Soc. 2013, 135, 3121–3135. 10.1021/ja310650n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dizdaroglu M.; Holwitt E.; Hagan M. P.; Blakely W. F. Formation of cytosine glycol and 5,6-dihydroxycytosine in deoxyribonucleic acid on treatment with osmium tetroxide. Biochem. J. 1986, 235, 531–536. 10.1042/bj2350531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood B. R. The importance of hydration and DNA conformation in interpreting infrared spectra of cells and tissues. Chem. Soc. Rev. 2016, 45, 1980–1998. 10.1039/c5cs00511f. [DOI] [PubMed] [Google Scholar]
- Swarts S. G.; Becker D.; Sevilla M.; Wheeler K. T. Radiation-induced DNA damage as a function of hydration. II. Base damage from electron-loss centers. Radiat. Res. 1996, 145, 304–314. 10.2307/3578986. [DOI] [PubMed] [Google Scholar]
- Chatgilialoglu C.; Krokidis M. G.; Masi A.. Oxidation of the C5′ Position in DNA and the Role of Purine 5′,8-Cyclo-2′-deoxynucleoside Lesions. DNA Damage, DNA Repair and Disease; Dizdaroglu M., Lloyd R. S., Eds.; Royal Society of Chemistry: London, 2020; Vol. 1, pp 117–139. [Google Scholar]
- Bowman M. K.; Becker D.; Sevilla M. D.; Zimbrick J. D. Track structure in DNA irradiated with heavy ions. Radiat. Res. 2005, 163, 447–454. 10.1667/rr3338. [DOI] [PubMed] [Google Scholar]
- Wallace S. S. Biological consequences of free radical-damaged DNA bases. Free Radical Biol. Med. 2002, 33, 1–14. 10.1016/s0891-5849(02)00827-4. [DOI] [PubMed] [Google Scholar]
- Fedeles B. I.; Essigmann J. M.. Mutational Spectra Provide Insight into the Mechanisms Bridging DNA Damage to Genetic Disease. DNA Damage, DNA Repair and Disease; Dizdaroglu M., Lloyd R. S., Eds.; Royal Society of Chemistry: London, 2020; Vol. 2, pp 214–253. [Google Scholar]
- Jasti V. P.; Das R. S.; Hilton B. A.; Weerasooriya S.; Zou Y.; Basu A. K. (5′S)-8,5′-Cyclo-2′-deoxyguanosine Is a Strong Block to Replication, a Potent pol V-Dependent Mutagenic Lesion, and Is Inefficiently Repaired inEscherichia coli. Biochemistry 2011, 50, 3862–3865. 10.1021/bi2004944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suman S.; Jaruga P.; Dizdaroglu M.; Fornace A. J. Jr.; Datta K. Heavy ion space radiation triggers ongoing DNA base damage by downregulating DNA repair pathways. Life Sci. Space Res. 2020, 27, 27–32. 10.1016/j.lssr.2020.07.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.