Abstract
In past 10 years, microRNAs (miRNAs) have gained scientific attention due to their importance in the pathophysiology of allergic diseases and their potential as biomarkers in liquid biopsies. They act as master post‐transcriptional regulators that control most cellular processes. As one miRNA can target several mRNAs, often within the same pathway, dysregulated expression of miRNAs may alter particular cellular responses and contribute, or lead, to the development of various diseases. In this review, we give an overview of the current research on miRNAs in allergic diseases, including atopic dermatitis, allergic rhinitis, and asthma. Specifically, we discuss how individual miRNAs function in the regulation of immune responses in epithelial cells and specialized immune cells in response to different environmental factors and respiratory viruses. In addition, we review insights obtained from experiments with murine models of allergic airway and skin inflammation and offer an overview of studies focusing on miRNA discovery using profiling techniques and bioinformatic modeling of the network effect of multiple miRNAs. In conclusion, we highlight the importance of research into miRNA function in allergy and asthma to improve our knowledge of the molecular mechanisms involved in the pathogenesis of this heterogeneous group of diseases.
Keywords: allergic disease, asthma, experimental models, microRNA, pollution
1. INTRODUCTION
The human body is constantly subjected to an onslaught of allergens and environmental irritants. These particles can trigger immune and inflammatory responses leading to a variety of alterations in gene expression. In recent years, the study of non‐coding RNAs has led to an increased understanding that gene regulation is more complex than previously imagined. 1 , 2 Among other mechanisms, small non‐coding microRNAs (miRNAs) have found themselves in the role of central regulators of post‐transcriptional gene expression. The details of miRNA molecular function, biogenesis, and processing have been thoroughly described in several reviews 1 , 2 , 3 , 4 and presented briefly in Figure 1. It should be highlighted that in mammalian cells miRNAs often initiate mRNA deadenylation followed by degradation of their target genes via imperfect binding to the 3′ untranslated regions of mRNAs. This imperfect binding leads to the suppression of multiple targets by one miRNA, while a single mRNA can be influenced by several miRNAs. There are several points to take into account when understanding miRNA nomenclature. 5 (a) Novel miRNAs are named sequentially and currently over 2500 are verified. There are naming exceptions for “historical” miRNAs such as let‐7 and lin‐4 which were first discovered in C. elegans. (b) miRNA clusters are areas where two or more miRNAs are transcribed from adjacent miRNA genes (eg, miR17~92). (c) miRNA strands are named ‐5p or ‐3p indicating if they originate from the 5′ or 3′ arm of the hairpin and either may be responsible for regulating cellular processes (Figure 1). Nowadays, technological advances such as real‐time PCR, microarray, and next‐generation sequencing have simplified the identification and validation of miRNAs, allowing for the exponential growth of investigation of miRNAs as regulatory molecules in numerous research areas.
FIGURE 1.
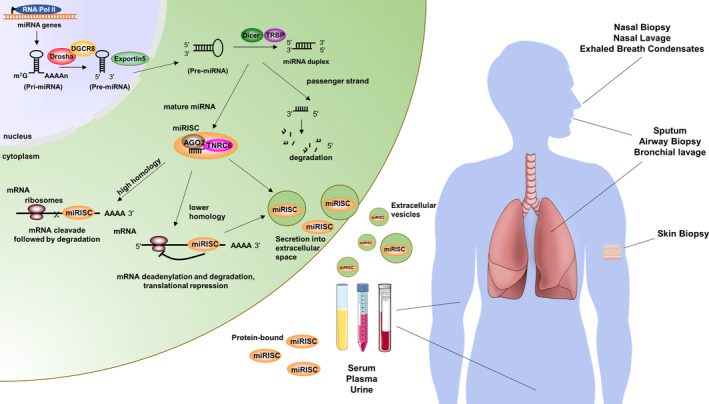
microRNA biogenesis and clinical sampling. (Left) miRNAs are transcribed in the nucleus by RNA polymerase II (RNA pol II) and processed by the enzymes Drosha and DiGeorge syndrome critical region 8 (DGCR8) from Pri‐ to Pre‐miRNA. Exportin5 acts to export Pre‐miRNAs to the cytoplasm where Dicer and the Dicer binding protein TRBP cut the hairpin to shorter duplexes. While the mature miRNA is incorporated into the miRNA induced silencing complex (miRISC) containing Ago2 and trinucleotide repeat‐containing gene 6 (TNRC6) proteins, the passenger strand is degraded. In the miRISC, the mature miRNA acts as a guide RNA for RISC proteins, of which Ago2 has the capacity to cleave mRNA if there is very high homology between the miRNA and the mRNA. If the homology is low, TNRC6 activity predominantly leads to deadenylation and degradation or translational repression of the target mRNA. In addition, miRNAs can be incorporated into different types of secretory vesicles and exit to the extracellular space. (Right) miRNAs are found in cells, tissues and fluids throughout the body. In lung diseases, miRNA levels in fluids, for example serum, plasma and urine (either protein‐bound, ie, free, or within extracellular vesicles), are often altered compared to healthy controls. This qualifies them to be used as non‐invasive biomarker for lung diseases. Other examples of clinical sampling that would allow for the identification of miRNAs in asthma, atopic dermatitis and allergic rhinitis are illustrated
Although miRNAs were first discovered nearly thirty years ago, their detailed role in the immune system has only begun to be elucidated in the past decade. While more thoroughly studied in cancer, recent research has reported alterations in miRNA expression in skin conditions and a variety of lung diseases, including, but not limited to: idiopathic pulmonary fibrosis, cystic fibrosis, chronic obstructive pulmonary disease, and asthma. 6 , 7 , 8 , 9 , 10 The use of experimental systems, such as cell culture and mouse models, has furthered our knowledge of the mechanistic role of miRNAs in airway hyper‐reactivity, allergy and immune responses. 2 , 4 , 11 , 12 , 13 Research into the role of miRNAs in allergy is expanding and many potential players have been identified in laboratory studies, but their actual role in human disease remains poorly understood.
This review highlights the recent steps toward a better understanding of the role of miRNAs in allergic diseases including atopic dermatitis (AD), allergic rhinitis (AR) and asthma. The importance of miRNA regulation in the pathogenesis of the aforementioned allergic diseases is supported by many studies, but also in other allergic diseases such as food allergy or chronic rhinosinusitis the evidence for a role of miRNAs is emerging. 4 , 29 Figure 2 provides an overview of miRNAs in cells and tissues that are associated with allergic diseases presented herein.
FIGURE 2.
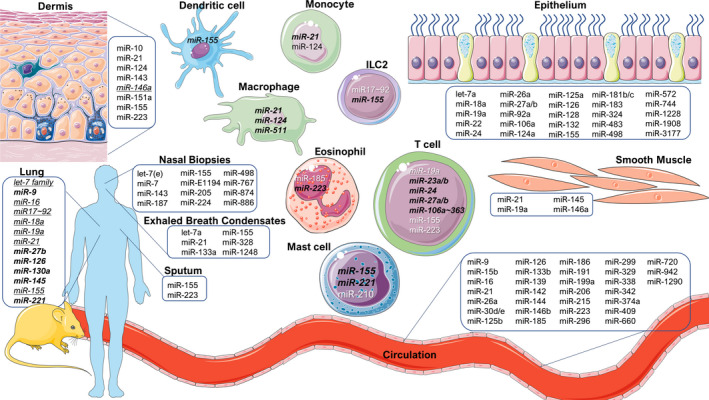
Overview of miRNAs discussed in this review. miRNAs are important regulators in allergic diseases. Herein, we provide an overview of the miRNAs described within the review and the cell types or organ systems where evidence of their actions has been reported. All miRNAs have been examined in human cells unless indicated inbold italics (mouse studies) or underlined italics (human and mouse)
2. HUMAN DISEASE
2.1. Atopic dermatitis
AD is a complex chronic inflammatory skin disease that is associated with skin barrier defects and activation of immune responses in the skin by environmental allergens and/or intrinsic factors. 30 Although type 2 inflammatory responses and elevated IgE are known as the main characteristics of AD, some patients actually develop stronger T helper cell (Th) Th17/Th22 responses. 31 Among other characteristic features, activation of keratinocytes plays an important role in AD. 30 , 32 Research on miRNAs in AD started with an array analysis of lesional skin samples from AD and psoriasis patients, representing, another inflammatory skin disease. The results of this early study suggested that alterations in miRNA levels in the skin of AD patients partially overlapped with that of psoriatic skin and included multiple miRNAs shown to be modulated in other inflammatory conditions. 33 For example, miR‐21 and miR‐146a were shown to be upregulated in the skin of psoriasis and AD patients. 33 miR‐146a was demonstrated to inhibit many pro‐inflammatory chemokines in keratinocytes through targeting multiple factors of the NF‐κB pathway 34 , 35 , 36 , 37 and miR‐146a‐deficient mice developed stronger inflammation in both AD and psoriasis models. 36 , 38 It has been shown that miR‐146a deficiency in mice leads to a defect in IgE production 39 , 40 and is linked to a Th1/Th17 skewing phenotype, 41 suggesting that miR‐146a is needed for the production of IgE and suppression of Th1/17‐cell‐mediated immune responses in mice. However, a negative relationship between miR‐146a and IgE levels was observed in serum samples from patients. 39 Therefore, the increased expression of miR‐146a in the case of allergic inflammation might have limited influence on type‐2 cell‐mediated immune responses in a subgroup of AD patients with increased IgE.
Another miRNA that may influence the development of AD through its function in the immune system is miR‐155. It was shown that miR‐155 is overexpressed in the skin of AD patients, likely due to infiltrating immune cells, and suggested that miR‐155 may influence the development of AD through the downregulation of cytotoxic T lymphocyte‐associated antigen 4 (CTLA4), a negative regulator of T‐cell activation. 37 In addition, miR‐155 expression was reported to positively correlate with AD severity, the number of Th17 cells, IL‐17 mRNA expression and IL‐17 plasma concentration, indicating that miR‐155 may influence AD pathogenesis through its effect Th17 cells. 42 , 43 The expression changes and effects in cell cultures of other miRNAs, including miR‐151a, ‐143, ‐124, and ‐10a, are reported and outlined in Table 1. Altogether, the studies of miRNAs in AD clearly demonstrate that miRNAs affect the severity of skin inflammation, modulate cellular responses of keratinocytes and specialized immune cells and thereby influence the pathogenesis of AD.
TABLE 1.
The functions of miRNAs in atopic dermatitis
| miRNA | Function | Targets | References |
|---|---|---|---|
| miR‐21 | Upregulated in AD | ND | 33 |
| miR‐146a | Upregulated in the skin and keratinocytes of AD patients; alleviates chronic inflammation in a mouse model of AD | IRAK1, CARD10, CCL5 | 34, 35, 36, 38, 39, 40, 175 |
| miR‐155 | May influence the development of AD by downregulating CTLA‐4 in T cells and by modulation of the development of Th17 cells | CTLA‐4, SOCS1 | 37, 42, 79 |
| miR‐151a | Altered in the blood plasma of AD patients, may contribute to Th2 skewing and pathogenesis of AD | IL12RB2 | 176 |
| miRNA‐143 | May reduce the influence of IL‐13 on epidermal keratinocytes | IL‐13Rα1 | 177 |
| miR‐124 | Suggested to decrease inflammation in chronic AD skin lesions | p65 | 178 |
| miR‐223 | Upregulated in whole blood of AD patients | ND | 148 |
| miR‐10a | Upregulated in AD skin, inhibits keratinocyte proliferation | MAP3K7, HAS3 | 179 |
ND—not determined in publications cited in the current table, may be described by other studies.
2.2. Allergic rhinitis
AR represents the most common allergic disease and is characterized by increased allergic symptoms along with circulating allergen‐specific IgE levels and/or positive skin prick test. This is triggered by various environmental allergens including pollen, molds, house dust mite and animal dander, resulting in a cascade of type 2 immune response in which type 2 cytokine production and eosinophil numbers are increased. AR, a condition of the upper airways, often coexists with inflammation of the lower airways. 44 , 45 Mucosal inflammation in AR and asthma shares many features, which has led to the “united airway concept” 46 and the idea that inflammation in AR can progressively extend to the lower airways. 47 Even though AR and asthma often co‐exist, many studies examining AR subjects aimed to uncover unique AR‐specific miRNA signatures (Table 2). Indeed, a subset of circulating miRNAs in plasma, miR‐206, ‐338‐3p, ‐329, and ‐26a, were found to be differentially expressed in patients with AR, but not in healthy individuals or those with asthma. Random forest model prediction suggested that a subset of six miRNAs allowed for high accuracy in distinguishing between these three groups. 48
TABLE 2.
miRNAs in allergic rhinitis
| miRNA | Function | Targets | Citations |
|---|---|---|---|
| let‐7 | Major regulatory mechanism for modulation IL‐13 secretion and thereby type‐2 inflammation | JAK1/STAT3, IL‐13, SOCS4 | 50, 140, 180 |
| miR‐206 | Regulator of the VEGF pathway | S100A7A | 48 |
| VEGF | |||
| miR‐338‐3p | Inhibitor of Wnt/β‐catenin signaling and inducer of epithelial‐mesenchymal transition | WNT/ β ‐Catenin | 48 |
| miR‐329 | Unknown | TGF‐β1 | 48 |
| miR‐26a | Modulation of TGF‐β‐dependent signaling pathways and repression of inflammatory responses by promoting regulatory T‐cell responses or through NF‐κB inhibition | SMAD2, SMAD3 | 48 |
| miR‐7 | Unknown | CMKLR1 | 49 |
| miR‐498 | Suppressing Th17 cell differentiation via STAT3 | STAT3 | 48, 50 |
| miR‐187 | Regulation of T‐cell response via CD276 | CD276 | 49, 50 |
| miR‐143 | Regulates memory T‐cell differentiation | TGF‐β1 | 49, 50 |
| miR‐886 | Regulates TGF pathway via SMAD3 | SMAD3, FoxO1 | 49, 50 |
| miR‐224 | Regulates TGF pathway via SMAD4 | SMAD4 | 49, 50 |
| miR‐155 | Important role in host defense, modulates IL‐13 pathway in macrophages determining the M2 phenotype | IL13Ra1 | 51, 52, 181, 182 |
| miR‐205 | Activation of ERK17 pathway | MICAL2 | 50 |
| miR‐572 | Regulates type‐1 cytokine expression | SOCS1 | 53 |
| miR‐1228 | Regulates type 2 responses | PPAR | 53 |
| miR‐483 | Inhibition of TGFβ1 | TGFβ1 | 53 |
| miR‐1908 | Inhibition of TGFβ1 |
SMAD2, SMAD3 MMP2 |
53 |
| miR‐126 | Counter‐regulation of IL4 effect | VEGF, IRS1 | 53 |
| miR‐92a | Regulation of IL4 effect | WNT5a | 53 |
| miR‐125a | Dampens TLR pathway via IL10, suppresses A20 | TLR, A20 | 53 |
| miR‐19a | Activates TGFβ signaling | TGFβ1 | 53 |
| miR‐106a | Regulation of autophagic activity | NOD1/2 | 53 |
| miR‐181c | Down‐regulates Osteopontin, modulating TGF | Osteopontin (SPP1) | 53 |
| miR18a | Regulating TGF pathway | CTGF | 48 |
In nasal biopsies, out‐of‐season AR patients displayed higher miR‐7 and miRPlus‐E1194 expression, whereas let‐7, miR‐498,‐187, ‐874, ‐143, ‐886, ‐224, and ‐767 were decreased compared to non‐allergic patients undergoing inferior turbinate surgery. 49 , 50 The reduced levels of let‐7e were confirmed by an additional study, which also showed increased levels of miR‐155, a miRNA involved in type 2 immune responses (see above 51 , 52 ), miR‐205 and miR‐498 in nasal biopsies of patients with current AR symptoms. 50 miR‐498 was also increased in the nasal mucosa of subjects suffering from perennial allergy, while miR‐18a expression was significantly lower in subjects with perennial allergy compared to subjects responding to seasonal allergens. 48
The diagnostic power of miRNAs was studied in relation to AR symptom severity (Total Nasal Symptoms Score) using microarray which revealed 3 down‐regulated (miR‐572, ‐1228‐, ‐483) as well as 9 up‐regulated miRNAs in nasal mucosa (miR‐1908, ‐126, ‐92a, ‐125a, 19a, 26a, 106a, ‐181c, ‐3177). Of the identified miRNAs, miR‐126, ‐19a, and ‐26a specifically and sensitively predicted AR disease activity in a receiver operating characteristic (ROC) analysis, 53 particularly when miR‐126, ‐19a, and 26a were used in combination. Taken together, several miRNAs were associated with AR, of which only let‐7 and miR‐26a were identified in independent studies.
2.3. Asthma
Asthma is a heterogeneous, chronic disease of the lower airways associated with airway hyper‐reactivity, bronchoconstriction, cough, wheeze and, in the majority of cases, inflammation. The most common and well‐studied form of asthma is the allergic type affecting both children and adults. Allergic asthma is associated with increased circulating allergen‐specific IgE levels and/or positive skin prick test and triggered by various allergens in addition to airway hyper‐reactivity. This allergen‐trigger leads to a cascading type 2 immune response in which type 2 cytokine production, eosinophil numbers and IgE levels are all increased. Given the vast heterogeneity of asthma sub‐phenotypes, this section will focus primarily on findings regarding the role of miRNAs in allergic asthma (Table 3).
TABLE 3.
miRNAs in asthma—studies using patient samples and cell cultures
| miRNA | Function or findings | Targets | Citations |
|---|---|---|---|
| miR‐15b, 126, ‐139,‐142, ‐186,‐191, ‐342, ‐374a, ‐409, ‐660, ‐942, ‐1290 | Circulating miRNAs (blood) correlating to lung function parameters in children | ND | 56 |
| miR‐16, ‐30d, ‐296 | Circulating miRNAs (blood) correlating to bronchial hyper‐responsiveness | ND | 57 |
| miR‐146a, ‐206, ‐720 | Circulating miRNAs (blood) used in combination as potential asthma prediction markers | ND | 58 |
| miR‐16, ‐125b, ‐133b, ‐206, ‐299 | Plasma miRNAs able to distinguish asthmatics from healthy individuals or those with allergic rhinitis | ND | 48 |
| let‐7a, miR‐21, ‐133a, ‐155, ‐328, ‐1248 | Decreased in exhaled breath condensates from asthmatic compared to healthy subjects | ND | 59 |
| miR‐21 | Dysregulated in circulation and lungs in allergic experimental murine models and human allergic asthmatics | ND | 9, 10, 11 |
| miR‐155 | Downregulated in the lymphocytes of allergic asthmatics during pollen season | ND | 60 |
| miR‐19a |
Increased in airway T cells. Reduction in smooth muscle cells leads to enhanced remodeling |
PTEN, A20 | 57, 61 |
| miR‐221 | Decreased levels in epithelial and sputum was associated with eosinophilic airway inflammation in asthma | ND | 64 |
| miR‐185 | Identified in circulating eosinophils as a distinguisher between healthy and asthmatic subjects. A potential predictor of asthma severity in blood sera | ND | 66 |
| miR‐16 | Negatively correlates to lung function parameters | ADRB2 | 70 |
| miR‐223, ‐513a and ‐625 | Downregulated in the blood of dust mite allergic asthmatics compared to healthy individuals | CBL, PPARGC1B, ESR1 | 55 |
| let‐7a | Abundant in the lungs and regulates IL‐13 expression | IL‐13 | 72 |
| miR‐1248 | Interacts with the 3′UTR to promote IL‐5 expression | IL‐5 | 73 |
| miR‐15a | Low levels in CD4+ T cells in pediatric asthma subject | VEGF | 74 |
| miR‐146a | Downregulated in bronchial brushing samples of asthma patients, inhibits IL‐8 and CXCL1 expression and neutrophil migration | IRAK1 | 153 |
| miR‐210 | Increases in human mast cells following IgE sensitization | ND | 183 |
| miR‐155, ‐146a, miR‐223, ‐374a | Serum miRNAs correlating to clinical parameters in asthma subgroups | ND | 65 |
ND—not determined in publications cited in the current table, may be described by other studies
Allergic asthma often begins in early life with up to half of adults reporting asthma symptoms in childhood. 54 Several studies examined the composition of miRNAs in the circulation and their potential as biomarkers. For example, 122 circulating miRNAs differentiated asthmatic from non‐asthmatic children. 55 The comprehensive inclusion of phenotypic characteristics in the Childhood Asthma Management Program (CAMP) studies allowed for the identification of miRNAs, through microarray analysis, that could potentially aid in the treatment of childhood asthma. 56 , 57 , 58 These miRNAs correlated to lung function parameters after stratification by sex (miR‐126, ‐139, ‐15b, ‐186, ‐342, ‐374a, ‐409, ‐660, ‐942, male associated and miR‐126, ‐1290, ‐142, 191, female associated) and to bronchial hyper‐responsiveness in response to methacholine challenge (miR‐296, ‐16, and ‐30d). Applying machine learning to miRNA expression and the clinical asthma score from the CAMP cohort, these studies suggested a combination of miRNAs as asthma prediction markers (miR‐146b, miR‐206 and miR‐720). 58
Studies in adult asthma have also identified numerous miRNAs that may assist in better identifying and understanding the disease. Significantly decreased expression of let‐7a, miR‐21, ‐133a, ‐155, ‐328, and ‐1248 were found in exhaled breath condensates from asthmatic individuals compared to healthy subjects. Furthermore, numerous type 2 mediators were predicted targets of these miRNAs, suggesting their role in asthma. 59 The dysregulation of miR‐21 in circulation and the airways has been commonly reported in allergic asthma, and thoroughly studied in humans and mice. 9 , 10 , 11 Certain miRNA expression was shown to be altered in a temporal manner with miR‐155 found to be downregulated in sputum lymphocytes from allergic asthmatics only during pollen season. 60 This raises the question as to which other miRNAs may be altered upon pollen exposure. miR‐19a, another miRNA often altered in asthma, has multiple roles in the asthmatic airway. 61 , 62 , 63 Increased expression of miR‐19a in airway T cells promoted type 2 cytokine production through direct targeting of Phosphatase and Tensin Homolog (PTEN) and TNF Alpha Induced Protein 3 (TNFAIP3) and reduced miR‐19a in the airway smooth muscle cells led to enhanced airway remodeling. 61 , 63 In a recent study, decreased epithelial and sputum miR‐221 were associated with eosinophilic airway inflammation in asthma. 64 Even out of the airways, miRNAs have shown potential as predictive markers in asthma. A study identified a set of plasma miRNAs, miR‐125b, ‐16, ‐299, ‐206, and ‐133b, that distinguished asthmatics from healthy individuals and subjects with AR. 48 Recent studies in blood serum used miRNAs to identify asthma subgroups 65 and another identified miR‐185, a circulating eosinophil derived miRNA, to be a predictor of asthma severity. 66
Numerous biomarker studies have been conducted to find both extracellular vesicle derived miRNAs from bronchoalveolar lavage (BAL) and cell‐specific miRNAs dysregulated in asthma. 67 , 68 , 69 Recently, more detailed studies have identified potential miRNA targets, suggesting that multiple signaling processes may be affected. A negative correlation between lung function parameters and miR‐16 in asthma was recently identified. 70 In silico analysis predicted Adrenoreceptor B‐2 (ADRB2), which is involved in bronchial smooth muscle contraction, as a target gene for miR‐16 and was later confirmed by luciferase assay. 70 Bioinformatic analysis of miRNA targets from the blood of house dust mite allergic asthmatic children revealed enrichment in the PI3K and NF‐κB pathways. More specifically, correlations were shown between their target miRNAs and three genes: the Cbl Proto‐Oncogene (CBL), PPARG Coactivator 1 Beta (PPARGC1B) and the estrogen receptor 1 (ESR1), suggesting that these pathways and genes have a role in asthma pathogenesis. 71 Additionally, miRNA expression in asthma has been correlated to expression and/or targeting of the type 2 cytokines, 59 , 61 IL‐13 72 and IL‐5, 73 as well as VEGF, 74 key molecules in asthma pathogenesis, strengthening the evidence for their role in the regulation of the disease. Having shown the alteration of a large set of miRNA in asthma calls for further investigation to mechanistically define their role(s) in asthma pathogenesis.
3. ENVIRONMENTAL FACTORS
3.1. miRNAs in the regulation of virus‐induced asthma exacerbation
Viruses affecting the respiratory system, such as human rhinoviruses (RVs), respiratory syncytial virus (RSV) and influenza, are known to cause serious illness and exacerbation in asthma patients. 75 , 76 , 77 When infecting human bronchial epithelial cells (HBECs), these viruses activate the NF‐κB pathway and interferon signaling in order to induce cellular responses, restrict virus replication and avoid tissue damage. It has been suggested that HBECs of asthmatic patients might have weakened interferon responses, resulting in increased viral propagation, enhanced activation of NF‐κB and immune responses and asthma exacerbations. 78 In this context, it can be envisioned that miRNAs targeting the NF‐κB pathway and influencing interferon signaling may have great potential to modulate cellular responses to respiratory viruses and influence the exacerbation of asthma. Accordingly, one of the earliest studies addressing the question of miRNA involvement in the regulation of viral responses showed an increase in viral replication of RV‐1B in HBECs when DICER was knocked down and, additionally, miR‐128 and ‐155 were inhibited. 79 Another study found that miR‐18a, ‐27a, ‐128, and ‐155 were downregulated in asthmatic HBECs and that simultaneous knockdown of these four miRNAs led to a significant increase in IL‐8 and IL‐6 expression. 80 Differences in the bronchial epithelium of asthmatic patients may also occur due to epigenetic changes. 81 miRNAs can influence genes involved in epigenetic regulation or modification and may also influence cellular responses to respiratory viruses. Indeed, a recent study demonstrated the upregulation of miR‐22 and downregulation of its target genes histone deacetylase (HDAC)4 and CD147 in response to influenza A virus H1N1 in bronchial epithelial cells from healthy subjects. However, cells from asthmatic patients were incapable of upregulating miR‐22 and showed increased and unchanged levels of HDAC4 and CD147, respectively. 82 Several additional studies suggest important functions for miRNAs in the regulation of cellular and immune responses to respiratory viruses (Table 4). Three miRNAs from different families (miR‐24, ‐124a, ‐744) all interfere with the p38MAPK pathway through the downstream kinases MK2 and Myc. MK2 and Myc are essential pro‐viral host factors and their downregulation by these miRNAs (or small interfering RNA [siRNAs]) confers broad‐spectrum antiviral activity against influenza A virus, RSV and adenovirus. 83 Recently more studies have utilized primary respiratory epithelial cultures, including air‐liquid interface cultures, clinical samples and in vivo mouse models 84 possibly leading to better clarification of the functions of miRNAs also during respiratory viral infections and virus‐induced asthma exacerbations.
TABLE 4.
miRNAs and respiratory viruses
| miRNA | Function | Targets | References |
|---|---|---|---|
| miR‐155 |
Inhibition results in increased replication of RV‐1B in HBECs. |
ND | 79, 83 |
| miR‐22 | May influence cellular responses to influenza A virus H1N1 in asthmatic HBECs |
HDAC4, CD147 (?) |
82 |
| miR‐155,‐27a, ‐18a, ‐128 |
Altered in asthmatic HBECs, simultaneous knockdown results in increased IL‐8 and IL‐6. Alter viral responses in bronchial epithelial cell line |
multiple | 79 |
| miRNA‐4776 | Downregulation of the NF‐κB inhibitor beta, increased. Influenza A virus survival in HBECs | NFKBIB | 184 |
| miR‐221 | Downregulated in response to RSV, inhibits viral replication and infectivity | NGF, TrKA | 185 |
| miR‐23b | Downregulates very low density lipoprotein receptor and thereby inhibits infection by minor group of RVs | VLDLR, LRP5 | 186 |
| miR‐136 | Increased in A549 human lung epithelial cells infected with H5N1 influenza A virus, upregulates IFN‐β | RIG‐I | 187 |
| miR‐29 | Induced in A549 cells by influenza A and PBMCs in influenza patients, induces COX2 and IFN‐λ | DNMT3A | 188 |
| miR‐29c | Induced by influenza in A549 cells, may contribute to virus‐mediated apoptosis, inhibits innate immune responses | BCL2L2 | 189, 190 |
| miR‐let‐7c | Upregulated in influenza infected A549 cells, may reduce virus replication | viral M1 | 191 |
| miR‐449b | Upregulated in influenza infected A549 cells, regulates antiviral cytokine signaling | HDAC1 | 192 |
| miR‐3145 | May inhibit influenza A virus replication | viral PB1 | 193 |
| miR‐485 | Prevents spurious activation of antiviral signaling, restricts influenza virus H5N1 infection | RIG‐I, viral PB1 | 194 |
| miR‐144 | Attenuates the host response to influenza virus by targeting the TRAF6‐IRF7 signaling axis | TRAF6 | 195 |
| miR‐324‐5p | Downregulated in A549 cells in response to infection with RNA viruses, enhanced type I and III interferons and interferon‐inducible genes | CUEDC2 | 196 |
|
miR‐24 miR‐124a miR‐744 |
Suppress influenza A (all) and RSV (miR‐124a, miR‐744) infection in A549 cells by inhibition of p38 MAPK expression and activation of MK2 |
P38MAPK MK2 |
83 |
| miR‐146a |
Increased in response to H3N2, targets TRAF6 in human nasal epithelial cells. Decreases IFN‐stimulated gene expression and enhances infection with influenza A virus in A549 cells |
TRAF6 TRAF6 |
|
| miRNA‐126a, miRNA‐16 and miRNA‐21 | Serum levels significantly lower during exacerbation visit as compared to follow‐up visit | ND | 199 |
ND—not determined in publications cited in the current table, may be described by other studies.
3.2. The role of air pollution in miRNA regulation
In addition to viral infections, air pollution and cigarette smoke exposure are important contributors to asthma development and/or exacerbations. 85 , 86 , 87 , 88 , 89 , 90 , 91 , 92 , 93 Air pollution is not only associated with aggravated type 2 responses, but can also lead to elevated neutrophil levels which are also a source of miRNAs. 94 Since exposure to air pollution alters miRNA expression both in the lungs and in blood (reviewed in Ref. 95), this could represent an important immunomodulatory mechanism in asthma. However, studies that investigate miRNA expression and function in association with air pollution and allergy or asthma are scarce (Table 5). In bronchial brushings from atopic individuals exposed to diesel exhaust and allergen, miR‐183, ‐324, and ‐132 expression was modulated by allergen exposure, but not by diesel exhaust. 96 Diesel exhaust exposure on the other hand increased expression of miR‐21, ‐30e, ‐215, and ‐144 in blood of mild asthmatics. Importantly, miR‐21 and miR‐144 expression was associated with increased oxidative stress markers and reduced antioxidant gene expression. 97 Increased miR‐155 in the serum of asthmatic children correlated with particulate matter level exposure. 98 Indirect exposure by maternal smoking reduced miR‐199a expression in cord blood. Interestingly, miR‐199a targets the receptor tyrosine kinase AXL, which is more methylated upon maternal smoking and the combination of maternal smoking and AXL methylation modifies the risk of childhood bronchitis symptoms. 99 Tobacco smoke exposure is also associated with increased miR‐223 expression in maternal and cord blood and with low numbers of regulatory T cells, which could be important in asthma development. 100 This miRNA was also identified in induced sputum of patients with severe asthma (both atopic and non‐atopic) and was associated with increased neutrophils. 101 In lung tissue of murine models with in utero smoke exposure combined with allergen challenge, the expression of miR‐221, ‐16, ‐155, ‐21, and ‐18a was increased, whereas miR‐130a expression was reduced compared to lungs challenged with allergen only. 102 , 103 Similarly, using miRNA arrays it was demonstrated that 133 miRNAs were dysregulated in fetal murine lungs upon maternal smoking. 104 Subsequent bioinformatic network analyses that included miRNAs and transcriptional regulators revealed insulin‐like growth factor (Igf‐1) as a major hub. Dysregulation of IgF‐1 was confirmed in PBMCs of healthy school‐aged children with early‐life smoke exposure. 104 In a murine cockroach asthma model, particulate matter exposure led to increased inflammatory responses, which were associated with elevated expression of miR‐206. This miRNA targets the antioxidant enzyme superoxide dismutase 1 (SOD1) leading to induction of oxidative stress, which may drive the pollution‐driven aggravated inflammatory response. 105 Expression analysis and functional experiments in epithelial cells (primary and cell lines) exposed to air pollution have revealed miRNA involvement in several processes that can be important in asthma, such as oxidative stress, apoptosis, autophagy, NF‐κB signaling and epithelial to mesenchymal transition (Figure 3). 106 , 107 , 108 , 109 , 110 , 111 , 112 , 113 , 114 , 115 , 116 , 117 , 118 miRNAs reported to be involved in chronic obstructive pulmonary disease (reviewed in Ref. 119, 120, 121) may also be important in asthma aggravated by air pollution. Although the actual involvement of these miRNAs in asthma remains to be further investigated, they could become interesting tools for exposure and risk assessment.
TABLE 5.
miRNA studies investigating the relation between pollution and allergy or asthma
| miRNA | Function or findings | Citations |
|---|---|---|
| Human data | ||
|
miR‐183 miR‐324 miR‐132 |
In controlled exposures in atopic subjects (exposure chamber), the miRNA expression was modulated by allergen exposure, but not additionally by diesel exposure | 96 |
|
miR‐21 (up) miR‐30e (up) miR‐215 (up) miR‐144 (up) |
In controlled exposures in asthma patients (exposure chamber), diesel exposure was associated with increased expression of miR‐21, miR‐30e, miR‐215, and miR‐144. miR‐144 and miR‐21 associated with systemic oxidative stress markers and negative correlation between miR‐144 and antioxidant genes |
97 |
| miR‐199a1 (down) | miR‐199a controls AXL (receptor kinase of the TAM (TYRO3, AXL, MERTK) family. Maternal smoking is associated with increased methylation of AXL and with reduced expression of miR‐199a. Combination of material smoking and increased AXL methylation alters the risk of childhood bronchitis symptoms | 99 |
| miR‐223 (up) | Prenatal tobacco exposure is associated with high miR‐223 expression in cord and maternal blood with low Treg numbers | 100 |
| Murine data | ||
|
miR‐221 (up) miR‐16 (up) miR‐130 (down) |
In a model cigarette‐aggravated allergic asthma (in utero side stream cigarette smoke, followed by Aspergillus fumigatus exposure), the altered miRNAs are associated with apoptosis and anti‐angiogenesis pathways | 102 |
|
miR‐155 (up) miR‐21 (up) miR‐18 (up) |
In a model cigarette‐aggravated allergic asthma (in utero side stream cigarette smoke, followed by Aspergillus fumigatus exposure), these miRNAs are positively associated with Type2 cytokines in bronchoalveolar lavage fluid | 103 |
FIGURE 3.
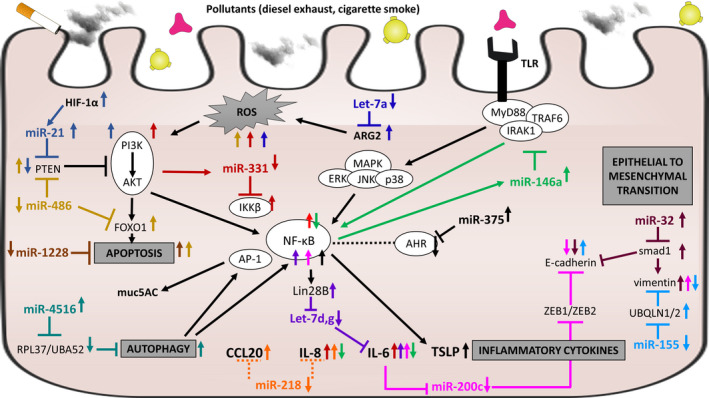
The effect of air pollution on miRNA networks and pathways in airway epithelial cells. Shown is a detailed schematic of miRNA action including known targets. Increased miRNA expression is indicated with upward red arrow, and decreased miRNA expression is indicated with downward blue arrow. Black arrows indicate a stimulatory effect on expression or process, and black line ending with perpendicular line indicates inhibitory effect. Smoke, pink and yellow figures represent air pollutants that may affect epithelial cells. Based on references 106, 107, 108, 109, 110, 111, 112, 113, 114, 115, 116, 117, 118
4. MECHANISTIC STUDIES
The availability of miRNA‐mimics, ‐inhibitors, and ‐knockout (KO) murine lines, in particular, have helped to delineate the impact of deregulated miRNA expression on disease pathology and revealed intricate interactions of altered miRNA‐regulation (Table 6). Mechanistic studies performed in murine models and human cell culture have broadened our understanding of miRNAs and have paved the way for future translational studies.
TABLE 6.
miRNAs and mouse models
| miRNA | Function | Target | Reference |
|---|---|---|---|
| miR‐21 | Induced by Th2 cytokines in DC and macrophages and promotes type 2‐ driven inflammation | IL‐12p35 | 130, 131, 132, 133 |
| miR‐126 | Induced in airway wall and promotes type 2 inflammation | TOM1 | 128, 129 |
| let‐7a‐e | Downregulated in CD4+ T cells and suppresses type 2 inflammation | IL‐13 | 140 |
| miR‐ 145 | Induced by allergen exposure and promotes type 2 inflammation | 139 | |
| miR‐155 |
Induced in ILC2 in type 2 inflamed airways and neutralization ameliorates experimental asthma phenotype. Involved in Th2‐mediated airway inflammation. Regulates mast cell activity |
S1pr1 PU.1(?) PI3Kγ pathway(?) |
41, 122, 123, 124, 125, 200 |
| miR‐23~27 | Type 2 cells lacking this miR‐cluster express elevated type 2 cytokines | Gene network regulating IL‐4 | 11, 138 |
| miR‐17‐92 | Mice deficient in this miR‐cluster develop an augmented experimental asthma phenotype | 126 | |
| miR‐19a | Upregulated in asthmatic airways and promotes experimental asthma | SOCS1/A20 | 61 |
| miR‐19b | Downregulated in asthmatic airways. Exogenous delivery of miR‐19b mimics ameliorates experimental asthma | TSLP | 127 |
|
miR network miR‐27b (up) miR‐206 (down) miR‐106b (down) miR‐203 (down) miR‐23b (up) |
A miR‐network is induced in lung‐resident type 2 cells and comprises a combination of induced miRs‐27b and ‐23b as well as silenced miR‐206, miR106b, and miR‐203. Antagonism of expression levels reduces type 2 cytokine expression | Fine tuning of multiple pathways, that suppress inhibitory signals and allow activation and survival of type 2 cells | 164 |
| miR‐1 | Downregulated by VEGF. Intranasal miR‐1 delivery inhibited inflammatory responses in experimental asthma models | Mpl | 201, 202 |
| miR‐221 | Influences effector functions and actin cytoskeleton in mast cells | ND | 203 |
ND—not determined in publications cited in the current table, may be described by other studies.
4.1. miRNAs in innate immune responses in allergic airway inflammation
The exposure to allergens, for example, house dust mite (HDM), is an important trigger for changes in lung‐specific miRNA expression. Allergen contact triggers epithelial release of IL‐33, which in turn tightly controls the activation and proliferation of innate lymphoid type 2 cells (ILC2). ILC2s provide early release of type2‐promoting IL‐5 and IL‐13 and initiate allergic airway inflammation. Studies in miRNA‐KO mice revealed the importance of miRNAs in these early pathogeneic events. Mice deficient in miR‐155 exposed to allergen had reduced levels of IL‐33 in the airways post allergen challenge and lower ILC2 numbers compared to wild‐type mice, revealing a critical role in inducing ILC2 proliferation. 122 In dendritic cells (DC), lack of miR‐155 led to reduced chemotaxis and type 2‐priming capacity, resulting in ameliorated hallmarks of experimental asthma. 41 , 123 , 124 , 125 A similar phenotype was observed for ILC2s recovered from miR‐17~92 cluster deficient mice. These cells were found to be defective in growth and cytokine expression in response to IL‐33 and thymic stromal lymphopoietin (Tslp). 126 However, further studies revealed the complexity within miRNA‐clusters, showing that individual family members can have opposing roles. For example, within the miR‐17~92 cluster, the family member mir‐19a was found to be elevated in allergic inflammation and shown to promote IL‐5 and IL‐13 production by targeting the known inhibitors SocS1 and Tnfaip3. 126 In contrast, miR‐19b was downregulated in allergic inflammation and shown to target Tslp. Treatment with miR‐19b was able to reduce allergic inflammation, providing evidence for a suppressive role and limiting type 2‐inflammation. 127 Another miRNA induced in the murine airway upon allergen contact and Toll‐like receptor (TLR) signaling was miR‐126. Inhibition of miR‐126 using antagomirs was sufficient to suppress the inflammatory response, implicating a prominent role in driving type 2 inflammation. 128 , 129 The inflammatory milieu in the lung induced the expression of miR‐21 in cells of the monocyte/macrophage lineage and structural cells. miR‐21 targets IL‐12p35 mRNA and thereby critically controls the type 1/type 2 balance in type 2‐high (Ovalbumin [OVA], HDM, Aspergillus fumigatus) and steroid‐insensitive experimental asthma. 130 , 131 , 132 , 133 Furthermore, it was shown that inhibition of miR‐9 in experimental allergic, steroid resistant asthma models restored the steroid sensitivity via targeting protein phosphatase 2 regulatory subunit B (B56) δ isoform (Ppp2r5d). 134 miR‐9 was also increased in the sputum of patients with neutrophilic asthma, which is often associated with steroid resistance. Additionally, miRNAs seem to control macrophage differentiation into their intrinsic sub‐phenotypes. Along this line, miR‐511 was increased in alternatively activated macrophages, but decreased in pro‐inflammatory macrophages. 135 A similar study identified an upregulation of miR‐124 in alternatively activated macrophages 136 and in CD14+CD16+ monocytes of patients with asthma compared to controls.
4.2. miRNAs controlling adaptive immunity in experimental asthma
In vitro studies in CD4+ T cells revealed a dynamic change of miRNA expression upon activation of cells and polarization into specialized CD4+ T‐cell subsets. 137 Some miRNAs involved in controlling the polarization process are encoded in the polycistronic clusters Mirc11 and Mirc22, comprising miRs‐23(a/b), ‐24 and ‐27(a/b). Bioinformatic analyses revealed several genes in a network upstream of IL‐4 to be among the targets for these miRNAs. In an acute model of experimental asthma, mice bearing CD4+ T cells deficient in these miRNAs developed an augmented type 2 response, including high type 2‐cytokine levels and elevated eosinophil numbers in BAL. 11 , 138 Conversely, miR‐145 expression was found induced in inflamed lungs and seemed to actively promote and sustain the inflammatory process. Indeed, blockade by antagomirs suppressed the production of IL‐5 and IL‐13 in the lungs and inhibited the inflammatory phenotype to an extent equal to dexamethasone. 139
Once established, the allergic phenotype is thought to stabilize and reinforce itself by IL‐13 production in the inflamed environment. Cellular control mechanisms, that restrict IL‐13 expression in the airways, seem to be suppressed in allergic airway inflammation and include the involvement of miRNAs. One example is the let‐7 family of miRNAs, 140 all of which were found to be downregulated in OVA‐induced experimental asthma. Exogenous delivery of let‐7 limited eosinophil recruitment and histopathological alterations and airway‐reactivity to methacholine. 140 The let‐7 miRNA family is also very abundant in the lung and their inhibition in vivo ameliorated murine experimental asthma. 72 However, in an additional study, let‐7a was shown to inhibit IL‐13 expression by directly targeting Il‐13 140 in vitro and exogenous administration of a let‐7 microRNA mimic alleviated asthmatic features in a mouse model of asthma. Additional examples that are both downregulated in OVA‐induced asthma lung tissue are miR‐133a and miR‐488 which directly target the genes IGF‐1 receptor (Igf1r) 141 and Tgfβ1, 142 respectively. Furthermore, overexpression of both miRNAs was able to reduce remodeling associated genes.
4.3. Cell‐based functional studies
Several studies investigated miRNA‐based molecular mechanisms ex vivo/in vitro in different cell types involved in asthma pathogenesis. 143 , 144 , 145 , 146 , 147 , 148 miR‐155 was shown to be induced by hyper‐stretch in human bronchial epithelial cells 149 and targets Src homology 2 domain–containing inositol 5‐phosphatase 1 (SHIP1) production and activates Janus Kinase (JNK) signaling leading to KC (the functional IL‐8 paralog) secretion in mouse models. miR‐181b was decreased in bronchial brushings and plasma from patients with asthma and inversely correlated with eosinophil counts in sputum. 150 Overexpressing this miRNA in a bronchial epithelial cell line (BEAS‐2B) confirmed the regulation of the target Secreted Phospho Protein 1 (SPP1) and reduced IL‐13 induced secretion of IL‐1β and CC‐Motif Chemokine Ligand 11 (CCL11). In this line, miR‐181b was induced following addition of dexamethasone. Further, miR‐27b has been described to be decreased in HDM induced experimental asthma, with a proposed function in the regulation of the PI3K‐AKT pathway via targeting Spleen Associated Tyrosine Kinase (SYK) and Epidermal Growth Factor Receptor (EGFR) in a bronchial epithelial cell line (16‐HBE). 55 In concordance to its effect in skin keratinocytes, 35 , 36 miR‐146a was shown to have anti‐inflammatory function in human lung alveolar epithelial cell line A549 151 , 152 and in HBECs. 68 , 153 , 154 , 155 , 156 , 157 Interestingly, it was recently shown that primary human airway epithelial cells secrete miRNAs in extracellular vesicles (EV) 154 as a means of intercellular communication by functionally transferring miRNAs to recipient cells. 155 The miRNA content of epithelial EVs was altered when the cells were treated with IL‐13, suggesting a differential regulation of target genes in recipient cells. Further, EVs isolated from BALF 68 or plasma 156 of patients with asthma contained different miRNAs compared to healthy individuals. This ability of miRNAs to be transferred between different cells adds another layer of complexity to their involvement in perpetuating anasthmatic response in the airways. 157
Besides epithelial cells, several studies have investigated miRNA‐regulated mechanisms in airway smooth muscle cells (hASMCs). In vitro stimulation of hASMCs with a cytokine cocktail (IL‐1β, TNF‐α, IFN‐γ) caused an increase in miR‐146a with the observed effect being stronger in asthmatic donor cells than healthy controls. 158 As inhibition of miR‐146a increases cyclooxygenase‐2 (COX‐2) levels and IL‐1β secretion by hASMCs, the authors suggested that miR‐146a may be an interesting anti‐inflammatory factor in asthma. In line with this study, upregulation of miR‐145 in hASMCs was demonstrated upon cytokine stimulation and was associated with enhanced migration and proliferation in vitro. 159 Inhibition of miR‐145 reversed this effect through the reduced expression of collagen type I and contractile protein MHC via targeting of Krüppel‐like factor 4 (KLF‐4). Finally, miR‐21 was shown to modulate hASMCs proliferation in vitro, via targeting PTEN, as identified by lentiviral overexpression experiments. 160 miR‐21 has been previously associated with asthma development, mainly due its targeting of, that is, IL‐12p35, 130 , 161 highlighting the multi‐functional roles of miRNAs in several cell types contributing synergistically to asthma pathology.
4.4. miRNA effects in gene networks
miRNA expression analyses from isolated cells, as well as in tissues from disease models, revealed simultaneously altered expression for several miRNAs. This implicates several parallel regulatory events which are not captured by traditional miRNA‐single target gene identification methodologies. Recently, network methods have been utilized to assess the outcome of miRNA‐regulation from a global perspective, revealing possible relationships between the miRNA‐targets and affected biological pathways. An example of such comprehensive regulatory miRNA‐mRNA networks has been simulated for in vitro differentiated Th17 cells. Compared to naive CD4+ T cells, Th17 cells expressed lower levels of miR‐106a, miR‐18b and miR‐363 all belonging to the miR‐106a~363 cluster. 162 Overexpression of the aforementioned miRNAs led to decreased expression of their confirmed target genes Nuclear Factor of Activated T cells (Nfat5), RAR related Orphan Receptor C (Rorc), and Rorα; entailing decreased Th17 differentiation and IL‐17 secretion, therefore identifying this miRNA cluster as a potential target for Th17‐mediated inflammation. Th17 cell differentiation is also controlled by the miR‐17~92 cluster, in particular by miR‐18a, 163 which targets Smad4, hypoxia‐inducible factor 1α (Hif1α), and Rorα. Thus, miR‐18a deficiency enhances Th17 differentiation in vitro and increases Th17 cells in tissue in experimental asthma models in vivo.
Another approach identified a distinct miRNA‐expression pattern in tissue‐resident type 2 cells in experimental allergic asthma. 164 Compared to naïve CD4+ T cells, type 2 cells in inflamed lungs displayed a strong downregulation of miRNAs 164 and this expression pattern changed with the transition from acute to chronic airway inflammation. Integrating gene and miRNA expression using a network approach, revealed distinct disease stage specific gene‐miRNA networks. 164 Pathogenic type 2 responses were predicted to result from combined and cumulative miRNA activities. Type 2 inflammation was predicted to result from elevated miR‐27b and miR‐23b, targeting immune regulatory Tgfb1 and Egfr pathways on the one hand and on the other hand, reduced expression of miR‐206, miR‐106b and miR‐203, allowing for the expression of genes involved in immune activation. Antagonizing this ex vivo miRNA‐expression pattern in vitro using miRNA‐inhibitors and mimics suppressed IL‐13 expression in Th2 cells and supported the in silico predictions of an intricate network of miRNA regulation. 164
Notably, we have mainly discussed the influence of miRNAs in the regulation of the immune response in allergic diseases. However, as individual miRNAs influence a wide range of targets and pathways, it is only logical to assume that several other pathways, such as, for example, metabolic pathways, might be affected at the same time and are also relevant for asthma pathogenesis.
5. CONCLUSION
miRNA research, thus far, has led to a breadth of information and long lists of potentially interesting miRNAs, but mechanistic studies of miRNA targeting and function are only beginning to emerge. Furthermore, it is unlikely that one miRNA alone holds the key to explain the pathology of asthma or allergic diseases. More likely, there are numerous players and complex networks of interactions that lead not only to disease pathogenesis, but also to heterogeneity, making mechanistic insight into the roles of miRNAs all the more important going forward. We propose that understanding common triggers that change miRNA expression in distinct cell populations, at defined disease stages and in specific phenotypes, together with assessing the net effects of miRNAs will help to decipher the pathophysiological consequences of altered miRNA expression in allergic diseases. Nonetheless, we have provided important evidence highlighting a crucial role of miRNA in the pathogenesis of asthma and allergic disease, making them interesting targets for clinical investigations. As asthma and allergy are very heterogeneous conditions and because current treatments are still inefficient in controlling severe forms of these diseases, more individualized and effective therapies are in a great demand. 165 , 166 , 167 Interest in novel therapeutics strategies to target single miRNAs 128 , 129 , 139 , 168 , 169 , 170 is increasing as well as the interest in using miRNA profiles as biomarkers for (lung) disease. 48 , 58 , 65 , 66 , 171 , 172 , 173 , 174 To implement the use of miRNAs as biomarkers, they should be specific, have the capacity to predict disease phenotypes, and be easily detectable in body fluids. Several studies mentioned in this review (eg, CAMP study) demonstrate that miRNAs indeed have this potential. We will address the challenges and potential pitfalls associated with the use of miRNAs as biomarkers in a future review.
CONFLICT OF INTEREST
Dr Weidner has nothing to disclose. Dr Bartel reports grants and personal fees from Bencard Allergie GmbH, outside the submitted work. Dr Kilic has nothing to disclose. Dr Zissler has nothing to disclose. Dr Renz has nothing to disclose. Dr Schwarze reports personal fees from MYLAN, personal fees from F2F events, outside the submitted work; and Industry support to educational activities of the Scottish Allergy and Respiratory Academy and of the Children's and Young people's Allergy Network Scotland. Industry support to EAACI, he is EAACI Secretary General 2019‐2021. Prof. Schmidt‐Weber reports grants from DFG, grants from BMBF, grants from EIT Health, and grants from German Center of lung research, outside the submitted work. Dr Maes reports grants from Ghent University, grants from Fund for Scientific Research in Flanders, during the conduct of the study; personal fees from GlaxoSmithKline, outside the submitted work; and is shareholder from Oryzon Genomics and Mendelion Lifesciences SL. Dr Rebane has nothing to disclose. Dr Krauss‐Etschmann has nothing to disclose. Dr Rådinger reports grants and personal fees from AstraZeneca outside the submitted work.
GLOSSARY
AGO2—a member of the Argonaute family of proteins that plays a central role in RNA silencing through the binding of small RNAs such as miRNAs.
DCGR8—a nuclear protein required for mRNA processing. DCGR8 binds to Drosha and forms a complex that cleaves the primary transcript known as pri‐miRNA into a characteristic stem‐loop structure known as a pre‐miRNA.
Drosha—a nuclear RNase III that cleaves primary miRNAs (pri‐miRNAs) to release hairpin‐shaped pre‐miRNAs.
Dicer—a cytoplasmic RNase III that cuts the pre‐miRNA to form mature miRNA.
pre‐miRNA hairpin—precursor‐miRNAs (pre‐miRNA) refer to the hairpin precursors of miRNAs formed by the cleavage of primary miRNAs by DCGR8 and Drosha.
miRNA duplex—the sense and antisense strand of the pre‐miRNA hairpin that is cleaved to a double stranded RNA. One strand will ultimately be degraded.
miRISC—The RNA‐induced silencing complex is a ribonucleoprotein complex comprised of proteins such as Dicer, Ago2 and TRBP and miRNA. The miRISC will target an mRNA for post‐transcriptional regulation.
Polycistronic clusters—a primary transcript encoding more than one miRNA or mRNA.
Antagomirs—a class of chemically engineered oligonucleotides that are used to silence endogenous miRNA.
Biomarker—A biological molecule found in fluid or tissue that allows for the identification of a specific condition/disease.
ACKNOWLEDGMENTS
We would like to thank the European Academy of Allergy and Clinical immunology for financing the EAACI task force "microRNAs in allergy and asthma."
Weidner J, Bartel S, Kılıç A, et al. Spotlight on microRNAs in allergy and asthma. Allergy.2021;76:1661–1678. 10.1111/all.14646
This review is an output from the EAACI task force “microRNAs in allergy and asthma.”
REFERENCES
- 1. Bartel DP. Metazoan microRNAs. Cell. 2018;173(1):20‐51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mehta A, Baltimore D. MicroRNAs as regulatory elements in immune system logic. Nat Rev Immunol. 2016;16(5):279‐294. [DOI] [PubMed] [Google Scholar]
- 3. Gebert LFR, MacRae IJ. Regulation of microRNA function in animals. Nat Rev Mol Cell Biol. 2019;20(1):21‐37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Johansson K, Weidner J, Radinger M. MicroRNAs in type 2 immunity. Cancer Lett. 2018;425:116‐124. [DOI] [PubMed] [Google Scholar]
- 5. Ambros V, Bartel B, Bartel DP, et al. A uniform system for microRNA annotation. RNA. 2003;9(3):277‐279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Alipoor SD, Adcock IM, Garssen J, et al. The roles of miRNAs as potential biomarkers in lung diseases. Eur J Pharmacol. 2016;791:395‐404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mestdagh P, Vandesompele J, Brusselle G, Vermaelen K. Non‐coding RNAs and respiratory disease. Thorax. 2015;70(4):388‐390. [DOI] [PubMed] [Google Scholar]
- 8. Ameis D, Khoshgoo N, Iwasiow BM, Snarr P, Keijzer R. MicroRNAs in lung development and disease. Paediatr Respir Rev. 2017;22:38‐43. [DOI] [PubMed] [Google Scholar]
- 9. Booton R, Lindsay MA. Emerging role of microRNAs and long noncoding RNAs in respiratory disease. Chest. 2014;146(1):193‐204. [DOI] [PubMed] [Google Scholar]
- 10. Dissanayake E, Inoue Y. MicroRNAs in allergic disease. Curr Allergy Asthma Rep. 2016;16(9):67. [DOI] [PubMed] [Google Scholar]
- 11. Pua HH, Ansel KM. MicroRNA regulation of allergic inflammation and asthma. Curr Opin Immunol. 2015;36:101‐108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rebane A. microRNA and allergy. Adv Exp Med Biol. 2015;888:331‐352. [DOI] [PubMed] [Google Scholar]
- 13. Weidner J, Malmhäll C, Rådinger M. microRNAs in asthma pathogenesis – from mouse to man. J Transl Genet Genom. 2019;3:2. 10.20517/jtgg.2018.30 [DOI] [Google Scholar]
- 14. D'Argenio V, Del Monaco V, Paparo L, et al. Altered miR‐193a‐5p expression in children with cow's milk allergy. Allergy. 2018;73(2):379‐386. [DOI] [PubMed] [Google Scholar]
- 15. Liu ZQ, Yang G, Geng XR, et al. Micro RNA‐17‐92 cluster mediates interleukin‐4‐suppressed IL‐10 expression in B cells. Am J Transl Res. 2016;8(5):2317‐2324. [PMC free article] [PubMed] [Google Scholar]
- 16. Yang LT, Li XX, Qiu SQ, et al. Micro RNA‐19a suppresses thrombospondin‐1 in CD35(+) B cells in the intestine of mice with food allergy. Am J Transl Res. 2016;8(12):5503‐5511. [PMC free article] [PubMed] [Google Scholar]
- 17. Larsen LF, Juel‐Berg N, Hansen A, et al. No difference in human mast cells derived from peanut allergic versus non‐allergic subjects. Immun Inflamm Dis. 2018;6(4):416‐427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Callejas‐Díaz B, Fernandez G, Fuentes M, et al. Integrated mRNA and microRNA transcriptome profiling during differentiation of human nasal polyp epithelium reveals an altered ciliogenesis. Allergy. 2020;75:2548‐2561. [DOI] [PubMed] [Google Scholar]
- 19. Cheng J, Chen J, Zhao Y, Yang J, Xue K, Wang Z. MicroRNA‐761 suppresses remodeling of nasal mucosa and epithelial‐mesenchymal transition in mice with chronic rhinosinusitis through LCN2. Stem Cell Res Ther. 2020;11(1):151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gu X, Yao X, Liu D. Up‐regulation of microRNA‐335‐5p reduces inflammation via negative regulation of the TPX2‐mediated AKT/GSK3beta signaling pathway in a chronic rhinosinusitis mouse model. Cell Signal. 2020;70:109596. [DOI] [PubMed] [Google Scholar]
- 21. Li L, Feng J, Zhang D, et al. Differential expression of miR‐4492 and IL‐10 is involved in chronic rhinosinusitis with nasal polyps. Exp Ther Med. 2019;18(5):3968‐3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li X, Li C, Zhu G, Yuan W, Xiao ZA. TGF‐beta1 induces epithelial‐mesenchymal transition of chronic sinusitis with nasal polyps through microRNA‐21. Int Arch Allergy Immunol. 2019;179(4):304‐319. [DOI] [PubMed] [Google Scholar]
- 23. Liu CC, Xia M, Zhang YJ, et al. Micro124‐mediated AHR expression regulates the inflammatory response of chronic rhinosinusitis (CRS) with nasal polyps. Biochem Biophys Res Commun. 2018;500(2):145‐151. [DOI] [PubMed] [Google Scholar]
- 24. Ma Z, Shen Y, Zeng Q, et al. MiR‐150‐5p regulates EGR2 to promote the development of chronic rhinosinusitis via the DC‐Th axis. Int Immunopharmacol. 2018;54:188‐197. [DOI] [PubMed] [Google Scholar]
- 25. Qing Xiang, Zhang Yongquan, Peng Ya, He Guangxiang, Liu An, Liu Huowang. Mir‐142‐3p Regulates Inflammatory Response by Contributing to Increased TNF‐α in Chronic Rhinosinusitis With Nasal Polyposis. Ear, Nose & Throat Journal. 2019;014556131984797 10.1177/0145561319847972 [DOI] [PubMed] [Google Scholar]
- 26. Xia G, Bao L, Gao W, Liu S, Ji K, Li J. Differentially expressed miRNA in inflammatory mucosa of chronic rhinosinusitis. J Nanosci Nanotechnol. 2015;15(3):2132‐2139. [DOI] [PubMed] [Google Scholar]
- 27. Xuan L, Luan G, Wang Y, et al. MicroRNAs regulating mucin type O‐glycan biosynthesis and transforming growth factor beta signaling pathways in nasal mucosa of patients with chronic rhinosinusitis with nasal polyps in Northern China. Int Forum Allergy Rhinol. 2019;9(1):106‐113. [DOI] [PubMed] [Google Scholar]
- 28. Zhang X‐H, Zhang Y‐N, Li H‐B, et al. Overexpression of miR‐125b, a novel regulator of innate immunity, in eosinophilic chronic rhinosinusitis with nasal polyps. Am J Respir Crit Care Med. 2012;185(2):140‐151. [DOI] [PubMed] [Google Scholar]
- 29. Zhang XH, Zhang YN, Liu Z. MicroRNA in chronic rhinosinusitis and allergic rhinitis. Curr Allergy Asthma Rep. 2014;14(2):415. [DOI] [PubMed] [Google Scholar]
- 30. Weidinger S, Novak N. Atopic dermatitis. Lancet. 2016;387(10023):1109‐1122. [DOI] [PubMed] [Google Scholar]
- 31. Czarnowicki T, He H, Krueger JG, Guttman‐Yassky E. Atopic dermatitis endotypes and implications for targeted therapeutics. J Allergy Clin Immunol. 2019;143(1):1‐11. [DOI] [PubMed] [Google Scholar]
- 32. Alexander H, Paller AS, Traidl‐Hoffmann C, et al. The role of bacterial skin infections in atopic dermatitis: expert statement and review from the International Eczema Council Skin Infection Group. Br J Dermatol. 2020;182:1331‐1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sonkoly E, Wei T, Janson PCJ, et al. MicroRNAs: novel regulators involved in the pathogenesis of psoriasis? PLoS One. 2007;2(7):e610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hermann H, Runnel T, Aab A, et al. miR‐146b probably assists miRNA‐146a in the suppression of keratinocyte proliferation and inflammatory responses in psoriasis. J Invest Dermatol. 2017;137(9):1945‐1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Meisgen F, Xu Landén N, Wang A, et al. MiR‐146a negatively regulates TLR2‐induced inflammatory responses in keratinocytes. J Invest Dermatol. 2014;134(7):1931‐1940. [DOI] [PubMed] [Google Scholar]
- 36. Rebane A, Runnel T, Aab A, et al. MicroRNA‐146a alleviates chronic skin inflammation in atopic dermatitis through suppression of innate immune responses in keratinocytes. J Allergy Clin Immunol. 2014;134(4):836‐847. [DOI] [PubMed] [Google Scholar]
- 37. Sonkoly E, Janson P, Majuri M‐L, et al. MiR‐155 is overexpressed in patients with atopic dermatitis and modulates T‐cell proliferative responses by targeting cytotoxic T lymphocyte‐associated antigen 4. J Allergy Clin Immunol. 2010;126(3):581‐589. [DOI] [PubMed] [Google Scholar]
- 38. Srivastava A, Nikamo P, Lohcharoenkal W, et al. MicroRNA‐146a suppresses IL‐17‐mediated skin inflammation and is genetically associated with psoriasis. J Allergy Clin Immunol. 2017;139(2):550‐561. [DOI] [PubMed] [Google Scholar]
- 39. Carreras‐Badosa G, Runnel T, Plaas M, et al. microRNA‐146a is linked to the production of IgE in mice but not in atopic dermatitis patients. Allergy. 2018;73(12):2400‐2403. [DOI] [PubMed] [Google Scholar]
- 40. Li F, Huang Y, Huang YY, et al. MicroRNA‐146a promotes IgE class switch in B cells via upregulating 14‐3‐3sigma expression. Mol Immunol. 2017;92:180‐189. [DOI] [PubMed] [Google Scholar]
- 41. Okoye IS, Czieso S, Ktistaki E, et al. Transcriptomics identified a critical role for Th2 cell‐intrinsic miR‐155 in mediating allergy and antihelminth immunity. Proc Natl Acad Sci USA. 2014;111(30):E3081‐E3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ma L, Xue HB, Wang F, Shu CM, Zhang JH. MicroRNA‐155 may be involved in the pathogenesis of atopic dermatitis by modulating the differentiation and function of T helper type 17 (Th17) cells. Clin Exp Immunol. 2015;181(1):142‐149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Moyle M, Cevikbas F, Harden JL, Guttman‐Yassky E. Understanding the immune landscape in atopic dermatitis: the era of biologics and emerging therapeutic approaches. Exp Dermatol. 2019;28(7):756‐768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Brożek JL, Bousquet J, Agache I, et al. Allergic Rhinitis and its Impact on Asthma (ARIA) guidelines‐2016 revision. J Allergy Clin Immunol. 2017;140(4):950‐958. [DOI] [PubMed] [Google Scholar]
- 45. Cruz AA, Popov T, Pawankar R, et al. Common characteristics of upper and lower airways in rhinitis and asthma: ARIA update, in collaboration with GA(2)LEN. Allergy 2007;62(Suppl 84):1‐41. [DOI] [PubMed] [Google Scholar]
- 46. Braunstahl GJ. United airways concept: what does it teach us about systemic inflammation in airways disease? Proc Am Thorac Soc. 2009;6(8):652‐654. [DOI] [PubMed] [Google Scholar]
- 47. Zissler UM, Ulrich M, Jakwerth CA, et al. Biomatrix for upper and lower airway biomarkers in patients with allergic asthma. J Allergy Clin Immunol. 2018;142(6):1980‐1983. [DOI] [PubMed] [Google Scholar]
- 48. Panganiban RP, Wang Y, Howrylak J, et al. Circulating microRNAs as biomarkers in patients with allergic rhinitis and asthma. J Allergy Clin Immunol. 2016;137(5):1423‐1432. [DOI] [PubMed] [Google Scholar]
- 49. Shaoqing YU, Ruxin Z, Guojun L, et al. Microarray analysis of differentially expressed microRNAs in allergic rhinitis. Am J Rhinol Allergy. 2011;25(6):e242‐e246. [DOI] [PubMed] [Google Scholar]
- 50. Suojalehto H, Lindström I, Majuri M‐L, et al. Altered microRNA expression of nasal mucosa in long‐term asthma and allergic rhinitis. Int Arch Allergy Immunol. 2014;163(3):168‐178. [DOI] [PubMed] [Google Scholar]
- 51. Kohlhaas S, Garden OA, Scudamore C, Turner M, Okkenhaug K, Vigorito E. Cutting edge: the Foxp3 target miR‐155 contributes to the development of regulatory T cells. J Immunol. 2009;182(5):2578‐2582. [DOI] [PubMed] [Google Scholar]
- 52. Rodriguez A, Vigorito E, Clare S, et al. Requirement of bic/microRNA‐155 for normal immune function. Science. 2007;316(5824):608‐611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jia M, Chu C, Wang M. Correlation of microRNA profiles with disease risk and severity of allergic rhinitis. Int J Clin Exp Pathol. 2018;11(3):1791‐1802. [PMC free article] [PubMed] [Google Scholar]
- 54. Simpson CR, Sheikh A. Trends in the epidemiology of asthma in England: a national study of 333,294 patients. J R Soc Med. 2010;103(3):98‐106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dong X, Zhong N, Fang Y, Cai Q, Lu M, Lu Q. MicroRNA 27b–3p modulates SYK in pediatric asthma induced by dust mites. Front Pediatr. 2018;6:301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kho AT, Sharma S, Davis JS, et al. Circulating microRNAs: association with lung function in asthma. PLoS One. 2016;11(6):e0157998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Davis JS, Sun M, Kho AT, et al. Circulating microRNAs and association with methacholine PC20 in the Childhood Asthma Management Program (CAMP) cohort. PLoS One. 2017;12(7):e0180329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kho AT, McGeachie MJ, Moore KG, Sylvia JM, Weiss ST, Tantisira KG. Circulating microRNAs and prediction of asthma exacerbation in childhood asthma. Respir Res. 2018;19(1):128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pinkerton M, Chinchilli V, Banta E, et al. Differential expression of microRNAs in exhaled breath condensates of patients with asthma, patients with chronic obstructive pulmonary disease, and healthy adults. J Allergy Clin Immunol. 2013;132(1):217‐219. [DOI] [PubMed] [Google Scholar]
- 60. Malmhall C, Johansson K, Winkler C, Alawieh S, Ekerljung L, Radinger M. Altered miR‐155 expression in allergic asthmatic airways. Scand J Immunol. 2017;85(4):300‐307. [DOI] [PubMed] [Google Scholar]
- 61. Simpson LJ, Patel S, Bhakta NR, et al. A microRNA upregulated in asthma airway T cells promotes TH2 cytokine production. Nat Immunol. 2014;15(12):1162‐1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Haj‐Salem I, Fakhfakh R, Berube JC, et al. MicroRNA‐19a enhances proliferation of bronchial epithelial cells by targeting TGFbetaR2 gene in severe asthma. Allergy. 2015;70(2):212‐219. [DOI] [PubMed] [Google Scholar]
- 63. Sun Q, Liu LI, Wang H, et al. Constitutive high expression of protein arginine methyltransferase 1 in asthmatic airway smooth muscle cells is caused by reduced microRNA‐19a expression and leads to enhanced remodeling. J Allergy Clin Immunol. 2017;140(2):510‐524 e513. [DOI] [PubMed] [Google Scholar]
- 64. Zhang K, Liang Y, Feng Y, et al. Decreased epithelial and sputum miR‐221‐3p associates with airway eosinophilic inflammation and CXCL17 expression in asthma. Am J Physiol Lung Cell Mol Physiol. 2018;315(2):L253‐L264. [DOI] [PubMed] [Google Scholar]
- 65. Weidner J, Ekerljung L, Malmhall C, Miron N, Radinger M. Circulating microRNAs correlate to clinical parameters in individuals with allergic and non‐allergic asthma. Respir Res. 2020;21(1):107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rodrigo‐Muñoz JM, Cañas JA, Sastre B, et al. Asthma diagnosis using integrated analysis of eosinophil microRNAs. Allergy. 2019;74(3):507‐517. [DOI] [PubMed] [Google Scholar]
- 67. Francisco‐Garcia AS, Garrido‐Martín EM, Rupani H, et al. Small RNA species and microRNA profiles are altered in severe asthma nanovesicles from broncho alveolar lavage and associate with impaired lung function and inflammation. Noncoding RNA 2019;5(4):51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Levänen B, Bhakta NR, Torregrosa Paredes P, et al. Altered microRNA profiles in bronchoalveolar lavage fluid exosomes in asthmatic patients. J Allergy Clin Immunol. 2013;131(3):894‐903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Solberg OD, Ostrin EJ, Love MI, et al. Airway epithelial miRNA expression is altered in asthma. Am J Respir Crit Care Med. 2012;186(10):965‐974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yu B, Yao L, Liu C, Tang L, Xing T. Upregulation of microRNA16 alters the response to inhaled betaagonists in patients with asthma though modulating expression of ADRB2. Mol Med Rep. 2019;19(5):4027‐4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Dong X, Xu M, Ren Z, et al. Regulation of CBL and ESR1 expression by microRNA‐223p, 513a–5p and 625–5p may impact the pathogenesis of dust mite‐induced pediatric asthma. Int J Mol Med. 2016;38(2):446‐456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Polikepahad S, Knight JM, Naghavi AO, et al. Proinflammatory role for let‐7 microRNAS in experimental asthma. J Biol Chem. 2010;285(39):30139‐30149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Panganiban RP, Pinkerton MH, Maru SY, Jefferson SJ, Roff AN, Ishmael FT. Differential microRNA epression in asthma and the role of miR‐1248 in regulation of IL‐5. Am J Clin Exp Immunol. 2012;1(2):154‐165. [PMC free article] [PubMed] [Google Scholar]
- 74. Nakano T, Inoue Y, Shimojo N, et al. Lower levels of hsa‐mir‐15a, which decreases VEGFA, in the CD4+ T cells of pediatric patients with asthma. J Allergy Clin Immunol. 2013;132(5):1224‐1227 e1212. [DOI] [PubMed] [Google Scholar]
- 75. Castillo JR, Peters SP, Busse WW. Asthma exacerbations: pathogenesis, prevention, and treatment. J Allergy Clin Immunol Pract. 2017;5(4):918‐927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Jartti T, Gern JE. Role of viral infections in the development and exacerbation of asthma in children. J Allergy Clin Immunol. 2017;140(4):895‐906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Schwarze J, Openshaw P, Jha A, et al. Influenza burden, prevention, and treatment in asthma – a scoping review by the EAACI Influenza in asthma task force. Allergy. 2018;73(6):1151‐1181. [DOI] [PubMed] [Google Scholar]
- 78. Edwards MR, Regamey N, Vareille M, et al. Impaired innate interferon induction in severe therapy resistant atopic asthmatic children. Mucosal Immunol. 2013;6(4):797‐806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Bondanese VP, Francisco‐Garcia A, Bedke N, Davies DE, Sanchez‐Elsner T. Identification of host miRNAs that may limit human rhinovirus replication. World J Biol Chem. 2014;5(4):437‐456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Martinez‐Nunez RT, Bondanese VP, Louafi F, et al. A microRNA network dysregulated in asthma controls IL‐6 production in bronchial epithelial cells. PLoS One. 2014;9(10):e111659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Clifford RL, Jones MJ, MacIsaac JL, et al. Inhalation of diesel exhaust and allergen alters human bronchial epithelium DNA methylation. J Allergy Clin Immunol. 2017;139(1):112‐121. [DOI] [PubMed] [Google Scholar]
- 82. Moheimani F, Koops J, Williams T, et al. Influenza A virus infection dysregulates the expression of microRNA‐22 and its targets; CD147 and HDAC4, in epithelium of asthmatics. Respir Res. 2018;19(1):145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. McCaskill JL, Ressel S, Alber A, et al. Broad‐spectrum inhibition of respiratory virus infection by microRNA mimics targeting p38 MAPK signaling. Mol Ther Nucleic Acids. 2017;7:256‐266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Tan KS, Lim RL, Liu J, et al. Respiratory viral infections in exacerbation of chronic airway inflammatory diseases: novel mechanisms and insights from the upper airway epithelium. Front Cell Dev Biol. 2020;8:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Balmes JR, Earnest G, Katz PP, et al. Exposure to traffic: lung function and health status in adults with asthma. J Allergy Clin Immunol. 2009;123(3):626‐631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Brunekreef B, Stewart AW, Anderson HR, et al. Self‐reported truck traffic on the street of residence and symptoms of asthma and allergic disease: a global relationship in ISAAC phase 3. Environ Health Perspect. 2009;117(11):1791‐1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Carlsten C, Dybuncio A, Becker A, Chan‐Yeung M, Brauer M. Traffic‐related air pollution and incident asthma in a high‐risk birth cohort. Occup Environ Med. 2011;68(4):291‐295. [DOI] [PubMed] [Google Scholar]
- 88. De Grove KC, Provoost S, Hendriks RW, et al. Dysregulation of type 2 innate lymphoid cells and TH2 cells impairs pollutant‐induced allergic airway responses. J Allergy Clin Immunol. 2017;139(1):246‐257 e244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Jacquemin B, Kauffmann F, Pin I, et al. Air pollution and asthma control in the Epidemiological study on the Genetics and Environment of Asthma. J Epidemiol Community Health. 2012;66(9):796‐802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Kunzli N, Bridevaux P‐O, Liu L‐JS, et al. Traffic‐related air pollution correlates with adult‐onset asthma among never‐smokers. Thorax. 2009;64(8):664‐670. [DOI] [PubMed] [Google Scholar]
- 91. Maes T, Provoost S, Lanckacker EA, et al. Mouse models to unravel the role of inhaled pollutants on allergic sensitization and airway inflammation. Respir Res. 2010;11:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. McCreanor J, Cullinan P, Nieuwenhuijsen MJ, et al. Respiratory effects of exposure to diesel traffic in persons with asthma. N Engl J Med. 2007;357(23):2348‐2358. [DOI] [PubMed] [Google Scholar]
- 93. Meng YY, Rull RP, Wilhelm M, Lombardi C, Balmes J, Ritz B. Outdoor air pollution and uncontrolled asthma in the San Joaquin Valley, California. J Epidemiol Community Health. 2010;64(2):142‐147. [DOI] [PubMed] [Google Scholar]
- 94. Wenzel SE. Asthma phenotypes: the evolution from clinical to molecular approaches. Nat Med. 2012;18(5):716‐725. [DOI] [PubMed] [Google Scholar]
- 95. Vrijens K, Bollati V, Nawrot TS. MicroRNAs as potential signatures of environmental exposure or effect: a systematic review. Environ Health Perspect. 2015;123(5):399‐411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Rider CF, Yamamoto M, Günther OP, et al. Controlled diesel exhaust and allergen coexposure modulates microRNA and gene expression in humans: effects on inflammatory lung markers. J Allergy Clin Immunol. 2016;138(6):1690‐1700. [DOI] [PubMed] [Google Scholar]
- 97. Yamamoto M, Singh A, Sava F, Pui M, Tebbutt SJ, Carlsten C. MicroRNA expression in response to controlled exposure to diesel exhaust: attenuation by the antioxidant N‐acetylcysteine in a randomized crossover study. Environ Health Perspect. 2013;121(6):670‐675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Liu Q, Wang W, Jing W. Indoor air pollution aggravates asthma in Chinese children and induces the changes in serum level of miR‐155. Int J Environ Health Res. 2019;29(1):22‐30. [DOI] [PubMed] [Google Scholar]
- 99. Gao LU, Liu X, Millstein J, et al. Self‐reported prenatal tobacco smoke exposure, AXL gene‐body methylation, and childhood asthma phenotypes. Clin Epigenetics. 2018;10(1):98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Herberth G, Bauer M, Gasch M, et al. Maternal and cord blood miR‐223 expression associates with prenatal tobacco smoke exposure and low regulatory T‐cell numbers. J Allergy Clin Immunol. 2014;133(2):543‐550. [DOI] [PubMed] [Google Scholar]
- 101. Maes T, Cobos FA, Schleich F, et al. Asthma inflammatory phenotypes show differential microRNA expression in sputum. J Allergy Clin Immunol. 2016;137(5):1433‐1446. [DOI] [PubMed] [Google Scholar]
- 102. Singh SP, Chand HS, Langley RJ, et al. Gestational exposure to sidestream (secondhand) cigarette smoke promotes transgenerational epigenetic transmission of exacerbated allergic asthma and bronchopulmonary dysplasia. J Immunol. 2017;198(10):3815‐3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Xiao R, Noel A, Perveen Z, Penn AL. In utero exposure to second‐hand smoke activates pro‐asthmatic and oncogenic miRNAs in adult asthmatic mice. Environ Mol Mutagen. 2016;57(3):190‐199. [DOI] [PubMed] [Google Scholar]
- 104. Dehmel S, Nathan P, Bartel S, et al. Intrauterine smoke exposure deregulates lung function, pulmonary transcriptomes, and in particular insulin‐like growth factor (IGF)‐1 in a sex‐specific manner. Sci Rep. 2018;8(1):7547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Wang L, Xu J, Liu H, Li J, Hao H. PM2.5 inhibits SOD1 expression by up‐regulating microRNA‐206 and promotes ROS accumulation and disease progression in asthmatic mice. Int Immunopharmacol. 2019;76:105871. [DOI] [PubMed] [Google Scholar]
- 106. Bleck B, Grunig G, Chiu A, et al. MicroRNA‐375 regulation of thymic stromal lymphopoietin by diesel exhaust particles and ambient particulate matter in human bronchial epithelial cells. J Immunol. 2013;190(7):3757‐3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Li J, Zhou Q, Liang Y, et al. miR‐486 inhibits PM2.5‐induced apoptosis and oxidative stress in human lung alveolar epithelial A549 cells. Ann Transl Med. 2018;6(11):209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Li X, Lv Y, Hao J, et al. Role of microRNA‐4516 involved autophagy associated with exposure to fine particulate matter. Oncotarget. 2016;7(29):45385‐45397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Liu L, Wan C, Zhang W, et al. MiR‐146a regulates PM1 ‐induced inflammation via NF‐kappaB signaling pathway in BEAS‐2B cells. Environ Toxicol. 2018;33(7):743‐751. [DOI] [PubMed] [Google Scholar]
- 110. Wang G, Zheng X, Tang J, et al. LIN28B/let‐7 axis mediates pulmonary inflammatory response induced by diesel exhaust particle exposure in mice. Toxicol Lett. 2018;299:1‐10. [DOI] [PubMed] [Google Scholar]
- 111. Yadav S, Singh N, Shah PP, et al. MIR155 regulation of ubiquilin1 and ubiquilin 2: implications in cellular protection and tumorigenesis. Neoplasia. 2017;19(4):321‐332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Yang D, Ma M, Zhou W, Yang B, Xiao C. Inhibition of miR‐32 activity promoted EMT induced by PM2.5 exposure through the modulation of the Smad1‐mediated signaling pathways in lung cancer cells. Chemosphere. 2017;184:289‐298. [DOI] [PubMed] [Google Scholar]
- 113. Zhou F, Li S, Jia W, et al. Effects of diesel exhaust particles on microRNA‐21 in human bronchial epithelial cells and potential carcinogenic mechanisms. Mol Med Rep. 2015;12(2):2329‐2335. [DOI] [PubMed] [Google Scholar]
- 114. Conickx G, Mestdagh P, Avila Cobos F, et al. MicroRNA profiling reveals a role for microRNA‐218‐5p in the pathogenesis of chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2017;195(1):43‐56. [DOI] [PubMed] [Google Scholar]
- 115. Lu L, Xu H, Yang P, et al. Involvement of HIF‐1alpha‐regulated miR‐21, acting via the Akt/NF‐kappaB pathway, in malignant transformation of HBE cells induced by cigarette smoke extract. Toxicol Lett. 2018;289:14‐21. [DOI] [PubMed] [Google Scholar]
- 116. Song L, Li D, Gu Y, Li X, Peng L. Let‐7a modulates particulate matter (</= 2.5 mum)‐induced oxidative stress and injury in human airway epithelial cells by targeting arginase 2. J Appl Toxicol. 2016;36(10):1302‐1310. [DOI] [PubMed] [Google Scholar]
- 117. Song L, Li D, Li X, et al. Exposure to PM2.5 induces aberrant activation of NF‐kappaB in human airway epithelial cells by downregulating miR‐331 expression. Environ Toxicol Pharmacol. 2017;50:192‐199. [DOI] [PubMed] [Google Scholar]
- 118. Zhao Y, Xu Y, Li Y, et al. NF‐kappaB‐mediated inflammation leading to EMT via miR‐200c is involved in cell transformation induced by cigarette smoke extract. Toxicol Sci. 2013;135(2):265‐276. [DOI] [PubMed] [Google Scholar]
- 119. De Smet EG, Mestdagh P, Vandesompele J, Brusselle GG, Bracke KR. Non‐coding RNAs in the pathogenesis of COPD. Thorax. 2015;70(8):782‐791. [DOI] [PubMed] [Google Scholar]
- 120. Huang X, Zhu Z, Guo X, Kong X. The roles of microRNAs in the pathogenesis of chronic obstructive pulmonary disease. Int Immunopharmacol. 2019;67:335‐347. [DOI] [PubMed] [Google Scholar]
- 121. Osei ET, Florez‐Sampedro L, Timens W, Postma DS, Heijink IH, Brandsma CA. Unravelling the complexity of COPD by microRNAs: it's a small world after all. Eur Respir J. 2015;46(3):807‐818. [DOI] [PubMed] [Google Scholar]
- 122. Johansson K, Malmhall C, Ramos‐Ramirez P, Radinger M. MicroRNA‐155 is a critical regulator of type 2 innate lymphoid cells and IL‐33 signaling in experimental models of allergic airway inflammation. J Allergy Clin Immunol. 2017;139(3):1007‐1016 e1009. [DOI] [PubMed] [Google Scholar]
- 123. Qiu L, Zhang Y, Do DC, et al. miR‐155 modulates cockroach allergen‐ and oxidative stress‐induced cyclooxygenase‐2 in asthma. J Immunol. 2018;201(3):916‐929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Zech A, Ayata CK, Pankratz F, et al. MicroRNA‐155 modulates P2R signaling and Th2 priming of dendritic cells during allergic airway inflammation in mice. Allergy. 2015;70(9):1121‐1129. [DOI] [PubMed] [Google Scholar]
- 125. Malmhall C, Alawieh S, Lu Y, et al. MicroRNA‐155 is essential for T(H)2‐mediated allergen‐induced eosinophilic inflammation in the lung. J Allergy Clin Immunol. 2014;133(5):1429‐1438, e1421–e1427. [DOI] [PubMed] [Google Scholar]
- 126. Singh PB, Pua HH, Happ HC, et al. MicroRNA regulation of type 2 innate lymphoid cell homeostasis and function in allergic inflammation. J Exp Med. 2017;214(12):3627‐3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Ye L, Mou Y, Wang J, Jin ML. Effects of microRNA‐19b on airway remodeling, airway inflammation and degree of oxidative stress by targeting TSLP through the Stat3 signaling pathway in a mouse model of asthma. Oncotarget. 2017;8(29):47533‐47546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Collison A, Herbert C, Siegle JS, Mattes J, Foster PS, Kumar RK. Altered expression of microRNA in the airway wall in chronic asthma: miR‐126 as a potential therapeutic target. BMC Pulm Med. 2011;11:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Mattes J, Collison A, Plank M, Phipps S, Foster PS. Antagonism of microRNA‐126 suppresses the effector function of TH2 cells and the development of allergic airways disease. Proc Natl Acad Sci USA. 2009;106(44):18704‐18709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Kim RY, Horvat JC, Pinkerton JW, et al. MicroRNA‐21 drives severe, steroid‐insensitive experimental asthma by amplifying phosphoinositide 3‐kinase‐mediated suppression of histone deacetylase 2. J Allergy Clin Immunol. 2017;139(2):519‐532. [DOI] [PubMed] [Google Scholar]
- 131. Lee HY, Lee HY, Choi JY, et al. Inhibition of MicroRNA‐21 by an antagomir ameliorates allergic inflammation in a mouse model of asthma. Exp Lung Res. 2017;43(3):109‐119. [DOI] [PubMed] [Google Scholar]
- 132. Lu TX, Hartner J, Lim EJ, et al. MicroRNA‐21 limits in vivo immune response‐mediated activation of the IL‐12/IFN‐gamma pathway, Th1 polarization, and the severity of delayed‐type hypersensitivity. J Immunol. 2011;187(6):3362‐3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Sawant DV, Wu H, Kaplan MH, Dent AL. The Bcl6 target gene microRNA‐21 promotes Th2 differentiation by a T cell intrinsic pathway. Mol Immunol. 2013;54(3‐4):435‐442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Li JJ, Tay HL, Maltby S, et al. MicroRNA‐9 regulates steroid‐resistant airway hyperresponsiveness by reducing protein phosphatase 2A activity. J Allergy Clin Immunol. 2015;136(2):462‐473. [DOI] [PubMed] [Google Scholar]
- 135. Karo‐Atar D, Itan M, Pasmanik‐Chor M, Munitz A. MicroRNA profiling reveals opposing expression patterns for miR‐511 in alternatively and classically activated macrophages. J Asthma. 2015;52(6):545‐553. [DOI] [PubMed] [Google Scholar]
- 136. Veremeyko T, Siddiqui S, Sotnikov I, Yung A, Ponomarev ED. IL‐4/IL‐13‐dependent and independent expression of miR‐124 and its contribution to M2 phenotype of monocytic cells in normal conditions and during allergic inflammation. PLoS One. 2013;8(12):e81774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Bronevetsky Y, Villarino AV, Eisley CJ, et al. T cell activation induces proteasomal degradation of Argonaute and rapid remodeling of the microRNA repertoire. J Exp Med. 2013;210(2):417‐432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Pua H, Steiner D, Patel S, et al. MicroRNAs 24 and 27 suppress allergic inflammation and target a network of regulators of T helper 2 cell‐associated cytokine production. Immunity. 2016;44(4):821‐832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Collison A, Mattes J, Plank M, Foster PS. Inhibition of house dust mite‐induced allergic airways disease by antagonism of microRNA‐145 is comparable to glucocorticoid treatment. J Allergy Clin Immunol. 2011;128(1):160‐167 e164. [DOI] [PubMed] [Google Scholar]
- 140. Kumar M, Ahmad T, Sharma A, et al. Let‐7 microRNA‐mediated regulation of IL‐13 and allergic airway inflammation. J Allergy Clin Immunol. 2011;128(5):1077‐1085 e1071–e1010. [DOI] [PubMed] [Google Scholar]
- 141. Shao Y, Chong L, Lin P, et al. MicroRNA‐133a alleviates airway remodeling in asthtama through PI3K/AKT/mTOR signaling pathway by targeting IGF1R. J Cell Physiol. 2019;234(4):4068‐4080. [DOI] [PubMed] [Google Scholar]
- 142. Yang ZC, Qu ZH, Yi MJ, et al. MiR‐448‐5p inhibits TGF‐beta1‐induced epithelial‐mesenchymal transition and pulmonary fibrosis by targeting Six1 in asthma. J Cell Physiol. 2019;234(6):8804‐8814. [DOI] [PubMed] [Google Scholar]
- 143. Huang H, Lu H, Liang L, et al. MicroRNA‐744 inhibits proliferation of bronchial epithelial cells by regulating Smad3 pathway via targeting transforming growth factor‐beta1 (TGF‐beta1) in severe asthma. Med Sci Monit. 2019;25:2159‐2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Jardim MJ, Dailey L, Silbajoris R, Diaz‐Sanchez D. Distinct microRNA expression in human airway cells of asthmatic donors identifies a novel asthma‐associated gene. Am J Respir Cell Mol Biol. 2012;47(4):536‐542. [DOI] [PubMed] [Google Scholar]
- 145. Liu D, Pan J, Zhao D, Liu F. MicroRNA‐223 inhibits deposition of the extracellular matrix by airway smooth muscle cells through targeting IGF‐1R in the PI3K/Akt pathway. Am J Transl Res. 2018;10(3):744‐752. [PMC free article] [PubMed] [Google Scholar]
- 146. Matsukura S, Osakabe Y, Sekiguchi A, et al. Overexpression of microRNA‐155 suppresses chemokine expression induced by Interleukin‐13 in BEAS‐2B human bronchial epithelial cells. Allergol Int. 2016;65(Suppl):S17‐S23. [DOI] [PubMed] [Google Scholar]
- 147. Qian FH, Deng X, Zhuang QX, Wei B, Zheng DD. miR6255p suppresses inflammatory responses by targeting AKT2 in human bronchial epithelial cells. Mol Med Rep. 2019;19(3):1951‐1957. [DOI] [PubMed] [Google Scholar]
- 148. Jia H‐Z, Liu S‐L, Zou Y‐F, et al. MicroRNA‐223 is involved in the pathogenesis of atopic dermatitis by affecting histamine‐N‐methyltransferase. Cell Mol Biol. 2018;64(3):103‐107. [DOI] [PubMed] [Google Scholar]
- 149. Kuo Y‐C, Li Y‐S, Zhou J, et al. Human mesenchymal stem cells suppress the stretch‐induced inflammatory miR‐155 and cytokines in bronchial epithelial cells. PLoS One. 2013;8(8):e71342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Huo X, Zhang K, Yi L, et al. Decreased epithelial and plasma miR‐181b‐5p expression associates with airway eosinophilic inflammation in asthma. Clin Exp Allergy. 2016;46(10):1281‐1290. [DOI] [PubMed] [Google Scholar]
- 151. Perry MM, Moschos SA, Williams AE, Shepherd NJ, Larner‐Svensson HM, Lindsay MA. Rapid changes in microRNA‐146a expression negatively regulate the IL‐1beta‐induced inflammatory response in human lung alveolar epithelial cells. J Immunol. 2008;180(8):5689‐5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Tsai M‐J, Tsai Y‐C, Chang W‐A, et al. Deducting microRNA‐mediated changes common in bronchial epithelial cells of asthma and chronic obstructive pulmonary disease – a next‐generation sequencing‐guided bioinformatic approach. Int J Mol Sci 2019;20(3).533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Kivihall A, Aab A, Soja J, et al. Reduced expression of miR‐146a in human bronchial epithelial cells alters neutrophil migration. Clin Transl Allergy. 2019;9:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Bartel S, La Grutta S, Cilluffo G, et al. Human airway epithelial extracellular vesicle miRNA signature is altered upon asthma development. Allergy. 2020;75(2):346‐356. [DOI] [PubMed] [Google Scholar]
- 155. Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome‐mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654‐659. [DOI] [PubMed] [Google Scholar]
- 156. Bahmer T, Krauss‐Etschmann S, Buschmann D, et al. RNA‐seq–based profiling of extracellular vesicles in plasma reveals a potential role of miR‐122‐5p in asthma. Allergy. 2021;76:366‐371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Bartel S, Deshane J, Wilkinson T, Gabrielsson S. Extracellular vesicles as mediators of cellular cross talk in the lung microenvironment. Front Med. 2020;7:326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Comer BS, Camoretti‐Mercado B, Kogut PC, Halayko AJ, Solway J, Gerthoffer WT. MicroRNA‐146a and microRNA‐146b expression and anti‐inflammatory function in human airway smooth muscle. Am J Physiol Lung Cell Mol Physiol. 2014;307(9):L727‐L734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Liu Y, Sun X, Wu Y, et al. Effects of miRNA‐145 on airway smooth muscle cells function. Mol Cell Biochem. 2015;409(1‐2):135‐143. [DOI] [PubMed] [Google Scholar]
- 160. Liu Y, Yang K, Shi H, et al. MiR‐21 modulates human airway smooth muscle cell proliferation and migration in asthma through regulation of PTEN expression. Exp Lung Res. 2015;41(10):535‐545. [DOI] [PubMed] [Google Scholar]
- 161. Lu TX, Munitz A, Rothenberg ME. MicroRNA‐21 is up‐regulated in allergic airway inflammation and regulates IL‐12p35 expression. J Immunol. 2009;182(8):4994‐5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Kästle M, Bartel S, Geillinger‐Kästle K, et al. microRNA cluster 106a–363 is involved in T helper 17 cell differentiation. Immunology. 2017;152(3):402‐413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Montoya MM, Maul J, Singh PB, et al. A distinct inhibitory function for miR‐18a in Th17 cell differentiation. J Immunol. 2017;199(2):559‐569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Kılıç Ayşe, Santolini Marc, Nakano Taiji, Schiller Matthias, Teranishi Mizue, Gellert Pascal, Ponomareva Yuliya, Braun Thomas, Uchida Shizuka, Weiss Scott T., Sharma Amitabh, Renz Harald. A systems immunology approach identifies the collective impact of 5 miRs in Th2 inflammation. JCI Insight. 2018;3 (11): 10.1172/jci.insight.97503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Muraro A, Lemanske RF Jr, Hellings PW, et al. Precision medicine in patients with allergic diseases: airway diseases and atopic dermatitis‐PRACTALL document of the European Academy of Allergy and Clinical Immunology and the American Academy of Allergy, Asthma & Immunology. J Allergy Clin Immunol. 2016;137(5):1347‐1358. [DOI] [PubMed] [Google Scholar]
- 166. Shrestha S, Miao R, Wang L, Chao J, Yuce H, Wei W. Burden of atopic dermatitis in the United States: analysis of healthcare claims data in the commercial, Medicare, and Medi‐Cal databases. Adv Ther. 2017;34(8):1989‐2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Zeiger RS, Tran TN, Schatz M, et al. Drivers of health care costs for adults with persistent asthma. J Allergy Clin Immunol Pract. 2018;6(1):265‐268 e264. [DOI] [PubMed] [Google Scholar]
- 168. Qin H‐B, Xu B, Mei J‐J, et al. Inhibition of miRNA‐221 suppresses the airway inflammation in asthma. Inflammation. 2012;35(4):1595‐1599. [DOI] [PubMed] [Google Scholar]
- 169. Sharma A, Kumar M, Ahmad T, et al. Antagonism of mmu‐mir‐106a attenuates asthma features in allergic murine model. J Appl Physiol (1985). 2012;113(3):459‐464. [DOI] [PubMed] [Google Scholar]
- 170. Tay HL, Kaiko GE, Plank M, et al. Antagonism of miR‐328 increases the antimicrobial function of macrophages and neutrophils and rapid clearance of non‐typeable Haemophilus influenzae (NTHi) from infected lung. PLoS Pathog. 2015;11(4):e1004549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Liao W, Dong J, Peh H, et al. Oligonucleotide therapy for obstructive and restrictive respiratory diseases. Molecules. 2017;22(1).139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Milger K, Götschke J, Krause L, et al. Identification of a plasma miRNA biomarker signature for allergic asthma: a translational approach. Allergy. 2017;72(12):1962‐1971. [DOI] [PubMed] [Google Scholar]
- 173. Rodrigo‐Muñoz JM, Rial MJ, Sastre B, et al. Circulating miRNAs as diagnostic tool for discrimination of respiratory disease: asthma, asthma‐chronic obstructive pulmonary disease (COPD) overlap and COPD. Allergy. 2019;74(12):2491‐2494. [DOI] [PubMed] [Google Scholar]
- 174. Wu C, Xu K, Wang Z, et al. A novel microRNA miR‐1165‐3p as a potential diagnostic biomarker for allergic asthma. Biomarkers. 2019;24(1):56‐63. [DOI] [PubMed] [Google Scholar]
- 175. Urgard E, Lorents A, Klaas M, et al. Pre‐administration of PepFect6‐microRNA‐146a nanocomplexes inhibits inflammatory responses in keratinocytes and in a mouse model of irritant contact dermatitis. J Control Release. 2016;235:195‐204. [DOI] [PubMed] [Google Scholar]
- 176. Chen XF, Zhang LJ, Zhang J, et al. MiR‐151a is involved in the pathogenesis of atopic dermatitis by regulating interleukin‐12 receptor beta2. Exp Dermatol. 2018;27(4):427‐432. [DOI] [PubMed] [Google Scholar]
- 177. Zeng YP, Nguyen GH, Jin HZ. MicroRNA‐143 inhibits IL‐13‐induced dysregulation of the epidermal barrier‐related proteins in skin keratinocytes via targeting to IL‐13Ralpha1. Mol Cell Biochem. 2016;416(1‐2):63‐70. [DOI] [PubMed] [Google Scholar]
- 178. Yang Z, Zeng B, Wang C, Wang H, Huang P, Pan Y. MicroRNA‐124 alleviates chronic skin inflammation in atopic eczema via suppressing innate immune responses in keratinocytes. Cell Immunol. 2017;319:53‐60. [DOI] [PubMed] [Google Scholar]
- 179. Vaher H, Runnel T, Urgard E, et al. miR‐10a‐5p is increased in atopic dermatitis and has capacity to inhibit keratinocyte proliferation. Allergy. 2019;74(11):2146‐2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Li L, Zhang S, Jiang X, Liu Y, Liu K, Yang C. MicroRNA‐let‐7e regulates the progression and development of allergic rhinitis by targeting suppressor of cytokine signaling 4 and activating Janus kinase 1/signal transducer and activator of transcription 3 pathway. Exp Ther Med. 2018;15(4):3523‐3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Martinez‐Nunez RT, Louafi F, Sanchez‐Elsner T. The interleukin 13 (IL‐13) pathway in human macrophages is modulated by microRNA‐155 via direct targeting of interleukin 13 receptor alpha1 (IL13Ralpha1). J Biol Chem. 2011;286(3):1786‐1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Suojalehto H, Toskala E, Kilpeläinen M, et al. MicroRNA profiles in nasal mucosa of patients with allergic and nonallergic rhinitis and asthma. Int Forum Allergy Rhinol. 2013;3(8):612‐620. [DOI] [PubMed] [Google Scholar]
- 183. Just J, Munk Ipsen P, Kruhøffer M, et al. Human mast cell sensitization with IgE increases miRNA‐210 expression. Int Arch Allergy Immunol. 2019;179(2):102‐107. [DOI] [PubMed] [Google Scholar]
- 184. Othumpangat S, Bryan NB, Beezhold DH, Noti JD. Upregulation of miRNA‐4776 in influenza virus infected bronchial epithelial cells is associated with downregulation of NFKBIB and increased viral survival. Viruses 2017;9(5).94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Othumpangat S, Walton C, Piedimonte G. MicroRNA‐221 modulates RSV replication in human bronchial epithelium by targeting NGF expression. PLoS One. 2012;7(1):e30030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Ouda R, Onomoto K, Takahasi K, et al. Retinoic acid‐inducible gene I‐inducible miR‐23b inhibits infections by minor group rhinoviruses through down‐regulation of the very low density lipoprotein receptor. J Biol Chem. 2011;286(29):26210‐26219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Zhao L, Zhu J, Zhou H, et al. Identification of cellular microRNA‐136 as a dual regulator of RIG‐I‐mediated innate immunity that antagonizes H5N1 IAV replication in A549 cells. Sci Rep. 2015;5:14991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Fang J, Hao Q, Liu L, et al. Epigenetic changes mediated by microRNA miR29 activate cyclooxygenase 2 and lambda‐1 interferon production during viral infection. J Virol. 2012;86(2):1010‐1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Guan Z, Shi N, Song Y, Zhang X, Zhang M, Duan M. Induction of the cellular microRNA‐29c by influenza virus contributes to virus‐mediated apoptosis through repression of antiapoptotic factors BCL2L2. Biochem Biophys Res Commun. 2012;425(3):662‐667. [DOI] [PubMed] [Google Scholar]
- 190. Zhang X, Dong C, Sun X, et al. Induction of the cellular miR‐29c by influenza virus inhibits the innate immune response through protection of A20 mRNA. Biochem Biophys Res Commun. 2014;450(1):755‐761. [DOI] [PubMed] [Google Scholar]
- 191. Ma Y‐J, Yang J, Fan X‐L, et al. Cellular microRNA let‐7c inhibits M1 protein expression of the H1N1 influenza A virus in infected human lung epithelial cells. J Cell Mol Med. 2012;16(10):2539‐2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Buggele WA, Krause KE, Horvath CM. Small RNA profiling of influenza A virus‐infected cells identifies miR‐449b as a regulator of histone deacetylase 1 and interferon beta. PLoS One. 2013;8(9):e76560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Khongnomnan K, Makkoch J, Poomipak W, Poovorawan Y, Payungporn S. Human miR‐3145 inhibits influenza A viruses replication by targeting and silencing viral PB1 gene. Exp Biol Med (Maywood). 2015;240(12):1630‐1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Ingle H, Kumar S, Raut AA, et al. The microRNA miR‐485 targets host and influenza virus transcripts to regulate antiviral immunity and restrict viral replication. Sci Signal. 2015;8(406):ra126. [DOI] [PubMed] [Google Scholar]
- 195. Rosenberger CM, Podyminogin RL, Diercks AH, et al. miR‐144 attenuates the host response to influenza virus by targeting the TRAF6‐IRF7 signaling axis. PLoS Pathog. 2017;13(4):e1006305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Kumar A, Kumar A, Ingle H, et al. MicroRNA hsa‐miR‐324‐5p suppresses H5N1 virus replication by targeting the viral PB1 and host CUEDC2. J Virol. 2018;92(19):e01057‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Deng Y, Yan Y, Tan KS, et al. MicroRNA‐146a induction during influenza H3N2 virus infection targets and regulates TRAF6 levels in human nasal epithelial cells (hNECs). Exp Cell Res. 2017;352(2):184‐192. [DOI] [PubMed] [Google Scholar]
- 198. Zhang F, Sun X, Zhu Y, Qin W. Downregulation of miR‐146a inhibits influenza A virus replication by enhancing the type I interferon response in vitro and in vivo. Biomed Pharmacother. 2019;111:740‐750. [DOI] [PubMed] [Google Scholar]
- 199. Wardzynska A, Pawelczyk M, Rywaniak J, Kurowski M, Makowska JS, Kowalski ML. Circulating microRNAs and T‐cell cytokine expression are associated with the characteristics of asthma exacerbation. Allergy Asthma Immunol Res. 2020;12(1):125‐136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Biethahn K, Orinska Z, Vigorito E, et al. miRNA‐155 controls mast cell activation by regulating the PI3Kgamma pathway and anaphylaxis in a mouse model. Allergy. 2014;69(6):752‐762. [DOI] [PubMed] [Google Scholar]
- 201. Takyar S, Vasavada H, Zhang J‐G, et al. VEGF controls lung Th2 inflammation via the miR‐1‐Mpl (myeloproliferative leukemia virus oncogene)‐P‐selectin axis. J Exp Med. 2013;210(10):1993‐2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Perry MM, Adcock IM, Chung KF. Role of microRNAs in allergic asthma: present and future. Curr Opin Allergy Clin Immunol. 2015;15(2):156‐162. [DOI] [PubMed] [Google Scholar]
- 203. Mayoral RJ, Deho L, Rusca N, et al. MiR‐221 influences effector functions and actin cytoskeleton in mast cells. PLoS One. 2011;6(10):e26133. [DOI] [PMC free article] [PubMed] [Google Scholar]


