Abstract
Background
PAOLA1 is a phase III study assessing olaparib maintenance therapy in advanced high-grade ovarian carcinoma patients responding to first-line platinum-taxane–based chemotherapy plus bevacizumab as standard of care. Randomization was stratified by treatment outcome and tumor BRCA1/2 status (tBRCA) at screening.
Methods
tBRCA was tested on formalin-fixed, paraffin-embedded tumor blocks on 5 French platforms using 2 next-generation sequencing methods based either on hybrid capture or amplicon technology. One of the exploratory objectives was to assess the concordance between germline (gBRCA) and tBRCA testing in French patients. gBRCA testing was performed on blood samples on the same platforms.
Results
From May 2015 to July 2017, tBRCA tests were performed for 1176 screened patients. Only 52 (4.4%) tumor samples were noncontributive. The median interval between reception of the tumor sample and availability of the tBRCA status result was 37 days (range = 8-260). A pathogenic variant was reported in 27.1% tumor samples (319 of 1176 screened patients). tBRCA and gBRCA testing were performed for 451 French patients with negative results for both tests in 306 patients (67.8%) and positive results for both tests in 85 patients (18.8%). Only 1 large genomic rearrangement of BRCA1 was detected, exclusively in the blood sample. Interestingly, tBRCA testing revealed 6.4% of pathogenic variant (29 of 451) not detected by gBRCA testing.
Conclusions
tBRCA testing is an appropriate tool with an acceptable turnaround time for clinical practice and a low failure rate, ensuring reliable identification of patients likely to benefit from poly(ADP-ribose) polymerase inhibitor therapy.
Epithelial ovarian cancer (EOC) is the sixth most common cancer among women worldwide and the leading cause of death due to gynecologic malignancies (1). Approximately 13%-31% of patients with early EOC and 75%-80% of those with advanced disease relapse after a median of 11-29 months and 18-24 months, respectively (2). For several decades, systemic therapy of ovarian cancer has consisted of chemotherapy, with the relatively recent addition of antiangiogenic strategies in combination with chemotherapy and in the maintenance setting (3). The benefit of bevacizumab, a humanized antivascular endothelial growth factor monoclonal antibody, has been demonstrated for advanced disease in combination with chemotherapy added to a maintenance phase (4,5). Recently, a major breakthrough was made with the approval of poly(ADP-ribose) polymerase inhibitors (PARPi) for the treatment of relapsing high-grade serous carcinoma patients responding to platinum-based chemotherapy in the SOLO2, ARIEL3, and NOVA trials (6-8) and as first-line treatment for mBRCA patients in the SOLO1 trial (9). Data from these clinical trials identified BRCA1 and/or BRCA2mutations, genome-wide loss of heterozygosity, and homologous repair deficiency as predictive biomarkers for PARPi therapy.
PAOLA1 (ENGOT-ov25) is a phase III, randomized, double-blind, placebo-controlled multicenter trial designed to assess the efficacy and safety of olaparib (tablet formulation) as maintenance therapy in patients with advanced high-grade serous or endometrioid ovarian cancer (HGOC) who have responded to first-line platinum-taxane–based chemotherapy plus bevacizumab concomitant with maintenance chemotherapy for up to 15 months. Stratification was performed on treatment outcome and tumor BRCA1/2 status (tBRCA) at screening (NCT02477644) (10). Of the 806 patients randomly assigned in the PAOLA1 study, 537 were assigned to receive olaparib plus bevacizumab, and 269 were assigned to receive placebo plus bevacizumab. Olaparib maintenance therapy improved median progression-free survival (hazard ratio [HR] for disease progression or death = 0.59, 95% confidence interval [CI] = 0.49 to 0.72; P < .001), which was more pronounced in patients with BRCA-mutated tumors (HR = 0.31, 95% CI = 0.20 to 0.47) compared with patients with wild-type BRCA tumors (HR = 0.71, 95% CI = 0.58 to 0.88) (10).
Determination of BRCA status at first-line treatment, the feasibility of tumor BRCA testing, and the concordance between germline and tumor BRCA status are therefore of interest with this type of clinical trial (11). Although BRCA1/2 gene sequencing has been performed routinely on blood samples for many years to detect hereditary predisposition, the search for theranostic BRCA1/2 tumor variants emerged with the arrival of PARPi and is far from trivial, as genetic tests on tumor blocks can encounter several pitfalls because of the small sample sizes, low tumor cell infiltration, and DNA degradation because of formalin fixation. DNA extraction and sequencing methods must therefore be adapted to ensure reliable tumor testing (12,13).
We report the experience of institutional platforms that performed prospective tBRCA testing for patients from all participating centers in the PAOLA1 trial, in parallel with germline BRCA (gBRCA) testing for French patients, particularly to determine the feasibility of tumor testing on formalin-fixed, paraffin-embedded (FFPE) samples, the compatibility with clinical practice, and the concordance between tBRCA and gBRCA testing for the subgroup of French patients.
Methods
Study Population
The randomized, double-blind, placebo-controlled PAOLA-1 trial was conducted in 11 countries (France, Germany, Italy, Austria, Spain, Belgium, Finland, Denmark, Monaco, Sweden, and Japan) according to the declaration of Helsinki guidelines. The trial was approved by the authorities of all participating countries, and signed informed consent was obtained from all patients.
A total of 1176 patients were potentially candidates for inclusion in the PAOLA1 trial, and tumor samples from all patients were tested for BRCA1/2 variants. However, 370 patients were screen failures (31%) because of the presence of clinical exclusion criteria (no response or progression during chemotherapy, toxic events, no bevacizumab therapy, abnormal laboratory test results, time frames, patient’s decision). Finally, 806 patients were randomly assigned between May 2015 and August 2017 (EudraCT No.: 2014-004027-52).
Tumor BRCA Testing
Tumor BRCA testing was centralized in 5 French national academic platforms based in Paris (Institut Curie and Assistance Publique-Hôpitaux de Paris-APHP), Villejuif (Institut Gustave Roussy), Caen (Centre François Baclesse), and Bordeaux (Institut Bergonié) selected by a national call for tenders from the French National Institute of Cancer (INCa) to which 10 platforms applied. Platforms were selected after examination of the mandatory technical document submitted by each platform (confidential documents) to ensure that all selected platforms provided a similar level of performance. All 5 platforms comply with the ISO15189 standard and have reached the requirements of the same external quality assessment. These platforms also included positive and negative controls in each run. Material and methods are detailed in Supplementary Table 1 (available online). The Institut Curie platform centralized analyses from parts of France and Germany. The APHP platform centralized analysis from parts of France, Finland, Denmark, and Sweden. The Institut Gustave Roussy platform centralized analysis from parts of France and Italy. The Institut Bergonié platform centralized analysis from parts of France, Monaco, and Spain. The François Baclesse platform centralized analysis from parts of France, Austria, Belgium, and Japan. Tumor samples were collected from local pathologists during the screening period. An adequately sized (minimum: 2 mm x 2 mm) archival paraffin-embedded tumor block, representative of the tumor obtained by surgical resection or core biopsy of the primary tumor or peritoneal carcinomatosis, was provided. Each participating center had to send either a tumor block or twenty 6 µm slides containing at least 30% of tumor cells. Participating centers were asked to send tumor samples about 2 months (at least 1 month) prior to random assignment to allow sufficient time to perform the analysis and to request another tumor block if the first block was unsuitable for analysis. DNA was extracted from FFPE tumor blocks according to local procedures at each of the 5 platforms. Tumor BRCA testing was performed on FFPE samples obtained from all centers. The 5 platforms met several times prior to initiation of inclusions to standardize their process of validation of pathogenic variants and variants of unknown significance. All loss-of-function variants (frameshift, nonsense, canonical splice site), as well as large genomic rearrangement or missense variants already classified as pathogenic in public databases (BRCA share, BRCA-UMD, ClinVar, COSMIC, Galaxy, ALAMUT) were considered to be pathogenic (class 5). These same public databases, sometimes completed by home-made databases (for 3 platforms), were used to classify missense variants as neutral variants or variants of unknown significance (VUS; class 3). Pathogenic variants (PV) were reported on a specific form that was sent to the study site and to the sponsor (ARCAGY Research). Variants of unknown significance were reported on a specific form that was sent only to ARCAGY Research and were classified as wild-type tBRCA status. Neutral and likely neutral variants were not reported.
Germline BRCA Testing
In compliance with European Union regulations for germline testing, blood BRCA tests were performed only on French patients. gBRCA testing was performed in parallel to tBRCA testing in French patients by the 5 same platforms selected by INCa. DNA was extracted from blood samples according to local procedures in each of the 5 platforms. All of the French platforms belong to the Groupe Génétique et Cancer French consortium, which have a national BRCA database, which helps to have a common approach in variants classifications. Material and methods are detailed in Supplementary Table 2 (available online).
Statistical Methods
Statistical analyses were performed using GraphPad Prism (version 5.01) software (GraphPad Software, Inc. California, USA). The lower and upper limits of the 95% confidence interval (CI) were calculated for each proportion, according to Robert Newcombe’s method. All statistical tests were two-sided and a P value of less than .05 was considered statistically signficant.
Results
Tumor BRCA Analysis
From May 2015 to July 2017, 1176 tests were performed. tBRCA status was assessed in 1124 samples with a median turnaround time of 37 days (range = 8-260; first quartile = 25; fourth quartile = 55) between reception of the tumor sample and availability of the BRCA result. The median turnaround time was reduced to 31 days (range = 8-109) during the second recruitment period (2016-2017), when tumor testing had been fully implemented on all platforms.
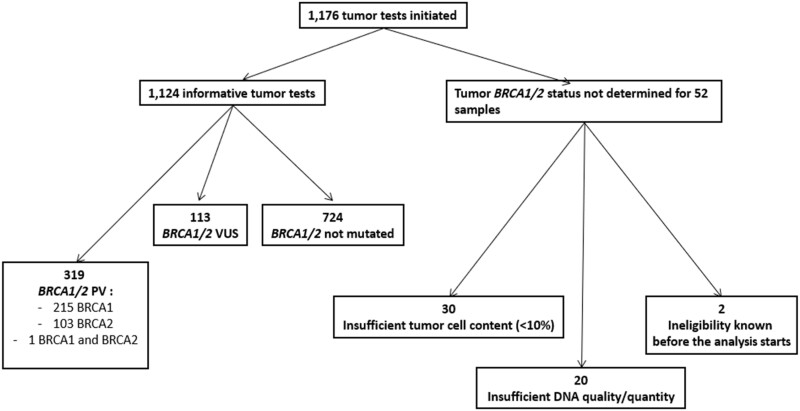
In this set of 1176 tests, results were reported as inconclusive for 52 samples (4.4%): 20 were considered to be noncontributive because of the insufficient quality or quantity of the extracted DNA to perform next-generation sequencing (NGS) analysis. For another 30 samples, the tumor cell content was insufficient (<10%) to initiate NGS analysis. Another 2 patients were not tested because of the presence of ineligibility criteria (an interval of less than 2 weeks between shipment of the tumor sample and randomization) prior to testing (Figure 1).
Figure 1.
Graphic summary of tumor testing results. PV = pathogenic variant; VUS = variant of unknown significance.
A PV was reported in 319 tumor samples (27.1% of the 1176 patients screened): 215 (67.3%) in BRCA1, 103 (32.2%) in BRCA2, and 1 in both genes. Two samples had a double PV in the BRCA2 gene, and 1 sample had a double PV in the BRCA1 gene (see Supplementary Table 3, available online, for details of tumor PV). The proportions of mutated tumor samples per country and per platform are detailed in Tables 1 and 2, respectively. The proportion of mutated tumors was statistically significantly higher for samples from Belgium (P = .004) than for the overall cohort (53%, 95% CI = 35% to 70%). The proportion of inconclusive results was lower for samples from Germany (P < .001) than for the overall cohort (0.5%, 95% CI = 0.1% to 1%) (Table 1). We did not detect any statistically significant difference in the proportion of mutated tumor samples reported by each platform according to the use of a hybrid capture or amplicon technique. The proportion of inconclusive tumor BRCA results was statistically significantly lower on the Institut Curie platform (P < .001) and higher on the Gustave Roussy platform (P < .001) than in the overall cohort (Table 2).
Table 1.
Tumor BRCA results per country (restricted to pathogenic variants only)a
| Center location | No. of BRCA tests performed on tumor | No. of tMut (%, 95% CI) | No. of tNeg (%, 95% CI) | No. of tBRCA unknown (%, 95% CI) | Screening failure for randomization No. (%, 95% CI) |
|---|---|---|---|---|---|
| France/Monaco | 498 | 126 (25.3, 21 to 29) | 338 (67.9, 63 to 71) | 34 (6.8, 5 to 9) | 169 (33.9, 30 to 38) |
| Belgium | 28 | 15 (53.6, 35 to 70) | 13 (46.4, 29 to 64) | 0 (0, 0 to 12) | 8 (28.5, 15 to 47) |
| Germany | 382 | 109 (28.5, 24 to 33) | 271 (70.9, 66 to 75) | 2 (0.5, 0.1 to 1) | 131 (34.2, 29 to 39) |
| Austria | 42 | 16 (38.1, 25 to 53) | 24 (57.1, 42 to 70) | 2 (4.7, 1 to 15) | 14 (33.3, 21 to 48) |
| Italy | 108 | 24 (22.2, 15 to 30) | 76 (70.4, 61 to 78) | 8 (7.4, 3 to 13) | 23 (21.2, 14 to 30) |
| Spain | 73 | 23 (31.5, 22 to 42) | 45 (61.6, 50 to 72) | 5 (6.8, 3 to 15) | 18 (24.6, 16 to 35) |
| Denmark | 6 | 0 (0, 0 to 39) | 6 (100, 61 to 100) | 0 (0, 0 to 39) | 0 (0, 0 to 39) |
| Finland | 12 | 1 (8.3, 1 to 35) | 11 (91.6, 64 to 98) | 0 (0, 0 to 24) | 5 (41.6, 19 to 68) |
| Sweden | 1 | 0 (0, 0 to 79) | 1 (100, 20 to 100) | 0 (0, 0 to 79) | 0 (0, 0 to 79) |
| Japan | 26 | 5 (19.2, 8 to 37) | 20 (76.9, 58 to 89) | 1 (3.8, 0.7 to 19) | 2 (7.6, 2 to 24) |
| Total | 1176 | 319 (27.1, 25 to 30) | 805 (68.4, 66 to 71) | 52 (4.4, 3 to 6) | 370 (31.4, 29 to 34) |
CI = confidence interval; tBCRA/2 = tumor BRCA1/2; tmut = tumor mutation; tNeg = tumor negative mutation.
Table 2.
Results per screening platform
| Screening center |
BRCA tests performed on tumor |
BRCA tests performed among French patients |
|||||
|---|---|---|---|---|---|---|---|
| No. of tBRCA tests performed | No. of unknown tBRCA tests (%, 95% CI) | No. of samples in which a mutation was detected (%, 95% CI) | No. tests performed |
Samples in which a mutation was detected |
|||
| On tumor | On blood | No. in tumor (%, 95% CI) | No. in blood (%, 95% CI) | ||||
| Institut Curie, Paris | 485 | 5 (1.1, 0.4 to 2) | 143 (29.4, 25 to 33) | 103 | 98 | 34 (33.0, 24 to 42) | 24 (24.4, 17 to 33) |
| Centre Baclesse, Caen | 196 | 10 (5.1, 2 to 9) | 62 (31.6, 25 to 38) | 100 | 90 | 26 (26.0, 18 to 35) | 15 (16.6, 10 to 25) |
| APHP, Paris | 124 | 3 (2.4, 0.8 to 7) | 23 (18.5, 12 to 26) | 105 | 104 | 22 (20.9, 14 to 29) | 16 (15.3, 9 to 23) |
| Institut Bergonié, Bordeaux | 174 | 10 (5.7, 3 to 10) | 49 (28.1, 22 to 35) | 98 | 91 | 26 (26.5, 18 to 36) | 21 (23.1, 15 to 32) |
| Gustave Roussy, Villejuif | 197 | 24 (12.1, 8 to 17) | 42 (21.3, 16 to 27) | 89 | 71 | 18 (20.2, 13 to 29) | 12 (16.9, 10 to 27) |
| Total | 1176 | 52 (4.4, 3 to 6) | 319 (27.1, 26 to 31) | 495 | 454a | 126 (25.4, 22 to 29) | 88 (19.3, 16 to 23) |
Including 3 patients without tBRCA testing. CI = confidence interval; tBRCA = tumor BRCA1/2.
A VUS was reported for 113 tumor samples (9.6% of the 1176 patients screened): in the BRCA1 gene in 43 (38.1%) cases, in the BRCA2 gene in 67 (59.2%) cases, and in both genes in 3 cases. Three samples had a double VUS in the BRCA1 gene (see Supplementary Table 4, available online, for details of tumor VUS). For 81 samples, the VUS reported were the only variant detected in BRCA1/2 genes. VUS were associated with a PV in the BRCA1 gene for 22 samples or a PV in the BRCA2 gene for 10 samples. The 6 samples in which 2 VUS were reported did not harbor a PV in the BRCA1/2 genes.
Germline BRCA Analysis
Between May 2015 and November 2017, 454 tests were performed exclusively on samples from French patients. All blood samples were contributive, and results were available after a median of 45 days (range = 10-456; first quartile = 32; fourth quartile = 64).
A germline PV was reported for 88 of 454 patients (19.3%) with 53 (60.2%) PV in the BRCA1 gene and 36 (40.9%) PV in the BRCA2 gene. One patient had 2 PV: 1 in the BRCA1 gene and the other in the BRCA2 gene (see Supplementary Table 5, available online, for details of germline PV). The proportions of germline PV on the 5 platforms ranged from 15.3% to 24.4%. Germline VUS were detected in 24 samples (5.2% of the patients screened). VUS were the only variant detected for 18 samples, whereas VUS were associated with a PV in 6 samples (see Supplementary Table 6, available online, for details of germline VUS).
Global results for gBRCA testing on each platform are presented in Table 2. No inconclusive results were observed for germline BRCA tests, and a similar proportion of mutated germline samples was reported on all platforms.
Concordance Between Tumor and Germline BRCA Testing in the Subgroup of French Patients
Tumor BRCA and germline BRCA testing were both performed for 451 French patients. gBRCA test results were available for 3 patients, in whom tBRCA testing was not performed (Table 3). In this group of 451 patients, tBRCA testing was inconclusive for 30 patients because of the inadequate DNA quantity/quality or tumor cell content. tBRCA and gBRCA results were consistent with negative results for both tests for 306 (67.8%) patients and positive results for both tests for 85 (18.8%) patients. For 1 patient, gBRCA testing was positive, whereas tBRCA testing was negative. The PV detected for this patient was a large genomic rearrangement (LGR) consisting of deletion of exons 1 and 2 of the BRCA1 gene. Interestingly, tBRCA testing revealed 29 of 451 PV (6.4%) not detected by gBRCA testing. The list of these variants, exclusively detected in the tumor, is presented in Supplementary Table 7 (available online). In 1 patient, a de novo BRCA1 PV was associated with a germline BRCA2 VUS (c.8958A>G, p. Ile2986Met). For 2 patients, de novo BRCA1 PV were associated with a tumor BRCA1 VUS (c.5194-8dup, p.? or c.2129 C > G, p. Thr710Ser).
Table 3.
Concordance between tBRCA and gBRCA testing in the French cohorta
| Testing result | gBRCA negative No. (%) | gBRCA positive No. (%) | Total No. (%) |
|---|---|---|---|
| tBRCA negative | 306 (67.8) | 1 (0.2) | 307 (68.1) |
| tBRCA positive | 29 (6.4) | 85 (18.8) | 114 (25.2) |
| Inconclusive tumor testing | 29 (6.4) | 1 (0.2) | 30 (6.6) |
| Total | 364 (80.7) | 87 (19.2) | 451 (100) |
gBRCA = germline BRCA; tBRCA = tumor BRCA1/2.
Discussion
During PAOLA-1 trial enrollment, 1176 tBRCA tests were performed on FFPE samples with a median turnaround time of 37 days and 4.4% of inconclusive results. The tBRCA testing failure rate was low and not different from that observed for other tests, such as KRAS or EGFR hotspot detection (mean failure rates of 5% and 8%, respectively, in 2013 for the 28 platforms certified by the French National Institute of Cancer; see the www.e-cancer.fr website for more details). Sequencing of the entire coding sequence of large suppressor genes such a BRCA1/2 is therefore feasible on fragmented DNA extracted from FFPE samples. However, we observed a difference in terms of the inconclusive result rate according to whether the comparison was based on platform or country. Between-country differences in sample formalin fixation technique cannot be excluded. The respective rates of neoadjuvant chemotherapy vs primary debulking surgery, which differ from one country to another, may also have influenced the percentage of tumor cells in the sample, the quality of DNA extracted, and the proportion of inconclusive results. As laboratories may have adapted their workflow to meet the randomization deadline (communicated at the time of reception of the tumor block), the turnaround time may therefore have been extended. All platforms routinely observed a turnaround time of less than 6 weeks, as recommended by INCa (see the www.e-cancer.fr website for more details). These performances demonstrate that tBRCA testing on FFPE samples is a reliable tool for routine clinical practice.
The proportion of mutated tumor samples in the PAOLA1 trial differed from one country to another, but no differences were observed between platforms. Between-country differences could be potentially related to differences in patient characteristics in recruitment groups, which may have been influenced by the different bevacizumab reimbursement policies in the participating countries.
The incidence of tBRCA PV in PAOLA1 (27.1%, 95% CI = 25% to 30%) was not statistically different from that observed in The Cancer Genome Atlas study (22.4%, 95% CI = 18% to 27%) (14). The phase III PRIMA trial exploring the benefit of niraparib maintenance therapy after first-line platinum-based chemotherapy reported a similar incidence of tBRCA mutation (30.4%, 95% CI = 27% to 34%) (15). Platinum-based chemotherapy sensitivity is known to be correlated with BRCA mutation and may have induced a bias. However, the phase III VELIA trial exploring the benefit of first-line veliparib without patient selection prior to inclusion also reported a 26.1% (95% CI = 24% to 29%) tBRCA mutation rate, suggesting that 25%-30% may represent the true tBRCA mutation rate in stage III-IV high-grade serous and endometrioid ovarian or fallopian tube or primaryperitoneal cancers (16).
In the subgroup of French patients, a high level of concordance was observed between tumor and germline BRCA test results. The only tBRCA- and gBRCA+ discordant case highlights the difficulty of detecting LGR in FFPE tumor samples but was responsible for failure in less than 1% of tumor BRCA tests. LGR are rare events, because the mutation profile for high-risk patients was 90.1% sequencing mutations vs 9.9% large rearrangements in the study by Judkins et al. (17). Moreover, improvement in the material quality, progress in bioinformatics tools, and NGS technologies should soon overcome this difficulty. We are therefore confident that a first tumor test will soon be able to rapidly provide a reliable result to initiate PARPi therapy and to refer patients for genetic counseling with no risk of false-negative results. This approach could facilitate focused germline testing and an overall reduction in genetic testing.
Although BRCA1 and BRCA2 gene mutations are the alterations most commonly observed, HGOC is also characterized by frequent genetic and epigenetic alterations of hazard ratio pathway genes (18). In the future, detection of homologous recombination deficiency (HRD) may predict those individuals most likely to derive a benefit from PARPi therapy in addition to tumor BRCA mutation (tBRCAm) (7,10,15). However, the limitations of the HRD tests used in the PAOLA1 trial are the proportion of samples with “unknown” status, the possibility of false-negatives, cost, and unavailability or lack of access to testing. In PAOLA1, the addition of maintenance olaparib provided a statistically significant progression-free survival benefit, which was substantial in patients with HRD-positive tumors, including those without BRCA mutation (10). Further studies are ongoing to identify the most reliable HRD biomarkers to predict sensitivity to PARPi. An ENGOT-led project is designed to explore new HRD tests [genotypic or phenotypic tests such as rad51 foci (19)] using tumor samples from the PAOLA1 trial.
In the French subgroup, PV were detected in tumors in 25.2% (114 of 451) of patients and in blood in 19.2% (87 of 451) of patients. Tumor BRCA testing therefore allowed the detection of 29 of 451 (6.4%) de novo somatic PV in this cohort. As a result of tumor BRCA testing instead of germline BRCA testing, an additional 25% (29 of [87 + 29]) of patients would be able to benefit from olaparib therapy in routine clinical practice. Jorge et al. (20) reported 20.9% (9 of 43) of somatic variants. Vos et al. (21) showed that the universal tumor BRCA1/2 workflow identified twice as many patients for PARPi therapy than conventional genetic predisposition testing of DNA from blood. These various studies therefore show that tumor testing can identify more patients likely to benefit from PARPi therapy. However, tumor testing should not overshadow germline testing for variant detection in other genes than BRCA1/2 (eg, RCA51C, RAD51D, MMR genes) and genetic counseling, which are essential for the prevention of second cancers and the surveillance of relatives.
In conclusion, tBRCA testing is a reliable tool for clinical trials with acceptable time frame and is now also useful to guide PARPi prescription in clinical practice. Knowledge of both tumor and germline BRCA1/2 status is essential at diagnosis and to ensure optimal care of EOC patients. Rapid and common statement between oncologists and geneticists for patients is essential to optimize therapeutic management and genetic counseling.
Funding
This work was supported by ARCAGY research, Institut National du Cancer, AstraZeneca, and F. Hoffmann-La Roche.
Notes
Role of the funders: Arcagy Research funding acquisition, Institut National du Cancer molecular platforms logistics, AstraZeneca funding acquisition, and F. Hoffmann-La Roche funding acquisition.
Disclosures: The authors have no potential conflicts of interest.
Author contributions: Céline Callens; Resources, Investigation, data curation, methodology, formal analysis, Writing- Original draft preparation. Dominique Vaur: Resources, Investigation, data curation, Writing- Reviewing and Editing. Isabelle Soubeyran: Resources, Investigation, data curation, Writing- Reviewing and Editing. Etienne RouleResources, Investigation, data curation, Writing- Reviewing and Editing. Pierre-Alexandre Just: Resources, Investigation, data curation, Writing- Reviewing and Editing. Erell Guillerm: Resources, Investigation, data curation, Writing- Reviewing and Editing. Lisa Golmard: Resources, Investigation, data curation, Writing- Reviewing and Editing. Nicolas Goardon: Resources, Investigation, data curation, Writing- Reviewing and Editing. Nicolas Sevenet: Resources, Investigation, data curation, Writing- Reviewing and Editing. Odile Cabaret: Resources, Investigation, data curation, Writing- Reviewing and Editing. Philipp Harter: Resources, Writing- Reviewing and Editing. Antonio Gonzalez-Martin: Resources, Writing- Reviewing and Editing. Keiichi Fujiwara: Resources, Writing- Reviewing and Editing. Sabrina Chiara Cecere: Resources, Writing- Reviewing and Editing. Nicoletta Colombo: Resources, Writing- Reviewing and Editing. Christian Marth: Resources, Writing- Reviewing and Editing. Ignace Vergote: Resources, Writing- Reviewing and Editing. Johanna Maenpaa: Resources, Writing- Reviewing and Editing. Eric Pujade-Lauraine: Data curation, funding acquisition, Methodology, Writing- Reviewing and Editing. Isabelle Ray-Coquard: Conceptualization, Resources, Writing- Original draft preparation, Reviewing and Editing, Supervision.
Acknowledgments: All investigators from the the following sites: In France: Centre Léon Bérard, Lyon; Gustave Roussy, Villejuif; Hôpital Tenon, Paris; Centre Eugène Marquis, Rennes; Groupe Confluent, Nantes; Hôpital Européen Georges Pompidou, Paris; Institut Curie, Paris; Institut Curie, Saint-Cloud; Centre Jean Perrin, Clermont-Ferrand; CH Lyon Sud, Lyon; Institut Bergonié, Bordeaux; Centre Jean Bernard, Le Mans; Centre CARIO - HPCA, Plérin sur mer; Institut Paoli Calmettes, Marseille; ICM Val d’Aurelle, Montpellier; ICO Centre René Gauducheau, Saint-Herblain; Hôpital les Diaconesses, Paris; Centre François Baclesse, Caen; Institut Claudius Regaud, Toulouse; ICO Paul Pain, Angers; Centre Oscar Lambret, Lille; Institut de Cancréoleogie de Lorraine, Vandoeuvre-les-Nancy; Hôpital Cochin, Paris; GHM de Grenoble; centre Georges-François Leclerc, Dijon; Clinique Pasteur, Toulouse; CHU de Lille; centre Azuréen de Cancérologie de Mougins; Institut Sainte-Catherine, Avignon; Hôpital Mont-de-Marsan; Centre Antoine Lacassagne, Nice; Hôpital Jean Minjoz, Besançon; Polyclinique Bordeaux Nord, Bordeaux; Clinique Francheville, Périgueux; Hôpital Michallon, Grenoble; CHR d’Orléans; Clinique Sainte-Anne, Strasbourg; Hôpitaux Universitaires de Strasbourg; CHD Les Oudairies, la Roche-sur-Yon; Groupe Hospitalier Saint-Joseph, Paris; CHU La Milétrie, Poitiers; Centre Paul Strauss, Strasbourg; Centre Henri Becquerel, Rouen.
In Germany: Kliniken Essen Mitte, Essen; Charité - Universitätsmedizin Berlin; Universitätsklinikum Carl Gustav Carus, Dresden; Universitätsklinikum Heidelberg; Universitätsklinikum Ulm; St. Vincentius Kliniken, Karlsruhe; Universitätsklinikum Essen; Klinkum Gütersloh; Klinikum der Universität München; Universitätsfrauenklinik Erlangen; Klinikum der Johann Wolfgang Goethe, Frankfurt; Universitätsklinikum Jena; Universitätsmedizin Greifswald; Gynäkologisch-Onkologische Praxis Hannover; Onkologie Ravensburg; Ortenau Klinikum, Offenburg; Universitätsklinikum Hamburg-Eppendorf, Hamburg; Universitätsfrauenklinik Regensburg; Städtisches Klinikum Dessau; Medizinische Hochschule Hannover; amO Wolfsburg; Klinikum Südstadt, Rostock; Klinikum Frankfurt Höchst, Frankfurt; Universitätsklinikum Schleswig-Holstein, Kiel; Klinikum rechts der Isar, München; Klinikum Kassel; HELIOS Klinikum Berlin-Buch; Universitätsklinikum Schleswig-Holstein, Lübeck; HELIOS Dr Horst Schmidt Kliniken, Wiesbaden; Johannes Wesling Klinikum, Minden; Marien Hospital Witten; Hochtaunus-Kliniken, Bad Homburg; Robert-Bosch-Krankenhaus, Stuttgart; Universitäts-Frauenklinik Tübingen; Universitätsklinikum Münster; Klinikum Esslingen; St. Elisabeth Krankenhaus, Köln; Universitätsfrauenklinik Köln; Albertinen Krankenhaus, Hamburg; Praxisklinik Krebsheilkunde für Frauen, Berlin; Klinikum Worms; Onkologie Bottrop; Klinikum Konstanz; Universitätsfrauenklinik Freiburg; Klinikum am Steinenberg, Reutlingen; Universitätsklinikum Halle; Sana Klinikum Offenbach; Klinikum Augsburg; Evangelisches Krankenhaus Düsseldorf; Universitäts-Frauenklinik Göttingen; HELIOS Klinikum Krefeld.
In Italy: Istituto Nazionale Tumori - IRCCS Pascale, Napoli; Policlinico Universitario Gemelli Università Cattolica del Sacro Cuore, Roma; Istituto Nazionale Tumori, Milano; Ospedale Santa Maria della Misericordia, Perugia; Policlinico S.Orsola-Malpighi, Bologna; Ospedale Senatore Antonio Perrino, Brindisi; Istituto Europeo di Oncologia, Milano; Spedali Civili-Università di Brescia; Arcispedale S. M. Nuova, Reggio Emilia.
In Spain: MD Anderson Cancer Center Madrid; H. U. Ramón y Cajal, Madrid; H. U. Reina Sofía, Cordoba; C.S. Parc Taulí, Sabadell; H.U. Central de Asturias, Oviedo; H. U. Miguel Servet, Zaragova; Complejo Hospitalario de Navarra, Pamplona; H.U. Arnau de Vilanova, Lleida; Instituto Valenciano de Oncología, València; H. de la Santa Creu i Sant Pau, Barcelona; H. General Universitario de Valencia; H. U. 12 de Octubre, Madrid; H. U. Fundación Alcorcón.
In Austria: Medical University of Vienna; Krankenhaus Hietzing, Vienna; Medical University of Innsbruck; Medical University of Graz; Landeskrankenhaus Salzburg; KH der Barmherzigen Brüder Graz.
In Belgium: UZ Gasthuisberg, Leuven; CHU Dinant Godinne, Yvoir; Clinique Sainte Elisabeth, Namur.
In Finland: Tampere University Hospital; Turku University Hospital.
In Denmark: Rigshospitalet, Copenhagen.
In Sweden: Linköping University Hospital.
In Monaco: Centre Hospitalier Princesse Grace.
In Japan: Hyogo Cancer Center; Saitama Medical University International Medical Center; University of Tsukuba Hospital, Ibaragi; Jichi Medical University Hospital, Tochigi; Ehime University Hospital; National Cancer Center Hospital, Tokyo; Kagoshima University Hospital.
All screening platform staff, especially Guillaume Bataillon, Virginie Moncoutier, Anne Salomon, Ivan Bieche, Samia Melaabi, Sabrina Croce, Isabelle Hostein, Ludovic Lacroix, Catherine Genestie, Karen Leroy, Florence Coulet, Pierre-Laurent Puig, Hélène Blons, Erell Guillerm, Odile Léopold, Eric Pasmant, Delfine Lafon, Françoise Bonnet, Natalie Jones, and Jennifer Chiron.
All operational staff of GINECO, MITO, GEICO, AGO-Austria, GOTIC, BGOG, MANGO, NSGO, and AGO who contributed to this trial. We thank Sylvie Mijonnet, Christine Montoto-Grillot, Aurélie Morvan, Sébastien Armanet, and Bénédicte Votan from ARCAGY for their assistance with trial coordination. We thank all women who participated in this study and their families.
Data Availability
All data generated or analyzed during this study are included in this published article.
ARCAGY-GINECO has a long history of academic data sharing for research purposes. The process is similar for every trial sponsored by ARCAGY-GINECO:
Researchers have to submit a request to the sponsor directly or through the principal investigator. The request should be written in a predefined format of a short synopsis indicating the objective of the research, the methodology intended to be used, including the statistical analysis plan, and the variables within the database required for the research.
A scientific board will review and approve the requests on a case-by-case basis.
Only encoded datasets will be used, which enables us to fulfill legal and ethical obligations to protect our patients while utilizing patient data in progressing medical research to its full potential in the best interests of public health.
A specific agreement between the sponsor and the researcher is requested for data transfer. This data transfer agreement details both parts responsibilities to ensure the required level of data integrity and legal and ethical obligations.
-
In the case of sharing encoded patient level data, please note that the full dataset may not be shared in view of the following:
Clinical consent for some countries prohibits secondary use of the data.
Patients may withdraw their consent for participation in the trial at any point.
Other aspects might also be taken into consideration to protect patient privacy (eg, review of rare clinical events where information is aggregated to a higher level before sharing).
Supplementary Material
References
- 1. Siegel R, Ma J, Zou Z, Jemal A.. Cancer statistics, 2014. CA A Cancer J Clin. 2014;64(1):9–29. [DOI] [PubMed] [Google Scholar]
- 2. Lorusso D, Tripodi E, Maltese G, et al. Spotlight on olaparib in the treatment of BRCA-mutated ovarian cancer: design, development and place in therapy. Drug Des Devel Ther. 2018;12:1501–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mirza MR, Pignata S, Ledermann JA.. Latest clinical evidence and further development of PARP inhibitors in ovarian cancer. Ann Oncol. 2018;29(6):1366–1376. [DOI] [PubMed] [Google Scholar]
- 4. Perren TJ, Swart AM, Pfisterer J, et al. A phase 3 trial of bevacizumab in ovarian cancer. N Engl J Med. 2011;365(26):2484–2496. [DOI] [PubMed] [Google Scholar]
- 5. Burger RA, Brady MF, Bookman MA, et al. Incorporation of bevacizumab in the primary treatment of ovarian cancer. N Engl J Med. 2011;365(26):2473–2483. [DOI] [PubMed] [Google Scholar]
- 6. Mirza MR, Monk BJ, Herrstedt J, et al. Niraparib maintenance therapy in platinum-sensitive, recurrent ovarian cancer. N Engl J Med. 2016;375(22):2154–2164. [DOI] [PubMed] [Google Scholar]
- 7. Coleman RL, Oza AM, Lorusso D, et al. Rucaparib maintenance treatment for recurrent ovarian carcinoma after response to platinum therapy (ARIEL3): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;390(10106):1949–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pujade-Lauraine E, Ledermann JA, Selle F, et al. Olaparib tablets as maintenance therapy in patients with platinum-sensitive, relapsed ovarian cancer and a BRCA1/2 mutation (SOLO2/ENGOT-Ov21): a double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol. 2017;18(9):1274–1284. [DOI] [PubMed] [Google Scholar]
- 9. Moore K, Colombo N, Scambia G, et al. Maintenance olaparib in patients with newly diagnosed advanced ovarian cancer. N Engl J Med. 2018;379(26):2495–2505. [DOI] [PubMed] [Google Scholar]
- 10. Ray-Coquard I, Pautier P, Pignata S, et al. Olaparib plus bevacizumab as first-line maintenance in ovarian cancer. N Engl J Med. 2019;381(25):2416–2428. [DOI] [PubMed] [Google Scholar]
- 11. Colombo N, Sessa C, Du Bois A, et al. ESMO-ESGO consensus conference recommendations on ovarian cancer: pathology and molecular biology, early and advanced stages, borderline tumours and recurrent disease. Ann Oncol. 2019;30(5):672–705. [DOI] [PubMed] [Google Scholar]
- 12. Capoluongo E, Ellison G, Lopez-Guerrero JA, et al. Guidance statement on BRCA1/2 tumor testing in ovarian cancer patients. Semin Oncol. 2017;44(3):187–197. [DOI] [PubMed] [Google Scholar]
- 13. Capoluongo E, Scambia G, Nabholtz JM.. Main implications related to the switch to BRCA1/2 tumor testing in ovarian cancer patients: a proposal of a consensus. Oncotarget. 2018;9(28):19463–19468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.The Cancer Genome Atlas. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gonzalez-Martin A, Pothuri B, Vergote I, et al. Niraparib in patients with newly diagnosed advanced ovarian cancer. N Engl J Med. 2019;381(25):2391–2402. [DOI] [PubMed] [Google Scholar]
- 16. Coleman RL, Fleming GF, Brady MF, et al. Veliparib with first-line chemotherapy and as maintenance therapy in ovarian cancer. N Engl J Med. 2019;381(25):2403–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Judkins T, Rosenthal E, Arnell C, et al. Clinical significance of large rearrangements in BRCA1 and BRCA2. Cancer. 2012;118(21):5210–5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Konstantinopoulos PA, Ceccaldi R, Shapiro GI, D’Andrea AD.. Homologous recombination deficiency: exploiting the fundamental vulnerability of ovarian cancer. Cancer Discov. 2015;5(11):1137–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cruz C, Castroviejo-Bermejo M, Gutierrez-Enriquez S, et al. RAD51 foci as a functional biomarker of homologous recombination repair and PARP inhibitor resistance in germline BRCA-mutated breast cancer. Ann Oncol. 2018;29(5):1203–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jorge S, McFaddin AS, Doll KM, et al. Simultaneous germline and somatic sequencing in ovarian carcinoma: mutation rate and impact on clinical decision-making. Gynecol Oncol. 2020;156(3):517–522. [DOI] [PubMed] [Google Scholar]
- 21. Vos JR, Fakkert IE, de Hullu JA, et al. Universal tumor DNA BRCA1/2 testing of ovarian cancer: prescreening PARPi treatment and genetic predisposition. J Natl Cancer Inst. 2020;112(2):161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.
ARCAGY-GINECO has a long history of academic data sharing for research purposes. The process is similar for every trial sponsored by ARCAGY-GINECO:
Researchers have to submit a request to the sponsor directly or through the principal investigator. The request should be written in a predefined format of a short synopsis indicating the objective of the research, the methodology intended to be used, including the statistical analysis plan, and the variables within the database required for the research.
A scientific board will review and approve the requests on a case-by-case basis.
Only encoded datasets will be used, which enables us to fulfill legal and ethical obligations to protect our patients while utilizing patient data in progressing medical research to its full potential in the best interests of public health.
A specific agreement between the sponsor and the researcher is requested for data transfer. This data transfer agreement details both parts responsibilities to ensure the required level of data integrity and legal and ethical obligations.
-
In the case of sharing encoded patient level data, please note that the full dataset may not be shared in view of the following:
Clinical consent for some countries prohibits secondary use of the data.
Patients may withdraw their consent for participation in the trial at any point.
Other aspects might also be taken into consideration to protect patient privacy (eg, review of rare clinical events where information is aggregated to a higher level before sharing).