Abstract
Background
Whole genome sequencing (WGS) surveillance and electronic health record data mining have the potential to greatly enhance the identification and control of hospital outbreaks. The objective was to develop methods for examining economic value of a WGS surveillance-based infection prevention (IP) program compared to standard of care (SoC).
Methods
The economic value of a WGS surveillance-based IP program was assessed from a hospital’s perspective using historical outbreaks from 2011–2016. We used transmission network of outbreaks to estimate incremental cost per transmission averted. The number of transmissions averted depended on the effectiveness of intervening against transmission routes, time from transmission to positive culture results and time taken to obtain WGS results and intervene on the transmission route identified. The total cost of an IP program included cost of staffing, WGS, and treating infections.
Results
Approximately 41 out of 89 (46%) transmissions could have been averted under the WGS surveillance-based IP program, and it was found to be a less costly and more effective strategy than SoC. The results were most sensitive to the cost of performing WGS and the number of isolates sequenced per year under WGS surveillance. The probability of the WGS surveillance-based IP program being cost-effective was 80% if willingness to pay exceeded $2400 per transmission averted.
Conclusions
The proposed economic analysis is a useful tool to examine economic value of a WGS surveillance-based IP program. These methods will be applied to a prospective evaluation of WGS surveillance compared to SoC.
Keywords: whole genome sequencing, electronic health record, healthcare-associated infections, economic evaluation, outbreak detection
Whole genome sequencing (WGS) surveillance can improve identification and control of hospital outbreaks. We have developed a method to assess economic value of a WGS surveillance-based infection prevention program. Our preliminary results suggest that it could be a cost-effective strategy.
Healthcare-associated infections (HAIs) are one of the most common complications of hospital care and are associated with a prolonged hospital stay, increased healthcare costs and poorer health outcomes [1–3]. The annual direct medical cost of HAIs in the United States is estimated to range from $28 to $45 billion [4].
Considering this significant public health burden, efforts to improve quality of care and reduce HAIs at the national level have increased [5]. The usual approach to detect an outbreak in hospitals is a multistep process [6]. First, systematic HAI surveillance is undertaken to identify patients who might be involved in an outbreak. If a cluster of infections is suspected by identifying presumptively related pathogens, the hospital’s infection prevention (IP) team may investigate possible epidemiological links between patients involved using the electronic health record (EHR) to identify plausible sources of transmission. Common routes of transmission include environmental contamination, transient (or less commonly, sustained) healthcare worker carriage, or a contaminated medical device or therapy [7]. For some outbreaks, there may be >1 transmission route. For example, transmission may start on a shared hospital unit and then continued via a contaminated device. As part of an IP investigation, bacterial whole genome sequencing (WGS) or another molecular typing method may confirm or refute that the outbreak is caused by a single strain. We refer to this approach as standard of care (SoC) where WGS is only done in reaction to a suspected outbreak, if available at the specific hospital. There is evidence that the SoC approach misses some outbreaks [8] or results in a delay in detecting an outbreak [9].
With the widespread use of the EHR in hospitals and a reduction in costs of bacterial WGS, it is possible to enhance the detection of hospital outbreaks. We can identify genetically related isolates through WGS and hence identify patients involved in the outbreak. Afterward, EHR can be used to identify probable transmission route(s) as it has near real-time information on the patient’s location, procedures, treatments and contact with healthcare personnel. We refer to this approach as WGS surveillance-based IP program.
Under WGS surveillance, WGS is performed on all clinical isolates of designated bacterial pathogens, which were selected based on (1) causing substantial morbidity and mortality, (2) being associated with antibiotic resistance, and (3) propensity to cause healthcare-associated outbreaks. In contrast, under SoC, WGS is performed on isolates of any bacterial species that are suspected to be involved in outbreak. Two features of this approach are that WGS surveillance generally requires sequencing to be performed on a much larger number of bacterial isolates than occurs under SoC and that WGS surveillance is anticipated to detect outbreaks that would not be detected by SoC.
Although IP programs are essential to patient care and safety, they also contribute to the operating costs of a hospital. Therefore, a hospital needs an effective and efficient IP program. A cost-effectiveness analysis can be used to evaluate whether additional expenditure on a WGS surveillance-based IP program is of benefit to a hospital. Even though a number of studies have examined economic value of IP programs [10–13], only Dymond et al [14] has examined cost-effectiveness of WGS-based surveillance. They concluded WGS-surveillance to be a cost saving and more effective approach than SoC for methicillin-resistant Staphylococcus aureus (MRSA) infections because they presumed 90% reduction in probability of MRSA acquisition under the WGS surveillance-based program. The present study contributes to knowledge by incorporating outbreak transmission networks, effectiveness of intervening against a variety of transmission routes and including outbreaks of high-impact organisms such as Klebsiella pneumoniae, and Pseudomonas aeruginosa into economic evaluation.
The objective of this study was to develop a method to comprehensively assess the cost-effectiveness and budget impact of a WGS surveillance-based IP program compared to the SoC using outbreaks that were detected by SoC over a 5-year period at our institution.
METHODS
The analysis was conducted from the perspective of University of Pittsburgh Medical Center (UPMC) Presbyterian Hospital, an adult medical/surgical tertiary care hospital [15]. The method for estimating economic benefit of IP program consisted of 3 steps: (i) building the transmission network for each outbreak, (ii) estimating the number of transmissions averted due to WGS surveillance-based IP program compared to SoC, as defined in the Introduction, and (iii) estimating number of lives saved and economic outcomes.
Building the Transmission Network
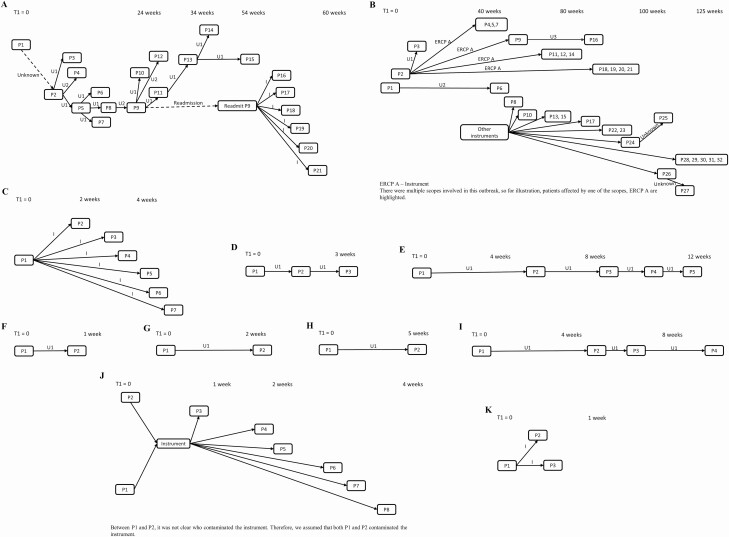
The transmission network of an outbreak comprised patients who were part of the outbreak and the epidemiological link connecting patients with the probable route(s) of transmission. Using WGS for selected bacterial isolates, patients with genetically closely related isolates were identified as described in prior studies [15, 16]. For these patients, we then identified common exposures through manual review of health records. If there was >1 patient who could have transmitted the infection to a patient, we assumed that the person with maximum duration of contact with the susceptible patient was the source. The date of acquiring infection was assumed as 5 days before the date of positive culture if the transmission route was a shared unit, and as procedure date, if the transmission route was an instrument. In this study, we have included 11 outbreaks that were detected by SoC at our hospital during 2011–16 (Table 1) (Figure 1A–K) [9, 17].
Table 1.
Outbreak-specific Inputs
| Variable | Value |
|---|---|
| Number of outbreaks by organism | 11 |
| Klebsiella pneumoniae | 3 |
| Acinetobacter baumannii | 2 |
| Clostridioides difficile | 4 |
| Pseudomonas aeruginosa | 1 |
| Pseudomonas putida | 1 |
| Total number of patients, N | 89 |
| Number of patients in each outbreak, median (range) | 4 (2–32) |
| Duration of outbreak in weeks, median (range) | 5.1 (0.6–125.1) |
| Total number of positive culturesa | 90 |
| Frequency of positive culture by source, n (%) | |
| Respiratory | 44 (49%) |
| Wound | 13 (14%) |
| Urine | 5 (6%) |
| Blood | 17 (19%) |
| Rectal swab | 1 (1%) |
| Stool | 10 (11%) |
aTotal number of positive cultures (n = 90) is one more than the number of patients (N = 89) because one patient had 2 positive cultures. The maximum number of transmissions that can be prevented would be 89; therefore, the patient having 2 positive cultures is counted once for estimating the number of transmissions averted but counted twice to estimate cost of treating infections.
Figure 1.
Transmission network of outbreaks (A–K) included in the economic analysis. Patients are represented as nodes (P1, P2, P3, etc.), arrows indicate transmission between patients, and transmission route is indicated above the arrows. The x-axis denotes time (not to scale) at which transmission occurred, and T1 represents start date of the outbreak. I, U1, U2, and U3 refer to Instrument, Unit 1, Unit 2, and Unit 3 as transmission routes, respectively. A, Klebsiella pneumoniae – A outbreak; B, Klebsiella pneumoniae – B outbreak; C, Klebsiella pneumoniae – C outbreak; D, Acinetobacter baumannii – A outbreak; E, Acinetobacter baumannii – B outbreak; F, Clostridioides difficile – A outbreak; G, Clostridioides difficile – B outbreak; H, Clostridioides difficile – C outbreak; I, Clostridioides difficile – D outbreak; J, Pseudomonas aeruginosa outbreak; K, Pseudomonas putida outbreak. Abbreviation: ERCP A, endoscopic retrograde cholangiopancreatography.
The effectiveness of intervention differs by transmission route (Table 2). The removal of a single contaminated instrument from service was assumed to have 100% effectiveness (relative risk, rr = 0) in halting transmission. If there were other instruments involved, the IP team would design an intervention that would affect all instruments, and the effectiveness of such an intervention was assumed 100%. The effectiveness of intervening to stop a unit-based outbreak would depend on the effectiveness of implemented interventions, such as improving or enhancing environmental cleaning, strengthening adherence to hand hygiene and personal protective equipment use by healthcare workers. We used estimates from 2 studies describing the effectiveness of unit-based interventions. First, Jayaraman et al [11] observed 48% reduction in the rate of infection with proactive IP program, which included deep cleaning of the intensive care unit, enhanced hand hygiene, and increased nurse-to-patient ratio. Second, Anderson et al [18] found that the risk of transmission reduced by 30% when room was decontaminated with ultraviolet C light and quaternary ammonium disinfectant compared to quaternary ammonium disinfectant alone. In the base case, we have used effectiveness value of 30% (rr = 0.70), but in a scenario analysis, we have used effectiveness value of 48% (rr = 0.52) to test the impact of our assumption. The effectiveness of intervening on a unit was assumed same for all types of inpatient units. For patients for whom we were not able to identify the transmission route, we have denoted the route as “Unknown” and to be conservative, we assigned an effectiveness value of 0%.
Table 2.
Data Inputs for Variables in the Model
| Variable | Mean | 95% CIa | Distribution | Source |
|---|---|---|---|---|
| Effectiveness related parameters | ||||
| Time from transmission to positive culture results under WGS surveillance-based IP program | 8 days | 6–10 days | Gamma | Assumption |
| Response time under WGS surveillance-based IP programb | 9 days | 7–11 days | Gamma | Assumption |
| Effectiveness (relative risk) of intervening against transmission routes | ||||
| Instrument | 0.00 | Not varied | Assumption | |
| Inpatient unit | 0.70 | .50–.98 | Lognormal | [18] |
| Unknown | 1.00 | Not varied | Assumption | |
| % colonized respiratory culturesc | 49% | 38%–60% | Beta | [19] |
| Attributable mortality risk due to infection | ||||
| Pneumonia | 0.143 | .142–.145 | Beta | [20] |
| Wound | 0.028 | .028–.029 | Beta | [20] |
| Urinary tract | 0.023 | .023–.024 | Beta | [20] |
| Bacteremia | 0.123 | .122–.125 | Beta | [20] |
| Clostridioides difficile | 0.030 | .029–.031 | Beta | [21] |
| Cost-related parametersd | ||||
| Annual salary of an IP professional | $89 398 | $86 236–$92 561 | Normal | [22] |
| Number of IP professionals in IP team | ||||
| SoC | 8 | Not varied | Unpublished data | |
| WGS surveillance-based | 8 | Not varied | Assumption | |
| % time spent on outbreak investigations | ||||
| SoC | 10% | 8%–12% | Beta | Unpublished data |
| WGS surveillance-based | 10% | 8%–12% | Beta | Assumption |
| Cost of performing WGS per isolate | ||||
| SoC | $70 | $57–$84 | Gamma | Unpublished data |
| WGS surveillance-based | $70 | $57–$84 | Gamma | Unpublished data |
| Number of isolates sequenced per year | ||||
| SoC | 76 | 62–92 | Gamma | Unpublished data |
| WGS surveillance-based | 1300 | 1058–1567 | Gamma | Unpublished data |
| Cost of treating infection | ||||
| Pneumonia (ICD10 J15.0) | $22 335 | $19 851–$24 964 | Gamma | [2,3] |
| Wound (ICD10 T81.4XXA) | $15 747 | $15 364–$16 135 | Gamma | [23] |
| Urinary tract (ICD10 N39.0) | $7284 | $7186–$7383 | Gamma | [23] |
| Bacteremia (ICD10 R78.81) | $11 805 | $11 242–$12 382 | Gamma | [23] |
| C. diff infection (ICD10 A04.7) | $9870 | $9664–$10 078 | Gamma | [23] |
Certain variables such as effectiveness against instrument, number of IP professionals in IP team is considered fixed and not varied in probabilistic sensitivity analysis.
Abbreviations: C. diff, Clostridioides difficile; CI, confidence interval; ICD, International Classification of Diseases; IP, infection prevention; SoC, standard of care; WGS, whole genome sequencing.
aThe 95% CI column represents confidence interval for parameters whose estimates are sourced from published studies, while it represents uncertainty range for parameters (eg, response time, cost of sequencing) whose estimates are either assumption-based or sourced from internal data (labelled as unpublished).
bResponse time was defined as time taken to obtain WGS results and intervene on the transmission route identified.
cIt was assumed that all positive cultures from wound, urine and blood represented infections, while 51% of positive respiratory cultures were assumed infections and remaining 49% were considered colonized.
dAll costs were adjusted to 2018 using medical component of consumer price index (CPI) obtained from Bureau of Labor Statistics.
Estimating the Number of Transmissions Averted
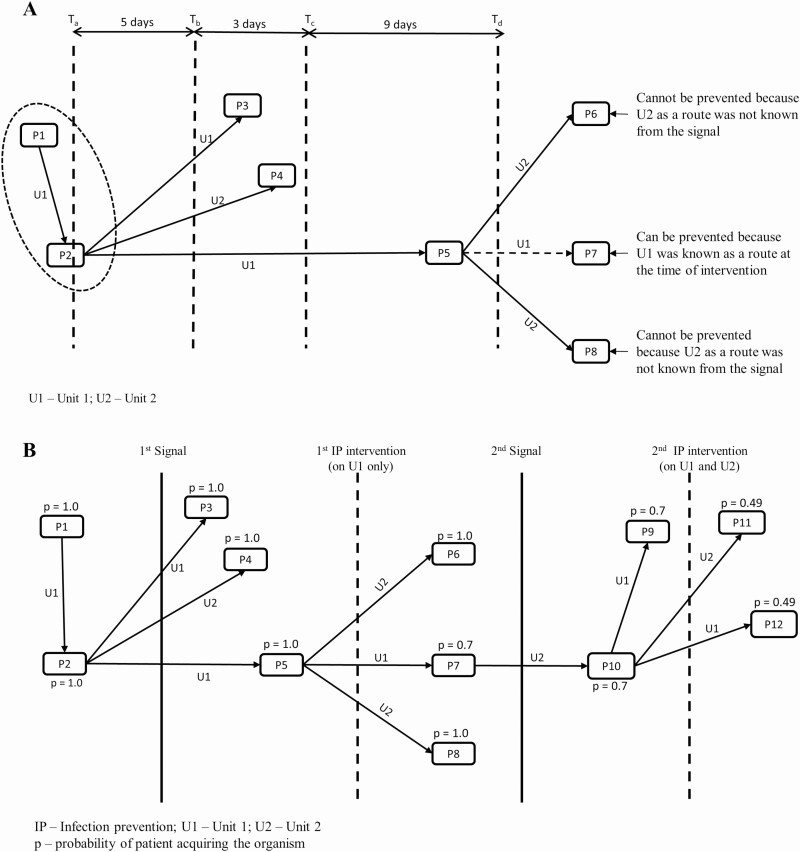
The number of transmissions averted depends on the effectiveness of intervening against transmission routes, time from transmission to receive positive culture results and response time, which was defined as total number of days taken to obtain WGS results and intervene on the transmission route identified. Figure 2 shows a pictorial representation of how we estimated expected number of transmissions under WGS surveillance-based program using a hypothetical infection transmission network. As shown in Figure 2A, the time from transmission to receive positive culture results was assumed 8 days (5 days to take the culture since transmission + 3 days to receive culture results) in the base case but was varied from 5 to 21 days in scenario analysis. The response time was assumed 9 days in the base case but was varied from 5 to 21 in scenario analysis. If IP team could have intervened at time Td (Figure 2A), transmission in P7 could have been prevented because, at the time of intervention, Unit 1 (U1) was known as the transmission route, but transmissions in P6 and P8 could not have been prevented because Unit 2 (U2) was not known as the transmission route. Mathematically, the probability of transmission was defined as:
Figure 2.
Conceptual diagram to estimate expected number of transmissions under WGS surveillance-based IP program. A, Healthcare associated transmission schematic. Hypothetical outbreak showing patients and the associated transmission routes. Patients are represented as nodes (P1, P2, P3, etc.), lines with arrows indicate the transmission between patients and transmission route is indicated above the arrows. At time Ta, patient P2 acquires organism and his/her sample is sent for culture at time Tb (ie, after 5 days of acquiring organism). At time Tc (ie, 3 days after Tb), the IP team would receive positive culture results confirming that patients P1 and P2 have the same species. After reviewing WGS results and health records, at time Td (ie, 9 days after Tc), the IP team would intervene on the transmission route identified. Assuming effectiveness value of 30% to stop unit-based outbreaks, the probability of P7 being infected would be 0.7, while the probability would be 1 for P6 and P8. B, Schematic depicting multiple signals if outbreak did not stop after intervention. Unit 1 (U1) would be the route identified from the first signal, and Unit 2 (U2) would be the route identified from the second signal. The probability of patient being infected was estimated assuming that the effectiveness of interventions to stop a unit-based outbreak would be 30%. Abbreviations: IP, infection prevention; WGS, whole genome sequencing.
| (1) |
Because the effectiveness of intervening against transmission routes can be <100%, some patients would still become infected despite the intervention. Therefore, the IP team would receive another outbreak signal, and a similar cycle of intervention would be repeated (Figure 2B). Consequently, IP team might become aware of new transmission routes as the outbreak progresses. We have assumed that the IP team would intervene on all transmission routes identified until the time of intervention. At each intervention point, we updated the probability of patient acquiring the organism using . The expected number of transmissions under the WGS surveillance-based IP program were then deducted from the corresponding number observed in SoC to estimate the number of transmissions averted.
Estimating Outcomes
Deaths Averted
We estimated expected number of deaths by applying infection-attributable mortality on only infected cases and not colonized. We assumed that all positive cultures from wound, urine, and blood represented infections. However, for respiratory cultures, it was not clear whether they were infections, so we assumed that 49% were colonized cases [19]. The mortality risk varies by the type of infection (eg, bacteremia vs wound infection) (Table 2) [20, 21].
Change in Costs
The total cost of an IP program included cost of staffing, WGS, and treating infections (Table 2). The staffing cost was based on salary and proportion of time spent on outbreak investigation by the IP professional staff involved. At our institution, there are 8 IP professionals who spend approximately 10% of their time on outbreak investigation activities in SoC. The salary was taken from Association for Professionals in Infection Control and Epidemiology’s MegaSurvey [22]. For WGS surveillance-based IP program, in the base case, the time commitment towards outbreak investigation was assumed same as that for SoC, that is, 10%.
The cost of WGS was based on the number of isolates sequenced over time. For SoC, 76 isolates were sequenced per year, whereas for WGS surveillance-based IP program, 1300 isolates per year are expected based on our current experience with WGS surveillance. The cost of WGS will vary according to the clinical setting and was considered to be $70 per isolate for both SoC and WGS surveillance-based IP programs based upon the method of Baym [24].
We used national data from AHRQ’s (Agency for Healthcare Research and Quality) Healthcare Cost and Utilization Project [23] to estimate attributable cost of treating infections. The cost was applied to infected cases assuming that colonized cases would not be treated. A scenario analysis was conducted where the proportion of colonized respiratory cases was varied from 5% to 95%. All cost-related inputs were adjusted to 2018 using the medical component of Consumer Price Index [25].
The primary cost-effectiveness measure was incremental cost per transmission averted. The budget impact measure was the change in total costs due to the WGS surveillance-based program compared to SoC. For cost-effectiveness, costs and number of transmissions averted were discounted at 3% to the start date of the earliest outbreak, that is, 2011, as per the guidelines for conducting cost-effectiveness analysis [26].
Sensitivity Analysis
We performed 3 types of sensitivity analysis: (i) scenario analyses as defined in previous sections, (ii) 1-way sensitivity analysis using 95% confidence interval for lower and upper bound values of model parameters to identify parameters driving cost-effectiveness (Table 2), and (iii) a probabilistic sensitivity analysis (PSA) [27] to assess uncertainty in costs and benefits of the WGS surveillance-based IP program. The PSA was based on 1000 simulations where, in each simulation, all but 3 model parameters were sampled randomly based on their probability distributions (Table 2). The exceptions were effectiveness of intervening against an instrument and unknown route, and the number of IP professionals in IP team, which were considered fixed and hence not varied in the PSA.
RESULTS
Base Case Results
The 11 outbreaks included 89 patients and each outbreak had 2–32 patients. Had WGS surveillance for outbreak detection been in place during the study period, there would have been approximately 41 fewer transmissions (including both colonization and infection) and 3.1 fewer deaths (Table 3 and Table 4).
Table 3.
Results: Number of Transmissions Averted Under WGS Surveillance-based infection prevention (IP) Program
| No. | Outbreak | SoC | WGS Surveillance | Transmissions averted |
|---|---|---|---|---|
| 1 | Klebsiella pneumoniae – A | 21.0 | 10.2 | 10.8 |
| 2 | K. pneumoniae – B | 32.0 | 7.0 | 25.0 |
| 3 | K. pneumoniae – C | 7.0 | 5.0 | 2.0 |
| 4 | Acinetobacter – A | 3.0 | 3.0 | 0.0 |
| 5 | Acinetobacter – B | 5.0 | 4.4 | 0.6 |
| 6 | Clostridioides difficile - A | 2.0 | 2.0 | 0.0 |
| 7 | C. difficile - B | 2.0 | 2.0 | 0.0 |
| 8 | C. difficile - C | 2.0 | 2.0 | 0.0 |
| 9 | C. difficile - D | 4.0 | 3.7 | 0.3 |
| 10 | Pseudomonas aeruginosa | 8.0 | 6.0 | 2.0 |
| 11 | Pseudomonas putida | 3.0 | 3.0 | 0.0 |
| Total | 89.0 | 48.3 | 40.7 |
Abbreviations: SoC, standard of care; WGS, whole genome sequencing.
Table 4.
Base Case Results
| SoC | WGS Surveillance | Change | |
|---|---|---|---|
| Budget impact resultsa | |||
| Number of transmissions | 89.0 | 48.3 | 40.7 averted |
| Number of deaths | 6.0 | 2.9 | 3.1 saved |
| Total costs | $1 468 778 | $1 456 962 | ($11 817) |
| IP program | $397 370 | $397 370 | $0 |
| WGS costs | $29 681 | $505 611 | $475 930 |
| Treating infections | $1 041 728 | $553 981 | ($487 747) |
| Cost-effectiveness resultsb | |||
| Number of transmissions | 81.2 | 43.3 | 37.9 averted |
| Total costs | $1 339 384 | $1 330 311 | ($9073) |
| IP program | $366 368 | $366 368 | $0 |
| WGS costs | $27 365 | $466 165 | $438 799 |
| Treating infections | $945 651 | $497 779 | ($447 872) |
Incremental cost per transmission averted for WGS surveillance-based IP program = $239 saved for each transmission averted, ie, less costly and more effective.
All costs and benefits are reported over 2011–2016 period; costs are in 2018 dollars.
Amount in parenthesis indicates savings.
Abbreviations: IP, infection prevention; SoC, standard of care; WGS, whole genome sequencing.
aFor budget impact, costs and outcomes were not discounted.
bFor cost effectiveness, costs and outcomes were discounted at 3% to the start date of earliest outbreak ie, 2011.
Had WGS surveillance been in place at the time of each outbreak and assuming the same number of outbreaks, it would have resulted in saving of $487 747 in infection treatment costs (~$11 900 per transmission averted) over the study period. However, the net savings would have been $11 817 because the cost of doing WGS surveillance increased by $475 930. The cost-effectiveness results indicated that the WGS surveillance-based IP program resulted in net saving of $9073 (discounted) and approximately 38 fewer transmissions (discounted), thereby making WGS surveillance-based IP program a less costly and more effective strategy than SoC (Table 4).
Sensitivity Analysis
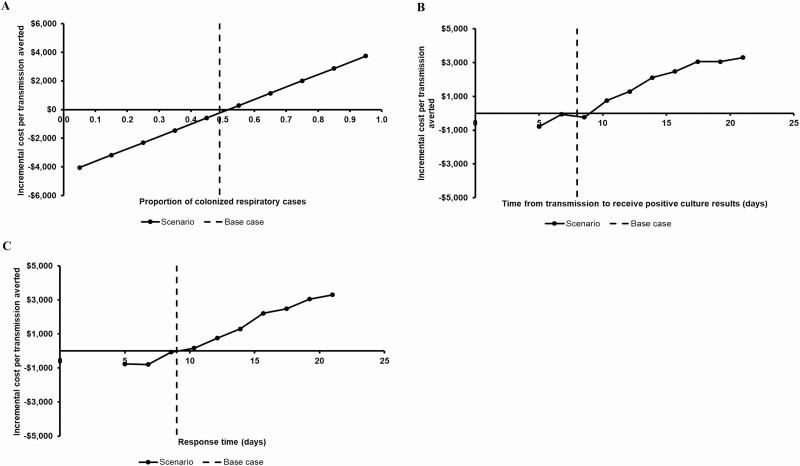
If the effectiveness of intervening against inpatient unit had been 48%, the WGS surveillance-based IP program would have resulted in net saving of $43 700 (vs $9073 in the base case) and 40 fewer transmissions (vs 38 in the base case) thereby making WGS surveillance even more favorable compared to SoC. We found that the WGS surveillance-based IP program became less favorable when the proportion of colonized respiratory cases increased (Figure 3A), time from transmission to receive positive culture results increased (Figure 3B), and response time increased (Figure 3C).
Figure 3.
Scenario analyses results (all other variables were held constant at base case values). A, Varying proportion of colonized respiratory positive cultures. The WGS surveillance-based infection prevention program became less favorable when proportion of colonized respiratory cases increased because cost savings in infection treatment costs decreased. The WGS surveillance-based program would be cost saving if the proportion of colonized respiratory cases was <55%. B, Varying time from transmission to receive positive culture results. The WGS surveillance-based infection prevention program became less favorable when time from transmission to receive positive culture results increased because it led to more delay in detecting an outbreak thereby making WGS surveillance less effective. C, Varying response time. The WGS surveillance-based infection prevention program became less favorable as response time increased because there would be more delay in intervening on transmission routes thereby making WGS surveillance less effective. Abbreviation: WGS, whole genome sequencing.
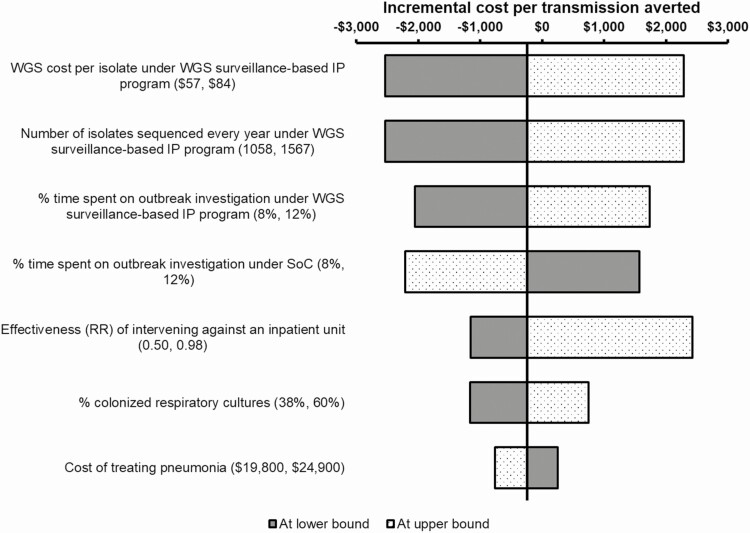
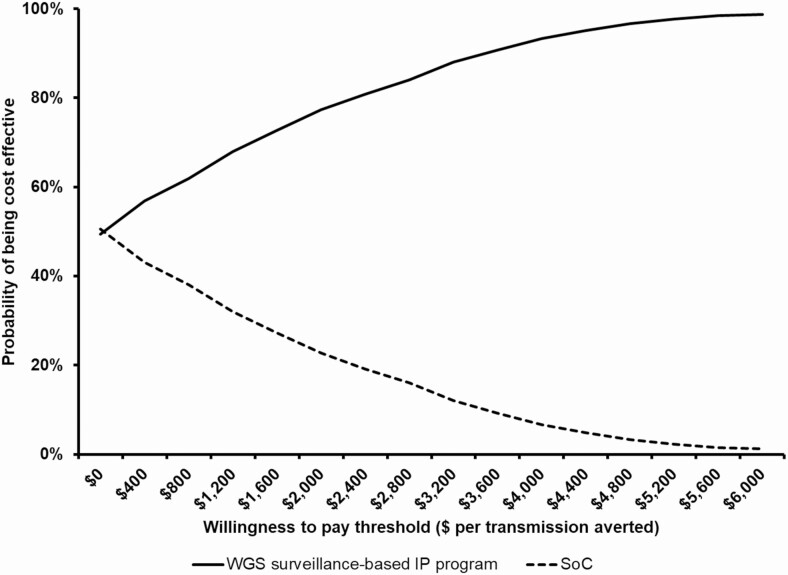
The results were most sensitive to the cost of performing WGS, the number of isolates sequenced per year, and amount of time spent on outbreak investigations under WGS surveillance (Figure 4). The PSA showed that the WGS surveillance-based program was cost saving and more effective than SoC in 49% of the simulations (data not shown). Based on the cost effectiveness acceptability curve, there was 80% chance that the WGS surveillance-based program would be cost-effective if willingness to pay exceeded $2400 per transmission averted (Figure 5).
Figure 4.
One-way sensitivity analysis results. The tornado diagram shows the most sensitive model parameters driving the cost effectiveness of WGS surveillance-based IP program. Each model parameter was varied 1 by 1 using 95% confidence interval as lower and upper bound values. The cost-effectiveness of WGS surveillance was most sensitive to cost of performing WGS, the number of isolates sequenced per year, and amount of time spent on outbreak investigations. Abbreviations: IP, infection prevention; RR, relative risk; SoC, standard of care; WGS, whole genome sequencing.
Figure 5.
Cost-effectiveness acceptability curve. The cost effectiveness acceptability curve shows the probability that the infection prevention program was cost effective for different levels of WTPs thresholds. If WTP is above $2400 per transmission averted, the WGS surveillance-based IP program has 80% chance of being cost-effective compared to SoC. Abbreviations: IP, infection prevention; SoC, standard of care; WGS, whole genome sequencing; WTP, willingness to pay.
DISCUSSION
This study proposes a robust method to evaluate economic benefit of a WGS surveillance-based IP program compared with a SoC approach that uses reactionary WGS in response to suspected outbreaks. The cost-effectiveness analysis showed that the WGS surveillance-based IP program would have saved 3 lives and was a less-costly and more-effective strategy than SoC. Our results are consistent with findings from other studies where IP programs were generally found cost-effective [10–14]. The cost-effectiveness of new IP programs has ranged from being a cost-saving program [10, 13, 14] to a more-costly program [11, 12]. However, the method we describe likely substantially underestimates the cost-effectiveness of a WGS surveillance-based IP program. This is because we have included only those outbreaks that were detected by SoC. Therefore, the present analysis is conservative and favors SoC as it does not take into account outbreaks that were likely missed by SoC [8].
In addition to cost-effectiveness, hospitals may be willing to invest in WGS surveillance given reductions in HAIs would reduce the financial penalty for falling in the worst-performing 25% hospitals with respect to hospital-acquired condition quality measures [28]. Also, hospital resources can be used more efficiently to provide better care to other patients because of freed up staff and bed-days due to transmissions averted. All these factors would further improve the value of a WGS surveillance-based IP program.
This study is methodologically similar to previous studies investigating the cost-effectiveness of infection prevention programs [10, 11, 13, 14]. Our method has several advantages. First, our model explicitly takes into account that the effectiveness of WGS surveillance is dependent not only on early outbreak detection but also on response time and effectiveness of interventions used to contain the outbreak. Second, the model has the flexibility to analyze customized infection outbreak networks. Third, the economic model is sufficiently transparent to understand how costs and benefits would change under WGS surveillance-based IP program.
This study also has limitations. First, as mentioned above, the method is based on outbreaks that were detected by SoC. As we and others have shown, WGS surveillance can detect previously unidentified outbreaks [6, 8]; therefore, the expected number of transmissions averted would be higher than estimated here. We are currently working on an analysis using prospective data to account for this limitation and determine the true cost-effectiveness of WGS surveillance. Second, WGS was done for only clinical isolates and not colonization isolates. Therefore, we are underestimating the number of transmissions. Third, we did not incorporate quality of life measures because the number of transmissions averted was the main outcome from hospital’s perspective, and there was significant variation in patient characteristics and decrement in quality of life due to an infection was not readily available. Both these factors complicated the selection of an appropriate quality of life value. Fourth, for some cases we were not able to identify the epidemiological link even when WGS showed isolates from patients to be genetically related. We accounted for this by considering the effectiveness of intervening against the unknown route as 0%. Fifth, we did not model recurrent infection; treatment was considered successful and a final result. Had we incorporated recurrence, the average cost of treating infection would increase thereby increasing the amount of savings.
This study has substantial implications. Hospitals can use this method to assess economic value and make a business case for their IP program. We are using this method to assess the economic benefit of Enhanced Detection System for Healthcare Associated Transmission (EDS-HAT), a new WGS surveillance-based IP program that is currently being developed and validated at UPMC [8, 15, 29]. EDS-HAT uses WGS surveillance and data mining of EHR to identify outbreaks and transmission routes. Our study contributes toward knowledge gaps regarding economic evaluation of WGS surveillance-based IP programs [6]. Our preliminary findings suggest that a WGS surveillance-based IP program would be a cost-effective strategy.
CONCLUSION
The proposed economic analysis is a useful tool to examine the potential cost-effectiveness and budget impact of any WGS surveillance-based IP program.
Notes
Acknowledgments. The authors are grateful for the expert technical assistance of Chinelo Ezeonwuka, Jessica Schlackman, and Joseph Penzelik.
Financial support . This study was funded in part by the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH) (R21Al109459 and R01AI127472). NIH played no role in data collection, analysis, or interpretation; study design; writing of the manuscript; or decision to submit for publication.
Potential conflicts of interest. G. M. S. serves as a scientific advisor to Infectious Disease (ID) Connect. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Stone PW. Economic burden of healthcare-associated infections: an American perspective. Expert Rev Pharmacoecon Outcomes Res 2009; 9:417–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Magill SS, Edwards JR, Bamberg W, et al. ; Emerging Infections Program Healthcare-Associated Infections and Antimicrobial Use Prevalence Survey Team . Multistate point-prevalence survey of health care-associated infections. N Engl J Med 2014; 370:1198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. AHRQ. AHRQ’s healthcare-associated infections program.2018. July 2018 [cited 21 December 2018]; Available at: https://www.ahrq.gov/professionals/quality-patient-safety/hais/index.html. Accessed 21 December 2018.
- 4. Scott R The direct medical costs of healthcare-associated infections in US hospitals and the benefits of prevention. Centers for Disease Control and Prevention, 2009. Available at: https://www.cdc.gov/HAI/pdfs/hai/Scott_CostPaper.pdf. Accessed 21 December 2018. [Google Scholar]
- 5. Jeeva RR, Wright D. Healthcare-associated infections: a national patient safety problem and the coordinated response. Med Care 2014; 52:S4–8. [DOI] [PubMed] [Google Scholar]
- 6. Peacock SJ, Parkhill J, Brown NM. Changing the paradigm for hospital outbreak detection by leading with genomic surveillance of nosocomial pathogens. Microbiology 2018; 164:1213–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sood G, Perl TM. Outbreaks in health care settings. Infect Dis Clin North Am 2016; 30:661–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sundermann AJ, Babiker A, Marsh JW, et al. . Outbreak of vancomycin-resistant Enterococcus faecium in interventional radiology: detection through whole genome sequencing-based surveillance. Clin Infect Dis 2020; 70:2336–43. doi:10.1093/cid/ciz666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Marsh JW, Krauland MG, Nelson JS, et al. . Genomic epidemiology of an endoscope-associated outbreak of Klebsiella pneumoniae carbapenemase (KPC)-producing K. pneumoniae. PLoS One 2015; 10:e0144310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hutton DW, Krein SL, Saint S, et al. . Economic evaluation of a catheter-associated urinary tract infection prevention program in nursing homes. J Am Geriatr Soc 2018; 66:742–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jayaraman SP, Jiang Y, Resch S, Askari R, Klompas M. Cost-effectiveness of a model infection control program for preventing multi-drug-resistant organism infections in critically ill surgical patients. Surg Infect (Larchmt) 2016; 17:589–95. [DOI] [PubMed] [Google Scholar]
- 12. Nelson RE, Jones M, Leecaster M, et al. . An Economic analysis of strategies to control Clostridium difficile transmission and infection using an agent-based simulation model. PLoS One 2016; 11:e0152248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cohen ER, Feinglass J, Barsuk JH, et al. . Cost savings from reduced catheter-related bloodstream infection after simulation-based education for residents in a medical intensive care unit. Simul Healthc 2010; 5:98–102. [DOI] [PubMed] [Google Scholar]
- 14. Dymond A, Davies H, Mealing S, et al. . Genomic surveillance of methicillin-resistant Staphylococcus aureus: a mathematical early modeling study of cost-effectiveness. Clin Infect Dis 2020; 70:1613–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sundermann AJ, Miller JK, Marsh LW, et al. . Automated data mining of the electronic health record for investigation of healthcare-associated outbreaks. Infect Control Hosp Epidemiol 2019; 40:314–9. doi:10.1017/ice.2018.343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Marsh JW, Mustapha MM, Griffith MP, et al. . Evolution of outbreak-causing carbapenem-resistant Klebsiella pneumoniae ST258 at a tertiary care hospital over 8 years. mBio 2019; 10:e01945-19. doi: 10.1128/mBio.01945-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Parr A, Querry A, Pasculle A, Morgan D, Muto C. Carbapenem-resistant Klebsiella pneumoniae cluster associated with gastroscope exposure among surgical intensive care unit patients at University of Pittsburgh Medical Center. Open Forum Infect Dis 2016; 3(suppl_1): 248. doi: 10.1093/ofid/ofw172.115 [DOI] [Google Scholar]
- 18. Anderson DJ, Chen LF, Weber DJ, et al. ; CDC Prevention Epicenters Program . Enhanced terminal room disinfection and acquisition and infection caused by multidrug-resistant organisms and Clostridium difficile (the Benefits of Enhanced Terminal Room Disinfection study): a cluster-randomised, multicentre, crossover study. Lancet 2017; 389:805–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Martin RM, Cao J, Brisse S, et al. . Molecular epidemiology of colonizing and infecting isolates of Klebsiella pneumoniae. mSphere 2016; 1:e00261–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Klevens RM, Edwards JR, Richards CL Jr, et al. . Estimating health care-associated infections and deaths in US hospitals, 2002. Public Health Rep 2007; 122:160–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. CDC. Nearly half a million Americans suffered from Clostridium difficile infections in a single year. US Department of Health and Human Services, 2015. Available at: https://www.cdc.gov/media/releases/2015/p0225-clostridium-difficile.html. Accessed 31 July 2018. [Google Scholar]
- 22. Landers T, Davis J, Crist K, Malik C. APIC MegaSurvey: methodology and overview. Am J Infect Control 2017; 45:584–8. [DOI] [PubMed] [Google Scholar]
- 23.HCUPnet, Healthcare Cost and Utilization Project. Rockville, MD: Agency for Healthcare Research and Quality, 2016. Available at: https://hcupnet.ahrq.gov/. Accessed 19 May 2019. [Google Scholar]
- 24. Baym M, Kryazhimskiy S, Lieberman TD, Chung H, Desai MM, Kishony R. Inexpensive multiplexed library preparation for megabase-sized genomes. PLoS One 2015; 10:e0128036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bureau of Labor Statistics. Medical care in US city average, all urban consumers, not seasonally adjusted. Washington, DC: Division of Consumer Prices and Price Indexes, 2019. [Google Scholar]
- 26. Sanders GD, Neumann PJ, Basu A, et al. . Recommendations for conduct, methodological practices, and reporting of cost-effectiveness analyses: second panel on cost-effectiveness in health and medicine. JAMA 2016; 316:1093–103. [DOI] [PubMed] [Google Scholar]
- 27. Doubilet P, Begg CB, Weinstein MC, Braun P, McNeil BJ. Probabilistic sensitivity analysis using Monte Carlo simulation: a practical approach. Med Decis Making 1985; 5:157–77. [DOI] [PubMed] [Google Scholar]
- 28. CMS.gov. Hospital-Acquired Condition Reduction Program (HACRP).2018. 07/30/2018 4:42 PM [cited 20 December 2018]; Available at: https://www.cms.gov/medicare/medicare-fee-for-service-payment/acuteinpatientpps/hac-reduction-program.html. Accessed 20 December 2018.
- 29. Miller JK, Chen J, Sundermann A, et al. . Statistical outbreak detection by joining medical records and pathogen similarity. J Biomed Inform 2019; 91:103126. [DOI] [PMC free article] [PubMed] [Google Scholar]