Thorough purification and cross‐linking of the NuRD complex from multiple angles led to a high‐density interaction network. The core is formed by MTA‐HDAC and RBBP proteins, to which MBD and GATAD2 proteins dock. The latter proteins associate with CHDs, which in turn recruit CDK2AP1 and its paralog CDK2AP2 via their C termini.
Keywords: CDK2AP2, cross‐linking, NuRD, xIP‐MS
Abstract
The multi‐subunit nucleosome remodeling and deacetylase (NuRD) complex consists of seven subunits, each of which comprises two or three paralogs in vertebrates. These paralogs define mutually exclusive and functionally distinct complexes. In addition, several proteins in the complex are multimeric, which complicates structural studies. Attempts to purify sufficient amounts of endogenous complex or recombinantly reconstitute the complex for structural studies have proven quite challenging. Until now, only substructures of individual domains or proteins and low‐resolution densities of (partial) complexes have been reported. In this study, we comprehensively investigated the relative orientation of different subunits within the NuRD complex using multiple cross‐link IP mass spectrometry (xIP‐MS) experiments. Our results confirm that the core of the complex is formed by MTA, RBBP, and HDAC proteins. Assembly of a copy of MBD and GATAD2 onto this core enables binding of the peripheral CHD and CDK2AP proteins. Furthermore, our experiments reveal that not only CDK2AP1 but also CDK2AP2 interacts with the NuRD complex. This interaction requires the C terminus of CHD proteins. Our data provide a more detailed understanding of the topology of the peripheral NuRD subunits relative to the core complex.
Database
Proteomics data are available in the PRIDE database under the accession numbers PXD017244 and PXD017378.
Abbreviations
- ADH
adipic acid dihydrazide
- BAC
bacterial artificial chromosome
- BAH domain
bromo‐adjacent homology domain
- BS3
bis(sulfosuccinimidyl) suberate sodium salt
- CDK2AP
cyclin‐dependent kinase 2‐associated protein
- CHD
chromodomain helicase DNA‐binding protein
- CR
conserved region
- DMTMM
4‐(4,6‐dimethoxy‐1,3,5‐triazin‐2‐yl)‐4‐methyl‐morpholinium chloride
- DOC‐1R
deleted in oral cancer 1‐related
- FRT‐TO
flippase recognition target–tetracycline operon
- GATAD
GATA‐zinc finger domain‐containing protein
- GFP
green fluorescent protein
- HDAC
histone deacetylase
- LFQ
label‐free quantification
- MBD
methyl‐CpG‐binding domain
- MTA
metastasis‐associated protein
- NuRD
nucleosome remodeling and deacetylase
- RBBP
retinoblastoma‐binding protein
- xIP‐MS
cross‐link IP mass spectrometry
- XL
cross‐linked peptide pair
- XL‐MS
cross‐linking followed by mass spectrometry
- ZNF687
zinc finger protein 687
Introduction
DNA in human cells is wrapped around an octamer of histone proteins to form nucleosomes. These nucleosomes protect DNA against (external) damaging factors, in addition to compacting the DNA. The assembly of nucleosomes and other proteins into a higher‐order structure called chromatin also plays an active role in regulating DNA accessibility and processes such as transcription and DNA replication. Chromatin accessibility is regulated by chromatin remodeling complexes that can reposition nucleosomes on DNA. These remodeler complexes can either positively or negatively affect DNA accessibility and thus gene expression. Examples of these multi‐subunit complexes are Ino80 and the Swi/Snf complex [1]. Some complexes combine chromatin remodeling activity with histone‐modifying activity. Modifications on the N‐terminal tails of histone proteins, such as lysine methylation or acetylation, are important for regulation of transcription by attracting effector or ‘reader’ proteins [2]. One of the few complexes that combines nucleosome remodeling activity with histone‐modifying activity is called the nucleosome remodeling and deacetylase (NuRD) complex [3]. The NuRD complex is conserved in the entire metazoan lineage, from sponges to humans [4]. However, both the subunits and the complex itself have evolved. For example, whereas in mammals the NuRD complex is best known for its ability to bind methylated DNA, the Drosophila genome is depleted of CpG methylation and Drosophila NuRD has a very low affinity for methyl‐CpG [5]. Additional functions of the complex probably evolved in conjunction with gene duplication events and the appearance of novel transcription factors. This may be why invertebrates have only one paralog for each of the seven NuRD subunits, while the mammalian genome encodes 2 or 3 paralogs for every subunit.
In this study, we investigated the overall architecture of the NuRD complex by performing the most comprehensive XL‐MS study performed for NuRD thus far. Our data reveal that the conserved region 2 (CR2) of GATAD2A is sufficient to bind CHD4 and CDK2AP1, whereas conserved region 1 (CR1) of GATAD2A is required for the interaction with the core NuRD complex. Furthermore, we identify a C‐terminal region of the CHD proteins, which likely serves as an interaction surface for CDK2AP1 and CDK2AP2. Finally, we show that the CDK2AP1 paralog CDK2AP2 is a bona fide NuRD complex subunit.
Results
Comprehensive topological analyses of the NuRD complex
To investigate the topology of the NuRD complex, we performed GFP affinity purifications followed by cross‐linking and mass spectrometry (xIP‐MS [6]) for multiple NuRD core subunits (Fig. 1A). In short, xIP‐MS reveals which proteins and which parts of those proteins are in close proximity, but not at atomic resolution, as the used cross‐linkers are about 0–15 Å in length. Since these cross‐linkers react with primary amines, such as those present in lysines, the flexibility of the side chains has to be taken into consideration as well, meaning that cross‐links can span a maximum distance of 35 Å between Cα atoms. To obtain a comprehensive interaction network, we performed xIP‐MS for six core NuRD complex subunits, namely MBD3, MTA1, CDK2AP1, RBBP7, CHD4, and HDAC1 and the substoichiometric interactor ZNF687 [7]. To obtain near‐endogenous expression of GFP‐tagged constructs in HeLa Kyoto cells, we used the bacterial artificial chromosome (BAC) system [8], since overexpression of a protein can cause large pools of the tagged protein to be unincorporated in the complex. For CHD4, we used CRISPaint to endogenously tag the CHD4 gene with a C‐terminal GFP, since no BAC construct or cell line was available [9]. Western blots using antibodies against endogenous NuRD subunits revealed that the transgenes are in all cases apart from CDK2AP1 expressed at lower levels compared with the endogenous proteins (Fig. 1A). For CHD4, no extra band is observed, possibly due to the size of the endogenous protein. MBD3‐GFP is very difficult to detect, probably due to its low expression in differentiated cells. We generated nuclear extracts from these stable cell lines and purified the GFP‐tagged proteins using GFP nanobodies immobilized on sepharose beads under stringent washing conditions (1 m NaCl, 1% NP40). A silver‐stained gel shows that in addition to the NuRD subunits, quite some other proteins are co‐purified, despite the stringent washing procedures (Fig. 1B). Whereas a part of these proteins may be specific interactors of HDAC1 and RBBP7 (which are in multiple protein complexes), many of them may also be nonspecific background proteins. To obtain a very dense dataset, we performed multiple replicate pull‐downs for each of the baits and performed cross‐linking with either BS3 or ADH + DMTMM (Table 1, Fig. 2A). Furthermore, to obtain more sequence information, we used multiple proteases for on‐bead digestion after cross‐linking, namely trypsin and chymotrypsin, resulting in 35 samples in total. Data were analyzed using the pLink2.0 software [10].
Fig. 1.
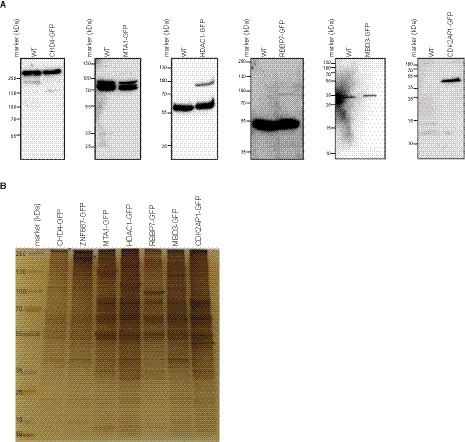
Expression analysis of the used cell lines. (A) Western blot analysis of 6 cell lines shows that the tagged proteins are expressed at much lower levels than the endogenous protein. The exception is CDK2AP1. Black triangle indicates the endogenous, untagged protein, while asterisk indicates the tagged protein (n = 1). (B) Silver‐stained gel to visualize the homogeneity of the purifications of NuRD via different bait proteins (n = 1). Asterisks indicate the tagged proteins.
Table 1.
Number of identified and filtered cross‐linked sites per condition. Ambiguous cross‐links were counted as a single cross‐link.
| Condition | Loop links | Cross‐links | % NuRD cross‐links | Filtered cross‐links | NuRD cross‐links |
|---|---|---|---|---|---|
| BS3‐trypsin | 293 | 248 | (197/248) 79.4% | 81 | (67/81) 82.7% |
| BS3‐chymotrypsin | 186 | 180 | (139/180) 77.2% | 45 | (39/45) 86.7% |
| ADH‐DMTMM‐trypsin | 443 | 166 | (153/166) 92.1% | 59 | (58/59) 98.3% |
| ADH‐DMTMM‐chymotrypsin | 99 | 74 | (46/74) 62.1% | 11 | (8/11) 72.7% |
Fig. 2.
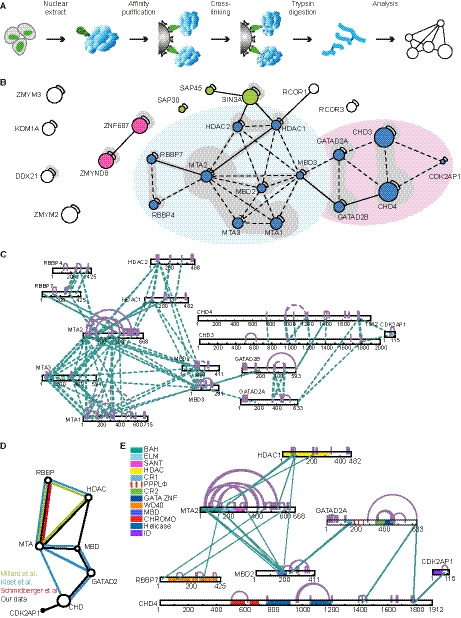
Cross‐linking mass spectrometry to unveil the NuRD complex conformation. (A) Schematic representation of the xIP‐MS workflow. (B) A schematic representation of the network of all identified cross‐links from our 35 affinity purifications. NuRD subunits are indicated in blue, and known interactors are shown in pink. The Sin3A co‐repressor complex is shown in green. No cross‐links between PRC2 and NuRD subunits were observed. Solid lines indicate unambiguous cross‐links, whereas dashed lines indicate ambiguous cross‐links. Gray shading indicates the amount of evidence per set of proteins connected. (C) Overview of the interaction network within the NuRD complex obtained in this study with 35 affinity purifications. Solid lines indicate unambiguous cross‐links, whereas dashed lines indicate ambiguous cross‐links. Purple lines indicate intraprotein cross‐links. (D) Comparison of this dataset to other datasets that have been published before. (E) Cross‐link network of all NuRD subunits with their functional domains indicated. The MTA2 and the MBD proteins are in the center of the network.
In total, these 35 samples resulted in 196 high‐confidence cross‐linked peptide pairs (Table S1). Since RBBP7 and HDAC1 are incorporated into many different protein complexes, the analyses of all purifications were performed with a database containing not only NuRD, but also SIN3A and PRC2 subunits. Between 72% and 98% of the identified cross‐links for each experiment were between or within NuRD subunits. In total, 169 high‐confidence (score < 10−5) cross‐links were assigned to NuRD complex subunits. Of these, 92 were unambiguous intraprotein cross‐links, of which 60 were cross‐links spanning less than 25 residues, whereas 32 spanned more than 25 residues and may contain information about the conformation of the subunit. In addition, 54 cross‐links were ambiguously assigned to different paralogs of the same subunit, although for all of these, intraprotein linking is a possible explanation that could also explain these cross‐links. Concerning interprotein cross‐links, we identified 7 unambiguous cross‐links and 16 cross‐links that were ambiguous either on one or on both sides. These 23 cross‐links provide information about the topology of the complex.
We used the xiNET viewer [11] to generate a network of all cross‐links with the NuRD complex subunits visualized in the center (Fig. 2B). Figure 2C shows an overview of the cross‐links within the NuRD complex. Although the absence of a cross‐link does not necessarily mean that an interaction does not exist, we did not identify any cross‐links between the GATAD2 proteins and the RBBP proteins or between the HDAC and RBBP proteins. In addition, the shape of the network suggests that the CHD proteins are peripheral subunits, which interact with the MBD/GATAD2 surface. Furthermore, we did not observe any cross‐links between CHD proteins and either HDACs or RBBPs. This is in agreement with multiple other studies [12, 13, 14, 15, 16]. Cross‐links from the smallest NuRD subunit, CDK2AP1, connect it to the C termini of both CHD3 and CHD4. Previous studies have not identified any cross‐links between CDK2AP1 and the NuRD complex (Fig. 2D).
It should be noted that although this cross‐linking network may suggest that both MBD2 and MBD3, GATAD2A and GATAD2B, and CHD3 and CHD4 can assemble together in NuRD complexes, this conclusion is not justified based on these experiments. NuRD complexes are purified from cells expressing all NuRD subunit paralogs, a heterogeneous mixture of distinct NuRD complexes is thus purified. Therefore, in a single HDAC or RBBP purification, both MBD2 and MBD3 are purified, for example. The—in theory—ambiguous cross‐links will in practice be derived from intraprotein cross‐links, based on previous work from others and us that has shown that MBD2/3, GATAD2A/B, and CHD3/4 define mutually exclusive NuRD complexes [7, 17, 18]. Conclusions regarding the stoichiometry of the complex based on this cross‐linking network cannot be made.
Previously, we have shown that the interaction between the Z3 module (ZMYND8, ZNF687, ZNF532, and ZNF592) and NuRD is mediated by the MYND domain of ZMYND8 and the PPPLՓ motifs in GATAD2A [ 7 ]. To further investigate the molecular interaction between Z3 and NuRD, we performed xIP‐MS experiments with ZNF687 as a bait. The ZNF687 purification resulted in 173 cross‐linked peptide spectra and 94 identified cross‐links. Most of these represent intraprotein ZNF687 and ZMYND8 cross‐links, as well as a few spectra mapping to CHD, GATAD2, and MBD subunits. Unfortunately, due to high stringency filtering, only cross‐links between ZNF687 and ZMYND8 remained.
To study these data in more detail, we reduced the ambiguous cross‐links to a single paralog of each subunit (Fig. 2E and Table S2). Most cross‐links were identified between MTA and the RBBP and HDAC proteins, but also the MBD of the MBD proteins are located in close proximity to the MTA BAH domain (Fig. 1E). From this MBD, a couple of cross‐links map to the GATAD2 proteins. The GATAD2 proteins are cross‐linked to the MBDs through their conserved region 1 (CR1) domains and to the CHD proteins through their C termini. This suggests a scaffold of MTA, RBBP, and HDAC proteins, on which an MBD, GATAD2, and finally a CHD and a CDK2AP protein can dock. This is in agreement with previous studies, although we did not identify any of the cross‐links identified by Schmidberger et al., and only 17 cross‐links overlapped with those identified by Kloet et al. [12, 19, 20]. The fact that so few overlapping and so many unique cross‐links are present in each dataset indicates that cross‐linking of large protein complexes is still fairly variable and inefficient, due to conformational changes in flexible parts.
To test the confidence of our cross‐links, we mapped the reduced set of BS3 and ADH/DMTMM cross‐links onto the known structures of single molecules of HDAC1 (PDB 4BKX), RBBP7 (PDB 3CFV), and a homology model of a residue 499–1281 of CHD4 using ucsf chimera and the plugin Xlink analyzer (Fig. 3A–C) [21, 22]. As the heterogeneity of the NuRD complex hinders structural studies of the complex, these are unfortunately the only larger structures available for NuRD complex subunits. All of the cross‐links that we mapped onto 3D models are in agreement with the published structures and did not exceed the distance restraint of 35 Å between Cα atoms, thus illustrating the quality of our datasets. When comparing the observed cross‐links to random solvent accessible surface distances between lysines using Jwalk [23], it is clear that the identified cross‐links did not violate the spatial restraints (Fig. 3D,E). These data also point out that most of the observed ambiguous cross‐links most likely represent short‐distance intraprotein cross‐links rather than interprotein cross‐links, except for the MTA subunits.
Fig. 3.
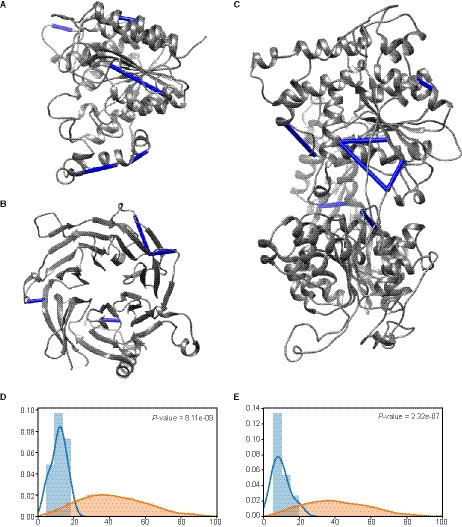
Validation of the observed XL lengths. (A–C) X‐ray (A, B) and homology (C) models for single molecules of HDAC1, RBBP7, and CHD4 with the intraprotein cross‐links indicated. All validated cross‐links have a Cα‐Cα length below 35 Å. Images were made using ucsf chimera with the Xlink Analyzer plugin [21, 22]. (D) Comparison of the observed Cα‐Cα cross‐link lengths (blue) to the random solvent accessible surface Cβ‐Cβ distance between lysines (orange) for BS3. (E) Comparison of the observed Cα‐Cα cross‐link lengths (blue) to the random solvent accessible surface Cβ‐Cβ distance between lysines (orange) for ADH + DMTMM.
In summary, we identified cross‐links in xIP‐MS pull‐downs of seven NuRD subunits and interactors, using either BS3 or ADH cross‐linking combined with digestion using different proteases. The resulting network shows a clear NuRD‐centered interactome, in which the core is formed by MTA, RBBP, and HDAC proteins.
GATAD2 proteins connect CHDs with the core NuRD complex
Our cross‐linking data suggest that whereas the GATAD2‐conserved region 1 (CR1) mediates the association with MBD, MTA, RBBP, and HDAC proteins, conserved region 2 (CR2) may be required for the interaction with the CHD proteins (Fig. 4A). To further investigate this, we generated several GFP‐tagged deletion constructs for GATAD2A (Fig. 4B). We then expressed these in HeLa Kyoto cells and performed GFP affinity purifications to test whether the GATAD2 CR2 is indeed the interaction domain that is required for the CHD proteins to interact with the complex. For full‐length GATAD2A, we observed interactions with all of the NuRD subunits tested. However, when looking at the interactions observed with either CR1 or CR2, these are complementary: CR1 associates with MTA, HDAC, RBBP, and MBD proteins, while CR2 associates with CHD and CDK2AP1 (Fig. 4C). These data are in agreement with Zhang et al., who showed that a core NuRD complex consisting of MTA, RBBP, HDAC, and MBD proteins is not sufficient to mediate an interaction with CHD4. GATAD2 protein had to be added to the reaction mix to facilitate an interaction between CHD4 and the core complex [20]. Interestingly, not only CHD4 but also CDK2AP1 associates specifically with GATAD2A‐CR2. This hints at an interaction between CDK2AP1 and CHD4, something which is also apparent from our cross‐linking data.
Fig. 4.
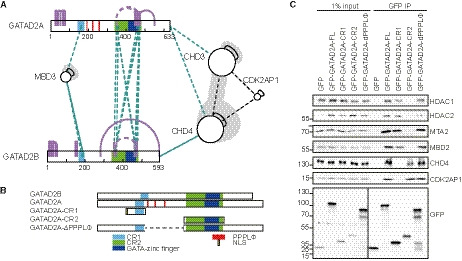
GATAD2 forms a bridge between the core of the NuRD complex and the CHD proteins. (A) Overview of the cross‐links identified between GATAD2A and GATAD2B and other NuRD subunits. Whereas MBD3 cross‐links to conserved region 1, CHD3 and CHD4 are the only proteins that cross‐link to conserved region 2. (B) Overview of the deletion constructs generated for GATAD2A. (C) Western blot analysis performed on GFP affinity purifications of the constructs in (B). Subunits of the core of the NuRD complex are enriched in purifications of full‐length GATAD2A and CR1, whereas CHD4 and CDK2AP1 are depleted from the CR1 affinity purification. CR2 exclusively co‐purifies CHD4 and CDK2AP1 (n = 2).
CDK2AP1 associates with CHD4
After being identified as a putative NuRD‐associated protein in 2006 [17], CDK2AP1 was formally identified as a NuRD subunit in 2010 [24]. The N‐terminal half of this ~ 14 kDa protein is intrinsically disordered, and the C‐terminal half forms a four‐helix bundle [25]. CDK2AP1 is an important tumor suppressor in oral cancer, although its exact function within the NuRD complex remains unclear. Despite its small size, the function of CDK2AP1 appears to be important, as this gene is conserved between mammals and fruit fly.
In our xIP‐MS experiments, we identified a few intermolecular cross‐links between CDK2AP1 and the C termini of CHD3 and CHD4 (Fig. 5A). The CDK2AP1 residues that formed the cross‐links reside on the edge of an alpha helix (Fig. 5B). This alpha helix is flanked by a positively charged cavity on the surface of CDK2AP1, which may accommodate some negatively charged amino acid side chains of the CHD proteins. To identify which residues are important for the interaction between CDK2AP1 and the CHDs, we aligned CHD3, CHD4, and CHD5 of human, mouse, zebrafish, and fruit fly. We identified a stretch in the C terminus that is highly conserved between the CHDs and across species (Fig. 5C). A few glutamic acids flank the cross‐linked residues in CHD3 and CHD4. These could potentially form part of the interaction surface with CDK2AP1.
Fig. 5.
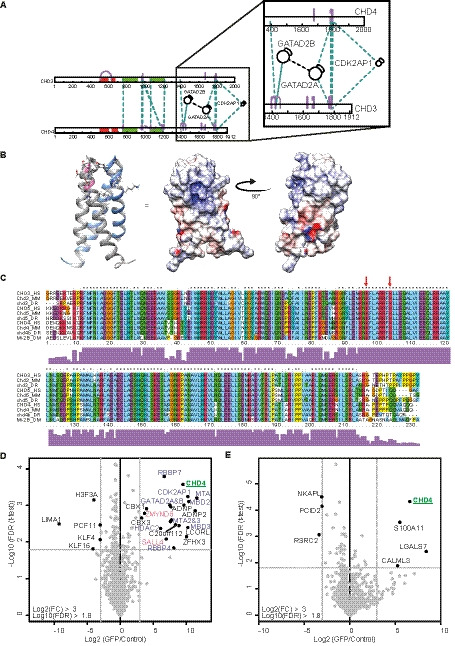
The C terminus of CHD proteins is required for the association with GATAD2 proteins and CDK2AP1. (A) Schematic overview of the cross‐links identified between CHD3 and CHD4 and other NuRD subunits. GATAD2A and GATAD2B mostly cross‐link to the C terminus. CDK2AP1 forms ambiguous cross‐links to the C terminus of both CHD3 and CHD4. (B) X‐ray structure of a dimer of (the C‐terminal parts of) CDK2AP1. One peptide is indicated in gray and the other in blue. Residues that cross‐link to the CHD proteins are indicated in pink. The surface potential of the CDK2AP1 dimer is also shown where blue is positively charged and red is negatively charged. Image is made using ucsf chimera [22]. (C) Sequence alignment of CHD3, CHD4, and CHD5 from human (HS), mouse (MM), zebrafish (DR), and fruit fly (DM) shows the presence of many glutamic acids around the sites of cross‐linking (indicated with red arrows). Used UniProt accession numbers are as follows: CHD3‐HS: Q12873‐1 (1696‐1925), CHD3‐MM: B1AR17‐1 (1622‐1851), CHD3‐DR: X1WBL0‐1 (1525‐1751), CHD5‐HS: Q8TDI0‐1 (1693‐1917), CHD5‐MM: A2A8L1‐1 (1691‐1915), CHD5‐DR: A0A0R4IJ89‐1 (1690‐1919), CHD4‐HS: Q14839‐1 (1685‐1912), CHD4‐MM: Q6PDQ2‐1 (1688‐1915), CHD4b‐DR: F1RBT2‐1 (1728‐1953), and Mi2‐DM: O97159‐1 (1716‐1945). Sequence alignment was generated using clustalx [36]. (D, E) Label‐free purifications of full‐length GFP‐CHD4 and CHD4‐ΔC‐GFP, respectively. Full‐length CHD4 co‐purifies all NuRD subunits in addition to a couple of new interactors (D). C‐terminal truncation of CHD4 leads to a loss of CDK2AP1 and of all other NuRD subunits (E).
To further investigate whether the C terminus of CHD4 contains a CDK2AP1 interaction domain, we expressed CHD41‐1627 with a C‐terminal GFP tag in HeLa FRT‐TO cells and performed a GFP purification followed by mass spectrometry. Unlike full‐length CHD4, regardless of whether it is tagged on the N terminus or C terminus (Fig. 5D), the truncated protein does not interact with CDK2AP1 nor any other NuRD subunits (Fig. 5E). This loss of interaction with CDK2AP1 is most likely caused by deletion of the direct interaction domain. Upon closer inspection of the cross‐links between GATAD2A and CHD proteins, it is clear that this interaction also requires the C terminus of the CHD proteins, which is in agreement with data published by the Mackay Lab [13, 26]. This may explain why the CHD41‐1627 protein fails to interact with any NuRD complex subunit.
The lost NuRD subunit paralog, CDK2AP2
As discussed above, multiple paralogs exist for all NuRD subunits. However, we never observed an interaction between NuRD and the CDK2AP1 paralog CDK2AP2 (or DOC‐1R), in contrast to previous observations [27]. Closer inspection of the amino acid sequence of CDK2AP1 and CDK2AP2 reveals that all tryptic peptides from CDK2AP2 that theoretically could be observed by mass spectrometry are shared with CDK2AP1 (Fig. 6A). MaxQuant assigns these so‐called shared peptides to the gene product that was identified with the most unique peptides. Since the unique peptides of CDK2AP2 are either too long or too small to be observed by mass spectrometry, all observed unique and shared peptides will therefore be assigned to CDK2AP1. To investigate whether CDK2AP2 associates with the NuRD complex, we cloned the cDNA of CDK2AP2 with either an N‐ or a C‐terminal GFP tag. We made a stably integrated dox‐inducible HeLa FRT‐TO cell line for both of them and performed GFP affinity purifications. As expected, both the N‐ and C‐terminal GFP fusions of CDK2AP2 enriched all subunits of the NuRD complex (Fig. 6B,C, respectively). In these pull‐downs, we only identified two shared CDK2AP1/2 peptides. For this reason, we also plotted the enrichment ratio and P‐value of GFP in the figure as representative of the bait. The cross‐links between CDK2AP1 and the CHD proteins that were discussed above could potentially belong to CDK2AP2 as well, since they lie within the conserved sequence. For the intraprotein cross‐links (CDK2AP1 75‐85, CDK2AP1 65‐104, and CDK2AP1 89‐92), one of the two peptides is always unique to CDK2AP1. The question therefore remains whether the two paralogs form heterodimers in the NuRD complex. In summary, CDK2AP1 and its paralog CDK2AP2 both assemble into the NuRD complex.
Fig. 6.
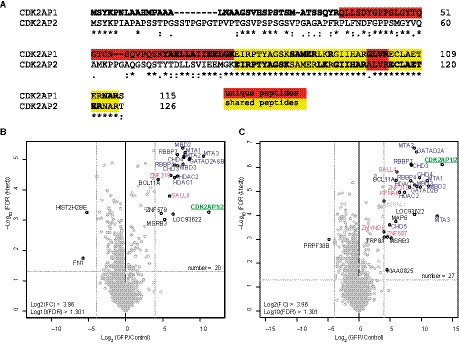
CDK2AP1 paralog CDK2AP2 also interacts with the NuRD complex. (A) Alignment of CDK2AP1 (O14519‐1) and CDK2AP2 (O75956‐1) shows a high degree of sequence conservation. Tryptic peptides are indicated with bold and regular font. Unique peptides are indicated in red and shared peptides in yellow. CDK2AP2 shares most peptides with CDK2AP1, but has 1 very short unique tryptic peptide. Alignment was made using Clustal Omega [37]. (B, C) Label‐free purifications of GFP‐CDK2AP1 (B) and CDK2AP1‐GFP (C) both show significant enrichment of all NuRD subunits. Some other identified proteins have been identified or suggested before to be CDK2AP2 interactors.
Conclusion
The NuRD complex is a highly conserved multi‐subunit protein complex, which has been difficult to study using conventional structural biology approaches such as X‐ray crystallography and NMR. The presence of multiple paralogs for each subunit, which often form multimers, complicates the purification of a homogenous native complex from mammalian cells. Many groups therefore focus on a few interacting subunits, or isolated structural domains. In this study, we have applied cross‐linking mass spectrometry on human NuRD purified from HeLa cells to obtain comprehensive topological information. We tagged and purified six different NuRD subunits and subjected the purified complexes to cross‐linking with two different cross‐linkers in combination with two different proteases. This resulted in the densest cross‐linking network for the NuRD complex to date. We identified many new cross‐links when comparing our data to that of Kloet et al., possibly due to the different proteins we used as a bait and use of a different mass spectrometer. In addition, we have used ADH/DMTMM, which allows cross‐linking of carboxylic acids and can thus be used to identify cross‐links in stretches of proteins that lack lysines. We believe that the cross‐linking network we generated, possibly in combination with the cross‐links identified by Kloet et al., can be instructive for modeling purposes or for mapping substructures resolved in single‐particle electron microscopy density maps. We validated some of the detected interactions using truncation mutants.
For clarity reasons, we have also generated a map of reduced cross‐links: In this case, all cross‐links on different paralogs of a single subunit have been projected onto a single paralog. Most of these comprised ambiguous cross‐links that could anyway belong to multiple paralogs and most likely originated from intrapeptide links instead of interpeptide links. However, some paralogs entail specific domains or motifs that may get lost in such a simplistic approach. This approach also assumes that all paralogs interact with other proteins in the same way. This may, however, be dependent on cellular context. Although we expect that expression levels will be one of the major regulatory mechanisms to incorporate different paralogs into the NuRD complex, post‐translational modifications could definitely also play a role, when situated close to an interaction surface. Further studies are clearly needed to shed further light on paralog‐specific interactions.
Our data revealed that CDK2AP1 binds to the C terminus of CHD4. Furthermore, we show that the CDK2AP1 paralog CDK2AP2 is a novel NuRD subunit. The expression levels of CDK2AP1 and CDK2AP2 in various cells and tissues may explain why loss of CDK2AP1 may be exclusively linked to oral cancers [28]. In these specific tumors, loss of CDK2AP1 may not be compensated for by expression of CDK2AP2. Further studies are, however, necessary to further investigate this. In summary, this study provides novel insights into the structural topology of the NuRD complex. Further efforts are still needed to resolve a holo‐structure of the NuRD complex at atomic resolution. This is not only relevant from a basic scientific perspective, but also relevant from a clinical point of view since perturbations of NuRD subunits are associated with multiple diseases.
Materials and methods
Cell lines
For the generation of the cross‐linking extracts, we used HeLa Kyoto cells stably transfected with BAC transgenes [8]. These constructs contain a GFP‐tagged gene under its endogenous promoter, ensuring endogenous expression levels. For CHD3 or CHD4, BAC lines were not available and we generated an endogenously TagGFP2‐tagged CHD4 cell line using CRISPaint [9] in the same HeLa Kyoto background.
To study the interactions of CHD4‐FL and deltaC, as well as CDK2AP2, we used HeLa Kyoto FRT cells, in which the pcDNA4‐TO‐GFP‐CHD4‐FL [29] or pcDNA5‐FRT‐TO construct with C‐terminally tagged CHD4‐dC or either N‐ or C‐terminally tagged CDK2AP2 was integrated.
Cells were grown in Dulbecco's modified Eagle's medium, supplemented with 10% fetal bovine serum and 100 U·mL−1 penicillin/streptomycin. HeLa Kyoto BAC lines were grown in the presence of 400 µg·mL−1 G418, while the HeLa Kyoto CRISPaint‐recombined cells were grown in the presence of 1 µg·mL−1 puromycin. HeLa Kyoto FRT‐TO cells were grown in the presence of 250 µg·mL−1 hygromycin.
CRISPaint
In short, we used the gRNA and frame selector suggested in Ref. [9] in combination with the TagGFP2‐T2A‐Puro universal donor plasmid (Addgene, constructs 80970, 66939, 66940, and 66941). The gRNA was cloned into a Cas9 containing plasmid without puromycin selection marker (Addgene, 64324) [30]. We transfected cells using polyethylenimine (PEI), and 48 h later, successfully transfected and genome‐edited cells were selected using 1 µg·mL−1 puromycin. After one week, monoclonal lines were obtained, which were then analyzed by western blot and fluorescence microscopy.
Label‐free affinity purification (LFQ)
Nuclear extracts were prepared as described earlier [31]. The affinity purifications are performed in triplicate, and so are the negative control purifications. For CHD4, the negative control purification consists of a GFP affinity purification on WT nuclear extract. For the CDK2AP2 purifications, blocked agarose beads were used as a negative control on the same nuclear extract. One mg of nuclear extract was incubated with 7.5 µL of GFP nano‐trap beads (Chromotek, Planegg‐Martinsried, Germany), in the presence of 50 µg·µL−1 ethidium bromide. After 1.5‐h incubation at 4 °C, beads were washed thoroughly (2× buffer C (1 m NaCl, 20 mm HEPES/KOH pH 7.9, 20% glycerol, 2 mm MgCl2, 0.2 mm EDTA, 0.25% NP40, 0.5 mm DTT, complete protease inhibitors (Roche, Basel, Switzerland)), 2× PBS with 0.25% NP40, and 2× PBS). Purified proteins were subjected to on‐bead digestion using trypsin (0.25 µg protease in 2 m urea, 100 mm Tris/HCl pH 8.5, 10 mm DTT) prior to STAGE tipping [32].
xIP‐MS
Nuclear extracts were prepared as described earlier [31]. One milliliter nuclear extract (5–10 mg of protein) was incubated with 20 µL of GFP nano‐trap beads (Chromotek), in the presence of 50 µg·µL−1 ethidium bromide. After 1.5‐h incubation at 4 °C, beads were washed thoroughly with a stringent washing buffer (2× buffer Cextra (1 m NaCl, 20 mm HEPES/KOH pH 7.9, 20% glycerol, 2 mm MgCl2, 0.2 mm EDTA, 1.0% NP40, 0.5 mm DTT, complete protease inhibitors (Roche)), 2× PBS with 1.0% NP40, and 2× PBS). Subsequently, proteins were cross‐linked using 2 mm BS3 or 43.4 mm DMTMM + 47.6 mm AHD in borate‐buffered saline at 37 °C for 1 h [14], which was quenched with 100 mm Tris/HCl pH 8.0 for 10 min. Proteins were subjected to on‐bead digestion using trypsin or chymotrypsin (0.25 µg protease in 2 m urea, 100 mm Tris/HCl pH 8.5, 10 mm DTT) prior to STAGE tipping [32].
Mass spectrometry
Cross‐linking samples were measured on a reverse‐phase Easy‐nLC 1000 online with a Thermo Q Exactive Mass Spectrometer or Tribrid Fusion Orbitrap. A 4‐hour gradient of buffer B (80% acetonitrile, 0.1% TFA) was used. Only peptides with a charge state of 3 or more were selected for HCD fragmentation.
The LFQ purifications were analyzed using either the LTQ Orbitrap Fusion or the LTQ Orbitrap Q Exactive, with online reverse‐phase nano‐HPLC during a 140‐min gradient of buffer B (80% acetonitrile, 0.1% TFA).
XL Mass spec data analysis
Thermo raw data files were converted to .mgf format using MSConvert (ProteoWizard, Palo Alto, California, USA). The data were analyzed using plink2.0 (from the pFind package) [10] using databases containing all NuRD subunits + ZMYND8, and ZNF687/592/532/512B with additional BHC, PRC2, and SIN3 subunits (NuRD_HDAC_RBBP_complexes.fasta). Data were filtered to have a score below 0.0001. Cross‐link networks are visualized using xinet (crosslinkviewer.org) [11].
Visualization on 3D structures
A homology model of a partial CHD4 (residue 499–1281) was generated using the structure known for yeast CHD1 (3MWY) and the automated online modeling server SWISS‐MODEL (https://swissmodel.expasy.org/interactive) [33].
Cross‐links were mapped onto single molecules extracted from existing X‐ray structures of HDAC1 (4BKX), RBBP7 (3CFV), and a homology model of residue 499 till 1281 of CHD4 using ucsf chimera 3D viewer and the Xlink Analyzer add‐on [21, 22].
The cross‐linked residues on CDK2AP1 are visualized on the PDB 2KW6, and the surface potential is calculated using the Delphi Webserver (http://compbio.clemson.edu/sapp/delphi_webserver/) [34, 35].
LFQ data analysis using MaxQuant and Perseus
RAW files were analyzed using maxquant 1.5.0.1 with carbamidomethyl as fixed modification and oxidation of methionines and acetylation of the N terminus as variable modifications. The database used was a SWISS‐Prot reviewed database downloaded in December 2015. After identification of proteins, data were filtered for reverse hits, contaminants, unique peptides (> 0), and valid values (3 in at least 1 group). Missing values were imputed, and a two‐samples t‐test was performed to identify specific outliers. Volcano plots were generated using r.
Interaction assays
Different mutants of GATAD2A were cloned into Clontech peGFP‐C3 backbone using HinDIII and BamHI restriction sites. Constructs were sequence‐verified and transiently transfected into HeLa Kyoto cells using PEI. Nuclear extracts were made, and 1 mg of extract was used per purification. After washing, proteins were eluted from the beads by boiling for 5 min in 2× SDS sample buffer. Proteins were separated using SDS/PAGE ranging from 6% to 15% and then blotted onto nitrocellulose membrane using the Bio‐Rad Turboblotting System. The membrane is blocked in 5% skimmed milk in TBST (50 mm Tris, 150 mm NaCl, 0.05% Tween) and sequentially incubated with primary and secondary antibodies. Results are visualized using enhanced chemiluminescence (Thermo Scientific, Waltham, Massachusetts, USA) on the LAS4000.
Silver stain
Proteins were purified as for xIP‐MS, except the beads were boiled in Laemmli buffer instead of cross‐linking. Proteins were then separated on a NuPAGE 4–12% Bis/Tris Gel (NP0335PK2; Invitrogen, Carlsbad, CA, USA) and visualized using the SilverQuest Staining Kit (Invitrogen, LC6070).
Used antibodies
The antibodies used are as follows: rabbit αGFP: ab‐290, Abcam; rabbit αCDK2AP1: in‐house generated, V41 [24]; rabbit αHDAC1: H51, SC‐7872, Santa Cruz; rabbit αHDAC2: SC‐7899, Santa Cruz; goat αMTA2: SC‐9447, Santa Cruz Biotechnologies, Inc., Dallas, Texas, USA; goat αMBD2: EB07538, Everest Biotech, Bicester, England, UK; rabbit αCHD4/Mi2beta: A301‐081A, Bethyl Laboratories, Inc., Montgomery, Texas, USA; rabbit αMTA1: A300‐911A, Bethyl; goat αRbAp46: SC‐8272, Santa Cruz; and rabbit αMBD3: A302‐528A, Bethyl.
PRIDE upload
All the raw mass spectrometry data, as well as output files from the analysis, have been uploaded on the PRIDE server under the accession numbers PXD017244 and PXD017378.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
CGS conceived the study and wrote the manuscript; CGS, MAB, and SK performed the experiments; and CGS and CG analyzed the data. MV and CGS supervised the study.
Peer Review
The peer review history for this article is available at https://publons.com/publon/10.1111/febs.15650.
Supporting information
Table S1. Overview of the filtered cross‐links mapping to the NuRD complex, per combination of cross‐linker and protease used.
Table S2. List of cross‐links reduced to a single paralog per subunit.
Acknowledgements
The authors would like to thank the Vermeulen group for nice discussions, especially Dr. M.M. Makowski. In addition, the authors are grateful to Dr. Kuhn and Dr. Hornung for sharing constructs via Addgene. We thank Dr. M. Luijsterburg for sharing the cDNA4TO‐GFP‐CHD4 construct. Molecular graphics and analyses were performed with UCSF Chimera, developed by the Resource for Biocomputing, Visualization, and Informatics, at the University of California, San Francisco, with support from NIH P41‐GM103311. CGS receives financial support from the Dutch Organisation of Science (NWO‐Veni 722.016.003). The Vermeulen Lab is part of the Oncode Institute, which is partly funded by the Dutch Cancer Society (KWF).
References
- 1. Clapier CR, Iwasa J, Cairns BR & Peterson CL (2017) Mechanisms of action and regulation of ATP‐dependent chromatin‐remodelling complexes. Nat Rev Mol Cell Biol 18, 407–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kouzarides T (2007) Chromatin modifications and their function. Cell 128, 693–705. [DOI] [PubMed] [Google Scholar]
- 3. Denslow SA & Wade PA (2007) The human Mi‐2/NuRD complex and gene regulation. Oncogene 26, 5433–5438. [DOI] [PubMed] [Google Scholar]
- 4. Cramer JM, Pohlmann D, Gomez F, Mark L, Kornegay B, Hall C, Siraliev‐Perez E, Walavalkar NM, Sperlazza MJ, Bilinovich S et al. (2017) Methylation specific targeting of a chromatin remodeling complex from sponges to humans. Sci Rep 7, 40674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hendrich B & Tweedie S (2003) The methyl‐CpG binding domain and the evolving role of DNA methylation in animals. Trends Genet 19, 269–277. [DOI] [PubMed] [Google Scholar]
- 6. Makowski MM, Willems E, Jansen PW & Vermeulen M (2016) Cross‐linking immunoprecipitation‐MS (xIP‐MS): topological analysis of chromatin‐associated protein complexes using single affinity purification. Mol Cell Proteomics 15, 854–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Spruijt CG, Luijsterburg MS, Menafra R, Lindeboom RG, Jansen PW, Edupuganti RR, Baltissen MP, Wiegant WW, Voelker‐Albert MC, Matarese F et al. (2016) ZMYND8 co‐localizes with NuRD on target genes and regulates poly(ADP‐ribose)‐dependent recruitment of GATAD2A/NuRD to sites of DNA damage. Cell Rep 17, 783–798. [DOI] [PubMed] [Google Scholar]
- 8. Poser I, Sarov M, Hutchins JR, Heriche JK, Toyoda Y, Pozniakovsky A, Weigl D, Nitzsche A, Hegemann B, Bird AW et al. (2008) BAC TransgeneOmics: a high‐throughput method for exploration of protein function in mammals. Nat Methods 5, 409–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schmid‐Burgk JL, Honing K, Ebert TS & Hornung V (2016) CRISPaint allows modular base‐specific gene tagging using a ligase‐4‐dependent mechanism. Nat Commun 7, 12338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yang B, Wu YJ, Zhu M, Fan SB, Lin J, Zhang K, Li S, Chi H, Li YX, Chen HF et al. (2012) Identification of cross‐linked peptides from complex samples. Nat Methods 9, 904–906. [DOI] [PubMed] [Google Scholar]
- 11. Combe CW, Fischer L & Rappsilber J (2015) xiNET: cross‐link network maps with residue resolution. Mol Cell Proteomics 14, 1137–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Low JK, Webb SR, Silva AP, Saathoff H, Ryan DP, Torrado M, Brofelth M, Parker BL, Shepherd NE & Mackay JP (2016) CHD4 is a peripheral component of the nucleosome remodeling and deacetylase complex. J Biol Chem 291, 15853–15866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Torrado M, Low JKK, Silva APG, Schmidberger JW, Sana M, Sharifi Tabar M, Isilak ME, Winning CS, Kwong C, Bedward MJ et al. (2017) Refinement of the subunit interaction network within the nucleosome remodelling and deacetylase (NuRD) complex. FEBS J 284, 4216–4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kloet SL, Baymaz HI, Makowski M, Groenewold V, Jansen PW, Berendsen M, Niazi H, Kops GJ & Vermeulen M (2015) Towards elucidating the stability, dynamics and architecture of the nucleosome remodeling and deacetylase complex by using quantitative interaction proteomics. FEBS J 282, 1774–1785. [DOI] [PubMed] [Google Scholar]
- 15. Millard CJ, Varma N, Saleh A, Morris K, Watson PJ, Bottrill AR, Fairall L, Smith CJ & Schwabe JW (2016) The structure of the core NuRD repression complex provides insights into its interaction with chromatin. eLife 5, e13941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schmidberger JW, Sharifi Tabar M, Torrado M, Silva AP, Landsberg MJ, Brillault L, AlQarni S, Zeng YC, Parker BL, Low JK et al. (2016) The MTA1 subunit of the nucleosome remodeling and deacetylase complex can recruit two copies of RBBP4/7. Protein Sci 25, 1472–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Le Guezennec X, Vermeulen M, Brinkman AB, Hoeijmakers WA, Cohen A, Lasonder E & Stunnenberg HG (2006) MBD2/NuRD and MBD3/NuRD, two distinct complexes with different biochemical and functional properties. Mol Cell Biol 26, 843–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hoffmeister H, Fuchs A, Erdel F, Pinz S, Grobner‐Ferreira R, Bruckmann A, Deutzmann R, Schwartz U, Maldonado R, Huber C et al. (2017) CHD3 and CHD4 form distinct NuRD complexes with different yet overlapping functionality. Nucleic Acids Res 45, 10534–10554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bode D, Yu L, Tate P, Pardo M & Choudhary J (2016) Characterization of two distinct nucleosome remodeling and deacetylase (NuRD) complex assemblies in embryonic stem cells. Mol Cell Proteomics 15, 878–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang W, Aubert A, Gomez de Segura JM, Karuppasamy M, Basu S, Murthy AS, Diamante A, Drury TA, Balmer J, Cramard J et al. (2016) The nucleosome remodeling and deacetylase complex NuRD is built from preformed catalytically active sub‐modules. J Mol Biol 428, 2931–2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kosinski J, von Appen A, Ori A, Karius K, Muller CW & Beck M (2015) Xlink Analyzer: software for analysis and visualization of cross‐linking data in the context of three‐dimensional structures. J Struct Biol 189, 177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC & Ferrin TE (2004) UCSF Chimera – a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- 23. Bullock JMA, Thalassinos K & Topf M (2018) Jwalk and MNXL web server: model validation using restraints from crosslinking mass spectrometry. Bioinformatics 34, 3584–3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Spruijt CG, Bartels SJ, Brinkman AB, Tjeertes JV, Poser I, Stunnenberg HG & Vermeulen M (2010) CDK2AP1/DOC‐1 is a bona fide subunit of the Mi‐2/NuRD complex. Mol Biosyst 6, 1700–1706. [DOI] [PubMed] [Google Scholar]
- 25. Ertekin A, Aramini JM, Rossi P, Leonard PG, Janjua H, Xiao R, Maglaqui M, Lee HW, Prestegard JH & Montelione GT (2012) Human cyclin‐dependent kinase 2‐associated protein 1 (CDK2AP1) is dimeric in its disulfide‐reduced state, with natively disordered N‐terminal region. J Biol Chem 287, 16541–16549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sharifi Tabar M, Mackay JP & Low JKK (2019) The stoichiometry and interactome of the nucleosome remodeling and deacetylase (NuRD) complex are conserved across multiple cell lines. FEBS J 286, 2043–2061. [DOI] [PubMed] [Google Scholar]
- 27. Buajeeb W, Zhang X, Ohyama H, Han D, Surarit R, Kim Y & Wong DT (2004) Interaction of the CDK2‐associated protein‐1, p12(DOC‐1/CDK2AP1), with its homolog, p14(DOC‐1R). Biochem Biophys Res Commun 315, 998–1003. [DOI] [PubMed] [Google Scholar]
- 28. Mohd‐Sarip A, Teeuwssen M, Bot AG, De Herdt MJ, Willems SM, Baatenburg de Jong RJ, Looijenga LHJ, Zatreanu D, Bezstarosti K, van Riet J et al. (2017) DOC1‐dependent recruitment of NURD reveals antagonism with SWI/SNF during epithelial‐mesenchymal transition in oral cancer cells. Cell Rep 20, 61–75. [DOI] [PubMed] [Google Scholar]
- 29. Luijsterburg MS, Acs K, Ackermann L, Wiegant WW, Bekker‐Jensen S, Larsen DH, Khanna KK, van Attikum H, Mailand N & Dantuma NP (2012) A new non‐catalytic role for ubiquitin ligase RNF8 in unfolding higher‐order chromatin structure. EMBO J 31, 2511–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chu VT, Weber T, Wefers B, Wurst W, Sander S, Rajewsky K & Kuhn R (2015) Increasing the efficiency of homology‐directed repair for CRISPR‐Cas9‐induced precise gene editing in mammalian cells. Nat Biotechnol 33, 543–548. [DOI] [PubMed] [Google Scholar]
- 31. Dignam JD, Lebovitz RM & Roeder RG (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res 11, 1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rappsilber J, Ishihama Y & Mann M (2003) Stop and go extraction tips for matrix‐assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem 75, 663–670. [DOI] [PubMed] [Google Scholar]
- 33. Arnold K, Bordoli L, Kopp J & Schwede T (2006) The SWISS‐MODEL workspace: a web‐based environment for protein structure homology modelling. Bioinformatics 22, 195–201. [DOI] [PubMed] [Google Scholar]
- 34. Sarkar S, Witham S, Zhang J, Zhenirovskyy M, Rocchia W & Alexov E (2013) DelPhi Web Server: a comprehensive online suite for electrostatic calculations of biological macromolecules and their complexes. Commun Comput Phys 13, 269–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Smith N, Witham S, Sarkar S, Zhang J, Li L, Li C & Alexov E (2012) DelPhi web server v2: incorporating atomic‐style geometrical figures into the computational protocol. Bioinformatics 28, 1655–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. [DOI] [PubMed] [Google Scholar]
- 37. Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J et al. (2011) Fast, scalable generation of high‐quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Overview of the filtered cross‐links mapping to the NuRD complex, per combination of cross‐linker and protease used.
Table S2. List of cross‐links reduced to a single paralog per subunit.


