Abstract
So far, more than 70 genes involved in the chronological lifespan (CLS) of Schizosaccharomyces pombe (fission yeast) have been reported. In this mini‐review, we arrange and summarize these genes based on the reported genetic interactions between them and the physical interactions between their products. We describe the signal transduction pathways that affect CLS in S. pombe: target of rapamycin complex 1, cAMP‐dependent protein kinase, Sty1, and Pmk1 pathways have important functions in the regulation of CLS extension. Furthermore, the Php transcription complex, Ecl1 family proteins, cyclin Clg1, and the cyclin‐dependent kinase Pef1 are important for the regulation of CLS extension in S. pombe. Most of the known genes involved in CLS extension are related to these pathways and genes. In this review, we focus on the individual genes regulating CLS extension in S. pombe and discuss the interactions among them.
Keywords: chronological lifespan, fission yeast, longevity, Schizosaccharomyces pombe, signal transduction, stationary phase
The microbial chronological lifespan is considered to be a model of cell lifespan after differentiation in higher organisms. In this review, factors extending the lifespan reported thus far in Schizosaccharomyces pombe and the relationships among them were summarized. According to the findings, the TORC1 pathway, PKA pathway, Sty1 pathway, Pmk1 pathway, Ecl1 family genes, Php complex, and cyclin‐dependent kinase Pef1 are considered to contribute to the extension of this yeast's chronological lifespan.

1. INTRODUCTION
The fission yeast Schizosaccharomyces pombe is a model organism of unicellular eukaryotes (Hayles and Nurse, 2018). This yeast is considered to have diverged from the budding yeast Saccharomyces cerevisiae hundreds of million years ago (Hayles and Nurse, 2018; Hedges, 2002; Sipiczki, 2000), and studies using these yeast species have contributed significantly to the understanding of various cellular processes. S. pombe has been actively used for research in multiple fields, including cell cycle studies, cellular morphology, sexual development, splicing, and chromosome structure. Furthermore, much information is accumulating regarding the stationary phase of cells, particularly chronological lifespan (CLS) (Hayles and Nurse, 2018; Lin and Austriaco, 2014; Ohtsuka and Aiba, 2017; Roux et al., 2010b).
There are two yeast lifespan fields of study: replicative lifespan (RLS) and CLS (Chen and Runge, 2012; Longo et al., 2012; Roux et al., 2010b). RLS is the number of divisions a cell can undergo, whereas CLS is the length of time a cell can survive. CLS of yeast corresponds with the survival period of cells that have entered the stationary phase. Both types of lifespans are relatively easy to measure in S. cerevisiae. In contrast, it is difficult to distinguish between mother and daughter cells in S. pombe and the number of RLS studies is significantly lower than that of CLS studies (Erjavec et al., 2008; Ohtsuka and Aiba, 2017). However, in S. cerevisiae, although several common factors are related to RLS and CLS, the exact relationship between these two lifespans has not been clarified yet (Longo et al., 2012).
Recently, microfluidic devices were established to study yeast lifespan, allowing the study of RLS in S. pombe (Nakaoka, 2017; Nakaoka and Wakamoto, 2017; Spivey et al., 2017). However, using this technique, at least two independent laboratories have reported that S. pombe is not related to RLS, that is, this yeast does not age replicatively and cellular aging and RLS can be unrelated in this yeast. Moreover, studies of the CLS of S. pombe are being conducted as actively as those of budding yeast. As described in detail later, some nutrient signaling pathways, including target of rapamycin (TOR) and cAMP‐dependent protein kinase (PKA) pathways, have been found to affect the CLS of S. pombe as well as the lifespans of other model organisms such as nematodes, flies, and mammals (Chen and Runge, 2009; Fontana et al., 2010; Lin and Austriaco, 2014; Rallis et al., 2013; Roux et al., 2009, 2010b).
Growth environment greatly influences the CLS of S. pombe. Both calorie restriction and restriction of specific nutrients, that is, dietary restriction, are established methods of delaying aging and prolonging lifespan, and their effects are conserved widely in many organisms including yeast, nematodes, flies, and mammals (Fontana and Partridge, 2015; Fontana et al., 2010; Lee and Longo, 2016; Ohtsuka and Aiba, 2017). In S. pombe, the limitation of nutrients such as nitrogen, sulfur, specific amino acids, and trace metals, as well as restriction of glucose as a carbon source, contributes to CLS extension (Lin and Austriaco, 2014; Ohtsuka and Aiba, 2017; Ohtsuka et al., 2017, 2019; Shimasaki et al., 2017; Su et al., 1996). CLS extension by calorie restriction is known to involve the PKA pathway and the stress‐dependent mitogen‐activated protein kinase (MAPK) Sty1 pathway in S. pombe (Roux et al., 2009; Zuin et al., 2010a, 2010b). Sty1 is a stress‐dependent MAPK, but it is also involved in the PKA pathway and nutrient signaling (Caspari, 1997; Madrid et al., 2004; Sansó et al., 2011; Stiefel et al., 2004). CLS extension due to the restriction of nutrients, such as sulfur, leucine, and zinc, depends on Ecl1 family genes and CLS regulators found in S. pombe, and CLS extensions by some types of dietary restriction may be associated with reduced translation including reduced ribosome level (Ohtsuka and Aiba, 2017). The relationship between translational repression and lifespan extension has been reported in other organisms including budding yeast and nematodes (Hansen et al., 2007; MacInnes, 2016; Steffen et al., 2008). Nitrogen restriction causes G1 arrest and G0 phase entry, where heterochromatin formation and autophagy have been implicated in the survival of S. pombe (Oya et al., 2019; Roche et al., 2016).
Moreover, studies on the effects of drugs on CLS in S. pombe suggest that supplementation with drugs such as acivicin, 3,3′‐diindolylmethane, mangosteen, monensin sodium, mycophenolic acid, nigericin sodium, prostaglandin J2, wortmannin, ribozinoindole‐1, diazaborine, actinomycin D, tschimganine, β‐hibitakanine, and Torin 1 can extend the CLS of S. pombe (Hibi et al., 2018; Ohtsuka and Aiba, 2017; Ohtsuka et al., 2017; Rodríguez‐López et al., 2020; Stephan et al., 2013). Ribozinoindole‐1 and diazaborine suppress rRNA maturation, and actinomycin D suppresses rRNA translation by acting on RNA polymerase (Cooper and Braverman, 1977; Hayashi et al., 2014; Kawashima et al., 2016; Loibl et al., 2014; Scala et al., 2016). The ionophores monensin and nigericin extend CLS by affecting vacuolar acidification (Stephan et al., 2013). Prostaglandin J2 reportedly exhibits antiaging properties by inhibiting mitochondrial mitosis (Stephan et al., 2013). Acivicin and mycophenolic acid inhibit guanosine monophosphate (GMP) synthesis, suggesting a relationship between GMP level and CLS (Stephan et al., 2013). Torin 1 inhibits TOR, which is related to the lifespan regulation (Rodríguez‐López et al., 2020).
Many studies report the relationship between aging and oxidative stress (Berlett and Stadtman, 1997; Fabrizio and Longo, 2003; Lu and Finkel, 2008; Muller et al., 2007). The well‐known free radical theory states that free radicals produced as a byproduct, mainly from mitochondria, oxidize cellular components such as DNA, proteins, and lipids, which cause aging. According to this theory, if free radicals cause aging, increased antioxidant activity should suppress aging and extend lifespan. However, in studies using model organisms, including S. pombe, increasing antioxidant activity has not always been shown to suppress aging and extend lifespan (Lam et al., 2010; Ohtsuka et al., 2012; Sadowska‐Bartosz and Bartosz, 2014; Selman et al., 2013).
Here, we summarize the relationship between gene groups and pathways among over 70 reported genes, each of which causes the CLS extension when it is overexpressed or deleted in S. pombe. Because the interactions of more than 70 longevity genes are extremely complicated, this review focuses only on the genes that cause longevity. Therefore, genes known to be involved in the CLS extension pathway, but not reported to cause the CLS extension by their activation or suppression, are not described in detail.
2. NOVEL CLS‐AFFECTING GENES: git5+, SPBC26H8.13c, nop14+, AND grx4+
We searched the DNA region of the S. pombe genome that causes the CLS extension by overexpression using a multicopy plasmid and found that overexpression of each DNA regions containing git5 +, SPBC26H8.13, nop14 +, or grx4 + caused CLS extension (Figure 1a,b). Interestingly, the deletion of git5 + also extended the CLS (Figure 1c). git5 + encodes the G protein subunit, which acts on glucose response and forms a heterotrimer with Gpa2 and Git11. Overexpression of git5 + alone in this heterotrimer may disrupt the precise regulation of this heterocomplex and, like Δgit5, suppress the glucose signaling pathway.
FIGURE 1.
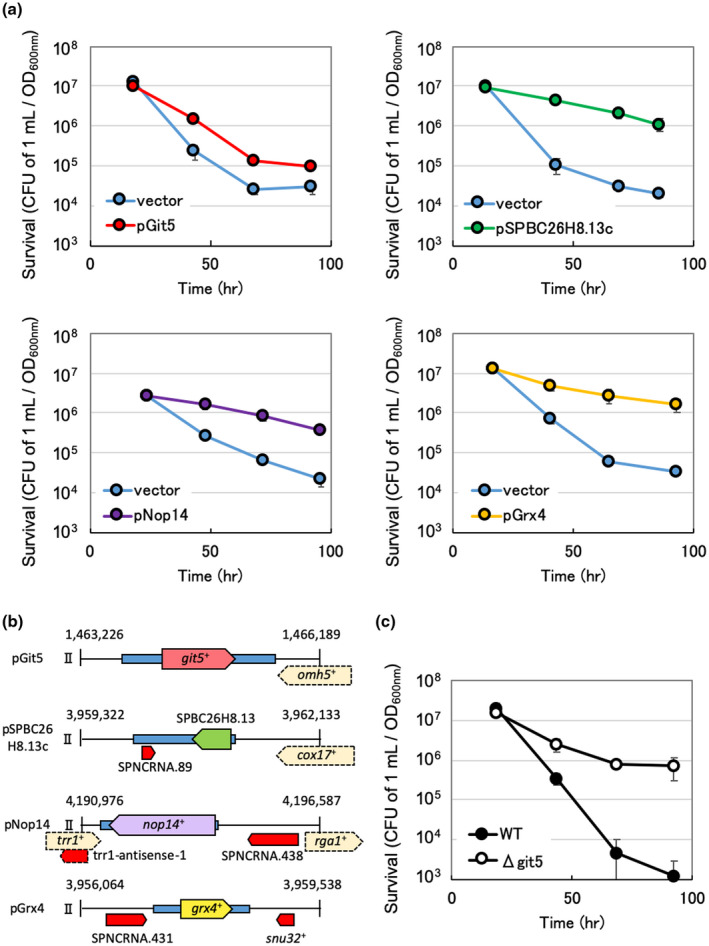
(a) The results of chronological lifespan (CLS) measurements. The strain of Schizosaccharomyces pombe used was JY333 and the plasmid vector was pLB‐Dblet. To determine cell viability, the cells were grown in SD liquid medium, sampled at each growth phase, and then, plated on YE agar plates using suitable dilutions. After incubation for several days as 30°C, the number of colonies derived from 1 ml of culture was counted. This number was divided by the cell turbidity at the sampling time. (b) The DNA fragments that were inserted into the plasmids are carried by the cells whose CLS were measured. (c) The results of CLS measurement of wild‐type JY333 and Δgit5 are shown
This review adds these four genes to the 77 previously reported genes involved in CLS extension and summarizes the function of a total of 81 genes involved in the regulation of CLS extension.
3. INTERACTIONS AMONG FACTORS THAT REGULATE CLS IN S. pombe
First, we summarized the physical and genetic interactions of 81 factors that are involved in CLS extension in S. pombe (Figure 2). Then, the factors that interact with many other factors involved in CLS extension were extracted and summarized as CLS‐regulated gene product groups and CLS‐regulated signal pathways (Figure 3). Some CLS regulatory genes encoded enzymes that are directly involved in energy metabolism (Figure 4). Below, we focus on the major signaling pathways and gene groups involved in CLS extension in S. pombe and discuss the regulation of CLS extension.
FIGURE 2.
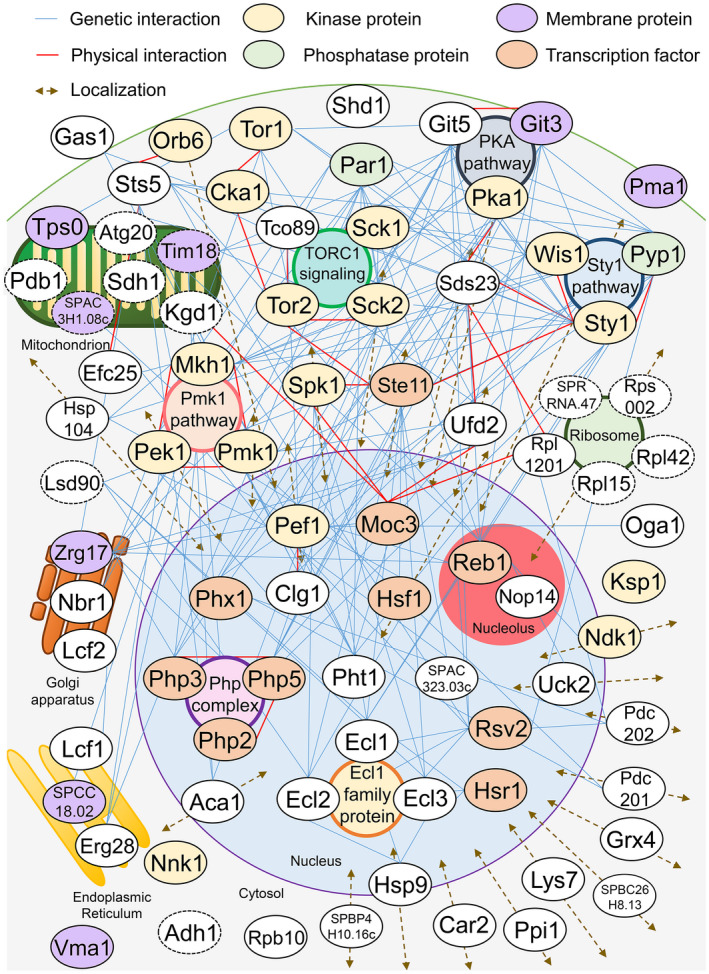
Factors that reportedly cause chronological lifespan extension in Schizosaccharomyces pombe. All the genetic and physical interactions reported so far are shown. Information on each factor's localization was based on the reports by Ding et al. (2000) and Matsuyama et al. (2006), in addition to those mentioned in the text. Dotted lines indicate factors with unknown localization. Studies in which two or more intracellular localization (e.g., nucleus and cytosol) were reported are indicated by double‐headed arrows
FIGURE 3.

A hypothetical model summarizing the representative signal pathways and factors involved in chronological lifespan regulation in Schizosaccharomyces pombe
FIGURE 4.
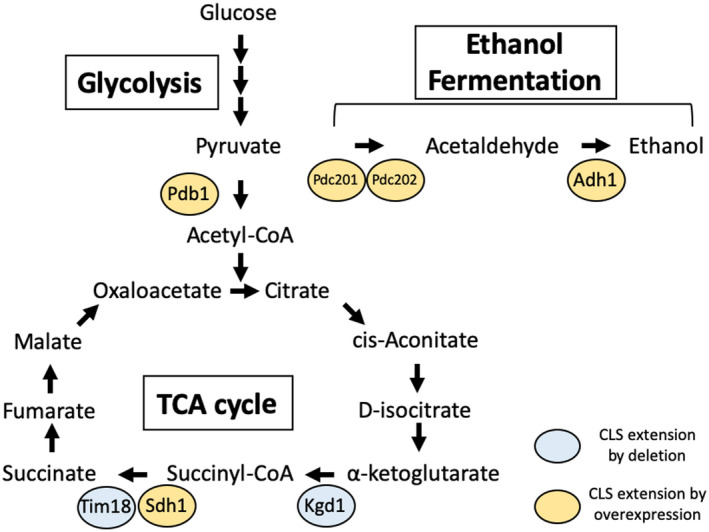
Model summarizing the chronological lifespan regulatory factors involved in energy metabolism
4. TOR COMPLEX 1 PATHWAY
In many model organisms, suppression of the TOR complex 1 (TORC1) pathway extends lifespan (Fontana et al., 2010; Lees et al., 2016), and similar phenomena have occurred in S. pombe (Rodríguez‐López et al., 2020). In S. pombe, tor2 +, which codes for a serine/threonine kinase of TORC1, is an essential gene. There is no analysis of the CLS of a tor2 + deletion strain, but its temperature‐sensitive (ts) mutant, tor2‐ts6, has an extended CLS (Ohtsuka et al., 2019). Furthermore, the deletion of tco89 +, which encodes the TORC1 subunit, extends CLS (Rallis et al., 2013). The serine/threonine kinases in the AGC (protein kinase A/protein kinase G/protein kinase C) kinase family, Sck1 and Sck2, which are orthologs of S. cerevisiae Sch9, are phosphorylated by Tor2 as targets of TORC1 (Nakashima et al., 2012). The deletion of S. cerevisiae SCH9 is known to extend the CLS (Madia et al., 2008) and, in S. pombe, the deletion of sck2 + can lead to larger CLS extension than that of sck1 + (Chen and Runge, 2009; Roux et al., 2006). CLS extension caused by Δsck2 occurs irrespective of the presence of Pka1, the loss of which also causes CLS extension, suggesting that the CLS extension mechanism by the TORC1 pathway and PKA pathway act in parallel (Roux et al., 2006). The regulation of CLS extension by the PKA pathway is described later.
Mass spectrometry analyses have identified Cka1 as a factor that interacts with the TOR complexes (Hayashi et al., 2007). The overexpression of cka1 + extends CLS (Roux et al., 2010a). cka1 + encodes a catalytic subunit of casein kinase 2 (CK2) (Nakazawa et al., 2019). Although it is unclear exactly how overexpression of cka1 + affects the TORC1 pathway, it is considered that CLS extension is involved (Roux et al., 2010a). Because CK2 represses the transcription of ribosomal proteins (Moreira‐Ramos et al., 2015), and the repression of ribosomes can extend CLS (Ohtsuka and Aiba, 2017), cka1 + overexpression may repress ribosomes, leading to CLS extension.
Tor2 phosphorylates the transcription factor Ste11, which is essential for sexual differentiation, and phosphorylation suppresses its functions including nuclear localization (Otsubo et al., 2017). Although ste11 + deletion does not affect CLS, its overexpression leads to CLS extension, albeit to a low extent (Ohtsuka et al., 2012). Therefore, Ste11 mainly regulates genes involved in sexual differentiation, but some Ste11‐regulated genes may contribute to CLS extension.
The deletion of zrg17 + extends CLS (Rallis et al., 2014). The cation diffusion facilitator (CDF) family protein Zrg17 forms a heteromer with Cis4, another CDF family protein, and is involved in Golgi membrane trafficking through the regulation of zinc homeostasis (Fang et al., 2008). Interestingly, synthetic genetic array analysis shows that zrg17 + performs positive genetic interactions, wherein the double‐mutant phenotype is weaker than anticipated, with sck2 + (Rallis et al., 2014). This suggests that CLS extension by zrg17 + deletion may be involved in the TORC1 pathway. Intriguingly, as mentioned above, it has also been reported that extracellular zinc concentration itself affects CLS (Shimasaki et al., 2017).
Overexpression of the stationary phase‐specific transcription factor Phx1 extends CLS and its deletion shortens CLS (Kim et al., 2012). Δsck2 longevity reportedly depends on phx1 + (Kim et al., 2014), suggesting that Phx1 contributes to CLS extension downstream of the TORC1 pathway. Furthermore, Phx1 is required for the stationary phase‐specific induction of pdc201 + and pdc202 +, which encode pyruvate decarboxylases. The overexpression of both pdc201 + and pdc202 + extends CLS, whereas their deletion decreases CLS (Kim et al., 2014).
The CLS extension of Δsck2 reportedly depends on Sty1 (Zuin et al., 2010a, 2010b). However, it is unclear how much of the CLS extension induced by the deletion of sck2 + depends on Sty1: under conditions wherein Δsck2 causes CLS extension, the comparison between survivals of sty1 + single‐deletion mutants and sck2 + and sty1 + double‐deletion mutants during the early stationary phase has not been reported. A comprehensive analysis indicated that tco89 +, which encodes the TORC1 subunit, and sty1 + have negative genetic interaction, wherein the double‐mutant phenotype is stronger than expected from the phenotypes associated with the single mutants (Ryan et al., 2012). Therefore, the TORC1 pathway may be involved in CLS extension in parallel with Sty1.
Current findings indicate that in addition to the signal pathway cascading from TORC1 (including Tor2 and Tco89) to Sck2, the transcription factor Phx1 (as the downstream factor) and its regulated genes, (pdc201 + and pdc202 +) contribute mainly to CLS extension by the TORC1 pathway in S. pombe (Figure 3). Furthermore, zrg17 +, which shows genetic interaction with sck2 +, may be involved in this pathway and affect CLS. Moreover, although its effect on CLS is not substantial, the transcription factor Ste11 can contribute to CLS extension in response to suppression of the TORC1 pathway. CK2 may also be involved in CLS regulation via this signaling pathway.
5. PKA AND Sty1 PATHWAYS
Similar to the TORC1 pathway, inhibition of the PKA pathway is reportedly associated with CLS extension in S. pombe as well as in other organisms, such as budding yeast and mice (Fabrizio et al., 2003; Fontana and Partridge, 2015; Fontana et al., 2010; Yan et al., 2007). In S. pombe, the deletion of the G protein‐coupled glucose receptor Git3 located in the plasma membrane extends CLS (Roux et al., 2009; Stephan et al., 2013). Similarly, the deletion of Git5, which encodes a heterotrimeric G protein β‐subunit that acts on this glucose receptor, also extends CLS (Figure 1). Signals from this glucose receptor are transmitted to PKA (Lin and Austriaco, 2014; Roux et al., 2010b). Besides, the deletion of pka1 +, which encodes the catalytic subunit of PKA (Gupta et al., 2011), also causes CLS extension (Roux et al., 2006). As described above, because Δpka1 and Δsck2 additively cause CLS extension (Roux et al., 2006), the TORC1 and PKA pathways are considered to contribute to CLS extension in parallel. This finding is consistent with the comprehensive analysis results that git3 + and git5 + indicate negative genetic interactions with sck2 + (Rallis et al., 2014; Ryan et al., 2012). However, similar to the TORC1 pathway, the longevity of Δpka1 mutant cells reportedly requires phx1 + (Kim et al., 2014). Therefore, although the upstream regions of the two signaling pathways differ, the downstream region may contain a common cell survival mechanism. However, git5 + reportedly indicates negative genetic interaction with pdc202 + (Ryan et al., 2012), which seems to act downstream of Phx1. Further analysis will clarify the relationship between these pathways in the regulation of CLS.
The PKA and Sty1 pathways have opposite effects on at least the regulation of CLS: inhibition of the PKA pathway extends CLS, whereas activation of the Sty1 pathway extends CLS (Roux et al., 2010b; Zuin et al., 2010a). Sty1 is a MAPK that responds to various stresses including heat, osmotic stress, oxidative stress, and nutritional stress in S. pombe (Vivancos et al., 2006). Deletion of sty1 + shortens CLS, whereas its overexpression extends CLS (Hibi et al., 2018; Ohtsuka et al., 2008). Consistent with this, although wis1 + encodes a MAPK kinase that phosphorylates Sty1, its activated mutation, wis1‐DD, which promotes Sty1 phosphorylation, also has a long CLS (Zuin et al., 2010a). Furthermore, the deletion of pyp1 +, which encodes tyrosine phosphatase and the product dephosphorylates Sty1 (Sansó et al., 2011), leads to the activation of Sty1 and extension of CLS (Zuin et al., 2010a). Thus, the Sty1 pathway plays a crucial role in regulating CLS in S. pombe. Because CLS extension by Δpyp1 mutant cells does not occur when phx1 + is absent (Kim et al., 2014), CLS extension by the activation of the Sty1 pathway may also depend on Phx1 (Figure 3).
As mentioned above, the PKA and Sty1 pathways (PKA–Sty1 pathway) are two pathways involved in CLS control, and there are many connections between them. Deletion of pka1 + promotes Sty1 phosphorylation, and its longevity also requires sty1 + (Zuin et al., 2010a; 2010b). This finding suggests that the longevity of Δpka1 mutant cells depends on sty1 + and that the activation of the Sty1 pathway is important for CLS extension by suppressing the PKA pathway. This finding is consistent with the report that pyp1 + overexpression suppresses the phenotypes that abnormally induce fbp1 +, encoding fructose‐1,6‐bisphosphatase, in git3 +, git5 +, and pka1 + mutants each (Santo et al., 1996). It is also consistent with the comprehensive analysis demonstrating that both git3 + and git5 + indicate positive genetic interactions with sty1 + (Ryan et al., 2012). However, git5 + reportedly indicates negative genetic interaction with pyp1 + and that deletion of both pka1 + and wis1 + causes synthetic growth defects (Jang et al., 2013; Ryan et al., 2012), suggesting that these pathways function in parallel. This finding may not rule out the possibility of a CLS extension mechanism via the repression of the PKA pathway that does not depend on the Sty1 pathway.
Sds23, which encodes a PP2A‐type phosphatase inhibitor, is thought to be involved in the PKA–Sty1 pathway and to regulate CLS in S. pombe. Sty1 reportedly interacts physically with Sds23 and directly phosphorylates it (Jang et al., 2013). Moreover, sds23 + overexpression inhibits stress sensitivity in Δsty1 mutant cells (Yakura et al., 2006), suggesting that Sds23 acts downstream of Sty1. Overexpression of sds23 + extends CLS, whereas its deletion reduces CLS (Roux et al., 2010a; Yakura et al., 2006). However, sds23 + reportedly indicates negative genetic interaction with pyp1 + (Ryan et al., 2012), suggesting that these factors may act in parallel. Meanwhile, another study, which used the yeast two‐hybrid assay and pull‐down assay, reports that Pka1 physically interacts with Sds23 (Jang et al., 2013). Although Jang et al. have shown that pka1 + is required for Sds23 phosphorylation during the stationary phase and that the catalytic subunit of PKA can phosphorylate Sds23, further studies are needed to elucidate these detailed molecular mechanisms. This is because Pka1 is inactivated by Cgs1, a regulatory subunit of PKA, during the nutrient‐poor stationary phase (Nishida et al., 2019). Additionally, since the deletion of sds23 + reportedly increases CLS under nitrogen depletion (Sideri et al., 2014), further analysis is required to understand the CLS regulation of sds23 + precisely.
Ecl1, which regulates CLS, is considered to contribute to CLS extension via the Sty1 pathway. Initially, ecl1 + was identified as a factor that complements short CLS of Δsty1 mutant cells (Ohtsuka et al., 2008); subsequently, ecl1 + is directly induced by Atf1, a transcription factor functioning downstream of Sty1 (Shimasaki et al., 2014).
Because the deletion of pka1 + induces ste11 + (Ohtsuka et al., 2008), ste11 + also contributes to CLS extension by inhibiting the PKA pathway. While the suppression of the PKA pathway induces CLS extension and ste11 +, the activation of Sty1 pathway also induces CLS extension and ste11 + (Shiozaki and Russell, 1996; Zuin et al., 2010a). Similarly, the overexpression of Sds23, the target of Sty1, induces ste11 + (Paul et al., 2009). Furthermore, two‐hybrid assay revealed that Sty1 physically interacts with Ste11 (Kjaerulff et al., 2005).
The deletion of tim18 +, which encodes the succinate dehydrogenase anchor subunit localized in the inner mitochondrial membrane, and may be involved in the tricarboxylic acid (TCA) cycle (Mercier et al., 2006), has been found to extend CLS (Rallis et al., 2014). tim18 + reportedly has a negative genetic interaction with sck2 + (involved in the TORC1 pathway) and positive genetic interaction with git3 + (involved in the PKA pathway) (Rallis et al., 2014; Ryan et al., 2012). These findings suggest that CLS extension by the deletion of tim18 + may occur in parallel with CLS extension via inhibition of the TORC1 pathway and may be involved in CLS extension via inhibition of the PKA pathway.
Thus, based on the findings obtained so far, it is likely that PKA and Sty1 pathways influence each other and contribute to CLS extension in S. pombe (Figure 3). Of note, CLS extension by glucose restriction is mediated through these pathways; low glucose levels inhibit the activity of Pka1 by suppressing signals from the receptor Git3 via G proteins, including Git5, and leads to the activation of MAPK Sty1 (Roux et al., 2006; Zuin et al., 2010a). When CLS extension occurs via the PKA–Sty1 pathway, the regulation of ecl1 +, sds23 +, ste11 +, and tim18 + may also contribute to CLS extension.
Although the activation of the Sty1 pathway contributes to CLS extension in S. pombe, the deletion of HOG1, the homolog of sty1 +, reportedly extends CLS of budding yeast (Garay et al., 2014; Zuin et al., 2010b). Moreover, in the presence of Sch9, Hog1 is reportedly phosphorylated under amino acid starvation and can contribute to longevity (Santos et al., 2013, 2016). Thus, the involvement of sty1 + and HOG1 in CLS regulation may be partially conserved depending on growth conditions such as the nutritional state of the environment. However, there are various differences between the two MAPKs: Hog1 is strongly activated by osmotic stress, but other stresses do not lead the same level of activation like of Sty1 and, unlike sty1 +, HOG1 does not appear to affect mating efficiency (Mutavchiev et al., 2016). Although the role of the PKA pathway in the conserved evolutionary regulation of lifespan is known, future studies should clarify whether the Sty1 pathway is conserved in longevity regulation.
6. Pmk1 PATHWAY
Inhibition of the Pmk1 pathway, which plays an important role in maintaining cell wall integrity, extends CLS in S. pombe (Figure 3). The Pmk1 pathway is composed of MAPK Pmk1, MAPK kinase Pek1, and MAPK kinase kinase Mkh1, and their deletion extends CLS (Imai et al., 2020). Because both pmk1 + and mkh1 + have negative genetic interactions with sck2 + (Rallis et al., 2014), the mechanisms of CLS extension by the Pmk1 and TORC1 pathways seem to function in parallel. Additionally, pmk1 + has a negative genetic interaction with git5 + (Ryan et al., 2012) and mkh1 + has negative genetic interactions with the genes involved in the Sty1 pathway, including wis1 +, pyp1 +, and sds23 + (Ryan et al., 2012; Sengar et al., 1997). Therefore, the mechanisms of CLS extension by the regulation of the Pmk1 and PKA–Sty1 pathways also seems to function in parallel. The transcription activity of the transcription factor Atf1, a target of Sty1, is reportedly regulated not only by Sty1 but also by Pmk1 (Zhou et al., 2012), suggesting that these pathways share common downstream factors, including Atf1 and factors regulated by Atf1. Moreover, although the overexpression of wis1 + and sty1 + does not complement all phenotypes of Δmkh1, they restore this mutant's β‐glucanase sensitivity (Sengar et al., 1997), indicating a connection between these pathways. Therefore, the mechanism of CLS extension regulated by the Pmk1 pathway may partially overlap with that of the PKA–Sty1 pathway. However, as described in detail later, in the absence of sty1 + or pmk1 +, the point mutation of gas1 + (gas1–287) does not extend CLS sufficiently, indicating that CLS extension by the gas1 + mutation is partially dependent on both sty1 + and pmk1 + (Imai et al., 2020). However, when both sty1 + and pmk1 + are deleted, CLS extension of the gas1 mutant disappears almost entirely (Imai et al., 2020). This finding suggests that the mechanisms of CLS regulation via the Pmk1 or Sty1 pathway are not the same but that these pathways regulate the CLS in parallel, at least in part.
Meanwhile, the deletion of SLT2, a budding yeast ortholog of pmk1 +, reportedly shortens CLS (Marek and Korona, 2013); thus, the effect of this MAPK on longevity does not seem to be the same, at least among these yeasts.
7. Ecl1 FAMILY GENES
The Ecl1 gene family is one of the most analyzed gene families in S. pombe CLS research (Ohtsuka and Aiba, 2017). S. pombe has three Ecl1 family genes, that is, ecl1 +, ecl2 +, and ecl3 +, whose overexpression extends CLS but triple deletion reduces CLS (Ohtsuka et al., 2008, 2009, 2011). Different signals induce these genes, but their gene products appear to have similar functions (Ohtsuka and Aiba, 2017). Ecl1 family gene‐dependent CLS extension occurs under conditions that induce Ecl1 family genes such as sulfur or leucine depletion (Ohtsuka et al., 2017, 2019). Furthermore, nitrogen depletion slightly induces ecl1 + (Miwa et al., 2011); oxidative stress induces ecl1 + via Atf1 (Shimasaki et al., 2014); and heat stress induces ecl2 + via Hsf1 (Ohtsuka et al., 2011), a heat shock transcription factor (Sakurai and Takemori, 2007). Overexpression of hsf1 + induces CLS extension and the expression of ste11 +, both of which are dependent on Ecl1 family genes (Ohtsuka et al., 2011). Heat shock transcription factor is also known to affect the lifespan of the nematode Caenorhabditis elegans; decreased hsf‐1 promotes tissue senescence and overexpression of extend lifespan (Hsu et al., 2003). Meanwhile, although the induction of Ecl1 family genes has not been observed, these genes are also required for CLS extension due to zinc limitation (Ohtsuka et al., 2015; Shimasaki et al., 2017). These findings indicate that Ecl1 family genes respond to environments that are disadvantageous for growth, such as nutrient depletion and stress, and contribute to cell survival.
Ecl1 family genes induce various genes that affect CLS including hsp9 +, hsr1 +, lsd90 +, spk1 +, ste11 +, and rsv2 +, whose overexpression leads to CLS extension, and some inductions depend on the transcription factor Prr1 (Ohtsuka et al., 2012). hsp9 + encodes a heat shock protein, and the involvement of heat shock proteins in lifespan and aging has been reported in other organisms, such as nematode and mammals (Fontana and Partridge, 2015; Fontana et al., 2010; Hsu et al., 2003; Walker and Lithgow, 2003). hsr1 + encodes a transcription factor that has low homology to Msn2 and Msn4, which are known to be involved in CLS in budding yeast (Wei et al., 2009). Lsd90 has been suggested to be involved in phospholipid metabolism including very long‐chain fatty acid metabolism (Yokoyama et al., 2008). Spk1, a MAPK involved in pheromone response, physically interacts with Ste11 and phosphorylates it in vitro (Kjaerulff et al., 2005). rsv2 + encodes a zinc finger transcription factor that induces stress‐related genes during spore formation (Mata et al., 2007). The mechanism of CLS extension regulated by rsv2 + may be involved in that of Pef1, a cyclin‐dependent kinase, because rsv2 + has positive genetic interactions with pef1 + (Roguev et al., 2008). rsv2 + has a positive genetic interaction with git5 + but causes synthetic growth defects with pyp1 + (Dixon et al., 2008; Roguev et al., 2008; Ryan et al., 2012), suggesting that the mechanism of CLS extension by rsv2 + may be related to that of the PKA pathway and not depend on the Sty1 pathway. Besides, rsv2 + reportedly has negative genetic interactions with mkh1 + and pdc202 + (Roguev et al., 2008; Ryan et al., 2012). Thus, the CLS extension by rsv2 + possibly occurs in parallel with the Pmk1 pathway and Pdc202. In S. cerevisiae, several studies of RPN4, a rsv2 + homolog, indicates different CLS results: one reports that the deletion of RPN4 increases RLS and another reports that its deletion decreases RLS (Kruegel et al., 2011; Longo et al., 2012; Schleit et al., 2013). Simultaneously, the loss of UBR2, which increases the Rpn4 level, extends RLS (Kruegel et al., 2011). Further research will be needed to clarify the precise CLS regulation of rsv2 +.
Ecl1 family genes also repress the expressions of many ribosomal proteins (Ohtsuka et al., 2012, 2017), some of which also depend on the transcription factor Prr1 (Ohtsuka et al., unpublished data). Furthermore, although many expressions of ribosomal proteins decrease during sulfur depletion, the repressions are dependent on Ecl1 family genes (Ohtsuka et al., 2017). CLS extension is also observed by the suppression of ribosomes by deletion of ribosomal proteins including rpl1201 +, rpl15 +, rpl42 +, and rps002 + (which encode ribosomal proteins), by the deletion of SPRRNA.47 (which encodes ribosomal RNA), and by drugs such as diazaborine or ribozionoindole‐1 (Chen et al., 2013; Ohtsuka et al., 2017). Therefore, CLS extension by Ecl1 family genes may be due to the suppression of ribosomes (Ohtsuka and Aiba, 2017). Meanwhile, because yeast two‐hybrid assay revealed that Rpl1201 physically interacts with Sds23 (Paul et al., 2009), CLS regulation by sds23 + may also be involved in ribosome regulation. Regulation of lifespan via ribosomes has been documented in various organisms including S. pombe (Hansen et al., 2007; MacInnes, 2016; Ohtsuka and Aiba, 2017; Rodríguez‐López et al., 2020; Steffen et al., 2008). In addition to Ecl1 family genes, the TORC1 pathway, including Sck2 ribosomal S6 kinase, and the PKA–Sty1 pathway, involved in Sds23, also affect ribosome regulation and control CLS. Ecl1 family genes, the TORC1 pathway, and the PKA–Sty1 pathway are closely related to response to dietary restriction of sulfur, nitrogen, and glucose, respectively, and all of these dietary restrictions lead to a significant reduction in ribosomes and extend CLS in S. pombe (Ohtsuka et al., 2017).
Consistent with the fact that Ecl1 family genes play an important role in CLS regulation in S. pombe, the S. cerevisiae ortholog ECL1 also functions in CLS regulation in budding yeast. However, unlike S. pombe, which has three Ecl1 family genes, S. cerevisiae has only one, that is, ECL1, the overexpression of which extends CLS and deletion shortens CLS (Azuma et al., 2009). Recently, Ecl1 family genes were found to be involved in CLS regulation as downstream factors of the general amino acid control (amino acid response in mammals) (Ohtsuka et al., 2019). Nevertheless, the orthologs of this gene have not been found in higher organisms.
Although the molecular mechanisms of Ecl1 family proteins are currently unknown, these genes respond to various stresses, particularly starvation, thereby contributing to the maintenance of cell survival and, consequently, CLS extension and sexual development, through the regulation of different other CLS‐related genes.
8. CYCLIN Clg1 AND CYCLIN‐DEPENDENT KINASE Pef1
The deletion of clg1 +, which encodes a cyclin‐like protein, extends CLS in S. pombe (Chen et al., 2013). Furthermore, the deletion of the cyclin‐dependent kinase Pef1 that interacts with Clg1 extends CLS (Chen et al., 2013). CLS extension by Δclg1 depends on cek1 +, which encodes the homologous protein of budding yeast protein kinase Rim15, although the deletion of cek1 + itself does not appear to have a significant effect on CLS in S. pombe (Chen et al., 2013).
CLS extension via Clg1 and Pef1 (Clg1–Pef1) suppression seems to function in parallel with the TORC1 pathway. clg1 + has a negative genetic interaction with sck2 +, which is involved in the TORC1 pathway (Rallis et al., 2014). pef1 + also has negative genetic interactions with tco89 + and sck2 + (Rallis et al., 2014; Ryan et al., 2012). Furthermore, CLS extension by Clg1–Pef1 suppression seems to function in parallel with the PKA–Sty1 pathway. clg1 + has a negative genetic interaction with pyp1 + (Ryan et al., 2012), and pef1 + has negative genetic interactions with git3 + and pyp1 + (Dixon et al., 2008; Ryan et al., 2012). Furthermore, it seems that CLS extension by Clg1–Pef1 suppression occurs in parallel with the Pmk1 pathway because clg1 + has negative genetic interactions with mkh1 +, pek1 +, and pmk1 + (Ryan et al., 2012), and pef1 + has negative genetic interactions with pek1 + and pmk1 + (Roguev et al., 2008; Ryan et al., 2012). Conversely, pef1 + has positive genetic interactions with pdc201 + and zrg17 + (Ryan et al., 2012); therefore, the mechanism of CLS extension by Clg1–Pef1 suppression may partially overlap with that of other pathways including TORC1 and PKA–Sty1 pathways.
Three genes, aca1 +, SPAC323.03c, and SPAC3H1.08c, whose deletions extend CLS (Rallis et al., 2014), reportedly have a positive genetic interaction with pef1 + (Roguev et al., 2008; Ryan et al., 2012). aca1 + is a homolog of the budding yeast gene MPR1 [which encodes an acetyltransferase of L‐azetidine‐2‐carboxylic acid, which is a toxic L‐proline analog (Nomura et al., 2003; Shichiri et al., 2001)] and acts to remove the intracellular oxidative stress (Du and Takagi, 2007). Based on sequence prediction, SPAC323.03c should encode a factor involved in peroxisome regulation, and SPAC3H1.08c encodes a mitochondrial calcium uniporter regulator. This indicates that the regulation of CLS extension by Clg1–Pef1 may be via the same pathway as that of aca1 +, SPAC323.03c, and SPAC3H1.08c. The negative interaction between aca1 + and tim18 + (Ryan et al., 2012) supports the hypothesis that aca1 + and Clg1–Pef1 function in parallel with the PKA–Sty1 pathway.
Additionally, pef1 + shows a synthetic growth defect with efc25 +, which encodes the Ras1 activator guanine nucleotide exchange factor (Dixon et al., 2008), and the deletion of efc25 + extends CLS (Chen et al., 2019). The amount of Efc25 protein is upregulated by the conserved NDR/LATS kinase Orb6 through the phosphorylation of efc25 mRNA‐binding protein Sts5 (Chen et al., 2019). Similar to efc25 +, the non‐phosphorylatable mutation at the Sts5 Ser‐86 site, sts5S86A, as well as downregulation of Orb6 also extends CLS (Chen et al., 2019). Consistent with these relationships between Clg1–Pef1 and Orb6–Sts5–Efc25, sts5 + shows a positive genetic interaction with aca1 + (Ryan et al., 2012). Based on these findings, CLS regulation by Orb6–Sts5–Efc25, which regulates the Ras1 GTPase activity, may be involved in the regulation of Clg1–Pef1. Moreover, sts5 + has positive genetic interactions with mkh1 + and pmk1 + (Ryan et al., 2012), and the cell morphology of sts5 mutant is complemented by wis1 deletion or pyp1 + overexpression (Toda et al., 1996), suggesting that CLS extension mechanism by sts5 + is also involved in the regulation of Pmk1 and Sty1 pathways. However, it has simultaneously been shown that efc25 + has a negative genetic interaction with mkh1 + (Ryan et al., 2012), but further studies will be needed to clarify the detailed mechanism. In addition, it has been reported that overexpression of spk1 +, a CLS regulator, complements the staurosporine sensitivity of sts5 mutant (Toda et al., 1991). Furthermore, because sts5 + has negative genetic interactions with sck2 + and zrg17 + (Rallis et al., 2014; Ryan et al., 2012), the CLS extension regulated by sts5 + may act in parallel with the TORC1 pathway.
Although the mechanism of CLS extension by Clg1–Pef1 is unclear, except the involvement of cek1 +, it is possible that the mechanism is related to the functions of other CLS‐regulated genes such as aca1 +, zrg17 +, SPAC323.03c, SPAC3H1.08c, orb6 +, sts5 +, and efc25 +.
9. Php COMPLEX
CCAAT‐binding factor (CBF) is a DNA‐binding transcription complex that binds to promoter regions containing the CCAAT sequence (Janoo et al., 2001). S. pombe CBF acts as a Php complex and comprises Php2, Php3, Php5, and its repressor Php4, and it plays an important role in various cellular regulations including iron response, TCA cycle, and respiration (Mercier et al., 2008). Deletion of php2 +, php3 +, and php5 +, but not of php4 +, causes CLS extension (Takuma et al., 2013).
Since both php3 + and php5 + have negative interactions with git3 + and git5 + (Ryan et al., 2012), the Php complex appears to function in parallel with the PKA–Sty1 pathway. Reports of the negative genetic interactions between php5 + and pyp1 +/sds23 + support this idea (Ryan et al., 2012). Moreover, since zrg17 +, which has positive genetic interactions with factors involved in the TORC1 pathway and Clg1–Pef1, has negative genetic interactions with php3 + and php5 + (Ryan et al., 2012), the Php complex may function in parallel with these pathways. However, the deletion of php2 + promotes Sty1 phosphorylation, and Sty1 is required for CLS extension by Δphp2 (Takuma et al., 2013). Thus, the suppression of the Php complex activates the Sty1 pathway and leads to CLS extension, probably indirectly, depending on certain conditions, such as an increase in ROS level due to abnormal expressions of mitochondrial components. Consistent with this, tim18 + expression, which has a positive genetic interaction with git3 +, is regulated by the Php complex (Mercier et al., 2006). Meanwhile, since php3 + has a positive genetic interaction with mkh1 + (Ryan et al., 2012), CLS regulation via the Php complex may also be related to the Pmk1 pathway.
In S. cerevisiae, the CBF Hap complex affects CLS. In contrast with the Php complex of S. pombe, deletion of HAP3, which encodes a component of the Hap complex, shortens CLS (Laschober et al., 2010). Besides, HAP4 overexpression extends CLS (Piper et al., 2006). The S. pombe Php complex, which carries Php4 as a repressor, may have a different regulatory system (Labbé et al., 2007), and the consistent effect to CLS by each CBF has not been observed, at least among budding yeast and S. pombe. Furthermore, although deletion of tim18 + extends CLS in S. pombe, the deletion of SDH4, one of its homologs, shortens CLS in budding yeast (Chang et al., 2015). Based on the findings to date, the effects of these CBFs on longevity are not consistent, but many respiratory mutants including CBF affect the lifespan. Strong inhibition of respiration shortens lifespan of C. elegans, whereas mild inhibition extends lifespan (Rea et al., 2007). Considering this, the difference in the effect of each factor on lifespan may be due to the difference in their effect on respiration in these yeasts. Further studies of these factors will contribute to the understanding of conserved regulation of lifespan and respiration.
10. OTHER GENES INVOLVED IN CLS
In addition to the TORC1 pathway, the PKA–Sty1 pathway, the Pmk1 pathway, Ecl1 family genes, Clg1–Pef1, and the Php complex, many genes are reportedly involved in CLS extension in S. pombe.
10.1. adh1 +
Overexpression of adh1 + extends CLS (Roux et al., 2010a). adh1 + encodes alcohol dehydrogenase, which reduces the acetaldehyde level, the last step in alcohol fermentation, and promotes the ethanol production (Sakurai et al., 2004). Efficient conversion of toxic acetaldehyde to ethanol, which can be used as a carbon source (Sakurai et al., 2004), may make a significant contribution to cell survival, particularly in the nutrient‐depleted stationary phase (Figure 4).
10.2. atg20 +
Deletion of atg20 +, which functions in organelle autophagy in S. pombe (Zhao et al., 2016), extends CLS (Rallis et al., 2014). However, according to a report by Rallis et al., the survival of Δatg20 cells was lower during the early stationary phase than that of wild‐type cells. Then, it increased after several days (Rallis et al., 2014), indicating the possibility of adaptive regrowth. Adaptive regrowth, which is often observed in short‐lived mutants, has been discussed as a phenomenon in which individual cells adapt to the environment and undergo regrowth during the stationary phase, thereby interfering with accurate CLS measurement (Fabrizio et al., 2004; Ohtsuka et al., 2011; Zambrano and Kolter, 1996). Since autophagy is required for lifespan control in various organisms (Ellis et al., 2018; Fontana and Partridge, 2015; Kapahi et al., 2017), results indicating a lack of autophagy factor extends CLS may mean either the existence of an unknown mechanism between CLS regulation and autophagy or regrowth of the short‐lived mutant due to autophagy deficiency. Further analysis will be needed for its determination. In the former case, because atg20 + has a positive genetic interaction with tim18 + (Ryan et al., 2012), CLS extension by Δatg20 may be involved in the PKA pathway or Php complex.
10.3. car2 +
Deletion of car2 +, which encodes ornithine transaminase and acts on amino acid metabolism (Bicho et al., 2010), extends CLS (Rallis et al., 2014). Since Car2 is required for the conversion of arginine to proline, glutamic acid, glutamine, and lysine, CLS extension by Δcar2 cells may be associated with the starvation of amino acids including lysine, which extends CLS (Ohtsuka et al., 2019).
10.4. erg28 +
Overexpression of erg28 + extends CLS (Ohtsuka et al., 2013). erg28 + encodes a protein conserved from yeast to humans, and the product is involved in sterol synthesis (Gachotte et al., 2001). Since erg28 + has negative genetic interactions with mkh1 + and pek1 + (Ryan et al., 2012), the mechanism of CLS extension may function in parallel with that of the Pmk1 pathway.
10.5. gas1 +
gas1 + encodes cell wall 1,3‐β‐glucanosyltransferase, and the point mutation gas1‐287 confer a long CLS (Imai et al., 2020). The CLS extension by gas1‐287 mutation depends on the Sty1 and Pmk1 pathways, both of which regulate CLS.
10.6. hsp104 +
The deletion of the heat shock protein Hsp104 extends CLS (Rallis et al., 2014). hsp104 + is induced by Hsf1 (Vjestica et al., 2013), whose overexpression causes CLS extension (Ohtsuka et al., 2011). Negative regulation of CLS by hsp104 + seems inconsistent with previous reports of the positive contributions of heat shock proteins to longevity (Fontana et al., 2010; Walker and Lithgow, 2003). Meanwhile, some heat shock proteins, including Hsp90 and Hsp70–Hsp40 chaperons, are also known to be inhibitors of Hsf1 activity (Vjestica et al., 2013). Thus, the deletion of hsp104 + might induce Hsf1 activation.
Because hsp104 + reportedly has negative genetic interactions with pyp1 + and mkh1 + (Ryan et al., 2012), hsp104 + in CLS may work in parallel with the Sty1 and Pmk1 pathways. In S. cerevisiae, the homolog HSP104 is reportedly required for survival under nutrient deprivation (Werner‐Washburne et al., 1993). Additionally, HSP104 overexpression contributes to the suppression of short RLS in SIR2 mutants and the deletion of HSP104 itself both decreases and increases RLS (Erjavec et al., 2007; Kaeberlein et al., 2005). Further research will be needed to gain an accurate understanding of the longevity conferred by Hsp104.
10.7. nop14 +
nop14 + is an ortholog of the budding yeast NOP14, which is involved in the maturation of the 40S ribosomal subunit (Granneman and Baserga, 2004; Pérez‐Fernández et al., 2007). As mentioned above, ribosome regulation has a significant influence on CLS, suggesting that the regulation of CLS via nop14 + may be involved in ribosome regulation.
10.8. kgd1 +
kgd1 + is a homolog of the budding yeast KGD1, which encodes the mitochondrial α‐ketoglutarate dehydrogenase complex subunit (Repetto and Tzagoloff, 1989). Its deletion reportedly causes CLS extension (Rallis et al., 2014). However, according to Rallis et al. (2014), the survival of kgd1 + (SPBC3h7.03c)‐deleted cells dropped sharply in the early stationary phase, stabilized, and was higher than that of wild‐type cells after several days as well as that of Δatg20 cells, suggesting adaptive regrowth. In S. cerevisiae, deletion of KGD1 decreases survival during stationary phase (Martinez et al., 2004). Further analysis will be needed to understand the role of Kgd1 in CLS regulation.
10.9. ksp1 +
Deletion of ksp1 +, an ortholog of budding yeast KSP1, extends CLS (Rallis et al., 2014). Since KSP1 is reported to be regulated by PKA and activates TORC1 (Umekawa and Klionsky, 2012), it may also be involved in these pathways and affect CLS.
10.10. lcf1 + and lcf2 +
Overexpression of lcf1 +, which encodes long‐chain fatty acyl‐CoA ligase, extends CLS (Oshiro et al., 2003), whereas its deletion shortens CLS (Fujita et al., 2007). S. cerevisiae has three homologs of lcf1 +: FAA1, FAA3, and FAA4. The deletion of FAA1 also reduces survival during the stationary phase when cultured at 37°C, as does Δlcf1 (Martinez et al., 2004). Meanwhile, the deletion of lcf2 +, a paralog of lcf1 +, extends CLS (Fujita et al., 2007). Although both lcf1 + and lcf2 + are involved in CLS regulation, their effects on CLS are different, partly because the catalytic levels of these two enzymes for each fatty acid are slightly different, depending on their substrates. Analysis using deletion strains demonstrated that Lcf1 mainly contributes to the catalytic reactions of three substrates: myristic acid, palmitic acid, and oleic acid. However, the only contribution of Lcf2 is to the catalysis of myristic acid (Fujita et al., 2007). Although the short CLS of Δlcf1 and the long CLS of Δlcf2 were both observed under the same conditions, a slight reduction in the activity of fatty acyl‐CoA ligase may be more advantageous than the intact condition for maintaining cell survival during the stationary phase under this condition.
10.11. lys7 +
Deletion of lys7 +, involved in lysine biosynthesis, extends CLS (Rallis et al., 2014). It is known that the availability of amino acids including leucine, arginine, histidine, and lysine has a remarkable influence on CLS of S. pombe (Ohtsuka et al., 2019). In S. pombe, the CLS of auxotrophic cells requiring leucine, lysine, or arginine is extended when cultured in media without a corresponding amino acid. In contrast, the CLS of cells requiring histidine decreases dramatically under histidine‐depleted conditions (Ohtsuka et al., 2019). Therefore, CLS extension by Δlys7 appears to be due to intracellular lysine restriction. The effects of amino acids on lifespan are reported in S. pombe and other organisms, such as budding yeast and animals. The restriction of specific amino acids, including asparagine, glutamate, and methionine, extends CLS in budding yeast, and a reduction of dietary amino acids, particularly tryptophan and methionine, extends lifespan in rodents (Dilova et al., 2007; Fontana and Partridge, 2015; Gallinetti et al., 2013; López‐Otín et al., 2016). Genes involved in amino acid metabolism and the related signal transductions are thought to be commonly involved in lifespan regulation in both S. pombe and other organisms.
10.12. moc3 +
The deletion of moc3 + extends CLS (Rallis et al., 2014). moc3 +, which encodes a Zn finger‐type protein localized in the nucleus, was identified as a factor that induces sexual differentiation even in the presence of cAMP, and is involved in stress response and sexual differentiation (Paul et al., 2009).
Since moc3 + has a negative genetic interaction with pef1 + (Ryan et al., 2012), its involvement in CLS may also be in parallel with that of Clg1–Pef1. Moreover, by yeast two‐hybrid assay, it has been reported that Moc3 physically interacts with Kgd1 (Vo et al., 2016), although each reported intracellular localization of Moc3 and Kgd1 is not the same, namely, nucleus and mitochondria, respectively. Similarly, since Moc3 physically interacts with the ribosomal protein Rpl1201 and MAPK Spk1 (Paul et al., 2009; Vo et al., 2016), the mechanism of CLS extension related to Moc3 may be involved in these factors.
10.13. nbr1 +
Deletion of nbr1 + (SPBP35G2.11c) extends CLS (Rallis et al., 2014). Nbr1 is a homolog of the mammalian autophagy receptor NBR1 and is distantly related to S. cerevisiae Atg19 (Zhao et al., 2016). Nbr1 mediates the transport of soluble hydrolases from the cytosol to the vacuole lumen (Liu et al., 2015).
10.14. ndk1 +
The deletion of ndk1 + extends CLS (Rallis et al., 2014). ndk1 + encodes a subunit of nucleoside–diphosphate kinases (Izumiya and Yamamoto, 1995). However, its mechanism of CLS extension has not been analyzed yet.
10.15. nnk1 +
Nonsense mutation of nnk1 +, a homolog of budding yeast NNK1, leads to CLS extension (Kurauchi et al., 2017). Since the budding yeast Nnk1 physically interacts with both Tor1 and Tor2 proteins (Breitkreutz et al., 2010), the mechanism of CLS regulation by nnk1 + may be associated with TOR. Moreover, the deletion of budding yeast NNK1 shortens CLS of S. cerevisiae (Garay et al., 2014). Since the deletion of nnk1 + is lethal in S. pombe, accurate comparison among these yeasts is difficult. Thus, the possibility of Nnk1 as a factor regulating lifespan beyond the species has not been verified.
10.16. oga1 +
Overexpression of oga1 + extends CLS (Ohtsuka et al., 2013). Oga1 is a homolog of budding yeast Stm1, which binds guanine–quadruplex nucleic acids and is involved in the TOR pathway and ribosome control (Ohtsuka et al., 2013; Van Dyke et al., 2006). Stm1 is reportedly important for maintaining survival during nutrient depletion, insdicating that Stm1 may act as a ribosome preservation factor under these conditions (Van Dyke et al., 2006, 2013). The TOR pathway and ribosome regulation are known to be involved in a conserved lifespan extension pathway in response to nutrient limitation (MacInnes, 2016; Ohtsuka and Aiba, 2017; Ohtsuka et al., 2017; Steffen et al., 2008). Thus, the mechanism of CLS extension by oga1 + may also be related to these processes. Since oga1 + has a negative genetic interaction with pef1 + (Ryan et al., 2012), CLS extension by oga1 + may function in parallel with Clg1–Pef1.
10.17. par1 +
The deletion of par1 + slightly extends CLS (Rallis et al., 2014). par1 + encodes a protein phosphatase 2A B′‐regulatory subunit, and its deletion causes abnormal septum formation and increases the septation index (Le Goff et al., 2001). Because elongated multinucleate multiseptated cells also appear in Δpar1 cells, this may also contribute to the high survival rate during the stationary phase. During colony formation, a connected cell population forms one colony regardless of the number of surviving cells among the population unless all cells in the population are dead. Cells with such a phenotype may not indicate accurate survival by CLS measurement using colony‐forming units.
Since par1 + has negative genetic interactions with mkh1 +, pek1 +, pmk1 +, pyp1 +, sds23 +, erg28 +, and zrg17 + (Ryan et al., 2012), the CLS regulation mechanism may occur in parallel with that of the Pmk1 and PKA–Sty1 pathways, Erg28, and Zrg17. Conversely, since par1 + has positive genetic interactions with sck2 + and nbr1 + (Rallis et al., 2014; Ryan et al., 2012), CLS extension by par1 + may be involved in the TORC1 pathway and Nbr1, which is involved in autophagy. In S. cerevisiae, deletion of RTS1, a homolog of par1 +, shortens CLS (Marek and Korona, 2013).
10.18. pdb1 +
Overexpression of pdb1 +, which encodes a subunit of pyruvate dehydrogenase, extends CLS (Ohtsuka et al., 2013). Although the CLS of pdb1 +‐deleted cells has not been reported in S. pombe, deletion of PDB1, which is a homolog of pdb1 + in S. cerevisiae, shortens CLS (Marek and Korona, 2013). Furthermore, in nematodes, dichloroacetate's activation of pyruvate dehydrogenase may lead to lifespan extension (Schaffer et al., 2011). Longevity regulation via pyruvate dehydrogenase may be conserved across species.
10.19. pdc201 + and pdc202 +
The transcription factor Phx1 may induce pdc201 + and pdc202 +, both of which encode pyruvate decarboxylase, and their individual induction contributes to CLS extension (Kim et al., 2014). As homologs of pdc201 + and pdc202 +, S. cerevisiae has three pyruvate decarboxylases: PCD1, PDC5, and PDC6. Unlike S. pombe pdc201 + and pdc202 +, deletion of PDC5 extends CLS (Garay et al., 2014). Therefore, at present, it is not easy to understand the regulation of CLS by pyruvate decarboxylase precisely across species.
10.20. pht1 +
The deletion of pht1 +, which encodes the histone H2A variant H2A.Z, extends CLS (Carr et al., 1994). Since pht1 + has negative genetic interactions with pmk1 +, sty1 +, pef1 +, sts5 +, par1 +, and moc3 + (Roguev et al., 2008; Ryan et al., 2012), the mechanism of CLS extension by pht1 + may be in parallel with the Pmk1 and Sty1 pathways, Clg–Pef1, Par1, and Moc3. In S. cerevisiae, deletion of HTZ1, a budding yeast homolog of pht1 +, also extends CLS (Garay et al., 2014). Furthermore, histone variants have been studied for their effects on aging in higher organisms (Contrepois et al., 2017; Re and Vinciguerra, 2017). CLS studies regulated by Pht1 may be useful in elucidating aging mechanisms across species.
10.21. pma1 +
Two loss‐of‐function mutations (D138N and A270D) of Pma1 extend CLS (Ito et al., 2010; Naito et al., 2014). pma1 + encodes P‐type proton ATPase (Kashiwazaki et al., 2011; Naito et al., 2014; Ulaszewski et al., 1987), and Pma1 mutations reduce glucose intake in addition to CLS extension, so its relationship with calorie restriction has been discussed (Ito et al., 2010). In S. cerevisiae, the functional decline of Pma1 reportedly extends RLS; furthermore, differences in Pma1 distribution between mother and daughter cells and the effects on vacuolar acidity and RLS have also been discussed previously (Henderson et al., 2014).
10.22. ppi1 +
Overexpression of ppi1 + extends CLS (Ohtsuka et al., 2013). ppi1 + encodes cyclophilin and has peptidyl‐prolyl cis/trans isomerase activity similar to the rapamycin‐acting protein FKBP (Siekierka et al., 1989; Skruzný et al., 2001; Van Dyke et al., 2013). The relationship between cyclophilin and aging is unclear (Nigro et al., 2013). Clarification of the regulatory mechanism of CLS extension by ppi1 + will contribute to the understanding of the relationship between cyclophilin and aging.
10.23. reb1 +
Deletion of reb1 +, which encodes RNA polymerase I transcription termination factor (Jaiswal et al., 2016), extends CLS (Rallis et al., 2014). However, Δreb1 cells have reportedly had lower survival during the early stationary phase than wild‐type cells (Rallis et al., 2014), so the possibility of adaptive regrowth cannot be ruled out. Since reb1 + has negative genetic interactions with git3 +, tim18 +, sty1 +, sds23 +, pef1 +, sts5 +, and zrg17 + (Roguev et al., 2008; Ryan et al., 2012), the CLS extension mechanism by Reb1 may work in parallel with the PKA–Sty1 pathway and Clg1–Pef1. Additionally, reb1 + has negative genetic interactions with par1 + and rsv2 + (Roguev et al., 2008; Ryan et al., 2012). Conversely, since reb1 + has a positive genetic interaction with ndk1 + (Ryan et al., 2012), these mechanisms of CLS regulation may work via the same pathways. Meanwhile, the deletions of the budding yeast reb1 + homologs REB1 and NSI1 reportedly shorten RLS (Ha et al., 2012; Kamei et al., 2015).
10.24. rpb10 +
Overexpression of rpb10 +, which encodes small subunits of RNA polymerase I, II, and III, extends CLS (Roux et al., 2010a). However, its interaction with other genes that cause CLS extension has not been reported; therefore, its mechanism of CLS extension is unknown.
10.25. sck1 +
Overexpression of sck1 + restores the phenotypes of git3, git5, and pka1 mutants (Jin et al., 1995), and sck1 + has positive genetic interactions with sty1 + and tim18 + (Ryan et al., 2012). Therefore, although the CLS extension by Δsck1 is weak, CLS regulation via sck1 + may be involved in the TORC1 pathway and PKA–Sty1 pathway. However, in contrast to this hypothesis, a negative genetic interaction between sck1 + and pyp1 + has also been reported (Ryan et al., 2012).
10.26. sdh1 +
Overexpression of sdh1 +, which encodes succinate dehydrogenase, extends CLS, and its deletion shortens CLS (Ohtsuka et al., 2013; Rallis et al., 2014). However, deletion of SDH1 (a budding yeast homolog of sdh1 +) has been reported to extend RLS (McCormick et al., 2015). In C. elegans, addition of TCA cycle metabolites related to succinate dehydrogenase (e.g., malate, fumarate, and succinate) activates nuclear translocation of the DAF‐16/FOXO transcription factor and suppresses oxidative stress (Edwards et al., 2013). Furthermore, the addition of malate or fumarate extends lifespan (Edwards et al., 2013). Similarly, in C. elegans, adding pyruvate and oxaloacetate also extends lifespan (Mouchiroud et al., 2011; Williams et al., 2009), similar to RNAi knockdown of aconitase or mitochondrial NAD+‐dependent isocitrate dehydrogenase (Hamilton et al., 2005; Rea et al., 2007). Thus, the knowledge regarding the TCA cycle and lifespan regulation is accumulating. It is unclear exactly how sdh1 + affects CLS of S. pombe, but there may be a conserved mechanism of lifespan regulation related to the TCA cycle.
10.27. shd1 +
The deletion of shd1 +, which encodes a cytoskeletal protein‐binding protein, extends CLS (Rallis et al., 2014); however, the detailed mechanism for CLS regulation by shd1 + remains unknown.
10.28. tor1 +
The relationship between Tor2, a component of TORC1, and Tor1, the catalytic subunit of S. pombe TORC2, is complicated. While they have the opposite effect on response to sexual differentiation (Laboucarié et al., 2017; Otsubo et al., 2017), there are reports that they share the same function in cell survival in adverse environments (Uritani et al., 2006; Weisman and Choder, 2001). Similarly, regarding CLS, deletion of tor1 + extends CLS in minimal (SD) medium, but shortens CLS in a complete (YE) medium (Ohtsuka et al., 2013; Rallis et al., 2013; Weisman and Choder, 2001). This indicates that the CLS of Δtor1 cells may be significantly affected by environmental and nutritional conditions. Since the deletion of tor1 + causes hypersensitivities to various stresses induced by the environment (Uritani et al., 2006; Weisman and Choder, 2001), the stresses caused by tor1 deletion may lead to the activation of stress response pathways such as the Sty1 pathway and then, cause CLS extension. Consistent with this idea, Tor1 functions upstream of Sty1 (Schonbrun et al., 2009) and CLS extension by Δtor1 may be involved in the Sty1 pathway. However, tor1 + reportedly has negative genetic interactions with git3 + and sds23 + (Ryan et al., 2012), making it difficult to understand exactly how Tor1 contributes to CLS regulation. Furthermore, since tor1 + has a negative genetic interaction with nbr1 + (Ryan et al., 2012), their involvement in CLS regulation may work in parallel.
10.29. tps0 +
Overexpression of tps0 +, which encodes mitochondrial lipid translocator protein, extends CLS (Ohtsuka et al., 2013). Since tps0 + has a negative genetic interaction with tor1 + (Ryan et al., 2012), these mechanisms of CLS extension may work in parallel.
10.30. uck2 +
The deletion of uck2 +, which encodes uracil phosphoribosyltransferase, extends CLS (Rallis et al., 2014). Since uck2 + has a negative genetic interaction with reb1 + (Ryan et al., 2012), CLS control of uck2 + may work in parallel with reb1 +.
10.31. ufd2 +
One study has reported that the deletion of ufd2 + (SPAC20H4.10), which encodes ubiquitin–protein ligase E4, extends CLS (Jang et al., 2013), whereas another study stated that the CLS of Δufd2 cells was almost the same as those of wild‐type cells (Rallis et al., 2014). The effect of ufd2 + on CLS may change depending on the culture conditions. Since ufd2 + has positive genetic interactions with git3 + and git5 + (Ryan et al., 2012), CLS extension by ufd2 + may be involved in the PKA pathway. This is consistent with the reports that Ufd2 physically interacts with Sds23 (Jang et al., 2013; Paul et al., 2009). Additionally, since ufd2 + has a positive genetic interaction with pmk1 + (Ryan et al., 2012), the CLS regulation mechanism by ufd2 + may be involved in both the PKA–Sty1 and Pmk1 pathways. Although the CLS extension regulated by ufd2 + may be environmentally dependent, ufd2 + interacts with many other CLS regulators. ufd2 + also has positive genetic interactions with sck2 + and par1 + (Rallis et al., 2014; Roguev et al., 2008), negative genetic interactions with pef1 + and zrg17 + (Roguev et al., 2008; Ryan et al., 2012), and the product Ufd2 physically interacts with Moc3 (Paul et al., 2009).
10.32. ure4 +
The deletion of ure4 +, which encodes an urease accessory protein, extends CLS (Rallis et al., 2014); however, the mechanism of CLS extension has not yet been elucidated.
10.33. vma1 +
Overexpression of vma1 +, which encodes the subunit A of vacuolar ATPase, extends CLS, and its deletion shortens CLS (Stephan et al., 2013). CLS regulation by vma1 + may be due to vacuolar acidification (Stephan et al., 2013). In S. cerevisiae, the deletion of VMA1, the homolog of vma1 +, also shortens CLS (Marek and Korona, 2013). Because vacuolar acidification is important for RLS in budding yeast (Henderson et al., 2014), this may be one of the evolutionarily conserved mechanisms of lifespan regulation.
10.34. SPAC323.03c
Deletion of SPAC323.03c extends CLS (Rallis et al., 2014). Since SPAC323.03c has a negative genetic interaction with par1 + (Ryan et al., 2012), the CLS extension mechanism may be parallel with that of par1 +.
10.35. SPBP4H10.16c
The deletion of SPBP4H10.16c, which may encode G‐patch RNA‐binding protein, extends CLS (Rallis et al., 2014). However, the deletion of WHI2, a budding yeast homolog of SPBP4H10.16c, decreases CLS (Burtner et al., 2011). An accurate understanding of this gene's role in CLS will require further study.
10.36. SPCC18.02
Overexpression of SPCC18.02, which should encode a transmembrane transporter protein, extends CLS (Ohtsuka et al., 2013). Since SPCC18.02 has a negative genetic interaction with mkh1 + (Ryan et al., 2012), CLS extension by SPCC18.02 is considered to be in parallel with the Pmk1 pathway.
11. CONCLUSIONS
While many model organisms, such as budding yeasts, nematodes, flies, and rodents, contribute to aging research, many CLS studies using S. pombe have also been conducted. In this review, we have summarized about more than 80 genes involved in CLS extension revealed in studies using S. pombe and have organized them based on information not only from CLS studies but also from non‐CLS studies including comprehensive interactome analysis. Many CLS regulatory genes have various interactions with many other CLS regulatory genes. Furthermore, among these CLS regulatory genes, we summarized the genes that have many interactions with each other and found that three pathways, namely, the TORC1, PKA–Sty1, and Pmk1 pathways, and three groups, namely, Ecl1 family genes, Clg1‐Pef1, and the Php complex, play central roles in CLS regulation in S. pombe.
Among the large volume of current results, some interactions were difficult to interpret. This may be because some of the CLS regulatory pathways function in parallel upstream, but have a common target downstream that regulates CLS extension. For example, in this review, the TORC1 and PKA–Sty1 pathways were described separately, but CLS extensions by both pathways are reportedly mediated by the common transcription factor Phx1 (Kim et al., 2014). Moreover, ribosome regulation may be important for CLS extension via the TORC1 pathway, Ecl1 family genes, and glucose restriction involved in the PKA–Sty1 pathway (Ohtsuka et al., 2017; Rodríguez‐López et al., 2020).
Although S. pombe is a unicellular organism, its signaling pathways that respond to nutrition and starvation function to mediate lifespans similarly to those in multicellular organisms (Fontana and Partridge, 2015; Fontana et al., 2010; Kapahi et al., 2017), suggesting evolutionarily conserved mechanisms to regulate lifespan. Furthermore, evolutionarily conserved mechanisms of lifespan extension other than those that respond to nutrition or starvation may exist. CLS research in S. pombe will also make a significant contribution to the elucidation of the aging mechanism in the same way as that of other model organisms.
CONFLICT OF INTEREST
None declared.
AUTHOR CONTRIBUTIONS
HO has made major contributions to (i) this study and writing of the manuscript. TS and HA have contributed to (i) the factual and logical confirmation; and (ii) revision of this manuscript.
ACKNOWLEDGMENTS
The authors are grateful to the scientists whose work provided the basis for this review and also thank Mr. Kanamaru and Mr. Hibi for discussions and Enago (www.enago.jp) for the English language review.
Ohtsuka H, Takafumi S, Aiba H. Genes affecting the extension of chronological life span in Schizosaccharomyces pombe (fission yeast). Mol Microbiol.2021;115:623–642. 10.1111/mmi.14627
Funding informationThis work was supported by a Grant‐in‐Aid for Young Scientists from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to HO) [JP19K15730] and by a Grant‐in‐Aid for Scientific Research (C) from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to HO) [JP16K07662]. Part of this work was also supported by the Institute for Fermentation, Osaka and The Asahi Glass Foundation (to HA) and a Grant‐in‐Aid for Scientific Research (B) from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to HA) [JP17H03792] and [JP20H02898]
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.
REFERENCES
- Azuma, K. , Ohtsuka, H. , Mita, S. , Murakami, H. and Aiba, H. (2009) Identification and characterization of an Ecl1‐family gene in Saccharomyces cerevisiae . Bioscience, Biotechnology, and Biochemistry, 73, 2787–2789. 10.1271/bbb.90599 [DOI] [PubMed] [Google Scholar]
- Berlett, B.S. and Stadtman, E.R. (1997) Protein oxidation in aging, disease, and oxidative stress. Journal of Biological Chemistry, 272, 20313–20316. 10.1074/jbc.272.33.20313 [DOI] [PubMed] [Google Scholar]
- Bicho, C.C. , de Lima Alves, F.D.L. , Chen, Z.A. , Rappsilber, J. and Sawin, K.E. (2010) A genetic engineering solution to the ‘arginine conversion problem’ in stable isotope labeling by amino acids in cell culture (SILAC). Molecular and Cellular Proteomics, 9, 1567–1577. 10.1074/mcp.M110.000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitkreutz, A. , Choi, H. , Sharom, J.R. , Boucher, L. , Neduva, V. , Larsen, B. , et al. (2010) A global protein kinase and phosphatase interaction network in yeast. Science, 328, 1043–1046. 10.1126/science.1176495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtner, C.R. , Murakami, C.J. , Olsen, B. , Kennedy, B.K. and Kaeberlein, M. (2011) A genomic analysis of chronological longevity factors in budding yeast. Cell Cycle, 10, 1385–1396. 10.4161/cc.10.9.15464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr, A.M. , Dorrington, S.M. , Hindley, J. , Phear, G.A. , Aves, S.J. and Nurse, P. (1994) Analysis of a histone H2A variant from fission yeast: evidence for a role in chromosome stability. Molecular and General Genetics, 245, 628–635. 10.1007/BF00282226 [DOI] [PubMed] [Google Scholar]
- Caspari, T. (1997) Onset of gluconate‐H+ symport in Schizosaccharomyces pombe is regulated by the kinases Wis1 and Pka1, and requires the gti1 + gene product. Journal of Cell Science, 110, 2599–2608. [DOI] [PubMed] [Google Scholar]
- Chang, Y.L. , Hsieh, M.H. , Chang, W.W. , Wang, H.Y. , Lin, M.C. , Wang, C.P. , et al. (2015) Instability of succinate dehydrogenase in SDHD polymorphism connects reactive oxygen species production to nuclear and mitochondrial genomic mutations in yeast. Antioxidants and Redox Signaling, 22, 587–602. 10.1089/ars.2014.5966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, B.R. , Li, Y. , Eisenstatt, J.R. and Runge, K.W. (2013) Identification of a lifespan extending mutation in the Schizosaccharomyces pombe cyclin gene clg1 + by direct selection of long‐lived mutants. PLoS One, 8, e69084. 10.1371/journal.pone.0069084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, B.R. and Runge, K.W. (2009) A new Schizosaccharomyces pombe chronological lifespan assay reveals that caloric restriction promotes efficient cell cycle exit and extends longevity. Experimental Gerontology, 44, 493–502. 10.1016/j.exger.2009.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, B.R. and Runge, K.W. (2012) Genetic approaches to aging in budding and fission yeasts: new connections and new opportunities. Sub‐Cellular Biochemistry, 57, 291–314. 10.1007/978-94-007-2561-4_13 [DOI] [PubMed] [Google Scholar]
- Chen, C. , Pino, M.R. , Haller, P.R. and Verde, F. (2019) Conserved NDR/LATS kinase controls RAS GTPase activity to regulate cell growth and chronological lifespan. Molecular Biology of the Cell, 30, 2598–2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contrepois, K. , Coudereau, C. , Benayoun, B.A. , Schuler, N. , Roux, P.F. , Bischof, O. , et al. (2017) Histone variant H2A.J accumulates in senescent cells and promotes inflammatory gene expression. Nature Communications, 8, 14995. 10.1038/ncomms14995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, H.L. and Braverman, R. (1977) The mechanism by which actinomycin D inhibits protein synthesis in animal cells. Nature, 269, 527–529. 10.1038/269527a0 [DOI] [PubMed] [Google Scholar]
- Dilova, I. , Easlon, E. and Lin, S.J. (2007) Calorie restriction and the nutrient sensing signaling pathways. Cellular and Molecular Life Sciences, 64, 752–767. 10.1007/s00018-007-6381-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, D.Q. , Tomita, Y. , Yamamoto, A. , Chikashige, Y. , Haraguchi, T. and Hiraoka, Y. (2000) Large‐scale screening of intracellular protein localization in living fission yeast cells by the use of a GFP‐fusion genomic DNA library. Genes to Cells, 5, 169–190. 10.1046/j.1365-2443.2000.00317.x [DOI] [PubMed] [Google Scholar]
- Dixon, S.J. , Fedyshyn, Y. , Koh, J.L. , Prasad, T.S. , Chahwan, C. , Chua, G. , et al. (2008) Significant conservation of synthetic lethal genetic interaction networks between distantly related eukaryotes. Proceedings of the National Academy of Sciences of the United States of America, 105, 16653–16658. 10.1073/pnas.0806261105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, X. and Takagi, H. (2007) N‐Acetyltransferase Mpr1 confers ethanol tolerance on Saccharomyces cerevisiae by reducing reactive oxygen species. Applied Microbiology and Biotechnology, 75, 1343–1351. 10.1007/s00253-007-0940-x [DOI] [PubMed] [Google Scholar]
- Edwards, C.B. , Copes, N. , Brito, A.G. , Canfield, J. and Bradshaw, P.C. (2013) Malate and fumarate extend lifespan in Caenorhabditis elegans . PLoS One, 8, e58345. 10.1371/journal.pone.0058345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, D.A. , Mustonen, V. , Rodríguez‐López, M. , Rallis, C. , Malecki, M. , Jeffares, D.C. , et al. (2018) Uncovering natural longevity alleles from intercrossed pools of aging fission yeast cells. Genetics, 210, 733–744. 10.1534/genetics.118.301262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erjavec, N. , Cvijovic, M. , Klipp, E. and Nyström, T. (2008) Selective benefits of damage partitioning in unicellular systems and its effects on aging. Proceedings of the National Academy of Sciences of the United States of America, 105, 18764–18769. 10.1073/pnas.0804550105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erjavec, N. , Larsson, L. , Grantham, J. and Nyström, T. (2007) Accelerated aging and failure to segregate damaged proteins in Sir2 mutants can be suppressed by overproducing the protein aggregation‐remodeling factor Hsp104p. Genes and Development, 21, 2410–2421. 10.1101/gad.439307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio, P. , Battistella, L. , Vardavas, R. , Gattazzo, C. , Liou, L.L. , Diaspro, A. , et al. (2004) Superoxide is a mediator of an altruistic aging program in Saccharomyces cerevisiae . Journal of Cell Biology, 166, 1055–1067. 10.1083/jcb.200404002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio, P. , Liou, L.L. , Moy, V.N. , Diaspro, A. , Valentine, J.S. , Gralla, E.B. , et al. (2003) SOD2 functions downstream of Sch9 to extend longevity in yeast. Genetics, 163, 35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio, P. and Longo, V.D. (2003) The chronological life span of Saccharomyces cerevisiae . Aging Cell, 2, 73–81. 10.1046/j.1474-9728.2003.00033.x [DOI] [PubMed] [Google Scholar]
- Fang, Y. , Sugiura, R. , Ma, Y. , Yada‐Matsushima, T. , Umeno, H. and Kuno, T. (2008) Cation diffusion facilitator Cis4 is implicated in Golgi membrane trafficking via regulating zinc homeostasis in fission yeast. Molecular Biology of the Cell, 19, 1295–1303. 10.1091/mbc.e07-08-0805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana, L. and Partridge, L. (2015) Promoting health and longevity through diet: From model organisms to humans. Cell, 161, 106–118. 10.1016/j.cell.2015.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana, L. , Partridge, L. and Longo, V.D. (2010) Extending healthy life span–from yeast to humans. Science, 328, 321–326. 10.1126/science.1172539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita, Y. , Mita, S. , Ohtsuka, H. and Aiba, H. (2007) Identification of a fatty acyl‐CoA synthetase gene, lcf2 +, which affects viability after entry into the stationary phase in Schizosaccharomyces pombe . Bioscience, Biotechnology, and Biochemistry, 71, 3041–3047. 10.1271/bbb.70442 [DOI] [PubMed] [Google Scholar]
- Gachotte, D. , Eckstein, J. , Barbuch, R. , Hughes, T. , Roberts, C. and Bard, M. (2001) A novel gene conserved from yeast to humans is involved in sterol biosynthesis. Journal of Lipid Research, 42, 150–154. [PubMed] [Google Scholar]
- Gallinetti, J. , Harputlugil, E. and Mitchell, J.R. (2013) Amino acid sensing in dietary‐restriction‐mediated longevity: roles of signal‐transducing kinases GCN2 and TOR. Biochemical Journal, 449, 1–10. 10.1042/BJ20121098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garay, E. , Campos, S.E. , González de la Cruz, J. , Gaspar, A.P. , Jinich, A. and Deluna, A. (2014) High‐resolution profiling of stationary‐phase survival reveals yeast longevity factors and their genetic interactions. PLoS Genetics, 10, e1004168. 10.1371/journal.pgen.1004168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman, S. and Baserga, S.J. (2004) Ribosome biogenesis: of knobs and RNA processing. Experimental Cell Research, 296, 43–50. 10.1016/j.yexcr.2004.03.016 [DOI] [PubMed] [Google Scholar]
- Gupta, D.R. , Paul, S.K. , Oowatari, Y. , Matsuo, Y. and Kawamukai, M. (2011) Multistep regulation of protein kinase A in its localization, phosphorylation and binding with a regulatory subunit in fission yeast. Current Genetics, 57, 353–365. 10.1007/s00294-011-0354-2 [DOI] [PubMed] [Google Scholar]
- Ha, C.W. , Sung, M.K. and Huh, W.K. (2012) Nsi1 plays a significant role in the silencing of ribosomal DNA in Saccharomyces cerevisiae . Nucleic Acids Research, 40, 4892–4903. 10.1093/nar/gks188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton, B. , Dong, Y. , Shindo, M. , Liu, W. , Odell, I. , Ruvkun, G. , et al. (2005) A systematic RNAi screen for longevity genes in C. elegans . Genes and Development, 19, 1544–1555. 10.1101/gad.1308205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, M. , Taubert, S. , Crawford, D. , Libina, N. , Lee, S.J. and Kenyon, C. (2007) Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans . Aging Cell, 6, 95–110. 10.1111/j.1474-9726.2006.00267.x [DOI] [PubMed] [Google Scholar]
- Hayashi, T. , Hatanaka, M. , Nagao, K. , Nakaseko, Y. , Kanoh, J. , Kokubu, A. , et al. (2007) Rapamycin sensitivity of the Schizosaccharomyces pombe tor2 mutant and organization of two highly phosphorylated TOR complexes by specific and common subunits. Genes to Cells, 12, 1357–1370. 10.1111/j.1365-2443.2007.01141.x [DOI] [PubMed] [Google Scholar]
- Hayashi, Y. , Kuroda, T. , Kishimoto, H. , Wang, C. , Iwama, A. and Kimura, K. (2014) Downregulation of rRNA transcription triggers cell differentiation. PLoS One, 9, e98586. 10.1371/journal.pone.0098586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayles, J. and Nurse, P. (2018) Introduction to fission yeast as a model system. Cold Spring Harbor Protocols, 2018, 323–333. 10.1101/pdb.top079749 [DOI] [PubMed] [Google Scholar]
- Hedges, S.B. (2002) The origin and evolution of model organisms. Nature Reviews. Genetics, 3, 838–849. 10.1038/nrg929 [DOI] [PubMed] [Google Scholar]
- Henderson, K.A. , Hughes, A.L. and Gottschling, D.E. (2014) Mother‐daughter asymmetry of pH underlies aging and rejuvenation in yeast. eLife, 3, e03504. 10.7554/eLife.03504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibi, T. , Ohtsuka, H. , Shimasaki, T. , Inui, S. , Shibuya, M. , Tatsukawa, H. , et al. (2018) Tschimganine and its derivatives extend the chronological life span of yeast via activation of the Sty1 pathway. Genes to Cells, 23, 620–637. 10.1111/gtc.12604 [DOI] [PubMed] [Google Scholar]
- Hsu, A.L. , Murphy, C.T. and Kenyon, C. (2003) Regulation of aging and age‐related disease by DAF‐16 and heat‐shock factor. Science, 300, 1142–1145. 10.1126/science.1083701 [DOI] [PubMed] [Google Scholar]
- Imai, Y. , Shimasaki, T. , Enokimura, C. , Ohtsuka, H. , Tsubouchi, S. , Ihara, K. , et al. (2020) gas1 mutation extends chronological lifespan via Pmk1 and Sty1 MAPKs in Schizosaccharomyces pombe . Bioscience, Biotechnology, and Biochemistry, 84, 330–337. 10.1080/09168451.2019.1676695 [DOI] [PubMed] [Google Scholar]
- Ito, H. , Oshiro, T. , Fujita, Y. , Kubota, S. , Naito, C. , Ohtsuka, H. , et al. (2010) Pma1, a P‐type proton ATPase, is a determinant of chronological life span in fission yeast. Journal of Biological Chemistry, 285, 34616–34620. 10.1074/jbc.M110.175562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumiya, H. and Yamamoto, M. (1995) Cloning and functional analysis of the ndk1 gene encoding nucleoside‐diphosphate kinase in Schizosaccharomyces pombe . Journal of Biological Chemistry, 270, 27859–27864. 10.1074/jbc.270.46.27859 [DOI] [PubMed] [Google Scholar]
- Jaiswal, R. , Choudhury, M. , Zaman, S. , Singh, S. , Santosh, V. , Bastia, D. , et al. (2016) Functional architecture of the Reb1‐Ter complex of Schizosaccharomyces pombe . Proceedings of the National Academy of Sciences of the United States of America, 113, E2267–E2276. 10.1073/pnas.1525465113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang, Y.J. , Won, M. and Yoo, H.S. (2013) Phosphorylations of Sds23/Psp1/Moc1 by stress‐activated kinase and cAMP‐dependent kinase are essential for regulating cell viability in prolonged stationary phase. Yeast, 30, 379–394. 10.1002/yea.2958 [DOI] [PubMed] [Google Scholar]
- Janoo, R.T. , Neely, L.A. , Braun, B.R. , Whitehall, S.K. and Hoffman, C.S. (2001) Transcriptional regulators of the Schizosaccharomyces pombe fbp1 gene include two redundant Tup1p‐like corepressors and the CCAAT binding factor activation complex. Genetics, 157, 1205–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, M. , Fujita, M. , Culley, B.M. , Apolinario, E. , Yamamoto, M. , Maundrell, K. , et al. (1995) sck1, a high copy number suppressor of defects in the cAMP‐dependent protein kinase pathway in fission yeast, encodes a protein homologous to the Saccharomyces cerevisiae SCH9 kinase. Genetics, 140, 457–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein, M. , Kirkland, K.T. , Fields, S. and Kennedy, B.K. (2005) Genes determining yeast replicative life span in a long‐lived genetic background. Mechanisms of Ageing and Development, 126, 491–504. 10.1016/j.mad.2004.10.007 [DOI] [PubMed] [Google Scholar]
- Kamei, Y. , Tai, A. , Dakeyama, S. , Yamamoto, K. , Inoue, Y. , Kishimoto, Y. , et al. (2015) Transcription factor genes essential for cell proliferation and replicative lifespan in budding yeast. Biochemical and Biophysical Research Communications, 463, 351–356. 10.1016/j.bbrc.2015.05.067 [DOI] [PubMed] [Google Scholar]
- Kapahi, P. , Kaeberlein, M. and Hansen, M. (2017) Dietary restriction and lifespan: lessons from invertebrate models. Ageing Research Reviews, 39, 3–14. 10.1016/j.arr.2016.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwazaki, J. , Yamasaki, Y. , Itadani, A. , Teraguchi, E. , Maeda, Y. , Shimoda, C. , et al. (2011) Endocytosis is essential for dynamic translocation of a syntaxin 1 orthologue during fission yeast meiosis. Molecular Biology of the Cell, 22, 3658–3670. 10.1091/mbc.E11-03-0255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima, S.A. , Chen, Z. , Aoi, Y. , Patgiri, A. , Kobayashi, Y. , Nurse, P. , et al. (2016) Potent, reversible, and specific chemical inhibitors of eukaryotic ribosome biogenesis. Cell, 167, 512–524.e14. 10.1016/j.cell.2016.08.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.Y. , Kim, E.J. , Lopez‐Maury, L. , Bähler, J. and Roe, J.H. (2014) A metabolic strategy to enhance long‐term survival by Phx1 through stationary phase‐specific pyruvate decarboxylases in fission yeast. Aging, 6, 587–601. 10.18632/aging.100682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.Y. , Kwon, E.S. and Roe, J.H. (2012) A homeobox protein Phx1 regulates long‐term survival and meiotic sporulation in Schizosaccharomyces pombe . BMC Microbiology, 12, 86. 10.1186/1471-2180-12-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjaerulff, S. , Lautrup‐Larsen, I. , Truelsen, S. , Pedersen, M. and Nielsen, O. (2005) Constitutive activation of the fission yeast pheromone‐responsive pathway induces ectopic meiosis and reveals Ste11 as a mitogen‐activated protein kinase target. Molecular and Cellular Biology, 25, 2045–2059. 10.1128/MCB.25.5.2045-2059.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruegel, U. , Robison, B. , Dange, T. , Kahlert, G. , Delaney, J.R. , Kotireddy, S. , et al. (2011) Elevated proteasome capacity extends replicative lifespan in Saccharomyces cerevisiae . PLoS Genetics, 7, e1002253. 10.1371/journal.pgen.1002253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurauchi, T. , Hashizume, A. , Imai, Y. , Hayashi, K. , Tsubouchi, S. , Ihara, K. , et al. (2017) Identification of a novel protein kinase that affects the chronological lifespan in fission yeast. FEMS Microbiology Letters, 364, fnw257. 10.1093/femsle/fnw257 [DOI] [PubMed] [Google Scholar]
- Labbé, S. , Pelletier, B. and Mercier, A. (2007) Iron homeostasis in the fission yeast Schizosaccharomyces pombe . BioMetals, 20, 523–537. 10.1007/s10534-006-9056-5 [DOI] [PubMed] [Google Scholar]
- Laboucarié, T. , Detilleux, D. , Rodriguez‐Mias, R.A. , Faux, C. , Romeo, Y. , Franz‐Wachtel, M. , et al. (2017) TORC1 and TORC2 converge to regulate the Saga co‐activator in response to nutrient availability. EMBO Reports, 18, 2197–2218 10.15252/embr.201744942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, Y.T. , Stocker, R. and Dawes, I.W. (2010) The lipophilic antioxidants α‐tocopherol and coenzyme Q10 reduce the replicative lifespan of Saccharomyces cerevisiae . Free Radical Biology and Medicine, 49, 237–244. 10.1016/j.freeradbiomed.2010.04.008 [DOI] [PubMed] [Google Scholar]
- Laschober, G.T. , Ruli, D. , Hofer, E. , Muck, C. , Carmona‐Gutierrez, D. , Ring, J. , et al. (2010) Identification of evolutionarily conserved genetic regulators of cellular aging. Aging Cell, 9, 1084–1097. 10.1111/j.1474-9726.2010.00637.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Goff, X. , Buvelot, S. , Salimova, E. , Guerry, F. , Schmidt, S. , Cueille, N. , et al. (2001) The protein phosphatase 2A B’‐regulatory subunit par1p is implicated in regulation of the S. pombe septation initiation network. FEBS Letters, 508, 136–142. 10.1016/s0014-5793(01)03047-2 [DOI] [PubMed] [Google Scholar]
- Lee, C. and Longo, V. (2016) Dietary restriction with and without caloric restriction for healthy aging. F1000Research, 5, 1–7. 10.12688/f1000research.7136.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees, H. , Walters, H. and Cox, L.S. (2016) Animal and human models to understand ageing. Maturitas, 93, 18–27. 10.1016/j.maturitas.2016.06.008 [DOI] [PubMed] [Google Scholar]
- Lin, S.J. and Austriaco, N. (2014) Aging and cell death in the other yeasts, Schizosaccharomyces pombe and Candida albicans . FEMS Yeast Research, 14, 119–135. 10.1111/1567-1364.12113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X.M. , Sun, L.L. , Hu, W. , Ding, Y.H. , Dong, M.Q. and Du, L.L. (2015) ESCRTs cooperate with a selective autophagy receptor to mediate vacuolar targeting of soluble cargos. Molecular Cell, 59, 1035–1042. 10.1016/j.molcel.2015.07.034 [DOI] [PubMed] [Google Scholar]
- Loibl, M. , Klein, I. , Prattes, M. , Schmidt, C. , Kappel, L. , Zisser, G. , et al. (2014) The drug diazaborine blocks ribosome biogenesis by inhibiting the AAA‐ATPase Drg1. Journal of Biological Chemistry, 289, 3913–3922. 10.1074/jbc.M113.536110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo, V.D. , Shadel, G.S. , Kaeberlein, M. and Kennedy, B. (2012) Replicative and chronological aging in Saccharomyces cerevisiae . Cell Metabolism, 16, 18–31. 10.1016/j.cmet.2012.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López‐Otín, C. , Galluzzi, L. , Freije, J.M.P. , Madeo, F. and Kroemer, G. (2016) Metabolic control of longevity. Cell, 166, 802–821. 10.1016/j.cell.2016.07.031 [DOI] [PubMed] [Google Scholar]
- Lu, T. and Finkel, T. (2008) Free radicals and senescence. Experimental Cell Research, 314, 1918–1922. 10.1016/j.yexcr.2008.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacInnes, A.W. (2016) The role of the ribosome in the regulation of longevity and lifespan extension. Wiley Interdisciplinary Reviews: RNA, 7, 198–212. 10.1002/wrna.1325 [DOI] [PubMed] [Google Scholar]
- Madia, F. , Gattazzo, C. , Wei, M. , Fabrizio, P. , Burhans, W.C. , Weinberger, M. , et al. (2008) Longevity mutation in SCH9 prevents recombination errors and premature genomic instability in a Werner/Bloom model system. Journal of Cell Biology, 180, 67–81. 10.1083/jcb.200707154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrid, M. , Soto, T. , Franco, A. , Paredes, V. , Vicente, J. , Hidalgo, E. , et al. (2004) A cooperative role for Atf1 and Pap1 in the detoxification of the oxidative stress induced by glucose deprivation in Schizosaccharomyces pombe . Journal of Biological Chemistry, 279, 41594–41602. 10.1074/jbc.M405509200 [DOI] [PubMed] [Google Scholar]
- Marek, A. and Korona, R. (2013) Restricted pleiotropy facilitates mutational erosion of major life‐history traits. Evolution; International Journal of Organic Evolution (N., Y), 67, 3077–3086. 10.1111/evo.12196 [DOI] [PubMed] [Google Scholar]
- Martinez, M.J. , Roy, S. , Archuletta, A.B. , Wentzell, P.D. , Anna‐Arriola, S.S. , Rodriguez, A.L. , et al. (2004) Genomic analysis of stationary‐phase and exit in Saccharomyces cerevisiae: gene expression and identification of novel essential genes. Molecular Biology of the Cell, 15, 5295–5305. 10.1091/mbc.e03-11-0856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata, J. , Wilbrey, A. and Bähler, J. (2007) Transcriptional regulatory network for sexual differentiation in fission yeast. Genome Biology, 8, R217. 10.1186/gb-2007-8-10-r217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama, A. , Arai, R. , Yashiroda, Y. , Shirai, A. , Kamata, A. , Sekido, S. , et al. (2006) ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe . Nature Biotechnology, 24, 841–847. 10.1038/nbt1222 [DOI] [PubMed] [Google Scholar]
- McCormick, M.A. , Delaney, J.R. , Tsuchiya, M. , Tsuchiyama, S. , Shemorry, A. , Sim, S. , et al. (2015) A comprehensive analysis of replicative lifespan in 4,698 single‐gene deletion strains uncovers conserved mechanisms of aging. Cell Metabolism, 22, 895–906. 10.1016/j.cmet.2015.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier, A. , Pelletier, B. and Labbé, S. (2006) A transcription factor cascade involving Fep1 and the CCAAT‐binding factor Php4 regulates gene expression in response to iron deficiency in the fission yeast Schizosaccharomyces pombe . Eukaryotic Cell, 5, 1866–1881. 10.1128/EC.00199-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier, A. , Watt, S. , Bähler, J. and Labbé, S. (2008) Key function for the CCAAT‐binding factor Php4 to regulate gene expression in response to iron deficiency in fission yeast. Eukaryotic Cell, 7, 493–508. 10.1128/EC.00446-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa, Y. , Ohtsuka, H. , Naito, C. , Murakami, H. and Aiba, H. (2011) Ecll, a regulator of the chronological lifespan of Schizosaccharomyces pombe, is induced upon nitrogen starvation. Bioscience, Biotechnology, and Biochemistry, 75, 279–283. 10.1271/bbb.100607 [DOI] [PubMed] [Google Scholar]
- Moreira‐Ramos, S. , Rojas, D.A. , Montes, M. , Urbina, F. , Miralles, V.J. and Maldonado, E. (2015) Casein kinase 2 inhibits HomolD‐directed transcription by Rrn7 in Schizosaccharomyces pombe . FEBS Journal, 282, 491–503. 10.1111/febs.13157 [DOI] [PubMed] [Google Scholar]
- Mouchiroud, L. , Molin, L. , Kasturi, P. , Triba, M.N. , Dumas, M.E. , Wilson, M.C. , et al. (2011) Pyruvate imbalance mediates metabolic reprogramming and mimics lifespan extension by dietary restriction in Caenorhabditis elegans . Aging Cell, 10, 39–54. 10.1111/j.1474-9726.2010.00640.x [DOI] [PubMed] [Google Scholar]
- Muller, F.L. , Lustgarten, M.S. , Jang, Y. , Richardson, A. and Van Remmen, H. (2007) Trends in oxidative aging theories. Free Radical Biology and Medicine, 43, 477–503. 10.1016/j.freeradbiomed.2007.03.034 [DOI] [PubMed] [Google Scholar]
- Mutavchiev, D.R. , Leda, M. and Sawin, K.E. (2016) Remodeling of the fission yeast Cdc42 cell‐polarity module via the Sty1 p38 stress‐activated protein kinase pathway. Current Biology, 26, 2921–2928. 10.1016/j.cub.2016.08.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito, C. , Ito, H. , Oshiro, T. , Ohtsuka, H. , Murakami, H. and Aiba, H. (2014) A new pma1 mutation identified in a chronologically long‐lived fission yeast mutant. FEBS Open Bio, 4, 829–833. 10.1016/j.fob.2014.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaoka, H. (2017) Live fast, die fast principle in a single cell of fission yeast. Microbial Cell, 4, 308–310 10.15698/mic2017.09.591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaoka, H. and Wakamoto, Y. (2017) Aging, mortality, and the fast growth trade‐off of Schizosaccharomyces pombe . PLoS Biology, 15, e2001109. 10.1371/journal.pbio.2001109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima, A. , Otsubo, Y. , Yamashita, A. , Sato, T. , Yamamoto, M. and Tamanoi, F. (2012) Psk1, an AGC kinase family member in fission yeast, is directly phosphorylated and controlled by TORC1 and functions as S6 kinase. Journal of Cell Science, 125, 5840–5849. 10.1242/jcs.111146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa, N. , Arakawa, O. , Ebe, M. and Yanagida, M. (2019) Casein kinase II– dependent phosphorylation of DNA topoisomerase II suppresses the effect of a catalytic topo II inhibitor, ICRF‐193, in fission yeast. Journal of Biological Chemistry, 294, 3772–3782. 10.1074/jbc.RA118.004955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigro, P. , Pompilio, G. , Capogrossi, M.C. and Cyclophilin, A. (2013) A key player for human disease. Cell Death and Disease, 4, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida, I. , Yokomi, K. , Hosono, K. , Hayashi, K. , Matsuo, Y. , Kaino, T. , et al. (2019) CoQ10 production in Schizosaccharomyces pombe is increased by reduction of glucose levels or deletion of pka1 . Applied Microbiology and Biotechnology, 103, 4899–4915. 10.1007/s00253-019-09843-7 [DOI] [PubMed] [Google Scholar]
- Nomura, M. , Nakamori, S. and Takagi, H. (2003) Characterization of novel acetyltransferases found in budding and fission yeasts that detoxify a proline analogue, azetidine‐2‐carboxylic acid. Journal of Biochemistry, 133, 67–74. 10.1093/jb/mvg003 [DOI] [PubMed] [Google Scholar]
- Ohtsuka, H. and Aiba, H. (2017) Factors extending the chronological lifespan of yeast: Ecl1 family genes. FEMS Yeast Research, 17, 10.1093/femsyr/fox066 [DOI] [PubMed] [Google Scholar]
- Ohtsuka, H. , Azuma, K. , Kubota, S. , Murakami, H. , Giga‐Hama, Y. , Tohda, H. , et al. (2012) Chronological lifespan extension by Ecl1 family proteins depends on Prr1 response regulator in fission yeast. Genes to Cells, 17, 39–52. 10.1111/j.1365-2443.2011.01571.x [DOI] [PubMed] [Google Scholar]
- Ohtsuka, H. , Azuma, K. , Murakami, H. and Aiba, H. (2011) hsf1 + extends chronological lifespan through Ecl1 family genes in fission yeast. Molecular Genetics and Genomics, 285, 67–77. 10.1007/s00438-010-0588-6 [DOI] [PubMed] [Google Scholar]
- Ohtsuka, H. , Ishida, M. , Naito, C. , Murakami, H. and Aiba, H. (2015) Sexual development of Schizosaccharomyces pombe is induced by zinc or iron limitation through Ecl1 family genes. Molecular Genetics and Genomics, 290, 173–185. 10.1007/s00438-014-0911-8 [DOI] [PubMed] [Google Scholar]
- Ohtsuka, H. , Kato, T. , Sato, T. , Shimasaki, T. , Kojima, T. and Aiba, H. (2019) Leucine depletion extends the lifespans of leucine‐auxotrophic fission yeast by inducing Ecl1 family genes via the transcription factor Fil1. Molecular Genetics and Genomics, 294, 1499–1509. 10.1007/s00438-019-01592-6 [DOI] [PubMed] [Google Scholar]
- Ohtsuka, H. , Mita, S. , Ogawa, Y. , Azuma, K. , Ito, H. and Aiba, H. (2008) A novel gene, ecl1 +, extends the chronological lifespan in fission yeast. FEMS Yeast Research, 8, 520–530. 10.1111/j.1567-1364.2008.00379.x [DOI] [PubMed] [Google Scholar]
- Ohtsuka, H. , Ogawa, S. , Kawamura, H. , Sakai, E. , Ichinose, K. , Murakami, H. , et al. (2013) Screening for long‐lived genes identifies Oga1, a guanine‐quadruplex associated protein that affects the chronological lifespan of the fission yeast Schizosaccharomyces pombe . Molecular Genetics and Genomics, 288, 285–295. 10.1007/s00438-013-0748-6 [DOI] [PubMed] [Google Scholar]
- Ohtsuka, H. , Ogawa, Y. , Mizuno, H. , Mita, S. and Aiba, H. (2009) Identification of Ecl family genes that extend chronological lifespan in fission yeast. Bioscience, Biotechnology, and Biochemistry, 73, 885–889. 10.1271/bbb.80804 [DOI] [PubMed] [Google Scholar]
- Ohtsuka, H. , Takinami, M. , Shimasaki, T. , Hibi, T. , Murakami, H. and Aiba, H. (2017) Sulfur restriction extends fission yeast chronological lifespan through Ecl1 family genes by downregulation of ribosome. Molecular Microbiology, 105, 84–97. 10.1111/mmi.13686 [DOI] [PubMed] [Google Scholar]
- Oshiro, T. , Aiba, H. and Mizuno, T. (2003) A defect in a fatty acyl‐CoA synthetase gene, lcf1 +, results in a decrease in viability after entry into the stationary phase in fission yeast. Molecular Genetics and Genomics, 269, 437–442. 10.1007/s00438-003-0841-3 [DOI] [PubMed] [Google Scholar]
- Otsubo, Y. , Nakashima, A. , Yamamoto, M. and Yamashita, A. (2017) TORC1‐dependent phosphorylation targets in fission yeast. Biomolecules, 7, 50. 10.3390/biom7030050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oya, E. , Durand‐Dubief, M. , Cohen, A. , Maksimov, V. , Schurra, C. , Nakayama, J. , et al. (2019) Leo1 is essential for the dynamic regulation of heterochromatin and gene expression during cellular quiescence. Epigenetics Chromatin, 12, 45. 10.1186/s13072-019-0292-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul, S.K. , Oowatari, Y. and Kawamukai, M. (2009) A large complex mediated by Moc1, Moc2 and Cpc2 regulates sexual differentiation in fission yeast. FEBS Journal, 276, 5076–5093. 10.1111/j.1742-4658.2009.07204.x [DOI] [PubMed] [Google Scholar]
- Pérez‐Fernández, J. , Román, A. , De Las Rivas, J. , Bustelo, X.R. and Dosil, M. (2007) The 90S preribosome is a multimodular structure that is assembled through a hierarchical mechanism. Molecular and Cellular Biology, 27, 5414–5429. 10.1128/MCB.00380-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper, P.W. , Harris, N.L. and MacLean, M. (2006) Preadaptation to efficient respiratory maintenance is essential both for maximal longevity and the retention of replicative potential in chronologically ageing yeast. Mechanisms of Ageing and Development, 127, 733–740. 10.1016/j.mad.2006.05.004 [DOI] [PubMed] [Google Scholar]
- Rallis, C. , Codlin, S. and Bähler, J. (2013) TORC1 signaling inhibition by rapamycin and caffeine affect lifespan, global gene expression, and cell proliferation of fission yeast. Aging Cell, 12, 563–573. 10.1111/acel.12080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rallis, C. , López‐Maury, L. , Georgescu, T. , Pancaldi, V. and Bähler, J. (2014) Systematic screen for mutants resistant to TORC1 inhibition in fission yeast reveals genes involved in cellular ageing and growth. Biology Open, 3, 161–171. 10.1242/bio.20147245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Re, O. and Vinciguerra, M. (2017) Histone macroH2A1: A chromatin point of intersection between fasting, senescence and cellular regeneration. Genes, 8, 1–14. 10.3390/genes8120367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea, S.L. , Ventura, N. and Johnson, T.E. (2007) Relationship between mitochondrial electron transport chain dysfunction, development, and life extension in Caenorhabditis elegans . PLoS Biology, 5, e259. 10.1371/journal.pbio.0050259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repetto, B. and Tzagoloff, A. (1989) Structure and regulation of KGD1, the structural gene for yeast α‐ketoglutarate dehydrogenase. Molecular and Cellular Biology, 9, 2695–2705. 10.1128/mcb.9.6.2695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche, B. , Arcangioli, B. , and Martinessen, R.A. (2016) RNA interference is essential for cellular quiescence. Science, 354, aah5651. 10.1126/science.aah5651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez‐López, M. , Gonzalez, S. , Hillson, O. , Tunnacliffe, E. , Codlin, S. , Tallada, V.A. , et al. (2020) The GATA transcription factor Gaf1 represses tRNAs, inhibits growth, and extends chronological lifespan downstream of fission yeast TORC1. Cell Reports, 30, 3240–3249.e4. 10.1016/j.celrep.2020.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roguev, A. , Bandyopadhyay, S. , Zofall, M. , Zhang, K. , Fischer, T. , Collins, S.R. , et al. (2008) Conservation and rewiring of functional modules revealed by an epistasis map in fission yeast. Science, 322, 405–410. 10.1126/science.1162609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux, A.E. , Arseneault, G. , Chartrand, P. , Ferbeyre, G. and Rokeach, L.A. (2010a) A screen for genes involved in respiration control and longevity in Schizosaccharomyces pombe . Annals of the New York Academy of Sciences, 1197, 19–27. 10.1111/j.1749-6632.2010.05198.x [DOI] [PubMed] [Google Scholar]
- Roux, A.E. , Chartrand, P. , Ferbeyre, G. and Rokeach, L.A. (2010b) Fission yeast and other yeasts as emergent models to unravel cellular aging in eukaryotes. Journals of Gerontology . Series A, Biological Sciences and Medical Sciences, 65, 1–8. 10.1093/gerona/glp152 [DOI] [PubMed] [Google Scholar]
- Roux, A.E. , Leroux, A. , Alaamery, M.A. , Hoffman, C.S. , Chartrand, P. , Ferbeyre, G. , et al. (2009) Pro‐aging effects of glucose signaling through a G protein‐coupled glucose receptor in fission yeast. PLoS Genetics, 5(3), e1000408. 10.1371/journal.pgen.1000408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux, A.E. , Quissac, A. , Chartrand, P. , Ferbeyre, G. and Rokeach, L.A. (2006) Regulation of chronological aging in Schizosaccharomyces pombe by the protein kinases Pka1 and Sck2. Aging Cell, 5, 345–357. 10.1111/j.1474-9726.2006.00225.x [DOI] [PubMed] [Google Scholar]
- Ryan, C.J. , Roguev, A. , Patrick, K. , Xu, J. , Jahari, H. , Tong, Z. , et al. (2012) Hierarchical modularity and the evolution of genetic interactomes across species. Molecular Cell, 46, 691–704. 10.1016/j.molcel.2012.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowska‐Bartosz, I. and Bartosz, G. (2014) Effect of antioxidants supplementation on aging and longevity. BioMed Research International, 2014, 404680. 10.1155/2014/404680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai, H. and Takemori, Y. (2007) Interaction between heat shock transcription factors (HSFs) and divergent binding sequences: binding specificities of yeast HSFs and human HSF1. Journal of Biological Chemistry, 282, 13334–13341. 10.1074/jbc.M611801200 [DOI] [PubMed] [Google Scholar]
- Sakurai, M. , Tohda, H. , Kumagai, H. and Giga‐Hama, Y. (2004) A distinct type of alcohol dehydrogenase, adh4 +, complements ethanol fermentation in an adh1‐deficient strain of Schizosaccharomyces pombe . FEMS Yeast Research, 4, 649–654. 10.1016/j.femsyr.2003.12.009 [DOI] [PubMed] [Google Scholar]
- Sansó, M. , Vargas‐Pérez, I. , García, P. , Ayté, J. and Hidalgo, E. (2011) Nuclear roles and regulation of chromatin structure by the stress‐dependent MAP kinase Sty1 of Schizosaccharomyces pombe . Molecular Microbiology, 82, 542–554. 10.1111/j.1365-2958.2011.07851.x [DOI] [PubMed] [Google Scholar]
- Santo, P.D. , Blanchard, B. and Hoffman, C.S. (1996) The Schizosaccharomyces pombe pyp1 protein tyrosine phosphatase negatively regulates nutrient monitoring pathways. Journal of Cell Science, 109, 1919–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos, J. , Leão, C. and Sousa, M.J. (2013) Ammonium‐dependent shortening of CLS in yeast cells starved for essential amino acids is determined by the specific amino acid deprived, through different signaling pathways. Oxidative Medicine and Cellular Longevity, 2013, 161986. 10.1155/2013/161986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos, J. , Leitão‐Correia, F. , Sousa, M.J. and Leão, C. (2016) Dietary restriction and nutrient balance in aging. Oxidative Medicine and Cellular Longevity, 2016, 4010357. 10.1155/2016/4010357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scala, F. , Brighenti, E. , Govoni, M. , Imbrogno, E. , Fornari, F. , Treré, D. , et al. (2016) Direct relationship between the level of p53 stabilization induced by rRNA synthesis‐inhibiting drugs and the cell ribosome biogenesis rate. Oncogene, 35, 977–989. 10.1038/onc.2015.147 [DOI] [PubMed] [Google Scholar]
- Schaffer, S. , Gruber, J. , Ng, L.F. , Fong, S. , Wong, Y.T. , Tang, S.Y. , et al. (2011) The effect of dichloroacetate on health‐ and lifespan in C. elegans . Biogerontology, 12, 195–209. 10.1007/s10522-010-9310-7 [DOI] [PubMed] [Google Scholar]
- Schleit, J. , Johnson, S.C. , Bennett, C.F. , Simko, M. , Trongtham, N. , Castanza, A. , et al. (2013) Molecular mechanisms underlying genotype‐dependent responses to dietary restriction. Aging Cell, 12, 1050–1061. 10.1111/acel.12130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonbrun, M. , Laor, D. , López‐Maury, L. , Bähler, J. , Kupiec, M. and Weisman, R. (2009) TOR complex 2 controls gene silencing, telomere length maintenance, and survival under DNA‐damaging conditions. Molecular and Cellular Biology, 29, 4584–4594. 10.1128/MCB.01879-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selman, C. , McLaren, J.S. , Collins, A.R. , Duthie, G.G. and Speakman, J.R. (2013) Deleterious consequences of antioxidant supplementation on lifespan in a wild‐derived mammal. Biology Letters, 9, 20130432. 10.1098/rsbl.2013.0432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengar, A.S. , Markley, N.A. , Marini, N.J. and Young, D. (1997) Mkh1, a MEK kinase required for cell wall integrity and proper response to osmotic and temperature stress in Schizosaccharomyces pombe . Molecular and Cellular Biology, 17, 3508–3519. 10.1128/mcb.17.7.3508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shichiri, M. , Hoshikawa, C. , Nakamori, S. and Takagi, H. (2001) A novel acetyltransferase found in Saccharomyces cerevisiae Sigma1278b that detoxifies a proline analogue, azetidine‐2‐carboxylic acid. Journal of Biological Chemistry, 276, 41998–42002. 10.1074/jbc.C100487200 [DOI] [PubMed] [Google Scholar]
- Shimasaki, T. , Ohtsuka, H. , Naito, C. , Azuma, K. , Tenno, T. , Hiroaki, H. , et al. (2017) Ecl1 is a zinc‐binding protein involved in the zinc‐limitation‐dependent extension of chronological life span in fission yeast. Molecular Genetics and Genomics, 292, 475–481. 10.1007/s00438-016-1285-x [DOI] [PubMed] [Google Scholar]
- Shimasaki, T. , Ohtsuka, H. , Naito, C. , Murakami, H. and Aiba, H. (2014) Ecl1 is activated by the transcription factor Atf1 in response to H2O2 stress in Schizosaccharomyces pombe . Molecular Genetics and Genomics, 289, 685–693. 10.1007/s00438-014-0845-1 [DOI] [PubMed] [Google Scholar]
- Shiozaki, K. and Russell, P. (1996) Conjugation, meiosis, and the osmotic stress response are regulated by Spc1 kinase through Atf1 transcription factor in fission yeast. Genes and Development, 10, 2276–2288. 10.1101/gad.10.18.2276 [DOI] [PubMed] [Google Scholar]
- Sideri, T. , Rallis, C. , Bitton, D.A. , Lages, B.M. , Suo, F. , Rodríguez‐López, M. , et al. (2014) Parallel profiling of fission yeast deletion mutants for proliferation and for lifespan during long‐term quiescence. G3: Genes|Genomes|Genetics, 5, 145–155. 10.1534/g3.114.014415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siekierka, J.J. , Hung, S.H.Y. , Poe, M. , Lin, C.S. and Sigal, N.H. (1989) A cytosolic binding protein for the immunosuppressant FK506 has peptidyl‐prolyl isomerase activity but is distinct from cyclophilin. Nature, 341, 755–757. 10.1038/341755a0 [DOI] [PubMed] [Google Scholar]
- Sipiczki, M. (2000) Where does fission yeast sit on the tree of life? Genome Biology, 1, REVIEWS1011. 10.1186/gb-2000-1-2-reviews1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skruzný, M. , Ambrozková, M. , Fuková, I. , Martínková, K. , Blahůsková, A. , Hamplová, L. , et al. (2001) Cyclophilins of a novel subfamily interact with SNW/SKIP coregulator in Dictyostelium discoideum and Schizosaccharomyces pombe . Biochimica et Biophysica Acta, 1521, 146–151. 10.1016/s0167-4781(01)00301-3 [DOI] [PubMed] [Google Scholar]
- Spivey, E.C. , Jones, S.K. , Rybarski, J.R. , Saifuddin, F.A. and Finkelstein, I.J. (2017) An aging‐independent replicative lifespan in a symmetrically dividing eukaryote. eLife, 6, 1–25. 10.7554/eLife.20340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen, K.K. , MacKay, V.L. , Kerr, E.O. , Tsuchiya, M. , Hu, D. , Fox, L.A. , et al. (2008) Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell, 133, 292–302. 10.1016/j.cell.2008.02.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan, J. , Franke, J. and Ehrenhofer‐Murray, A.E. (2013) Chemical genetic screen in fission yeast reveals roles for vacuolar acidification, mitochondrial fission, and cellular GMP levels in lifespan extension. Aging Cell, 12, 574–583. 10.1111/acel.12077 [DOI] [PubMed] [Google Scholar]
- Stiefel, J. , Wang, L. , Kelly, D.A. , Janoo, R.T. , Seitz, J. , Whitehall, S.K. , et al. (2004) Suppressors of an adenylate cyclase deletion in the fission yeast Schizosaccharomyces pombe . Eukaryotic Cell, 3, 610–619. 10.1128/EC.3.3.610-619.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, S.S.Y. , Tanaka, Y. , Samejima, I. , Tanaka, K. and Yanagida, M. (1996) A nitrogen starvation‐induced dormant G0 state in fission yeast: the establishment from uncommitted G1 state and its delay for return to proliferation. Journal of Cell Science, 109, 1347–1357. [DOI] [PubMed] [Google Scholar]
- Takuma, K. , Ohtsuka, H. , Azuma, K. , Murakami, H. and Aiba, H. (2013) The fission yeast php2 mutant displays a lengthened chronological lifespan. Bioscience, Biotechnology, and Biochemistry, 77, 1548–1555. 10.1271/bbb.130223 [DOI] [PubMed] [Google Scholar]
- Toda, T. , Niwa, H. , Nemoto, T. , Dhut, S. , Eddison, M. , Matsusaka, T. , et al. (1996) The fission yeast sts5 + gene is required for maintenance of growth polarity and functionally interacts with protein kinase C and an osmosensing MAP‐kinase pathway. Journal of Cell Science, 109, 2331–2342. http://www.ncbi.nlm.nih.gov/pubmed/8886983 [DOI] [PubMed] [Google Scholar]
- Toda, T. , Shimanuki, M. and Yanagida, M. (1991) Fission yeast genes that confer resistance to staurosporine encode an AP‐1‐like transcription factor and a protein kinase related to the mammalian ERK1/MAP2 and budding yeast FUS3 and KSS1 kinases. Genes and Development, 5, 60–73. [DOI] [PubMed] [Google Scholar]
- Ulaszewski, S. , Van Herck, J.C. , Dufour, J.P. , Kulpa, J. , Nieuwenhuis, B. and Goffeau, A. (1987) A single mutation confers vanadate resistance to the plasma membrane H+‐ATPase from the yeast Schizosaccharomyces pombe . Journal of Biological Chemistry, 262, 223–228. [PubMed] [Google Scholar]
- Umekawa, M. and Klionsky, D.J. (2012) Ksp1 kinase regulates autophagy via the target of rapamycin complex 1 (TORC1) pathway. Journal of Biological Chemistry, 287, 16300–16310. 10.1074/jbc.M112.344952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uritani, M. , Hidaka, H. , Hotta, Y. , Ueno, M. , Ushimaru, T. and Toda, T. (2006) Fission yeast Tor2 links nitrogen signals to cell proliferation and acts downstream of the Rheb GTPase. Genes to Cells, 11, 1367–1379. 10.1111/j.1365-2443.2006.01025.x [DOI] [PubMed] [Google Scholar]
- Van Dyke, N. , Baby, J. and Van Dyke, M.W. (2006) Stm1p, a ribosome‐associated protein, is important for protein synthesis in Saccharomyces cerevisiae under nutritional stress conditions. Journal of Molecular Biology, 358, 1023–1031. 10.1016/j.jmb.2006.03.018 [DOI] [PubMed] [Google Scholar]
- Van Dyke, N. , Chanchorn, E. and Van Dyke, M.W. (2013) The Saccharomyces cerevisiae protein Stm1p facilitates ribosome preservation during quiescence. Biochemical and Biophysical Research Communications, 430, 745–750. 10.1016/j.bbrc.2012.11.078 [DOI] [PubMed] [Google Scholar]
- Vivancos, A.P. , Jara, M. , Zuin, A. , Sansó, M. and Hidalgo, E. (2006) Oxidative stress in Schizosaccharomyces pombe: different H2O2 levels, different response pathways. Molecular Genetics and Genomics, 276, 495–502. 10.1007/s00438-006-0175-z [DOI] [PubMed] [Google Scholar]
- Vjestica, A. , Zhang, D. , Liu, J. and Oliferenko, S. (2013) Hsp70‐Hsp40 chaperone complex functions in controlling polarized growth by repressing Hsf1‐driven heat stress‐associated transcription. PLoS Genetics, 9, e1003886. 10.1371/journal.pgen.1003886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo, T.V. , Das, J. , Meyer, M.J. , Cordero, N.A. , Akturk, N. , Wei, X. , et al. (2016) A proteome‐wide fission yeast interactome reveals network evolution principles from yeasts to human. Cell, 164, 310–323. 10.1016/j.cell.2015.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, G.A. and Lithgow, G.J. (2003) Lifespan extension in C. elegans by a molecular chaperone dependent upon insulin‐like signals. Aging Cell, 2, 131–139. 10.1046/j.1474-9728.2003.00045.x [DOI] [PubMed] [Google Scholar]
- Wei, M. , Fabrizio, P. , Madia, F. , Hu, J. , Ge, H. , Li, L.M. , et al. (2009) Tor1/Sch9‐regulated carbon source substitution is as effective as calorie restriction in life span extension. PLoS Genetics, 5, e1000467. 10.1371/journal.pgen.1000467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisman, R. and Choder, M. (2001) The fission yeast TOR homolog, tor1 +, is required for the response to starvation and other stresses via a conserved serine. Journal of Biological Chemistry, 276, 7027–7032. 10.1074/jbc.M010446200 [DOI] [PubMed] [Google Scholar]
- Werner‐Washburne, M. , Braun, E. , Johnston, G.C. and Singer, R.A. (1993) Stationary phase in the yeast Saccharomyces cerevisiae . Microbiological Reviews, 57, 383–401. 10.1128/MMBR.57.2.383-401.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, D.S. , Cash, A. , Hamadani, L. and Diemer, T. (2009) Oxaloacetate supplementation increases lifespan in Caenorhabditis elegans through an AMPK/FOXO‐dependent pathway. Aging Cell, 8, 765–768. 10.1111/j.1474-9726.2009.00527.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakura, M. , Ishikura, Y. , Adachi, Y. and Kawamukai, M. (2006) Involvement of Moc1 in sexual development and survival of Schizosaccharomyces pombe . Bioscience, Biotechnology, and Biochemistry, 70, 1740–1749. 10.1271/bbb.60088 [DOI] [PubMed] [Google Scholar]
- Yan, L. , Vatner, D.E. , O'Connor, J.P. , Ivessa, A. , Ge, H. , Chen, W. , et al. (2007) Type 5 adenylyl cyclase disruption increases longevity and protects against stress. Cell, 130, 247–258. 10.1016/j.cell.2007.05.038 [DOI] [PubMed] [Google Scholar]
- Yokoyama, K. , Nakagawa, M. , Satoh, M. , Saitoh, S. , Dohmae, N. , Harada, A. , et al. (2008) Expression of a novel 90‐kDa protein, Lsd90, involved in the metabolism of very long‐chain fatty acid‐containing phospholipids in a mitosis‐defective fission yeast mutant. Journal of Biochemistry, 143, 369–375. 10.1093/jb/mvm232 [DOI] [PubMed] [Google Scholar]
- Zambrano, M.M. and Kolter, R. (1996) GASPing for life in stationary phase. Cell, 86, 181–184. 10.1016/s0092-8674(00)80089-6 [DOI] [PubMed] [Google Scholar]
- Zhao, D. , Liu, X.M. , Yu, Z.Q. , Sun, L.L. , Xiong, X. , Dong, M.Q. , et al. (2016) Atg20‐ and Atg24‐family proteins promote organelle autophagy in fission yeast. Journal of Cell Science, 129, 4289–4304. 10.1242/jcs.194373 [DOI] [PubMed] [Google Scholar]
- Zhou, X. , Ma, Y. , Kato, T. and Kuno, T. (2012) A measurable activation of the bZIP transcription factor Atf1 in a fission yeast strain devoid of stress‐activated and cell integrity mitogen‐activated protein kinase (MAPK) activities. Journal of Biological Chemistry, 287, 23434–23439. 10.1074/jbc.C111.338715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuin, A. , Carmona, M. , Morales‐Ivorra, I. , Gabrielli, N. , Vivancos, A.P. , Ayté, J. , et al. (2010a) Lifespan extension by calorie restriction relies on the Sty1 MAP kinase stress pathway. EMBO Journal, 29, 981–991. 10.1038/emboj.2009.407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuin, A. , Castellano‐Esteve, D. , Ayté, J. and Hidalgo, E. (2010b) Living on the edge: stress and activation of stress responses promote lifespan extension. Aging, 2, 231–237. 10.18632/aging.100133 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.


