Abstract
Background
Genome-wide association studies suggest that the combined effects of breast cancer (BC)-associated single nucleotide polymorphisms (SNPs) can improve BC risk stratification using polygenic risk scores (PRSs). The performance of PRSs in genome-wide association studies–independent clinical cohorts is poorly studied in individuals carrying mutations in moderately penetrant BC predisposition genes such as CHEK2.
Methods
A total of 760 female CHEK2 mutation carriers were included; 561 women were affected with BC, of whom 74 developed metachronous contralateral BC (mCBC). For PRS calculations, 2 SNP sets covering 77 (SNP set 1, developed for BC risk stratification in women unselected for their BRCA1/2 germline mutation status) and 88 (SNP set 2, developed for BC risk stratification in female BRCA1/2 mutation carriers) BC-associated SNPs were used. All statistical tests were 2-sided.
Results
Both SNP sets provided concordant PRS results at the individual level (r = 0.91, P < 2.20 × 10−16). Weighted cohort Cox regression analyses revealed statistically significant associations of PRSs with the risk for first BC. For SNP set 1, a hazard ratio of 1.71 per SD of the PRS was observed (95% confidence interval = 1.36 to 2.15, P = 3.87 × 10−6). PRSs identify a subgroup of CHEK2 mutation carriers with a predicted lifetime risk for first BC that exceeds the surveillance thresholds defined by international guidelines. Association of PRS with mCBC was examined via Cox regression analysis (SNP set 1 hazard ratio = 1.23, 95% confidence interval = 0.86 to 1.78, P = .26).
Conclusions
PRSs may be used to personalize risk-adapted preventive measures for women with CHEK2 mutations. Larger studies are required to assess the role of PRSs in mCBC predisposition.
Personalized risk prediction is essential for optimized decision making in clinical management for women with a breast cancer (BC) family history (1-5). Genome-wide association studies (GWAS) identified germline BC susceptibility loci, that is, single nucleotide polymorphisms (SNPs), which were shown to modify BC risk in addition to germline mutations in established high-risk BC predisposition genes such as BRCA1 and BRCA2 (6-9). The effects conferred by each of the BC-associated SNPs are low but can be combined into polygenic risk scores (PRSs), which could achieve a clinically useful degree of BC risk discrimination for women with or without a family history of BC and for women carrying pathogenic BRCA1 or BRCA2 mutations (10-12). For female BRCA1/BRCA2 mutation carriers, the effect of an 88-SNP–based PRS on BC risk was evaluated by Kuchenbaecker et al. (12) in a GWAS dataset of 23 463 European mutation carriers. For BRCA1 mutation carriers, the calculated BC risks at the age of 60 years with a PRS in the highest and lowest deciles of the PRS distribution were 57% and 38%, respectively, and for BRCA2 mutation carriers 40% and 28%, respectively (12). In a GWAS sample of 33 673 patients with BC and 33 381 control women of European descent, Mavaddat et al. (11) demonstrated that a 77-SNP–based PRS stratifies BC risk in women unselected for their BRCA1/2 germline mutation status. A study focusing on 369 CHEK2 c.1100delC germline mutation carriers (285 with BC; 84 without BC) and 33 624 noncarriers (17 640 with BC; 15 984 female controls) and the 77 BC-associated SNPs described by Mavaddat et al. (11) revealed that the PRS was associated with an odds ratio of 1.59 (95% confidence interval [CI] = 1.21 to 2.09) per SD for BC (13), similar to the odds ratio estimated in noncarriers. Of note, both SNP sets described by Mavaddat et al. (11) (subsequently referred to as SNP set 1) and Kuchenbaecker et al. (12) (subsequently referred to as SNP set 2) do have 55 SNPs in common, and 13 SNPs unique to SNP set 1 are in linkage disequilibrium to at least 1 out of 13 SNPs unique to SNP set 2, and vice versa. From the remaining 9 SNPs exclusive to SNP set 1, 4 were reported as specific for estrogen receptor (ER)-positive BC and 1 for ER-negative BC, respectively, by Kuchenbaecker et al. (12).
In many European countries, CHEK2 is the third-most frequently mutated BC risk gene (14), with c.1100delC (p.Thr367Metfs*15) being by far the most frequently observed pathogenic mutation. CHEK2 germline mutations confer estimated lifetime risks (LTRs) for BC of approximately 20% (15) and a 3.5-fold increased risk for a second BC compared with noncarriers (16). The performance of PRSs in GWAS-independent clinical cohorts is poorly studied for moderate penetrant risk genes, and thus the clinical implementation of PRSs is pending for these individuals. Moreover, it remains elusive whether PRSs may predict the risk for secondary contralateral BC (CBC) in addition to that for unilateral BC. To address these issues, we analyzed a clinical cohort of women at increased risk for both unilateral BC and CBC because of carrying a pathogenic CHEK2 germline variant.
Methods
Study Cohort of Female CHEK2 Germline Mutation Carriers
Female individuals carrying monoallelic protein-truncating germline variants (PTVs) in the CHEK2 gene (MIM +604373, transcript NM_007194.3) were eligible for this investigation. All individuals were identified through diagnostic germline testing in families recruited between January 1997 and June 2018 by 17 centers of the German Consortium for Hereditary Breast and Ovarian Cancer (GC-HBOC). All families met the inclusion criteria of the GC-HBOC for germline testing of the respective index patient (17) (Supplementary Table 1, available online). Demographic data, disease characteristics, family history, and medical history were documented in a central registry at the Institute for Medical Informatics, Statistics, and Epidemiology, University of Leipzig (Supplementary Table 2, available online). Written informed consent was obtained from all individuals, and ethical approval was granted by the Ethics Committee of the University of Cologne (07-048). All PTVs were classified as likely deleterious (class 4) or deleterious (class 5) based on the 5-tier variant classification system suggested by the International Agency for Research on Cancer, Unclassified Genetic Variants Working Group and in accordance with the regulations proposed by the international Evidence-Based Network for the Interpretation of Germline Mutant Alleles Consortium (18); class 4/5 PTVs were referred to as mutations. For this investigation, we excluded 1) CHEK2 mutation carriers who were part of previous GWAS studies aimed at identifying SNPs associated with BC risk [iCOGs study (19); OncoArray study (20)], and 2) CHEK2 mutation carriers who also carried BRCA1/2 mutations. This resulted in a sample of 769 female individuals.
Combined PRS-SNP Genotyping by Next-Generation Sequencing
PRSs were computed based on an SNP set comprising 77 SNPs developed using data on women unselected for germline mutation status [Mavaddat et al. (11) SNP set 1] and an SNP set comprising of 88 SNPs developed for BC risk stratification for female BRCA1/2 mutation carriers [Kuchenbaecker et al. (12) SNP set 2] (12). For SNP genotyping, we used a customized 48.48-amplicon–based target enrichment panel (Access Array, Fluidigm, San Francisco, CA). Variants that could not be covered due to technical limitations were replaced by adjacent SNPs in linkage disequilibrium (Supplementary Table 3, available online). Subsequent parallel next-generation sequencing of the barcoded amplicons of 384 samples was performed by using an Illumina NextSeq500 sequencing device (Illumina, San Diego, CA) (Supplementary Methods, available online). All DNA samples were analyzed at the Center for Familial Breast and Ovarian Cancer, University Hospital Cologne, Germany.
Quality Control of PRS-SNP Genotyping Results
For SNP set 1, 6 of 77 SNPs were excluded (rs1045485, rs7726159, rs12662670, rs13281615, rs8170, rs2363956; Supplementary Table 4, available online). For SNP set 2, 6 of 88 SNPs were excluded due to SNP call rates below 0.95 (rs12048493, rs56963355, rs9257408, rs13281615, rs494406, rs146699004; Supplementary Table 4, available online). SNP rs132390, located approximately 500 kb upstream of the CHEK2 gene and considered in both SNP sets, was described to be associated with the CHEK2 c.1100delC mutation (6,21). Consistently, the rs132390 minor allele frequency (MAF) in the subgroup of 557 CHEK2 c.1100delC mutation carriers was significantly increased compared with 203 individuals carrying other CHEK2 mutations (MAF = 0.35 vs MAF = 0.04, Fisher’s exact test P < 2.2 × 10−16). Thus, rs132390 unlikely defines an independent BC risk allele and was therefore excluded from the PRSs. In summary, these analyses resulted in an effective set of 70 SNPs for SNP set 1 and 81 SNPs for SNP set 2.
PRS Computation
For each sample , an individual PRS was derived via
where β is the per-allele log odds ratio and is the number of effect alleles in person for locus. Missing genotypes were imputed to the average observed genotype in the sample (see Supplementary Methods, available online). Values of , the theoretically expected mean PRSs with respect to the final SNP sets, were derived as described by Kuchenbaecker et al. (12) using the European subset of 1000 Genomes data (Supplementary Methods, available online).
Statistical Analysis
Analyses were conducted using the GenABEL v1.8 utilities (22) under R v3.6 and PLINK v1.9 (23). Quality controls, including checks for duplicate samples and ethnicity outliers (Supplementary Methods, available online), resulted in a curated data set comprising 760 CHEK2 mutation carriers out of 578 families as input for the PRS computation. Differences between obtained PRS values were assessed using Welsh’s t test. All statistical tests were 2-sided, with P less than .05 considered statistically significant.
PRSs for both SNP sets were standardized to have mean 0 and variance 1. To account for the nonrandom sampling of CHEK2 mutation carriers with respect to their disease status, the association of standardized PRSs with BC risk was analyzed using a weighted cohort Cox regression (24) with time to first BC diagnosis as the outcome (Supplementary Methods, available online). The weighted cohort approach aimed to correct the bias towards CHEK2 mutation carriers affected with BC in the sample by assignment of adapted weights to individuals with and without BC per age group, such that the observed weighted incidence rate agrees with the expected population-based incidence rates of CHEK2 mutation carriers (15,25). Observations were censored at age of first BC diagnosis, ovarian cancer (OC) diagnosis, prophylactic mastectomy, or age at last observation, whichever appeared at earliest. Analyses were adjusted for year of birth and counseling center of origin (Supplementary Methods, available online).
Age-specific PRS analyses, that is, examination of the proportional hazard assumption in age-stratified Cox regression models, were performed as described by Zhang et al. (26). Absolute age-specific cumulative risks of developing BC at different percentiles of the standardized PRS were calculated using age-specific hazard ratios (HRs) per SD of the PRS as described previously (12), based on UK incidences for women with CHEK2 c.1100delC mutation using recently published relative risk estimates (15,25).
CBCs occurring within a year of the first BC diagnosis were defined as synchronous CBC (sCBC) and those detected after 1 year as metachronous CBC (mCBC).
A Cox regression from time from 1 year after BC diagnosis to mCBC as the outcome was applied to individuals with BC, but not with sCBC, to evaluate the association of PRSs and age at first BC diagnosis with mCBC (Supplementary Methods, available online). To prevent a bias towards genetic testing due to mCBC, patients who entered the study after the occurrence of mCBC were excluded. Patients were censored at the age at mCBC diagnosis, OC diagnosis, prophylactic mastectomy of the healthy breast, or last observation.
Results
The study sample consisted of 769 female CHEK2 mutation carriers. After genotype quality control, 760 individuals were included, of whom 557 carried the c.1100delC mutation and 203 carried other PTVs (Supplementary Table 5, available online).
A total of 561 mutation carriers were diagnosed with BC. This included 460 patients diagnosed with unilateral BC (mean age [range] at diagnosis of 46.2 years [23-78 years]), 74 patients with mCBC (mean age at first BC diagnosis was 41.2 years [25-64 years] and mean age at mCBC diagnosis was 49.5 years [31-79 years]), and 27 patients with sCBC (mean age at diagnosis was 46.6 years [30-57 years]). Ten patients, all with unilateral BC, also developed OC; 6 of these were affected by OC before BC. A total of 199 mutation carriers had not been diagnosed with BC. Key characteristics of the study sample are summarized in Supplementary Table 2 (available online).
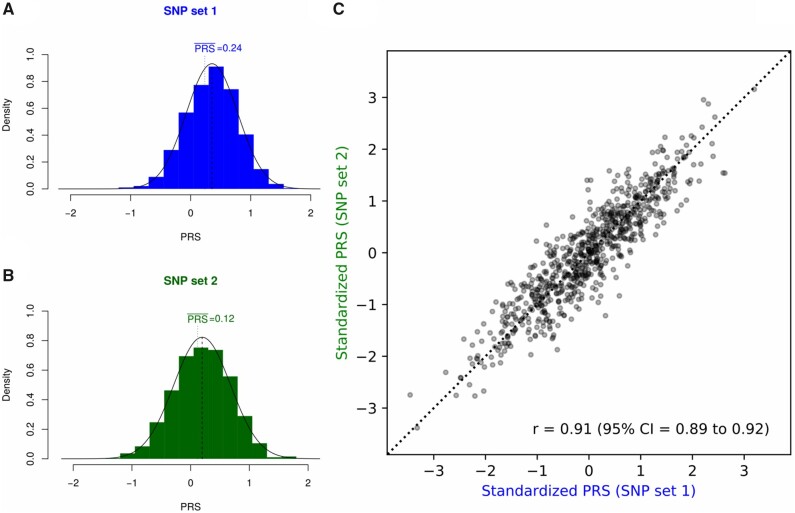
The distribution of the raw PRSs for both SNP set 1 and SNP set 2 in the overall study sample is shown in Figure 1. For SNP set 1, the PRS ranged from −1.12 to 1.72 (mean = 0.36) and for SNP set 2 from −1.44 to 1.73 (mean = 0.20). For both SNP sets, the mean PRS in the overall study sample was statistically significantly increased compared with the theoretically expected value (SNP set 1: P = 2.95 × 10−14 and SNP set 2: P = 1.49 × 10−5; 2-sided t test; Figure 1, A and B), reflecting the enrichment for BC and BC family history in the sample. Standardized PRSs from both SNP sets showed statistically significant correlation at the individual level (Pearson’s correlation coefficient r = 0.91, P < 2.20 × 10−16; Figure 1, C). Due to the high concordance between both SNP sets, results shown hereafter are restricted to SNP set 1; results for SNP set 2 are shown in the Supplementary Tables 6-8 (available online).
Figure 1.
Empirical distributions of the polygenic risk score (PRS) for single nucleotide polymorphism (SNP) set 1 (A) and SNP set 2 (B) in the overall cohort of 760 CHEK2 mutation carriers. Values of theoretically expected mean are indicated with vertical lines. (C) Standardized PRS values per individual for SNP set 1 and SNP set 2.
Weighted cohort Cox regression analysis revealed a statistically significant association of the standardized PRS with BC risk (HR = 1.71, 95% CI = 1.36 to 2.15, P = 3.87 × 10−6) (Table 1). In addition, we tested whether the association between the c.1100delC mutation and BC differed from the association between other PTVs in CHEK2 and BC, and whether the PRS association was consistent across carriers of different CHEK2 mutations by including an interaction term between the PRS and c.1100delC carrier status. The association with BC did not differ between c.1100delC and other PTVs (HR = 0.86, 95% CI = 0.53 to 1.41, P = .56), nor did the association between PRS and BC differ statistically significantly between carriers of c.1100delC and carriers of other PTVs in CHEK2 (interaction HR = 0.67, 95% CI = 0.42 to 1.08, P = .10) (Table 1). Testing for the violation of the proportional hazards assumption pointed towards an interaction of the PRS with age (Table 2). The PRS showed stronger associations with first BC in younger age groups, and associations were attenuated for ages older than 50 years (Table 2).
Table 1.
Hazard ratios per SD of the standardized PRS for first breast cancera
| Variable | SNP set 1 |
|
|---|---|---|
| HR (95% CI) | P | |
| Weighted cohort Cox regression | ||
| PRS | 1.71 (1.36 to 2.15) | 3.87 × 10−6 |
| Weighted cohort Cox regression with PRS × c.1100delC interaction | ||
| PRS | 2.29 (1.56 to 3.38) | 2.7 x 10−5 |
| c.1100delC | 0.86 (0.53 to 1.41) | .56 |
| PRS × c.1100delC interaction term | 0.67 (0.42 to 1.08) | .10 |
95% Confidence intervals and P values (2-sided Wald test) for the association of PRS with BC risk in 760 female CHEK2 mutation carriers, with and without inclusion of an interaction term for the PRS with c.1100delC carrier status. CI = confidence interval; HR = hazard ratio; PRS = polygenic risk score; SNP = single nucleotide polymorphisms.
Table 2.
Age-specific hazard ratios per SD of the standardized PRSa
| Variable | No. at risk | No. of events | SNP set 1 |
|
|---|---|---|---|---|
| HR (95% CI) | P | |||
| Age category, y | ||||
| ≤40 | 760 | 163 | 1.43 (1.04 to 1.97) | .03 |
| 41-50 | 503 | 254 | 2.32 (1.69 to 3.20) | 2.62 × 10−7 |
| 51-60 | 204 | 96 | 1.59 (1.07 to 2.35) | .02 |
| >60 | 71 | 42 | 1.34 (0.78 to 2.30) | .29 |
| PRS | — | — | — | .15 |
| Global | — | — | — | 9.50 × 10−91 |
95% Confidence intervals and P values (2-sided Wald test) for the association of PRS with breast cancer risk, and 2-sided P values for testing for the proportional hazards assumption of the Cox model stratified by age groups. CI = confidence interval; HR = hazard ratio; PRS = polygenic risk score; SNP = single nucleotide polymorphism.
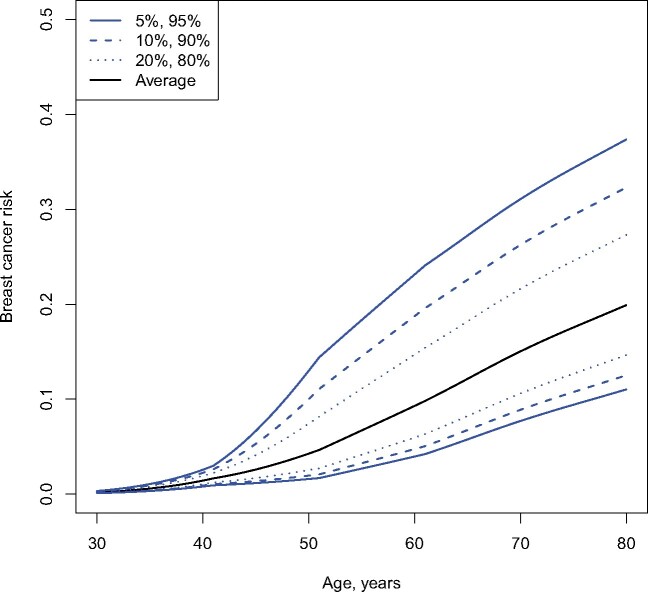
Age-specific hazard ratio estimates served as input for computation of absolute cumulative BC risks by PRS percentile (Figure 2). CHEK2 mutation carriers at the 10th percentile of the PRS had a risk of 2% of developing BC by the age of 50 years and a 13% risk by the age of 80 years; mutation carriers at the 90th percentile of the PRS had an 11% BC risk by the age of 50 years and 33% by the age of 80 years. The high concordance of absolute cumulative BC risk predictions based on both SNP set 1 and SNP set 2 (Supplementary Figure 2, available online), along with the high concordance of standardized PRSs at the individual level (Figure 1, C), suggest a similar clinical utility for both SNP sets.
Figure 2.
Predicted cumulative breast cancer risks by percentile of the polygenic risk scores in CHEK2 mutation carriers.
In addition to that for unilateral BC, we tested whether the PRS predicts the risk for CBC in a sample of 528 patients, including 34 individuals with mCBC. The mean interval between the first and the second BC was 8.3 years (range = 1-30 years). Cox regression analyses with time from 1 year after first BC diagnosis to mCBC as the outcome pointed towards an association of the PRS with mCBC, although a level of statistical significance was reached only for the association of first BC age with mCBC risk (HR per SD of the PRS = 1.23, 95% CI = 0.86 to 1.78, P = .26; age at first BC diagnosis HR = 0.95, 95% CI = 0.92 to 0.98, P = 3.67 × 10−4) (Table 3).
Table 3.
Hazard ratios per SD of the standardized PRS for mCBCa
| Variable | SNP set 1 |
|
|---|---|---|
| HR (95% CI) | P | |
| PRS | 1.23 (0.86 to 1.78) | .26 |
| Age at first breast cancer diagnosis, y | 0.95 (0.92 to 0.98) | 3.67 × 10−4 |
95% Confidence intervals and P values (2-sided Wald test) for a Cox model with time from 1 year after first BC diagnosis to mCBC as the outcome, including 528 CHEK2 mutation carriers with BC but not sCBC. The standardized PRSs do not show statistically significant association with mCBC. A statistically significant negative association with age at first diagnosis was observed. BC = breast cancer; CI = confidence interval; HR = hazard ratio; mCBC = metachronous contralateral breast cancer; PRS = polygenic risk score; sCBC = synchronous contralateral breast cancer; SNP = single nucleotide polymorphisms.
Discussion
Although the clinical management of women carrying pathogenic mutations in high-risk BC genes, such as BRCA1 and BRCA2, is well-established, the clinical management of women carrying mutations in moderately penetrant BC genes, such as CHEK2, is less standardized and may vary between countries (27). The clinical decision whether preventive measures are offered is mainly based on the estimated LTR and family history of cancer. We demonstrate that PRSs are suited to identify women with CHEK2 mutations in a risk category that exceeds, for example, the surveillance thresholds for BC according to the UK National Institute for Health and Care Excellence (NICE) and US National Comprehensive Cancer Network (NCCN) guidelines. The NICE guidelines generally consider an annual mammography for women with an estimated LTR greater than 30%, starting at the age of 30 years (28). The NCCN recommends annual mammography for patients with a LTR greater than 20% starting earliest at the age of 30 years or at an age that is 10 years younger than the age at the earliest BC diagnosis in the family, whichever is later (29). For CHEK2 germline mutation carriers, the NCCN suggests an annual breast screening by magnetic resonance imaging starting at the age of 40 years, depending on the family history (30). We demonstrate that the PRSs may be useful in identifying those women with risks exceeding the general surveillance thresholds of 20% or 30% LTR, respectively. In our study sample, on the basis of the PRS alone, approximately 10% of women with CHEK2 mutations fall into a risk category that exceeds the surveillance thresholds for BC according to the NICE recommendations (112 of 760), and approximately 50% fall into a risk category that exceeds the surveillance thresholds for BC according to NCCN guidelines (400 of 760).
Differences in the cumulative risk of mutation carriers within the highest decile of the PRS distribution were more than fivefold increased compared with the lowest decile at the age of 50 years (>11% vs <2%), whereas this effect was attenuated by the age of 80 years (>33% vs <13%). The stronger association of the PRS in younger age groups is in line with results published for BRCA1/2 mutation carriers (12). These data suggest that especially women younger than 50 years may benefit from a PRS-based risk prediction due to the higher levels of risk discrimination.
A study of women unselected for their germline mutation status demonstrated that a PRS based on 67 polymorphic loci associated with BC is associated with the development of CBC (31). A recent study considering 99 969 women enrolled in the FinnGen study reported an association of PRS greater than the 90th percentile with mCBC risk in 5979 Finnish individuals with BC, including 202 CHEK2 c.1100delC mutation carriers, for a PRS including millions of loci (32). We found no statistically significant evidence of association between the PRS and mCBC in our study. However, our analysis was based on small numbers of mCBC cases, and the estimates were associated with wide confidence intervals; therefore, effects of the PRSs cannot be excluded either.
In conclusion, PRSs have the potential to improve personal risk prediction accuracy for first and CBC in CHEK2 mutation carriers. Our GWAS-independent study was not restricted to the most common pathogenic CHEK2 variant c.1100delC, and we demonstrated that women with other PTVs in the CHEK2 gene may benefit from the PRS-based BC risk prediction. The recently extended Breast and Ovarian Analysis of Disease Incidence and Carrier Estimation Algorithm (BOADICEA) model (25) implemented in the CanRisk tool (www.canrisk.org) includes the effects of CHEK2 mutations, family history, and the BC PRS. Our results support the use of such tools in a clinical setting for providing more personalized BC risks for CHEK2 mutation carriers on the basis of the combined effects of family history and PRS.
This study has limitations. Our study sample consists of individuals who met the GC-HBOC inclusion criteria for germline testing. Therefore, a strong bias towards a familial BC background exists. Antoniou et al. (24) have shown that weighted cohort approaches based on values that deviate statistically significantly from true incidences can, in extreme cases, lead to estimates outside the true confidence intervals; therefore, we must also point out that the incidences used here are based on CHEK2 c.1100delC carriers only. Moreover, PRS analyses were restricted to individuals of European descent, and our results may not be applicable to other populations.
Because CHEK2 c.1100delC mutation carriers are more likely predisposed to ER-positive disease compared with noncarriers (13,16), the question arises whether an ER-positive–specific PRS may be more suitable for CHEK2 mutation carriers. Here, the overall PRS was used due to ER-negative mutation carriers in our cohort (52 of 441 first BC cases with known receptor status were ER-negative [11.79%]) and a high correlation between the ER-positive BC and the overall BC PRS at the individual level (r = 0.96, 95% CI = 0.95 to 0.96).
Larger studies based on more recently defined 313- or 3820-SNP–based PRSs (10) will provide more precise estimates of the association of PRS with BC and mCBC.
Funding
This work was supported by the German Cancer Aid (#110837, #70111850) and the Ministry for Innovation, Science and Research of the State of North Rhine-Westphalia (#323–8.0302.16.02–132142).
Notes
Role of the funder: The funders were not involved in study design, data collection and analysis, decision to publish, or prepare the manuscript.
Disclosures: The authors have nothing to disclose.
Prior presentations: European Society Human Genetics (ESHG) meeting 2019, Gothenburg (oral presentation by Julika Borde).
Acknowledgements: We thank all individuals who participated in this study. Computations were partially run on the German Research Foundation (DFG)-funded high-performance computing cluster CHEOPS, which is provided and maintained by the computing center of the University of Cologne (RRZK). We thank Stefanie Mueller for her greatly appreciated advice.
Author contributions: Conceptualization, J.B., C.Er., R.K.S., K.K., E.H.; Methodology, J.B., C.Er., J.Ha., N.W.L., E.P.R., O.G., A.C.A, M.K.S, K.K., E.H., R.K.S.; Software, J.B., C.Er., K.K.; Formal Analysis, J.B., C.Er.; Investigation, J.B., C.Er., K.W.L.; Resources, B.W., D.N., K.W.L., G.S., J.Ha., A.S.Q., N.W.L., J.Ho., E.P-R., N.A., A.R., A.G., J.He., U.F., V.D., A.M., M.K., S.W.G., B.H.F.W., C.S., A.E.V., A.L., C.En., M.K.S., A.C.A., R.K.S., E.H.; Data Curation, J.B., C.Er., C.En., E.H.; Writing—Original Draft, J.B., C.Er., K.K., R.K.S., E.H.; Writing—review & editing, J.B., C.Er., B.W., D.N., K.W.L., G.S., J.Ha., A.S.Q., N.W.L., J.Ho., E.P.R., N.A., A.R., A.G., J.He., U.F., V.D., A.M., M.K., S.W.G., B.H.F.W., C.S., A.E.V., O.G., A.L., C.En., M.K.S., A.C.A., R.K.S., K.K., E.H.; Visualization, C.Er.; Supervision, R.K.S., K.K., E.H.; Project Administration, R.K.S., E.H.; Funding Acquisition, R.K.S.
Data Availability
All relevant data are shown in the main manuscript and the Supplementary Materials.
Supplementary Material
References
- 1. Bleyer A, Welch HG.. Effect of three decades of screening mammography on breast-cancer incidence. N Engl J Med. 2012;367(21):1998–2005. [DOI] [PubMed] [Google Scholar]
- 2. Cuzick J, Sestak I, Bonanni B, et al. Selective oestrogen receptor modulators in prevention of breast cancer: an updated meta-analysis of individual participant data. Lancet. 2013;381(9880):1827–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Howell A, Astley S, Warwick J, et al. Prevention of breast cancer in the context of a national breast screening programme. J Intern Med. 2012;271(4):321–330. [DOI] [PubMed] [Google Scholar]
- 4. Pashayan N, Duffy SW, Chowdhury S, et al. Polygenic susceptibility to prostate and breast cancer: implications for personalised screening. Br J Cancer. 2011;104(10):1656–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Burton H, Chowdhury S, Dent T, et al. Public health implications from COGS and potential for risk stratification and screening. Nat Genet. 2013;45(4):349–351. [DOI] [PubMed] [Google Scholar]
- 6. Michailidou K, Hall P, Gonzalez-Neira A, et al. The Breast and Ovarian Cancer Susceptibility Collaboration. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet. 2013;45(4):353–361.361e1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kuchenbaecker KB, Ramus SJ, Tyrer J, et al. EMBRACE. Identification of six new susceptibility loci for invasive epithelial ovarian cancer. Nat Genet. 2015;47(2):164–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Michailidou K, Beesley J, Lindstrom S, et al. ; BOCS. Genome-wide association analysis of more than 120,000 individuals identifies 15 new susceptibility loci for breast cancer. Nat Genet. 2015;47(4):373–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Easton DF, Pooley KA, Dunning AM, et al. ; The SEARCH orators. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447(7148):1087–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mavaddat N, Michailidou K, Dennis J, et al. Polygenic risk scores for prediction of breast cancer and breast cancer subtypes. Am J Hum Genet. 2019;104(1):21–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mavaddat N, Pharoah PD, Michailidou K, et al. Prediction of breast cancer risk based on profiling with common genetic variants. J Natl Cancer Inst. 2015;107(5):djv036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kuchenbaecker KB, McGuffog L, Barrowdale D, et al. Evaluation of polygenic risk scores for breast and ovarian cancer risk prediction in BRCA1 and BRCA2 mutation carriers. J Natl Cancer Inst. 2017;109(7):999–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *13. Muranen TA, Greco D, Blomqvist C, et al. ; on behalf of the Breast Cancer Association Consortium. Genetic modifiers of CHEK21100delC-associated breast cancer risk. Genet Med. 2017;19(5):599–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lilyquist J, Ruddy KJ, Vachon CM, et al. Common genetic variation and breast cancer risk-past, present, and future. Cancer Epidemiol Biomarkers Prev. 2018;27(4):380–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *15. Schmidt MK, Hogervorst F, van Hien R, et al. Age- and tumor subtype-specific breast cancer risk estimates for CHEK21100delC carriers. J Clin Oncol. 2016;34(23):2750–2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *16. Weischer M, Nordestgaard BG, Pharoah P, et al. CHEK21100delC heterozygosity in women with breast cancer associated with early death, breast cancer-specific death, and increased risk of a second breast cancer. J Clin Oncol. 2012;30(35):4308–4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kast K, Rhiem K, Wappenschmidt B, et al. Prevalence of BRCA1/2 germline mutations in 21 401 families with breast and ovarian cancer. J Med Genet. 2016;53(7):465–471. [DOI] [PubMed] [Google Scholar]
- 18.ENIGMA. Evidence-Based Network for the Interpretation of Germline Mutant Alleles. https://enigmaconsortium.org/. Accessed December 2019.
- 19. Sakoda LC, Jorgenson E, Witte JS.. Turning of COGS moves forward findings for hormonally mediated cancers. Nat Genet. 2013;45(4):345–348. [DOI] [PubMed] [Google Scholar]
- 20. Amos CI, Dennis J, Wang Z, et al. The OncoArray Consortium: a network for understanding the genetic architecture of common cancers. Cancer Epidemiol Biomarkers Prev. 2017;26(1):126–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bahcall O. Shared susceptibility loci for breast, prostate and ovarian cancers. Nat Genet. 2019; doi: 10.1038/ngicogs.2. [Google Scholar]
- 22. Aulchenko YS, Ripke S, Isaacs A, et al. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23(10):1294–1296. [DOI] [PubMed] [Google Scholar]
- 23. Chang CC, Chow CC, Tellier LC, et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Antoniou AC, Goldgar DE, Andrieu N, et al. A weighted cohort approach for analysing factors modifying disease risks in carriers of high-risk susceptibility genes. Genet Epidemiol. 2005;29(1):1–11. [DOI] [PubMed] [Google Scholar]
- 25. Lee A, Mavaddat N, Wilcox AN, et al. BOADICEA: a comprehensive breast cancer risk prediction model incorporating genetic and nongenetic risk factors. Genet Med. 2019;21(8):1708–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang Z, Reinikainen J, Adeleke KA, et al. Time-varying covariates and coefficients in Cox regression models. Ann Transl Med. 2018;6(7):121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kleiblova P, Stolarova L, Krizova K, et al. Identification of deleterious germline CHEK2 mutations and their association with breast and ovarian cancer. Int J Cancer. 2019;145(7):1782–1797. [DOI] [PubMed] [Google Scholar]
- 28.National Collaborating Centre for Cancer. National Institute for Health and Clinical Excellence: Guidance. In: Familial Breast Cancer: Classification and Care of People at Risk of Familial Breast Cancer and Management of Breast Cancer and Related Risks in People with a Family History of Breast Cancer. Cardiff, UK: National Collaborating Centre for Cancer; 2013. [PubMed] [Google Scholar]
- 29.National Comprehensive Cancer Network. NCCN clinical practice guidelines in oncology: breast cancer screening and diagnosis, Version 1.2019, May 17. http://www.nccn.org/professionals/physician gls/breast-screening.pdf
- 30.NCCN Clinical Practice Guidelines in Oncology. Genetic/Familial High-Risk Assessment: Breast and Ovarian. Version 1-2018, 2017, October 3. nccn.org.
- 31. Robson ME, Reiner AS, Brooks JD, et al. Association of common genetic variants with contralateral breast cancer risk in the WECARE Study. J Natl Cancer Inst. 2017;109(10):djx051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mars N, Widen E, Kerminen S, et al. Polygenic risk, susceptibility genes, and breast cancer over the life course. medRxiv. 2020;11(1):6383. doi: 10.1101/2020.04.17.20069229:2020.04.17.20069229. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are shown in the main manuscript and the Supplementary Materials.