Abstract
Background
The HIV Prevention Trials Network (HPTN) 075 study evaluated the feasibility of enrolling and retaining men who have sex with men (MSM) and transgender women (TGW) from Kenya, Malawi, and South Africa. During the study follow-up, 21 participants acquired human immunodeficiency virus (HIV) (seroconverters). We analyzed HIV subtype diversity, drug resistance, transmission dynamics, and HIV superinfection data among MSM and TGW enrolled in HPTN 075.
Methods
HIV genotyping and drug resistance testing were performed for participants living with HIV who had viral loads >400 copies/mL at screening (prevalent cases, n = 124) and seroconverters (n = 21). HIV pol clusters were identified using Cluster Picker. Superinfection was assessed by a longitudinal analysis of env and pol sequences generated by next-generation sequencing.
Results
HIV genotyping was successful for 123/124 prevalent cases and all 21 seroconverters. The major HIV subtypes were A1 (Kenya) and C (Malawi and South Africa). Major drug resistance mutations were detected in samples from 21 (14.6%) of 144 participants; the most frequent mutations were K103N and M184V/I. Phylogenetic analyses identified 11 clusters (2–6 individuals). Clusters included seroconverters only (n = 1), prevalent cases and seroconverters (n = 4), and prevalent cases only (n = 6). Superinfections were identified in 1 prevalent case and 2 seroconverters. The annual incidence of superinfection was higher among seroconverters than among prevalent cases, and was higher than the rate of primary HIV infection in the cohort.
Conclusions
This report provides important insights into HIV genetic diversity, drug resistance, and superinfection among MSM and TGW in sub-Saharan Africa. These findings may help to inform future HIV prevention interventions in these high-risk groups.
Keywords: HIV drug resistance, phylogenetic analysis, men who have sex with men, transgender women, sub-Saharan Africa
Human immunodeficiency virus (HIV) drug resistance, transmission dynamics, subtype diversity, and superinfection data among men who have sex with men and transgender women in a clinical trial in sub-Saharan Africa may inform future HIV prevention interventions in these high-risk groups.
Sub-Saharan Africa has a high human immunodeficiency virus (HIV) burden, accounting for the majority of global HIV infections [1]. Men who have sex with men (MSM) are disproportionally affected by HIV, compared to the general African population. The median reported HIV prevalences among MSM are 19% in Western and Central Africa and 13% in Eastern and Southern Africa [1]. The HIV acquisition risk among African MSM in part reflects high rates of bisexual concurrency, with mature and widespread generalized epidemics [2]. Data on HIV among transgender women (TGW) in Africa and elsewhere are limited [3]. Violence, stigma, criminalization, and discrimination against transgender individuals and those in same-sex sexual relationships limit access to health services for HIV education, prevention, and treatment in many settings.
The prevalence of pretreatment resistance to antiretroviral (ARV) drugs in first-line treatment regimens exceeds 10% in some African countries [4]. The prevalence of HIV drug resistance is likely to be higher among treatment-experienced individuals, and to increase with broader use of ARV drugs for HIV prevention and treatment. Data on antiretroviral treatment (ART) and HIV drug resistance are limited among African MSM and TGW [5, 6]. These data are needed to guide interventions for reducing HIV incidence in these high-risk populations. Phylogenetic studies may also provide important information for understanding the HIV epidemic among African MSM and TGW. Those studies can identify transmission clusters and transmission chains, as well as the demographic, clinical, and behavioral characteristics associated with HIV transmission and acquisition. Previous studies in MSM have demonstrated associations of HIV transmission with recent infection [7], with discrete introductions of HIV into MSM populations [8, 9].
The HIV Prevention Trials Network (HPTN) 075 study evaluated the feasibility of enrolling and retaining MSM and TGW from sub-Saharan Africa for future HIV prevention studies. A high HIV prevalence (30.1%) was observed among those screened for the study [10]. Among those who were living with HIV at screening, 34.4% were on ART and 28.4% were virally suppressed [10]. During the study follow-up, 21 participants acquired HIV, corresponding to annual HIV incidences of 1.3% to 14.4% across the 4 study sites [10, 11]. In this study, we analyzed HIV strains from MSM and TGW screened for participation in HPTN 075. This provided data on HIV subtype diversity, drug resistance, transmission dynamics, and HIV superinfection in these high-risk groups.
METHODS
Study Cohort
HPTN 075 (NCT03201510) was conducted in Kisumu, Kenya; Blantyre, Malawi; Cape Town, South Africa; and Soweto, South Africa. The study enrolled participants living both with and without HIV who were 18–44 years old and reported that they were biologically male at birth. Those who reported that they were on ART or were in HIV care were not eligible for enrollment. Participants were followed quarterly for 12 months and were offered local standard-of-care clinical services. Participants living with HIV were referred for HIV care and treatment. Preexposure prophylaxis (PrEP) was available at study sites near the end of the study. Participants living without HIV were referred for PrEP, but none reported using PrEP during the study.
Human Immunodeficiency Virus Laboratory Testing
Individuals were tested for HIV infection at screening and enrollment; participants who were living without HIV at enrollment were tested for HIV infection at follow-up visits [5]. ARV drug testing had been performed previously for those who were living with HIV at screening [5]; testing was performed in this study for participants who acquired HIV infection during the study (seroconverters; at the first visit with documented HIV infection). HIV genotyping was performed using samples from individuals living with HIV at screening; samples from seroconverters at the first visit with documented HIV infection; and samples from follow-up visits for a subset of enrolled participants. Genotyping was performed using the ViroSeq HIV-1 Genotyping System v2.0 (Abbott Diagnostics, Des Plaines, IL) for samples with HIV viral loads ≥ 400 copies/mL. This system generates HIV pol sequences (1302 base pairs). HIV drug resistance was assessed using ViroSeq system software v3.0.
Human Immunodeficiency Virus Subtyping
HIV subtypes were identified using automated tools: REGA v3.0 [12], COntext-based Modeling for Expeditious Typing (COMET) [13], and the recombinant identification program (RIP) [14]. HIV subtypes were also identified by constructing phylogenetic trees that included subtype reference sequences from the Los Alamos National Laboratory (LANL) HIV Sequence Database [15]. HIV subtypes were assigned if the same result was obtained with at least 3 subtyping methods.
Clustering Analysis
Phylogenetic trees were constructed using HIV pol sequences from study samples and subtype and country-specific background sequences from the LANL HIV Sequence Database using the following search criteria: subtype, A1 or C; genomic region, 2252–3554 base pairs; and sampling country (Kenya, Malawi, and South Africa). For each study participant, 1 background sequence was included (selected from all available background sequences in the LANL database). Sequences with >5% nucleotide ambiguity were excluded from the analysis. The recombination detection program (RPD4) [16] was used to identify potential recombination breakpoints; sequences with evidence of recombination were excluded from analysis. Multiple pairwise sequence alignment was performed using multiple alignment using fast Fourier transform (MAFFT) v6.864 [17]. Phylogenetic trees were constructed using the randomized accelerated maximum-likelihood (RAxML) v8.2.10 approach, accessed through the Cyber Infrastructure for Phylogenetic Research (CIPRES) Science Gateway [18]. HIV pol clusters were identified using Cluster Picker [19], using a maximum genetic distance threshold of 4.5% and a bootstrap support value threshold of ≥90% [20]. Recent transmission clusters were identified using a maximum genetic distance of 1.5%. Phylogenetic trees were visualized using Interactive Tree of Life (iTOL) software v4 [21]. Genetic distances among pol sequences were calculated using the “ape” package [22] in R.
Statistical Analysis
A logistic regression analysis was used to identify associations between phylogenetic clustering with sociodemographic and clinical characteristics. Individual characteristics of participants were collected at the screening visit.
Analysis of Human Immunodeficiency Virus Superinfection
HIV superinfection was analyzed for a subset of the individuals included in the phylogenetic analysis. Next-generation sequencing (NGS) of HIV env and pol was performed using samples collected at the screening visit (for those who were living with HIV at enrollment); at the first visit where the participant had documented HIV infection (for seroconverters); and at the end-of-study visit (12 months, for all participants). NGS was performed using the MiSeq System (Illumina, Inc., San Diego, CA) with methods adapted from a previous study [23]. In brief, a 1-step reverse-transcription (RT)/polymerase chain reaction (PCR) was used to amplify a region of HIV gp41 (env; HXB2 coordinates: 7938–8256); if amplification failed, an alternate region was amplified (HXB2 coordinates: 7950–8299) with separate steps for RT and PCR. The same RT-PCR procedure was used to amplify a region of HIV reverse transcriptase (pol; HXB2 coordinates: 2696–3252). Sequences with ≥99% similarity were merged into a single consensus sequence.
Identification of Human Immunodeficiency Virus Superinfection
Phylogenetic trees were constructed using NGS env study sequences, HIV subtyping reference sequences, and background sequences for the corresponding genomic region. A unique set of background sequences was evaluated for each participant using Basic Local Alignment Search Tool (BLAST); for each study sequence, the 10 closest unique sequences were selected for analysis. Potential superinfection cases were identified if the phylogenetic tree included study sequences that were phylogenetically separated by reference or background branches from other study sequences. Distinct groups of study sequences representing <2% of the sequenced reads in a sample and supported by a bootstrap value <80% were not considered to be indicative of potential superinfection.
To confirm superinfection and determine the timing of superinfection, HIV genotyping and NGS were performed for samples collected between screening and the end of the study. Phylogenetic trees were constructed using env and pol NGS sequences for each participant; trees included study sequences from all visits. Superinfection was confirmed if the introduction of a new viral strain was observed in multiple samples, and if the maximum genetic distance between the original and new viral strains was great enough to rule out natural evolutionary drift (≥5% per year). The incidence of superinfection was calculated as cases per 100 person-years (py) of follow-up.
Nucleotide Sequence Accession Numbers
HIV pol consensus sequences generated in this study were submitted to GenBank (accession numbers: MG597249-MG597259 and MT185157-MT185334); env and pol NGS data are available on request.
Ethical Considerations
HPTN 075 was approved by Institutional Review Boards (IRBs) at each study site. Written informed consent for study participation was obtained at 3 of the 4 sites; oral consent was obtained at the fourth site, at the direction of the local IRB.
RESULTS
Human Immunodeficiency Virus Sequences Used for Analysis
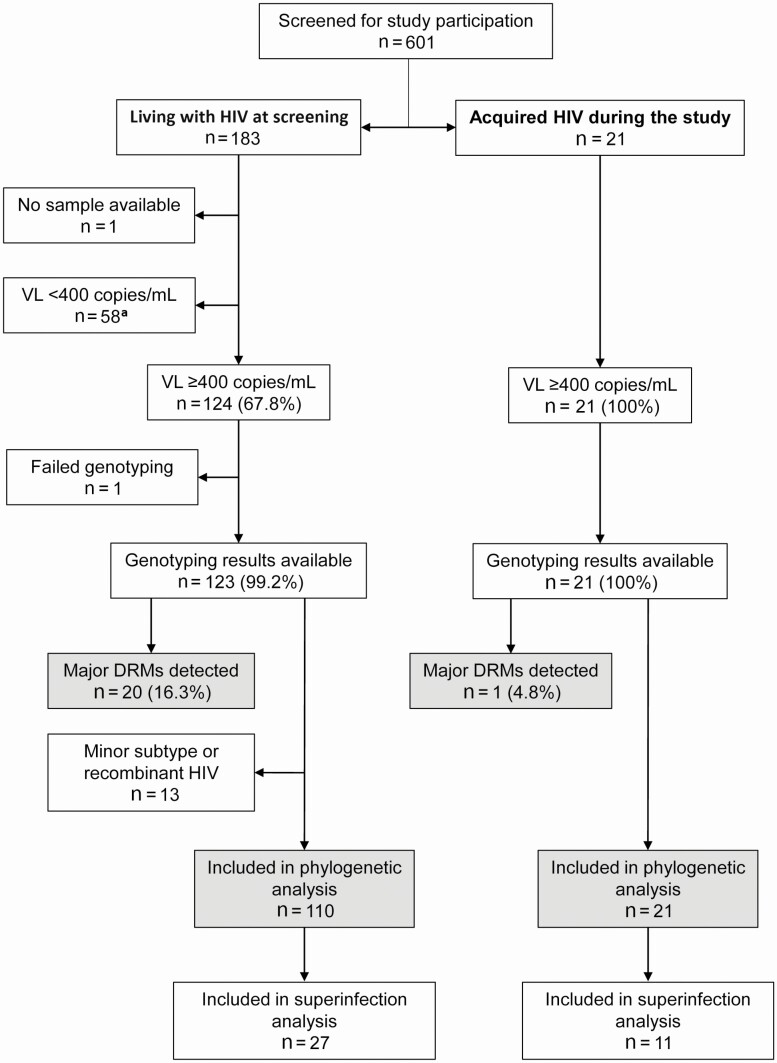
HIV genotyping results were obtained for 123 (99.2%) of 124 individuals who were living with HIV at screening and for all 21 seroconverters (Figure 1). The major HIV subtypes identified were A1 in Kenya (11/17, 64.7%); C in Malawi (17/19, 89.5%); and C in South Africa (103/108, 95.4%, Supplementary Table 1). We also identified 8 intersubtype recombinant sequences.
Figure 1.
Study cohort. The figure shows the number of individuals included in each step of the analysis. Shaded boxes show the number of participants who had major DRMs detected, and the number of participants who were included in the phylogenetic analysis. Data are shown separately for individuals who were living with HIV at the screening visit and those who acquired HIV during the study (seroconverters). The 144 participants included in the analysis of HIV drug resistance (those with genotyping results available) included 17 from Kenya; 19 from Malawi; 44 from Cape Town, South Africa; and 64 from Soweto, South Africa. The participant group included 41 aged 18–20, 55 aged 21–25, and 48 aged 26–44. Of the 144 participants, 48 (33.3%) identified as transgender women. aOf the individuals who were living with HIV at screening, 58 had viral loads <400 copies/mL; ARV drugs were detected in samples from 52 of those 58 individuals. Abbreviations: ARV, antiretroviral; DRM, drug-resistance mutation; HIV, human immunodeficiency virus; mL, milliliter; VL, viral load.
Human Immunodeficiency Virus Drug Resistance
Major HIV drug resistance mutations (DRMs) were detected in samples from 21 of the 144 persons with genotyping data (14.6%; 95% confidence interval [CI], 9.3–21.4%), including 1 seroconverter (Figure 1; Supplementary Table 2). DRMs detected conferred resistance to non-nucleoside reverse transcriptase inhibitors (NNRTIs; 19 cases) and nucleoside/nucleotide reverse transcriptase inhibitors (NRTIs; 8 cases); 6 of those cases had resistance to both ARV drug classes. Resistance to protease inhibitors was not detected. The most frequent NNRTI DRM detected was K103N (n = 14). The most frequent NRTI DRMs detected were M184V/I (n = 6) and K65R (n = 4). The seroconverter had a single NNRTI DRM (K103N).
Clustering Analysis
HIV pol sequences were obtained for 144 persons (Figure 1). We excluded 13 sequences from analysis: 8 intersubtype recombinant sequences and 5 sequences with subtypes that differed from the major subtype in the corresponding country (2 subtype C sequences from Kenya; 1 subtype D sequence from Malawi; 2 subtype B sequences from South Africa). The final data set included 131 sequences: 11 subtype A1 sequences from Kenya, 17 subtype C sequences from Malawi, and 103 subtype C sequences from South Africa. The analysis included country-specific background pol sequences (150 from Kenya; 264 from Malawi; and 710 from South Africa). The median pol gene diversity was higher among study sequences (6.3–7.0%) than among background sequences (4.1–4.6%; Supplementary Table 1).
A phylogenetic tree is shown for each country (Supplementary Figure 1). Most study sequences formed sporadic tree branches dispersed among the background sequences with no evidence of large clusters (>10 individuals). Overall, 11 clusters were detected that included study sequences (1 from Kenya, 1 from Malawi, and 9 from South Africa; Supplementary Table 3). In each cluster, sequences were from individuals enrolled at the same study site. These included 7 sequence pairs, 3 clusters of 3 sequences, and 1 cluster of 6 sequences. Of the 11 clusters, 6 included only individuals living with HIV at study screening, 1 cluster included seroconverters only, and 4 clusters included both individuals living with HIV at screening and seroconverters. Of the 39 sequences in clusters, 8 were from seroconverters. This included 1 cluster with 2 seroconverters and a nonseroconverter; 3 clusters with 1 seroconverter and 1 nonseroconverter; and 1 cluster of 3 seroconverters. Of the 3 seroconverters whose baseline sequences were clustered together, 2 were superinfected during the study follow-up.
Major DRMs were detected in only 1 cluster; in that case, K103N was detected in both individuals in a 2-sequence cluster. Of the 11 clusters, 6 met the more stringent criteria for recent transmission clusters (genetic distance ≤ 1.5%, Table 1). These included the cluster of 3 seroconverters from Cape Town, South Africa; 2 pairs from Cape Town, South Africa, and Kenya (both included a seroconverter and a participant living with HIV at screening); 1 cluster of 3 individuals from Cape Town, South Africa (all living with HIV at screening); 1 cluster of 3 individuals from Malawi (all living with HIV at screening); and 1 pair from Soweto, South Africa (both living with HIV at screening; Table 1). Clustering was not statistically associated with any sociodemographic or clinical characteristic evaluated, except for self-report of a positive result for the participant’s last HIV test (Supplementary Table 4); as compared to participants who reported a positive test, those who reported that their last HIV test was negative were less likely to be in a phylogenetic cluster (68.8% vs. 31.3%, respectively; P = .009). An HIV clustering analysis was also performed for the 13 cases with minor subtypes and recombinant HIV. None of those sequences grouped with any other study or background sequence. Individual characteristics of participants in pol sequence clusters are shown in Supplementary Table 3.
Table 1.
Human immunodeficiency virus pol sequence clusters identified in study samples
| Cluster Type | Cluster Size | Study Site | Genetic Distance, % |
|---|---|---|---|
| Seroconverters only | 3a | Cape Town, South Africa | 1.0 |
| Mixed | 3a,b,c | Kenya | 2.9 |
| 2a | Cape Town, South Africa | .6 | |
| 2a | Soweto, South Africa | 1.1 | |
| 2 | Soweto, South Africa | 3.7 | |
| All living with HIV at screening | 3a | Malawi | 1.2 |
| 2 | Cape Town, South Africa | 3.3 | |
| 6a,d | Cape Town, South Africa | 3.8 | |
| 2 | Soweto, South Africa | 2.7 | |
| 2 | Soweto, South Africa | 2.7 | |
| 2e | Soweto, South Africa | 1.6 |
The table shows the cluster types, cluster sizes, and study sites for 11 pol clusters identified in phylogenetic trees, using a maximum genetic distance threshold of 4.5% and a branch support threshold of ≥90% (see Figure 2).
Abbreviation: HIV, human immunodeficiency virus.
aRecent transmission clusters (identified using a maximum genetic distance of 1.5%).
bOf the 3 participants in this cluster, 2 seroconverted during the study.
cOf the 3 participants in this cluster, 2 met criteria for a recent transmission cluster (1 living with HIV at screening and 1 who seroconverted); a maximum genetic distance between those participants was 1.2%.
dOf the 6 participants in this cluster, 3 met criteria for a recent transmission cluster (all living with HIV at screening); a maximum genetic distance between those participants was 0.8%.
eThe drug resistance mutation, K103N, was identified in both participants in this cluster.
Human Immunodeficiency Virus Superinfection Analysis
HIV superinfection was analyzed for 38 (29%) of the 131 participants included in the phylogenetic analysis (Figure 1; Supplementary Figure 2). This included 27 (38.0%) of 72 participants who were living with HIV at screening and were enrolled in the study (the remaining 44 participants included 7 with no 12-month sample, 37 with viral loads <400 copies/mL at 12 months, and 1 with a 12-month sample that failed analysis). HIV superinfection was also analyzed for 11 (52.4%) of the 21 seroconverters (the remaining 10 seroconverters included 3 who seroconverted at 12 months, 6 with viral loads <400 copies/mL at 12 months, and 1 with no 12-month sample).
Potential superinfection was identified in 1 (3.7%) of the 27 participants who were living with HIV at screening and in 2 (18.2%) of the 11 seroconverters. In all 3 cases, superinfection was confirmed with additional testing (env and pol NGS analysis of longitudinal samples). The incidence of superinfection among seroconverters (30.3/100 py) was higher than that among participants who were living with HIV at enrollment (3.6/100 py; P = .08) and was significantly higher than the rate of primary HIV infection in the HPTN 075 cohort (6.96/100 py; P = .046).
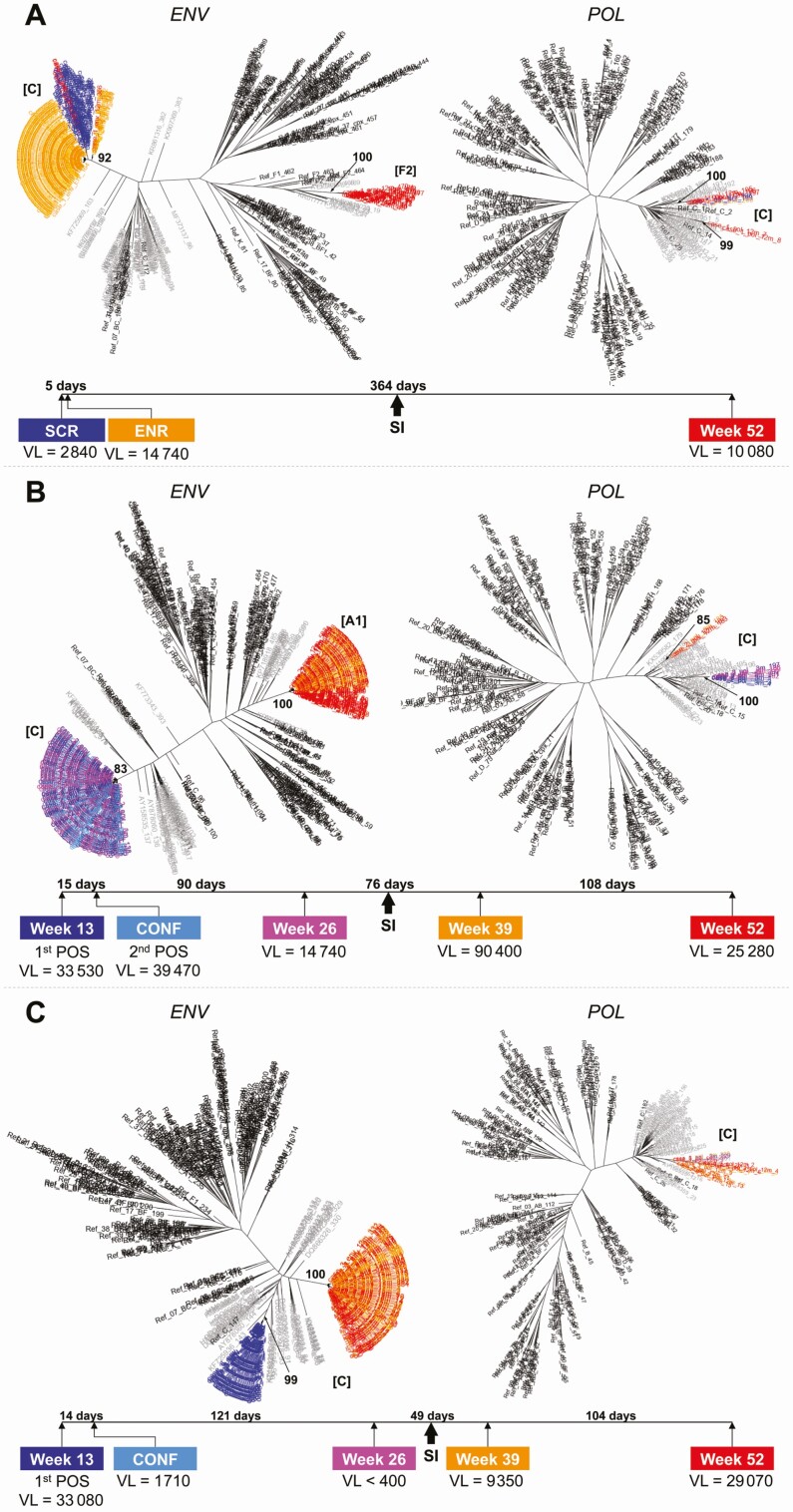
In the superinfection case where the participant was living with HIV at enrollment, subtype C was present at enrollment, while an intersubtype recombinant strain (pol subtype C, env subtype F2) was detected 364 days later; both strains were present at the follow-up visit (Figure 2A). The genetic distance between the original strain and the new strain was 17.8% in env and 8.2% in pol. In the 2 cases of superinfection involving seroconverters, the viral strain present at seroconversion shifted entirely to a new strain (Figure 2B and C). In 1 case, the original subtype C strain was replaced with an intersubtype recombinant strain (pol subtype C, env subtype A1) after 76 days (Figure 2B); the genetic distance between the original strain and the new strain was 19.2% in env and 6.9% in pol. In the final case, the original subtype C strain was replaced with a different subtype C strain after 184 days (Figure 2C); the genetic distance between the original strain and the new strain was 13.4% in env, and the phylogenetic tree generated using pol sequences did not show evidence of superinfection.
Figure 2.
Phylogenetic trees of HIV env and pol sequences from participants living with HIV superinfection. A–C, The figure shows maximum-likelihood trees of consensus HIV sequences from a longitudinal next-generation sequencing analysis of HIV env and pol for 3 superinfection cases. Bootstrap values are indicated for each cluster (1000 replicates). VL values are expressed as copies/mL. HIV subtypes are shown in brackets for each cluster. Abbreviations: CONF, confirmatory visit; ENR, enrollment sample; ENV, envelope gene; HIV, human immunodeficiency virus; POL, polymerase gene; POS, seropositive sample for HIV; SCR, screening sample; SI, HIV superinfection; VL, viral load.
The superinfecting strains in the 3 cases did not cluster with each other or with any other sequence from the study cohort. In 2 superinfection cases, the participants were viremic at all study visits; in the third case, the participant was virally suppressed prior to superinfection (Figure 2). This participant may have started ART after study enrollment. This participant was viremic at both visits after the superinfection event. HIV drug resistance testing was performed for all postsuperinfection samples in all 3 cases; none of the superinfecting strains had HIV drug resistance.
DISCUSSION
We evaluated HIV subtype, drug resistance, phylogenetics, and superinfection data in MSM and TGW in sub-Saharan Africa. Major DRMs were detected in 16.3% of those who were living with HIV at screening (95% CI, 10.2–24.0%) and in 4.8% of the seroconverters (95% CI, .1–23.8%). Approximately 1 in 5 study sequences (20.4%) clustered with 1–5 other sequences from the study cohort. Phylogenetic analyses showed that HIV sequences from MSM and TGW in HPTN 075 were interspersed with sequences from a public database, suggesting that HIV strains circulated between the general population and these high-risk groups.
The highest subtype diversity was observed in Kenya and Soweto, South Africa; B and C subtypes and recombinant strains were detected at both sites. Intersubtype recombinant strains were also detected in Malawi. The subtypes and strains identified in this report were similar to those reported previously from these regions [24–26]. HIV genetic diversity was similar across the 3 countries, with the highest diversity in South Africa and the lowest diversity in Malawi. Overall, the level of genetic diversity in the study population was similar to those levels observed for background sequences from each of the 3 countries.
ART scale-up in Africa is likely to increase the prevalences of transmitted and acquired drug resistance. Previous reports found pretreatment prevalences of HIV drug resistance in sub-Saharan Africa at rates ranging from 2.9–19.4%, mostly reflecting NNRTI resistance [4]. The frequency of HIV drug resistance among MSM and TGW in HPTN 075 was in this range; these individuals reported that they were not on ART or in care, but may have had prior exposure to ARV drugs. Drug resistance was assessed in 21 seroconverters; 1 had drug resistance (NNRTI, K103N) with no ARV drugs detected. This may represent a case of transmitted drug resistance. The detection of NNRTI resistance in this cohort suggests that ART regimens with alternate backbones may be preferable for ART in this population. The presence of circulating HIV strains with the K65R and M184V mutations could compromise the use of current PrEP regimens in this population.
Phylogenetic analyses revealed clustering of sequences from approximately 1 in 5 participants (29/129, 20.4%). The clusters included individuals with prevalent and incident infections. The relatively low frequency of clustering observed may reflect the low sampling density in the study. HIV clustering results from studies with sampling densities <10% are not considered adequate for assessing HIV transmission dynamics [27]. The presence of clustered infections highlights the need for frequent HIV testing and for implementation of prevention interventions in individuals living both with and without HIV in this population.
The incidence of superinfection has been reported to be similar to those of primary HIV infection in some cohorts [28–31], including a study among MSM [32]; other studies have reported incidences of superinfection to be lower than those of primary infection [29, 33–36]. In this cohort of African MSM and TGW, the incidence of superinfection was higher than the incidence of primary infection; the incidence of superinfection among seroconverters in this study (18.2%) was also higher than rates reported previously among seroconverters (eg, 2.6% [33], 4.7% [28]). The overall rate of superinfection in the HPTN 075 cohort (7.9%) was similar to a relatively high rate reported among MSM in the United States (8.5%) [37] and was lower than the rate of 15.6% reported among MSM in China [38]. In 2 of the 3 superinfection cases in this report, the superinfection strain was an intersubtype recombinant. Subtype C/A1 recombinants were reported previously in Malawi [39] and Tanzania [40, 41]. Subtype C/F1 recombinants were previously reported among MSM in South Africa [42]. Further studies are needed to explore whether intersubtype recombinant strains have a selective advantage for superinfection (eg, by escaping the antibodies produced in response to a prior infection with a single subtype strain). While we did not observe superinfection with drug-resistant strains, HIV superinfection with resistant strains can lead to ART failure [43]. Superinfection can also impact disease progression, if more pathogenic strains are transmitted [44]. This highlights the importance of limiting ongoing HIV exposure in persons living with HIV.
The relatively small number of MSM and TGW who were available for analysis in this study may have limited our ability to identify transmitted drug resistance and clusters; the stringent criteria used for cluster identification may also have limited the number of clusters identified. Other limitations in the superinfection analysis include potential biases introduced during NGS amplification and the relatively small amplicon size. Some cases of superinfection may also not have been identified because of the relatively short follow-up period (52 weeks for prevalent cases, and shorter periods for seroconverters).
This report provides new insights into HIV genetic diversity, drug resistance, and superinfection among MSM and TGW in sub-Saharan Africa. These findings may help inform future studies evaluating HIV prevention interventions in these high-risk populations.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Author contributions. M. V. S., P. J. P., and Y. Z. performed human immunodeficiency virus (HIV) genotyping. M. V. S. performed the phylogenetic analysis and HIV subtyping. P. J. P. performed the next-generation sequencing and the superinfection analysis. Y. Z. processed the next-generation sequencing data. V. C. was the HIV Prevention Trials Network (HPTN) Laboratory Center QAQC Representative for HPTN 075. X. G. was a data analyst for HPTN 075. E. L. H. was a HPTN Leadership and Operations Center Clinical Research Manager for HPTN 075. L. M. was a data manager for HPTN 075. A. O., N. K., R. P., and K. D. were Site Principal Investigators for HPTN 075. Y. Q. C. was a Statistician for HPTN 075. T. G. M. S. was the Protocol Chair for HPTN 075. S. H. E. was the HPTN 075 Virologist and designed the study. M. V. S., P. J. P., and S. H. E. analyzed the data. M. V. S., P. J. P., Y. Z., and S. H. E. prepared the manuscript. All of the authors contributed to manuscript preparation and reviewed the manuscript before publication.
Acknowledgments. The authors thank the HIV Prevention Trials Network (HPTN) 075 study team and participants for providing the samples and data used in this study. They thank the laboratory staff at study sites and at the HPTN Laboratory Center for their assistance with sample management, processing, and testing.
Financial support. This work was supported by the Division of AIDS of the US National Institute of Allergy and Infectious Diseases; and by the Office of AIDS Research of the US National Institutes of Health (grant numbers UM1-AI068613 to S. H. E.; UM1-AI068617 to Donnell; and UM1-AI068619 to Cohen/El-Sadr). Additional support was provided by the Human Immunodeficiency Virus Center for Clinical and Behavioral Research (grant number P30-MH43520 to Remien).
Potential conflicts of interest. S. H. E. has collaborated on research studies with investigators from Abbott Diagnostics, which has provided reagents for collaborative research studies. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Presented in part: Conference on Retroviruses and Opportunistic Infections, Boston, Massachusetts, 8-11 March 2020. Abstract #0194.
References
- 1. Joint United Nations Programme on Human Immunodeficiency Virus/AIDS (UNAIDS). The gap report. 2014. Available at: https://www.unaids.org/en/resources/documents/2014/20140716_UNAIDS_gap_report. Accessed 5 March 2020.
- 2. Beyrer C, Baral SD, Walker D, Wirtz AL, Johns B, Sifakis F. The expanding epidemics of HIV type 1 among men who have sex with men in low- and middle-income countries: diversity and consistency. Epidemiol Rev 2010; 32:137–51. [DOI] [PubMed] [Google Scholar]
- 3. Poteat T, Scheim A, Xavier J, Reisner S, Baral S. Global epidemiology of HIV infection and related syndemics affecting transgender people. J Acquir Immune Defic Syndr 2016; 72(Suppl 3):S210–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gupta RK, Gregson J, Parkin N, et al. HIV-1 drug resistance before initiation or re-initiation of first-line antiretroviral therapy in low-income and middle-income countries: a systematic review and meta-regression analysis. Lancet Infect Dis 2018; 18:346–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang Y, Fogel JM, Guo X, et al. Antiretroviral drug use and HIV drug resistance among MSM and transgender women in sub-Saharan Africa. AIDS 2018; 32:1301–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stannah J, Dale E, Elmes J, Staunton R, Beyrer C, Mitchell KM, Boily M-C. HIV testing and engagement with the HIV treatment cascade among men who have sex with men in Africa: a systematic review and meta-analysis. Lancet HIV 2019; 6:e769–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fisher M, Pao D, Brown AE, et al. Determinants of HIV-1 transmission in men who have sex with men: a combined clinical, epidemiological and phylogenetic approach. AIDS 2010; 24:1739–47. [DOI] [PubMed] [Google Scholar]
- 8. Hué S, Pillay D, Clewley JP, Pybus OG. Genetic analysis reveals the complex structure of HIV-1 transmission within defined risk groups. Proc Natl Acad Sci USA 2005; 102:4425–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen JH, Wong KH, Chan KC, To SW, Chen Z, Yam WC. Phylodynamics of HIV-1 subtype B among the men-having-sex-with-men (MSM) population in Hong Kong. PLoS One 2011; 6:e25286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sandfort TGM, Dominguez K, Kayange N, et al. HIV testing and the HIV care continuum among sub-Saharan African men who have sex with men and transgender women screened for participation in HPTN 075. PLoS One 2019; 14:e0217501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sandfort TGM, Guo X, Hamilton EL, et al. HIV incidence among men who have sex with men and transgender women in sub-Saharan Africa: findings from the multi-country HPTN 075 cohort study. Madrid, Spain: Human Immunodeficiency Virus Research for Prevention Meeting (HIVR4P), 2018. [Google Scholar]
- 12. Pineda-Peña AC, Faria NR, Imbrechts S, et al. Automated subtyping of HIV-1 genetic sequences for clinical and surveillance purposes: performance evaluation of the new REGA version 3 and seven other tools. Infect Genet Evol 2013; 19:337–48. [DOI] [PubMed] [Google Scholar]
- 13. Struck D, Lawyer G, Ternes AM, Schmit JC, Bercoff DP. COMET: adaptive context-based modeling for ultrafast HIV-1 subtype identification. Nucleic Acids Res 2014; 42:e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Siepel AC, Halpern AL, Macken C, Korber BT. A computer program designed to screen rapidly for HIV type 1 intersubtype recombinant sequences. AIDS Res Hum Retroviruses 1995; 11:1413–6. [DOI] [PubMed] [Google Scholar]
- 15. Los Alamos National Laboratory. HIV sequence database.2018. Available at: https://www.hiv.lanl.gov/content/index. Accessed 5 March 2020.
- 16. Martin DP, Murrell B, Golden M, Khoosal A, Muhire B. RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol 2015; 1:vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform 2008; 9:286–98. [DOI] [PubMed] [Google Scholar]
- 18. Miller MA, Schwartz T, Pickett BE, et al. A RESTful API for access to phylogenetic tools via the CIPRES science gateway. Evol Bioinform Online 2015; 11:43–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rose R, Lamers SL, Dollar JJ, et al. Identifying transmission clusters with cluster picker and HIV-TRACE. AIDS Res Hum Retroviruses 2017; 33:211–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. de Oliveira T, Kharsany AB, Gräf T, et al. Transmission networks and risk of HIV infection in KwaZulu-Natal, South Africa: a community-wide phylogenetic study. Lancet HIV 2017; 4:e41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 2016; 44:W242–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Paradis E, Claude J, Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 2004; 20:289–90. [DOI] [PubMed] [Google Scholar]
- 23. Courtney CR, Mayr L, Nanfack AJ, et al. Contrasting antibody responses to intrasubtype superinfection with CRF02_AG. PLoS One 2017; 12:e0173705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hassan AS, Esbjörnsson J, Wahome E, et al. HIV-1 subtype diversity, transmission networks and transmitted drug resistance amongst acute and early infected MSM populations from Coastal Kenya. PLoS One 2018; 13:e0206177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wilkinson E, Rasmussen D, Ratmann O, Stadler T, Engelbrecht S, de Oliveira T. Origin, imports and exports of HIV-1 subtype C in South Africa: a historical perspective. Infect Genet Evol 2016; 46:200–8. [DOI] [PubMed] [Google Scholar]
- 26. Bbosa N, Kaleebu P, Ssemwanga D. HIV subtype diversity worldwide. Curr Opin HIV AIDS 2019; 14:153–60. [DOI] [PubMed] [Google Scholar]
- 27. Novitsky V, Moyo S, Lei Q, DeGruttola V, Essex M. Impact of sampling density on the extent of HIV clustering. AIDS Res Hum Retroviruses 2014; 30:1226–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Redd AD, Mullis CE, Serwadda D, et al. The rates of HIV superinfection and primary HIV incidence in a general population in Rakai, Uganda. J Infect Dis 2012; 206:267–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Redd AD, Quinn TC, Tobian AA. Frequency and implications of HIV superinfection. Lancet Infect Dis 2013; 13:622–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Piantadosi A, Chohan B, Chohan V, McClelland RS, Overbaugh J. Chronic HIV-1 infection frequently fails to protect against superinfection. PLoS Pathog 2007; 3:e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Piantadosi A, Ngayo MO, Chohan B, Overbaugh J. Examination of a second region of the HIV type 1 genome reveals additional cases of superinfection. AIDS Res Hum Retroviruses 2008; 24:1221–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Smith DM, Wong JK, Hightower GK, et al. Incidence of HIV superinfection following primary infection. JAMA 2004; 292:1177–8. [DOI] [PubMed] [Google Scholar]
- 33. Redd AD, Mullis CE, Wendel SK, et al. Limited HIV-1 superinfection in seroconverters from the CAPRISA 004 Microbicide Trial. J Clin Microbiol 2014; 52:844–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gonzales MJ, Delwart E, Rhee SY, Tsui R, Zolopa AR, Taylor J, Shafer RW. Lack of detectable human immunodeficiency virus type 1 superinfection during 1072 person-years of observation. J Infect Dis 2003; 188:397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rachinger A, Manyenga P, Burger JA, et al. Low incidence of HIV-1 superinfection even after episodes of unsafe sexual behavior of homosexual men in the Amsterdam Cohort Studies on HIV Infection and AIDS. J Infect Dis 2011; 203:1621–8. [DOI] [PubMed] [Google Scholar]
- 36. Ronen K, McCoy CO, Matsen FA, et al. HIV-1 superinfection occurs less frequently than initial infection in a cohort of high-risk Kenyan women. PLoS Pathog 2013; 9:e1003593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wagner GA, Pacold ME, Kosakovsky Pond SL, et al. Incidence and prevalence of intrasubtype HIV-1 dual infection in at-risk men in the United States. J Infect Dis 2014; 209:1032–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Luan H, Han X, Yu X, et al. Dual infection contributes to rapid disease progression in men who have sex with men in China. J Acquir Immune Defic Syndr 2017; 75:480–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sturdevant CB, Dow A, Jabara CB, et al. Central nervous system compartmentalization of HIV-1 subtype C variants early and late in infection in young children. PLoS Pathog 2012; 8:e1003094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nofemela A, Bandawe G, Thebus R, et al. Defining the human immunodeficiency virus type 1 transmission genetic bottleneck in a region with multiple circulating subtypes and recombinant forms. Virology 2011; 415:107–13. [DOI] [PubMed] [Google Scholar]
- 41. Billings E, Sanders-Buell E, Bose M, et al. HIV-1 genetic diversity among incident infections in Mbeya, Tanzania. AIDS Res Hum Retroviruses 2017; 33:373–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Middelkoop K, Rademeyer C, Brown BB, et al. Epidemiology of HIV-1 subtypes among men who have sex with men in Cape Town, South Africa. J Acquir Immune Defic Syndr 2014; 65:473–80. [DOI] [PubMed] [Google Scholar]
- 43. Pingen M, Nouwen JL, Dinant S, et al. Therapy failure resulting from superinfection by a drug-resistant HIV variant. Antivir Ther 2012; 17:1621–5. [DOI] [PubMed] [Google Scholar]
- 44. Gottlieb GS, Nickle DC, Jensen MA, et al. HIV type 1 superinfection with a dual-tropic virus and rapid progression to AIDS: a case report. Clin Infect Dis 2007; 45:501–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.