Abstract
We describe a proteomics analysis to determine the molecular differences between normothermically perfused (normothermic machine perfusion, NMP) human kidneys with urine recirculation (URC) and urine replacement (UR). Proteins were extracted from 16 kidney biopsies with URC (n = 8 donors after brain death [DBD], n = 8 donors after circulatory death [DCD]) and three with UR (n = 2 DBD, n = 1 DCD), followed by quantitative analysis by mass spectrometry. Damage‐associated molecular patterns (DAMPs) were decreased in kidney tissue after 6 hours NMP with URC, suggesting reduced inflammation. Vasoconstriction was also attenuated in kidneys with URC as angiotensinogen levels were reduced. Strikingly, kidneys became metabolically active during NMP, which could be enhanced and prolonged by URC. For instance, mitochondrial succinate dehydrogenase enzyme levels as well as carbonic anhydrase were enhanced with URC, contributing to pH stabilization. Levels of cytosolic and the mitochondrial phosphoenolpyruvate carboxykinase were elevated after 24 hours of NMP, more prevalent in DCD than DBD tissue. Key enzymes involved in glucose metabolism were also increased after 12 and 24 hours of NMP with URC, including mitochondrial malate dehydrogenase and glutamic‐oxaloacetic transaminase, predominantly in DCD tissue. We conclude that NMP with URC permits prolonged preservation and revitalizes metabolism to possibly better cope with ischemia reperfusion injury in discarded kidneys.
Keywords: ischemia reperfusion injury (IRI), kidney (allograft) function / dysfunction, kidney biology, kidney transplantation / nephrology, organ perfusion and preservation, translational research / science
Short abstract
The authors perform a proteomics analysis to identify molecular differences between normothermically machine perfused and discarded human kidneys with urine recirculation and urine replacement using lactated Ringers.
Abbreviations
- CIT
cold ischemia time
- DBD
donation after brain death
- DCD
donation after circulatory death
- HMP
hypothermic machine perfusion
- IRR
intrarenal resistance
- NMP
normothermic machine perfusion
- SCS
static cold storage
- TPN
total parenteral nutrition
- URC
urine recirculation
- UR
urine replacement
- WIT
warm ischemia time
1. INTRODUCTION
Proteomic profiling by mass spectrometry (MS) can rapidly identify global protein changes in cells and tissues, providing insights into pathways and interaction networks. 1 , 2 , 3 MS has been widely used in medical research and in the field of transplantation, contributing to the knowledge of organ function and failure at a molecular level. 4 , 5 , 6 However, none of the numerous protein biomarkers, which have been described in the recent literature, has been implemented in the clinical routine yet to provide specific diagnosis for a defined phenotype of kidney allograft disease. 6 Most of the available data on kidney proteomics are on the detection of acute kidney allograft rejection markers earlier than biopsy could prove it. For instance, interferon‐gamma induced chemokines CXCL9 and CXCL10, recruiting T cells to inflammatory sites, are clinically the most promising urine protein biomarkers at present. 7 , 8 Kaisar et al described the ability of proteomic analysis on biopsies to differentiate between outcomes after deceased donor kidney transplantation. Their results suggested that the outcome‐related protein abundance was strongly linked with profibrotic and antioxidative proteins as well as proteins involved in apoptosis, 9 also observed in transcriptomics studies. 7 , 8 Besides the desire to improve prediction of outcome and rejection after kidney transplantation using ‐omics, there is little available on the topic of ex vivo normothermic perfusion (NMP). The purpose of applying proteomics to NMP tissue samples was to determine the molecular effects of normothermically perfused discarded human kidneys.
In light of all the relevant circumstances necessary for developing a transportable normothermic kidney perfusion device, we analyzed and published URC in discarded human kidneys and in experimental porcine kidney work previously. 9 , 10 , 11 Urine recirculation (URC) should help to facilitate an automated stand‐alone kidney NMP system which could be clinically applicable without the requirement to replace the excreted urine volume actively. Furthermore, URC enables stable hemodynamics, perfusate volume, and homeostasis. 9 , 10 Compared to urine replenishment with Ringer's lactate, it is possible to perfuse discarded human kidneys for up to 24 hours normothermically when the urine is recirculated. 9 , 10 , 11
We aimed to profile kidney proteomes, which are biologically distinct from others, and explore the impact of URC to control perfusion homeostasis and perfusate volume. This was compared to biopsies taken from kidneys perfused with urine replacement (UR), using Ringer's lactate, as described by other research groups before. 12 , 13 , 14 , 15
2. METHODS
Human kidneys were recovered for the purpose of transplantation but deemed untransplantable during post‐recovery assessment at the donor hospital or at the transplant center. After transport to Oxford, normothermic perfusions were performed at the Institute of Biomedical Engineering (IBME), University of Oxford. 16 Donor characteristics, warm (WIT) and cold ischemia times (CIT) and perfusion parameters were collated and analyzed. The study was evaluated and approved by the national ethics review committee of the United Kingdom (REC reference 12/EE/0273 IRAS project ID 106793).
2.1. Kidney biopsy preparation, protein extraction and digestion
Biopsies were taken at six different time points; zero biopsy at start of back table preparation, 1, 6, 12, 18, and 24 hours after perfusion start and at the end of perfusion, respectively. Core needle biopsy specimens, 18 gauge, were taken and stored at −80°C until use. The kidney biopsy retrieval protocol and –omics workflow are illustrated in Figure 1. Detailed description in Data S1.
Figure 1.
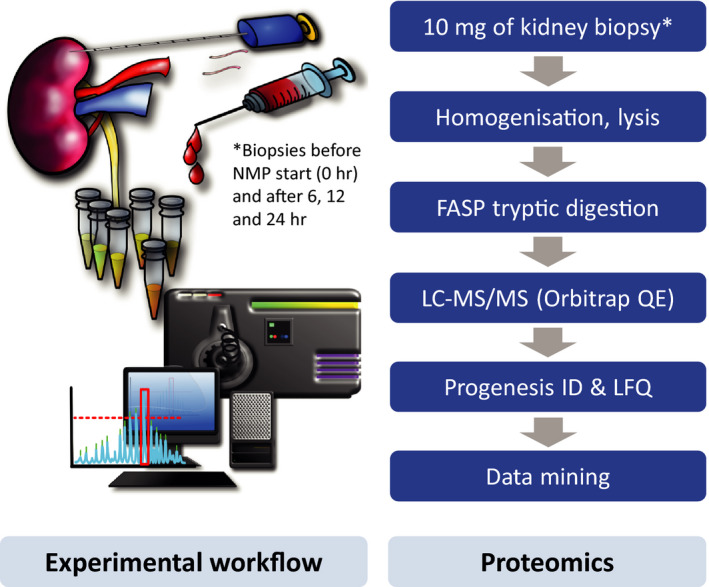
Kidney biopsy retrieval and proteomics workflows used in this study. Discarded kidneys were subjected to normothermic perfusion (NMP) either with urine recirculation (URC) or urine replacement (UR), and kidney cortex tissue biopsies collected at time points between 0 and 24 hours. Tissue material was homogenized, lysed, digested with trypsin using a filter‐assisted sample preparation (FASP) protocol followed by liquid chromatography tandem mass spectrometry (LC‐MS/MS) label‐free quantitative analysis (LFQ) and data processing using MaxQuant and Perseus software
2.2. Protein identification and quantitation by mass spectrometry
Samples were analyzed using liquid chromatography tandem mass spectrometry (LC‐MS/MS) as described previously. 17 In brief, a Dionex Ultimate 3000 nano‐ultra performance liquid chromatography system coupled to an Orbitrap Fusion Lumos Tribrid Mass Spectrometer (Thermo Scientific) acquiring data in electron spray ionization (ESI) positive mode was used with a 75‐µm‐inner diameter x 50 cm C18 EASY‐Spray column (2 µm particle size, Thermofisher Scientific, United Kingdom). Peptides were separated with a 60 minutes gradient of 0% solvent A to 40% solvent B (solvent A: 99.9% H2O, 0.1% formic acid, 5% DMSO; solvent B: 94.9% ACN, 0.1% formic acid, 5% DMSO) at 250 nl/min. MS survey acquisition was set with a resolution of 120 000 FWHM with a recording window between 400 and 1500 m/z. A maximum of 20 MS/MS scans were triggered in data‐dependent acquisition (DDA) mode.
2.3. Proteomics data analysis
Protein identification. Raw LC‐MS/MS data were uploaded to Andromeda/Maxquant version 1.5.2.8 for MS/MS spectra matching against complete human data set (UPR_HumanSapiens, retrieved at 15/02/2017, 92 954 entries) with trypsin specificity. 18 Two missed cleavages, NQ deamination and methionine oxidation were selected as variable modification, carbamidomethylation of cysteine was selected as fixed modification. For identification, the FDR (false discovery rate) at the peptide spectrum matches (PSM) and protein level were set to 0.01. Unmodified unique+razor peptides were chosen for label‐free quantification (LFQ) with minute ratio count set as 2. Match between run was selected to increased number of proteins with quantitative value. PANTHER classification system was used to perform gene ontology enrichment analysis (over represented or under represented). KEGG (Kyoto Encyclopaedia of Genes and Genomes) was used for canonical pathway assignment and STRING (v 11.0) for visualization.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD021507.
Quantitative analysis. Quantitative MS data were uploaded to the Perseus software package (v1.5.5.3) 19 for data visualization and statistical analysis. Label‐free quantitation (LFQ) values were first log2 transformed, then plotted in histograms to check normality of the data. Scatter plot display was used to display data in Figure 3. Hierarchical clustering analysis and heat maps were used to group samples based on the abundance of identified proteins after Z‐score normalization and subtraction of mean values across columns (Figure S2). Two‐sample t tests combined with Permutation FDR to correct for multiple testing were applied to detected differentially expressed proteins between comparisons. Statistical FDR set as 0.05, s0 as 0.1 in the volcano plot used to visualize differentially expressed proteins in Figure 2.
Figure 3.
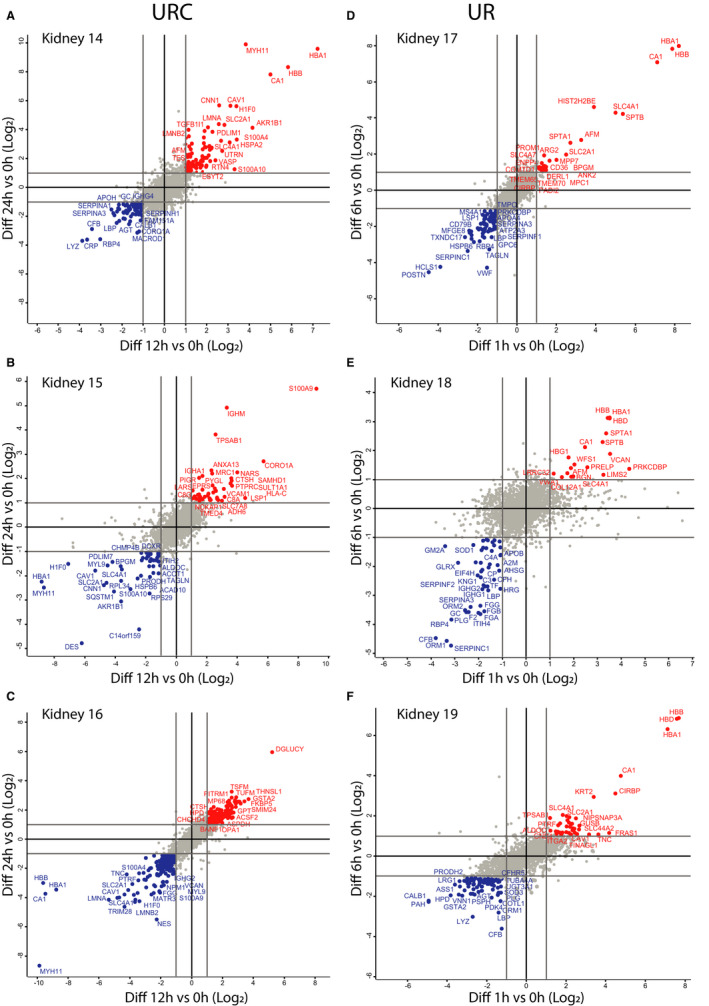
NMP with URC or UR affects kidney proteome dynamics over time. Kidneys were subjected to NMP either with URC for 12 and 24 hours (A‐C) or with UR for 1 and 6 hours (D‐F), followed by quantitative proteomics analysis. Scatter plots show 24 hours/0 hours (y‐axis) versus 12 hours/0 hours (X‐axis) abundant ratios (A‐C, for URC kidneys) or 6 hours/0 hours (y‐axis) versus 1 hours/0 hours (X‐axis) abundant ratios (D‐F, for UR kidneys). Proteins elevated over NMP time are labeled in red, whereas proteins with reduced levels are highlighted in blue
Figure 2.
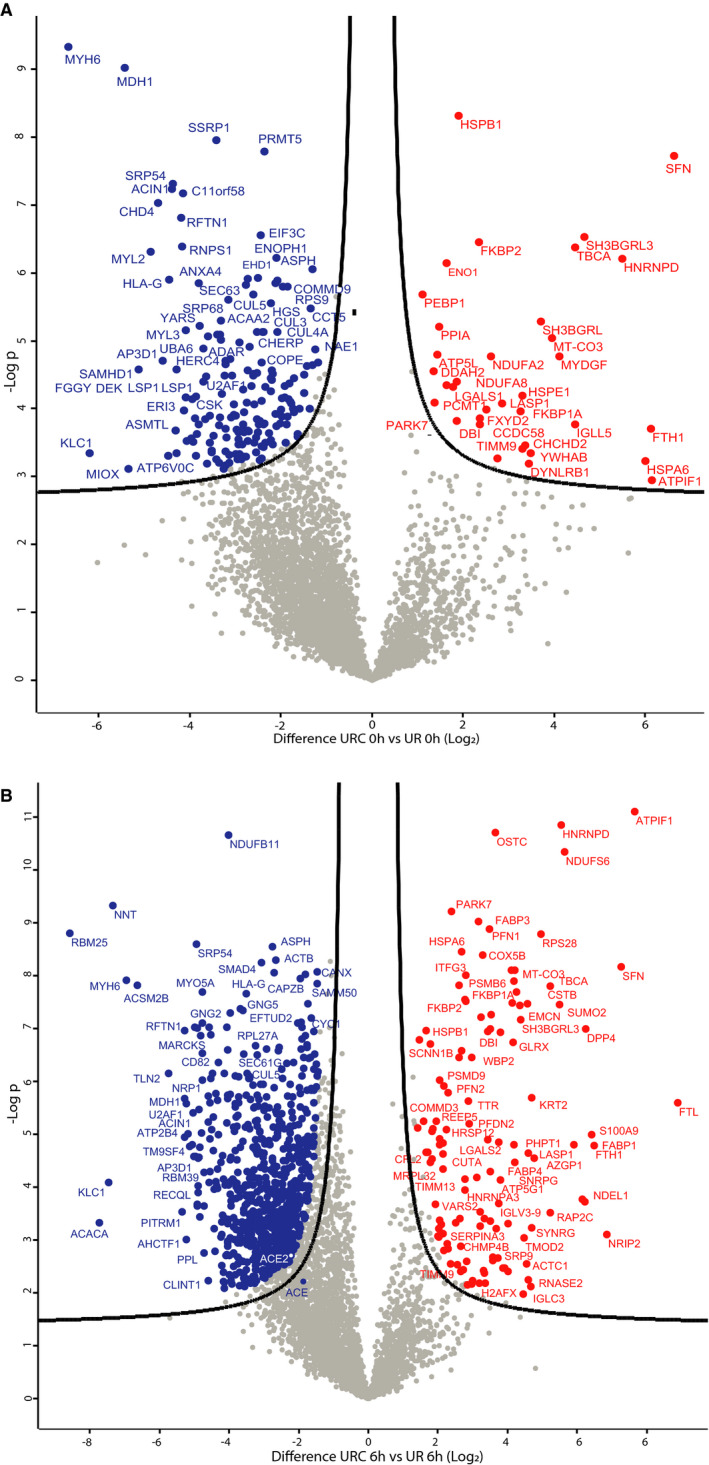
NMP with URC or UR leads to different Kidney tissue proteomes. A. Volcano plot showing kidney proteomes of time zero URC versus UR biopsies. In red are proteins whose abundances were increased and in blue proteins with decreased abundance levels in URC T0 hours kidney tissues as compared to UR T0 hours. B. Volcano plot showing the proteome of kidneys subjected to 6 hours NMP either with URC or UR. In red are proteins whose abundances were increased, in blue proteins with decreased abundance levels in URC T6 hours kidney tissue as compared to UR T6 hours
2.4. Western blot validation of differentially expressed proteins
Twenty microgram of homogenized renal cortex proteins was denatured at 95°C for 5 minutes in Laemmli buffer and loaded onto 4%‐12% pre‐cast SDS‐PAGE gels (Bio‐Rad). Detailed description in Data S1.
2.5. Statistical analysis
For statistical evaluation of the data (excluding proteomics), unpaired t tests (parametric) and Mann–Whitney tests (nonparametric) were performed using GraphPad Prism 7. Correlation analysis was performed using IBM SPSS Statistics Version 23. A value of P < 0.05 was considered significant.
3. RESULTS
Overall, biopsies of 19 kidneys were analyzed using an optimized proteomics workflow (Figure 1). Protein was extracted from 16 kidneys with URC (n = 8 organs from brain dead donors [DBD], n = 8 from donors after circulatory death [DCD]), and three with UR (n = 2 DBD, n = 1 DCD). The mean cold ischemia time (CIT) for kidneys perfused with URC was 32.8 ± 30.6 hours (median 19.9 [10.4‐108]hours); the mean warm ischemia time (WIT) was 18.5 ± 8.3 minutes. For the UR group, mean CIT was 20.5 ± 1.9 hours (median 21.1 [18.4‐22] hours) and the only DCD in this group had 11 minutes WIT. The mean arterial flow of URC kidneys was 370 ± 139.5 ml/min; corresponding mean intrarenal resistance (IRR) was 0.27 ± 0.08 mmHg/ml/min. Kidneys perfused with UR had a mean arterial flow of 319 ± 198.2 ml/min; their mean IRR was 0.44 ± 0.4 mmHg/ml/min. Table 1 displays donor and kidney demographics. Kidney parenchyma did not deteriorate over time in any of the perfused organs. Table S3 shows histology and KIM‐1 immunohistochemistry results, the tubular condition, of kidneys before perfusion start and at the end of NMP.
Table 1.
Kidneys undergoing proteomics after normothermic machine perfusion
| Kidneys with urine recirculation (URC) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Overall (n = 16) | Age in years | Sex | GFRa (ml/min/1.73 m2) | Hypertension | DBD (n = 8) | DCD (n = 8) | Reason for discard | CIT in hours | CIT >24 hours | NMP time in hours | Median arterial flow in ml/min | Lactate @6 hours NMP in mmol/l | Urine in ml/hour |
| Kidney 1 | 70 | male | 59 | no | X | patchy perfusion | 15.0 | 16.2 | 330 | 14.1 | 86.3 | ||
| Kidney 2 | 61 | male | 92 | no | X | esophageal cancer | 19.1 | 24.3 | 330 | 15.1 | 41.0 | ||
| Kidney 3 | 70 | male | 125 | no | X | patchy perfusion | 32.0 | X | 12.3 | 410 | 19.9 | 3.3 | |
| Kidney 4 | 58 | male | 118 | no | X | huge cyst lower pole | 10.4 | 18.6 | 330 | 2.6 | 20.0 | ||
| Kidney 5 | 73 | male | 45 | yes | X | patchy perfusion | 20.7 | 24.1 | 250 | 19.3 | 5.8 | ||
| Kidney 6 | 51 | female | 70 | no | X | renal neoplasia | 11.4 | 24.2 | 320 | 12.9 | 44.9 | ||
| Kidney 7 | 48 | male | 232 | no | X | patchy perfusion, long CIT | 51.2 | X | 15.0 | 250 | 20.0 | 111.8 | |
| Kidney 8 | 44 | female | 100 | no | X | patchy perfusion, long CIT | 36.1 | X | 24.1 | 305 | 11.2 | 16.7 | |
| Kidney 9 | 44 | female | 92 | no | X | patchy perfusion, cortical necrosis | 104.1 | X | 24.0 | 325 | 17.2 | 11.0 | |
| Kidney 10 | 49 | male | 178 | no | X | patchy perfusion | 25.3 | X | 24.3 | 510 | 10.6 | 25.4 | |
| Kidney 11 | 67 | female | 63 | no | X | patchy perfusion | 18.5 | 24.0 | 260 | 13.4 | 17.2 | ||
| Kidney 12 | 38 | male | 58 | yes | X | suspicion of pancreatic cancer | 29.2 | X | 24.4 | 365 | 15.1 | 26.3 | |
| Kidney 13 | 68 | male | 101 | no | X | patchy perfusion | 108.1 | X | 24.0 | 514 | 6.6 | 6.5 | |
| Kidney 14 | 70 | female | 119 | yes | X | stenosis of renal artery | 17.5 | 24.1 | 413 | 8.2 | 53.5 | ||
| Kidney 15 | 47 | female | 29 | no | X | vascular damage, patchy perfusion | 12.7 | 24.1 | 230 | 20.0 | 471.9 | ||
| Kidney 16 | 76 | female | 152 | yes | X | long CIT | 15.4 | 24.0 | 787 | 5.6 | 51.0 | ||
| Kidneys without urine recirculation (UR) | |||||||||||||
| Overall (n = 3) | Age in years | Sex |
GFR a (ml/min/1.73 m2 ) |
Hypertension | DBD (n = 2) | DCD (n = 1) | Reason for discard | CIT in hours | CIT >24 hours | NMP time in hours | Median arterial flow in ml/min | Lactate @6 hours NMP in mmol/l | Urine in ml/hour |
| Kidney 17 | 74 | female | 90 | yes | X | lesion on partner kidney | 22.0 | 6.2 | 486 | 4.5 | 74.2 | ||
| Kidney 18 | 78 | female | 61 | yes | X | vascular damage | 18.4 | 9.4 | 100 | 19.7 | 102.2 | ||
| Kidney 19 | 71 | female | 77 | no | X | organ size | 21.1 | 8.2 | 371 | 17.8 | 84.4 | ||
Abbreviations: DBD, donation after brain death; DCD, donation after circulatory death; CIT, cold ischemia time; NMP, normothermic machine perfusion.
MDRD (modification of diet in renal disease) GFR (glomerular filtration rate) calculator.
Urine recirculation achieved a stabilization of urine flow, which was not observed for kidneys under UR (Figure S1). The perfusion protocol appears to have global effects on total kidney tissue proteomes, separating the overall profiles of URC and UR kidneys, whereas separations of DCD versus DBD and lengths of CIT were less prominent (Figure S2). Different protein abundance profiles were visualized in groups stratified for perfusion types: URC versus UR. Figure 2A‐B illustrates the comparison of URC and UR kidneys. These volcano plots revealed significant differences already between protein abundance in the zero biopsies, as compared to the analysis performed after 6 hours of NMP. For instance, enzymes involved in energy metabolism such as ATP synthase (ATPF1), NADH dehydrogenase (NDUFS6), and oligosaccharyltransferase (OSTC), are emerging in the profiles of URC kidneys after 6 hours NMP (Figure 2B). To examine the differences between perfusion conditions at an individual kidney level, scatter plots of kidneys 14‐16 (URC) and 17‐19 (UR) were generated (Figure 3A‐F). We observed considerable variation between kidneys but noted also even more profound changes beyond 6 hours of NMP with URC, reflecting a revitalization of metabolism, in particular energy homeostasis. Perhaps not unexpectedly, kidney tissue proteome profiles at NMP and URC endpoints (beyond 6 up to 24 hours) were distinct from NMP and UR (6 hours).
3.1. Enzymes involved in glucose and succinate metabolism increased after URC
Due to the fact that all NMP kidneys became metabolically active and consumed a different amount of glucose, in particular beyond the 6‐hour time point, we first analyzed results specifically for proteins involved in glucose metabolism to discover potential differences in their up‐ and/or downregulation.
The comparison of biopsy samples from kidneys with a long CIT (>24 hours; kidneys 3, 7‐10, 12, and 13; Table 1) with kidneys normothermically perfused after a shorter CIT (<24 hours; kidneys 1 and 2, 4‐6, 11, 14‐16; Table 1) revealed that the following enzymes stayed twofold, or less, downregulated throughout the time of perfusion (albeit not significantly) in the longer CIT cohort: glycolysis enzymes hexokinase (HK), phosphofructokinase (PFK) and pyruvate kinase (PK), glucose‐6‐phosphatase (G6P), fructose‐1,6‐bisphosphatase (FBP), phosphoenolpyruvate carboxykinase (PCK) and pyruvate carboxylase (PC), the key enzymes of gluconeogenesis.
When stratifying the proteomics analysis for donor type, DCD (n = 8) versus DBD (n = 8) URC kidneys, we could detect interesting trends of all mentioned enzymes involved in glucose metabolism, including mitochondrial malate dehydrogenase (MDH) and glutamic‐oxaloacetic transaminase (GOT). All stated enzymes above were downregulated in the DCD zero biopsies compared to the DBD zero biopsies; FBP isoenzyme 1 was significantly (P< 0.05) downregulated in DCD kidneys compared to DBD organs. After 12 and 24 hours of NMP, all but one enzyme were more upregulated in DCD tissue compared to DBD kidney biopsies. The mitochondrial version of PCK, PCK2, was significantly downregulated (P < 0.05) after 12 hours of NMP in DCD kidney biopsies compared to DBD. Interestingly, both the cytosolic (PCK1) and the mitochondrial (PCK2) enzyme were more upregulated after 24 hours of NMP in DCD as compared to DBD tissue.
In DCD kidneys, succinate dehydrogenase (SDH; subunits A and B) were significantly upregulated in the zero biopsies as compared to DBD kidneys. After 12 and 24 hours of NMP the same proteins were significantly upregulated in DCD biopsies. Fumarate hydratase (FH), an enzyme which can reversibly catalyze the hydration/dehydration of fumarate to malate, was also significantly upregulated in the zero biopsies of DCD kidneys as compared to DBD tissue and vice versa after 12 hours of NMP.
3.2. Proteins with altered abundance in kidney NMP with URC or UR
Damage‐associated molecular pattern (DAMPs) proteins, known to be contributing to ischemia reperfusion injury (IRI), were significantly downregulated after 6 hours of kidney NMP and URC as compared to NMP and UR (Figure 3, Table S1).
Furthermore, the abundance of mitochondrial proteins such as d‐glutamate cyclase (DGLUCY) 20 and succinate dehydrogenase subunits were significantly altered in URC biopsies, which could play a role regarding succinate accumulation and ROS production 21 : FH (fumarate hydratase), SDHB (succinate dehydrogenase complex, subunit B), SDHC (succinate dehydrogenase complex, subunit C), SDHD (succinate dehydrogenase complex, subunit D) (Figure 3A‐C vs. D‐F).
Kidneys perfused with URC expressed significantly more SLC4A1, a band 3 anion transport protein, which acts as a chloride/bicarbonate exchanger and is able to maintain the correct acid levels (pH) in the body. Furthermore, this protein mediates the chloride–bicarbonate exchange in the kidney which is required for normal acidification of the urine. 1 , 2 , 3
Generally, protein profiles in kidneys with URC were exactly the opposite to the ones without, independent of their statistical significance. Figure 4A‐D displays the comparison of up‐ and downregulated proteins with and without urine recirculation, emphasizing the four most striking clusters of metabolic activity: (1) acid–base balance and pH stability, (2) glucose metabolism, (3) anti‐inflammation, and (4) the renin–angiotensin system.
Figure 4.
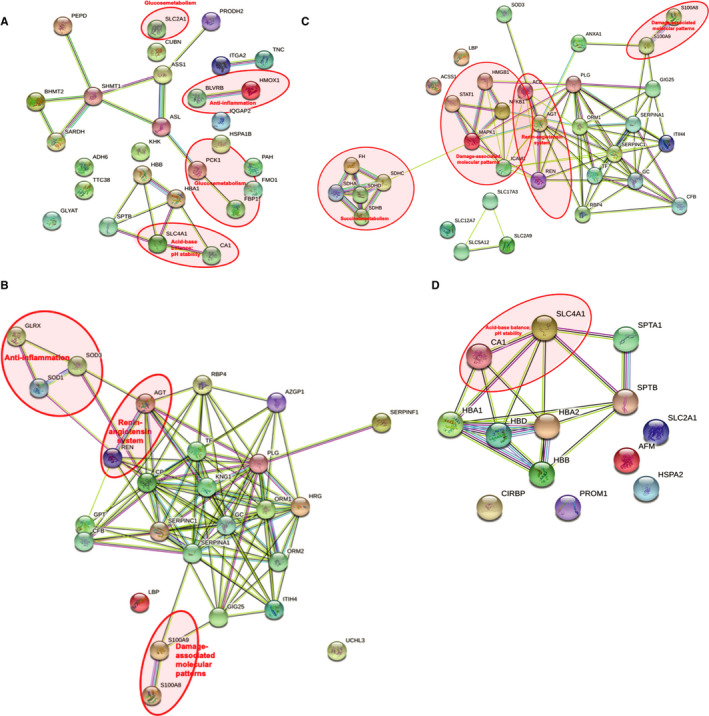
Prolonged NMP with URC drives metabolic homeostasis. (A) Upregulated cellular pathways after 6 hours NMP with URC. Acid–base balance and pH stability: CA1 carbonic anhydrase 1; SLC4A1 band 3 anion transport protein. Glucose metabolism: PCK1 phosphoenolpyruvate carboxykinase; FBP1 fructose‐1,6‐bisphosphatase 1; SLC2A1 solute carrier family 2, facilitated glucose transporter member 1. Anti‐inflammation: BLVRB flavin reductase (NADPH); broad specificity oxidoreductase that catalyzes the NADPH‐dependent reduction of a variety of flavins, such as riboflavin, FAD or FMN, biliverdins, methemoglobin and PQQ (pyrroloquinoline quinone); contributes to heme catabolism and metabolizes linear tetrapyrroles; can also reduce the complexed Fe(3+) iron to Fe(2+) in the presence of FMN and NADPH. HMOX1 heme oxygenase 1; cleaves the heme ring at the alpha methene bridge to form biliverdin; biliverdin is subsequently converted to bilirubin by biliverdin reductase; exhibits cytoprotective effects since excess of free heme sensitizes cells to undergo apoptosis. (B) Upregulated cellular pathways after 6 hours NMP with UR. Renin–angiotensin system: AGT angiotensinogen; a potent regulator of blood pressure, body fluid, and electrolyte homeostasis. REN renin; to generate angiotensin I from angiotensinogen in the plasma, initiating a cascade of reactions that produce an elevation of blood pressure and increased sodium retention by the kidney. Damage‐associated molecular patterns (DAMPs): S100A8 protein S100‐A8; a calcium‐ and zinc‐binding protein which plays a prominent role in the regulation of inflammatory processes and immune response. It can induce neutrophil chemotaxis and adhesion. Predominantly found as calprotectin (S100A8/A9) which has a wide plethora of intra‐ and extracellular functions. S100A9 protein S100‐A9; a calcium‐ and zinc‐binding protein which plays a prominent role in the regulation of inflammatory processes and immune response. It can induce neutrophil chemotaxis, adhesion, can increase the bactericidal activity of neutrophils by promoting phagocytosis via activation of SYK, PI3 K/AKT, and ERK1/2 and can induce degranulation of neutrophils by a MAPK‐dependent mechanism. Anti‐inflammation: GLRX glutaredoxin 1; has a glutathione‐disulfide oxidoreductase activity in the presence of NADPH and glutathione reductase. SOD1 superoxide dismutase; destroys radicals which are normally produced within the cells and which are toxic to biological systems. SOD3 extracellular superoxide dismutase; protects the extracellular space from toxic effect of reactive oxygen intermediates by converting superoxide radicals into hydrogen peroxide and oxygen. (C) Downregulated cellular pathways after 6 hours NMP with URC. Renin–angiotensin system: AGT angiotensinogen; ACE angiotensin‐converting enzyme; converts angiotensin I to angiotensin II by release of the terminal His‐Leu, this results in an increase in the vasoconstrictor activity of angiotensin. Also able to inactivate bradykinin, a potent vasodilator. Damage‐associated molecular patterns (DAMPs): HMGB1 high‐mobility group box 1; promotes host inflammatory response to sterile and infectious signals and is involved in the coordination and integration of innate and adaptive immune responses. NFKB1 nuclear factor NF‐kappa‐B p105; a pleiotropic transcription factor; related to many biological processes such as inflammation, immunity, differentiation, cell growth, tumorigenesis, and apoptosis. MAPK1 mitogen‐activated protein kinase 1; serine/threonine kinase which acts as an essential component of the MAP kinase signal transduction pathway. ICAM1 intercellular adhesion molecule 1; ligands for the leukocyte adhesion protein LFA‐1 (integrin alpha‐L/beta‐2). STAT1 signal transducer and activator of transcription 1‐alpha/beta; signal transducer and transcription activator that mediates cellular responses to interferons (IFNs), cytokine KITLG/SCF and other cytokines and other growth factors. S100A8 protein S100‐A8. Succinate metabolism: SDHA/B/C/D succinate dehydrogenase subunits; A= flavoprotein subunit; B= iron–sulfur protein subunit; C and D= membrane anchoring subunits; involved in complex II of the mitochondrial electron transport chain and is responsible for transferring electrons from succinate to ubiquinone (coenzyme Q). FH fumarate hydratase; class II fumarase/aspartate family. (D) Downregulated cellular pathways after 6 hours NMP with UR. Acid–base balance and pH stability: CA1 carbonic anhydrase 1; SLC4A1 band 3 anion transport protein. NMP=normothermic machine perfusion of the kidney. UR=urine replacement. URC=urine recirculation. String Version 11.0; © String Consortium 2020. 22 Lines: line color indicates the type of interaction evidence; Nodes: colored—query proteins and first shell of interactors; white—second shell of interactors; empty—proteins of unknown 3D structure; filled—some 3D structure is known or predicted
Angiotensinogen (AGT) was upregulated in kidneys with UR and downregulated in kidneys with URC. Angiotensin I converting enzyme (ACE) was significantly downregulated in kidneys with URC, and serpin peptidase inhibitor (SERPINA3) was significantly upregulated in kidneys with URC after 6 hours (Figures 3 and 4), contributing to stabilizing urine flow rates during prolonged NMP (Figure S1). 1 , 2 , 3
Carbonic anhydrase (CA1) was downregulated in kidneys with UR. 1 , 2 , 3 , 23 In kidneys perfused with URC it was possible to maintain physiological pH values throughout the perfusion period without giving any supplements. In kidneys without urine recirculation, UR group, the pH changed significantly over time in both directions—alkalotic and acidotic.
3.3. Correlation of proteins with perfusion and organ characteristics
Several correlations were found between proteins of kidneys perfused with URC and UR with perfusion parameters (e.g. arterial flow, URC, blood gases, NMP time) and kidney factors (e.g. donor age) (Table S2).
3.4. NMP‐ and URC‐dependent prolonged induction of enzymes contributing to metabolic homeostasis
In normothermically perfused kidneys with URC, proteins differentially expressed included antioxidants, stress proteins, anion exchangers, and enzymes like anhydrases and carboxykinases. We selected seven upregulated proteins in the group of kidneys with urine recirculation and validated the abundance profiles of these proteins by performing Western Blots on kidney tissue material. These included ICAM‐1, SLC4A1 (band 3 anion transport protein), HMOX1 (hem oxygenase 1), flavin reductase (NADPH, gene BLVRB), CA1, PCK, and HSPA4 (Figure 5A‐G). The signal intensity was quantified and normalized to beta‐actin (Figure 6A‐G). The number of selected proteins was limited by the available biopsy material. Strikingly, most effects were noticeable beyond 6 hours NMP. For instance, ICAM‐1 increased significantly over time, reaching its peak 16 hours after perfusion start. Similarly, SLC4A1 and HMOX1 increased significantly from zero biopsy up to 18‐24 hours after NMP initiation. BLVRB reached its peak 16 hours after NMP start compared to the zero biopsy. PCK got expressed throughout the perfusion without any significant differences, the lowest amount of protein could be detected in biopsies taken 16 hours after NMP start; CA1 changed most significantly within the first 6 hours of NMP, followed by an additional increase in the time frame between 12 and 18 hours after NMP start; HSPA4 did not show significant changes between time points after NMP start. We conclude that prolonged NMP (>6 hours), facilitated by URC, permits a more effective metabolic homeostasis.
Figure 5.
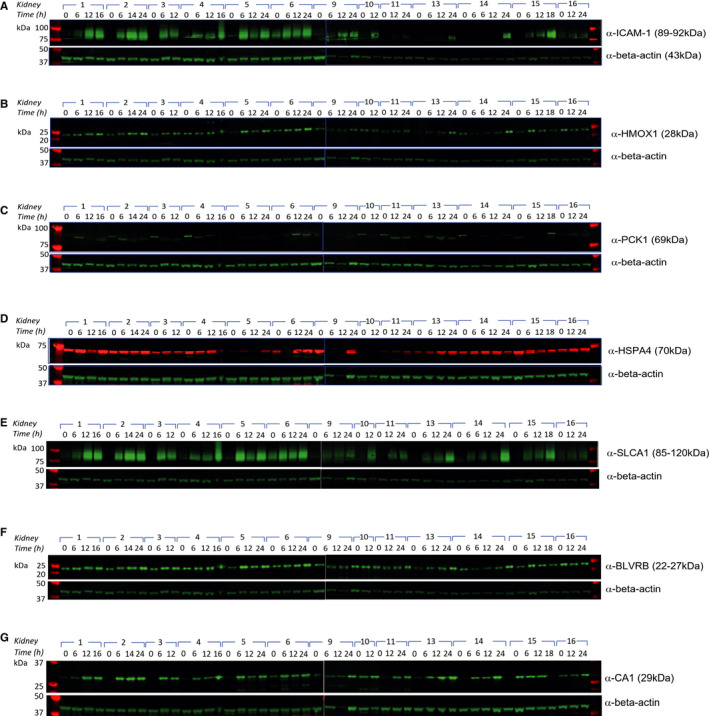
Altered protein profiles associated with prolonged NMP and URC. Western blotting of discarded kidney tissue extracts collected at different time points during NMP with URC: (A) α‐ICAM‐1, (B) α‐HMOX1, (C) α‐PCK1, (D) α‐HSPA4, (E) α‐SLC4A1, (F) α‐BLVRB, (G) α‐CA1. Beta‐actin was used as loading control
Figure 6.
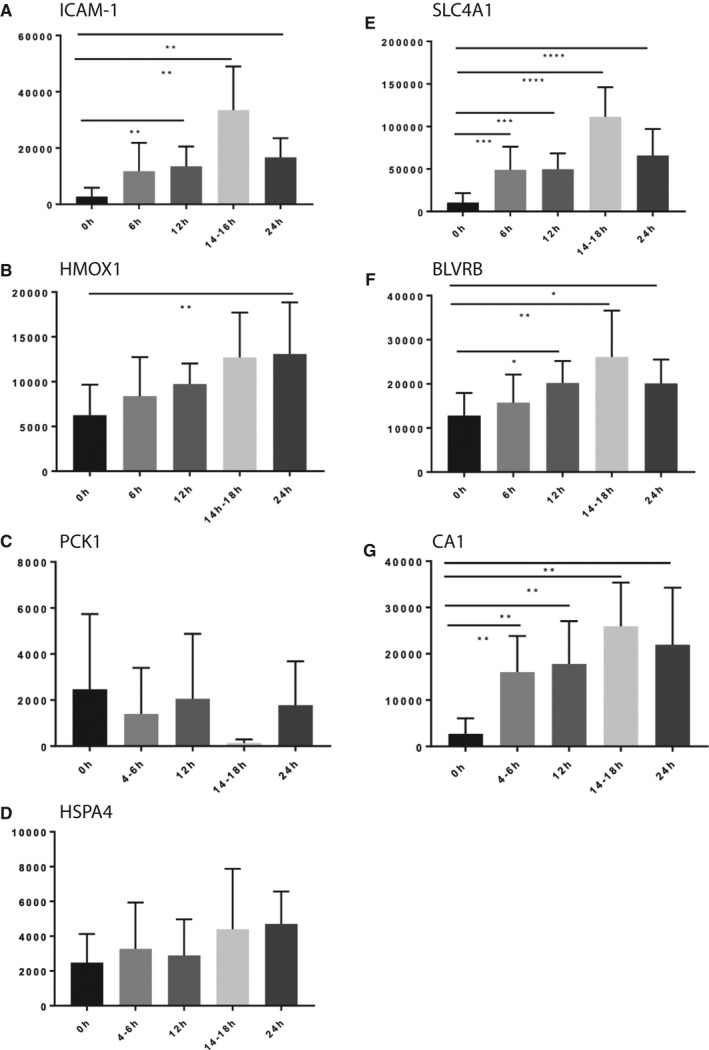
Prolonged NMP leads to increased levels of proteins contributing to metabolic homeostasis. Bar graphs show quantitative levels of proteins at times zero (0 hours), 6 hours, 12 hours, 14 hours‐18 hours, and 24 hours, respectively. Protein levels of ICAM‐1 (A), HMOX1 (B), PCK1 (C), HSPA4 (D), SLC4A1 (E), BLVRB (F), and CA1 (G) derived from Western blotting analysis are shown, normalized to beta‐actin
4. DISCUSSION
Proteomics technology was applied to uncover the molecular profiles of kidney biopsies after NMP up to 24 hours, facilitated by urine recirculation (URC). Remarkably, prolonged NMP (>6 hours), not readily achievable without urine recirculation (UR), resulted in a more optimal metabolic homeostasis and showed signs of tissue damage recovery. For instance, we detected the downregulation of potential markers promoting ischemia reperfusion injury (IRI) during and after kidney NMP on a fully closed circuit by recirculating the urine. The aim of our series of normothermic perfusions was rather focusing on the development of a clinically applicable perfusion device for kidneys and to learn about and to understand processes going on during NMP. Therefore, we have not used whole blood to reperfuse NMP kidneys after a certain time of preservation mimicking IRI in the preclinical experimental setting.
Recirculation of the urine has been applied to a non‐transplant model of kidney NMP up to 24 hours by our group before. 9 , 10 Volume control and balanced perfusate homeostasis as well as the possibility to have a fully automated and transportable kidney perfusion device, represent the three main advantages of URC in kidney NMP. 9 , 10 Generally, normothermic organ preservation has been an emerging field for the last two decades. Kidney NMP has been implemented as a clinical tool in the United Kingdom not only to reduce delayed graft function rates but also to enlarge the donor pool by rescuing discarded kidneys. 12 , 13 , 24 , 25 , 26 Several transplant centers around the globe have reported their first experiences with short‐term kidney NMP (up to 1 to 2 hours) in the clinical setting, but with the exception of the group around Nicholson et al publications are still missing. The first results of a prospective randomized trial comparing 1 hour NMP with static cold storage (SCS) in DCD kidneys is expected to be presented end of 2020. 13 Short‐term kidney NMP appears to be quite common in the world of clinical transplantation, but long‐term ex vivo kidney preservation going beyond 12 hours is still a subject of experimental work. Selzner et al are leading the field of porcine kidney perfusion, reaching overall preservation times of 24 hours. 14 , 15 , 27 , 28 , 29 In their recent publication, they describe the clear superiority of porcine DCD grafts preserved normothermically over grafts preserved by hypothermic perfusion or SCS in an auto‐transplant model. 29 Although promising and clearly leading the way towards long‐term kidney NMP, the necessity of replacing the lost urine volume in a perfusion system constitutes a limitation for a straightforward use in the clinical world.
In our perfusion system of kidney NMP up to 24 hours, we experienced the application of URC as extremely helpful, facilitating long‐term NMP even after CIT beyond 50 hours without harming kidney parenchyma. 9 , 10 Keeping kidneys going normothermically by replacing excreted urine 1:1 using Ringer's lactate, as demonstrated by other research groups, was not feasible in our setting and led to renal artery flows <50 ml/min after 6 to 8 hours of NMP and/or severely alkalotic or acidotic perfusate (Figures S1 and S3). 9 , 10 The heterogenous cohort of perfused discarded human kidneys in this study, which were deemed untransplantable by all transplant centers in the United Kingdom at the time and perfused after variable durations of SCS, were not considered for clinical transplantation, however, we wanted to investigate any differences and similarities on the protein level of such preserved organs. Despite the low number of NMP kidneys with UR, which demands cautious interpretation of statistical significance, the most striking findings were the extent of differences (Figure 2) and the complete reversed up‐ and downregulation of proteins in kidneys perfused with and without urine URC (Figure 3). In all kidneys perfused with UR the proteins extracted after 6 hours NMP, such as DAMPs, proteins involved in succinate metabolism and vasoconstriction, were all upregulated compared to kidneys perfused with URC (Table S1). DAMPs can be released by injured tissue or activated cells, following a stimulus and are recognized by cellular receptors as a universal damage‐associated molecular pattern, able to initiate repair, provoke remodeling and innate immune signaling. 30 Reduction of DAMPs by using URC might not have an immediate impact on the kidney while being perfused on the circuit. However, it might cause less reperfusion injury when such kidneys are reperfused in a potential organ recipient in future transplant studies. A similar explanation applies to downregulated proteins involved in the succinate metabolism after NMP with URC. Studies on ischemic tissue and reperfusion showed succinate accumulation as a consequence of IRI. 31 , 32 Chouchani et al described in an ischemic model of the heart that succinate accumulation is a metabolic signature of ischemia and is responsible for mitochondrial ROS production during reperfusion. Succinate accumulation is caused by reverse function of succinate dehydrogenase driven by fumarate overflow. 31 In our biopsies, several subunits of the succinate dehydrogenase complex were significantly downregulated after six NMP hours with URC as compared to UR. A hallmark of perfused kidneys with UR was the significant upregulation of proteins involved in vasoconstrictive processes when compared to kidneys perfused with URC. Angiotensinogen (AGT) 33 which was upregulated in kidneys with UR and downregulated in kidneys with URC, is an interesting target due to its function and the fact that kidneys with UR developed increasing intrarenal resistance during NMP, leading to a deterioration of the arterial flow from hour 6 onwards. Angiotensinogen is part of the renin–angiotensin system regulating blood pressure, fluid balance, and salts in the human body. Angiotensinogen is converted to angiotensin I, which is converted to angiotensin II, a vasoconstrictor. 17 In addition to this finding, the comparison of the entire biopsy cohort of kidneys with and without URC revealed significant differences in other proteins involved in the renin–angiotensin system: ACE was significantly downregulated in kidneys with URC. Serpin peptidase inhibitor (SERPINA3) can inhibit the conversion of angiotensin‐1 to the active angiotensin‐2 indirectly and was significantly upregulated in kidneys with URC after 6 hours (Figure 2B). 22 , 34 , 35 Moreover, sodium content of the perfusate changed over time. Perfusate of kidneys undergoing NMP with UR had significantly higher sodium levels with 159.6 ± 4.63 mmol/l9 as compared to URC kidneys with physiological levels of 141.3 ± 2.71 mmol/l, P < 0.01. Hypernatremia is a well‐known factor inducing vasoconstriction of renal vasculature, but if this is triggered in our cohort by different protein up‐ or downregulations induced by different ways of handling urine and perfusate volume, cannot be answered at the current stage of investigations.
Carbonic anhydrase (CA1) may also contribute to an increased performance of kidneys perfused with urine recirculation (URC), and was found to be downregulated in kidneys with UR. We were able to maintain physiological pH values throughout the perfusion period only in kidneys perfused with URC. In contrast, urine replacement, caused either acidosis or alkalosis of the perfusate in our experiments. This could be due to a lack of presence or activity of CA1. 22 , 34 , 35 , 36 The most important function of this enzyme is to maintain the acid–base balance in human blood and tissue. In addition, band 3 anion transport protein SLC4A1 was also significantly upregulated over time in kidneys perfused with URC (Figures 3,5,6). This protein is crucial to maintain the correct acid levels in the human body, mediated by the chloride–bicarbonate exchange in the kidney, and it is required for the normal acidification of the urine. 22 , 34 , 35
The profile of enzymes involved in gluconeogenesis and glycolysis could also help to explain the difficulty to interpret lactate levels in kidney NMP, which is an easy task in liver NMP. In humans, lactate metabolism plays a major role in the native kidney and the renal cortex appears to be the most important lactate‐consuming organ after the liver. 18 Bartlett et al showed years ago that renal dysfunction impacts lactate and glucose metabolism. It is most likely that lactate levels are related to active glucose metabolism, glycolysis, and gluconeogenesis in the kidney. Also, there seems to be a cortico‐medullary glucose‐lactate recycling system: the medulla consumes glucose by active glycolysis and generates lactate. The cortex has the ability to take up the lactate released by the medulla and uses it for oxidation and gluconeogenesis. 19 A publication by Hui et al demonstrated that glucose is essential in feeding the Krebs‐cycle via circulating lactate. 37 In their experimental setting in mice they detected strongly labeled lactate in the kidney after infusion of 13 C‐labeled glucose, although intermediates of the Krebs cycle were more labeled after lactate infusion. An explanation for this pattern in the kidney is the heterogeneity of kidney cells; the presence of glycolytic cells producing lactate from glucose and oxidative cells using lactate to make Krebs cycle intermediates. 37
Having in mind that proteomics technology will not be able to instantly advise us on decision making to transplant or not to transplant a kidney or to reallocate an organ to a better‐matching recipient, and was not developed for this purpose, it is still offering important information by providing protein profiles able to be correlated with perfusion, donor, and transplant characteristics. Interestingly, levels of CA1, SLC4A1, and Tenascin‐C (TNC) increased in accordance with higher arterial flow through the kidneys. Tenascin‐C was fairly recently reported to be protective against acute kidney injury (AKI). 38 In an ex vivo model of AKI TNC is induced at injured sites of the organ, recruits Wnt ligands and creates an optimal microenvironment for tubular repair and regeneration 38 In addition, levels of SOD3 were lower the higher the arterial flow was. This underlines the importance of reaching high and physiological renal plasma flow and, moreover, should recommend arterial flow as one of the most essential perfusion characteristics to assess a kidney ex vivo. Catalase expression increased the longer the kidney was on the circuit with urine recirculation. As expected, ICAM‐1 was elevated the higher the arterial pressure was, but considerably reduced the longer the kidney underwent NMP. FAM3C, which promotes epithelial to mesenchymal transition, 1 , 2 , 3 was significantly associated with donor age, CIT, and intrarenal resistance, but correlated inversely with the time on the NMP device. This would suggest FAM3C to have a positive effect of longer term kidney NMP on the epithelial structures and proposes the implementation of such technology in the clinical setting for extended criteria donor organs in particular.
In conclusion, proteomics applied on consecutively taken biopsies of NMP kidneys revealed clear differences between URC and UR. Ameliorated metabolic processes due to prolonged NMP and URC could lead to enhanced kidney organ function recovery and render kidneys more amenable to transplantation.
DISCLOSURE
The authors of this manuscript have conflicts of interest to disclose as described by the American Journal of Transplantation. In addition to being full‐time academics at the University of Oxford, PJF and CCC receive consultancy payments as nonexecutive medical and technical directors of OrganOx Limited, and are shareholders.
OPEN RESEARCH BADGES
This article has earned an Open Data badge for making publicly available the digitally‐shareable data necessary to reproduce the reported results. The data is available at http://proteomecentral.proteomexchange.org/cgi/GetDataset.
Supporting information
ACKNOWLEDGMENTS
Special thanks to Stefan Scheidl, MD, Department of Visceral, Transplant and Thoracic Surgery, Medical University of Innsbruck, Innsbruck, Austria, for designing, creating, and providing Figure 1. Honglei Huang, PhD, was supported by a National Institute of Health Research (NIHR) grant to Rutger J. Ploeg.
[Correction added on 11 December 2020, after first online publication: the author name 'Maria L. L. Faro' has been corrected to 'Maria L. Lo Faro'.]
DATA AVAILABILITY STATEMENT
Data available on request from the authors.
REFERENCES
- 1. Sigdel TK, Lee S, Sarwal MM. Profiling the proteome in renal transplantation. Proteomics Clin Appl. 2011;5(5–6):269‐280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sigdel TK, Salomonis N, Nicora CD, et al. The identification of novel potential injury mechanisms and candidate biomarkers in renal allograft rejection by quantitative proteomics. Mol Cell Proteomics MCP. 2014;13(2):621‐631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Marx D, Metzger J, Olagne J, et al. Proteomics in Kidney Allograft Transplantation‐Application of molecular pathway analysis for kidney allograft disease phenotypic biomarker selection. Proteomics Clin Appl. 2019;13(2):e1800091. [DOI] [PubMed] [Google Scholar]
- 4. Eikmans M, Gielis EM, Ledeganck KJ, Yang J, Abramowicz D, Claas FFJ. Non‐invasive biomarkers of acute rejection in kidney transplantation: novel targets and strategies. Front Med. 2018;5:358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hricik DE, Nickerson P, Formica RN, et al. Multicenter validation of urinary CXCL9 as a risk‐stratifying biomarker for kidney transplant injury. Am J Transplant. 2013;13:2634‐2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kaisar M, van Dullemen L, Charles P, et al. Subclinical changes in deceased donor kidney proteomes are associated with 12‐month allograft function posttransplantation—a preliminary study. Transplantation. 2019;103(2):323‐328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Perco P, Heinzel A, Leierer J, et al. Validation of systems biology derived molecular markers of renal donor organ status associated with long term allograft function. Sci Rep. 2018;8:6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kirita Y, Chang‐Panesso M, Humphreys BD. Recent insights into kidney injury and repair from transcriptomic analyses. Nephron. 2019;143(3):162‐165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Weissenbacher A, Lo Faro L, Boubriak O, et al. Twenty‐four‐hour normothermic perfusion of discarded human kidneys with urine recirculation. Am J Transplant. 2019;19:178‐192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Weissenbacher A, Voyce D, Ceresa CDL, et al. Urine recirculation improves hemodynamics and enhances function in normothermic kidney perfusion. Transplant Direct. 2020;6(4):e541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Weissenbacher A. Normothermic kidney preservation. University of Oxford; 2018. [Google Scholar]
- 12. Hosgood SA, Nicholson ML. The first clinical case of intermediate ex vivo normothermic perfusion in renal transplantation. Am J Transplant. 2014;14(7):1690‐1692. [DOI] [PubMed] [Google Scholar]
- 13. Hosgood SA, Saeb‐Parsy K, Wilson C, Callaghan C, Collett D, Nicholson ML. Protocol of a randomised controlled, open‐label trial of ex vivo normothermic perfusion versus static cold storage in donation after circulatory death renal transplantation. BMJ Open. 2017;7(1):e012237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kaths JM, Hamar M, Echeverri J, et al. Normothermic ex vivo kidney perfusion for graft quality assessment prior to transplantation. Am J Transplant. 2018;18:580‐589. [DOI] [PubMed] [Google Scholar]
- 15. Kaths JM, Cen JY, Chun YM, et al. Continuous normothermic ex vivo kidney perfusion is superior to brief normothermic perfusion following static cold storage in donation after circulatory death pig kidney transplantation. Am J Transplant. 2017;17:957‐969. [DOI] [PubMed] [Google Scholar]
- 16. Katane M, Ariyoshi M, Tateishi S, et al. Structural and enzymatic properties of mammalian d‐glutamate cyclase. Arch Biochem Biophys. 2018;654:10‐18. [DOI] [PubMed] [Google Scholar]
- 17. Satou R, Shao W, Navar LG. Role of stimulated intrarenal angiotensinogen in hypertension. Ther Adv Cardiovasc Dis. 2015;9(4):181‐190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bellomo R. Bench‐to‐bedside review: Lactate and the kidney. Crit Care. 2002;6:322‐326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bartlett S, Espinal J, Janssens P, Ross BD. The influence of renal function on lactate and glucose metabolism. Biochem J. 1984;219(1):73‐78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huang H, van Dullemen LFA, Akhtar MZ, et al. Proteo‐metabolomics reveals compensation between ischemic and non‐injured contralateral kidneys after reperfusion. Sci Rep. 2018;8(1):8539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tyanova S, Temu T, Cox J. The MaxQuant computational platform for mass spectrometry‐based shotgun proteomics. Nat Protoc. 2016;11(12):2301‐2319. [DOI] [PubMed] [Google Scholar]
- 22. Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome‐wide experimental datasets. Nucleic Acids Res. 2019;47:D607‐D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Perseus . http://www.biochem.mpg.de/5111810/Perseus Accessed August 25, 2020.
- 24. Hosgood SA, Nicholson ML. First in man renal transplantation after ex vivo normothermic perfusion. Transplantation. 2011;92(7):735‐738. [DOI] [PubMed] [Google Scholar]
- 25. Hosgood SA, Thompson E, Moore T, Wilson CH, Nicholson ML. Normothermic machine perfusion for the assessment and transplantation of declined human kidneys from donation after circulatory death donors. Br J Surg. 2018;105(4):388‐394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hosgood SA, Nicholson ML. Ex vivo normothermic perfusion of declined human kidneys after inadequate in situ perfusion. Am J Transplant. 2014;14(2):490‐491. [DOI] [PubMed] [Google Scholar]
- 27. Kaths JM, Echeverri J, Linares I, et al. Normothermic ex vivo kidney perfusion following static cold storage‐brief, intermediate, or prolonged perfusion for optimal renal graft reconditioning? Am J Transplant. 2017;17(10):2580‐2590. [DOI] [PubMed] [Google Scholar]
- 28. Kaths JM, Echeverri J, Goldaracena N, et al. Eight‐Hour continuous normothermic ex vivo kidney perfusion is a safe preservation technique for kidney transplantation: a new opportunity for the storage, assessment, and repair of kidney grafts. Transplantation. 2016;100(9):1862‐1870. [DOI] [PubMed] [Google Scholar]
- 29. Urbanellis P, Hamar M, Kaths JM, et al. Normothermic ex‐vivo kidney perfusion improves early DCD graft function compared to hypothermic machine perfusion and static cold storage. Transplantation. 2020;104:947‐955. [DOI] [PubMed] [Google Scholar]
- 30. Seong S‐Y, Matzinger P. Hydrophobicity: an ancient damage‐associated molecular pattern that initiates innate immune responses. Nat Rev Immunol. 2004;4(6):469‐478. [DOI] [PubMed] [Google Scholar]
- 31. Chouchani ET, Pell VR, Gaude E, et al. Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS. Nature. 2014;515(7527):431‐435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Martin JL, Gruszczyk AV, Beach TE, Murphy MP, Saeb‐Parsy K. Mitochondrial mechanisms and therapeutics in ischaemia reperfusion injury. Pediatr Nephrol. 2019;34:1167‐1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Reference GH .AGT gene. Genetics Home Reference. https://ghr.nlm.nih.gov/gene/AGTAccessed August 25, 2020.
- 34. UniProt . https://www.UniProt.org/ Accessed August 25, 2020.
- 35. STRING: functional protein association networks. https://string‐db.org/cgi/input.pl Accessed August 25, 2020.
- 36. Carbonic anhydrase. In: Wikipedia. 2018. Accessed August 25, 2020.
- 37. Hui S, Ghergurovich JM, Morscher RJ, et al. Glucose feeds the TCA cycle via circulating lactate. Nature. 2017;551(7678):115‐118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen S, Fu H, Wu S, et al. Tenascin‐C protects against acute kidney injury by recruiting Wnt ligands. Kidney Int. 2019;95(1):62‐74. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data available on request from the authors.


