To the Editor,
Type 2 (T2) inflammation in asthma encompasses the presence of eosinophilic inflammation and/or selected cytokines including interleukin (IL)‐13. Some chronic obstructive pulmonary disease (COPD) patients have increased airway eosinophil numbers, 1 which is associated with greater effects of inhaled corticosteroids (ICS). 2 Studies of T2 inflammation in COPD have used hypothesis free, open platform approaches. 3 , 4 We used these results, plus data from T2 asthma studies, 5 to test a restricted panel of biomarkers (chloride channel accessory 1 [CLCA1], periostin [POSTN], serpin family B member 2 [SERPINB2], C‐C motif chemokine ligand 26 [CCL26], IL‐13 and cystatin SN [CST1]) to further investigate the nature of T2 inflammation in eosinophilic COPD.
We used two cohorts: bronchial brushings and sputum were obtained from 17 eosinophilhigh and 20 eosinophillow COPD patients (bronchoscopy cohort previously reported 1 : eosinophilhigh defined as blood eosinophils > 250 eosinophils µL−1 and eosinophillow defined as blood eosinophils < 150 eosinophils µL−1) and sputum only was obtained from 15 eosinophilhigh and 18 eosinophillow COPD patients (validation cohort: eosinophilhigh defined as > 150 eosinophils µL−1 blood and > 3% sputum eosinophils and eosinophillow defined as < 150 eosinophils µL−1 blood and < 3% sputum eosinophils). Full details of inclusion criteria and clinical characteristics are in the Appendix S1 and Table 1. Both cohorts were generally well matched. RNA sequencing examined gene expression in the bronchoscopy cohort, while RT‐qPCR was used in the validation cohort. Full details of methods and analysis are in the Appendix S1.
TABLE 1.
Clinical Characteristics of the Bronchoscopy and Validation cohorts
| Characteristic | Bronchoscopy Cohort | Validation Cohort | ||||
|---|---|---|---|---|---|---|
| Eosinophillow | Eosinophilhigh | P value | Eosinophillow | Eosinophilhigh | P value | |
| Number | 17 | 20 | N/A | 18 | 15 | N/A |
| Age (y) | 62 ± 6 | 62 ± 4 | .7 | 64 ± 8 | 67 ± 9 | .3 |
| Gender: Male (%) | 65 | 70 | .7 | 44 | 73 | .1 |
| BMI (kg/m2) | 27 ± 5 | 25 ± 4 | .2 | 25 ± 5 | 28 ± 4 | .06 |
| Current smokers (%) | 42 | 60 | .2 | 61 | 20 | .01 |
| Pack‐years history | 43 ± 15 | 38 ± 14 | .3 | 48 ± 19 | 45 ± 25 | .7 |
| ICS use (%) | 82 | 55 | .08 | 39 | 93 | .02 |
| ICS dose | 0 (0‐2000) | 800 (0‐2000) | .009 | 1000 (0‐2000) | 600 (0‐2000) | .35 |
| Post‐bronchodilator FEV1 (L) | 1.9 ± 0.4 | 1.9 ± 0.4 | .9 | 1.7 ± 0.6 | 1.8 ± 0.5 | .6 |
| Postbronchodilator FEV1 (% predicted) | 64 ± 12 | 66 ± 11 | .6 | 67 ± 17 | 66 ± 16 | .8 |
| Postbronchodilator FVC (L) | 3.8 ± 1.3 | 3.6 ± 0.9 | .5 | 3.2 ± 1 | 3.3 ± 0.7 | .7 |
| Postbronchodilator FEV1/FVC ratio | 50 ± 10 | 53 ± 7 | .3 | 53 ± 12 | 55 ± 11 | .7 |
| Reversibility (mL) | 207 ± 189 | 217 ± 154 | .9 | 166 ± 142 | 145 ± 147 | .7 |
| FeNO50 (ppm) | 16 ± 8.4 | 23 ± 15 | .2 | 9.2 ± 5.2 | 14.9 ± 8.1 | .02 |
| Atopy (% positive) | 0 | 0 | N/A | 6 | 13 | .4 |
| Total SGRQ | 42 ± 17 | 37 ± 21 | .4 | 56 ± 16 | 55 ± 16 | .9 |
| mMRC | 1.8 ± 1.0 | 1.3 ± 1.2 | .2 | 3.6 ± 0.7 | 3.7 ± 0.6 | .6 |
| CAT | 19 ± 9 | 17 ± 8 | .4 | 22 ± 7 | 22 ± 6 | 1 |
| Exacerbation rate, 12 mo prior | 0.8 (0‐3) | 0.7 (0‐3) | .6 | 0.8 (0‐4) | 1.6 (0‐4) | .04 |
| Blood Eosinophils | 10 ± 3 | 432 ± 144 | <.0001 | 9 ± 3 | 350 ± 240 | <.0001 |
| Sputum Cell Counts | ||||||
| Total (×106/g) | 5.6 (0.1‐26.3) | 5.9 (0.5‐32.4) | .9 | 1.9 (0.2‐11.4) | 2.4 (0.8‐12.6) | .2 |
| Neutrophil (×106/g) | 3.2 (0.1‐20.8) | 3.6 (0.2‐29.6) | .9 | 3.9 (0.5‐24.4) | 5.4 (0.4‐16.6) | .8 |
| Macrophage (×106/g) | 1.3 (0.01‐4.6) | 1.2 (0.2‐7.7) | .6 | 1.4 (0.3‐3.5) | 1.3 (0.4‐4.5) | .5 |
| Eosinophil (×106/g) | 0.02 (0.0‐0.2) | 0.3 (0.01‐4.2) | <.001 | 0.04 (0.00 ‐ 0.4) | 0.3 (0.1‐1.3) | <.001 |
| Lymphocyte (×106/g) | 0.02 (0.0‐0.1) | 0.02 (0.0‐0.2) | .4 | 0.04 (0.0‐0.6) | 0.01 (0.0‐0.2) | .4 |
| Epithelial (×106/g) | 0.04 (0.0‐0.7) | 0.03 (0.0‐1.0) | .7 | 0.1 (0.02‐0.6) | 0.2 (0.04‐1.6) | .07 |
| Eosinophils (%) | 0.5 (0‐2.5) | 4.5 (0.3‐70) | <.0001 | 0.5 (0‐2) | 6.5 (2.8‐16.5) | <.0001 |
| Neutrophils (%) | 81 (17‐91) | 63 (15‐91) | .06 | 67 ± 20 | 65 ± 18 | .8 |
| Macrophages (%) | 17 (5‐82) | 23 (4‐63) | .6 | 30 ± 19 | 23 ± 15 | .3 |
| Lymphocytes (%) | 0.5 (0‐2.3) | 0.3 (0‐2.5) | .3 | 0.3 (0‐2.8) | 0.3 (0‐3.5) | .8 |
| Epithelial (%) | 1 (0‐8) | 1 (0‐8) | .85 | 2 (0.3‐10) | 3 (0.3‐17) | .08 |
Data presented as %, mean ± standard deviation, or median (range).
Abbreviations: BMI, body mass index; ICS, inhaled corticosteroids; FEV1, forced expiratory volume in 1 s; FVC, forced vital capacity; FeNO50, fractional exhaled nitric oxide at 50 mL−1 s flow rate; PPM, parts per million; SGRQ, St George's Respiratory Questionnaire; mMRC, modified Medical Research Council; CAT, COPD Assessment Test.
In the bronchoscopy cohort, bronchial epithelial gene expression of CLCA1, CCL26, IL‐13 and CST1 was significantly higher (P < .05) in eosinophilhigh versus eosinophillow COPD patients (Figure 1) with fold change differences of 1.9, 2.2, 2.9 and 2.5, respectively. POSTN and SERPINB2 expression trended higher in eosinophilhigh versus eosinophillow COPD patients but did not achieve statistical significance (P = .06 and P = .07, respectively; Figure S1).
FIGURE 1.
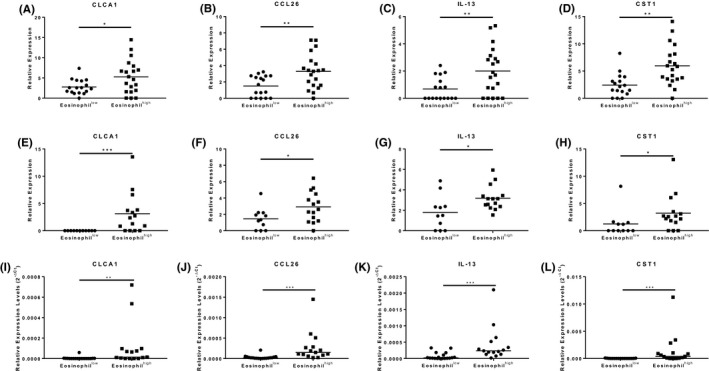
T2 gene expression in bronchial brushings and sputum from the bronchoscopy and validation cohorts. RNA sequencing was used to examine the expression of CLCA1 (A, E, and I), CCL26 (B, F and J), IL‐13 (C, G and K), CST1 (D, H and L), in RNA isolated from bronchial brushings (A‐D) and sputum cells (E‐H) from the bronchoscopy cohort and sputum cells from the validation cohort (I‐L). Data presented as individual data points with mean (A‐H) or median (I‐L) where *, ** and *** = significant difference (P < .05, P < .01 and P < .001, respectively). For A‐H, gene expression was normalized to the transcript length for each gene. For I‐L, gene expression was normalized to GAPDH
In the bronchoscopy and validation cohorts, sputum gene expression of CLCA1, CCL26, IL‐13 and CST1 was significantly higher (P < .05) in eosinophilhigh versus eosinophillow COPD patients (Figure 1); fold change differences were 3.1, 2.0, 1.8 and 3.5 for the bronchoscopy cohort and 28.2, 9.4, 6.0 and 79.3 for the validation cohort, respectively. Expression of POSTN and SERPINB2 was not different between groups (Figure S1). ICS use and smoking status did not influence gene expression (data not shown). There was a significant positive correlation between a four‐gene mean and sputum eosinophil counts in both cohorts (rho 0.6 to 0.7 and P < .001; Figure S1), but not other cell types (data not shown).
We report that eosinophilic COPD is associated with a profile of T2 inflammation in both bronchial epithelium and sputum samples; this was consistently observed in sputum samples from two independent cohorts. We tested six genes associated with T2 inflammation in asthma and observed increased expression of four of these genes (CLCA1, CCL26, IL‐13 and CST1) in eosinophilhigh compared to eosinophillow COPD patients. The strengths of this study include a validation cohort, and different lung samples (bronchial epithelium and sputum). The positive signals were > 1.8‐fold higher (gene expression) in the eosinophilhigh group. Different techniques (RNA sequencing and RT‐qPCR) were used to analyse sputum samples, but the conclusions and relative fold changes showed the same pattern, adding confidence to our observations. The association between eosinophils and IL‐13 expression levels suggests pathways linking eosinophilic inflammation to airway remodelling and mucus secretion, 6 discussed further in the Appendix S1.
These results indicate a wider profile of T2 inflammation in eosinophilic COPD that extends to mechanisms including IL‐13 driven pathways. While T2 inflammation is well recognized in asthma, we now provide further evidence of T2 inflammation in a subset of COPD patients. The components of T2 inflammation described here may represent therapeutic targets.
CONFLICT OF INTEREST
AH has received personal fees from Chiesi. AB, SW, NJ, GL, UK and TS have no conflicts of interest. T‐HP, SS, CM and PN are employees of AstraZeneca. DS has received personal fess from AstraZeneca, Boehringer Ingelheim, Chiesi, Cipla, GlaxoSmithKline, Glenmark, Menarini, Mundipharma, Novartis, Peptinnovate, Pfizer, Pulmatrix, Therevance and Verona.
Supporting information
Appendix S1
ACKNOWLEDGEMENTS
This research was co‐funded by the NIHR Manchester Biomedical Research Centre and the North West Lung Centre Charity, Manchester. This report is independent research, and the views expressed in this publication are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health. Data included in this study were obtained from samples collected from a previous bronchoscopy study funded by Medimmune.
REFERENCES
- 1. Kolsum U, Damera G, Pham TH, et al. Pulmonary inflammation in patients with chronic obstructive pulmonary disease with higher blood eosinophil counts. J Allergy Clin Immunol. 2017;140(4):1181‐1184. [DOI] [PubMed] [Google Scholar]
- 2. Singh D, Bafadhel M, Brightling CE, et al. Blood Eosinophil Counts in Clinical Trials for Chronic Obstructive Pulmonary Disease. Am J Respir Crit Care Med. 2020;202(5):660‐671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Christenson SA, Steiling K, van den Berge M, et al. Asthma‐COPD overlap. Clinical relevance of genomic signatures of type 2 inflammation in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2015;191(7):758‐766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. George L, Taylor AR, Esteve‐Codina A, et al. Blood eosinophil count and airway epithelial transcriptome relationships in COPD versus asthma. Allergy. 2020;75(2):370‐380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Woodruff PG, Modrek B, Choy DF, et al. T‐helper type 2‐driven inflammation defines major subphenotypes of asthma. Am J Respir Crit Care Med. 2009;180(5):388‐395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Atherton HC, Jones G, Danahay H. IL‐13‐induced changes in the goblet cell density of human bronchial epithelial cell cultures: MAP kinase and phosphatidylinositol 3‐kinase regulation. Am J Physiol Lung Cell Mol Physiol. 2003;285(3):L730‐L739. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1


