Abstract
Background
Oral food challenges have demonstrated that diagnosis of almond allergy based on extract‐sIgE tests displays low specificity. Molecular allergy diagnosis is expected to improve accuracy, but its value in diagnosing almond allergy remains unknown. The aim of this study was to identify relevant almond allergens and examine their ability to improve almond allergy diagnosis.
Methods
IgE‐reactive proteins were purified from almond kernels. IgE binding to almond extract and the allergens was analyzed by quantitative ELISA using sera from 18 subjects with a proven almond allergy. The control group consisted of sera from 18 subjects allergic to peanut and/or tree nuts but tolerant to almond.
Results
Three IgE‐binding proteins were identified: legumin (Pru du 6), alpha‐hairpinin (Pru du 8), and mandelonitrile lyase (Pru du 10). Positive IgE (≥0.35 kU/L) to almond extract showed 94% sensitivity but only 33% specificity. IgE to Pru du 6 maintained high sensitivity (83%) and provided superior specificity (78%). Sera from almond‐allergic subjects had significantly higher IgE levels to almond extract (P < .0001) and Pru du 6 (P < .0001) than sera from tolerant donors. Sensitization to Pru du 6 was highly specific for almond allergy, while frequencies of sensitization to legumins from peanut, walnut, hazelnut, and cashew were similar in both groups. IgE to Pru du 8 and Pru du 10 was less sensitive (41% and 67%), but showed specificities of 100% and 61%.
Conclusion
The use of almond allergens markedly increases the diagnostic specificity compared to the extract. Pru du 6 is a potential new molecular marker for almond allergy.
Keywords: almond allergens, diagnostic specificity, food allergy, molecular allergy diagnosis, tree nut allergy
Three allergens were identified in almond kernels: Pru du 6, Pru du 8, and a new allergen, Pru du 10, belonging to a novel family of allergenic proteins. Sensitization profiles of almond‐allergic individuals were dominated by almond‐specific allergens, particularly Pru du 6. The use of almond allergens for IgE tests markedly increased the diagnostic specificity compared to the extract.
Abbreviations: FPLC, fast protein liquid chromatography; LC‐MS/MS: liquid chromatography tandem mass spectrometry; sIgE: specific IgE

Abbreviations
- FPLC
fast protein liquid chromatography
- LC‐MS/MS
liquid chromatography‐tandem mass spectrometry
- sIgE
specific IgE
1. INTRODUCTION
Almond, along with walnut, hazelnut, and cashew, is considered an important cause of tree nut allergy in children and adults. Recent survey data in the United States reported almond and cashew with prevalences of 0.7% each as the most common tree nut allergies in children and adults. 1 , 2 Australian studies reported almond allergy prevalences of 0.2% in adolescents 3 and 0.3% in children. 4 The proportion of patients with peanut allergy who have concurrent almond sensitization is high with rates from 45% to 71%, though only up to 11% reported allergic symptoms. 5 , 6 , 7 Almond sensitization among those with tree nut allergy was 54% and 44%, yet the prevalence of food‐challenge‐defined almond allergy was ≤2%. 8 , 9
The main challenge in managing patients with suspected allergies to peanut and tree nuts is to distinguish between concurrent allergy and asymptomatic co‐sensitization due to multitude of food sources to be avoided. Current data from oral food challenges (OFCs) have revealed that routine almond allergy diagnosis based on clinical history confirmed by extract‐based tests such as skin prick test (SPT) and serum‐specific IgE (sIgE) test is poor in predicting almond allergy. 10 , 11 From 590 patients who were referred for an OFC for suspected almond allergy, 97% were sensitized to almond extract, but only 5% failed almond challenge. 10 Similarly, of 400 patients who underwent almond‐OFCs, 96% were sensitized to almond, yet 4% failed the challenge. 11 In both cohorts, neither almond sensitization nor prior history of almond reaction were predictive of challenge failure. Consequently, OFC remains the diagnostic standard for almond allergy to prevent unnecessary elimination diets.
In recent years, molecular allergy diagnosis using single allergens to quantify sIgE levels in serum has been shown to be a valuable tool to elucidate distinct sensitization profiles and more accurately reflect clinical reactivity, avoiding the need for a challenge. Allergens, such as Ara h 2, 12 Jug r 1, 13 Cor a 14, 14 and Ana o 3, 15 were described as superior in predicting allergic reactions to peanut, walnut, hazelnut, and cashew, compared to the extracts. In contrast to peanut and other tree nuts, the value of almond allergens in diagnosing almond allergy is unknown. Prior to this study, there were five almond allergens registered in the WHO/IUIS database of allergens: Pru du 3 (nonspecific lipid‐transfer protein), 16 Pru du 4 (profilin), 17 Pru du 5 (60S ribosomal protein), 18 Pru du 6 (legumin), 19 , 20 and Pru du 8 (α‐hairpinin antimicrobial protein). 21 Pru du 6, also known as amandin, is a highly abundant storage protein which is frequently recognized by IgE from almond sensitized patients. 19 Almond profilin Pru du 4 seems to be relevant for patients with grass pollinosis. 17 The other three allergens have been identified by cDNA cloning from immature almonds but their presence in mature almond kernels was not investigated. 18 , 22
The aim of this study was to (a) identify and purify IgE‐binding proteins from almond kernels, (b) establish the allergen sensitization profiles of patients with proven almond allergy, and (c) investigate the ability of individual almond allergens to discriminate between almond‐allergic and almond‐tolerant patients with peanut and other tree nuts allergies.
2. MATERIALS AND METHODS
A more detailed version of the Methods section can be found in the supplement.
2.1. Patient's sera
Eighteen patients with almond allergy were included in the study: 14 sera were collected from patients with a positive double‐blind, placebo‐controlled food challenge (DBPCFC) to almond and four from patients with a convincing history of almond allergy. DBPCFCs were performed at the Sean N. Parker Center for Allergy and Asthma Research at Stanford University using standardized methodology according to validated guidelines 23 , 24 , 25 and as previously described. 26 The inclusion criteria for DBPCFC were suspected almond allergy, defined as an sIgE ≥ 0.35 kU/L and/or a positive skin prick test (SPT) to almond. The blood was drawn before the challenge. A challenge was considered positive if objective symptoms occurred within 4 hours of exposure. Inclusion criteria for four almond‐allergic subjects enrolled at the Department of Dermatology, University Hospital St. Poelten, were a history of an allergic reaction after ingestion of almond and sIgE or positive SPT to almond.
To compare allergen recognition profiles between almond‐allergic and almond‐tolerant patients and to analyze the diagnostic specificity of the almond allergens, a control group consisting of 18 subjects was included. In the tolerant group, peanut and/or nut allergic patients were included who reported regular consumption of almonds without any allergic symptoms. The inclusion criteria were a clinically proven history of a peanut and/or tree nut allergy but tolerance of almond and sIgE to peanut and/or tree nuts (Tables 2 and S1). An additional negative control group consisted of four atopic subjects with no history of food allergy and negative sIgE tests to food allergens. Allergen‐sIgE levels were measured by ImmunoCAP (Thermo Fisher Scientific) or ALEX (Macro Array Diagnostics GmbH). The use of clinical data and serum samples for this study was approved by the local ethics committees (Stanford Institutional Review Board, the Ethics Committee of Lower Austria, and the Ethics Committee of the Medical University of Vienna), and signed informed consent was obtained from all patients.
TABLE 2.
Almond‐tolerant (19‐36) and atopic (37‐40) patients: clinical characteristics and IgE sensitization pattern as determined by quantitative ELISA
| Pat No. | Almond allergy a | Sex/Age | Symptoms to almond | Other food allergies | sIgE (kU/L) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Extract | Pru du 6 | Pru du 8 | Pru du 10 | Bet v 1 b | Phl p 12 b | Pru p 3 b | |||||
| 19 | No | F/31 | None | Peanut, hazelnut, walnut, poppy seed | 4.2 | ≤0.35 | ≤0.35 | ≤0.35 | 17.1 | ≤0.35 | ≤0.35 |
| 20 | No | F/22 | None | Peanut, hazelnut, lupine, peach, kiwi | 1.4 | 0.9 | ≤0.35 | 3.3 | ≤0.35 | ≤0.35 | ≤0.35 |
| 21 | No | M/7 | None | Peanut, hazelnut, walnut, milk | 3.2 | 11.3 | 0.41 | 1.8 | 21.3 | ≤0.35 | ≤0.35 |
| 22 | No | M/18 | None | Peanut | ≤0.35 | 1.0 | ≤0.35 | ≤0.35 | 4.3 | ≤0.35 | ≤0.35 |
| 23 | No | M/17 | None | Hazelnut, walnut, milk, egg | ≤0.35 | ≤0.35 | ≤0.35 | ≤0.35 | 25.4 | ≤0.35 | ≤0.35 |
| 24 | No | M/20 | None | Hazelnut, macadamia | 0.4 | ≤0.35 | ≤0.35 | 0.6 | ≤0.35 | ≤0.35 | ≤0.35 |
| 25 | No | F/27 | None | Peanut, walnut | 2.0 | 1.6 | ≤0.35 | 0.9 | 6.0 | ≤0.35 | ≤0.35 |
| 26 | No | F/9 | None | Peanut, hazelnut, walnut, cashew | 4.1 | ≤0.35 | ≤0.35 | 1.4 | ≤0.35 | ≤0.35 | ≤0.35 |
| 27 | No | F/37 | None | Hazelnut walnut, | 0.8 | ≤0.35 | ≤0.35 | 0.7 | 4.4 | ≤0.35 | 16.3 |
| 28 | No | M/28 | None | Peanut | 0.6 | ≤0.35 | ≤0.35 | ≤0.35 | 3.1 | ≤0.35 | ≤0.35 |
| 29 | No | F/31 | None | Peanut, hazelnut, walnut | ≤0.35 | ≤0.35 | ≤0.35 | ≤0.35 | 7.9 | ≤0.35 | ≤0.35 |
| 30 | No | M/10 | None | Peanut, apple | ≤0.35 | ≤0.35 | ≤0.35 | 0.5 | 6.7 | ≤0.35 | ≤0.35 |
| 31 | No | F/26 | None | Peanut, hazelnut, walnut | 0.8 | ≤0.35 | ≤0.35 | ≤0.35 | 2.0 | ≤0.35 | ≤0.35 |
| 32 | No | F/21 | None | Peanut, hazelnut, walnut | ≤0.35 | ≤0.35 | ≤0.35 | ≤0.35 | 15.3 | ≤0.35 | ≤0.35 |
| 33 | No | F/42 | None | Walnut | 0.45 | ≤0.35 | ≤0.35 | ≤0.35 | ≤0.35 | ≤0.35 | 3.4 |
| 34 | No | M/18 | None | Peanut | ≤0.35 | ≤0.35 | ≤0.35 | ≤0.35 | 0.9 | ≤0.35 | ≤0.35 |
| 35 | No | F/48 | None | Hazelnut, walnut | 0.4 | ≤0.35 | ≤0.35 | ≤0.35 | ≤0.35 | ≤0.35 | ≤0.35 |
| 36 | No | F/30 | None | Peanut, hazelnut | 0.5 | ≤0.35 | ≤0.35 | ≤0.35 | 0.5 | ≤0.35 | ≤0.35 |
| 37 | No | F/48 | None | None | ≤0.35 | ≤0.35 | ≤0.35 | ≤0.35 | 0.6 | ≤0.35 | ≤0.35 |
| 38 | No | M/29 | None | None | ≤0.35 | ≤0.35 | ≤0.35 | ≤0.35 | 25.5 | ≤0.35 | ≤0.35 |
| 39 | No | M/32 | None | None | ≤0.35 | ≤0.35 | ≤0.35 | ≤0.35 | 2.1 | ≤0.35 | ≤0.35 |
| 40 | No | M/50 | None | None | ≤0.35 | ≤0.35 | ≤0.35 | ≤0.35 | ≤0.35 | ≤0.35 | ≤0.35 |
Based on a detailed questionnaire, all subjects consumed almonds regularly without allergic symptoms.
IgE specific to Bet v 1, Phl p 12, and Pru p 3 was measured as representatives of an almond Bet v 1‐homologue, almond profilin, and almond nsLTP, respectively.
2.2. Purification and characterization of almond allergens
IgE‐binding proteins were purified from almonds using different protein extraction conditions and diverse chromatographic techniques. Protein identification was performed by Nano‐LC ESI‐Orbitrap MS/MS and N‐terminal sequencing. Purity of the proteins was analyzed by SDS‐PAGE, 2D electrophoresis, and Coomassie Brilliant Blue staining. Secondary structure content was determined by CD‐spectroscopy. To demonstrate IgE binding, immunoblotting and quantitative ELISA were performed. The detailed protocols are described in the supplement.
2.3. Statistical analysis
The Mann‐Whitney U test was used for comparing allergen‐specific IgE levels of patients with and without almond allergy. Chi‐square test was applied to assess statistically significant differences between the percentages of sera from the two groups reacting with individual allergens or extracts. Receiver operating characteristics (ROC) curves were generated to evaluate the diagnostic capacity of the test for specific IgE levels. An optimal cutoff point for sIgE to the tested allergens was obtained using the Youden index (sensitivity + specificity − 1). Paired correlation was calculated by using the Pearson test. P‐values < .05 were considered significant.
3. RESULTS
3.1. Patients' characteristics
Sera from 18 patients with clinically relevant allergic reactions following ingestion of almonds were included in this study (Table 1). The most common symptoms elicited by almond were urticaria (n = 10), rash (n = 4), and angioedema (n = 6). Most of the patients (56%) had a history of immediate reaction to peanut, 44% to cashew, 39% to hazelnut, and 44% to walnut. As control group, 18 sera from patients with allergy to at least one tree nut or peanut were used (Table 2). At the time of inclusion, all control patients indicated to consume almond regularly without allergic symptoms. Similar to the almond‐allergic group, most of the patients had peanut (72%) and hazelnut allergy (67%) followed by walnut allergy (61%). The total IgE levels and sIgE levels to peanut, hazelnut, walnut, Brazil nut, and macadamia did not differ significantly between the two groups (Table S1).
TABLE 1.
Almond‐allergic (1‐18) patients: clinical characteristics and IgE sensitization pattern as determined by quantitative ELISA
| Pat No. | Almond allergy confirmed by | Sex/Age | Symptoms to almond | Other food allergies | sIgE (kU/L) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Extract | Pru du 6 | Pru du 8 | Pru du 10 | Bet v 1 | Phl p 12 a | Pru p 3 a | |||||
| 1 | DBPCFC | M/7 | Pruritus, rash, angioedema, nasal congestion | Peanut, cashew, egg, milk | 7.1 | 6.7 | ≤0.35 | ≤0.35 | 0.6 | 0 | ≤0.35 |
| 2 | DBPCFC | F/12 | Urticaria | Hazelnut | 183.8 | 148.1 | 13.4 | 75.7 | 25.5 | 0 | ≤0.35 |
| 3 | DBPCFC | M/8 | Rash | Peanut, cashew, hazelnut, walnut | 125.9 | 69.4 | 159.1 | 33.1 | 2.1 | 0.7 | 3.2 |
| 4 | DBPCFC | F/4 | Rash | Peanut, milk, wheat | 7.5 | 11.5 | ≤0.35 | 0.9 | ≤0.35 | ≤0.35 | ≤0.35 |
| 5 | DBPCFC | F/5 | Angioedema | Peanut, egg, walnut | 6.3 | 5.9 | ≤0.35 | ≤0.35 | 1.0 | ≤0.35 | ≤0.35 |
| 6 | DBPCFC | F/10 | Urticaria | Cashew, hazelnut, walnut | 0.4 | ≤0.35 | ≤0.35 | ≤0.35 | 21.8 | ≤0.35 | ≤0.35 |
| 7 | DBPCFC | M/6 | Urticaria, rash | Peanut, cashew, hazelnut, walnut | 8.0 | 6.5 | ≤0.35 | 1.0 | ≤0.35 | 1.7 | 1.1 |
| 8 | DBPCFC | F/11 | Urticaria | Peanut, cashew, hazelnut, walnut | 317.2 | 181.7 | 44.5 | 24.2 | ≤0.35 | ≤0.35 | ≤0.35 |
| 9 | DBPCFC | F/7 | Urticaria | Egg | 2.7 | 2.0 | 1.3 | ≤0.35 | ≤0.35 | ≤0.35 | ≤0.35 |
| 10 | DBPCFC | F/12 | Urticaria | Cashew, hazelnut, walnut | 12.6 | 3.6 | ≤0.35 | 14.4 | 14.2 | ≤0.35 | ≤0.35 |
| 11 | DBPCFC | F/9 | Throat tightness, abdominal pain | None | 27.4 | 8.7 | 1.5 | 4.0 | 3.6 | ≤0.35 | ≤0.35 |
| 12 | DBPCFC | M/12 | Urticaria, angioedema | None | 2.2 | 1.1 | 1.3 | ≤0.35 | 6.0 | 1.5 | ≤0.35 |
| 13 | DBPCFC | M/15 | Urticaria | None | 21.1 | 6.0 | 54.1 | 2.0 | 33.4 | ≤0.35 | ≤0.35 |
| 14 | DBPCFC | M/9 | Urticaria | None | 73.7 | 81.1 | 0.5 | 95.1 | 18.7 | ≤0.35 | ≤0.35 |
| 15 | positive history | Angioedema | Peanut, hazelnut, walnut, cashew, pecan, | 38.2 | 8.7 | ≤0.35 | 0.9 | ≤0.35 | ≤0.35 | 9.6 | |
| 16 | positive history | M/4 | Urticaria | Peanut, cashew, pistachio | 19.3 | 22.9 | ≤0.35 | 3.3 | 7.0 | ≤0.35 | ≤0.35 |
| 17 | positive history | F/18 | Angioedema, oropharyngeal pruritus and mild erythema | Peanut | 1.6 | ≤0.35 | ≤0.35 | 0.4 | ≤0.35 | 3.1 | 3.4 |
| 18 | positive history | F/48 | Angioedema, oropharyngeal pruritus and mild erythema | Peanut, walnut | ≤0.35 | ≤0.35 | ≤0.35 | ≤0.35 | 2.6 | ≤0.35 | 2.1 |
IgE specific to Bet v 1, Phl p 12, and Pru p 3 was measured as representatives of an almond Bet v 1‐homologue, almond profilin, and almond nsLTP, respectively.
3.2. Identification of novel almond allergens
SDS‐PAGE analysis of almond extract showed multiple protein bands in the range of 10‐100 kDa (Figure 1, lanes 1‐2). Immunoblotting of the extract with a pool of sera from almond‐allergic patients (Table 1, sera 4, 5, 10 and 16) showed several IgE‐binding signals in this range (Figure 2A, lanes 1‐2). After purification, IgE‐binding proteins were identified as Pru du 6, Pru du 8, and mandelonitrile lyase 2, a novel plant food allergen (Figure 1, lanes 3‐8 and Table S2). The complete inhibition of IgE binding after preincubation of the serum pool with the mixture of the three proteins indicated that the IgE‐binding bands in the extracts represent the isolated proteins (Figure 2A, lane 3). Pru du 6 migrates as a band of around 60 kDa and dissociates under reducing conditions into two subunits of 20 and 40 kDa (Figure 1, lanes 3–4). As shown in 2D‐PAGE (Figure S1), Pru du 6 occurs in different processed forms indicated by multiple spots of similar molecular masses but varying isoelectric points (pIs). Pru du 8 migrates at 13 kDa under reducing and at 26 kDa under nonreducing conditions (Figure 1, lanes 5‐6 and Figure S1) indicating dimer formation. The newly identified mandelonitrile lyase 2 migrates at about 60 kDa and a pI of 5.5 (Figure 1, lanes 7‐8 and Figure S1). This allergen was designated Pru du 10 by the WHO/IUIS Allergen Nomenclature Sub‐Committee.
FIGURE 1.
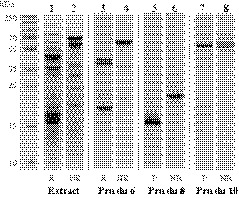
Coomassie‐stained 15% SDS‐PAGE of almond protein extract and purified allergens separated under reducing (R) and nonreducing (NR) conditions
FIGURE 2.
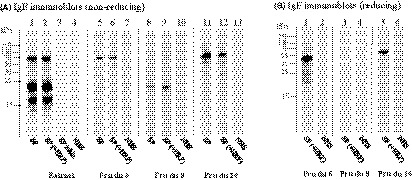
IgE immunoblots of almond extract and allergens under reducing and nonreducing conditions. IgE reactivity of pooled sera (SP) from 4 almond‐allergic patients (sera: 4, 5, 10, and 16). To inhibit CCD‐specific antibodies, serum pool was preincubated with HRP (+HRP). IgE inhibition of the serum pool (SP inhib.) was performed using a mix of Pru du 6, Pru du 8, and Pru du 10. As a negative control, a pool of 2 sera (NHS pool) from atopic subjects (sera: 43 and 44) was used
The three proteins showed high purity (>95%) and no cross‐contamination with other allergens, as determined by high‐accurate tandem mass spectrometry (Table S2). A clear IgE reactivity to Pru du 6 and Pru du 10 was observed under nonreducing (Figure 2A, lines 5‐6 and 11‐12) as well as reducing conditions (Figure 2B, lines 1 and 5). However, IgE reactivity to Pru du 8 was only observed under nonreducing conditions (Figure 2A, line 8 and Figure 2B, line 3). No considerable reduction of IgE binding to Pru du 10, a glycoprotein, was observed after inhibition of IgE antibodies specific to CCDs using HRP, indicating involvement of protein residues in the IgE binding (Figure 2A, lines 11‐12). Similarly, IgE binding to the other allergens was not altered upon preincubation of sera with HRP (Figure 2A,B).
Changing protein extraction conditions such as the use of different buffers, pH values, salt concentration, and water‐alcohol ratios did not result in detection of other IgE‐binding proteins (eg, vicilins and 2S albumins), typically present in other tree nuts.
3.3. Sensitization profiles to individual almond allergens differ between almond‐allergic and almond‐tolerant subjects
IgE reactivity to almond allergens differed substantially between the almond‐allergic and almond‐tolerant group (Tables 1 and 2, Figure 3 and Table S1). Almond‐allergic patients had significantly more frequently (P < .001) IgE specific to Pru du 6 than patients who tolerate almond. Sixteen of 18 (89%) sera from almond‐allergic subjects had specific IgE levels of ≥0.35 kUA/L, while only 22% (4 of 18) of sera of the control group were positive to the allergen. Rates of IgE binding to other allergenic legumins from peanut (Ara h 3), walnut (Jug r 4), hazelnut (Cor a 9), and cashew (Ana o 2) were similar among almond‐allergic and asymptomatic patients indicating that IgE binding to Pru du 6 was specific for almond allergy (Figure 4).
FIGURE 3.
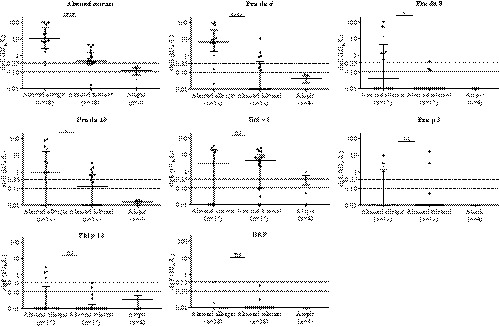
Specific IgE concentrations (kUA/L) to almond extract and individual purified allergens in sera from almond‐allergic, almond‐tolerant, and other atopic subjects. Horizontal bars indicate medians ± interquartile ranges. IgE values ≤ 0.01 kUA/L were set to 0.01 kUA/L. Dotted lines indicate the 0.10 and 0.35 kUA/L cutoff levels. Differences between the patient groups were analyzed by nonparametric Mann‐Whitney U test. *P ≤ .05, ***P ≤ .001, ****P ≤ .0001
FIGURE 4.
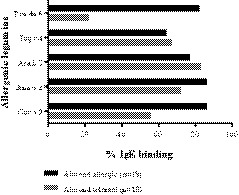
Percentages of sensitization of almond‐allergic and almond‐tolerant subjects to Pru du 6 homologous from walnut (Jug r 4), peanut (Ara h 3), cashew (Ana o 2), and hazelnut (Cor a 9)
Sensitization to Pru du 8 was observed in 44% of the almond‐allergic patients and to the newly identified allergen Pru du 10 in 67%. Only one serum from the control group reacted with Pru du 8, and 7 sera with Pru du 10. Sixteen almond‐allergic patients (89%) were sensitized to at least one almond allergen and six (33%) to all three allergens. The two sera negative to the three almond allergens were positive to Bet v 1 and the Bet v 1‐homologous allergens Cor a 1.04, Mal d 1, and Gly m 4. One of these patients also had IgE specific to Pru p 3, as a representative of almond nsLTP (Tables 1 and 2). The IgE‐binding frequency to Bet v 1 was 67% and in the control group 72%. Sensitization to Phl p 12, as a representative of almond profilin, was observed in 28% of almond‐allergic and 6% of almond‐tolerant subjects. No serum from the atopic control subjects reacted with almond allergens.
There was a strong correlation (R 2 = .9) between the IgE reactivity to almond extract and sIgE reactivity to the three almond allergens, expressed as the sum of sIgE values of the three individual allergens (Figure S2), indicating that IgE to these allergens represents the majority of almond‐specific IgE.
When specific IgE antibody levels to individual allergens were investigated, sera from almond‐allergic subjects had significantly higher IgE specific to Pru du 6 (median, 8.7 kUA/L; range, 1.1‐181.7, P < .0001), Pru du 8 (median, 7.4 kUA/L; range, 0.5‐159.0 kUA/L, P = .013), and Pru du 10 (median, 3.6 kUA/L; range, 0.4‐95.1, P = .031) compared to the tolerant group (Figure 3 and Table S1). The median levels of sIgE to Bet v 1 and Pru p 3 were similar between reactive and nonreactive patients. IgE specific to cashew extract (median, 17.3 kUA/L, P = .0030) and Ana o 3 (median, 20.9 kUA/L; P = .0073) was significantly higher in the group of allergic patients owing to the fact that most patients had co‐existing cashew allergy. There was no difference in the quantity of IgE specific to other extracts or allergens listed in Table S1.
3.4. Using single almond allergens increases diagnostic specificity
Seventeen (94%) of 18 patients with almond allergy and 12 (67%) of 18 almond‐tolerant patients had sIgE ≥0.35 kUA/L to almond extract. The median level of sIgE to almond extract (12.6 kUA/L; range, 0.4‐317.2 kUA/L, P < .0001) was significantly higher in sera from patients allergic to almond compared with tolerant patients (Tables 1, 2 and Table S1). Positive sIgE to almond extract was highly sensitive (94%), but not specific (33%). At the same threshold level, Pru du 6 maintained high sensitivity (83%), but provided superior specificity (78%). Pru du 8 and Pru du 10 were less sensitive (44% and 67%), but with specificities of 100% and 61%. The cutoff almond‐sIgE level that predicted clinical reactivity with the highest sum of sensitivity and specificity was 5.2 kUA/L, with a sensitivity of 72% and a specificity of 100% (Table 3). The optimal cutoff point of IgE to Pru du 6 was 1.8 kUA/L, with sensitivity and specificity of 77% and 94%, respectively. The calculated optimal cutoff point for Pru du 8 was 0.35 kUA/L with a sensitivity of 44% and specificity of 100%, and for Pru du 10 0.75 kUA/L with a sensitivity of 61% and specificity of 78% (Table 3). Based on the calculations of areas under the ROC curves, sIgE to almond extract and Pru du 6 had the highest accuracy in discriminating between allergic and tolerant individuals: area (95% CI), almond extract (0.89), Pru du 6 (0.87), Pru du 10 (0.71), and Pru du 8 (0.70) (Figure 5).
TABLE 3.
Sensitivity and specificity of different cutoff sIgE levels for almond extract, Pru du 6, Pru du 8, and Pru du 10
| Test cutoff (kUA/L) | Sensitivity (%) | Specificity (%) | Youden index |
|---|---|---|---|
| Almond sIgE | |||
| ≥0.1 | 100 | 11 | 0.11 |
| ≥0.35 a | 94 | 33 | 0.28 |
| ≥5.2 b | 72 | 100 | 0.72 |
| Pru du 6 sIgE | |||
| ≥0.1 | 89 | 56 | 0.50 |
| ≥0.35 a | 83 | 78 | 0.61 |
| ≥1.8 b | 77 | 94 | 0.72 |
| Pru du 8 sIgE | |||
| ≥0.1 | 45 | 83 | 0.29 |
| ≥0.35 a , b | 44 | 100 | 0.44 |
| Pru du 10 sIgE | |||
| ≥0.1 | 72 | 50 | 0.22 |
| ≥0.35 a | 67 | 61 | 0.28 |
| ≥0.75 b | 61 | 78 | 0.38 |
Clinically used cutoff value.
Calculated optimal cutoff points.
FIGURE 5.
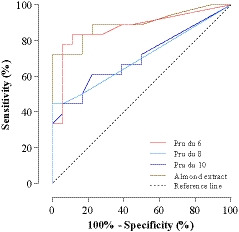
Receiver operating characteristic curves for sIgE to almond extract, Pru du 6, Pru du 8, and Pru du 10 with regard to discrimination between almond allergy and tolerance of almond
4. DISCUSSION
Recent studies have demonstrated that diagnosis of almond allergy by extract‐based tests poorly differentiate between sensitized and allergic individuals. Thus, the diagnosis is heavily reliant on OFCs. Molecular diagnostics, which measure allergen‐specific IgE, offer the potential to improve the diagnostic accuracy, but no information about diagnostic performance of almond allergens is available. In this study, we identified relevant allergens in almond kernels and determined allergen sensitization profiles of almond‐allergic and almond‐tolerant patients. Furthermore, we evaluated for the first time the diagnostic value of molecular diagnosis of almond allergy in particular with regard to specificity compared with that of an extract‐based diagnosis.
Our results showed a difference in the allergen compositions of almond compared with peanut and other tree nuts. Allergies to peanut and tree nuts are typically associated with sensitization to seed storage proteins (2S albumins, legumins, and vicilins) and additionally often to one or several less prominent allergens. With exception of almond legumin (Pru du 6), which was also previously detected as an important allergen, 19 2S albumin and vicilin, typically abundant in other nuts, could not be detected in almond kernels. Although recently a vicilin‐related cDNA was isolated from immature almonds and a recombinant protein produced has been shown to bind IgE, the presence of its natural counterpart in mature almonds was not confirmed. 22 In line with our results, neither a vicilin nor a 2S albumin was identified by mass spectrometric proteome analysis of the almond kernel. 27
Instead, we purified two almond‐specific allergens, Pru du 10 and a 13 kDa IgE‐binding protein which was identified as a C‐terminal fragment of previously reported almond allergen Pru du 8. Recombinant Pru du 8 obtained by cDNA cloning from immature almonds is a 26 kDa protein belonging to the α‐hairpinin family. 28 Our results indicate that this sequence codes for a preproprotein which in mature almond undergoes post‐translational proteolytic processing, as has been shown for a homologous protein from wheat. 28 Interestingly, IgE‐binding peptides from the α‐hairpinin protein family have also been identified in peanut 29 and walnut, 30 but in contrast to almond those peptides originate from the N‐terminal region of vicilin preproproteins.
Another interesting finding of this study is the identification of almond mandelonitrile lyase 2 as a novel allergen, which was designated Pru du 10. Mandelonitrile lyase 2, a key enzyme for a process known as cyanogenesis, is the most studied almond protein. It is a flavoprotein belonging to the glucose‐methanol‐choline oxidoreductase family. Although N‐glycosylation sites were previously identified by crystallography, 31 our results showed that glycan moieties are not involved in IgE reactivity to Pru du 10.
As a next step, we used the panel of purified almond allergens to establish sensitization profiles of 18 almond‐allergic subjects. We showed that 94% of almond‐allergic patients had IgE to at least one of those three allergens and that most patients displayed IgE reactivity to Pru du 6 (83%).
The main finding of our study was that sensitization to almond legumin Pru du 6 was highly specific for almond allergy. Notably, sensitization to other allergenic legumins from peanut, walnut, hazelnut, and cashew was similar in almond‐allergic and almond‐tolerant groups, which may qualify Pru du 6 as a specific biomarker for symptomatic almond allergy. The degree of sequence identity between Pru du 6 and the other legumins is low, ranging from 41% to 51%, 32 but future inhibition experiments will be necessary to evaluate the extent of cross‐reactivity between allergens from almond and other nuts.
Furthermore, we evaluated the diagnostic value of molecular diagnosis of almond allergy with regard to specificity compared with that of an extract‐based diagnosis. Because there is currently no commercially available diagnostic tool, measurement of sIgE to almond was performed using an in‐house developed quantitative ELISA. In line with other studies, 7 , 8 , 9 , 10 , 11 we found a high sensitization frequency to almond extract not only among almond‐allergic, but also almond‐tolerant patients and that almond‐allergic patients had significantly higher median sIgE levels to almond extract. Our data demonstrate that positive sIgE to almond extract was sensitive (94%), but not specific (33%). At the same threshold level, Pru du 6 maintained high sensitivity (83%) but provided superior specificity (78%). These results clearly demonstrate that the use of single almond components increases the diagnostic specificity compared to extract.
This observed improvement is in accordance with other component‐resolved diagnosis studies showing that measurement of specific IgE to allergens from hazelnut (Cor a 14), walnut (Jug r 1), or cashew (Ana o 3) improved specificity compared with whole extracts. 13 , 15 , 33
According to the above‐mentioned studies and current state of knowledge, 2S albumins seem to be most important and potent allergens not only in peanut, 12 but also in hazelnut, walnut, and cashew. Compared to almond allergy, those allergies are common and often severe. One could speculate that the distinct allergen composition of almond, in particular the absence of a 2S albumin, might give rise to usually uncommon and mild reactions to almond.
The limitations of this study are its monocentric retrospective design and the rather small size of the sample consisting mainly of children. Our findings need to be replicated in other populations, different geographic areas, and larger cohorts. For example, in birch endemic areas, almond allergy is often associated with birch pollinosis which may be caused by a cross‐reacting Bet v 1‐homologue. 34 , 35 Although evidence for a Bet v 1‐homologue in almond was provided at the mRNA level, to our knowledge no one has yet confirmed presence of a corresponding protein in almond kernels. In our study, we also could not detect it by immunoblot at the protein level using sera from almond‐allergic patients containing Bet v 1‐specific IgE. Identification of a Bet v 1‐homologue in almond requires further studies.
Further, interpretation of our findings is limited by the fact that the two groups, almond‐allergic and almond‐tolerant subjects, were not comparable by age. The almond‐allergic group consisted mainly of children, while the almond‐tolerant group included mostly adolescents and adults. Although the results of a previous study using sera of adult almond‐allergic patients identified Pru du 6 as a major allergen, 19 validation in adults should be the focus of future studies on almond allergy. However, despite these limitations, as being an exploratory in‐depth analysis of almond allergens and sensitization profiles of almond‐allergic subjects, our results will be of much help for subsequent studies specifically aimed at establishing predictive protocols in even larger cohorts.
In conclusion, we found marked differences in allergen composition of almond compared to other tree nuts. Our study provides the first comprehensive evaluation of the sensitization profiles of almond‐allergic patients using a panel of allergens isolated from almond kernels. We showed that the sensitization profile of almond‐allergic individuals is dominated by sensitization to almond‐specific allergens, particularly Pru du 6, which may potentially assist in effectively discriminating almond‐allergic from tolerant patients.
CONFLICT OF INTEREST
Dr. Chinthrajah reports grants from NIAID, CoFAR, Aimmune, DBV Technologies, Astellas, Regeneron, an Advisory member for Alladapt, Genentech, Novartis, and receives personal fees from Before Brands outside the submitted work. Dr. Nadeau reports grants from National Institute of Allergy and Infectious Diseases (NIAID), grants from National Heart, Lung, and Blood Institute (NHLBI), grants from National Institute of Environmental Health Sciences (NIEHS), grants from Food Allergy Research & Education (FARE), other from World Allergy Organization (WAO), other from Cour Pharma, other from Before Brands, other from Alladapt, other from Latitude, other from IgGenix, other from Immune Tolerance Network (ITN), other from National Institutes of Health (NIH) clinical research centers, outside the submitted work; In addition, Dr. Nadeau has a patent Inhibition of Allergic Reaction to Peanut Allergen using an IL‐33 Inhibitor pending, a patent Special Oral Formula for Decreasing Food Allergy Risk and Treatment for Food Allergy pending, a patent Basophil Activation Based Diagnostic Allergy Test pending, a patent Granulocyte‐based methods for detecting and monitoring immune system disorders pending, a patent Methods and Assays for Detecting and Quantifying Pure Subpopulations of White Blood Cells in Immune System Disorders pending, a patent Mixed Allergen Compositions and Methods for Using the Same pending, and a patent Microfluidic Device and Diagnostic Methods for Allergy Testing Based on Detection of Basophil Activation pending. The rest of the authors declare that they have no relevant conflict of interest related to this study.
AUTHOR CONTRIBUTIONS
SK performed experiments, analyzed data, and prepared the manuscript. MB designed and supervised the study. CH, SC, SBS, DK, LEK, and AJL undertook patient recruitment, data collection and were involved in data analysis. HB, KCN, MB, and CH critically revised the manuscript.
Supporting information
Appendix S1
ACKNOWLEDGMENTS
This work was supported by funds of the Oesterreichische Nationalbank (Austrian Central Bank), Anniversary Fund, project number: 17560. The protein identification was performed using resources of the VetCore Facility (Proteomics) of the University of Veterinary Medicine Vienna.
Kabasser S, Hafner C, Chinthrajah S, et al. Identification of Pru du 6 as a potential marker allergen for almond allergy. Allergy.2021;76:1463–1472. 10.1111/all.14613
REFERENCES
- 1. Gupta RS, Warren CM, Smith BM, et al. The public health impact of parent‐reported childhood food allergies in the United States. Pediatrics. 2018;142:e20181235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gupta RS, Warren CM, Smith BM, et al. Prevalence and severity of food allergies among US adults. JAMA Netw Open. 2019;2:e185630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sasaki M, Koplin JJ, Dharmage SC, et al. Prevalence of clinic‐defined food allergy in early adolescence: the SchoolNuts study. J Allergy Clin Immunol. 2018;141:391‐398. [DOI] [PubMed] [Google Scholar]
- 4. McWilliam V, Peters R, Tang MLK, et al. Patterns of tree nut sensitization and allergy in the first 6 years of life in a population‐based cohort. J Allergy Clin Immunol. 2019;143:644‐650. [DOI] [PubMed] [Google Scholar]
- 5. Uotila R, Kukkonen AK, Blom WM, et al. Component‐resolved diagnostics demonstrates that most peanut‐allergic individuals could potentially introduce tree nuts to their diet. Clin Exp Allergy. 2018;48:712‐721. [DOI] [PubMed] [Google Scholar]
- 6. Maloney JM, Rudengren M, Ahlstedt S, Bock SA, Sampson HA. The use of serum‐specific IgE measurements for the diagnosis of peanut, tree nut, and seed allergy. J Allergy Clin Immunol. 2008;122:145‐151. [DOI] [PubMed] [Google Scholar]
- 7. Mustafa SS, Vadamalai K, Bingemann T, Mortezavi M, Aranez V, Ramsey A. Real‐world tree nut consumption in peanut‐allergic individuals. Ann Allergy, Asthma Immunol. 2020;124:277‐282. [DOI] [PubMed] [Google Scholar]
- 8. Elizur A, Appel MY, Nachshon L, et al. NUT co reactivity ‐ ACquiring knowledge for elimination recommendations (NUT CRACKER) study. Allergy Eur J Allergy Clin Immunol. 2018;73:593‐601. [DOI] [PubMed] [Google Scholar]
- 9. Couch C, Franxman T, Greenhawt M. Characteristics of tree nut challenges in tree nut allergic and tree nut sensitized individuals. Ann Allergy, Asthma Immunol. 2017;118:591‐596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Virkud YV, Chen YC, Stieb ES, et al. Analysis of oral food challenge outcomes in IgE‐mediated food allergies to almond in a large cohort. J Allergy Clin Immunol Pract. 2019;7:2359‐2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Baker MG, Kattan JD. Review of 400 consecutive oral food challenges to almond. Ann Allergy, Asthma Immunol. 2019;122:189‐192. [DOI] [PubMed] [Google Scholar]
- 12. Nicolaou N, Murray C, Belgrave D, Poorafshar M, Simpson A, Custovic A. Quantification of specific IgE to whole peanut extract and peanut components in prediction of peanut allergy. J Allergy Clin Immunol. 2011;127:684‐685. [DOI] [PubMed] [Google Scholar]
- 13. Sato S, Yamamoto M, Yanagida N, et al. Jug r 1 sensitization is important in walnut‐allergic children and youth. J Allergy Clin Immunol Pract. 2017;5:1784‐1786. [DOI] [PubMed] [Google Scholar]
- 14. Masthoff LJN, Mattsson L, Zuidmeer‐Jongejan L, et al. Sensitization to Cor a 9 and Cor a 14 is highly specific for a hazelnut allergy with objective symptoms in Dutch children and adults. J Allergy Clin Immunol. 2013;132(2):393‐399. [DOI] [PubMed] [Google Scholar]
- 15. Lange L, Lasota L, Finger A, et al. Ana o 3‐specific IgE is a good predictor for clinically relevant cashew allergy in children. Allergy Eur J Allergy Clin Immunol. 2017;72:598‐603. [DOI] [PubMed] [Google Scholar]
- 16. Buhler S, Tedeschi T, Faccini A, et al. Isolation and full characterisation of a potentially allergenic lipid transfer protein (LTP) in almond. Food Addit Contam ‐ Part A Chem Anal Control Expo Risk Assess. 2015;32:648‐656. [DOI] [PubMed] [Google Scholar]
- 17. Tawde P, Venkatesh YP, Wang F, Teuber SS, Sathe SK, Roux KH. Cloning and characterization of profilin (Pru du 4), a cross‐reactive almond (Prunus dulcis) allergen. J Allergy Clin Immunol. 2006;118:915‐922. [DOI] [PubMed] [Google Scholar]
- 18. Abolhassani M, Roux KH. cDNA cloning, expression and characterization of an allergenic 60s ribosomal protein of almond (Prunus dulcis). Iran J Allergy, Asthma Immunol. 2009;8:77‐84. [PubMed] [Google Scholar]
- 19. Willison LAN, Tripathi P, Sharma G, Teuber SS, Sathe SK, Roux KH. Cloning, expression and patient IgE reactivity of recombinant Pru du 6, an 11S globulin from almond. Int Arch Allergy Immunol. 2011;156:267‐281. [DOI] [PubMed] [Google Scholar]
- 20. Sathe SK, Wolf WJ, Roux KH, Teuber SS, Venkatachalam M, Sze‐Tao KWC. Biochemical characterization of amandin, the major storage protein in almond (Prunus dulcis L.). J Agric Food Chem. 2002;50:4333‐4341. [DOI] [PubMed] [Google Scholar]
- 21. Che H, Zhang Y, Jiang S, et al. Almond (Prunus dulcis) allergen Pru du 8, the first member of a new family of food allergens. J Agric Food Chem. 2019;67:8626‐8631. [DOI] [PubMed] [Google Scholar]
- 22. Che H, Zhang Y, Lyu SC, Nadeau KC, McHugh T. Identification of almond (Prunus dulcis) vicilin as a food allergen. J Agric Food Chem. 2019;67:425‐432. [DOI] [PubMed] [Google Scholar]
- 23. Allan Bock S, Sampson HA, Atkins FM, et al. Double‐blind, placebo‐controlled food challenge (DBPCFC) as an office procedure: a manual. J Allergy Clin Immunol. 1988;82:986‐997. [DOI] [PubMed] [Google Scholar]
- 24. Boyce JA, Assa'ad A, Burks AW, et al. Guidelines for the diagnosis and management of food allergy in the United States: summary of the NIAID‐sponsored expert panel report. J Allergy Clin Immunol. 2010;126:1105‐1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sampson HA, Aceves S, Bock SA, et al. Food allergy: a practice parameter update ‐ 2014. J Allergy Clin Immunol. 2014;134:1016‐1025. [DOI] [PubMed] [Google Scholar]
- 26. Sindher S, Long AJ, Purington N, et al. Analysis of a large standardized food challenge data set to determine predictors of positive outcome across multiple allergens. Front Immunol. 2018;9:2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li S, Geng F, Wang P, Lu J, Ma M. Proteome analysis of the almond kernel (Prunus dulcis). J Sci Food Agric. 2016;96:3351‐3357. [DOI] [PubMed] [Google Scholar]
- 28. Utkina LL, Andreev YA, Rogozhin EA, et al. Genes encoding 4‐Cys antimicrobial peptides in wheat Triticum kiharae Dorof. et Migush.: multimodular structural organization, instraspecific variability, distribution and role in defence. FEBS J. 2013;280:3594‐3608. [DOI] [PubMed] [Google Scholar]
- 29. Aalberse RC, Mueller GA, Derksen NIL, et al. Identification of the amino‐terminal fragment of Ara h 1 as a major target of the IgE‐binding activity in the basic peanut protein fraction. Clin Exp Allergy. 2020;50:401‐405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Downs ML, Semic‐Jusufagic A, Simpson A, et al. Characterization of low molecular weight allergens from English walnut (Juglans regia). J Agric Food Chem. 2014;62(48):11767‐11775. [DOI] [PubMed] [Google Scholar]
- 31. Dreveny I, Gruber K, Glieder A, Thompson A, Kratky C. The hydroxynitrile lyase from almond: a lyase that looks like an oxidoreductase. Structure. 2001;8:803‐815. [DOI] [PubMed] [Google Scholar]
- 32. Smeekens JM, Bagley K, Kulis M. Tree nut allergies: allergen homology, cross‐reactivity, and implications for therapy. Clin Exp Allergy. 2018;48:762‐772. [DOI] [PubMed] [Google Scholar]
- 33. Buyuktiryaki B, Cavkaytar O, Sahiner UM, et al. Cor a 14, Hazelnut‐specific IgE, and SPT as a reliable tool in hazelnut allergy diagnosis in eastern Mediterranean children. J Allergy Clin Immunol Pract. 2016;4:265‐272. [DOI] [PubMed] [Google Scholar]
- 34. Uotila R, Kukkonen AK, Pelkonen AS, Mäkelä MJ. Cross‐sensitization profiles of edible nuts in a birch‐endemic area. Allergy Eur J Allergy Clin Immunol. 2016;71:514‐521. [DOI] [PubMed] [Google Scholar]
- 35. Rentzos G, Johanson L, Goksör E, Telemo E, Lundbäck B, Ekerljung L. Prevalence of food hypersensitivity in relation to IgE sensitisation to common food allergens among the general adult population in West Sweden. Clin Transl Allergy. 2019;9:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1


