Abstract
Randomized controlled trials and Mendelian randomization studies are two study designs that provide randomized evidence in human biological and medical research. Both exploit the power of randomization to provide unconfounded estimates of causal effect. However, randomized trials and Mendelian randomization studies have very different study designs and scientific objectives. As a result, despite sometimes being referred to as “nature's randomized trial,” a Mendelian randomization study cannot be used to replace a randomized trial but instead provides complementary information. In this review, we explain the similarities and differences between randomized trials and Mendelian randomization studies, and suggest several ways that Mendelian randomization can be used to directly inform and improve the design of randomized trials illustrated with practical examples. We conclude by describing how Mendelian randomization studies can employ the principles of trial design to be framed as “naturally randomized trials” that can provide a template for the design of future randomized trials evaluating therapies directed against genetically validated targets.
A randomized trial provides the highest level of evidence for human medical and biological research aiming to assess treatment effects, because it exploits the power and elegance of randomization (Collins et al. 2020). In a randomized trial, a group of eligible participants are randomly divided into two groups. If the sample size is large enough, randomization should equally distribute all known and unknown risk factors for an outcome between the two groups. Participants in one group are then given the intervention under study, while participants in the other group are given a comparator. Because randomization has created two groups that are, on average, nearly identical in every way, any differences in outcomes that occur between the two groups during follow-up can be reasonably inferred to have been caused by the intervention being evaluated.
A Mendelian randomization study is another study design that exploits the power and elegance of randomization in human medical and biological research. In a Mendelian randomization study, a genetic variant associated with the exposure of interest is used as an instrumental variable. Because genetic variants are randomly allocated at the time of gamete formation and conception according to Mendel's laws, the genetic variant can be used as an instrument to randomly divide the study population into groups with higher or lower levels of the exposure under study (Davey Smith and Ebrahim 2003). If the sample size is large enough, and if the genetic variant is not associated with potential confounders, then quasi-random allocation of this variant should have created groups that are, on average, nearly identical in every way except for levels of the exposure under study. Therefore, any differences in outcomes that occur between the groups later in life can be reasonably inferred to have been caused by differences in the exposure under study, assuming that the general assumptions of Mendelian randomization hold true (Davey Smith and Ebrahim 2003). Further details of these assumptions, and the difference between within-family and population studies, are provided in Richmond and Davey Smith (2020) and Davey Smith et al. (2020).
Although randomized trials and Mendelian randomization studies both rely on randomization to make causal inferences, they have very different study designs and scientific objectives. As a result, despite sometimes being referred to as “nature's randomized trial” (Hingorani and Humphries 2005), a Mendelian randomization study cannot be used to replace a randomized trial but instead provides complementary information. In this review, we begin by describing the similarities and differences between randomized trials and Mendelian randomization studies. We then suggest several ways that Mendelian randomization can be used to directly inform and improve the design of randomized trials, illustrated with practical examples. We conclude by describing how a Mendelian randomization study can borrow from the principles of trial design to create a “naturally randomized trial,” which can then be used as a template to design a subsequent randomized trial evaluating an intervention directed against a genetically validated target.
DESIGN OF RANDOMIZED TRIALS
A randomized trial is designed to answer a very specific question. For example, does intervention A reduce the risk of outcome X as compared to intervention B, by at least an increment of Y (defined as the minimal clinically important difference) in the population under study?
The design of a randomized trial is meticulously planned and memorialized in a written protocol that must be registered prior to initiating the trial (Chan et al. 2013). The protocol describes each step of the trial design in detail. First, the target population is defined. This population is usually the group of persons who may potentially benefit from the intervention under study. A series of inclusion and exclusion criteria are then enumerated to define the specific members of the target population who are eligible to participate in the trial, and procedures are established to recruit eligible members of the target population into the trial.
Next, the intervention to be studied is selected and carefully defined, including the specific dose or protocol for delivering the intervention. A comparator intervention is selected and carefully described. The comparator intervention may be an inactive placebo, usual care, or an alternative active intervention. Then, the allowable background therapies for enrolled participants are specified in detail (Schulz et al. 2010).
A group of primary, secondary, and tertiary outcomes are then selected, and a detailed set of criteria for adjudicating the occurrence of each event is defined (Pocock et al. 2002; Pocock 2006). Frequently, several related outcomes are combined into a single primary composite outcome to increase the statistical power of the study (because the power of a trial is determined primarily by the number of outcome events that occur rather than the total sample size).
Next, a randomization scheme is selected (Schulz and Grimes 2002). If practical, protocols are described to blind both trial participants and trial investigators from the interventions being compared to minimize the potential for bias (Psaty and Prentice 2010). An independent clinical events committee is then established to blindly adjudicate all possible outcome events that occur during the prospective follow-up of the trial; and a data safety and monitoring board is established to monitor the safety of the intervention under investigation (Montgomery et al. 2003). The Data and Safety Monitoring Board is also empowered to potentially stop the trial if any evidence of harm accumulates that exceeds a prespecified threshold of statistical evidence, or if the statistical thresholds for futility or overwhelming efficacy have been satisfied (Ellenberg et al. 2003).
Finally, the required sample size for the trial is determined. To calculate a sample size, several parameters must be explicitly estimated. First, the expected event rate of the outcome among participants receiving the comparator intervention must be estimated. Next, the minimal increment of difference in the primary outcome that is clinically meaningful to detect is explicitly stated (which is generally the smallest increment of difference in an outcome that will change clinical practice). An a priori definition of such a minimal clinically important difference is of relevance both in the context of superiority trials and in other trial designs, such as noninferiority trials (Treadwell et al. 2012). Of note, this review focuses primarily on ways Mendelian randomization can be used to inform the design of the more usual superiority trial. Finally, the desired level of statistical power needed to reliably detect the minimal clinically important difference is selected. These values are then entered into a mathematical formula to calculate the required sample size and the number of events that must accrue during follow-up to achieve the desired statistical power (Wittes 2002). Because of the time and expense required to conduct a randomized trial, the sample size calculation is a critical component of trial design. It is very important that the trial be large enough and of sufficient duration to accrue the required numbers of cases to both reliably reject the null hypothesis if a clinically important difference in outcomes exists between the two interventions being compared or to provide an informative null result if the interventions being compared do not result in any clinically meaningful differences in the outcomes under study.
DESIGN OF MENDELIAN RANDOMIZATION STUDIES
A Mendelian randomization study is also designed to answer a very specific question. For example, is the observed association between an exposure (E) and an outcome (X) likely to be causal?
A Mendelian randomization study uses genetic variants associated with the exposure of interest as an instrumental variable to explicitly introduce a randomization scheme into an observational study. This permits causal inferences about the association between the exposure and an outcome (Davey Smith and Hemani 2014). A unique feature of genetic variants that make them potentially powerful instrumental variables is that they are allocated randomly at gamete formation and conception under Mendel's Laws. Although strictly speaking alleles are randomly allocated within families (Davies et al. 2019; Davey Smith et al. 2020; Hwang et al. 2020), the distribution of alleles within a population is approximately random (Davey Smith and Ebrahim 2003; Davey Smith et al. 2007). As a result, genetic variants can be used as both instruments of randomization and instruments of effect. This is in contrast to a randomized trial, where the instrument of randomization into groups is distinct from the intervention under study.
The first step in the design of a Mendelian randomization study is to identify a study population that has measured the exposure and outcome of interest, and for which genetic information is available to permit construction of the genetic instrumental variable. Ideally, this would be a single large population in which the exposure, outcome, and genetic information has been measured for each participant so that the study can be performed using individual-level data (with selection of the variants to be included in the genetic instrument and their weights derived using data from an independent study population) (Haycock et al. 2016). Alternatively, a Mendelian randomization study can be performed by combining summary data from a genome-wide association study of the exposure conducted in one population, with summary data from a genome-wide association study of the outcome conducted in another independent population. However, these types of “two-sample” Mendelian randomization study designs that rely exclusively on summary-level data can narrow the range of clinically relevant questions that can be studied with regard to alternative outcome definitions or specific populations to be studied (Burgess et al. 2015; Davies et al. 2018).
Next, the genetic instrument is defined. This can consist of a single genetic variant that is strongly associated with a quantitatively “large” increment of difference in the exposure under study, or an instrument constructed by combining several independently inherited variants associated with “smaller” effects on the exposure of interest into a genetic score (Lawlor et al. 2008; Burgess et al. 2016; Dudbridge 2020). The variants are then aligned by defining the allele associated with a higher (or lower) level of the exposure under study as the “exposure allele” for each variant included in the instrument. While in a conventional Mendelian randomization study, a genetic instrument is typically used as a quantitative trait to estimate the causal effect for a given difference in the exposure, an alternative approach can be undertaken to more closely approximate the design of a randomized controlled trial (RCT). To do so, the genetic variant or score can be used as an instrument to randomly divide the study population into groups with higher (or lower) levels of the exposure under study. Because each allele included in the instrument is randomly allocated at gamete formation and conception under Mendel's Laws, and independent of all other variants included in the instrument by design, the number of exposure-increasing (or -decreasing) alleles in the instrument that a person inherits should also be approximately random. Therefore, either a single genetic variant or a genetic score can be used as a valid instrument of randomization and effect.
Finally, the association between the genetic instrument and the outcome of interest is measured. If the assumptions of Mendelian randomization are satisfied (which is only true under a number of assumptions as detailed in the literature; see Richmond and Davey Smith 2020), and if the genetic instrument is associated with the outcome of interest, then one can conclude that random allocation to higher (or lower) levels of the exposure arising due to the genetic instrument is associated with the outcome under study. As a result, one can conclude that the observed association between the exposure and outcome may be causal. Sensitivity analyses can then be conducted to test the robustness of the association to the assumptions of instrumental variable analysis (Bowden et al. 2016). Some examples of sensitivity analyses include the use of Mendelian randomization approaches that are robust to potential violation of the genetic instrumental variable arising from unbalanced horizontal pleiotropy (Hemani et al. 2018). By contrast, if the genetic instrument is not associated with the outcome under study, then one can conclude that the observed association between the exposure and outcome is unlikely to be causal but instead may be explained by reverse causation, residual confounding, or other biases that commonly occur in nonrandomized observational studies; or that the effect of the genetic instrument on the exposure was too small to permit a reliable quantitative assessment of the association between the exposure and outcome under study.
SIMILARITIES AND DIFFERENCES BETWEEN RANDOMIZED TRIALS AND MENDELIAN RANDOMIZATION STUDIES
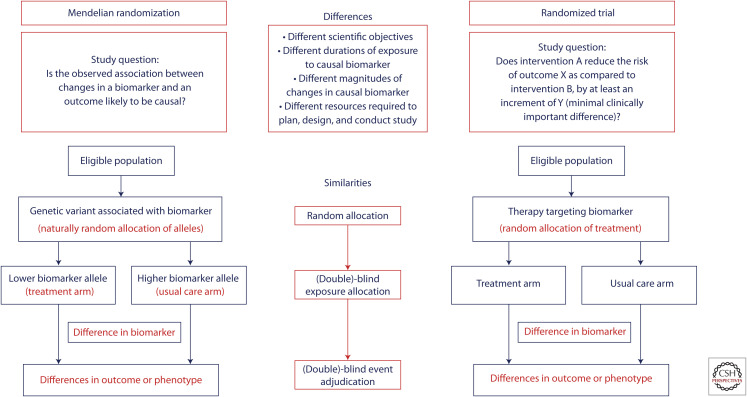
The defining feature of both randomized trials and Mendelian randomization studies is that they rely on randomization to make causal inferences (Fig. 1). Indeed, randomization is the only research technique that can equally distribute all measured and unmeasured variables between groups to permit an unbiased comparison. However, randomization by itself is not enough to ensure an unbiased comparison in either randomized trials or in Mendelian randomization studies.
Figure 1.
Structural similarities between Mendelian randomization and randomized trials.
The effect of randomization can potentially be undermined by an inadequate sample size (Peto and Baigent 1998). Despite a valid randomization scheme, a small sample size can lead to imbalances in the distribution of variables by chance alone among the groups being compared in both randomized trials and Mendelian randomization studies. For example, in Mendelian randomization studies evaluating large-effect, loss-of-function variants in drug targets (e.g., PCSK9, ANGPTL3, etc.), very few participants will have one of these large-effect variants because they are typically rare. As a result, the exposed group typically experiences only a relatively few events leading to imprecise estimates of effect with wide confidence intervals around the point estimates and potential imbalances in the distribution of risk factors by chance alone. Although these imbalances can be partially adjusted for in statistical models, such imbalances can potentially produce spurious results in both trials and Mendelian randomization studies.
In addition, both study designs are vulnerable to spurious results when allocation to the intervention or exposure under study is not adequately blinded from study participants, study investigators, or treating physicians (Psaty and Prentice 2010). Knowledge of which intervention a study participant is receiving in a randomized trial can lead to differences in the intensity of surveillance for the outcome of interest or differences in the administration of background therapies to participants in the groups being compared—either of which can bias the results of the study. Analogously, the presence of elevated levels of an exposure believed to cause a disease among persons who inherit a genetic variant with a large effect (e.g., a low-density lipoprotein [LDL] receptor mutation causing familial hypercholesterolemia) can result in more intense surveillance for the outcome under study or more aggressive treatments to prevent the disease as compared to persons without the mutation—thus potentially biasing the results of a Mendelian randomization study.
Therefore, both randomized trials and Mendelian randomization studies require a rigorous study design in addition to randomization to produce valid study results. Despite these similarities, however, there are important conceptual differences between a randomized trial and a Mendelian randomization study.
First, they are designed to test very different scientific questions. A Mendelian randomization study is designed specifically to assess whether the effect of an exposure on an outcome is likely to be causal, without any consideration of the “clinical significance” of the magnitude of the potential causal effect. By contrast, a randomized trial is not a simple test of causality. Instead, a randomized trial is specifically designed to evaluate whether the effect of an intervention on an outcome is quantitatively large enough to be clinically significant or to change clinical practice. Indeed, unlike a Mendelian randomization study, a randomized trial is designed to test both the null hypothesis that the intervention has no effect on the outcome of interest, and the alternative hypothesis that the intervention has a clinically important effect on the outcome (Feinstein and Concato 1998).
Second, most genetic variants or genetic scores used as instrumental variables in a Mendelian randomization study have a relatively small quantitative impact on the exposure under study. In addition, the effect of the genetic variants used to instrument the exposure are present since conception. As a result, a Mendelian randomization study generally evaluates the effect of lifetime (including intrauterine) exposure to a small increment of difference in the exposure of interest on the risk of an outcome (Davey Smith and Ebrahim 2004; Ference et al. 2012). By contrast, the intervention evaluated in a randomized trial generally has a large quantitative impact on the intermediate biomarkers that determine the therapeutic effect of the intervention. This effect can be an order of magnitude larger than the effect of a genetic variant or score that instruments the same biomarker used in a Mendelian randomization study. Furthermore, the intervention in a randomized trial is delivered for only a relatively short duration of time, typically between several months to 5 years (with the median duration of phase III trials being 40 months) (Martin et al. 2017), and in response to a specific clinical indication. As a result, unlike a Mendelian randomization study, a randomized trial evaluates the relatively short-term effect of an intervention with a quantitatively “large” effect on an intermediate biomarker (i.e., the effect of a therapeutic intervention on a biomarker is typically far greater than the effect of genetic variants instrumenting the intervention on the same biomarker). Such a pharmacological intervention is typically initiated in response to some triggering event such as an acute insult, or to slow the progression of a disease, to prevent a recurrent event, or to reduce the risk of an outcome among persons who cross a specific threshold of risk for that outcome.
Third, a Mendelian randomization study is generally a study of convenience that relies on the availability of existing study samples. Indeed, it is rare that a new study will be initiated for the purposes of performing a Mendelian randomization analysis because very large sample sizes are needed to reliably assess the causal effect of a small genetically instrumented increment of difference in an exposure on the risk of an outcome. By contrast, each randomized trial is a new interventional prospective cohort study that is designed to address a very specific question about the potential clinical efficacy of a therapeutic intervention. These studies are expensive and labor intensive, and can take a decade or more to design and complete. Because of this resource-intensive effort, randomized trials can be designed to investigate more precisely defined questions as compared to what can be explored in a typical Mendelian randomization study.
USING MENDELIAN RANDOMIZATION TO INFORM THE DESIGN OF RANDOMIZED TRIALS
Clearly randomized trials and Mendelian randomization studies have very different study designs and scientific objectives. As a result, Mendelian randomization studies cannot be used to replace or substitute for randomized trials. Instead, Mendelian randomization studies can provide complementary information or provide a biological perspective for interpreting the results of randomized trials.
However, recognizing that a Mendelian randomization study is merely a way to introduce a randomization scheme into an observational study, it becomes clear that Mendelian randomization can be designed specifically to provide information that is needed to optimally design a randomized trial. Below are several questions that must be considered when designing a randomized trial, with examples of how Mendelian randomization studies can be used to directly answer each of these questions and thus directly inform the design of the trial (Table 1).
Table 1.
Potential uses of Mendelian randomization studies to resolve uncertainties in the design of randomized trials
| Uncertainty when designing a randomized trial | Potential use of Mendelian randomization to resolve the uncertainty | Implications |
|---|---|---|
| 1. Is the target of the intervention under study in the causal pathway of the outcome? | Mendelian randomization study to assess whether the biomarker altered by a therapy has a causal effect on the outcome of interest. | If the targeted biomarker does not have a causal effect on the risk of disease, the trial is unlikely to show a clinical benefit. |
| 2. Which biomarker(s) in the causal pathway altered by the therapy determines the causal dose–response? | Multivariable Mendelian randomization study to assess which of several biologically related biomarkers altered by a therapy is likely to be responsible for the causal dose–response. | The trial should be powered; and inclusion criteria developed based on expected changes that can be achieved in the causal biomarker with the therapy under study. Changes in this biomarker should be measured during the trial. |
| 3. How much must the causal biomarker be changed to produce the minimal clinically important difference (MCID)? | Mendelian randomization study to assess the shape and magnitude of the causal dose–response curve; assess whether the association between a biomarker altered by the therapy and an outcome has a cumulative or threshold effect; and to determine how much the causal biomarker must be changed to produce the MCID in a short-term trial. | The trial should enroll participants with a high enough baseline level of the causal biomarker to achieve a large enough absolute reduction in the biomarker in response to treatment to produce the MCID. Enrolling participants with lower levels of the causal biomarker raises the risk that the causal biomarker will not be lowered enough to produce the MCID, thus leading to an underpowered study and the potential for a null result. |
| 4. What are the optimal inclusion and exclusion criteria for the trial? | Additional Mendelian randomization studies to those described above designed to evaluate differential benefits within subgroups defined by age, sex, comorbidities, etc. | The inclusion and exclusion criteria for the trial depend substantially on the identity of the causal biomarker, and much it must be altered to produce the MCID. Evaluating a therapy that does not produce a large enough change in the causal biomarker to produce an MCID, or enrolling participants who are unlikely to achieve a large enough change in the causal biomarker in response to therapy will lead to an underpowered trial. |
| 5. What background therapies should be allowed? | Factorial Mendelian randomization studies to assess the separate and combined effect of genetic variants or scores designed to instrument the therapy and any essential standard of care therapies that trial participants may require. | Identification of essential background therapies that may attenuate the effect of the therapy under study can help choose the appropriate clinical indication for the therapy to study in the trial, select appropriate permissible background therapy, or alter inclusion and exclusion criteria to enroll participants who cannot or will not take the recommended background therapy. |
| 6. Which outcomes should be included in the primary composite outcome? | Mendelian randomization studies evaluating the effect of the causal biomarker on each of several outcomes separately and combined into composites to assess for consistency of the magnitude of effect on the risk of the outcome per unit change in the causal biomarker. | The primary composite outcome should include only those outcomes for which reliable quantitative evidence exists to suggest a consistent magnitude of clinical effect per unit change in the causal biomarker. |
| 7. What potential adverse events are most likely? | PheWAS Mendelian randomization studies to assess for potential “on-target” adverse events, and to assess the quantitative magnitude and potential clinical relevance of any potential adverse effects. | Care should be taken to avoid speculation for associations with weak statistical evidence or that lack biological plausibility. |
| 8. Are there specific subgroups who are likely to benefit most from the intervention? | Factorial and stratified Mendelian randomization studies, alone and nested with machine and deep learning algorithms, to identify factors that modify the effect of the causal biomarker on the risk of disease. | Persons who experience a greater relative increased risk per unit change in a biomarker may be more vulnerable to that biomarker, and therefore may derive a larger relative risk reduction per unit change in the biomarker in response to therapy. Enriching a trial with “vulnerable” participants can lead to smaller trials that produce larger proportional (and absolute) reductions in risk per unit change in the causal biomarker. This information can further inform inclusion and exclusion criteria or inform the adaptive trial designs. |
-
Is the target of the intervention under study in the causal pathway of the outcome?
Mendelian randomization can help to determine whether the target of an intervention has a causal effect on the risk of an outcome. Obviously, if the target of an intervention does not have a causal effect on an outcome, then a randomized trial evaluating the effect of that intervention on the outcome is unlikely to be successful. This is the motivation behind the current trend of developing new interventions directed against “genetically validated targets.” Indeed, it has been estimated that the success rate of novel therapies being evaluated in randomized trials can double from 5%–10% to 10%–20% for interventions directed against targets that have genetic evidence of a causal effect on an outcome (Nelson et al. 2015). Remarkably, however, this means that potentially ∼80%–90% of interventions directed against genetically validated targets will fail somewhere between phase I and phase III clinical trials. It is clear therefore that evidence from a Mendelian randomization study suggesting that the target of an intervention has a causal effect on an outcome is not sufficient to ensure that an intervention directed against that exposure will have a clinically significant impact on the outcome.
-
Which biomarker in the causal pathway determines the causal dose–response?
Frequently an intervention directed against a specific biomarker target may alter the level of several other biologically related biomarkers in the same pathway. In this case, the challenge is determining which one or more of the related biomarkers are responsible for the causal dose–response. If only one trait is responsible for the causal dose–response relationship, then this has implications regarding the biomarker(s) that should be investigated in clinical trials. While in many cases it may be reasonable to assume that most traits on the causal pathway show dose–response relationships with disease, there will be particular situations where this may not be the case. Multivariable Mendelian randomization can help address this issue (Sanderson et al. 2019; Sanderson 2020). For example, several Mendelian randomization studies have suggested that plasma triglyceride levels may be causally associated with the risk of cardiovascular disease (Sarwar et al. 2010; Nordestgaard and Varbo 2014; White et al. 2016). However, several randomized trials of triglyceride-lowering therapies have failed to consistently demonstrate that lowering plasma triglycerides reduces the risk of cardiovascular events (Keech et al. 2005; Ginsberg et al. 2010). Recent multivariable Mendelian randomization studies have helped resolve this issue by revealing that triglycerides may not have a causal effect on the risk of cardiovascular events (Ference et al. 2019a; Richardson et al. 2020). Instead, the causal effect of genetic variants and pharmacologic interventions that reduce plasma triglyceride levels is likely determined by the absolute reduction in the circulating concentration of apoB-containing lipoprotein particles that carry triglycerides, rather than by the decrease in triglyceride content carried by those particles as measured by changes in plasma triglyceride concentration. As a result, the relevant biomarker to measure in the trial to assess the potential clinical efficacy of a triglyceride-lowering therapy is the reduction in plasma apoB levels, not necessarily the reduction in triglyceride levels.
-
How much must the causal biomarker be changed to produce the minimal clinically important difference?
Once the biomarker that determines the causal dose–response of the intervention under study is identified, the critical question then becomes how much must levels of the causal biomarker be changed to produce the desired minimal clinically important difference in the associated outcome? Surprisingly, this question is frequently ignored in the design of randomized trials (Cook et al. 2018). Without an estimate of how much a causal biomarker must be changed to produce the minimally important clinical difference in the outcome under study, it is impossible to reliably estimate whether the therapy under study is potent enough to lower the causal biomarker by this amount. It is also impossible to estimate whether the participants enrolled in the trial have baseline levels of the biomarker high enough to achieve therapeutic reductions in the biomarker that are large enough to produce the desired clinical benefit.
Mendelian randomization can help answer this critical question. For example, Mendelian randomization studies consistently demonstrate that Lp(a) has a causal effect on the risk of cardiovascular events (Clarke et al. 2009; Kamstrup et al. 2009). However, randomized trials of several therapies that reduce plasma Lp(a) levels by 30%–40% have consistently failed to show that lowering Lp(a) reduces risk of cardiovascular events (Boden et al. 2001; Barter et al. 2007; Landray et al. 2014). A recent Mendelian randomization study has helped reconcile these inconsistent findings by demonstrating that large absolute decreases in plasma Lp(a) are required to produce clinically meaningful reductions in cardiovascular risk (Burgess et al. 2018). Because plasma Lp(a) levels are extremely right-skewed, most members of the general population have very low absolute plasma Lp(a) levels. As a result, the failure of these trials can be explained by the fact that even relatively large proportional reductions in Lp(a) did not produce large enough absolute reductions in Lp(a) to produce a clinically meaningful reduction in cardiovascular events because the overwhelming majority of participants enrolled in these trials had very low baseline Lp(a) levels.
-
What are the optimal inclusion and exclusion criteria for the trial?
Determining how much the causal biomarker must be altered by an intervention to produce the minimal clinically important difference in an outcome directly informs the optimal inclusion and exclusion criteria for a trial. For example, continuing the example from above, only participants with very high plasma Lp(a) levels are likely to achieve a large enough absolute reduction in Lp(a) levels to experience a clinically significant reduction in the risk of cardiovascular events when treated with an Lp(a)-lowering therapy. Therefore, trials of an Lp(a)-lowering therapy should only include participants with very elevated levels of Lp(a), because they are the only ones likely to experience a large enough absolute reduction in Lp(a) to achieve a clinically meaningful reduction in the risk of cardiovascular events. Optimal inclusion criteria can be further informed by evaluating whether the association between the causal biomarker and the outcome under study differs among relevant subgroups defined by sex, age, or the presence of specific comorbidities (thus explicitly testing the fourth instrumental variable assumption of homogeneity).
-
What background therapies should be allowed?
Factorial Mendelian randomization can be used to assess whether the effect of an intervention on an outcome is likely to be modified by the coadministration of background therapies (Ference et al. 2015). This information will, in turn, further inform the optimal inclusion and exclusion criteria for the trial. For example, a recent Mendelian randomization study demonstrated that when genetic variants that mimic CETP inhibitors are combined with genetic variants that mimic statins (which are required standard of care background therapy for persons with cardiovascular disease), the expected reductions in plasma LDL-C remain unchanged but the corresponding reduction in plasma apoB becomes attenuated. Furthermore, the observed reduction in cardiovascular events was proportional to the attenuated absolute reduction in apoB, not the change in LDL-C (Ference et al. 2017). These findings were subsequently confirmed in a large randomized trial (HPS3/TIMI55-REVEAL Collaborative Group et al. 2017), and in a post hoc analysis of another large randomized trial (Riesmeyer et al. 2018). The implication is that CETP inhibitors should produce much greater reductions in cardiovascular events when used alone because this would avoid the attenuation in apoB reduction that occurs when CETP inhibitors are used in combination with a statin. As a result, any future trials of this class of medications should evaluate CETP inhibitors as monotherapy, and should include only participants with elevated lipid levels who are unable or unwilling to take statins.
-
Which outcomes should be included in the primary composite outcome?
Randomized trials frequently combine multiple outcomes into a composite primary outcome to increase the number of events that will accrue during follow-up to increase the statistical power of the trial. However, it is important to include only those outcomes in the composite that are similarly affected by the intervention under study to avoid diluting the observed relative impact of the intervention. Mendelian randomization can be used to compare the expected effect of the intervention on a variety of related outcomes per unit change in the causal biomarker altered by the intervention. This information can then be used to select those outcomes for which the expected effect of the therapy is approximately similar per unit change in causal biomarker altered by the intervention. These outcomes can be included in the primary composite outcome. All other outcomes should be excluded from the primary composite outcome to avoid attenuating the effect of the therapy under study toward the null.
-
What potential adverse events are most likely?
Mendelian randomization can be also used to identify potential adverse events that may be associated with the intervention under study, characterize the frequency of these potential adverse events, and estimate their clinical relevance. For example, several Mendelian randomization studies have reported that genetic variants that mimic the effect of statins, or other therapies that reduce LDL-C through up-regulation of the LDL receptor, are associated with an increased risk of diabetes (Swerdlow et al. 2015; Ference et al. 2016; Lotta et al. 2016; Schmidt et al. 2017). This finding was confirmed in some but not all randomized trials (Baigent et al. 2010; Sabatine et al. 2017; de Carvalho et al. 2018). Importantly, however, both the Mendelian randomization studies and the randomized trials have demonstrated that the reduction in LDL-C with a statin far outweighs the slight increased risk of developing diabetes on the risk of cardiovascular events. Specifically, in both Mendelian randomization studies and in randomized trials, the proportional reduction in cardiovascular events is the same per unit lower LDL-C among persons with and without diabetes, thus suggesting that any increased risk in diabetes does not substantially attenuate the clinical benefit (Baigent et al. 2010; Ference et al. 2015, 2016). This information can be used to further refine the design of the trial by excluding groups of participants who are most likely to experience an adverse event in response to the intervention.
-
Are there specific subgroups who are likely to benefit most from the intervention?
Factorial Mendelian randomization, and Mendelian randomization studies nested within machine and deep learning algorithms, can potentially identify subgroups of persons who are differentially susceptible to the deleterious effects of causal biomarkers. Persons who are more vulnerable to the causal effects of a biomarker are likely to be at greater risk of the outcome and to derive a greater “relative benefit” from the same reduction in that biomarker in response to an intervention as compared to persons who are less vulnerable to the biomarker. This information can then be used to further refine the inclusion and exclusion criteria for a trial by enrolling only those persons who are the most likely to benefit from an intervention. Alternatively, this information can be used to inform the design of so-called population-enrichment, adaptive clinical trial designs (Bhatt and Mehta 2016). In this approach, the trial can be adapted to preferentially enroll participants identified as vulnerable to the causal biomarker if an interim analysis indicates that the vulnerable participants are benefiting more from the therapy than the less vulnerable participants.
DESIGNING MENDELIAN RANDOMIZATION STUDIES AS NATURALLY RANDOMIZED TRIALS
Just as the design of randomized trials can be improved by using information from Mendelian randomization studies, the design of Mendelian randomization studies can be improved by employing the principles of trial design. Indeed, a Mendelian randomization study can be framed as a “naturally randomized trial” by using the same protocol as a randomized trial (Ference 2018). The results of a Mendelian randomization study framed as a “naturally randomized trial” can then be used to either anticipate the results of an ongoing randomized trial or serve as a template for the design of a future randomized trial evaluating a genetically validated target.
A “naturally randomized trial” is simply a Mendelian randomization study designed as if it were a randomized trial. Indeed, a naturally randomized trial can be conducted using a similar protocol to that used to conduct a randomized trial evaluating the effect of a therapeutic intervention. This study protocol should be memorialized prior to the initiation of the study. In addition, it should specify the same inclusion and exclusion criteria, the same definition of the same primary composite outcome, assess the same key biomarker and safety outcomes, and use the same threshold for the magnitude of the minimal clinically important difference that would be used in a randomized trial evaluating the intervention of interest. The distinction is that rather than evaluating the therapeutic intervention of interest using this protocol, a “naturally randomized trial” evaluates the effect of a genetic variant or genetic score designed as an instrumental variable to mimic the effect of the intervention. The genetic variant or score is then used as the instrument of randomization and effect to conduct the “naturally randomized trial.” We note that the change in exposure required to achieve the minimal clinically important difference may well differ between Mendelian randomization and therapeutic trials, and that other nuances should be considered including time-dependent effects (Holmes et al. 2017; Ference 2018, Ference et al. 2019b; Holmes 2019).
The results of Mendelian randomization studies framed as “naturally randomized trials” have already been used to accurately anticipate the results of several recent landmark studies in cardiovascular medicine, each of which reported unexpected results. For example, despite being called an “expensive placebo” (Mascitelli and Goldstein 2016), a Mendelian randomization study accurately anticipated that lowering plasma LDL-C with ezetimibe would reduce the risk of cardiovascular events proportional to the absolute reduction in plasma LDL-C regardless of whether used alone or in combination with a statin (Ference et al. 2015). This is precisely what the landmark IMPROVE-IT trial showed (Cannon et al. 2015). In addition, despite an almost irrational exuberance that led many to assume that lowering LDL-C by inhibiting PCSK9 could reduce the risk of cardiovascular events by 50% or more (Cohen et al. 2006; Robinson et al. 2015; Sabatine et al. 2015), a Mendelian randomization analysis showed that genetic variants in the PCSK9 and HMG-CoA reductase genes (the genes that encode for the targets of PCSK9 inhibitors and statins, respectively) have biologically equivalent effects on the risk of cardiovascular disease per unit change in LDL-C, and therefore treatment with a PCSK9 inhibitor should reduce the risk of cardiovascular events by the same amount as statins for the same reduction in LDL-C and the same duration of therapy (Ference et al. 2016). This is precisely what the landmark FOURIER and ODYSSEY trials showed (Sabatine et al. 2017; Schwartz et al. 2018). Indeed, PCSK9 inhibitors and statins appear to have the same effect on the risk of cardiovascular disease per unit change in LDL-C during each year of treatment, precisely as predicted by the Mendelian randomization naturally randomized trials (Ference et al. 2018). Finally, even though three previous large phase III cardiovascular outcomes trials evaluating CETP inhibitors were stopped prematurely for safety or futility (Barter et al. 2007; Schwartz et al. 2012; Lincoff et al. 2017), a Mendelian randomization study accurately predicted that when added to treatment with a statin, treatment with the CETP inhibitor anacetrapib would likely produce large absolute reductions in LDL-C but only small corresponding absolute reductions in apoB, and that the observed clinical benefit would be proportional to the attenuated changes in apoB not the larger changes in LDL-C (Ference et al. 2017). This is precisely what the 30,000 participant REVEAL trial showed (HPS3/TIMI55-REVEAL Collaborative Group et al. 2017). The results of this Mendelian randomization study not only accurately anticipated the results of this study, it also provided a biological explanation for the observed results, and has motivated the development of additional agents in this class.
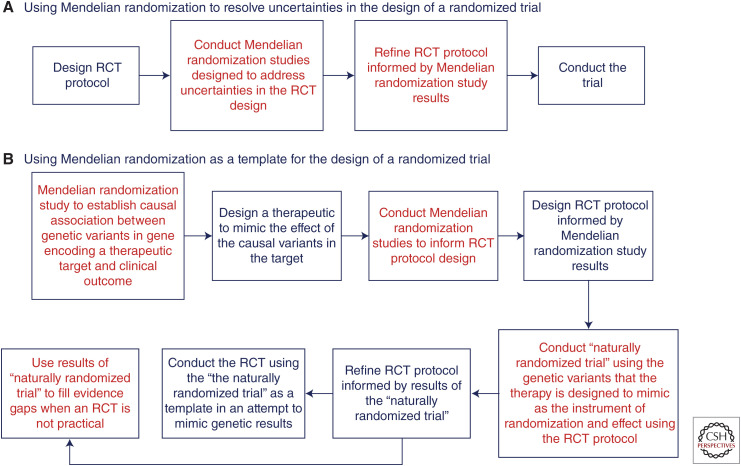
However, more than simply anticipating the results of randomized trials, a Mendelian randomization study framed as a naturally randomized trial can also serve as a template to design a future randomized trial, particularly trials that are evaluating therapies directed against a genetically validated target (Fig. 2).
Figure 2.
Using Mendelian randomization to (A) resolve uncertainties, and (B) as a template for the design of randomized trials. (RCT) Randomized controlled trial.
As a wider array of therapeutic technologies become available, including small interfering RNAs, many therapies are now being designed to explicitly recapitulate the phenotype of genetic variants that are associated with a lower risk of disease (Nishikido and Ray 2018; Tsimikas 2018). The goal of these therapies is to mimic the effect of the causal genetic variants on both the outcome of interest, and the intermediate biomarker that determines the causal effect. Importantly, the potential clinical relevance for such a therapy can be conveniently evaluated by a portfolio of related Mendelian randomization studies, which can then serve to directly inform the clinical development program for the therapy.
First, a series of Mendelian randomization studies can be designed to directly inform the clinical trial protocol. These studies would be designed to determine, for example, which of a series of related biomarkers altered by the genetic variant that a therapy is designed to mimic is responsible for the causal effect, to characterize the shape and magnitude of the causal dose–response curve for this biomarker, to determine whether the causal biomarker has a cumulative or threshold effect on the outcome, to estimate how much the causal biomarker must be altered to produce a predefined minimally clinically important difference in the outcome, and to determine the optimal inclusion criteria, primary composite outcome definition, and permissible background therapy among the groups being compared.
Next, this information could be used to construct the protocol for the randomized trial designed to evaluate the therapy that is mimicking the causal genetic variant. A “naturally randomized trial” could then be conducted according to the protocol informed by the earlier Mendelian randomization studies, using the genetic variant (or score) that the therapy is attempting to mimic as the instrument of randomization and effect.
Based on the results of the “naturally randomized trial,” the protocol for the trial could be revised and optimized as needed. Finally, the definitive randomized trial evaluating the therapy mimicking the causal genetic variant can be conducted using the revised protocol as a template in an explicit attempt to recapitulate the results of the “naturally randomized trial” (Fig. 2).
Indeed, as the biopharmaceutical industry increasingly focuses on developing therapies that attempt to recapitulate the phenotypes of genetic variants that are associated with the risk of various outcomes, the potential clinical benefit of these therapies will likely be initially evaluated in Mendelian randomization studies framed as “naturally randomized trials” (Ference 2019). These studies can serve as a template to optimize the design of the trial evaluating the therapies directed against genetically validated targets to minimize the potential that the trial will produce an unexpectedly null result.
CONCLUDING REMARKS
Both randomized trials and Mendelian randomization studies use the power and elegance of randomization to make unconfounded causal inferences. However, these two study designs address different scientific questions. As a result, Mendelian randomization studies cannot replace randomized trials. Instead, Mendelian randomization studies can provide complementary information to randomized trials addressing the same question. In addition, Mendelian randomization studies can be designed to provide information that is needed to inform each step in the design of a randomized trial. Indeed, Mendelian randomization studies can adopt the principles of trial design to conduct “naturally randomized trials,” which may eventually be used as templates to optimize the design of randomized trials evaluating novel therapies directed against genetically validated targets. Therefore, the clinical trial and Mendelian randomization communities may benefit from working together more closely toward the common goal of using Mendelian randomization to directly inform and improve the design of randomized trials, particularly when evaluating therapies either directed against a genetically validated target or therapies designed to mimic the effect of a causal genetic variant (Holmes et al. 2020).
Footnotes
Editors: George Davey Smith, Rebecca Richmond, and Jean-Baptiste Pingault
Additional Perspectives on Combining Human Genetics and Causal Inference to Understand Human Disease and Development available at www.perspectivesinmedicine.org
REFERENCES
*Reference is also in this collection.
- ACCORD Study Group; Ginsberg HN, Elam MB, Lovato LC, Crouse JR III, Leiter LA, Linz P, Friedewald WT, Buse JB, Gerstein HC, et al. 2010. Effects of combination lipid therapy in type 2 diabetes mellitus. N Engl J Med 362: 1563–1574. 10.1056/NEJMoa1001282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baigent C, Blackwell L, Emberson J, Holland LE, Reith C, Bhala N, Peto R, Barnes EH, Keech A, Simes J, et al. 2010. Efficacy and safety of more intensive lowering of LDL cholesterol: a meta-analysis of data from 170,000 participants in 26 randomised trials. Lancet 376: 1670–1681. 10.1016/S0140-6736(10)61350-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barter PJ, Caulfield M, Eriksson M, Grundy SM, Kastelein JJP, Komajda M, Lopez-Sendon J, Mosca L, Tardif JC, Water DD, et al. 2007. Effects of torcetrapib in patients at high risk for coronary events. N Engl J Med 357: 2109–2122. 10.1056/NEJMoa0706628 [DOI] [PubMed] [Google Scholar]
- Bhatt DL, Mehta C. 2016. Adaptive designs for clinical trials. N Engl J Med 375: 65–74. 10.1056/NEJMra1510061 [DOI] [PubMed] [Google Scholar]
- Boden WE, Probstfield JL, Anderson T, Chaitman BR, Desvignes-Nickens P, Koprowicz K, McBride R, Teo K, Weintraub W, AIM-HIGH Investigators. 2001. Niacin in patients with low HDL cholesterol levels receiving intensive statin therapy. N Engl J Med 365: 2255–2267. [DOI] [PubMed] [Google Scholar]
- Bowden J, Davey Smith G, Haycock PC, Burgess S. 2016. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol 40: 304–314. 10.1002/gepi.21965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S, Scott RA, Timpson NJ, Davey Smith G, Thompson SG, EPIC-InterAct Consortium. 2015. Using published data in Mendelian randomization: a blueprint for efficient identification of causal risk factors. Eur J Epidemiol 30: 543–552. 10.1007/s10654-015-0011-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S, Dudbridge F, Thompson SG. 2016. Combining information on multiple instrumental variables in Mendelian randomization: comparison of allele score and summarized data methods. Statist Med 35: 1880–1906. 10.1002/sim.6835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S, Ference BA, Staley JR, Freitag DF, Mason AM, Nielsen SF, Willeit P, Young R, Surendran P, Karthikeyan S, et al. 2018. European prospective investigation into cancer and nutrition–cardiovascular disease (EPIC-CVD) consortium. Association of LPA variants with risk of coronary disease and the implications for lipoprotein(a)-lowering therapies: a Mendelian randomization analysis. JAMA Cardiol 3: 619–627. 10.1001/jamacardio.2018.1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon CP, Blazing MA, Giugliano RP, McCagg A, White JA, Theroux P, Darius H, Lewis BS, Ophuis TO, Jukema JW, et al. 2015. Ezetimibe added to statin therapy after acute coronary syndromes. N Engl J Med 372: 2387–2397. 10.1056/NEJMoa1410489 [DOI] [PubMed] [Google Scholar]
- Chan AW, Tetzlaff JM, Altman DG, Laupacis A, Gøtzsche PC, Krleža-Jerić K, Hróbjartsson A, Mann H, Dickersin K, Berlin JA, et al. 2013. SPIRIT 2013 statement: defining standard protocol items for clinical trials. Ann Intern Med 158: 200–207. 10.7326/0003-4819-158-3-201302050-00583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke R, Peden JF, Hopewell JC, Kyriakou T, Goel A, Heath SC, Parish S, Barlera S, Franzosi MG, Rust S, et al. 2009. Genetic variants associated with Lp(a) lipoprotein level and coronary disease. N Engl J Med 361: 2518–2528. 10.1056/NEJMoa0902604 [DOI] [PubMed] [Google Scholar]
- Cohen JC, Boerwinkle E, Mosley TH Jr, Hobbs HH. 2006. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med 354: 1264–1272. 10.1056/NEJMoa054013 [DOI] [PubMed] [Google Scholar]
- Collins R, Bowman L, Landray M, Peto R. 2020. The magic of randomization versus the myth of real-world evidence. N Engl J Med 382: 674–678. 10.1056/NEJMsb1901642 [DOI] [PubMed] [Google Scholar]
- Cook JC, Julious SA, Sone W, Hampson LV, Hewitt C, Berlin JA, Ashby D, Emsley R, Fergusson DA, Walters SJ, et al. 2018. DELTA2 guidance on choosing the target difference and undertaking and reporting the sample size calculation for a randomized controlled trial. BMJ 363: k3750. 10.1136/bmj.k3750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey Smith G, Ebrahim S. 2003. “Mendelian randomization”: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol 32: 1–22. 10.1093/ije/dyg070 [DOI] [PubMed] [Google Scholar]
- Davey Smith G, Ebrahim S. 2004. Mendelian randomization: prospects, potentials, and limitations. Int J Epidemiol 33: 30–42. 10.1093/ije/dyh132 [DOI] [PubMed] [Google Scholar]
- Davey Smith G, Hemani G. 2014. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet 23: R89–R98. 10.1093/hmg/ddu328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey Smith G, Lawlor DA, Harbord R, Timpson NJ, Day I, Ebrahim S. 2007. Clustered environments and randomized genes: a fundamental distinction between conventional and genetic epidemiology. PLoS Med 4: 1985–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey Smith G, Holmes MV, Davies NM, Ebrahim S. 2020. Mendel's laws, Mendelian randomization and causal inference in observational data: substantive and nomenclatural issues. Eur J Epidemiol 35: 99–111. 10.1007/s10654-020-00622-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies NM, Holmes MV, Davey Smith G. 2018. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ 362: k601. 10.1136/bmj.k601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies NM, Howe LJ, Brumpton B, Havdahl A, Evans DM, Davey Smith G. 2019. Within family Mendelian randomization studies. Hum Mol Genet 28: R170–R179. 10.1093/hmg/ddz204 [DOI] [PubMed] [Google Scholar]
- de Carvalho LSF, Campos AM, Sposito AC. 2018. Proprotein convertase subtilisin/kexin type 9 (PCSK9) inhibitors and incident type 2 diabetes: a systematic review and meta-analysis with over 96,000 patient-years. Diabetes Care 41: 364–367. 10.2337/dc17-1464 [DOI] [PubMed] [Google Scholar]
- *.Dudbridge F. 2020. Polygenic Mendelian randomization. Cold Spring Harb Perspect Med 10.1101/cshperspect.a039586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenberg SS, Fleming TR, DeMets DL. 2003. Data monitoring committees in clinical trials: a practical perspective. Wiley, Chichester, UK. [Google Scholar]
- Feinstein AR, Concato J. 1998. The quest for “power”: contradictory hypotheses and inflated sample sizes. J Clin Epidemiol 51: 537–545. 10.1016/S0895-4356(98)00029-8 [DOI] [PubMed] [Google Scholar]
- Ference BA. 2018. How to use Mendelian randomization to anticipate the results of randomized trials. Eur Heart J 39: 360–362. 10.1093/eurheartj/ehx462 [DOI] [PubMed] [Google Scholar]
- Ference BA. 2019. Using genetic variants in the targets of lipid lowering therapies to inform drug discovery and development: current and future treatment options. Clin Pharmacol Ther 105: 568–581. 10.1002/cpt.1163 [DOI] [PubMed] [Google Scholar]
- Ference BA, Yoo W, Alesh I, Mahajan N, Mirowska KK, Mewada A, Kahn J, Afonso L, Williams KA Sr, Flack JM. 2012. Effect of long-term exposure to lower low-density lipoprotein cholesterol beginning early in life on the risk of coronary heart disease: a Mendelian randomization analysis. J Am Coll Cardiol 60: 2631–2639. 10.1016/j.jacc.2012.09.017 [DOI] [PubMed] [Google Scholar]
- Ference BA, Majeed F, Penumetcha R, Flack JM, Brook RD. 2015. Effect of naturally random allocation to lower low-density lipoprotein cholesterol on the risk of coronary heart disease mediated by polymorphisms in NPC1L1, HMGCR, or both. J Am Coll Cardiol 65: 1552–1561. 10.1016/j.jacc.2015.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ference BA, Robinson JG, Brook RD, Catapano AL, Chapman MJ, Neff DR, Voros S, Giugliano RP, Davey Smith G, Fazio S, et al. 2016. Variation in PCSK9 and HMGCR and risk of cardiovascular disease and diabetes. N Engl J Med 375: 2144–2153. 10.1056/NEJMoa1604304 [DOI] [PubMed] [Google Scholar]
- Ference BA, Kastelein JJP, Ginsberg HN, Chapman MJ, Nicholls SJ, Ray KK, Packard CJ, Laufs U, Brook RD, Oliver-Williams C, et al. 2017. Association of genetic variants related to CETP inhibitors and statins with lipoprotein levels and cardiovascular risk. JAMA 318: 947–956. 10.1001/jama.2017.11467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ference BA, Cannon CP, Landmesser U, Lüscher TF, Catapano AL, Ray KK. 2018. Reduction of low density lipoprotein-cholesterol and cardiovascular events with proprotein convertase subtilisin-kexin type 9 (PCSK9) inhibitors and statins: an analysis of FOURIER, SPIRE, and the cholesterol treatment trialists collaboration. Eur Heart J 39: 2540–2545. 10.1093/eurheartj/ehx450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ference BA, Kastelein JJP, Ray KK, Ginsberg HN, Chapman MJ, Packard CJ, Laufs U, Oliver-Williams C, Wood AM, Butterworth AS, et al. 2019a. Association of triglyceride-lowering LPL variants and LDL-C-lowering LDLR variants with risk of coronary heart disease. JAMA 321: 364–373. 10.1001/jama.2018.20045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ference BA, Ray KK, Catapano AL, Ference TB, Burgess S, Neff DR, Oliver-Williams C, Wood AM, Butterworth AS, Di Angelantonio E, et al. 2019b. Mendelian randomization study of ACLY and cardiovascular disease. N Engl J Med 380: 1033–1042. 10.1056/NEJMoa1806747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haycock PC, Burgess S, Wade KH, Bowden J, Relton C, Davey Smith G. 2016. Best (but oft-forgotten) practices: the design, analysis, and interpretation of Mendelian randomization studies. Am J Clin Nutr 103: 965–978. 10.3945/ajcn.115.118216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemani G, Bowden J, Davey Smith G. 2018. Evaluating the potential role of pleiotropy in Mendelian randomization studies. Hum Mol Genet 27: R195–R208. 10.1093/hmg/ddy163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hingorani A, Humphries S. 2005. Nature's randomised trials. Lancet 366: 1906–1908. 10.1016/S0140-6736(05)67767-7 [DOI] [PubMed] [Google Scholar]
- Holmes MV. 2019. Human genetics and drug development. N Engl J Med 380: 1076–1079. 10.1056/NEJMe1901565 [DOI] [PubMed] [Google Scholar]
- Holmes MV, Ala-Korpela M, Davey Smith G. 2017. Mendelian randomization in cardiometabolic disease: challenges in evaluating causality. Nat Rev Cardiol 14: 577–590. 10.1038/nrcardio.2017.78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes MV, Richardson TG, Ference BA, Davies N, Davey Smith G. 2020. Integrating genomics with biomarkers and therapeutic targets to invigorate cardiovascular drug development. Nat Rev Cardiol (in press). [DOI] [PubMed] [Google Scholar]
- HPS3/TIMI55-REVEAL Collaborative Group; Bowman L, Hopewell JC, Chen F, Wallendszus K, Stevens W, Collins R, Wiviott SD, Cannon CP, Braunwald E, et al. 2017. Effects of anacetrapib in patients with atherosclerotic vascular disease. N Engl J Med 377: 1217–1227. 10.1056/NEJMoa1706444 [DOI] [PubMed] [Google Scholar]
- *.Hwang LD, Davies NM, Warrington NM, Evans DM. 2020. Integrating family-based and Mendelian randomization designs. Cold Spring Harb Perspect Med 10.1101/cshperspect.a039503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamstrup PR, Tybjaerg-Hansen A, Steffensen R, Nordestgaard BG. 2009. Genetically elevated lipoprotein(a) and increased risk of myocardial infarction. JAMA 301: 2331–2339. 10.1001/jama.2009.801 [DOI] [PubMed] [Google Scholar]
- Keech A, Simes RJ, Barter P, Best J, Scott R, Taskinen MR, Forder P, Pillai A, Davis T, Glasziou P, et al. 2005. Effects of long-term fenofibrate therapy on cardiovascular events in 9795 people with type 2 diabetes mellitus (the FIELD study): randomised controlled trial. Lancet 366: 1849–1861. 10.1016/S0140-6736(05)67667-2 [DOI] [PubMed] [Google Scholar]
- Landray MJ, Haynes R, Hopewell JC, Parish S, Aung T, Tomson J, Wallendszus K, Craig M, Liang J, Collins R, et al. 2014. Effects of extended-release niacin with laropiprant in high-risk patients. N Engl J Med 371: 203–212. 10.1056/NEJMoa1300955 [DOI] [PubMed] [Google Scholar]
- Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G. 2008. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med 27: 1133–1163. 10.1002/sim.3034 [DOI] [PubMed] [Google Scholar]
- Lincoff AM, Nicholls SJ, Riesmeyer JS, Barter PJ, Brewer HB, Fox KAA, Gibson CM, Granger C, Menon V, Montalescot G, et al. 2017. Evacetrapib and cardiovascular outcomes in high-risk vascular disease. N Engl J Med 376: 1933–1942. 10.1056/NEJMoa1609581 [DOI] [PubMed] [Google Scholar]
- Lotta LA, Sharp SJ, Burgess S, Perry JRB, Stewart ID, Willems SM, Luan J, Ardanaz E, Arriola L, Balkau B, et al. 2016. Association between low-density lipoprotein cholesterol-lowering genetic variants and risk of type 2 diabetes: a meta-analysis. JAMA 316: 1383–1391. 10.1001/jama.2016.14568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin L, Hutchens M, Hawkins C. 2017. Clinical trial cycle times continue to increase despite industry efforts. Nat Rev Drug Discov 16: 157. 10.1038/nrd.2017.21 [DOI] [PubMed] [Google Scholar]
- Mascitelli L, Goldstein MR. 2016. Might ezetimibe be considered an expensive placebo? Int J Cardiol 207: 48–49. 10.1016/j.ijcard.2016.01.150 [DOI] [PubMed] [Google Scholar]
- Montgomery AA, Peters TJ, Little P. 2003. Design, analysis and presentation of factorial randomised controlled trials. BMC Med Res Methodol 3: 26. 10.1186/1471-2288-3-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MR, Tipney H, Painter JL, Shen J, Nicoletti P, Shen Y, Floratos A, Sham PC, Li MJ, Wang J, et al. 2015. The support of human genetic evidence for approved drug indications. Nat Genet 47: 856–860. 10.1038/ng.3314 [DOI] [PubMed] [Google Scholar]
- Nishikido T, Ray KK. 2018. Inclisiran for the treatment of dyslipidemia. Expert Opin Investig Drugs 27: 287–294. 10.1080/13543784.2018.1442435 [DOI] [PubMed] [Google Scholar]
- Nordestgaard BG, Varbo A. 2014. Triglycerides and cardiovascular disease. Lancet 384: 626–635. 10.1016/S0140-6736(14)61177-6 [DOI] [PubMed] [Google Scholar]
- Peto R, Baigent C. 1998. Trials: the next 50 years large scale randomised evidence of moderate benefits. BMJ 317: 1170–1171. 10.1136/bmj.317.7167.1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pocock SJ. 2006. Current controversies in data monitoring for clinical trials. Clin Trials 3: 513–521. 10.1177/1740774506073467 [DOI] [PubMed] [Google Scholar]
- Pocock SJ, Assmann SE, Enos LE, Kasten LE. 2002. Subgroup analysis, covariate adjustment and baseline comparisons in clinical trial reporting: current practice and problems. Stat Med 21: 2917–2930. 10.1002/sim.1296 [DOI] [PubMed] [Google Scholar]
- Psaty BM, Prentice RL. 2010. Minimizing bias in randomized trials: the importance of blinding. JAMA 304: 793–794. 10.1001/jama.2010.1161 [DOI] [PubMed] [Google Scholar]
- Richardson TG, Sanderson E, Palmer TM, Ala-Korpela M, Ference BA, Davey Smith G, Holmes MV. 2020. Evaluating the relationship between circulating lipoprotein lipids and apolipoproteins with risk of coronary heart disease: a multivariable Mendelian randomisation analysis. PLoS Med 17: e1003062. 10.1371/journal.pmed.1003062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesmeyer JS, Ruotolo G, Weerakkody G, Wolski K, McErlean E, Menon V, Lincoff MA, Nissen SE, Nicholls SJ 2018. Evacetrapib alone or in combination with different statin doses results in differential effect on lipoproteins and MACE rates: the ACCELERATE trial. Scientific Sessions, American Heart Association (AHA), Chicago, November 10. [Google Scholar]
- *.Richmond R, Davey Smith G. 2020. Mendelian randomization: concepts and scope. Cold Spring Harb Perspect Med 10.1101/cshperspect.a040501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JG, Farnier M, Krempf M, Bergeron J, Luc G, Averna M, Stroes ES, Langslet G, Raal FJ, El Shahawy M, et al. 2015. Efficacy and safety of alirocumab in reducing lipids and cardiovascular events. N Engl J Med 372: 1489–1499. 10.1056/NEJMoa1501031 [DOI] [PubMed] [Google Scholar]
- Sabatine MS, Giugliano RP, Wiviott SD, Raal FJ, Blom DJ, Robinson J, Ballantyne CM, Somaratne R, Legg J, Wasserman SM, et al. 2015. Open-label study of long-term evaluation against LDL cholesterol (OSLER) investigators. Efficacy and safety of evolocumab in reducing lipids and cardiovascular events. N Engl J Med 372: 1500–1509. 10.1056/NEJMoa1500858 [DOI] [PubMed] [Google Scholar]
- Sabatine MS, Giugliano RP, Keech AC, Honarpour N, Wiviott SD, Murphy SA, Kuder JF, Wang H, Liu T, Wasserman SM, et al. 2017. Evolocumab and clinical outcomes in patients with cardiovascular disease. N Engl J Med 376: 1713–1722. 10.1056/NEJMoa1615664 [DOI] [PubMed] [Google Scholar]
- *.Sanderson E. 2020. Multivariable Mendelian randomization and mediation. Cold Spring Harb Perspect Med 10.1101/cshperspect.a038984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson E, Davey Smith G, Windmeijer F, Bowden J. 2019. An examination of multivariable Mendelian randomization in the single sample and two-sample summary data settings. Int J Epidemiol 48: 713–727. 10.1093/ije/dyy262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt AF, Swerdlow DI, Holmes MV, Patel RS, Fairhurst-Hunter Z, Lyall DM, Hartwig FP, Horta BL, Hyppönen E, Power C, et al. 2017. PCSK9 genetic variants and risk of type 2 diabetes: a Mendelian randomisation study. Lancet Diabetes Endocrinol 5: 97–105. 10.1016/S2213-8587(16)30396-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz KF, Grimes DA. 2002. Generation of allocation sequences in randomised trials: chance, not choice. Lancet 359: 515–519. 10.1016/S0140-6736(02)07683-3 [DOI] [PubMed] [Google Scholar]
- Schulz KF, Altman DG, Moher D; CONSORT Group. 2010. CONSORT 2010 statement: updated guidelines for reporting parallel group randomised trials. BMJ 340: c332. 10.1136/bmj.c332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz GG, Olsson AG, Abt M, Ballantyne CM, Barter PJ, Brumm J, Chaitman BR, Holme IM, Kallend D, Leiter LA, et al. 2012. Effects of dalcetrapib in patients with a recent acute coronary syndrome. N Engl J Med 367: 2089–2099. 10.1056/NEJMoa1206797 [DOI] [PubMed] [Google Scholar]
- Schwartz GG, Steg PG, Szarek M, Bhatt DL, Bittner VA, Diaz R, Edelberg JM, Goodman SG, Hanotin C, Harrington RA, et al. 2018. Alirocumab and cardiovascular outcomes after acute coronary syndrome. N Engl J Med 379: 2097–2107. 10.1056/NEJMoa1801174 [DOI] [PubMed] [Google Scholar]
- Swerdlow DI, Preiss D, Kuchenbaecker KB, Holmes MV, Engmann JE, Shah T, Sofat R, Stender S, Johnson PC, Scott RA, et al. 2015. HMG-coenzyme A reductase inhibition, type 2 diabetes, and bodyweight: evidence from genetic analysis and randomised trials. Lancet 385: 351–361. 10.1016/S0140-6736(14)61183-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treadwell JR, Uhl S, Tipton K, Singh S, Santaguida L, Sun X, Berkman N, Viswanathan M, Coleman C, Shamliyan T, et al. 2012. Assessing equivalence and noninferiority. Agency for Healthcare Research and Quality (US), Rockville, MD. [PubMed] [Google Scholar]
- Triglyceride Coronary Disease Genetics Consortium and Emerging Risk Factors Collaboration; Sarwar N, Sandhu MS, Ricketts SL, Butterworth AS, Di Angelantonio E, Boekholdt SM, Ouwehand W, Watkins H, Samani NJ, et al. 2010. Triglyceride-mediated pathways and coronary disease: collaborative analysis of 101 studies. Lancet 375: 1634–1639. 10.1016/S0140-6736(10)60545-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsimikas S. 2018. RNA-targeted therapeutics for lipid disorders. Curr Opin Lipidol 29: 459–466. 10.1097/MOL.0000000000000549 [DOI] [PubMed] [Google Scholar]
- White J, Swerdlow DI, Preiss D, Fairhurst-Hunter Z, Keating BJ, Asselbergs FW, Sattar N, Humphries SE, Hingorani AD, Holmes MV. 2016. Association of lipid fractions with risks for coronary artery disease and diabetes. JAMA Cardiol 1: 692–699. 10.1001/jamacardio.2016.1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittes J. 2002. Sample size calculations for randomized controlled trials. Epidemiol Rev 24: 39–53. 10.1093/epirev/24.1.39 [DOI] [PubMed] [Google Scholar]