Abstract
Plant fitness is largely dependent on the root, the underground organ, which, besides its anchoring function, supplies the plant body with water and all nutrients necessary for growth and development. To exploit the soil effectively, roots must constantly integrate environmental signals and react through adjustment of growth and development. Important components of the root management strategy involve a rapid modulation of the root growth kinetics and growth direction, as well as an increase of the root system radius through formation of lateral roots (LRs). At the molecular level, such a fascinating growth and developmental flexibility of root organ requires regulatory networks that guarantee stability of the developmental program but also allows integration of various environmental inputs. The plant hormone auxin is one of the principal endogenous regulators of root system architecture by controlling primary root growth and formation of LR. In this review, we discuss recent progress in understanding molecular networks where auxin is one of the main players shaping the root system and acting as mediator between endogenous cues and environmental factors.
The evolution of land plants is characterized by the appearance of specialized organs such as leaves, flowers, and roots. In particular, roots have developed to anchor the plant body to the ground, mediate absorption of water and nutrients from the soil, and serve as storage organs for water and photo-assimilates (Raven and Edwards 2001; Osmont et al. 2007; Pires and Dolan 2012; Bellini et al. 2014).
Contrary to animals, where organ formation occurs primarily during embryogenesis, plants can produce new organs along their life cycle. Hence, plant roots are either of embryonic origin, such as primary roots (PRs), or formed postembryonically from parental roots or nonroot tissues (i.e., lateral roots [LRs] or adventitious roots [ARs], respectively). The root system in dicotyledonous plants is mainly based on the PR that sprouts to numerous lateral branches, which is known as a taproot or allorhizic system. Unlike dicots, monocotyledonous plants form fibrous or homorhizic root systems consisting of a dense mass of ARs that arise from the stem (Atkinson et al. 2014).
Arabidopsis thaliana, with one of the simplest and therefore the best anatomically described root systems within dicotyledonous plant species, became the preferred model to study molecular mechanisms regulating root branching. In the PR of Arabidopsis, concentric single layers of epidermis, cortex, and endodermis surround the pericycle, the tissue giving rise to LRs (Dolan et al. 1993). The pericycle consists of two cell populations, the xylem- and the phloem-pole pericycle cells (XPP and PPP, respectively). Typically, cells adjacent to xylem can acquire characteristics of founder cells (FCs) and reenter the cell cycle necessary for the initiation of LR primordia (LRP) organogenesis (Beeckman et al. 2001; Casimiro et al. 2003; Himanen et al. 2004; Parizot et al. 2008). LRPs, formed through a series of anti- and periclinal divisions classified in seven developmental stages, traverse through surrounding tissues until they emerge from the parental root (Malamy and Benfey 1997). Ultimately, the apical meristem of LRs, functionally analogous to that of the PR, is established. This simple, well-described root system allowed genetic and molecular screens for key factors controlling LR organogenesis. As a result, core genetic networks that regulate different phases of LR development have been identified and they are constantly updated for new components.
The plant hormone auxin is one of the principal regulators of LR organogenesis. Research focused on the mechanisms underlying root branching has corroborated the role of auxin as a trigger and a major coordinator of this postembryonic developmental process and provided molecular insights into auxin-orchestrated pathways (Torrey 1950; Blakely and Evans 1979; Blakely et al. 1988; Laskowski et al. 1995; Reed et al. 1998; Dubrovsky et al. 2008; Du and Scheres 2018; Israeli et al. 2020; Xuan et al. 2020). Intriguingly, besides its role as activator and regulator steering specific phases of LR organogenesis, auxin acts as an integrator of various endo- and exogenous inputs, thereby providing developmental plasticity and flexibility of the root system to adapt to fluctuating environmental conditions (Sun et al. 2009; Kapulnik et al. 2011; Duan et al. 2013).
Here we discuss recent advances in understanding pathways that, under the coordination of auxin, control different aspects of LR organogenesis and we cover views on the role of auxin as an integrator of environmental inputs to coordinate adaptive root responses.
ENDOGENOUS MECHANISMS DETERMINING RHIZOTACTIC PATTERNING
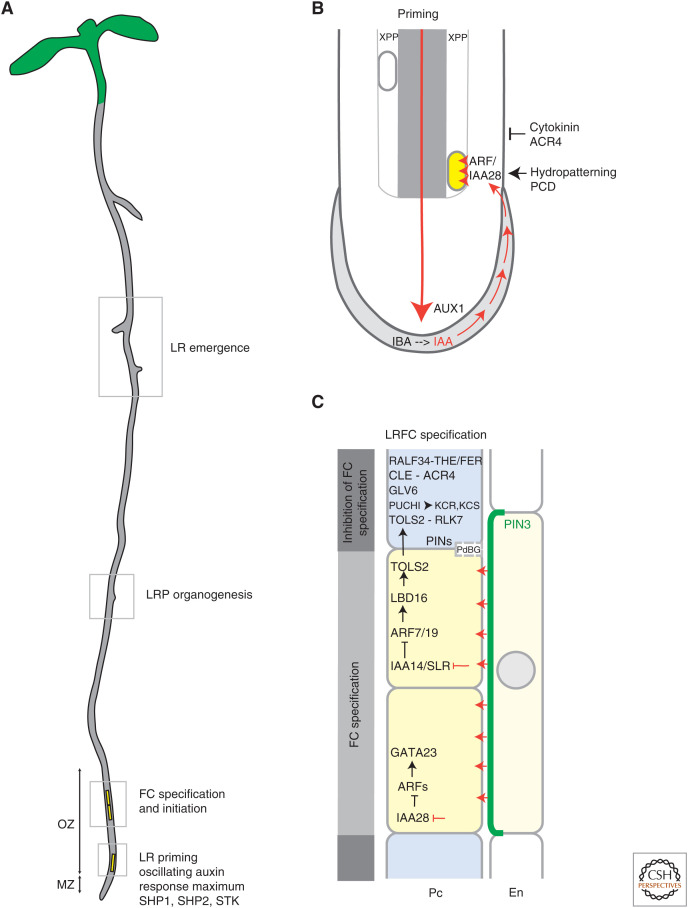
In Arabidopsis, a zone above the root apical meristem from the transition zone (TZ) to elongation zone (EZ) spatially defines a developmental window where LR initiation takes place (Casimiro et al. 2001; Dubrovsky et al. 2006). This region, also named the oscillation zone (OZ), is characterized by recurrent pulses of the DIRECT REPEAT5 (DR5) auxin-sensitive reporter and oscillatory expression of numerous genes (Ulmasov et al. 1997; De Smet et al. 2007; Moreno-Risueno et al. 2010). The oscillating auxin maxima and coregulated genes such as MADS-box transcription factors SHATTERPROOF1 (SHP1), SHP2, and SEEDSTICK (STK) define prebranch sites, where XPP cells are specified to FCs and subsequently LRs are initiated (Moreno-Risueno et al. 2010; Van Norman et al. 2013). In the XPP cells, auxin, through the signaling module encompassing AUXIN/INDOLE-3-ACETIC ACID repressor IAA28 and transcriptional activator AUXIN RESPONSE FACTOR7 (ARF7), triggers expression of the GATA23 transcription factor that controls FC identity (Fig. 1; De Rybel et al. 2010). Although the molecular principle of this endogenous mechanism working as a “root clock” is not fully understood, several recent studies provide important insights into pathways that contribute to regulation of the oscillation-based rhizotactic patterning.
Figure 1.
Lateral root priming and founder cell specification. (A) Schematic representation of the Arabidopsis seedling. Specific steps during lateral root (LR) development including priming, LR primordia (LRP) specification and initiation, and LR emergence are highlighted along the primary root (PR) in gray boxes. Pericycle cells involved in early stages of LR development are depicted in yellow. (B) Priming of pericycle founder cells (FCs) starts in the basal meristem and is driven by auxin reflux from the LR cap (LRC) (red arrows) to epidermal root cells. In the LRC, auxin flow is reinforced by the conversion of indole-3-butyric (IBA) to indole-3-acetic acid (IAA) (black dashed line). Factors promoting or repressing LR priming are included in the scheme (black arrows and bar-headed lines, respectively). Auxin priming of a xylem-pole pericycle (XPP) cell is highlighted in yellow. (C) Specification of FCs triggers the inhibition of FC establishment and LR initiation in flanking XPP cells. Polar localization of PIN3 (marked in green) regulates active auxin flow from the endodermis to the FC. Auxin signaling promoting or repressing expression of the indicated modules is given in red (arrows or bar-headed lines, respectively). Factors involved in FC specification and inhibition in the neighboring cells and their signaling pathways are depicted (black arrows and bar-headed lines). Auxin accumulates in the FC (yellow), while the neighboring FC flanking cells have a higher cytokinin content (blue). (Pc) Pericycle, (En) endodermis, (MZ) meristematic zone, (OZ) oscillation zone.
Inhibition of auxin transport by N-1-naphthylphthalamic acid (NPA) dims the oscillation of auxin response and the initiation of LRs, thus hinting at a role of polar auxin transport (PAT) in control of LR positioning (Xuan et al. 2016). Analyses of mutants defective in AUXIN RESISTANT1 (AUX1)/LIKE AUX1 (LAXs) auxin influx- and PIN-FORMED (PIN) or ABCB/PGP auxin efflux transporters further corroborate contributions of PAT to LR initiation (Fig. 1B; Marchant et al. 2002; Benková et al. 2003; Laskowski et al. 2008; Mravec et al. 2008). Intriguingly, targeted expression of AUX1 in the LR cap (LRC) and epidermal cells restored the aberrant LR initiation in the aux1 mutant, suggesting that the LRC-derived auxin might act as a trigger of LR organogenesis (De Smet et al. 2007; Xuan et al. 2016). A role of the LRC as a source of auxin essential for root branching is further supported by Naxillin, a non-auxin-like molecule that promotes LRC-specific conversion of the auxin precursor indole-3-butyric acid (IBA) to the active indole-3-acetic acid (IAA) and thereby stimulates LR formation. Accordingly, the ibr1ibr3ibr10 and ech2ibr1ibr3ibr10 mutants defective in enzymes of the IBA-to-IAA conversion pathway have a reduced number of initiated LRs (De Rybel et al. 2012; Xuan et al. 2015). Importantly, the LRC is not the only source of auxin required for establishment of prebranch sites, but it might accommodate a pathway that steers oscillation and thereby the spatiotemporal pattern of LR initiation. Programmed cell death (PCD) of the cells at the most distal end of the LRC displays oscillations and, consequently, auxin pulses are released to surrounding root tissues and define the LR spacing along the main root (Xuan et al. 2016; van den Berg and ten Tusscher 2018).
To maintain the acropetal pattern of root branching (i.e., younger LRs are positioned distally from older LRs), initiation of LRs must be restricted to a developmental window. Several processes contribute to LR spacing by suppressing ectopic initiation and thereby reinforcing and stabilizing the rhizotactic pattern. Enhanced expression of the cytokinin-sensitive two-component signaling sensor (TCS) reporter in pericycle cells adjacent to FCs and LRPs, suggests that cytokinin forms an inhibitory field that restricts the initiation of ectopic LRPs in proximity to existing ones (Fig. 1B). Consistently, inhibition of cytokinin biosynthesis or signaling resulted in LRP clustering (Bielach et al. 2012; Chang et al. 2015). In parallel, ARABIDOPSIS CRINKLY4 (ACR4), encoding a plasma membrane (PM)-localized receptor-like protein kinase (RLK), contributes to regulation of LR spacing. Defective rhizotactic patterning and accumulation of ectopic LRPs in acr4 mutants is in line with the inhibitory function of ACR4 during the initiation phase (De Smet et al. 2008; Chang et al. 2015). Putative ligands of ACR4 are signaling peptides of the CLAVATA3/ESR-RELATED (CLE) family and in particular CLE40 has been proposed to act as a direct ligand for ACR4 in the root (Stahl et al. 2013; Berckmans et al. 2020). However, quantitative binding assays revealed that ACR4 cannot directly interact with CLE40, and it may require additional components, like the ACR4 peptide coreceptor CLAVATA1 (CLV1) in the signaling complex (Okuda et al. 2020).
Recently, a peptide-signaling cascade involving TARGET OF LBD SIXTEEN (TOLS2) and its receptor RLK7 was identified as a local inhibitory mechanism controlling LR spacing. TOLS2 peptide, whose expression is auxin-dependent in FCs, moves to neighboring cells where it inhibits FC specification in ectopic positions through interaction with the RLK7 receptor (Toyokura et al. 2019). Activation of RLK7 promotes expression of PUCHI, a member of the AP2/EREBP transcription factor family, previously shown to control cell proliferation during LRP formation. Typically, loss of PUCHI activity results in abnormally enlarged flank cells and clustering of LRPs (Hirota et al. 2007). Interestingly, PUCHI was found to target transcription of genes involved in the synthesis of very long chain fatty acids (VLCFAs) such as a 3-Ketoacyl-CoA Synthase (KCS), a 3-Ketoacyl-CoA Reductase (KCR), and PASTICCINO2 (PAS2) (Fig. 1C; Trinh et al. 2019). Mutants perturbed in VLCFA biosynthesis showed similar LR initiation defects as observed in the puchi mutant, which suggests that PUCHI-regulated VLCFA biosynthesis is involved in control of spacing between LRPs. How VLCFAs suppress the initiation of LRs awaits further study. In addition, expression of RAPID ALKALINISATION FACTOR34 (RALF34) peptide was detected in pericycle cells flanking LRPs (Fig. 1C). RALF34, through activation of the THESEUS1 (THE1) and related FERONIA (FER) receptors, controls pathways involved in repression of LR initiation (Murphy et al. 2016; Gonneau et al. 2018). The downstream mechanism by which the RALF34-THE1/FER module regulates LR initiation has not yet been uncovered. Because swelling of the FCs is crucial for the onset of LR initiation (Vermeer et al. 2014; Ramakrishna et al. 2019; Jourquin et al. 2020), a similar mechanism to the known RALF1-FER signaling in PRs may involve the activity of the proton exporters, preventing acidification of the apoplast and triggering cell wall loosening and cell expansion (Fendrych et al. 2016; Haruta et al. 2018).
It has become evident that a rhizotactic pattern is controlled by multiple, mutually coordinated pathways. The root clock mechanism that drives oscillatory gene expression and steers the regulatory circuits to specify prebranch sites and LRP initiation is complemented by pathways that suppress LR formation outside of this developmental window.
AUXIN COORDINATES THE ESTABLISHMENT OF A DEVELOPMENTAL NICHE FOR LR INITIATION
Typically, the LR developmental program at the prebranching sites is triggered by auxin accumulating in two longitudinally adjacent FCs (i.e., two per cell file). Besides the longitudinal bicellular type of initiation, the longitudinal unicellular type has been reported in which a single FC in a cell file gives rise to the entire LRP. Long-term live-imaging experiments demonstrated that after specification of single FC its immediate neighboring pericycle cells are recruited and participate in LRP formation (von Wangenheim et al. 2016; Torres-Martínez et al. 2020).
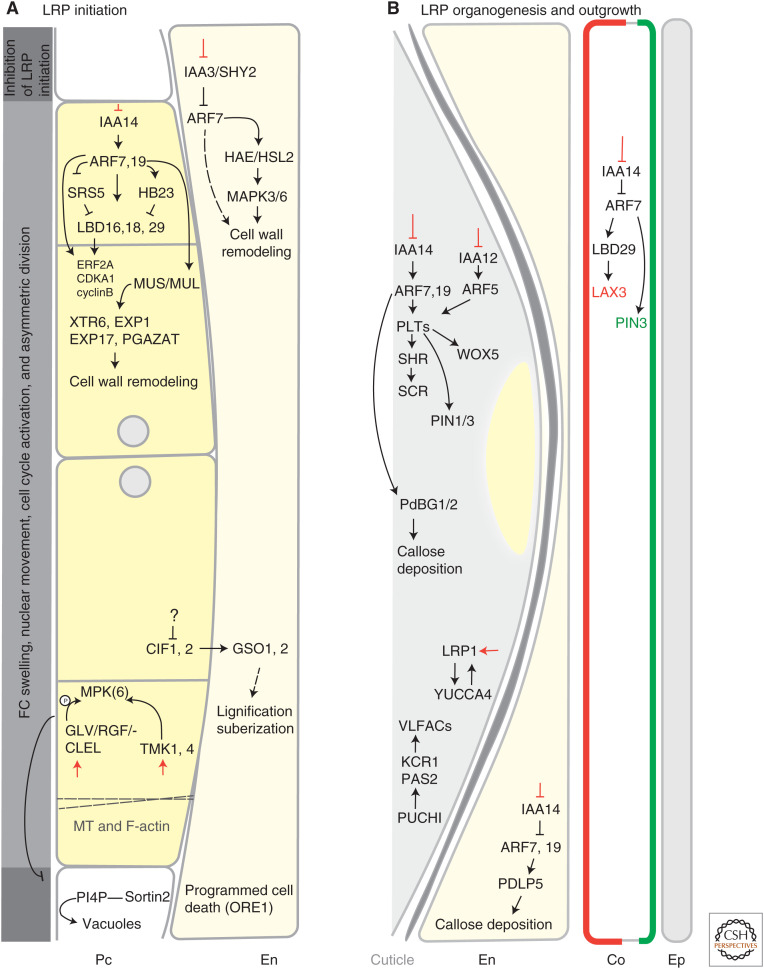
The PIN3 auxin efflux carrier reinforces reflux of auxin from surrounding endodermal to XPP cells (Fig. 1C) and thereby allows accumulation of auxin above a threshold level required for FC specification (Dubrovsky et al. 2008; Marhavý et al. 2013). Radial swelling of pairs of FCs and the migration of their nuclei toward the shared cell wall followed by asymmetric anticlinal divisions (perpendicular to the PR growth axis) are hallmarks of initiated LR organogenesis (Malamy and Benfey 1997; Dubrovsky et al. 2000). During the early phases of LR initiation, FCs and adjacent endodermal cells undergo complex morphocytological changes that encompass modulation of actin and microtubule cytoskeleton dynamics (Vilches Barro et al. 2019), changes of biomechanical cell wall properties (Swarup et al. 2008; Vermeer et al. 2014; Ramakrishna et al. 2019), and specification of symplastic fields of cell-to-cell connectivity (Fig. 2A; Hernández-Hernández et al. 2019). These processes, essential for the proper LR developmental program, are controlled through a few auxin-regulated signaling modules, such as SOLITARY ROOT(SLR)/IAA14-ARF7 and downstream acting transcription factors LATERAL ORGAN BOUNDARIES-DOMAIN16 (LBD16) and LBD18, BODENLOS (BDL)/IAA12-ARF5, or IAA3-ARF7 (Fukaki et al. 2002; Okushima et al. 2005, 2007; Vanneste et al. 2005; De Smet et al. 2007, 2010; Orosa-Puente et al. 2018). Importantly in FCs, the cell cycle is reactivated through transcriptional regulation of the E2Fa transcription factor by auxin-responsive IAA14-ARF7-LBD16 and LBD18 modules (Fig. 2A; Berckmans et al. 2011; Sanz et al. 2011; Lee et al. 2015). It is noteworthy that attempts to trigger LR initiation through either tissue-specific activation of cell-cycle machinery in XPP cells (Vanneste et al. 2005), or release of mechanical constraints imposed on XPP cells by endodermal cell ablation (Marhavý et al. 2016) demonstrate that a simple transition of XPP cells to a meristematically active stage is not sufficient to launch an LR developmental program. Hence, auxin, through spatiotemporal coordination of all the above-discussed pathways, establishes a developmental niche for LR initiation.
Figure 2.
Lateral root primordia development: from initiation to outgrowth. (A) Swelling of founder cells (FCs), nuclear movement, cytoskeleton rearrangements, and first asymmetric cell divisions defines the onset of lateral root primordia (LRP) development. Auxin signaling (red bar-headed lines) promotes the degradation of Aux/IAA repressing factors such as IAA14 and IAA3 and thereby stimulates the expression of downstream ARF, LBD transcription factors, and cell cycle and cell wall remodeling genes. Different factors are acting in the neighboring cells in which ectopic LRP initiation is inhibited. The initial cells of LRP accumulate higher auxin concentrations and are reported in yellow. (B) LRP organogenesis and outgrowth is regulated through several auxin signaling modules. The auxin signaling cascade and expression of specific factors involved are indicated in the relevant cell layers. At the surface of the developing LRP, a cuticle layer is formed to prevent adhesion when penetrating the overlaying tissues. In the cortex, LAX3 and PIN3 localization are marked in red and green, respectively. (MT) Microtubule, (Pc) pericycle, (En) endodermis, (Co) cortex, (Ep) epidermis.
New molecular players involved in the regulation of LR initiation and contributing to the plasticity of root branching have been identified recently. AtHB23, a homeodomain-leucine zipper (HD-Zip) I transcription factor, represses the expression of LBD16 and thereby suppresses the initiation of LRPs (Fig. 2A). Interestingly, expression of this TF differs in LRPs of secondary and tertiary order, suggesting that AtHB23 might act in the upstream pathway that fine-tunes the core “LR initiation module” as the root system branches and expands (Perotti et al. 2019). Similarly, SHI-RELATED SEQUENCE5 (SRS5), a member of the SHORT-INTERNODES family, represses the transcription of LBD16. SRS5 is under negative control of ARF7 and ARF19 and might act in a negative feedback loop repressing LR initiation (Fig. 2B; Yuan et al. 2020). Intriguingly, the activity of the IAA14-ARF7-LBD16 regulatory module that controls asymmetric cell divisions of FCs is complemented by GOLVEN/ROOT GROWTH FACTOR/CLE-like (GLV/RGF/CLEL) signaling peptides (Fig. 2A). Two members of the GLV gene family, GLV6 and GLV10, act as inhibitors of asymmetric cell divisions and signal through RGF1 INSENSITIVE receptors and MITOGEN-ACTIVATED PROTEIN KINASE6 (MPK6) to restrict the number of initial asymmetric cell divisions that take place during LR initiation (Fernandez et al. 2020).
ROLE OF CELL-TO-CELL TRANSPORT IN LRP ORGANOGENESIS
The LR developmental program starts with the asymmetric division of FCs. Although the subsequent rounds of divisions are less deterministic and do not follow a strictly stereotypical pattern, this division process results in a highly conserved tissue patterning and shape of developing LRP (Malamy and Benfey 1997; Lucas et al. 2013; von Wangenheim et al. 2016). LRPs originate from XPP cells embedded deeply within the parental PR, implying that significant parts of LR organogenesis take place while the primordium is surrounded by several tissue layers, each of them exhibiting specific biomechanical properties (Malamy 2005; Richter et al. 2009; Naseer et al. 2012; Vermeer et al. 2014; Banda et al. 2019). Hence, LR formation is governed by a tight cross talk between regulatory circuits acting in the body of the primordium and adjacent tissues. Remarkably, auxin and its distribution driven by PAT plays an instructive role in the proper formation of LRP as well as in the coordination of its emergence through surrounding tissues. In the LRP, PIN, and AUX1/LAX auxin transporters (Marchant et al. 2002; Benková et al. 2003; Marhavý et al. 2013; Péret et al. 2013) occupy distinct expression domains and polarize to specific membranes, thereby determining a graded auxin distribution with maximum at the tip of the primordium (Figs. 1C and 2B). Hence, numerous factors and pathways that control expression or polar membrane localization of the auxin transporters, including regulators of vesicular trafficking such as ARF GTPases and ARF guanine-nucleotide exchange factors (ARF-GEFs) (Geldner 2003), and hormones such as cytokinin (Marhavý et al. 2011, 2014), gibberellins (Löfke et al. 2013), and salicylic acid (Tan et al. 2020), play an important role in steering LRP organogenesis.
Auxin released from LRP to adjacent tissues activates pathways that promote cell separation and thereby facilitate emergence of the growing primordium (Swarup et al. 2008; Banda et al. 2019). Directional transport of auxin from the tip of the LRP toward neighboring tissues is driven by coordinated activities of LAX3 and PIN3 influx and efflux transporters, respectively, acting in few cells in direct contact with the emerging primordia (Fig. 2B). The remarkably specific spatiotemporal expression pattern of both transporters is the result of tightly coordinated consecutive expression. Auxin, through an ARF7-LBD29 transcriptional cascade, induces expression of LAX3, followed by enhancement of PIN3-mediated efflux (Péret et al. 2013; Porco et al. 2016).
PAT appears to play an essential role in auxin distribution during LRP organogenesis and, recently, novel factors that fine-tune the distribution of auxin at the subcellular level have been identified (Mravec et al. 2009; Barbez et al. 2012). Among them TOB1, a vacuolar transporter of the auxin precursor IBA, plays a role in LRP organogenesis. TOB1 expression is cytokinin-responsive and expands from LRP-flanking regions to the whole primordia and outer layers of the parental root. Mutations in TOB1 result in enhanced LRP development and while detailed mechanisms underlying TOB1 function await further investigation, TOB1 appears to act as an integrator of cytokinin signaling and auxin homeostasis in the regulation of LRP organogenesis (Michniewicz et al. 2019). The role of vacuoles in LR formation is also supported by a recent study of Sortin2, a chemical identified in screening for molecules affecting protein trafficking (Norambuena et al. 2008). Sortin2, through phosphatidylinositol 4-kinases PI4KIIIb1 and PI4KIIIb2, affects the abundance of phosphatidylinositol 4-phosphate (PI4P) at the PM and endosomal compartments, thereby promoting protein trafficking to the vacuoles. Intriguingly, the effect of Sortin2 on subcellular trafficking correlates with enhanced LR organogenesis independently of the canonical SCFTIR1/AFB auxin-mediated signaling. Whether Sortin2-mediated protein trafficking to the vacuoles modulates the activity of factors acting downstream of auxin signaling or involves different auxin-independent programs still needs to be resolved (Pérez-Henríquez et al. 2012; Rubilar-Hernández et al. 2019).
In parallel to cell-to-cell transport of auxin driven by PM transporters, symplastic plasmodesmata (PD)-dependent transport of molecules is critical for proper LRP organogenesis. Two related callose-degrading enzymes plasmodesmal-localized β-1,3 glucanase1 (PdBG1) and PdBG2, both transcriptionally regulated by auxin in an SLR/IAA14-dependent manner, control callose deposition in LRPs (Fig. 2B). Mutations in these genes result in defective primordia, in line with a role of symplastic transport in the regulation of LRP organogenesis (Benitez-Alfonso et al. 2013). Interestingly, in cells overlying LRPs, the auxin-inducible PLASMODESMATA-LOCATED PROTEIN5 (PDLP5) governs a temporary symplastic isolation of primordia via reversible callose accumulation. Interference with PDLP5 affects the outgrowth of LRP (Fig. 2B; Sager et al. 2020). Thus, auxin-controlled PD-dependent movement of molecules in both primordia and adjacent tissues contributes to fine-tuning of LRP organogenesis and outgrowth (Benitez-Alfonso et al. 2013; Sager et al. 2020).
AUXIN ORCHESTRATES GENETIC NETWORKS IN LRP AND ADJACENT TISSUES TO REGULATE LRP DEVELOPMENT
Auxin in LRP and adjacent cells triggers and coordinates essential pathways for the proper organogenesis and emergence. Importantly, recent works show that tissues overlaying LRPs are not just biomechanical barriers that need to be crossed during emergence from the parental root, but they play an active role in shaping and pattering the LRP (Lucas et al. 2008; Vermeer et al. 2014).
In the primordium, auxin signals transduced through specific modules, including SLR/IAA14-ARF7, ARF19, and BDL/IAA12-ARF5, activate multiple downstream pathways. Among them, the auxin-controlled PLETHORA (PLT) transcription factors, which further define expression of PIN1 and PIN3 transporters, as well as the SHORT ROOT (SHR), SCARECROW (SCR), and WUSCHEL-RELATED HOMEOBOX5 (WOX5) transcription factors steer the cell differentiation and patterning of LRPs (Du and Scheres 2017). Recently, LATERAL ROOT PRIMORDIA1 (LRP1), a member of the SHORT INTERNODES/STYLISH (SHI/STY) family, whose expression is regulated by histone deacetylation in an auxin-dependent manner, was recognized as a novel negative regulator of LRP organogenesis. LRP1 promotes auxin biosynthesis through the regulation of YUCCA4 (YUC4) and thus contributes to LRP formation (Fig. 2B; Singh et al. 2020).
Strikingly, in parallel to the well-established Aux/IAA-ARF-mediated signaling, a new noncanonical auxin transduction pathway acting through transmembrane kinases (TMKs) has been proposed. TMKs, in response to auxin, activate the mitogen-activated protein kinase (MAPK) pathway and thereby control the cell-division patterns during LRP organogenesis (Huang et al. 2019).
In outer tissues, auxin, through signaling modules such as SHORT HYPOCOTYL2 (SHY2)/IAA3-ARF7 in the endodermis and SLR/IAA14-ARF7 and ARF19 in the cortex and epidermis, regulates distinct pathways that enable separation of cells with different cell wall properties. For example, in the walls of endodermal cells, Casparian strips, enriched for suberin and lignin, form an extracellular hydrophobic barrier between the outer and inner root tissues to control water and nutrient uptake (Doblas et al. 2017; Nakayama et al. 2017). Interestingly, the expression of the CASPARIAN STRIP INTEGRITY FACTOR1 (CIF1) and CIF2, peptides, which through SCHENGEN3/GASSHO1 (SGN3/GSO1) and GSO2 leucine-rich repeat (LRR)-RLK receptors regulate lignification and suberization of the endodermal tissues, is suppressed in cells adjacent to LRPs (Fig. 2A). Although the mechanisms that control local down-regulation of CIFs and the role of auxin in this process need to be examined, the findings support the concept of a local, tissue-specific pathway engaged in cell separation during LRP outgrowth (Ghorbani et al. 2015; Nakayama et al. 2017). Degradation of pectins, which are major components of plant cell walls, is facilitated by the INFLORESCENCE DEFICIENT IN ABSCISSION (IDA) peptide. IDA interacts with the HAESA (HAE)/HAESA-LIKE2 (HSL2) LRR-RLK and activates the MKK4/MKK5–MPK3/MPK6 cascade, which promotes expression of a specific set of the cell wall remodeling enzymes and thereby enhances LR emergence (Fig. 2A; Kumpf et al. 2013; Zhu et al. 2019b).
Whereas initially major attention was given to mechanisms associated with separation of cells overlying LRP, current studies point out that in LRPs, auxin-controlled cell wall remodeling also has an important patterning function. MUSTACHE (MUS) and MUSTACHES-LIKE (MUL) genes, encoding inactive LRR-RLKs, are expressed in early-stage LRPs in an auxin/ARF7-ARF19-dependent manner. RNA-seq data suggest that MUS and MUL control LRP development, likely via regulating cell wall biosynthesis and remodeling genes such as XYLOGLUCAN ENDOTRANSGLYCOSYLASE6 (XTR6), EXPANSIN1 (EXP1), EXP17, and POLYGALACTURONASE ABSCISSION ZONE A. THALIANA (PGAZAT) (Xun et al. 2020).
In addition, recent findings hint at PCD as a parallel regulatory pathway to cell wall remodeling–driven cell separation during LR emergence. A subset of LRP-overlying cells display induction of PCD marker genes and their cell death was detected by electron and other microscopic techniques. Cell-death-deficient mutants lacking the positive cell death regulator ORESARA1/ANAC092 (ORE1) exhibit a delayed outgrowth of LRPs and these defects were restored by physical or genetic elimination of cells overlying the LRPs (Escamez et al. 2020).
Tight contact of developing LRPs and surrounding tissues raises an important question about the mechanisms that prevent adhesion of newly formed organs and parental root tissues. Intriguingly, a layer comprised of an insoluble polyester (cutin) and soluble lipids (waxes), called cuticle, was detected from the early stages on the surface of developing LRPs (Berhin et al. 2019). Mutants defective in cuticle formation exhibit deformations of LRPs, suggesting that, like in the shoot apical meristem and other plant organs, cuticle forms a separating layer, which prevents adhesion of adjacent tissues and thereby facilitates penetration of LRP toward the root surface (Fig. 2B; Nawrath et al. 2013; Ingram and Nawrath 2017; Berhin et al. 2019).
DISTINCT REGULATORY PATHWAYS ARE INVOLVED IN THE REGULATION OF LR DEVELOPMENT
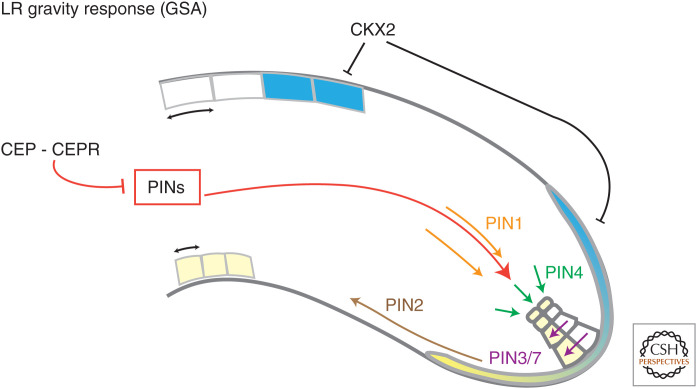
The outgrowth of LRs and their maturation encompasses a number of events including the establishment of a connection of the vascular system with that of the parental root and the formation of an apical meristem at the proximal end of the LR. The apical meristem, analogous to that of the PR, is essential for production of new cells and to sustain growth of LRs. Although many regulatory pathways are shared with PRs, some genetic programs are specific to LRs. An obvious example of such a nonshared pathway is the SLR/IAA14-ARF7-ARF19 signaling module. Mutations in components of this module severely affect LR formation, while the PR remains mostly unaffected (Fukaki et al. 2002). Similarly, expression and functional analyses of AtHB23 indicate that specialized pathways have developed to control PRs and different-order LRs (Perotti et al. 2019). Genetically distinct pathways controlling roots of either embryonic or postembryonic origin provide crucial advantages for plant survival under conditions that limit growth of the PR. Furthermore, radial expansion of the root system, essential for maximizing soil exploration, relies on diversification of gravity-sensing mechanisms in LR and PR. Whereas PRs are highly responsive to gravity and they rapidly realign with changed gravity vector (Friml et al. 2002; Zhu et al. 2019a), attenuated gravity sensing in LRs allows their horizontal growth (Rosquete et al. 2013; Waidmann and Kleine-Vehn 2020). Reduced gravity-driven bending of LRs is the result of partially suppressed activity of the PAT machinery, which mediates root gravity response through rapid relocation of auxin to the lower gravi-stimulated side of the root (Rosquete et al. 2013; Ogura et al. 2019). Moreover, in LRs, auxin-driven gravity sensing is counterbalanced by a cytokinin-dependent mechanism. Local increase of cytokinin signaling at the upper LR flank leads to growth inhibition and attenuation of downward gravitropic bending of LRs (Fig. 3). Interestingly, a genome-wide association study revealed that activity of Cytokinin Oxidase 2 (CKX2) which catalyzes the degradation of cytokinins and thereby controls levels of cytokinins at the tip of LR, is an important factor determining gravity set point angle (GSA) of LRs (Fig. 3; Waidmann et al. 2019). Notably, regulation of the LR GSA is not fully root-autonomous and can be fine-tuned by systemic signals that coordinate the development of root and shoot organs. Recently, interaction of CARBOXY-TERMINALLY ENCODED PEPTIDE (CEP) with the CEP RECEPTOR1 (CEPR1) in shoot was shown to control the GSA of LRs by reducing rootward auxin transport and/or by altering auxin levels in shoots (Chapman et al. 2020).
Figure 3.
Auxin and cytokinin define the gravitropic set point angle of the lateral root (LR) upon emergence. PIN-driven auxin flow to LR columella cells (yellow) is depicted as a red central arrow. Asymmetric cytokinin distribution in the LR cap (LRC) cell and in the upper side of LR is highlighted in blue. Interactions among factors interfering with the gravitropic angle are indicated (bar-headed lines). Differential cell elongation between the upper and lower side of LR is represented as double-headed black arrows.
ROOT SYSTEM ARCHITECTURE SHAPING BY ENVIRONMENTAL CUES
In heterogeneous soil environments, the root system is exposed to numerous cues such as mechanical obstacles, the availability of water and patches of nutrients, the level of oxygen, and light passing through the soil. The root continuously perceives these variables and translates them to adaptive developmental responses, including modulation of the branching pattern. Bending of PRs triggered either by gravity or deflection from obstacles are examples of an environmentally driven mechanism that significantly contribute to modulation of the LR initiation pattern (Ditengou et al. 2008; Lucas et al. 2008). Although the molecular details remain unclear, in silico models suggest that mechanical deformation of tissues during curve formation leads to local increases of auxin activity, which stimulates the initiation of LR (Laskowski et al. 2008). Alternatively, root bending could trigger a transient increase of Ca2+ within the pericycle that might act as a signal to translate mechanical forces to initiate LR development (Richter et al. 2009).
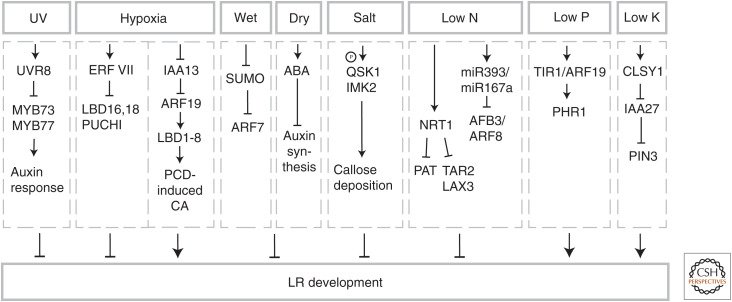
Light is one of the environmental factors essential for plant survival. It affects not only the development and physiology of the shoot, but also the underground root system. Importantly, root cells also possess photoreceptors that can be activated by light. Hence, roots not only receive information about light conditions through signaling molecules traveling from the shoot, but they also directly perceive light passing through the soil (Lee et al. 2016; Yang et al. 2020). For example, light perceived by the photoreceptor PHYTOCHROME B (PHYB) in roots activates the ELONGATED HYPOCOTYL5 (HY5) transcription factor and thereby modulates growth and gravitropic responses of the PR (Lee et al. 2016). Recently, another root-based mechanism of light-regulated responses has been described. Ultraviolet B (UV-B) light is an inherent component of sunlight that markedly affects root branching. At the molecular level, UV-B light activates the UVB-RESISTANCE8 (UVR8) receptor and triggers its relocation to the nucleus, where it inhibits the DNA-binding activities of MYB73/MYB77 transcription factors. As a result, transcription of the auxin-responsive genes is suppressed and consequently LR development is inhibited (Fig. 4; Yang et al. 2020).
Figure 4.
Environmental factors regulating lateral root (LR) development. Schematic representation of environmental cues, signaling pathways, and molecular mechanism regulating LR organogenesis. Factors promoting or repressing LR developments are indicated as black arrows or bar-headed lines, respectively.
The soil environment displays temporary oxygen deprivations due to seasonal rainfalls, flooding, or snow melting. Plants suffering from this drop in the soil oxygen content react by adaptive responses including inhibition of LR development. In-depth analyses revealed that in planta, the lack of active circulation may lead to the formation of internal hypoxic areas. These hypoxic niches, characterized by the expression of hypoxia marker genes, including a subgroup of ERF-VII transcription factors, were detected at specific stages of LRP development both in aerobic and enhanced at low oxygen conditions. ERF-VII transcription factors bind to promoters of the auxin-induced genes LBD16, LBD18, and PUCHI and suppress their expression, resulting in inhibition of LRP development (Shukla et al. 2019). Further insights into mechanisms associated with the regulation of oxygen levels and their impact on the root system were obtained from rice. In rice, lysigenous aerenchyma, a gas space created by cortical cell death, is established constitutively under aerobic conditions to enhance internal oxygen transport and its formation increases under oxygen-deficient conditions. In the dominant-negative mutant of the IAA13 auxin signaling repressor, reduced aerenchyma formation correlated with defects in LR formation. This hints at a role for auxin signaling in the coordination of both processes. Consistently, LBD1-8, a transcriptional target of IAA13 and ARF19, is expressed in cortex and LRP. Restoration of LBD1-8 expression recovered aerenchyma and LR formation in the iaa13 background, implying that auxin signaling coregulates aerenchyma and LR formation (Yamauchi et al. 2019).
Roots, as they grow through heterogenic soil environments, are exposed to wet soil aggregates or airy pores, where they experience a local transient water deficit. Recent advances in bioimaging techniques, which allowed monitoring of root growth in the natural soil environments, revealed that, while LR organogenesis is enhanced at the root side exposed to moisture, it is suppressed in airy microenvironments (Bao et al. 2014; Orman-Ligeza et al. 2018; Orosa-Puente et al. 2018; von Wangenheim et al. 2020). Intriguingly, both processes, hydro-patterning (promoting effect of moisture on LR organogenesis) and xero-branching (LR organogenesis inhibition in airy microenvironments) appear to be regulated by specific molecular pathways. Hydro-patterning is primarily driven through differential posttranslational control of the sumoylation pattern of ARF7 and its impact on auxin signaling. ARF7 is sumoylated on the airy side of the root, resulting in an interaction with the IAA3 repressor that inhibits LR initiation. On the water-exposed side of the root, ARF7 is not sumoylated, enabling the transcription factor to activate expression of genes involved in LR initiation (Orosa-Puente et al. 2018). Intriguingly, local heterogeneity in moisture also affects later phases of LRP organogenesis, namely the LR outgrowth angle. Typically, LR initiate from multiple adjacent XPP files, where the central cell file forms the tip of the primordium and contributes most of the cell mass to developing LRP (von Wangenheim et al. 2017). Exposure to water gradient might change proportion with which the individual XPP files contribute to formation of LRP and promote leaning of LR toward the water reach area (von Wangenheim et al. 2020).
Studies of xero-branching in cereals and Arabidopsis point at an indispensable role of abscisic acid (ABA) signaling when roots are not in contact with water. Endogenous ABA accumulation in the root correlates with attenuated auxin biosynthesis and consequently decreased LRP initiation (Deak and Malamy 2005; Orman-Ligeza et al. 2018).
When roots are exposed to osmotic stress in the presence of high soil salinity, the PM-located LRR-RLKs Qian Shou kinase (QSK1) and inflorescence meristem kinase2 (IMK2) rapidly relocate to the PD and through the regulation of callose-deposition fine-tune LRP organogenesis (Grison et al. 2019).
LATERAL ROOT DEVELOPMENT AS A FORAGING STRATEGY
The root system is highly responsive to nutrient availability and distribution within the soil. This is particularly true for major nutrients like nitrogen (N), phosphorus (P), and potassium (K), which are limiting nutrients for plant growth (Kellermeier et al. 2013; Guan 2017; Crombez et al. 2019; Vidal et al. 2020).
Nitrogen is an essential element that plants can acquire from various inorganic or organic sources in the soil. Fluctuations in both concentrations and the form of N sources available in the soil have prominent effects on root systems. Deficiency in N severely interferes with root growth and development, whereas low availability of nitrogen enhances root branching to promote the exploitation of this macronutrient. High levels of N might trigger production of stress hormones such as ethylene that lead to inhibition of PR and LR elongation growth (Tian et al. 2009; Gruber et al. 2013; Jia and von Wirén 2020). Interestingly, the local nitrate-rich zones enhance LR outgrowth (Remans et al. 2006). These complex adaptive responses of roots to N sources and heterogeneity in availability are regulated by a combination of systemic and local signaling (Ruffel 2018). The impact of available N on the root system is closely interconnected with the activity of plant hormones including auxin. A number of studies have demonstrated that auxin biosynthesis and transport are altered in response to different N regimes in various plant species (Gutiérrez et al. 2007; Ma et al. 2014; Krouk 2016; Maghiaoui et al. 2020). At the level of auxin signaling, auxin receptor AFB3 and auxin response factor AUXIN RESPONSE FACTOR 8 (ARF8) with associated miR393 and miRNA167s, respectively, were identified as N-responsive modules that control root system architecture in response to N availability in Arabidopsis (Gifford et al. 2008; Vidal et al. 2010). The NITRATE TRANSPORTER1.1 (NRT1.1), a principal component of nitrate transport and sensing, acts as an important integrator of nitrate and auxin pathways in Arabidopsis (Krouk et al. 2010; Bouguyon et al. 2015; Sun et al. 2017; Naz et al. 2019; Vidal et al. 2020). NRT1.1 is a nitrate transceptor with a dual transport and sensing function, which in the absence or at low nitrate availability displays auxin transport activity. The ability of NRT1.1 to transport auxin has a critical role in adjustment of LRP development to nitrate levels (Fig. 4). At low nitrate, NRT1 negatively affects LRP development by promoting auxin transport out of the primordia tip, whereas at the optimal level of this macronutrient, accumulation of auxin promotes LRP development (Krouk et al. 2010; Bouguyon et al. 2015). Intriguingly, NRT1, whose abundance is tightly controlled by nitrate, not only fine-tunes flow of auxin in LRPs but also controls both auxin biosynthesis by repressing expression of TRYPTOPHAN AMINOTRANSFERASE-RELATED2 (TAR2), and auxin influx into tissues overlaying LRP by attenuating LAX3 expression (Maghiaoui et al. 2020). Hence, NRT1.1 acts as a regulatory hub that senses and controls nitrate levels in the plant body and through multiple pathways controls activity of auxin, thereby translating N status of the plant into adaptive developmental responses including the root system architecture.
Although P is a major component in soil nutrients, only a small fraction is bioavailable for plants in the form of phosphate (Pi). Pi starvation triggers a series of adaptive responses in plants (Crombez et al. 2019). A Pi shortage fosters the formation of a shallow root system by attenuating PR growth and promoting branching (Naumann et al. 2019). Increased LR formation in Pi-deprived Arabidopsis seedlings is concomitant with increased expression of the TIR1 auxin receptor and ARF19 (Pérez-Torres et al. 2008). Both ARF7 and ARF19 positively regulate PHOSPHATE STARVATION RESPONSE1 (PHR1), an MYB-CC transcription factor, which is one of the main positive regulators of Pi-response genes (Bustos et al. 2010; Huang et al. 2018). Expression of PHR1 can partially rescue the LR-lacking phenotype of arf7/arf19 mutants, suggesting a compensatory feedback mechanism acting independently of LBD16 and LBD29 in response to low Pi (Huang et al. 2018). Other factors like light, ethylene, and strigolactones can coordinate PHR1 expression and the phosphate response, suggesting a high degree of cross talk among hormone signaling pathways (Sun et al. 2014; Liu et al. 2017; Crombez et al. 2019). In particular, PHR1 activation leads to a reduction in nitrate uptake and represents a direct link between phosphorus and nitrogen regulation (Maeda et al. 2018).
In contrast to Pi, potassium is highly soluble and K-deficiency inhibits LR organogenesis by interfering with auxin signaling and the expression of PIN3 in LRP (Kellermeier et al. 2013; Shahzad et al. 2020). Characteristic regulation of LR development in low K is achieved by RNA-directed DNA methylation (RdDM) (Matzke and Mosher 2014; Shahzad et al. 2020). The chromatin remodeling factor CLASSY (CLSY1) is a component of the RdDM complex, which is involved in silencing of a specific subset of transposable elements (TEs) and genes (Zhou et al. 2018). The promoter region of IAA27, an auxin signaling repressor and negative regulator of root branching, contains several of these TEs, and accordingly displays hypomethylation in RdDM mutants. Hence, a DNA-methylation-mediated transcriptional repression by CLSY1 has been proposed as an alternative pathway promoting LR development in low K by silencing IAA27 expression when the auxin-dependent degradation is impaired (Shahzad et al. 2020).
CONCLUSION AND PERSPECTIVES
Roots are underground organs that anchor and provide the plant body with water and nutrients essential for sustained growth and development. Developmental plasticity of the root system, based on the ability of genetic programs that control LR organogenesis to integrate environmental cues, has become a successful strategy supporting plant survival in very different habitats. Current research is providing exciting insights into mechanisms that act in the core of these unique root system features. Particularly, advances in monitoring root growth in real time and in natural soil environments have brought attention to some, so far, neglected factors that significantly impact the root system (Orman-Ligeza et al. 2018; von Wangenheim et al. 2020). Heterogeneity of soils with diverse microenvironments differing in water and oxygen content, nutrient composition, temperature, or light forms a complex universe that continuously shapes the root system and determines the fitness of the whole plant. In this dynamic soil environment, our view on auxin as a key endogenous orchestrator of root branching gains new perspectives and is challenged by intriguing open questions. How would relatively few auxin signaling modules integrate such a variety of environmental signals and translate them to coordinate diverse processes during LR organogenesis: the regulation of the cell cycle and divisions, actin and microtubule dynamics, cell wall remodeling, tissue patterning, and others. What are the molecular principles underlying specificity of the auxin action? Identification of a novel noncanonical TMK-mediated auxin transduction pathway (Huang et al. 2019), acting in parallel to well-established auxin signaling modules in LRP formation represents an exciting possibility of parallel pathways acting in the context of varying environmental conditions. Arabidopsis is a suitable model for investigation of basic developmental and molecular principles in root system architecture, but anatomical studies on a broader range of plant species point at significant diversities in LRP organogenesis (Xiao et al. 2019). This demonstrates that studying the molecular mechanisms controlling root branching in other plant species may provide important insights into evolution of root system architecture and environmental responses. Moreover, the rapidly progressing area of soil microbiota research and interaction with the plant's roots will reveal more genetic pathways that feed into modules controlling LR organogenesis.
In conclusion, the root system architecture is the result of a fine balance between the endogenous plant developmental program and exogenous inputs perceived from the environment. In this scenario, auxin is standing as a master regulator of LR organogenesis and future research will provide more elements to fully understand its primary role in shaping root architecture.
ACKNOWLEDGMENTS
We apologize to all the authors whose scientific work could not be cited and discussed because of space restrictions. We thank Dr. Inge Verstraeten (IST Austria) and Dr. Juan Carlos Montesinos-Lopez (ETH Zürich) for helpful suggestions. This work was supported by the DOC Fellowship Programme of the Austrian Academy of Sciences (25008) to C.A.
Footnotes
Editors: Dolf Weijers, Karin Ljung, Mark Estelle, and Ottoline Leyser
Additional Perspectives on Auxin Signaling available at www.cshperspectives.org
REFERENCES
- Atkinson JA, Rasmussen A, Traini R, Voss U, Sturrock C, Mooney SJ, Wells DM, Bennett MJ. 2014. Branching out in roots: uncovering form, function, and regulation. Plant Physiol 166: 538–550. 10.1104/pp.114.245423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banda J, Bellande K, von Wangenheim D, Goh T, Guyomarc'h S, Laplaze L, Bennett MJ. 2019. Lateral root formation in Arabidopsis: a well-ordered LRexit. Trends Plant Sci 24: 826–839. 10.1016/j.tplants.2019.06.015 [DOI] [PubMed] [Google Scholar]
- Bao Y, Aggarwal P, Robbins NE, Sturrock CJ, Thompson MC, Tan HQ, Tham C, Duan L, Rodriguez PL, Vernoux T, et al. 2014. Plant roots use a patterning mechanism to position lateral root branches toward available water. Proc Natl Acad Sci 111: 9319–9324. 10.1073/pnas.1400966111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbez E, Kubeš M, Rolčík J, Béziat C, Pěnčík A, Wang B, Rosquete MR, Zhu J, Dobrev PI, Lee Y, et al. 2012. A novel putative auxin carrier family regulates intracellular auxin homeostasis in plants. Nature 485: 119–122. 10.1038/nature11001 [DOI] [PubMed] [Google Scholar]
- Beeckman T, Burssens S, Inzé D. 2001. The peri-cell-cycle in Arabidopsis. J Exp Bot 52: 403–411. [DOI] [PubMed] [Google Scholar]
- Bellini C, Pacurar DI, Perrone I. 2014. Adventitious roots and lateral roots: similarities and differences. Annu Rev Plant Biol 65: 639–666. 10.1146/annurev-arplant-050213-035645 [DOI] [PubMed] [Google Scholar]
- Benitez-Alfonso Y, Faulkner C, Pendle A, Miyashima S, Helariutta Y, Maule A. 2013. Symplastic intercellular connectivity regulates lateral root patterning. Dev Cell 26: 136–147. 10.1016/j.devcel.2013.06.010 [DOI] [PubMed] [Google Scholar]
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J. 2003. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602. 10.1016/S0092-8674(03)00924-3 [DOI] [PubMed] [Google Scholar]
- Berckmans B, Vassileva V, Schmid SPC, Maes S, Parizot B, Naramoto S, Magyar Z, Kamei CLA, Koncz C, Bögre L, et al. 2011. Auxin-dependent cell cycle reactivation through transcriptional regulation of Arabidopsis E2Fa by lateral organ boundary proteins. Plant Cell 23: 3671–3683. 10.1105/tpc.111.088377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berckmans B, Kirschner G, Gerlitz N, Stadler R, Simon R. 2020. CLE40 signaling regulates root stem cell fate. Plant Physiol 182: 1776–1792. 10.1104/pp.19.00914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berhin A, de Bellis D, Franke RB, Buono RA, Nowack MK, Nawrath C. 2019. The root cap cuticle: a cell wall structure for seedling establishment and lateral root formation. Cell 176: 1367–1378.e8. 10.1016/j.cell.2019.01.005 [DOI] [PubMed] [Google Scholar]
- Bielach A, Podlesáková K, Marhavý P, Duclercq J, Cuesta C, Müller B, Grunewald W, Tarkowski P, Benková E. 2012. Spatiotemporal regulation of lateral root organogenesis in Arabidopsis by cytokinin. Plant Cell 24: 3967–3981. 10.1105/tpc.112.103044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakely L, Evans TA. 1979. Cell dynamics studies on the pericycle of radish seedling roots. Plant Sci Lett 14: 79–83. 10.1016/0304-4211(79)90158-5 [DOI] [Google Scholar]
- Blakely LM, Blakely RM, Colowit PM, Elliott DS. 1988. Experimental studies on lateral root formation in radish seedling roots. II: Analysis of the dose-response to exogenous auxin. Plant Physiol 87: 414–419. 10.1104/pp.87.2.414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouguyon E, Brun F, Meynard D, Kubeš M, Pervent M, Leran S, Lacombe B, Krouk G, Guiderdoni E, Zažímalová E, et al. 2015. Multiple mechanisms of nitrate sensing by Arabidopsis nitrate transceptor NRT1.1. Nat Plants 1: 15015. 10.1038/nplants.2015.15 [DOI] [PubMed] [Google Scholar]
- Bustos R, Castrillo G, Linhares F, Puga MI, Rubio V, Pérez-Pérez J, Solano R, Leyva A, Paz-Ares J. 2010. A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet 6: e1001102. 10.1371/journal.pgen.1001102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casimiro I, Marchant A, Bhalerao RP, Beeckman T, Dhooge S, Swarup R, Graham N, Inzé D, Sandberg G, Casero PJ, et al. 2001. Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell 13: 843–852. 10.1105/tpc.13.4.843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casimiro I, Beeckman T, Graham N, Bhalerao R, Zhang H, Casero P, Sandberg G, Bennett MJ. 2003. Dissecting Arabidopsis lateral root development. Trends Plant Sci 8: 165–171. 10.1016/S1360-1385(03)00051-7 [DOI] [PubMed] [Google Scholar]
- Chang L, Ramireddy E, Schmülling T. 2015. Cytokinin as a positional cue regulating lateral root spacing in Arabidopsis. EXBOTJ 66: 4759–4768. 10.1093/jxb/erv252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman K, Ivanovici A, Taleski M, Sturrock CJ, Ng JLP, Mohd-Radzman NA, Frugier F, Bennett MJ, Mathesius U, Djordjevic MA. 2020. CEP receptor signalling controls root system architecture in Arabidopsis and Medicago. New Phytol 226: 1809–1821. 10.1111/nph.16483 [DOI] [PubMed] [Google Scholar]
- Crombez H, Motte H, Beeckman T. 2019. Tackling plant phosphate starvation by the roots. Dev Cell 48: 599–615. 10.1016/j.devcel.2019.01.002 [DOI] [PubMed] [Google Scholar]
- Deak KI, Malamy J. 2005. Osmotic regulation of root system architecture: osmotic regulation of lateral root formation. Plant J 43: 17–28. 10.1111/j.1365-313X.2005.02425.x [DOI] [PubMed] [Google Scholar]
- De Rybel B, Vassileva V, Parizot B, Demeulenaere M, Grunewald W, Audenaert D, Van Campenhout J, Overvoorde P, Jansen L, Vanneste S, et al. 2010. A novel Aux/IAA28 signaling cascade activates GATA23-dependent specification of lateral root founder cell identity. Curr Biol 20: 1697–1706. 10.1016/j.cub.2010.09.007 [DOI] [PubMed] [Google Scholar]
- De Rybel B, Audenaert D, Xuan W, Overvoorde P, Strader LC, Kepinski S, Hoye R, Brisbois R, Parizot B, Vanneste S, et al. 2012. A role for the root cap in root branching revealed by the non-auxin probe naxillin. Nat Chem Biol 8: 798–805. 10.1038/nchembio.1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I, Tetsumura T, De Rybel B, Frey NF, Laplaze L, Casimiro I, Swarup R, Naudts M, Vanneste S, Audenaert D, et al. 2007. Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development 134: 681–690. 10.1242/dev.02753 [DOI] [PubMed] [Google Scholar]
- De Smet I, Vassileva V, De Rybel B, Levesque MP, Grunewald W, Van Damme D, Van Noorden G, Naudts M, Van Isterdael G, De Clercq R, et al. 2008. Receptor-like kinase ACR4 restricts formative cell divisions in the Arabidopsis root. Science 322: 594–597. 10.1126/science.1160158 [DOI] [PubMed] [Google Scholar]
- De Smet I, Lau S, Voss U, Vanneste S, Benjamins R, Rademacher EH, Schlereth A, De Rybel B, Vassileva V, Grunewald W, et al. 2010. Bimodular auxin response controls organogenesis in Arabidopsis. Proc Natl Acad Sci 107: 2705–2710. 10.1073/pnas.0915001107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditengou FA, Teale WD, Kochersperger P, Flittner KA, Kneuper I, van der Graaff E, Nziengui H, Pinosa F, Li X, Nitschke R, et al. 2008. Mechanical induction of lateral root initiation in Arabidopsis thaliana. Proc Natl Acad Sci 105: 18818–18823. 10.1073/pnas.0807814105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doblas VG, Geldner N, Barberon M. 2017. The endodermis, a tightly controlled barrier for nutrients. Curr Opin Plant Biol 39: 136–143. 10.1016/j.pbi.2017.06.010 [DOI] [PubMed] [Google Scholar]
- Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B. 1993. Cellular organisation of the Arabidopsis thaliana root. Development 119: 71. [DOI] [PubMed] [Google Scholar]
- Du Y, Scheres B. 2017. PLETHORA transcription factors orchestrate de novo organ patterning during Arabidopsis lateral root outgrowth. Proc Natl Acad Sci 114: 11709–11714. 10.1073/pnas.1714410114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Scheres B. 2018. Lateral root formation and the multiple roles of auxin. J Exp Bot 69: 155–167. 10.1093/jxb/erx223 [DOI] [PubMed] [Google Scholar]
- Duan L, Dietrich D, Ng CH, Chan PMY, Bhalerao R, Bennett MJ, Dinneny JR. 2013. Endodermal ABA signaling promotes lateral root quiescence during salt stress in Arabidopsis seedlings. Plant Cell 25: 324–341. 10.1105/tpc.112.107227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovsky JG, Doerner PW, Colón-Carmona A, Rost TL. 2000. Pericycle cell proliferation and lateral root initiation in Arabidopsis. Plant Physiol 124: 1648–1657. 10.1104/pp.124.4.1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovsky JG, Gambetta GA, Hernández-Barrera A, Shishkova S, González I. 2006. Lateral root initiation in Arabidopsis: developmental window, spatial patterning, density and predictability. Ann Bot 97: 903–915. 10.1093/aob/mcj604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovsky JG, Sauer M, Napsucialy-Mendivil S, Ivanchenko MG, Friml J, Shishkova S, Celenza J, Benkova E. 2008. Auxin acts as a local morphogenetic trigger to specify lateral root founder cells. Proc Natl Acad Sci 105: 8790–8794. 10.1073/pnas.0712307105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escamez S, André D, Sztojka B, Bollhöner B, Hall H, Berthet B, Voß U, Lers A, Maizel A, Andersson M, et al. 2020. Cell death in cells overlying lateral root primordia facilitates organ growth in Arabidopsis. Curr Biol 30: 455–464.e7. 10.1016/j.cub.2019.11.078 [DOI] [PubMed] [Google Scholar]
- Fendrych M, Leung J, Friml J. 2016. TIR1/AFB-Aux/IAA auxin perception mediates rapid cell wall acidification and growth of Arabidopsis hypocotyls. eLife 5: e19048. 10.7554/eLife.19048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez AI, Vangheluwe N, Xu K, Jourquin J, Claus LAN, Morales-Herrera S, Parizot B, De Gernier H, Yu Q, Drozdzecki A, et al. 2020. GOLVEN peptide signalling through RGI receptors and MPK6 restricts asymmetric cell division during lateral root initiation. Nat Plants 6: 533–543. 10.1038/s41477-020-0645-z [DOI] [PubMed] [Google Scholar]
- Friml J, Wiśniewska J, Benková E, Mendgen K, Palme K. 2002. Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415: 806–809. 10.1038/415806a [DOI] [PubMed] [Google Scholar]
- Fukaki H, Tameda S, Masuda H, Tasaka M. 2002. Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J 29: 153–168. 10.1046/j.0960-7412.2001.01201.x [DOI] [PubMed] [Google Scholar]
- Geldner N. 2003. Partial loss-of-function alleles reveal a role for GNOM in auxin transport-related, post-embryonic development of Arabidopsis. Development 131: 389–400. 10.1242/dev.00926 [DOI] [PubMed] [Google Scholar]
- Ghorbani S, Lin YC, Parizot B, Fernandez A, Njo MF, Van de Peer Y, Beeckman T, Hilson P. 2015. Expanding the repertoire of secretory peptides controlling root development with comparative genome analysis and functional assays. EXBOTJ 66: 5257–5269. 10.1093/jxb/erv346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford ML, Dean A, Gutierrez RA, Coruzzi GM, Birnbaum KD. 2008. Cell-specific nitrogen responses mediate developmental plasticity. Proc Natl Acad Sci 105: 803–808. 10.1073/pnas.0709559105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonneau M, Desprez T, Martin M, Doblas VG, Bacete L, Miart F, Sormani R, Hématy K, Renou J, Landrein B, et al. 2018. Receptor kinase THESEUS1 is a rapid alkalinization factor 34 receptor in Arabidopsis. Curr Biol 28: 2452–2458.e4. 10.1016/j.cub.2018.05.075 [DOI] [PubMed] [Google Scholar]
- Grison MS, Kirk P, Brault ML, Wu XN, Schulze WX, Benitez-Alfonso Y, Immel F, Bayer EM. 2019. Plasma membrane-associated receptor-like kinases relocalize to plasmodesmata in response to osmotic stress. Plant Physiol 181: 142–160. 10.1104/pp.19.00473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber BD, Giehl RFH, Friedel S, von Wirén N. 2013. Plasticity of the Arabidopsis root system under nutrient deficiencies. Plant Physiol 163: 161–179. 10.1104/pp.113.218453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan P. 2017. Dancing with hormones: a current perspective of nitrate signaling and regulation in Arabidopsis. Front Plant Sci 8: 1697. 10.3389/fpls.2017.01697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez RA, Lejay LV, Dean A, Chiaromonte F, Shasha DE, Coruzzi GM. 2007. Qualitative network models and genome-wide expression data define carbon/nitrogen-responsive molecular machines in Arabidopsis. Genome Biol 8: R7. 10.1186/gb-2007-8-1-r7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta M, Tan LX, Bushey DB, Swanson SJ, Sussman MR. 2018. Environmental and genetic factors regulating localization of the plant plasma membrane H+-ATPase. Plant Physiol 176: 364–377. 10.1104/pp.17.01126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Hernández V, Benítez M, Boudaoud A. 2019. Interplay between turgor pressure and plasmodesmata during plant development. J Exp Bot 71: 768–777. 10.1093/jxb/erz434 [DOI] [PubMed] [Google Scholar]
- Himanen K, Vuylsteke M, Vanneste S, Vercruysse S, Boucheron E, Alard P, Chriqui D, Van Montagu M, Inzé D, Beeckman T. 2004. Transcript profiling of early lateral root initiation. Proc Natl Acad Sci 101: 5146–5151. 10.1073/pnas.0308702101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota A, Kato T, Fukaki H, Aida M, Tasaka M. 2007. The auxin-regulated AP2/EREBP gene PUCHI is required for morphogenesis in the early lateral root primordium of Arabidopsis. Plant Cell 19: 2156–2168. 10.1105/tpc.107.050674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang KL, Ma GJ, Zhang ML, Xiong H, Wu H, Zhao CZ, Liu CS, Jia HX, Chen L, Kjorven JO, et al. 2018. The ARF7 and ARF19 transcription factors positively regulate PHOSPHATE STARVATION RESPONSE1 in Arabidopsis roots. Plant Physiol 178: 413–427. 10.1104/pp.17.01713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R, Zheng R, He J, Zhou Z, Wang J, Xiong Y, Xu T. 2019. Noncanonical auxin signaling regulates cell division pattern during lateral root development. Proc Natl Acad Sci 116: 21285–21290. 10.1073/pnas.1910916116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram G, Nawrath C. 2017. The roles of the cuticle in plant development: organ adhesions and beyond. J Exp Bot 68: 5307–5321. 10.1093/jxb/erx313 [DOI] [PubMed] [Google Scholar]
- Israeli A, Reed JW, Ori N. 2020. Genetic dissection of the auxin response network. Nat Plants 6: 1082–1090. 10.1038/s41477-020-0739-7 [DOI] [PubMed] [Google Scholar]
- Jia Z, von Wirén N. 2020. Signaling pathways underlying nitrogen-dependent changes in root system architecture: from model to crop species. J Exp Bot 71: 4393–4404. 10.1093/jxb/eraa033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jourquin J, Fukaki H, Beeckman T. 2020. Peptide-receptor signaling controls lateral root development. Plant Physiol 182: 1645–1656. 10.1104/pp.19.01317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapulnik Y, Delaux PM, Resnick N, Mayzlish-Gati E, Wininger S, Bhattacharya C, Séjalon-Delmas N, Combier JP, Bécard G, Belausov E, et al. 2011. Strigolactones affect lateral root formation and root-hair elongation in Arabidopsis. Planta 233: 209–216. 10.1007/s00425-010-1310-y [DOI] [PubMed] [Google Scholar]
- Kellermeier F, Chardon F, Amtmann A. 2013. Natural variation of Arabidopsis root architecture reveals complementing adaptive strategies to potassium starvation. Plant Physiol 161: 1421–1432. 10.1104/pp.112.211144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krouk G. 2016. Hormones and nitrate: a two-way connection. Plant Mol Biol 91: 599–606. 10.1007/s11103-016-0463-x [DOI] [PubMed] [Google Scholar]
- Krouk G, Lacombe B, Bielach A, Perrine-Walker F, Malinska K, Mounier E, Hoyerova K, Tillard P, Leon S, Ljung K, et al. 2010. Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev Cell 18: 927–937. 10.1016/j.devcel.2010.05.008 [DOI] [PubMed] [Google Scholar]
- Kumpf RP, Shi CL, Larrieu A, Sto IM, Butenko MA, Peret B, Riiser ES, Bennett MJ, Aalen RB. 2013. Floral organ abscission peptide IDA and its HAE/HSL2 receptors control cell separation during lateral root emergence. Proc Natl Acad Sci 110: 5235–5240. 10.1073/pnas.1210835110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski MJ, Williams ME, Nusbaum HC, Sussex IM. 1995. Formation of lateral root meristems is a two-stage process. Development 121: 3303. [DOI] [PubMed] [Google Scholar]
- Laskowski M, Grieneisen VA, Hofhuis H, ten Hove CA, Hogeweg P, Marée AFM, Scheres B. 2008. Root system architecture from coupling cell shape to auxin transport. PLoS Biol 6: e307. 10.1371/journal.pbio.0060307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HW, Cho C, Kim J. 2015. Lateral organ boundaries Domain16 and 18 act downstream of the AUXIN1 and LIKE-AUXIN3 auxin influx carriers to control lateral root development in Arabidopsis. Plant Physiol 168: 1792–1806. 10.1104/pp.15.00578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Ha JH, Kim SG, Choi HK, Kim ZH, Han YJ, Kim JI, Oh Y, Fragoso V, Shin K, et al. 2016. Stem-piped light activates phytochrome B to trigger light responses in Arabidopsis thaliana roots. Sci Signal 9: ra106. 10.1126/scisignal.aaf6530 [DOI] [PubMed] [Google Scholar]
- Liu Y, Xie Y, Wang H, Ma X, Yao W, Wang H. 2017. Light and ethylene coordinately regulate the phosphate starvation response through transcriptional regulation of PHOSPHATE STARVATION RESPONSE1. Plant Cell 29: 2269–2284. 10.1105/tpc.17.00268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löfke C, Zwiewka M, Heilmann I, Van Montagu MCE, Teichmann T, Friml J. 2013. Asymmetric gibberellin signaling regulates vacuolar trafficking of PIN auxin transporters during root gravitropism. Proc Natl Acad Sci 110: 3627–3632. 10.1073/pnas.1300107110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas M, Godin C, Jay-Allemand C, Laplaze L. 2008. Auxin fluxes in the root apex co-regulate gravitropism and lateral root initiation. J Exp Bot 59: 55–66. 10.1093/jxb/erm171 [DOI] [PubMed] [Google Scholar]
- Lucas M, Kenobi K, von Wangenheim D, Voss U, Swarup K, De Smet I, Van Damme D, Lawrence T, Peret B, Moscardi E, et al. 2013. Lateral root morphogenesis is dependent on the mechanical properties of the overlaying tissues. Proc Natl Acad Sci 110: 5229–5234. 10.1073/pnas.1210807110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Li J, Qu B, He X, Zhao X, Li B, Fu X, Tong Y. 2014. Auxin biosynthetic gene TAR2 is involved in low nitrogen-mediated reprogramming of root architecture in Arabidopsis. Plant J 78: 70–79. 10.1111/tpj.12448 [DOI] [PubMed] [Google Scholar]
- Maeda Y, Konishi M, Kiba T, Sakuraba Y, Sawaki N, Kurai T, Ueda Y, Sakakibara H, Yanagisawa S. 2018. A NIGT1-centred transcriptional cascade regulates nitrate signalling and incorporates phosphorus starvation signals in Arabidopsis. Nat Commun 9: 1376. 10.1038/s41467-018-03832-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maghiaoui A, Bouguyon E, Cuesta C, Perrine-Walker F, Alcon C, Krouk G, Benkova E, Nacry P, Gojon A, Bach L. 2020. The Arabidopsis NRT1.1 transceptor coordinately controls auxin biosynthesis and transport to regulate root branching in response to nitrate. J Exp Bot 71: 4480–4494. 10.1093/jxb/eraa242 [DOI] [PubMed] [Google Scholar]
- Malamy JE. 2005. Intrinsic and environmental response pathways that regulate root system architecture. Plant Cell Environ 28: 67–77. 10.1111/j.1365-3040.2005.01306.x [DOI] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN. 1997. Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124: 33.–. [DOI] [PubMed] [Google Scholar]
- Marchant A, Bhalerao R, Casimiro I, Eklöf J, Casero PJ, Bennett M, Sandberg G. 2002. AUX1 promotes lateral root formation by facilitating indole-3-acetic acid distribution between sink and source tissues in the Arabidopsis seedling. Plant Cell 14: 589–597. 10.1105/tpc.010354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marhavý P, Bielach A, Abas L, Abuzeineh A, Duclercq J, Tanaka H, Pařezová M, Petrášek J, Friml J, Kleine-Vehn J, et al. 2011. Cytokinin modulates endocytic trafficking of PIN1 auxin efflux carrier to control plant organogenesis. Dev Cell 21: 796–804. 10.1016/j.devcel.2011.08.014 [DOI] [PubMed] [Google Scholar]
- Marhavý P, Vanstraelen M, De Rybel B, Zhaojun D, Bennett MJ, Beeckman T, Benková E. 2013. Auxin reflux between the endodermis and pericycle promotes lateral root initiation. EMBO J 32: 149–158. 10.1038/emboj.2012.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marhavý P, Duclercq J, Weller B, Feraru E, Bielach A, Offringa R, Friml J, Schwechheimer C, Murphy A, Benková E. 2014. Cytokinin controls polarity of PIN1-dependent auxin transport during lateral root organogenesis. Curr Biol 24: 1031–1037. 10.1016/j.cub.2014.04.002 [DOI] [PubMed] [Google Scholar]
- Marhavý P, Montesinos JC, Abuzeineh A, Van Damme D, Vermeer JEM, Duclercq J, Rakusová H, Nováková P, Friml J, Geldner N, et al. 2016. Targeted cell elimination reveals an auxin-guided biphasic mode of lateral root initiation. Genes Dev 30: 471–483. 10.1101/gad.276964.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke MA, Mosher RA. 2014. RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat Rev Genet 15: 394–408. 10.1038/nrg3683 [DOI] [PubMed] [Google Scholar]
- Michniewicz M, Ho CH, Enders TA, Floro E, Damodaran S, Gunther LK, Powers SK, Frick EM, Topp CN, Frommer WB, et al. 2019. TRANSPORTER of IBA1 links auxin and cytokinin to influence root architecture. Dev Cell 50: 599–609.e4. 10.1016/j.devcel.2019.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Risueno MA, Van Norman JM, Moreno A, Zhang J, Ahnert SE, Benfey PN. 2010. Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science 329: 1306–1311. 10.1126/science.1191937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mravec J, Kubes M, Bielach A, Gaykova V, Petrasek J, Skupa P, Chand S, Benkova E, Zazimalova E, Friml J. 2008. Interaction of PIN and PGP transport mechanisms in auxin distribution-dependent development. Development 135: 3345–3354. 10.1242/dev.021071 [DOI] [PubMed] [Google Scholar]
- Mravec J, Skůpa P, Bailly A, Hoyerová K, Křeček P, Bielach A, Petrášek J, Zhang J, Gaykova V, Stierhof YD, et al. 2009. Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature 459: 1136–1140. 10.1038/nature08066 [DOI] [PubMed] [Google Scholar]
- Murphy E, Vu LD, Van den Broeck L, Lin Z, Ramakrishna P, van de Cotte B, Gaudinier A, Goh T, Slane D, Beeckman T, et al. 2016. RALFL34 regulates formative cell divisions in Arabidopsis pericycle during lateral root initiation. J Exp Bot 67: 4863–4875. 10.1093/jxb/erw281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama T, Shinohara H, Tanaka M, Baba K, Ogawa-Ohnishi M, Matsubayashi Y. 2017. A peptide hormone required for Casparian strip diffusion barrier formation in Arabidopsis roots. Science 355: 284–286. 10.1126/science.aai9057 [DOI] [PubMed] [Google Scholar]
- Naseer S, Lee Y, Lapierre C, Franke R, Nawrath C, Geldner N. 2012. Casparian strip diffusion barrier in Arabidopsis is made of a lignin polymer without suberin. Proc Natl Acad Sci 109: 10101–10106. 10.1073/pnas.1205726109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumann C, Müller J, Sakhonwasee S, Wieghaus A, Hause G, Heisters M, Bürstenbinder K, Abel S. 2019. The local phosphate deficiency response activates endoplasmic reticulum stress-dependent autophagy. Plant Physiol 179: 460–476. 10.1104/pp.18.01379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C, Schreiber L, Franke RB, Geldner N, Reina-Pinto JJ, Kunst L. 2013. Apoplastic diffusion barriers in Arabidopsis. The Arabidopsis Book 11: e0167. 10.1199/tab.0167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naz L, Guo L, Chen F. 2019. Overexpression of nitrate transporter OsNRT2.1 enhances nitrate-dependent root elongation. Genes (Basel) 10: 290. 10.3390/genes10040290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norambuena L, Zouhar J, Hicks GR, Raikhel NV. 2008. Identification of cellular pathways affected by Sortin2, a synthetic compound that affects protein targeting to the vacuole in Saccharomyces cerevisiae. BMC Chem Biol 8: 1. 10.1186/1472-6769-8-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura T, Goeschl C, Filiault D, Mirea M, Slovak R, Wolhrab B, Satbhai SB, Busch W. 2019. Root system depth in Arabidopsis is shaped by EXOCYST70A3 via the dynamic modulation of auxin transport. Cell 178: 400–412.e16. 10.1016/j.cell.2019.06.021 [DOI] [PubMed] [Google Scholar]
- Okuda S, Hothorn LA, Hothorn M. 2020. Crystal structures of Arabidopsis and Physcomitrella CR4 reveal the molecular architecture of CRINKLY4 receptor kinases. bioRxiv doi:10.1101/2020.08.10.245050 [Google Scholar]
- Okushima Y, Overvoorde PJ, Arima K, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Lui A, Nguyen D, et al. 2005. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell 17: 444–463. 10.1105/tpc.104.028316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M. 2007. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19: 118–130. 10.1105/tpc.106.047761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orman-Ligeza B, Morris EC, Parizot B, Lavigne T, Babé A, Ligeza A, Klein S, Sturrock C, Xuan W, Novák O, et al. 2018. The xerobranching response represses lateral root formation when roots are not in contact with water. Curr Biol 28: 3165–3173.e5. 10.1016/j.cub.2018.07.074 [DOI] [PubMed] [Google Scholar]
- Orosa-Puente B, Leftley N, von Wangenheim D, Banda J, Srivastava AK, Hill K, Truskina J, Bhosale R, Morris E, Srivastava M, et al. 2018. Root branching toward water involves posttranslational modification of transcription factor ARF7. Science 362: 1407–1410. 10.1126/science.aau3956 [DOI] [PubMed] [Google Scholar]
- Osmont KS, Sibout R, Hardtke CS. 2007. Hidden branches: developments in root system architecture. Annu Rev Plant Biol 58: 93–113. 10.1146/annurev.arplant.58.032806.104006 [DOI] [PubMed] [Google Scholar]
- Parizot B, Laplaze L, Ricaud L, Boucheron-Dubuisson E, Bayle V, Bonke M, De Smet I, Poethig SR, Helariutta Y, Haseloff J, et al. 2008. Diarch symmetry of the vascular bundle in Arabidopsis root encompasses the pericycle and is reflected in distich lateral root initiation. Plant Physiol 146: 140–148. 10.1104/pp.107.107870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péret B, Middleton AM, French AP, Larrieu A, Bishopp A, Njo M, Wells DM, Porco S, Mellor N, Band LR, et al. 2013. Sequential induction of auxin efflux and influx carriers regulates lateral root emergence. Mol Syst Biol 9: 699. 10.1038/msb.2013.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Henríquez P, Raikhel NV, Norambuena L. 2012. Endocytic trafficking towards the vacuole plays a key role in the auxin receptor SCFTIR-independent mechanism of lateral root formation in A. thaliana. Mol Plant 5: 1195–1209. 10.1093/mp/sss066 [DOI] [PubMed] [Google Scholar]
- Pérez-Torres CA, López-Bucio J, Cruz-Ramírez A, Ibarra-Laclette E, Dharmasiri S, Estelle M, Herrera-Estrella L. 2008. Phosphate availability alters lateral root development in Arabidopsis by modulating auxin sensitivity via a mechanism involving the TIR1 auxin receptor. Plant Cell 20: 3258–3272. 10.1105/tpc.108.058719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perotti MF, Ribone PA, Cabello JV, Ariel FD, Chan RL. 2019. AtHB23 participates in the gene regulatory network controlling root branching, and reveals differences between secondary and tertiary roots. Plant J 100: 1224–1236. 10.1111/tpj.14511 [DOI] [PubMed] [Google Scholar]
- Pires ND, Dolan L. 2012. Morphological evolution in land plants: new designs with old genes. Phil Trans R Soc B 367: 508–518. 10.1098/rstb.2011.0252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porco S, Larrieu A, Du Y, Gaudinier A, Goh T, Swarup K, Swarup R, Kuempers B, Bishopp A, Lavenus J, et al. 2016. Lateral root emergence in Arabidopsis is dependent on transcription factor LBD29 regulation of auxin influx carrier LAX3. Development 143: 3340–3349. 10.1242/dev.136283 [DOI] [PubMed] [Google Scholar]
- Ramakrishna P, Ruiz Duarte P, Rance GA, Schubert M, Vordermaier V, Vu LD, Murphy E, Vilches Barro A, Swarup K, Moirangthem K, et al. 2019. EXPANSIN A1-mediated radial swelling of pericycle cells positions anticlinal cell divisions during lateral root initiation. Proc Natl Acad Sci 116: 8597–8602. 10.1073/pnas.1820882116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven JA, Edwards D. 2001. Roots: evolutionary origins and biogeochemical significance. J Exp Bot 52: 381–401. 10.1093/jxb/52.suppl_1.381 [DOI] [PubMed] [Google Scholar]
- Reed RC, Brady SR, Muday GK. 1998. Inhibition of auxin movement from the shoot into the root inhibits lateral root development in Arabidopsis. Plant Physiol 118: 1369–1378. 10.1104/pp.118.4.1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remans T, Nacry P, Pervent M, Filleur S, Diatloff E, Mounier E, Tillard P, Forde BG, Gojon A. 2006. The Arabidopsis NRT1.1 transporter participates in the signaling pathway triggering root colonization of nitrate-rich patches. Proc Natl Acad Sci 103: 19206–19211. 10.1073/pnas.0605275103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter GL, Monshausen GB, Krol A, Gilroy S. 2009. Mechanical stimuli modulate lateral root organogenesis. Plant Physiol 151: 1855–1866. 10.1104/pp.109.142448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosquete MR, von Wangenheim D, Marhavý P, Barbez E, Stelzer EHK, Benková E, Maizel A, Kleine-Vehn J. 2013. An auxin transport mechanism restricts positive orthogravitropism in lateral roots. Curr Biol 23: 817–822. 10.1016/j.cub.2013.03.064 [DOI] [PubMed] [Google Scholar]
- Rubilar-Hernández C, Osorio-Navarro C, Cabello F, Norambuena L. 2019. PI4KIIIβ activity regulates lateral root formation driven by endocytic trafficking to the vacuole. Plant Physiol 181: 112–126. 10.1104/pp.19.00695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffel S. 2018. Nutrient-related long-distance signals: common players and possible cross-talk. Plant Cell Physiol 59: 1723–1732. 10.1093/pcp/pcy152 [DOI] [PubMed] [Google Scholar]
- Sager R, Wang X, Hill K, Yoo BC, Caplan J, Nedo A, Tran T, Bennett MJ, Lee JY. 2020. Auxin-dependent control of a plasmodesmal regulator creates a negative feedback loop modulating lateral root emergence. Nat Commun 11: 364. 10.1038/s41467-019-14226-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz L, Dewitte W, Forzani C, Patell F, Nieuwland J, Wen B, Quelhas P, De Jager S, Titmus C, Campilho A, et al. 2011. The Arabidopsis D-type cyclin CYCD2;1 and the inhibitor ICK2/KRP2 modulate auxin-induced lateral root formation. Plant Cell 23: 641–660. 10.1105/tpc.110.080002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahzad Z, Eaglesfield R, Carr C, Amtmann A. 2020. Cryptic variation in RNA-directed DNA-methylation controls lateral root development when auxin signalling is perturbed. Nat Commun 11: 218. 10.1038/s41467-019-13927-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla V, Lombardi L, Iacopino S, Pencik A, Novak O, Perata P, Giuntoli B, Licausi F. 2019. Endogenous hypoxia in lateral root primordia controls root architecture by antagonizing auxin signaling in Arabidopsis. Mol Plant 12: 538–551. 10.1016/j.molp.2019.01.007 [DOI] [PubMed] [Google Scholar]
- Singh S, Yadav S, Singh A, Mahima M, Singh A, Gautam V, Sarkar AK. 2020. Auxin signaling modulates LATERAL ROOT PRIMORDIUM 1 (LRP 1) expression during lateral root development in Arabidopsis. Plant J 101: 87–100. 10.1111/tpj.14520 [DOI] [PubMed] [Google Scholar]
- Stahl Y, Grabowski S, Bleckmann A, Kühnemuth R, Weidtkamp-Peters S, Pinto KG, Kirschner GK, Schmid JB, Wink RH, Hülsewede A, et al. 2013. Moderation of Arabidopsis root stemness by CLAVATA1 and ARABIDOPSIS CRINKLY4 receptor kinase complexes. Curr Biol 23: 362–371. 10.1016/j.cub.2013.01.045 [DOI] [PubMed] [Google Scholar]
- Sun J, Xu Y, Ye S, Jiang H, Chen Q, Liu F, Zhou W, Chen R, Li X, Tietz O, et al. 2009. Arabidopsis ASA1 is important for jasmonate-mediated regulation of auxin biosynthesis and transport during lateral root formation. Plant Cell 21: 1495–1511. 10.1105/tpc.108.064303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Tao J, Liu S, Huang S, Chen S, Xie X, Yoneyama K, Zhang Y, Xu G. 2014. Strigolactones are involved in phosphate- and nitrate-deficiency-induced root development and auxin transport in rice. J Exp Bot 65: 6735–6746. 10.1093/jxb/eru029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun CH, Yu JQ, Hu DG. 2017. Nitrate: a crucial signal during lateral roots development. Front Plant Sci 8: 485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup K, Benková E, Swarup R, Casimiro I, Péret B, Yang Y, Parry G, Nielsen E, De Smet I, Vanneste S, et al. 2008. The auxin influx carrier LAX3 promotes lateral root emergence. Nat Cell Biol 10: 946–954. 10.1038/ncb1754 [DOI] [PubMed] [Google Scholar]
- Tan S, Abas M, Verstraeten I, Glanc M, Molnár G, Hajný J, Lasák P, Petřík I, Russinova E, Petrášek J, et al. 2020. Salicylic acid targets protein phosphatase 2A to attenuate growth in plants. Curr Biol 30: 381–395.e8. 10.1016/j.cub.2019.11.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian QY, Sun P, Zhang WH. 2009. Ethylene is involved in nitrate-dependent root growth and branching in Arabidopsis thaliana. New Phytol 184: 918–931. 10.1111/j.1469-8137.2009.03004.x [DOI] [PubMed] [Google Scholar]
- Torres-Martínez HH, Hernández-Herrera P, Corkidi G, Dubrovsky JG. 2020. From one cell to many: morphogenetic field of lateral root founder cells in Arabidopsis thaliana is built by gradual recruitment. Proc Natl Acad Sci 117: 20943–20949. 10.1073/pnas.2006387117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrey JG. 1950. The induction of lateral roots by indoleacetic acid and root decapitation. Am J Bot 37: 257–264. 10.1002/j.1537-2197.1950.tb12192.x [DOI] [Google Scholar]
- Toyokura K, Goh T, Shinohara H, Shinoda A, Kondo Y, Okamoto Y, Uehara T, Fujimoto K, Okushima Y, Ikeyama Y, et al. 2019. Lateral inhibition by a peptide hormone-receptor cascade during Arabidopsis lateral root founder cell formation. Dev Cell 48: 64–75.e5. 10.1016/j.devcel.2018.11.031 [DOI] [PubMed] [Google Scholar]
- Trinh D-C, Lavenus J, Goh T, Boutté Y, Drogue Q, Vaissayre V, Tellier F, Lucas M, Voß U, Gantet P, et al. 2019. PUCHI regulates very long chain fatty acid biosynthesis during lateral root and callus formation. Proc Natl Acad Sci 116: 14325–14330. 10.1073/pnas.1906300116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. 1997. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg T, ten Tusscher KH. 2018. Lateral root priming synergystically arises from root growth and auxin transport dynamics. bioRxiv 10.1101/361709 [DOI] [Google Scholar]
- Vanneste S, De Rybel B, Beemster GTS, Ljung K, De Smet I, Van Isterdael G, Naudts M, Iida R, Gruissem W, Tasaka M, et al. 2005. Cell cycle progression in the pericycle is not sufficient for SOLITARY ROOT/IAA14-mediated lateral root initiation in Arabidopsis thaliana. Plant Cell 17: 3035–3050. 10.1105/tpc.105.035493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Norman JM, Xuan W, Beeckman T, Benfey PN. 2013. To branch or not to branch: the role of pre-patterning in lateral root formation. Development 140: 4301–4310. 10.1242/dev.090548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeer JEM, von Wangenheim D, Barberon M, Lee Y, Stelzer EHK, Maizel A, Geldner N. 2014. A spatial accommodation by neighboring cells is required for organ initiation in Arabidopsis. Science 343: 178–183. 10.1126/science.1245871 [DOI] [PubMed] [Google Scholar]
- Vidal EA, Araus V, Lu C, Parry G, Green PJ, Coruzzi GM, Gutierrez RA. 2010. Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc Natl Acad Sci 107: 4477–4482. 10.1073/pnas.0909571107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal EA, Alvarez JM, Araus V, Riveras E, Brooks M, Krouk G, Ruffel S, Lejay L, Crawford N, Coruzzi GM, et al. 2020. Nitrate 2020: thirty years from transport to signaling networks. Plant Cell 32: 2094–2119. 10.1105/tpc.19.00748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilches Barro A, Stöckle D, Thellmann M, Ruiz-Duarte P, Bald L, Louveaux M, von Born P, Denninger P, Goh T, Fukaki H, et al. 2019. Cytoskeleton dynamics are necessary for early events of lateral root initiation in Arabidopsis. Curr Biol 29: 2443–2454.e5. 10.1016/j.cub.2019.06.039 [DOI] [PubMed] [Google Scholar]
- von Wangenheim D, Fangerau J, Schmitz A, Smith RS, Leitte H, Stelzer EHK, Maizel A. 2016. Rules and self-organizing properties of post-embryonic plant organ cell division patterns. Curr Biol 26: 439–449. 10.1016/j.cub.2015.12.047 [DOI] [PubMed] [Google Scholar]
- von Wangenheim D, Hauschild R, Friml J. 2017. Light sheet fluorescence microscopy of plant roots growing on the surface of a gel. J Vis Exp 119: 55044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wangenheim D, Banda J, Schmitz A, Boland J, Bishopp A, Maizel A, Stelzer EHK, Bennett M. 2020. Early developmental plasticity of lateral roots in response to asymmetric water availability. Nat Plants 6: 73–77. 10.1038/s41477-019-0580-z [DOI] [PubMed] [Google Scholar]
- Waidmann S, Kleine-Vehn J. 2020. Asymmetric cytokinin signaling opposes gravitropism in roots. J Integr Plant Biol 62: 882–886. 10.1111/jipb.12929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waidmann S, Ruiz Rosquete M, Schöller M, Sarkel E, Lindner H, LaRue T, Petřík I, Dünser K, Martopawiro S, Sasidharan R, et al. 2019. Cytokinin functions as an asymmetric and anti-gravitropic signal in lateral roots. Nat Commun 10: 3540. 10.1038/s41467-019-11483-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao TT, van Velzen R, Kulikova O, Franken C, Bisseling T. 2019. Lateral root formation involving cell division in both pericycle, cortex and endodermis is a common and ancestral trait in seed plants. Development 146: dev182592. 10.1242/dev.182592 [DOI] [PubMed] [Google Scholar]
- Xuan W, Audenaert D, Parizot B, Möller BK, Njo MF, De Rybel B, De Rop G, Van Isterdael G, Mähönen AP, Vanneste S, et al. 2015. Root cap-derived auxin pre-patterns the longitudinal axis of the Arabidopsis root. Curr Biol 25: 1381–1388. 10.1016/j.cub.2015.03.046 [DOI] [PubMed] [Google Scholar]
- Xuan W, Band LR, Kumpf RP, Van Damme D, Parizot B, De Rop G, Opdenacker D, Moller BK, Skorzinski N, Njo MF, et al. 2016. Cyclic programmed cell death stimulates hormone signaling and root development in Arabidopsis. Science 351: 384–387. 10.1126/science.aad2776 [DOI] [PubMed] [Google Scholar]
- Xuan W, De Gernier H, Beeckman T. 2020. The dynamic nature and regulation of the root clock. Development 147: dev181446. 10.1242/dev.181446 [DOI] [PubMed] [Google Scholar]
- Xun Q, Wu Y, Li H, Chang J, Ou Y, He K, Gou X, Tax FE, Li J. 2020. Two receptor-like protein kinases, MUSTACHES and MUSTACHES-LIKE, regulate lateral root development in Arabidopsis thaliana. New Phytol 227: 1157–1173. 10.1111/nph.16599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi T, Tanaka A, Inahashi H, Nishizawa NK, Tsutsumi N, Inukai Y, Nakazono M. 2019. Fine control of aerenchyma and lateral root development through AUX/IAA- and ARF-dependent auxin signaling. Proc Natl Acad Sci 116: 20770–20775. 10.1073/pnas.1907181116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Zhang L, Chen P, Liang T, Li X, Liu H. 2020. UV-b photoreceptor UVR8 interacts with MYB73/MYB77 to regulate auxin responses and lateral root development. EMBO J 39: e101928. 10.15252/embj.2019101928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan T, Xu H, Li J, Lu Y. 2020. Auxin abolishes SHI-RELATED SEQUENCE 5-mediated inhibition of lateral root development in Arabidopsis. New Phytol 225: 297–309. 10.1111/nph.16115 [DOI] [PubMed] [Google Scholar]
- Zhou M, Palanca AMS, Law JA. 2018. Locus-specific control of the de novo DNA methylation pathway in Arabidopsis by the CLASSY family. Nat Genet 50: 865–873. 10.1038/s41588-018-0115-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Gallemí M, Pospíšil J, Žádníková P, Strnad M, Benková E. 2019a. Root gravity response module guides differential growth determining both root bending and apical hook formation in Arabidopsis. Development 146: dev175919. 10.1242/dev.175919 [DOI] [PubMed] [Google Scholar]
- Zhu Q, Shao Y, Ge S, Zhang M, Zhang T, Hu X, Liu Y, Walker J, Zhang S, Xu J. 2019b. A MAPK cascade downstream of IDA–HAE/HSL2 ligand–receptor pair in lateral root emergence. Nat Plants 5: 414–423. 10.1038/s41477-019-0396-x [DOI] [PubMed] [Google Scholar]