Here, Dimitrova-Paternoga et al. present the high-resolution crystal structure of Khc–aTm1 (Drosophila kinesin-1, also called kinesin heavy chain [Khc], in complex with a putative cargo adaptor, the atypical tropomyosin [aTm1]), which mediates transport of oskar mRNA to the posterior pole of the Drosophila oocyte. They show that aTm1 binds to an evolutionarily conserved cargo binding site on Khc, demonstrate that Khc binds RNA directly, and show that aTm1 plays a stabilizing role in the interaction of Khc with RNA, which distinguishes aTm1 from classical motor adaptors.
Keywords: kinesin, kinesin adaptor, kinesin–atypical tropomyosin complex, mRNA transport, oskar mRNA
Abstract
Kinesin-1 carries cargos including proteins, RNAs, vesicles, and pathogens over long distances within cells. The mechanochemical cycle of kinesins is well described, but how they establish cargo specificity is not fully understood. Transport of oskar mRNA to the posterior pole of the Drosophila oocyte is mediated by Drosophila kinesin-1, also called kinesin heavy chain (Khc), and a putative cargo adaptor, the atypical tropomyosin, aTm1. How the proteins cooperate in mRNA transport is unknown. Here, we present the high-resolution crystal structure of a Khc–aTm1 complex. The proteins form a tripartite coiled coil comprising two in-register Khc chains and one aTm1 chain, in antiparallel orientation. We show that aTm1 binds to an evolutionarily conserved cargo binding site on Khc, and mutational analysis confirms the importance of this interaction for mRNA transport in vivo. Furthermore, we demonstrate that Khc binds RNA directly and that it does so via its alternative cargo binding domain, which forms a positively charged joint surface with aTm1, as well as through its adjacent auxiliary microtubule binding domain. Finally, we show that aTm1 plays a stabilizing role in the interaction of Khc with RNA, which distinguishes aTm1 from classical motor adaptors.
Intracellular transport is an essential cellular function that is ensured by a vast cytoskeletal network and three major classes of motor proteins, namely kinesins, dyneins, and myosins (Hirokawa et al. 2009; de Lanerolle 2012; Olenick and Holzbaur 2019). Whereas myosins are responsible for short-range transport along randomly oriented actin filaments, kinesins and dyneins provide long-range directional transport on a polarized microtubule network (Titus 2018). Kinesin-1, a microtubule plus end-directed motor, was the first kinesin identified and is well characterized (Vale et al. 1985; Hirokawa et al. 2009). It transports diverse cargos including proteins, organelles, mRNA nucleoprotein complexes (mRNPs), and viruses (Chudinova and Nadezhdina 2018; Garcin and Straube 2019; Banerjee et al. 2020) and is essential for numerous processes such as endoplasmic reticulum (ER)-Golgi transport, mitochondrial distribution in axons and dendrites, cell migration, and embryonic axis formation (Hirokawa et al. 2009).
Kinesin-1 harbors an N-terminal motor domain, followed by a long coiled-coil stalk that mediates homodimerization, and a flexible tail with regulatory function (Fig. 1A; Seeger and Rice 2013). The two motor domains are powered by ATP hydrolysis, which elicits conformational changes that allow the kinesin molecule to move processively along microtubules (Qin et al. 2020). The kinesin-1 stalk associates with cargos either directly or via adaptor proteins that bind to the C-terminal region of the domain (Cross and Dodding 2019). Although a large portion of kinesin-1 transport is mediated by the adaptor kinesin light chain (Klc), the transport of mRNA cargos such as mRNP granules in dendrites or oskar in the Drosophila oocyte appears to be independent of Klc (Diefenbach et al. 1998; Kanai et al. 2004; Loiseau et al. 2010; Cross and Dodding 2019).
Figure 1.
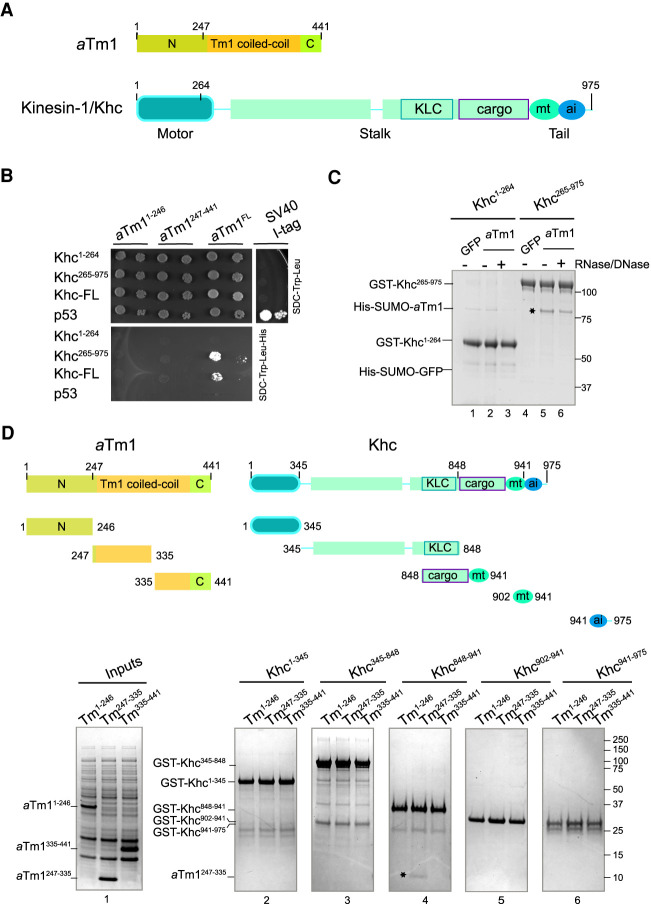
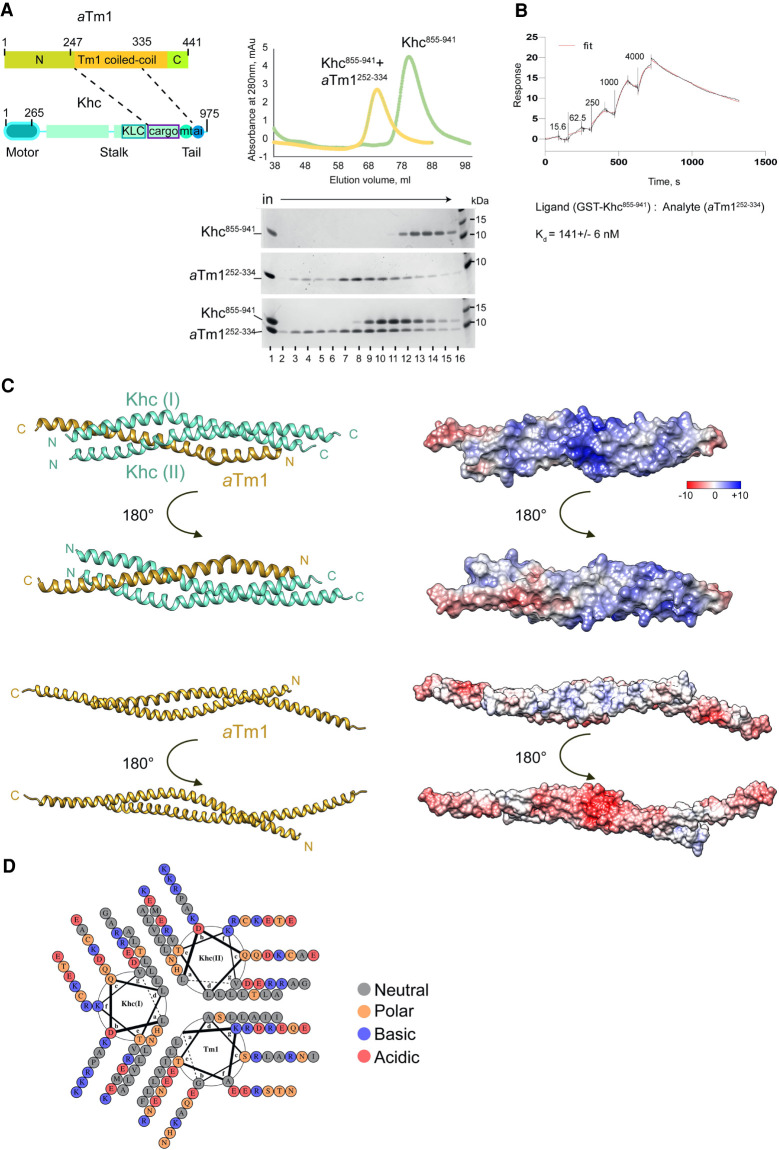
aTm1 interacts with the alternative cargo binding domain of Khc. (A) Schematic representation of aTm1 and kinesin heavy chain (Khc). aTm1 has a central coiled-coil domain, flanked by mostly unstructured N- and C-terminal regions. Khc comprises an N-terminal motor-domain (amino acids 1–345), followed by a long coiled-coil stalk (amino acids 345–902) and an unstructured tail (amino acids 902–975). The stalk includes the kinesin light chain (Klc) binding region (amino acids 792–839) and an alternative cargo binding region (amino acids 848–902). The tail comprises an ATP-independent microtubule binding domain (amino acids 910–938) and an IAK (isoleucine-alanine-lysine) motif (amino acids 938–950), responsible for the autoinhibition of Khc (Williams et al. 2014). Although in many studies the alternative cargo binding region has been considered part of the tail, based on its structural and functional features, we categorize it as part of the coiled-coil stalk. (B) Yeast two-hybrid analysis shows that aTm1 and Khc interact. The Khc variants were tagged with the GAL4 binding domain, and the aTm1 variants were fused to the GAL4 activation domain. Yeast cells were cotransformed with different combinations of the above-mentioned constructs and spotted in two 10-fold dilutions on SDC−Leu−Trp and SDC−Leu−Trp−His plates. (C) A GST (glutathione S-transferase) pull-down assay reveals direct binding of aTm1 to kinesin: GST-tagged Khc1–264 and Khc265–975 were immobilized on glutathione beads and mixed with E.coli lysates containing recombinantly expressed His6-SUMO-aTm1-FL and His6-SUMO-GFP (Supplemental Fig S1A). The eluates were analyzed by SDS-PAGE. The asterisk indicates the interaction of aTm1FL with Khc265–975. (D) aTm1247–335 interacts with the alternative cargo binding domain of Khc848–941. In vitro binding of GST-tagged Khc truncations (schematic top left) to different aTm1 truncations (schematic top right), premixed with E.coli whole-cell lysate. The asterisk indicates the interaction of aTm1247–334 with Khc848–941.
In Drosophila, localization of oskar mRNA to the posterior pole of the developing oocyte drives abdominal patterning and germline formation in the embryo (Ephrussi and Lehmann 1992). The directional transport of RNA is mediated by both the dynein and kinesin transport machineries (Clark et al. 2007; Zimyanin et al. 2008). In the germline syncytium, the oocyte is transcriptionally quiescent and relies on mRNAs provided by 15 interconnected nurse cells for its growth and development (Roth and Lynch 2009). Upon transcription in the nurse cells, oskar mRNA is transported into the oocyte on the microtubule cytoskeleton by the minus end-directed dynein motor complex (Clark et al. 2007). Within the oocyte, the microtubule plus end-directed motor kinesin-1 or kinesin heavy chain (Khc) in Drosophila (Lawrence et al. 2004) transports oskar to the posterior pole where the mRNA is localized (for review, see Trcek and Lehmann 2019). oskar mRNA transport depends on a region of Khc downstream from the Klc binding site that includes an alternative cargo binding domain and an ATP-independent microtubule binding site (Williams et al. 2014).
Moreover, kinesin-mediated oskar transport requires a number of cis and trans factors. One of these factors is a unique isoform of the actin binding protein tropomyosin 1, known as Tm1-I/C, which we refer to here as atypical or aTm1 for simplicity (Loiseau et al. 2010; Veeranan-Karmegam et al. 2016; Gáspár et al. 2017a). In the absence of aTm1, oskar mRNA fails to localize to the posterior pole of the oocyte, as kinesin recruitment to oskar mRNA requires aTm1 function (Erdélyi et al. 1995; Gáspár et al. 2017a). Furthermore, the fact that tethering of the Khc motor domain to oskar restores posterior localization of the mRNA in a Tm1 protein-null background suggested that aTm1 is an essential adaptor for Khc-mediated oskar transport (Gáspár et al. 2017a).
Here, we show that aTm1 homodimers adopt an unusual antiparallel coiled-coil conformation. Moreover, aTm1 binds to an alternative conserved cargo binding region in Khc, forming a tripartite coiled-coil complex with a positively charged surface. Finally, we show in vivo that the function of aTm1 is ensured by its Khc interacting domain and that this atypical Tm1 functions in stabilizing the interaction of Khc with RNA rather than as a classical adaptor.
Results
aTm1 binds to the alternative cargo binding site of Khc
It was previously reported that aTm1 is necessary for the recruitment of Khc to oskar mRNA and that the two proteins physically interact (Veeranan-Karmegam et al. 2016; Gáspár et al. 2017a). To elucidate the molecular basis of the aTm1-Khc interaction, we first performed a yeast two-hybrid analysis, which confirmed that aTm1 (Tm1-FL) binds both full-length Khc (Khc-FL) and Khc265–975 (Fig. 1B). Because the interaction of Khc and aTm1 was formerly tested by coimmunoprecipitation (IP) experiments from S2 cells or Drosophila ovarian lysates, neither of which is performed in a nucleic acid-free environment (Veeranan-Karmegam et al. 2016; Gáspár et al. 2017a), we also probed the interaction by pull-down of recombinant Khc and aTm1 proteins in the presence of DNase and RNase (Fig. 1C; inputs in Supplemental Fig. S1A). Binding of aTm1 to Khc was preserved under these conditions (Fig. 1C, right panel, lanes 5,6), indicating the specific and direct interaction of aTm1 with Khc.
To further narrow down the aTm1 and Khc interacting regions, we performed GST pull-down assays on shorter fragments of the proteins (Fig. 1D; Supplemental Fig. S1B,C). This revealed that aTm1247–335 binds Khc848–941 (Fig. 1D, panel 4), which comprises the alternative cargo binding region and the ATP-independent microtubule binding region of Khc (Fig. 1D; Seiler et al. 2000; Williams et al. 2014). Veeranan-Karmegam et al. (2016) have previously suggested that aTm1 interacts with residues 914–936 of Khc. However, the fact that Khc902–941 did not interact with aTm1 in our assays (Fig. 1D, panel 5) indicates that aTm1247–335 binds to the conserved alternative cargo binding site of Khc (residues 848–902) (Supplemental Fig. S1D).
The alternative cargo binding region of Khc is essential for oskar mRNA localization
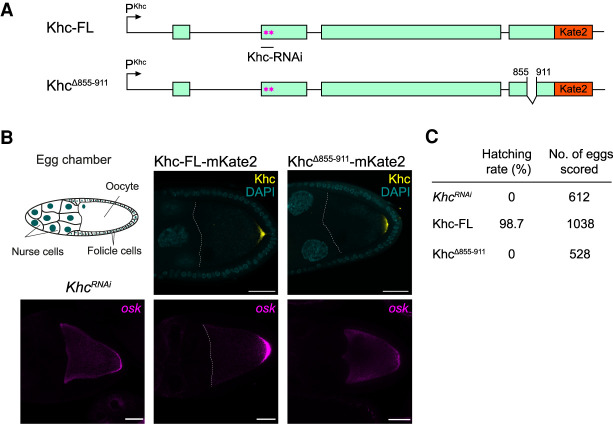
The alternative cargo binding region in Khc was originally identified in Neurospora crassa kinesin (Nkin), where deletion of this site abolished the ability of the protein to rescue the Nkin null mutant phenotype (Seiler et al. 2000). In Drosophila, deletion of the entire C terminus of Khc, which includes the alternative cargo binding region and the regulatory tail, also led to an oskar mRNA localization defect among others (Williams et al. 2014). To investigate the relevance of this cargo binding region for oskar mRNA transport, we expressed RNAi-resistant Khc-FL-mKate2 and Khc855–911Δ-mKate2 (wherein the cargo binding domain is deleted) fusion proteins in a khc-RNAi background (Fig. 2A; Supplemental Fig. S2). Khc855–911Δ localized to the posterior pole of the oocyte (Fig. 2B, upper right panel), indicating that Khc motor function and polarity of the microtubule cytoskeleton were preserved. However, oskar mRNA failed to localize at the posterior pole (Fig. 2B, bottom right panel) and the eggs produced failed to hatch (Fig. 2C), demonstrating that the alternative cargo binding region of Drosophila Khc, and likely its interaction with aTm1, play a major role in oskar mRNA transport.
Figure 2.
oskar mRNA localization depends on the alternative cargo binding domain of Khc. (A) Schematic of Khc-FL and KhcΔ855–911 transgenic constructs resistant to khc-RNAi used in this study. The pink stars indicate silent point mutations in the region targeted by khc-RNAi; KhcΔ855–911 lacks the alternative cargo binding domain. (B) Deletion of the alternative cargo binding region of Khc leads to oskar mRNA mislocalization. (Top middle and right panels) Khc-FL and KhcΔ855–911 variants tagged with mKate2 were expressed in a khc-RNAi background. Khc variants and khc-RNAi were expressed in the germline, which includes nurse cells and the oocyte (see schematic top left). (Bottom panels) oskar mRNA was visualized by single-molecule (sm)FISH. Scale bar, 25 µm. (C) Hatching rates of eggs from flies expressing Khc-FL or KhcΔ855–911 transgenes in Khc knockdown background.
The Khc interaction domain of aTm1 is essential for oskar localization
The region of aTm1 (residues 247–335) that interacts with Khc consists of a coiled coil that is also present in the classical Tm1-A isoform (residues 282–335), preceded by a unique N-terminal sequence predicted to be at least partially helical (Fig. 1D). To understand the functional significance of the aTm1–Khc interaction, we generated GFP-tagged aTm1 truncations in which the Khc interaction site was either present or absent and tested them in vivo. We expressed the constructs in Tm1eg9 mutant flies, in which aTm1 is not present and oskar mRNA fails to localize at the posterior of the oocyte (Supplemental Fig. S3A–C, first panel; Erdélyi et al. 1995; Gáspár et al. 2017a).
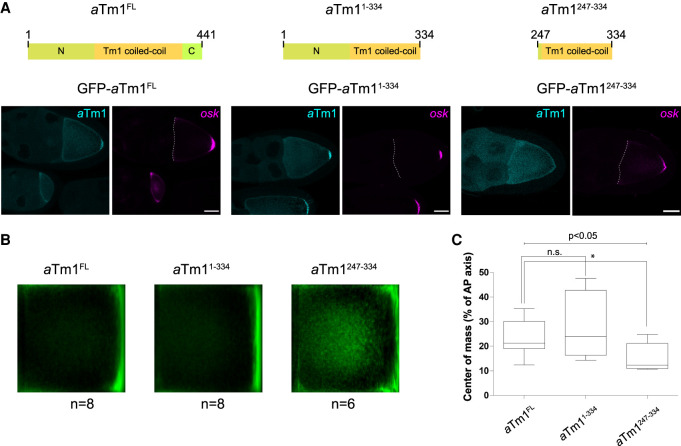
Expression of transgenic full-length aTm1 (aTm1FL) and aTm11–334 restored oskar localization to the posterior pole of Tm1eg9 oocytes (Fig. 3A–C), suggesting that the aTm1 C-terminal region is nonessential for this function. Expression of the minimal Khc interaction region of aTm1 (aTm1247–334) also promoted oskar localization in Tm1eg9 oocytes. Although the RNA was not as concentrated at the posterior pole as upon aTm1FL or aTm11–334 expression (Fig. 3B,C; analysis was performed as in Gaspar et al. 2014), the female progeny developed ovaries and were fertile. Neither aTm11–247 nor aTm1335–441 could restore oskar localization in Tm1eg9 oocytes (Supplemental Fig. S3C). These data show that the core domain of aTm1 (residues 247–334) identified as the Khc binding domain is essential for oskar mRNA localization.
Figure 3.
aTm1247–334 rescues oskar mRNA localization in Tm1eg9/Tm1eg9 mutant. (A) Confocal images of Tm1eg9/Tm1eg9 egg chambers expressing GFP-tagged aTm1FL, aTm11–334, and aTm1247–334 variants. oskar mRNA was visualized by smFISH. Scale bar, 25 µm. (B) Mean oskar distribution (green) in stage 9 oocytes from the flies in A. n indicates the number of oocytes analyzed. (C) Position of oskar mRNA center of mass relative to the geometric center of the oocyte represented in box plots with minimum and maximum whiskers. The analysis was performed as in Gaspar et al. (2014).
Interestingly, despite supporting oskar mRNA localization, GFP-aTm1247–334, which lacks a major portion of the unique N-terminal region of aTm1, does not accumulate at the posterior pole of the oocyte, in contrast to aTm1FL or aTm11–334. This suggests that the aTm1 N-terminal domain interacts with components of the pole plasm or with oskar mRNA itself, resulting in aTm1 accumulation at the oocyte posterior.
aTm1 coiled-coil dimers adopt an antiparallel configuration
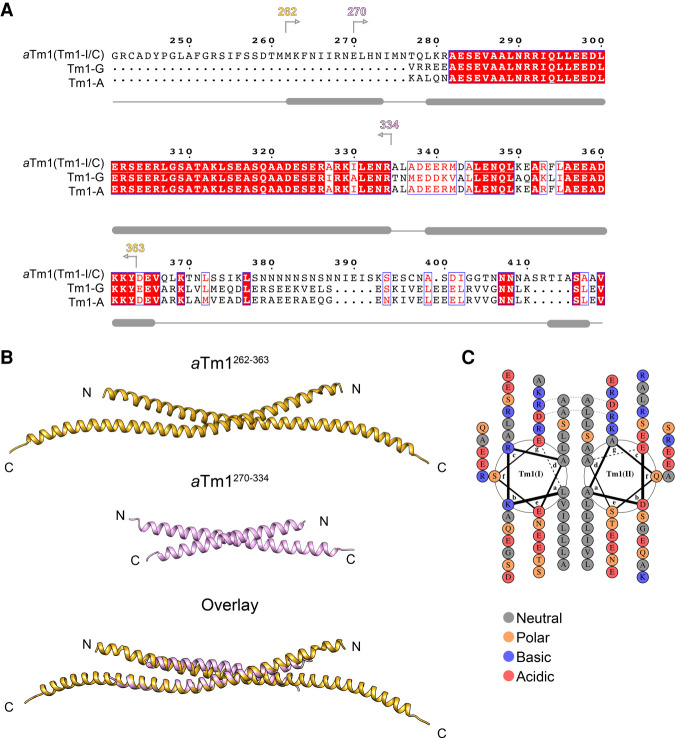
To gain insight into the molecular basis of aTm1–Khc interaction, we used crystallography to determine the structure of aTm1 alone and in complex with Khc. The classical actin-binding tropomyosins (Tm1-PG and Tm1-PA in Drosophila) are highly conserved in eukaryotes (Fig. 4A), where they form long parallel coiled coils that polymerize in a head-to-tail fashion along the actin filaments (Parry and Squire 1973). Residues 81–165 of Tm1-PG are present in aTm1 whose coiled-coil region is further extended by 20 unique residues at the N terminus (aTm1262–366) (Fig. 4A), which we included in our constructs.
Figure 4.
aTm1 forms an antiparallel coiled coil. (A) Multiple sequence alignment of three Tm1 isoforms from Drosophila melanogaster. The shared coiled-coil region of aTm1 extends N-terminally into a unique predicted α helix (the predicted secondary structure is indicated under the alignment where regions with thick gray bars indicate α helices). Tm1-PG and Tm1-PA are two classical actin-binding nonmuscle isoforms that lack this N-terminal extension. The boundaries of the crystallized constructs of aTm1 are indicated above the alignment. The alignment was done by ESPript 3 (Robert and Gouet 2014). (B) aTm1 coiled-coil region crystallized in an antiparallel conformation. Crystal structures of aTm1262–363 (top panel) and aTm1270–334 (middle panel) and their superimposition (bottom panel). Residues 261–358 are resolved in the crystal structure of the aTm1262–363 construct. Residues 277–319 in the first coil and residues 273–330 in the second coil are resolved in aTm1270–334. (C) The coiled coil of aTm1 is stabilized primarily by hydrophobic interactions. Helical wheel diagram of aTm1262–363 as generated by DrawCoil 1.0 (https://grigoryanlab.org/drawcoil). The dashed lines connecting Arg292 and Asp299 indicate salt bridges between these residues.
We performed crystallization trials with different aTm1 truncations (Fig. 4A) and obtained crystals for aTm1262–363 and aTm1270–334 constructs that diffracted to a resolution of 2.45 Å and 2.3 Å, respectively (Fig. 4B). The structures were solved by ab initio molecular replacement with Ample and were refined to an Rwork/Rfree of 0.25/0.29 and of 0.23/0.27 for Tm1262–363 and Tm1270–334, respectively (see Supplemental Table S1 for structural statistics; Bibby et al. 2012). Residues 261–358 were fitted into the density of the aTm1262–363 crystal structure, whereas residues 277–319 in the first coil and residues 273–330 in the second coil were resolved in aTm1270–334.
Both truncations crystallized as antiparallel coiled coils with an average backbone RMSD of 1 Å between residues 278 and 328 (Fig. 4B). A helical wheel diagram of aTm1262–363 in Figure 4C demonstrates that the two α helices are held together primarily by hydrophobic residues at the a and d positions of the heptad repeats, which is typical for coiled-coil structures. The interaction is additionally stabilized by two symmetric salt bridges on the surface from Arg292 and Asp299 of chain A to Asp299 and Arg292 of chain B, respectively.
The unusual antiparallel coiled-coil conformation of aTm1 distinguishes it from the canonical Tm1 isoforms, which form parallel coiled coils, and might be an additional factor attributing to it specific functions.
aTm1–Khc interaction domains form a triple coiled coil of two kinesin chains and one aTm1 chain
We next reconstituted, biophysically characterized, and crystallized the minimal aTm1–Khc complex. To do so, the initially identified protein boundaries (Fig. 1D) were further refined to improve protein solubility and yields, after which individually purified aTm1252–334 and Khc855–941 were mixed and analyzed by gel filtration (Fig. 5A). Upon addition of aTm1, the Khc peak shifted to a higher apparent molecular weight, indicating complex formation. Interestingly, the aTm1 peak fractions shifted to a later retention time (larger elution volume) indicative of compaction of the protein in the complex upon binding (Fig. 5A, middle gel [lanes 7–9] and lower gel [lanes 8–10]). Also, although we mixed the proteins in a 1:1 ratio before injection onto the column (Fig. 5A, right panel, lowest gel, lane 1), the ratio of the bands in the complex peak (Fig. 5A, right panel, lowest gel, lanes 8–12), as well as the presence of an additional aTm1 peak (Fig. 5A, right panel, lowest gel, lanes 3–7) indicate that Khc is in excess in the complex. To determine the affinity of the aTm1–Khc interaction, we performed surface plasmon resonance (SPR) experiments using an anti-GST antibody coated chip allowing immobilization of GST-Khc848–941. aTm1252–334 was added stepwise (fivefold increasing concentration at each step), and the resulting sensorgram could be fitted to a standard 1:1 kinetic model with a dissociation constant (Kd) of 141 ± 6 nM (Fig. 5B).
Figure 5.
The aTm1–Khc complex forms a tripartite coiled coil. (A) Reconstitution of an aTm1–Khc complex, consisting of the minimal interaction regions of the two proteins (left schematic). Size exclusion chromatography (S200 resin) of Khc855–941, aTm1252–334, and a 1:1 mixture of both proteins. The UV trace of aTm1 alone could not be recorded because of the very low extinction coefficient of this truncation. Peak fractions were analyzed by gel electrophoresis in the bottom panel. (B) Binding of aTm1252–334 to Khc848–941 analyzed by surface plasmon resonance (SPR). GST-tagged Khc848–941 was used as a ligand, and increasing concentrations (indicated above the sensogram in nanomoles) of aTm1 were added as analyte. Ligand to analyte was fitted using a standard 1:1 kinetic model with a Kd = 141 ± 6 nM. (C) Crystal structure of the aTm1252–334/Khc855–941 complex. Two different helical views (left panels) and the corresponding electrostatic views (right panels) are presented. The two Khc chains (aquamarine) are parallel and in register, whereas the single aTm1 chain (gold) is in an antiparallel conformation. The crystal structures consist of aTm1 (residues 259–324), KhcI (residues 855–923), and KhcII (residues 855–916). The bottom two left and right panels represent ribbon and electrostatic views of aTm1262–363 for comparison. (D) The triple coiled coil of the aTm1–Khc complex is stabilized primarily by hydrophobic interactions; helical wheel diagram generated by DrawCoil 1.0 (https://grigoryanlab.org/drawcoil).
The aTm1252–334–Khc855–942 complex crystallized in a P21 space group. A data set was collected to a resolution of 2.3 Å and a model obtained by ab initio molecular replacement with Arcimboldo (Rodríguez et al. 2009). The structure was refined to a final Rwork/Rfree of 0.24/0.28, incorporating aTm1 (residues 259–324), KhcI (residues 855–923), and KhcII (residues 855–916) in 1:2 (aTm1:Khc) stoichiometry (Fig. 5C). The trimeric structure comprises two parallel, in-register Khc chains and a single, antiparallel Tm1 chain, with the Khc C-terminal residues in close proximity to the aTm1 N-terminal region. This arrangement suggests that the coiled coil of aTm1 opens to accommodate the coiled-coil stalk of Khc. A possible explanation for the mechanism of aTm1–Khc complex formation would be a lower dimerization propensity of the aTm1 homotypic coiled coil. To estimate the Kd of the aTm1 homodimer, we performed an isothermal titration calorimetry (ITC) experiment whereby we fitted the heat changes upon dilution of aTm1252–334 (Supplemental Fig. S4A), and obtained a Kd of 10 µM. Thus, aTm1 has a higher affinity to Khc (Kd in the nanomolar range) (Fig. 5B) than to another aTm1, which explains why aTm1 forms a complex with the Khc homodimer.
Superimposition of the aTm1 homodimer on the Khc–aTm1 complex structure revealed no major changes of the aTm1 helix except for a kink caused by the looping out of T277 (Supplemental Fig. S4B). As crystals of Khc truncations were only obtained in the presence of aTm1, we could not evaluate the effect of aTm1 binding on Khc conformation.
The triple coiled coil is held together by van der Waals forces between mostly hydrophobic residues at “a” and “d” positions of each helix (Fig. 5D). Additionally, the surface of the complex is characterized by a positively charged patch, not present in the aTm1262–363 homodimer, formed by the polar side chains of K884, K887, and R888 of Khc (Fig. 5C, right panels), important for RNA binding (see below).
Mutational analysis of aTm1–Khc interaction contacts
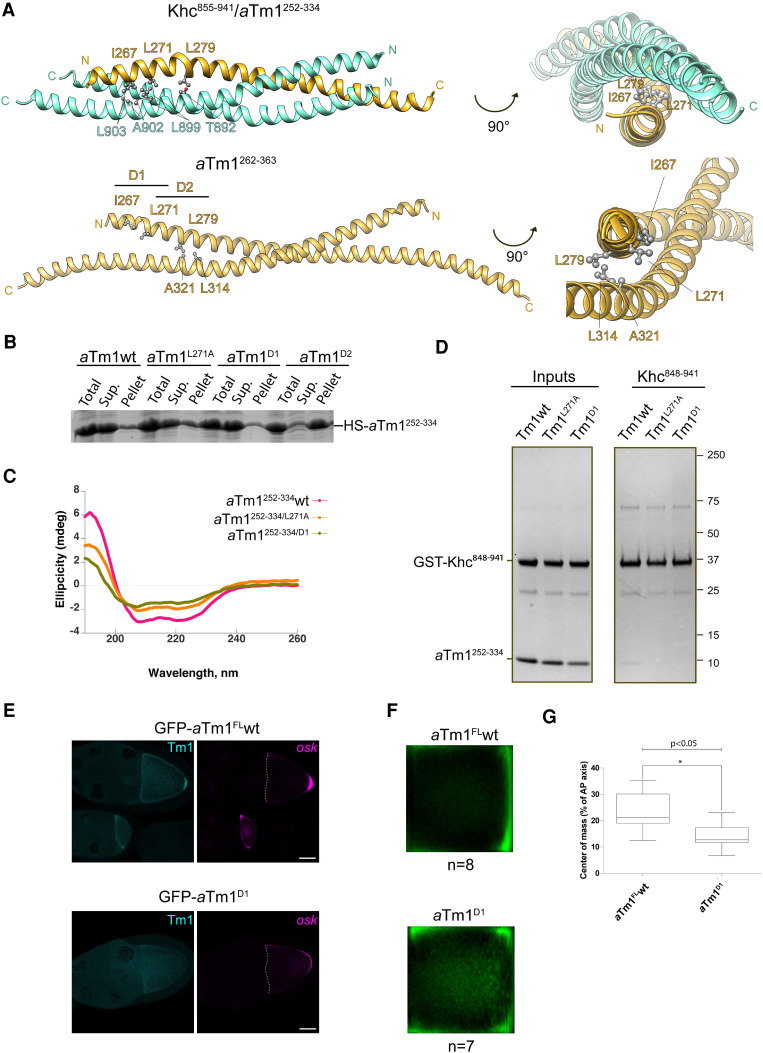
To verify the interaction surface of aTm1 and Khc, we generated single point mutants that would be expected to disrupt complex formation. We first mutated two amino acid residues in the unique region of the aTm1 coiled coil, L271 at the hydrophobic core of the complex, and Q278, whose polar side chain is positioned near the hydrophobic core due to a kink in aTm1 (Supplemental Figs. S4B, S5A). Mutating aTm1 L271 to the smaller nonpolar residue alanine or the bulkier, charged arginine only mildly affected the interaction with Khc, whereas the Q278A mutation had no effect (Supplemental Fig. S5B, left and middle panels). This might be due to the extensive hydrophobic core of the aTm1–Khc complex. To achieve disruption of the interaction, we designed two triple mutants—aTm1I267A/L271A/L279A (aTm1T1) at the unique N-terminal extension of aTm1, and aTm1L296A/L300A/L307A (aTm1T2) in the middle of the aTm1 coiled coil—and a combination of the two (aTm1Hexa) (Fig. 6A; Supplemental Fig. S5C). Both the aTm1T1 and aTm1Hexa mutations fully abolished interaction with Khc, as determined by in vitro binding assays (Supplemental Fig. S5D). However, although we changed the target residues to alanines in order to preserve the helical propensity of aTm1, circular dichroism (CD) spectroscopy showed that the mutations affected substantially the structure of the aTm1 homodimers (Supplemental Fig. S5E).
Figure 6.
Mutational analysis of the aTm1–Khc interaction. (A) Designing double point mutations at the hydrophobic core of aTm1–Khc coiled coil. Ribbon views of aTm1–Khc complex (top panels) or aTm1 homodimer (bottom panels) in two different orientations. Three residues—I267, L271, and L279—at the hydrophobic core of the trimeric complex are highlighted. In aTm1D1, residues I267 and L271 are mutated to alanine. In aTm1D2, residues L271 and L279 are mutated to alanine. (B) aTm1D2 has significantly reduced solubility. Total, supernatant, and pellet fractions of E. coli lysate with recombinantly expressed His-SUMO tagged aTm1252–334 wild-type and mutant variants were analyzed on SDS-PAGE and stained with Coomassie. (C) aTm1D1 has the characteristic coiled-coil fold. (Magenta) CD analysis of aTm1wt, (orange) aTm1L271A , (green) aTm1D1. (D) aTm1D1 mutation causes strong reduction of the interaction with Khc. Pull-down assays of GST-tagged Khc848–942 with different variants of aTm1252–334. (Left panel) Purified aTm1 variants were mixed in excess with Khc848–942, immobilized on GSH beads. (Right panel) After incubation and extensive washing, the eluates were analyzed on SDS-PAGE and stained with Coomassie. (E) Tm1D1 causes oskar mRNA localization defects in vivo. (Left panels) Egg chambers (stage 9) from flies expressing GFP-aTm1-FL and GFP-aTm1D1 in the Tm1eg9 /Tm1/eg9 background. (Right panels) oskar mRNA was detected by smFISH. Scale bar, 25 µm. (F) Mean oskar distribution (green) in stage 9 oocytes from the flies in E. n corresponds to the number of analyzed oocytes. (G) Position of oskar mRNA center of mass relative to the geometric center of the oocyte, represented in box plot with minimum and maximum whiskers. GFP-aTm1FL wt panels and quantification in E–G are the same as in Figure 3.
As the triple mutant T2 showed residual binding to Khc, we focused on T1 and designed two double mutants, aTm1I267A/L271A (aTm1D1) and aTm1 L271A/L279A (aTm1D2) (Fig. 6A). The mutations introduced in aTm1D2 substantially reduced the solubility of aTm1252–334 since most of the recombinantly expressed protein precipitated after centrifugation of the E.coli lysate (Fig. 6B). This might be due to the fact that L279 lies at the junction of the two aTm1 chains in close proximity to L314 and A321 of the second chain and is therefore essential for the stability of the aTm1 coiled coil. This again demonstrates how the fact that aTm1 uses the same surface to form a homodimer and to interact with Khc renders it difficult to generate an aTm1 mutant that has no effect on the structure of aTm1 yet disrupts its interaction with Khc. Thus, even the two mutations in aTm1D1 appear to destabilize the coiled coil to some extent, as a SEC-MALS (size exclusion chromatography multiangle light scattering) measurement indicates that aTm1D1 behaves as a mixture of dimers and monomers (Supplemental Fig. S5F). Nonetheless, the aTm1D1 mutations did not affect solubility of the protein and had only a moderate effect on its secondary structure as assessed by CD spectroscopy (Fig. 6B,C). More importantly, aTm1D1 interaction with Khc was notably reduced in our GST pull-down assays and as evaluated by SPR (Fig. 6D; Supplemental Fig. S5G).
To assess aTm1D1 function in vivo, we generated a GFP-tagged aTm1D1 transgene and tested its ability to rescue oskar mRNA localization in tm1eg1 oocytes. smFISH revealed that the bulk of oskar mRNA was unlocalized, with only trace amounts detected at the posterior pole (Fig. 6E; Supplemental Fig. S5H), further confirming the importance of the aTm1–Khc interaction for oskar mRNA transport.
aTm1 stabilizes the interaction of Khc with RNA
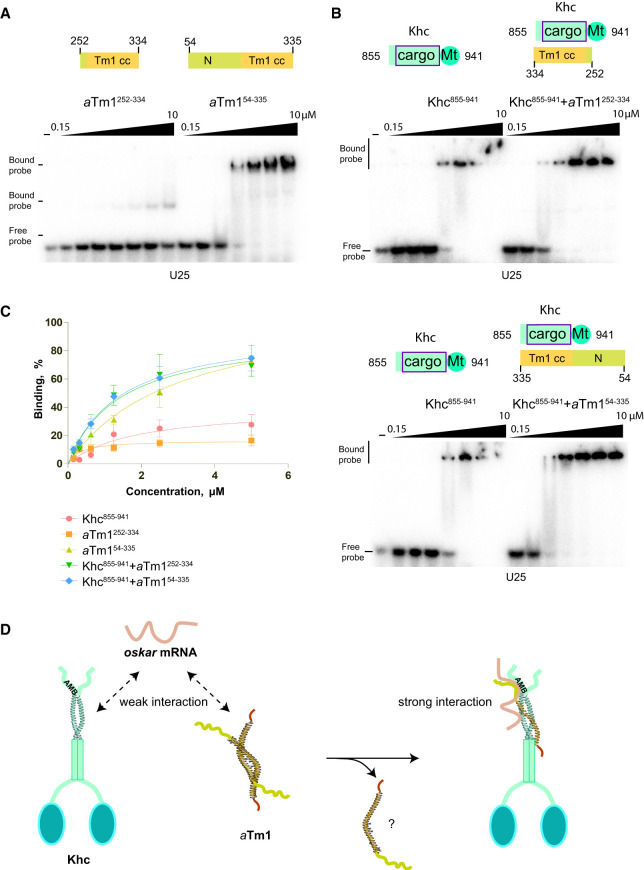
Previous findings suggest a direct interaction of aTm1 with the oskar 3′ UTR, supporting the notion that aTm1 might function as a Khc adaptor for RNA (Gáspár et al. 2017a). We therefore tested whether aTm1 interacts with poly (U) RNA in vitro. Considering the rescue of the Tm1eg9/eg9 loss-of-function mutant by aTm11–334 (Fig. 3), we focused our analysis on this portion of the protein. Since recombinantly expressed aTm11–335 was unstable and gave rise to two proteolytic products (Supplemental Fig. S6A), we generated further truncations of aTm1 to probe its interaction with RNA. Previously we showed by NMR, which allows detection of stable as well as transient interactions, that Tm11–246, which encompasses most of the low complexity N-terminal region of aTm1, interacts directly with U25 RNA (Vaishali et al. 2021). Moreover, electrophoretic mobility shift assays (EMSAs) showed that the C-terminal region of aTm11–334, Tm1252–334, binds oligo-(U25) only weakly, with a Kd >10 µM, as compared with Tm154–335, which demonstrated significantly higher affinity for RNA (Fig. 7A). These findings suggest that a major RNA binding activity of aTm1 lies within the N-domain (residues 1–246).
Figure 7.
aTm1 as a kinesin modulator. (A) aTm1-N domain binds to RNA. Electrophoretic mobility shift assay (EMSA) of aTm1252–334 and aTm154–335 variants. 32P-labeled oligo-(U25) was incubated with twofold increasing concentrations (0.15, 0.3 ….10 µM) of recombinantly purified proteins in 15-µL reactions as indicated. The samples were then loaded on 6% native gel and visualized by autoradiography. Free probe was run in the first lane. (B) Khc binds to RNA alone and better in the presence of aTm1. Electrophoretic mobility shift assay (EMSA) of Khc855–942 alone and in the presence of aTm1252–334 (left gel) or of aTm154–335 (right gel). For the complex, aTm1 was added to Khc in 1:2 molar ratio prior to performing serial dilutions. (C) Nitrocellulose filter binding assay of aTm1 and Khc variants alone and in complex. 32P-labeled oligo-(U25) probe was incubated with increasing concentrations of proteins and protein complexes. The mixture was then passed through a nitrocellulose filter, and the radioactive probe left on the filter was measured as a fraction of the total probe that was added to the reaction. (D) Hypothetical model of aTm1 as a modulator of Khc interaction with RNA. aTm1 and Khc each bind RNA weakly on their own. To accommodate the Khc coiled coil, the aTm1 homodimer opens, allowing formation of a tripartite coiled coil composed of one aTm1 and two Khc chains. The second aTm1 might be rearranged and promote formation of higher order structures. Binding of aTm1 to Khc might elicit conformational changes in Khc or, alternatively, stabilize the Khc RNA binding surface and thus allow the Khc-aTm1 complex to interact with higher affinity and/or specificity with RNA. Since the region with which Khc interacts with RNA incorporates both the alternative cargo binding domain and the adjacent auxiliary microtubule binding domain, the binding of aTm1 to Khc might regulate the kinetic activity of Khc.
As deletion of the N domain of aTm1 had only a mild effect on oskar localization (Fig. 3; Supplemental Fig. S3), we suspected the presence of an additional RNA binding moiety within the Khc–aTm1 complex. Indeed, as mentioned above, the structure of the aTm1–Khc complex displays a joint positively charged surface (Fig. 5C). We therefore performed EMSAs with Khc855–941 alone or in complex with aTm1. Khc on its own bound RNA with high affinity (high nanomolar range) and, at higher concentrations, appeared to form oligomers with RNA that failed to enter the gel (Fig. 7B). Upon addition of Tm1252–334 or Tm154–335 to Khc in a 1:2 ratio, the affinity increased and the protein–RNA complexes could be resolved (Fig. 7B). The affinity increase for Tm154–335, as expected, has a stronger effect, as the N domain contributes to RNA binding. As an independent method to assess the interaction of aTm1–Khc with RNA, we performed filter binding assays, which overall confirmed the EMSA results showing that the interaction of Khc with RNA in solution is enhanced almost to the same extent by Tm1252–334 and Tm154–334 (Fig. 7C). Together, these data show that aTm1 prevents formation of Khc–RNA aggregates and that the aTm1 and Khc synergize in RNA binding.
To ascertain that the interaction of Khc with RNA is not due to stickiness of the Khc855–941 truncation, we also performed EMSAs with the full-length Khc, alone or in the presence of Tm1252–334 or Tm154–335. This confirmed that Khc-FL binds to RNA and that its affinity for RNA increases when aTm1 is present (Supplemental Fig. S6B).
The Khc855–941 fragment used in our RNA binding and crystallization experiments contains both the aTm1 binding site (residues 855–909) and the ATP-independent microtubule binding site (AMB, residues 910–938). To determine which of the two regions mediates RNA binding, we tested two further truncations from which the AMB was either partially or fully deleted (Khc855–920 and Khc855–909). Both proteins, and especially Khc855–909, showed reduced binding to RNA (Supplemental Fig. S6C). To test for involvement of the positively charged patch on the surface of aTm1–Khc in RNA binding, we mutated Lys884, Lys887, and Arg888 of Khc to alanines (Khc855–941/TA) (Supplemental Fig. S6D). This reduced the binding of Khc to RNA but had no effect on protein folding as measured by CD spectroscopy or on binding to aTm1 as assessed by a GST pull-down assay (Supplemental Fig. S6E–G). These experiments show that both the aTm1 binding site and the AMB contribute to RNA binding, most likely by formation of an extended RNA interaction surface. These data are consistent with the finding of Williams et al. (2014) that Khc1–910, which lacks the auxiliary microtubule binding domain, does not support oskar mRNA localization.
Taken together, our findings demonstrate that Khc can bind RNA directly and that aTm1 acts in stabilizing rather than in mediating the Khc–RNA interaction, distinguishing aTm1 from a classical kinesin adaptor.
Discussion
In recent decades, mechanisms of mRNA transport and the composition of mRNP transport complexes have been extensively studied, yet only a few adaptor proteins linking RNAs to the transport machineries have been identified. These include She2p/She3p for myosin-based RNA transport in S. cerevisiae (Takizawa and Vale 2000), and Egalitarian/BicD proteins for dynein-based transport in Drosophila (Dienstbier and Li 2009; Dienstbier et al. 2009). The atypical tropomyosin isoform aTm1 was recently proposed to function as an adaptor for Khc-mediated transport of oskar mRNA in Drosophila (Veeranan-Karmegam et al. 2016; Gáspár et al. 2017a). Classical Tm1 isoforms form parallel coiled coils that polymerize in head-to-tail fashion along actin filaments and are known to regulate different functions of the actin cytoskeleton (Parry and Squire 1973; Gunning et al. 2015). aTm1, which is present in Drosophilidae, has a shortened coiled-coil region flanked on both sides by unique, intrinsically disordered N-terminal and C-terminal sequences. Crystal structures of the aTm1 coiled coil presented in this study reveal an antiparallel conformation with a coiled-coil region extended by 20 amino acids beyond the conserved region (Fig. 4). Coiled coils play vital roles in nearly all biological processes and are among the most studied structural motifs (Truebestein and Leonard 2016). A few point mutations in the hydrophobic core are sufficient to cause a switch in the orientation of the coiled coil or the number of coils involved in the interaction (Mason and Arndt 2004; Malashkevich et al. 2015). Therefore, it is likely that the unique extension of the aTm1 coiled-coil region causes the antiparallel topology of the two chains. Importantly, two point mutations in this region have a dramatic effect on aTm1's interaction with Khc, which suggests that the unique sequence contributes to defining the specificity of aTm1 for Khc, both at the secondary and tertiary structure level (Fig. 6). Additionally, the switched orientation of the aTm1 coiled coil prevents interaction of the protein with actin and thus prevent its interference with the function of the classical actin-binding tropomyosin isoforms (Cho et al. 2016).
Although we could not determine a structure of the Khc dimer on its own, we assume that its conformation does not change significantly when bound to aTm1. This is corroborated by rotary shadowing negative stain EM images of Khc, which show that the motor domains are symmetrically attached to a long rod, a conformation that can be achieved only when Khc is a parallel in-register coiled coil (Hirokawa et al. 1989). The aTm1 homodimeric coiled coil, on the other hand, must open to allow binding to the kinesin homodimer, thus posing the question of what happens to the second aTm1 chain (Fig. 7D). It might form an individual complex with another Khc homodimer or, alternatively, rearrange such that it remains bound to the portion of the aTm1 coiled coil not in complex with Khc. This arrangement can further provide a platform for the formation of higher-order structures.
The interaction of Tm1 with Khc involves the alternative cargo binding region of Khc, which lies immediately downstream from the Klc binding domain (Fig. 1). This region was initially identified as cargo binding in the fungus N. crassa and was subsequently shown to be the major binding site for other adaptors and their respective cargos (Diefenbach et al. 2002, 2004; Setou et al. 2002; Kanai et al. 2004; Glater et al. 2006). We have shown that deletion of this region in Drosophila Khc results in oskar mislocalization and female sterility (Fig. 2), a phenotype more severe than that of complete loss of aTm1, in which case the eggs still hatch and develop into adult sterile flies. This suggests that the alternative cargo binding region of Khc might have functions beyond oskar mRNA localization also in Drosophila. For example, it has been shown that the mitochondrial adaptor Milton binds also to the alternative cargo binding domain, suggesting that in KhcΔ855–911 mitochondrial transport to and within the oocyte might also be affected (Cox and Spradling 2006). Moreover, Milton along with SNAP25, which is an adaptor for vesicular transport, and the ribosome receptor p180 appear to bind Khc also via coiled-coil interactions, suggesting a binding mechanism similar to the aTm1–Khc interaction described here (Diefenbach et al. 2002, 2004; Glater et al. 2006; Randall et al. 2013).
Functionally, coiled coils allow the formation of long rigid structures, especially when they involve more than two chains, and they may play the role of “molecular rulers,” spacing the different functional regions of a protein (Truebestein and Leonard 2016). Thus, with the Klc binding site and the alterative cargo binding region adjacent to one another and potentially prone to steric hindrance, the coiled-coil structure might provide space for the simultaneous binding of cargos to these two regions. It will be interesting in the future to determine whether different cargos can indeed be cotransported by a single Khc molecule.
Retrograde transport by different kinesins along microtubules comprises a major fraction of the intracellular transport of mRNP granules. Surprisingly however, there are to date scarce mechanistic details of how the cargo might be recognized and how it can be attached to Khc. Only recently, the transport of β-actin and β2Β-tubulin mRNAs was reconstituted in vitro, whereby APC (adenomatous polyposis coli) links the mRNAs to kinesin-2 through the cargo adaptor KAP3 (Baumann et al. 2020). Another study suggests that the transport of some axonal mRNPs depends on KLC1, which also implies a more classical adaptor-based transport mechanism (Fukuda et al. 2021). Here, we have shown that the N domain (residues 1–246) of aTm1 binds RNA, whereas the region immediately adjacent to it (residues 247–334) interacts with the alternative cargo binding region of Khc (Figs. 1, 7). Therefore, it is appealing to imagine aTm1 as an adaptor for Khc as well. However, deletion of the N domain in vivo resulted in only minor oskar mRNA mislocalization, and sole expression of the Khc binding domain almost restored fully the accumulation of oskar at the posterior pole of the oocyte (Fig. 3). As demonstrated, Khc binds to RNA also on its own, and its RNA affinity increases in the presence of aTm1 (Fig. 7). Moreover, the Khc-RNA interaction appears to be mediated by both the aTm1 binding surface and the adjacent ATP-independent microtubule binding region (Supplemental Fig. S6). This explains the strong effect of deletion of the ATP-independent microtubule binding region on oskar mRNA localization (Williams et al. 2014). Therefore, aTm1 appears to act as a stabilizer of the Khc interaction with RNA rather than as a classical adaptor (Fig. 7).
It is possible that aTm1, upon binding, induces a conformation change of Khc, leading to increased RNA binding. This is supported by the structure reported here, as part of the AMB site of Khc appears to acquire a helical conformation (resides 909–920), although predicted to be disordered. Despite the lack of an isolated Khc structure for comparison, it can be speculated that the binding of aTm1 stabilizes the helical conformation of the alternative cargo binding domain that extends into the AMB site and thus enables a higher-affinity interaction of Khc with RNA. Interestingly, downstream from AMB (residues 938–950) (Williams et al. 2014) is the Khc IAK autoinhibitory motif, which interacts with the motor domain of Khc and keeps the protein inactive in the absence of cargo (Kaan et al. 2011). Because of the antiparallel topology of the aTm1 chain within the aTm1–Khc complex, the N domain of aTm1 is in proximity to the IAK motif of Khc. Thus, besides enabling the binding of cargo to Khc, aTm1 might also play a role in release of the autoinhibited conformation of Khc. This might explain the observation that, upon deletion of the N domain of aTm1, most but not all oskar localizes to the oocyte posterior pole (Fig. 3; Supplemental Fig. S3).
Our high-resolution structure of the atypical Tm1 (aTm1) suggests that the unique antiparallel coiled-coil conformation of its dimers might underlie the specific functions of this isoform. Together with our high-resolution structure of the complex of aTm1 with Khc, our findings show that aTm1 stabilizes the interaction of Khc with RNA. Whether aTm1 binding increases Khc's specificity for oskar mRNPs alone or through additional protein-protein or protein-RNA interactions in vivo remains to be investigated. Future identification of the aTm1 interactome and of the oskar mRNA sequence or structure to which the aTm1–Khc complex binds will be essential to elucidate the full mechanism whereby this RNA transport complex recognizes its different cargos.
Materials and methods
Plasmid construction
Cloning techniques including digestion, ligation, and DNA electrophoresis were performed essentially as described in Sambrook et al. (1989) unless indicated otherwise. Mutagenesis was performed according to Liu and Naismith (2008). All GST-tagged Khc constructs (Khc1–264, Khc265–975, Khc1–345, Khc345–848, Khc848–941, Khc941–975, Khc902–941, Khc559–941, Khc623–941, and Khc786–941) were cloned between BamHI and EcoRI sites of the pGEX6p-1 vector (Amersham). All His6-SUMO tagged Tm1 (Tm1-FL, Tm11–335, Tm1247–378, Tm11–246, Tm1247–334, and Tm1335–441) and Khc (Khc855–941, Khc855–909, and Khc855–920) truncations were cloned between BamHI and SacI sites of the pETM11-His6-SUMO plasmid (derived from pBR322, H. Besir, Protein Expression and Purification Core Facility, EMBL Heidelberg). The simple modular architecture research tool (SMART) was used for designing the truncation boundaries (Letunic and Bork 2018).
For yeast two-hybrid assays, Khc-FL, Khc265–975, and Khc265–975 were cloned between NdeI and BamHI sites of the pG4BDN22 vector, whereas Tm1-FL, Tm11–247, and Tm1247–441 were cloned between NdeI and BamHI sites of the pG4ADHAN111 vector (Thoms et al. 2015).
The Tm1 integrase constructs were based on pUASp2-emGFP-Tm1RI (Gáspár et al. 2017a). Essentially, the aTm1 open reading frame (ORF) and 3′ UTR were cloned between BamHI and BglII sites of the pUASp vector (Rørth 1998). emGFP was inserted downstream from the pUAS promoter in-frame with the Tm1 coding sequence. To generate pUASp-attB-ΔK10-Tm1 constructs, the region of pUASp2-emGFP-Tm1 containing the pUAS promoter, emGFP, Tm1-FL, or truncations’ ORFs, and Tm1 3′ UTR region were PCR-amplified and inserted between NotI and XbaI sites of pUASp-attB-ΔK10 plasmid using In-Fusion cloning according to the manufacturer's instructions (Takara).
To generate the pKhcp-attB-ΔK10-Khc-FL construct, we amplified the 2R:16266960–16273471 region from fly genomic DNA and inserted it between Eco147I and BamHI sites of pUASp-attB-ΔK10 vector, removing the pUAS promoter. We then performed site-directed mutagenesis using the following oligos: 5′-GTCTACCTGTTTGATAAAGTCTTCAAACCGAATGCATC-3′ (fwd) and 5′-TTTATCAAACAGGTAGACCTTGCCCTGCAGACGATG-3′ (rev) to change the region targeted by Khc-RNAi (Bloomington Stock 35409). The mKate2 coding sequence was added in-frame at the C terminus of the Khc ORF using the ligation-free cloning approach. To generate the pUASp-attB-ΔK10-KhcΔ855–911 truncation, we performed inverse PCR with oligos starting at the boundaries of the truncation, followed by ligation of the product and transformation in E.coli.
Khc-FL was cloned with a HisSUMO-SNAP-tag between BamHI and HindIII sites of the pFastBacDual vector. All plasmids used in this study are summarized in Supplemental Table S2.
Protein expression and purification
GST- and His6-SUMO-tagged proteins were expressed in BL21-CodonPlus(DE3)-RIL cells (Stratagene) by isopropyl β-D-1-thiogalactopyranoside (IPTG) induction for 16 h at 18°C. Cells were grown in Luria-Bertani (LB) medium (0.5% [w/v] yeast extract [MP], 1% [w/v] tryptone [MP], 0.5% [w/v] NaCl at pH 7.2) supplemented with antibiotics (100 µg/mL ampicillin or 10 µg/mL kanamycin and 34 µg/mL chloramphenicol). After harvesting, the pellets were lysed by a microfluidizer processor (Microfluidics) in 500 mM NaCl, 20 mM Tris-HCl (pH 7.5), 5% glycerol, 0.01% NP-40, and 40 mM imidazole (only in the case of His6-tagged proteins) buffer supplemented with protease inhibitor cocktail (Roche) and 5 mM β-mercaptoethanol. The lysates were subsequently cleared by centrifugation at 18,000 rpm for 20 min.
For GST-tagged proteins, the lysates were incubated with gluthatione sepharose 4B (GE Healthcare) for 1 h at 4°C. After washing, the beads were directly used for binding assays (see below).
His6-SUMO tagged proteins were affinity-purified by a His TRAP Ni column (GE Healthcare) and eluted over an imidazole gradient (40–600 mM). After cleavage of the tag by Senp2 protease, the proteins were passed one more time over a His TRAP Ni column to remove the tag and protease. Finally, the eluates were further separated on a Superdex 200 16/600 column in a 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, and 0.5 mM TCEP buffer for crystallization and binding assays and 20 mM HEPES-Na (pH 7.5), 150 mM NaCl, and 0.5 mM TCEP for surface plasmon resonance (SPR) and CD spectroscopy.
His6-SUMO-SNAP-tagged Khc-FL was expressed in 1 L of SF-21 insect cells at 1 × 106 cells/mL shaking culture infected with 10 mL of P1 virus stock. After expression for 3 d at 27.5°C, cells were harvested by centrifugation at 1000g for 20 min at 4°C. The pellet was lysed in a dounce tissue grinder in lysis buffer (20 mM Tris/HCl at pH 7.5, 500 mM NaCl, 1 mM MgCl2, 0.1 mM ATP, 2 mM DTT, 5% glycerol, 0.2% Tween-20). The lysate was cleared by centrifugation and the soluble protein fraction was affinity-purified on a HisTrap Excel column (GE Healthcare). After elution in a 0–300 mM imidazole gradient, the HisSUMO-SNAP-fusion tag was cleaved by 3C protease digest upon dialysis in 25 mM HEPES/KOH (pH 7.3), 150 mM KCl, 1 mM MgCl2, 0.1 mM ATP, and 2 mM DTT for 16 h before further purification by anion exchange chromatography on a HiTrap Q HP column (GE Healthcare), followed by size exclusion chromatography (SEC) on a Superdex 200 10/300 Increase column (GE Healthcare).
For expression of seleno-methionine (SeMet)-labeled Tm1252–334, we used the B834(DE3) methionine auxotrophic E. coli strain. After transformation, the cells were grown in minimal M9 medium containing 50 µg/mL methionine overnight at 37°C. The bacteria were then scaled up to the volume of the expression culture and grown to OD600 = 1 in minimal medium supplemented with methionine. The cells were then harvested, resuspended in minimal medium without methionine, and starved for 4–8 h before the addition of 50 µg/mL SeMet to the culture. After 30 min, protein expression was induced with 200 mM IPTG for 16 h at 18°C (for a detailed protocol see https://www.embl.de/pepcore/pepcore_services/protein_expression/ecoli/seleno).
Yeast two-hybrid assay
The yeast two-hybrid analysis was performed according to James et al. (1996). The yeast two-hybrid strain PJ69-4a was transformed with the indicated aTm1 or Khc fragments cloned in pAS or pG4ADHAN vectors (Clontech Laboratories, Inc.), and plated on SDC−TRP−LEU or SDC−TRP−LEU−HIS plates. A positive interaction was monitored by growth on SDC−TRP−LEU−HIS plates. pTD1-1 (Clontech Laboratories, Inc.), which contains pGAL4-AD SV40 large T-antigen, and pVA3-1 (Clontech Laboratories, Inc.), which contains pGAL4-BD-tagged murine p53, were used as positive controls.
GST pull-down assays
Proteins for pull-down assays were purified in 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 5% glycerol, and 0.01% NP-40 binding buffer. Baits were purified on gluthatione sepharose beads (see above). Lysates from E. coli expressing the protein of interest, E. coli lysates mixed with prepurified bait proteins or pure proteins were added with bait protein five times in excess of the prey protein, immobilized on GSH beads. After incubation on a turning wheel at 4°C and extensive washing, the proteins were eluted in sample buffer (200 mM Tris-HCl at pH 6.8, 8% [w/v] SDS, 40% glycerol, 0.4% bromophenol blue, 100 mM DTT) for 30 sec at 90°C. The eluates were analyzed by SDS-PAGE and stained with Coomassie.
Crystallization, data collection, and structure determination
Tm1262–363 was concentrated to 36 mg/mL and mixed in 1:1 ratio (100 nL:100 nL) with 0.1 M Tris-HCl (pH 8) and 40% MPD reservoir solution at 20°C. Rod-shaped crystals were obtained by sitting drop vapor diffusion method between day 21 and 35. Crystals were harvested in mother liquor without additional cryoprotectant. Tm1270–334 produced needle-like crystals 1 d after setting the hanging drops at 20 mg/mL in 1:1 ratio with 0.1 M NH4Ac (pH 4.6) and 15% (w/v) PEG 4000 reservoir solution. The crystals were harvested in mother liquor supplemented with 25% glycerol.
Tm1252–334/Khc855–942 complex crystals were obtained after mixing the SeMet-labeled Tm1252–334 and unlabeled Khc855–942 proteins in 1:1 ratio followed by further purification by size exclusion chromatography. The peak fractions corresponding to the complex were concentrated to 4.5 mg/mL and mixed in 1:1 ratio (100 nL:100 nL) with 0.2 M NaAc, 20% PEG 3350. Thin plate-shaped crystals were obtained after 1 d, and crystals were harvested in mother liquor supplemented with 25% glycerol.
Diffraction data sets were recorded at ID-29 beamline at the European Synchrotron Radiation Facility (ESRF), Grenoble, France. Data were processed with XDS (Kabsch 2010), and the structures were solved by ab initio molecular replacement with Ample (Bibby et al. 2012) or Archimboldo (Rodríguez et al. 2009). Model building and several cycles of refinement were performed with Coot (Emsley et al. 2010) and with the Phenix suite (Liebschner et al. 2019), respectively.
The crystal structures are deposited at the Protein Data Bank under the following accession codes: Tm1262–363 (7BJG), Tm1270–334 (7BJN), and Tm1252–334/Khc855–942 complex (7BJS).
Circular dichroism spectroscopy
The proteins were purified in 20 mM HEPES/NaOH (pH 7.5), 150 mM NaCl, and 0.5 mM TCEP for circular dichroism (CD) spectroscopy. Prior to measurement, the salt was diluted to 50 mM NaCl. The measurements were performed at a 20 μM concentration in a 0.2-mm cuvette at 20°C using a Jasco J-815 CD spectrometer. The monitored wavelength range was from 240 to 190 nm in 0.1-nm steps with an average of five points per wavelength. The data was plotted using GraphPad Prism 5.
Surface plasmon resonance (SPR)
Surface plasmon resonance (SPR) measurements were conducted using a BIAcore T200 instrument (GE Healthcare) in 20 mM HEPES-Na, pH 7.5, 150 mM NaCl, 0.5 mM TCEP, 0.005% Tween20, and 0.05% BSA at 25°C. Anti-GST antibody from the GST Capture kit was covalently immobilized to a CM5 sensor chip according to the supplier‘s instructions (GE Healthcare), and GST-tagged Khc848–941 or GST were site-directed affinity-captured to the sample and reference flow cell, respectively. The binding of Tm1252–334 at 15.625, 62.5, 250, 1000, and 4000 nM was measured in a single cycle kinetic with 90 sec injections and a final dissociation for 600 sec at a flow rate of 30 µL/min. The interaction was analyzed either as steady state affinity or by a kinetic fit of the sensorgrams using a 1:1 binding model.
Size exclusion chromatography multiangle light scattering (SEC-MALS)
One-hundred microliters of Tm1 WT or Tm1 D1 (2.0 mg/mL) was injected onto a Superdex 200 10/300 GL gel filtration column (Cytiva) in 10 mM HEPES (pH 7.5), 150 mM NaCl, and 5 mM TCEP buffer at room temperature. The column was coupled to a MALS system (MiniDAWN and Optilab, Wyatt Technology). Data were analyzed using the Astra 7 software (Wyatt Technology). Measurements were performed in triplicate.
Isothermal titration calorimetry (ITC)
Experiments were performed using a Microcal PEAQ-ITC (Malvern Instrument GmbH). The protein was dialyzed overnight at 4°C against the ITC buffer (20 mM HEPES at pH 7.5, 150 mM NaCl, 5 mM TCEP). The Tm1 WT protein in the syringe, at a concentration of 1 mM, was injected into the cell containing the buffer. Titrations were performed at 20°C or 25°C and consisted of 13 or 25 injections. The data were fitted using the dissociation fitting model of the MicroCal PEAQ-ITC analysis software (Malvern).
Fly stocks and husbandry
Tm1 and Khc transgenic flies were generated by site-specific integration of the respective pUASp-attB-ΔK10 or pKhcp-attB-ΔK10 constructs (see above) with Φc31 integrase in VK18 {vas-phi-ZH2A, PBac(y[+]-attP-9A)VK00018, Bellen laboratory} fly line. emGFP-Tm1-FL, truncations, and point mutations were driven by one copy of oskGal4 ((Telley et al. 2012); FBtp0083699) in the Tm1eg9/Tm1eg9 background (FBal0049223) (Erdélyi et al. 1995). Khc-FL-mKate2 and Khc855–911Δ-mKate2 were constitutively expressed by the Khc endogenous promoter in the UASp-Khc RNAi Trip Line GL00330 (Staller et al. 2013) background where the expression of Khc-RNAi was driven by oskGal4.
All fly stocks were grown at 21°C–25°C in vials on standard cornmeal agar. Prior to dissection, freshly hatched flies were fed with dried yeast for 1–2 d.
Single-molecule fluorescent in situ hybridization (smFISH)
Forty-two probes against the oskar mRNA coding region and 3′ UTR were labeled with Atto633 according to Gáspár et al. (2017b). smFISH was performed essentially as in Gáspár et al. (2017a). In short, two to three pairs of Drosophila ovaries were fixed with 2% (v/v) PFA and 0.05% (v/v) Triton X-100 in PBS (pH 7.4) for 20 min on an orbital shaker, followed by two washes with PBT (PBS + 0.1% [v/v] Triton X-100 at pH 7.4) for 10 min each. Ovaries were then prehybridized in 100 µL of hybridization buffer (300 mM NaCl, 30 mM sodium citrate at pH 7.0, 15% [v/v] ethylene carbonate, 1 mM EDTA, 50 µg/mL heparin, 100 µg/mL salmon sperm DNA, 1% [v/v] Triton X-100) for 30 min at 42°C, to which 100 µL of probe mixture (25 nM per individual oligonucleotide in hybridization buffer) was added for an additional 2–3 h at 42°C. After hybridization, the following washes were carried out to remove excess probes: hybridization buffer, hybridization buffer to PBT in a 1:1 mixture, PBT (10 min at 42°C each), and PBT at room temperature. The samples were then mounted in 80% (v/v) 2,2-thiodiethanol (TDE) in PBS and viewed using a Leica TCS SP8 confocal microscope with a 63× 1.4 NA objective. To detect GFP-tagged Tm1 variants, the native fluorescence of GFP was visualized. Because the mKate2 tag was not visible after smFISH, we imaged the ovaries directly after fixation in 2% PFA/PBT for 20 min. The images were analyzed in ImageJ/Fiji. Analysis of the oskar mRNA center of mass distribution was performed according to Gáspár et al. (2014) and plotted with GraphPad Prism 5.
Immunological techniques
For Western blots, dissected ovaries were lysed in 2× SDS sample buffer (100 mM Tris-HCl at pH 6.8, 4% [w/v] SDS, 20% glycerol, 0.2% bromophenol blue, 50 mM DTT) by manual grinding and heated for 10 min at 85°C. After separation by SDS-PAGE, the lysates were blotted on Immobilon-P PVDF membrane (Millipore) by wet blot transfer for 2 h at 4°C and 80 V. After blocking in 5% milk/PBS-Tween, incubation with primary antibody for 16 h and secondary horse radish peroxidase (HPR)-conjugated antibody for 1 h, the blot was developed with Immobilon Western chemiluminiscent HPR substrate (Millipore).
The anti-Khc antibody was raised against the Khc345–559 truncation in rabbits.
Electrophoretic mobility shift assays (EMSAs)
The U25 RNA synthetic probe (IDT) was 5′ end-labeled with ATP, [γ-32P] (Hartmann Analytic) using T4 polynucleotide kinase (Thermo Fisher) and subsequently purified using Illustra microspin G25 columns (GE Healthcare). Recombinant Tm1 or/and Khc truncations (concentrations indicated in the figure legends) were mixed with 2.5 nM probe in 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 5% glycerol, and 0.5 mM TCEP binding buffer in 15-µL reactions and incubated for 45 min on ice. The samples were subsequently separated on 6% native 0.5× TBE polyacrylamide gel for 1 h at 100 V. The gel was then dried and exposed overnight to a storage phosphor screen (GE), which was visualized with a Typhoon Trio Imager (GE Healthcare).
Filter binding assays
For the filter binding assays, 2 CPS/µL (counts per second) radiolabeled probe was mixed with protein or protein mixture in a 100-µL reaction volume. After incubation for 45 min on ice, the mixture was filtered through Whatman 0.45-μm pore size nitrocellulose filters. Only protein-RNA complexes were retained on the filters and detected by scintillation counting. Data fits were performed with GraphPad Prism 5 using a single site saturation binding mode.
Supplementary Material
Acknowledgments
We thank the European Synchrotron Radiation Facility Grenoble, beamline ID-29, and beamline scientist Montseratt Soler Lopez for support; Brice Marciano and the EMBL Crystallography Facility for assisting with crystallization screens; Alessandra Reversi for Drosophila transgenesis, crosses, and help with immunostainings; the EMBL Proteomics Core Facility for mass spectrometry; and the Advanced Light and Fluorescence Microscopy (ALFM) facility at the Centre for Structural Systems Biology (CSSB), Hamburg, for the use of their microscopes. We are highly indebted to Klaus Schmitt and the Laboratory Animal Resources at EMBL for making the anti-Khc antibody. We are very grateful to the CCP4/APS School in Macromolecular Crystallography (2019) for assistance with structure refinement. We are highly indebted to Thomas Pils at Biochemie Zentrum Heidelberg (BZH) for access and assistance in the isotope laboratory. We thank Sruthi Raja, Eva Boberlin, and Joscha Weis for assistance with protein purifications and crystallization screens. We thank Imre Gáspár and Zeljko Durdjevic for their help with Drosophila genetics. Additionally, we are very grateful to Imre Gáspár for his critical reading of the manuscript. L.D.-P. and P.K.A.J. were supported by fellowships from the EMBL Interdisciplinary Postdoc (EIPOD) Programme under Marie Curie Cofund Actions MSCA-COFUND-FP (664726). J.H. gratefully acknowledges support by an Emmy-Noether Fellowship (HE 7291). L.D.-P. and this research were supported by a Priority Program SPP1935 (EP37/3-1, 3-2) grant of the Deutsche Forschungsgemeinschaft (DFG) to A.E. and J.H. We also gratefully acknowledge the support of the EMBL.
Author contributions: L.D.-P., J.H., and A.E. conceived the study and wrote the manuscript. P.K.A.J. helped with data collection and solved the crystal structures. V. and A.C. helped with cloning, protein purifications, and fly husbandry. K.L. performed the ITC and SEC-MALS experiments. P.S. carried out the biacore experiments. K.P. measured the CD spectra. S.H. provided the full-length Khc. L.D-P. carried out the rest of the experiments and data analysis. C.L. provided laboratory space and resources. All authors commented on the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.348443.121.
Freely available online through the Genes & Development Open Access option.
Competing interest statement
The authors declare no competing interests.
References
- Banerjee A, Kulkarni S, Mukherjee A. 2020. Herpes simplex virus: the hostile guest that takes over your home. Front Microbiol 11: 733. 10.3389/fmicb.2020.00733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann S, Komissarov A, Gili M, Ruprecht V, Wieser S, Maurer SP. 2020. A reconstituted mammalian APC-kinesin complex selectively transports defined packages of axonal mRNAs. Sci Adv 6: eaaz1588. 10.1126/sciadv.aaz1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibby J, Keegan RM, Mayans O, Winn MD, Rigden DJ. 2012. AMPLE: a cluster-and-truncate approach to solve the crystal structures of small proteins using rapidly computed ab initio models. Acta Crystallogr D Biol Crystallogr 68: 1622–1631. 10.1107/S0907444912039194 [DOI] [PubMed] [Google Scholar]
- Cho A, Kato M, Whitwam T, Kim JH, Montell DJ. 2016. An atypical Tropomyosin in Drosophila with intermediate filament-like properties. Cell Rep 16: 928–938. 10.1016/j.celrep.2016.06.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chudinova EM, Nadezhdina ES. 2018. Interactions between the translation machinery and microtubules. Biochemistry (Mosc) 83: S176–s189. 10.1134/S0006297918140146 [DOI] [PubMed] [Google Scholar]
- Clark A, Meignin C, Davis I. 2007. A dynein-dependent shortcut rapidly delivers axis determination transcripts into the Drosophila oocyte. Development 134: 1955–1965. 10.1242/dev.02832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox RT, Spradling AC. 2006. Milton controls the early acquisition of mitochondria by Drosophila oocytes. Development 133: 3371–3377. 10.1242/dev.02514 [DOI] [PubMed] [Google Scholar]
- Cross JA, Dodding MP. 2019. Motor-cargo adaptors at the organelle-cytoskeleton interface. Curr Opin Cell Biol 59: 16–23. 10.1016/j.ceb.2019.02.010 [DOI] [PubMed] [Google Scholar]
- de Lanerolle P. 2012. Nuclear actin and myosins at a glance. J Cell Sci 125: 4945–4949. 10.1242/jcs.099754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diefenbach RJ, Mackay JP, Armati PJ, Cunningham AL. 1998. The C-terminal region of the stalk domain of ubiquitous human kinesin heavy chain contains the binding site for kinesin light chain. Biochemistry 37: 16663–16670. 10.1021/bi981163r [DOI] [PubMed] [Google Scholar]
- Diefenbach RJ, Diefenbach E, Douglas MW, Cunningham AL. 2002. The heavy chain of conventional kinesin interacts with the SNARE proteins SNAP25 and SNAP23. Biochemistry 41: 14906–14915. 10.1021/bi026417u [DOI] [PubMed] [Google Scholar]
- Diefenbach RJ, Diefenbach E, Douglas MW, Cunningham AL. 2004. The ribosome receptor, p180, interacts with kinesin heavy chain, KIF5B. Biochem Biophys Res Commun 319: 987–992. 10.1016/j.bbrc.2004.05.069 [DOI] [PubMed] [Google Scholar]
- Dienstbier M, Li X. 2009. Bicaudal-D and its role in cargo sorting by microtubule-based motors. Biochem Soc Trans 37: 1066–1071. 10.1042/BST0371066 [DOI] [PubMed] [Google Scholar]
- Dienstbier M, Boehl F, Li X, Bullock SL. 2009. Egalitarian is a selective RNA-binding protein linking mRNA localization signals to the dynein motor. Genes Dev 23: 1546–1558. 10.1101/gad.531009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66: 486–501. 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ephrussi A, Lehmann R. 1992. Induction of germ cell formation by oskar. Nature 358: 387–392. 10.1038/358387a0 [DOI] [PubMed] [Google Scholar]
- Erdélyi M, Michon AM, Guichet A, Glotzer JB, Ephrussi A. 1995. Requirement for Drosophila cytoplasmic tropomyosin in oskar mRNA localization. Nature 377: 524–527. 10.1038/377524a0 [DOI] [PubMed] [Google Scholar]
- Fukuda Y, Pazyra-Murphy MF, Silagi ES, Tasdemir-Yilmaz OE, Li Y, Rose L, Yeoh ZC, Vangos NE, Geffken EA, Seo HS, et al. 2021. Binding and transport of SFPQ-RNA granules by KIF5A/KLC1 motors promotes axon survival. J Cell Biol 220: e202005051. 10.1083/jcb.202005051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcin C, Straube A. 2019. Microtubules in cell migration. Essays Biochem 63: 509–520. 10.1042/EBC20190016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gáspár I, Yu YV, Cotton SL, Kim DH, Ephrussi A, Welte MA. 2014. Klar ensures thermal robustness of oskar localization by restraining RNP motility. J Cell Biol 206: 199–215. 10.1083/jcb.201310010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gáspár I, Sysoev V, Komissarov A, Ephrussi A. 2017a. An RNA-binding atypical tropomyosin recruits kinesin-1 dynamically to oskar mRNPs. EMBO J 36: 319–333. 10.15252/embj.201696038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gáspár I, Wippich F, Ephrussi A. 2017b. Enzymatic production of single-molecule FISH and RNA capture probes. RNA 23: 1582–1591. 10.1261/rna.061184.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glater EE, Megeath LJ, Stowers RS, Schwarz TL. 2006. Axonal transport of mitochondria requires milton to recruit kinesin heavy chain and is light chain independent. J Cell Biol 173: 545–557. 10.1083/jcb.200601067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunning PW, Hardeman EC, Lappalainen P, Mulvihill DP. 2015. Tropomyosin—master regulator of actin filament function in the cytoskeleton. J Cell Sci 128: 2965–2974. [DOI] [PubMed] [Google Scholar]
- Hirokawa N, Pfister KK, Yorifuji H, Wagner MC, Brady ST, Bloom GS. 1989. Submolecular domains of bovine brain kinesin identified by electron microscopy and monoclonal antibody decoration. Cell 56: 867–878. 10.1016/0092-8674(89)90691-0 [DOI] [PubMed] [Google Scholar]
- Hirokawa N, Noda Y, Tanaka Y, Niwa S. 2009. Kinesin superfamily motor proteins and intracellular transport. Nat Rev Mol Cell Biol 10: 682–696. 10.1038/nrm2774 [DOI] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144: 1425–1436. 10.1093/genetics/144.4.1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaan HY, Hackney DD, Kozielski F. 2011. The structure of the kinesin-1 motor-tail complex reveals the mechanism of autoinhibition. Science 333: 883–885. 10.1126/science.1204824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. 2010. XDS. Acta Crystallogr D Biol Crystallogr 66: 125–132. 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai Y, Dohmae N, Hirokawa N. 2004. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron 43: 513–525. 10.1016/j.neuron.2004.07.022 [DOI] [PubMed] [Google Scholar]
- Lawrence CJ, Dawe RK, Christie KR, Cleveland DW, Dawson SC, Endow SA, Goldstein LS, Goodson HV, Hirokawa N, Howard J, et al. 2004. A standardized kinesin nomenclature. J Cell Biol 167: 19–22. 10.1083/jcb.200408113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P. 2018. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res 46: D493–D496. 10.1093/nar/gkx922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebschner D, Afonine PV, Baker ML, Bunkóczi G, Chen VB, Croll TI, Hintze B, Hung LW, Jain S, McCoy AJ, et al. 2019. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr D Struct Biol 75: 861–877. 10.1107/S2059798319011471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Naismith JH. 2008. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol 8: 91. 10.1186/1472-6750-8-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiseau P, Davies T, Williams LS, Mishima M, Palacios IM. 2010. Drosophila PAT1 is required for kinesin-1 to transport cargo and to maximize its motility. Development 137: 2763–2772. 10.1242/dev.048108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malashkevich VN, Higgins CD, Almo SC, Lai JR. 2015. A switch from parallel to antiparallel strand orientation in a coiled-coil X-ray structure via two core hydrophobic mutations. Biopolymers 104: 178–185. 10.1002/bip.22631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason JM, Arndt KM. 2004. Coiled coil domains: stability, specificity, and biological implications. Chembiochem 5: 170–176. 10.1002/cbic.200300781 [DOI] [PubMed] [Google Scholar]
- Olenick MA, Holzbaur ELF. 2019. Dynein activators and adaptors at a glance. J Cell Sci 132. 10.1242/jcs.227132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry DA, Squire JM. 1973. Structural role of tropomyosin in muscle regulation: analysis of the x-ray diffraction patterns from relaxed and contracting muscles. J Mol Biol 75: 33–55. 10.1016/0022-2836(73)90527-5 [DOI] [PubMed] [Google Scholar]
- Qin J, Zhang H, Geng Y, Ji Q. 2020. How kinesin-1 utilize the energy of nucleotide: the conformational changes and mechanochemical coupling in the unidirectional motion of kinesin-1. Int J Mol Sci 21: 6977. 10.3390/ijms21186977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall TS, Moores C, Stephenson FA. 2013. Delineation of the TRAK binding regions of the kinesin-1 motor proteins. FEBS Lett 587: 3763–3769. 10.1016/j.febslet.2013.09.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert X, Gouet P. 2014. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42: W320–W324. 10.1093/nar/gku316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez DD, Grosse C, Himmel S, González C, de Ilarduya IM, Becker S, Sheldrick GM, Usón I. 2009. Crystallographic ab initio protein structure solution below atomic resolution. Nat Methods 6: 651–653. 10.1038/nmeth.1365 [DOI] [PubMed] [Google Scholar]
- Rørth P. 1998. Gal4 in the Drosophila female germline. Mech Dev 78: 113–118. 10.1016/S0925-4773(98)00157-9 [DOI] [PubMed] [Google Scholar]
- Roth S, Lynch JA. 2009. Symmetry breaking during Drosophila oogenesis. Cold Spring Harb Perspect Biol 1: a001891. 10.1101/cshperspect.a001891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Seeger MA, Rice SE. 2013. Intrinsic disorder in the kinesin superfamily. Biophys Rev 5: 233–247. 10.1007/s12551-012-0096-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler S, Kirchner J, Horn C, Kallipolitou A, Woehlke G, Schliwa M. 2000. Cargo binding and regulatory sites in the tail of fungal conventional kinesin. Nat Cell Biol 2: 333–338. 10.1038/35014022 [DOI] [PubMed] [Google Scholar]
- Setou M, Seog DH, Tanaka Y, Kanai Y, Takei Y, Kawagishi M, Hirokawa N. 2002. Glutamate-receptor-interacting protein GRIP1 directly steers kinesin to dendrites. Nature 417: 83–87. 10.1038/nature743 [DOI] [PubMed] [Google Scholar]
- Staller MV, Yan D, Randklev S, Bragdon MD, Wunderlich ZB, Tao R, Perkins LA, Depace AH, Perrimon N. 2013. Depleting gene activities in early Drosophila embryos with the ‘maternal-Gal4-shRNA’ system. Genetics 193: 51–61. 10.1534/genetics.112.144915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa PA, Vale RD. 2000. The myosin motor, Myo4p, binds Ash1 mRNA via the adapter protein, She3p. Proc Natl Acad Sci 97: 5273–5278. 10.1073/pnas.080585897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telley IA, Gáspár I, Ephrussi A, Surrey T. 2012. Aster migration determines the length scale of nuclear separation in the Drosophila syncytial embryo. J Cell Biol 197: 887–895. 10.1083/jcb.201204019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoms M, Thomson E, Baßler J, Gnädig M, Griesel S, Hurt E. 2015. The exosome is recruited to RNA substrates through specific adaptor proteins. Cell 162: 1029–1038. 10.1016/j.cell.2015.07.060 [DOI] [PubMed] [Google Scholar]
- Titus MA. 2018. Myosin-driven intracellular transport. Cold Spring Harb Perspect Biol 10: a021972. 10.1101/cshperspect.a021972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trcek T, Lehmann R. 2019. Germ granules in Drosophila. Traffic 20: 650–660. 10.1111/tra.12674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truebestein L, Leonard TA. 2016. Coiled-coils: the long and short of it. Bioessays 38: 903–916. 10.1002/bies.201600062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishali V, Dimitrova-Paternoga L, Haubrich K, Sun M, Ephrussi A, Hennig J. 2021. Validation and classification of RNA binding proteins identified by mRNA interactome capture. bioRxiv 10.1101/2021.02.02.429302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale RD, Reese TS, Sheetz MP. 1985. Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell 42: 39–50. 10.1016/S0092-8674(85)80099-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeranan-Karmegam R, Boggupalli DP, Liu G, Gonsalvez GB. 2016. A new isoform of Drosophila non-muscle Tropomyosin 1 interacts with kinesin-1 and functions in oskar mRNA localization. J Cell Sci 129: 4252–4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams LS, Ganguly S, Loiseau P, Ng BF, Palacios IM. 2014. The auto-inhibitory domain and ATP-independent microtubule-binding region of kinesin heavy chain are major functional domains for transport in the Drosophila germline. Development 141: 176–186. 10.1242/dev.097592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimyanin VL, Belaya K, Pecreaux J, Gilchrist MJ, Clark A, Davis I, St Johnston D. 2008. In vivo imaging of oskar mRNA transport reveals the mechanism of posterior localization. Cell 134: 843–853. 10.1016/j.cell.2008.06.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.