In this study, Tsai et al. set out to study how cellular m6A recognition proteins regulate HIV gene expression. Using genetic and genomic approaches, the authors report that m6A regulates HIV gene expression by at least two distinct mechanisms. First, m6A assures the optimal alternative splicing of HIV-1 transcripts by recruiting the nuclear m6A reader YTHDC1. Second, while they confirm previous reports that m6A can destabilize cellular mRNAs via recruitment of the cytoplasmic m6A reader YTHDF2, they found that YTHDF2 binding to m6A sites on HIV-1 transcripts instead enhances the stability of these viral mRNAs.
Keywords: RNA, epitranscriptomic regulation, YTHDF2, YTHDC1, HIV-1, splicing, RNA stability
Abstract
Previous work has demonstrated that the epitranscriptomic addition of m6A to viral transcripts can promote the replication and pathogenicity of a wide range of DNA and RNA viruses, including HIV-1, yet the underlying mechanisms responsible for this effect have remained unclear. It is known that m6A function is largely mediated by cellular m6A binding proteins or readers, yet how these regulate viral gene expression in general, and HIV-1 gene expression in particular, has been controversial. Here, we confirm that m6A addition indeed regulates HIV-1 RNA expression and demonstrate that this effect is largely mediated by the nuclear m6A reader YTHDC1 and the cytoplasmic m6A reader YTHDF2. Both YTHDC1 and YTHDF2 bind to multiple distinct and overlapping sites on the HIV-1 RNA genome, with YTHDC1 recruitment serving to regulate the alternative splicing of HIV-1 RNAs. Unexpectedly, while YTHDF2 binding to m6A residues present on cellular mRNAs resulted in their destabilization as previously reported, YTHDF2 binding to m6A sites on HIV-1 transcripts resulted in a marked increase in the stability of these viral RNAs. Thus, YTHDF2 binding can exert diametrically opposite effects on RNA stability, depending on RNA sequence context.
Eukaryotic mRNAs are subject to a range of covalent modifications at the nucleotide level, collectively referred to as epitranscriptomic modifications. The most common modification of mammalian mRNAs is methylation of the N6 position of adenosine (m6A), which comprises ∼0.4% of all adenosines (Desrosiers et al. 1974; Dominissini et al. 2012; Meyer et al. 2012). m6A residues are deposited on mRNAs by a “writer” complex, minimally consisting of METTL3, METTL14, and WTAP, which adds m6A to some, but not all, copies of the RNA motif 5′-RRACH-3′ (R = G/A, H = A/U/C) (Bokar et al. 1997; Liu et al. 2014; Ping et al. 2014; Shi et al. 2019). Cells also express two demethylases or “erasers,” called FTO and ALKBH5, which have been proposed to dynamically regulate m6A levels (Jia et al. 2011; Zheng et al. 2013). Finally, the phenotypic effects of m6A on mRNA are largely conferred by m6A binding proteins or readers. These include the nuclear m6A reader YTHDC1, which has been reported to regulate cellular mRNA splicing and nuclear export, and the cytoplasmic m6A readers YTHDF1, YTHDF2, and YTHDF3, which are thought to regulate the stability, and possibly also translation, of m6A-containing mRNAs (Wang et al. 2014, 2015; Xiao et al. 2016; Roundtree et al. 2017). Recent reports suggest that the cytosolic readers YTHDF1, 2, and 3 confer redundant functions, with YTHDF2 the dominant reader due to its higher expression level (Lasman et al. 2020; Zaccara and Jaffrey 2020).
Whereas m6A is the most common epitranscriptomic modification on cellular mRNAs, analysis of the genomic RNAs encoded by retroviruses revealed an even higher level of m6A on these transcripts, with m6A comprising as much as ∼2.4% of the adenosines found on murine leukemia virus (MLV) genomic RNAs (Courtney et al. 2019a). This high prevalence suggests that m6A might be promoting some aspect(s) of the viral replication cycle. Indeed, addition of m6A has now been shown to boost the replication of a diverse range of viruses, including not only MLV but also influenza A virus (IAV), SV40, human metapneumovirus (HMPV), enterovirus 71, and respiratory syncytial virus (RSV) (Tsai and Cullen 2020). In the case of IAV, HMPV, and RSV, the presence of m6A residues on viral transcripts was also shown to boost pathogenicity in vivo. Similarly, for HIV-1, knockdown of the m6A writers METTL3 and METTL14 inhibited HIV-1 gene expression, while knockdown of the m6A erasers FTO or ALKBH5 had the opposite effect (Lichinchi et al. 2016a; Tirumuru et al. 2016). However, the mechanism underlying this positive effect has remained controversial. One group (Lichinchi et al. 2016a) reported that a key adenosine residue in the HIV-1 Rev response element (RRE) is methylated to m6A, which then promotes Rev binding. However, we and others (Kennedy et al. 2016; Tirumuru et al. 2016; Chu et al. 2019; Courtney et al. 2019b) have been unable to confirm the presence of any m6A residues on the RRE.
While the addition of m6A to viral transcripts has therefore been reported to increase viral gene expression in most instances, exceptions exist. In particular, m6A addition has been reported to inhibit the replication of several flavivirus species and to destabilize hepatitis B virus transcripts (Gokhale et al. 2016; Lichinchi et al. 2016b; Imam et al. 2018). This effect is similar to what has been reported for a range of cellular mRNAs, which generally are destabilized by addition of m6A residues (Wang et al. 2014; Ke et al. 2017; Park et al. 2019; Zaccara and Jaffrey 2020). However, here also exceptions exist, with m6A being reported to stabilize specific cellular mRNAs during hypoxic shock (Fry et al. 2017).
While we have previously reported (Kennedy et al. 2016) that the cytoplasmic m6A readers YTHDF1, YTHDF3, and especially YTHDF2 all promote HIV-1 gene expression and replication, another group (Tirumuru et al. 2016) has argued that these same proteins bind to HIV-1 genomic RNAs and are then packaged into virions where they inhibit reverse transcription. In contrast, a third group has reported that while YTHDF3, but not YTHDF2, is selectively incorporated into HIV-1 virions, it is then inactivated by cleavage by the viral protease. Only when cleavage by the HIV-1 protease was inhibited was YTHDF3 found to exert an inhibitory effect (Jurczyszak et al. 2020).
Here, we have sought to more fully define how m6A residues interact with cellular readers to positively affect HIV-1 mRNA function and report that m6A regulates HIV-1 gene expression by at least two distinct mechanisms. On the one hand, m6A assures the optimal alternative splicing of HIV-1 transcripts by recruiting the nuclear m6A reader YTHDC1. On the other hand, while we confirm previous reports that m6A can destabilize cellular mRNAs via recruitment of the cytoplasmic m6A reader YTHDF2 (Wang et al. 2014; Park et al. 2019; Zaccara and Jaffrey 2020), we found that YTHDF2 binding to m6A sites on HIV-1 transcripts instead enhances the stability of these viral mRNAs. Thus, recruitment of YTHDF2 to m6A residues on mRNAs can have disparate effects on mRNA stability depending on as yet undefined differences in sequence context.
Results
Removal of m6A residues reduces the level of HIV-1 RNA expression
Previously, it has been demonstrated that knockdown of the m6A writers METTL3 and METTL14 using RNA interference (RNAi) inhibits HIV-1 Gag expression while knockdown of the m6A erasers ALKBH5 or FTO increases HIV-1 Gag expression (Lichinchi et al. 2016a; Tirumuru et al. 2016), yet the molecular basis for this positive effect of the m6A modification has remained unclear. As an alternative complementary approach, we overexpressed the m6A eraser ALKBH5, which has been previously demonstrated to globally reduce m6A levels on RNA (Zheng et al. 2013; Lu et al. 2020). A lentiviral ALKBH5 expression vector was used to transduce the CD4+ T-cell line CEM, and three single cell clones were isolated, each expressing an approximately threefold higher level of ALKBH5 than the parental cells (Fig. 1A,B). To confirm m6A depletion on mRNAs expressed in these ALKBH5-overexpressing cells (+ALKBH5), we quantified the m6A content of purified poly(A)+ RNA extracted from HIV-1-infected parental or +ALKBH5 CEM cells at 3 d postinfection (dpi) using an m6A ELISA assay. Isolation of poly(A)+ RNA by oligo-dT purification successfully removed >90% of rRNA (Supplemental Fig. S1), and we observed that poly(A)+ RNA isolated from the +ALKBH5 CEM cells contained ∼50% less m6A than WT CEM cells (Fig. 1C).
Figure 1.

Global depletion of m6A in T cells suppresses HIV-1 RNA and protein expression. Three separate clones of the CD4+ CEM T-cell line overexpressing ALKBH5 (A1, A3, and A12) were compared with WT CEM cells (Ctrl). (A) HIV-1 Gag expression was analyzed by Western blot at 2 dpi after single cycle HIV-1 infection of + ALKBH5 CEM cells or control cells (one WT and the others expressing the irrelevant GFP-targeted Cas9 [ΔG]). Also analyzed were ALKBH5 (the overexpressed epitope tagged ALKBH5 runs slightly slower than endogenous ALKBH5) and GAPDH, as a loading control. (B) ALKBH5 band intensities from A were quantified and are shown relative to control cells, set at 1. (C) The m6A content of poly(A)+ mRNAs from Ctrl and +ALKBH5 cells was quantified by ELISA (n = 5). (D) Quantification of the HIV-1 Gag band intensities (p24 + p55) from A. (E) Aliquots of the samples from C were analyzed for viral RNA expression levels by qRT-PCR. The m6A-free cellular transcript (NONO) served as a negative control. Statistical analyses by two-tailed Student's t-test. Error bars indicate SD. (**) P < 0.01.
We next infected WT and +ALKBH5 CEM cells with HIV-1 and then limited this infection to a single cycle by addition of the reverse transcriptase (RT) inhibitor nevirapine at 16 h postinfection (hpi), followed by harvest at 48 hpi for analysis of viral gene expression. In all three +ALKBH5 clones, reduced m6A addition resulted in a ∼60% drop in viral Gag protein expression (Fig. 1A,D) while cellular GAPDH, which is translated from an mRNA lacking m6A residues (Dominissini et al. 2012), was unaffected. We also saw a similar ∼60% drop in the level of both unspliced and spliced HIV-1 transcripts but again saw no change in the level of an m6A-free host mRNA, NONO (Fig. 1E; Dominissini et al. 2012). These data indicate that removal of m6A residues from HIV-1 mRNAs upon ALKBH5 overexpression selectively decreases HIV-1 gene expression at the RNA level without globally impacting cellular mRNA expression.
YTHDF2 binding stabilizes HIV-1 mRNAs
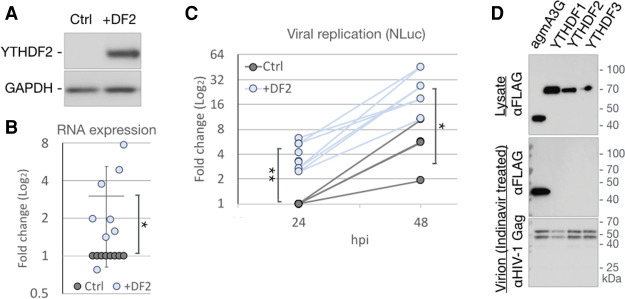
Whereas we have previously reported that the cytoplasmic m6A readers YTHDF1, YTHDF2, and YTHDF3 increase HIV-1 gene expression and replication, others have argued that they exert an inhibitory effect (Kennedy et al. 2016; Tirumuru et al. 2016). As it has recently been reported that all three YTHDF proteins function by similar mechanisms (Zaccara and Jaffrey 2020), and YTHDF2 is the most highly expressed variant in T cells (Kennedy et al. 2016), we focused on this protein. We first sought to confirm that YTHDF2 can indeed enhance viral replication by transfecting 293T cells with a plasmid expressing the HIV-1 receptor CD4, along with either an empty plasmid (Ctrl) or a plasmid expressing YTHDF2 (+DF2), and confirmed that this results in YTHDF2 overexpression (Fig. 2A). Control and YTHDF2-overexpressing cells were then infected with a fully replication competent HIV-1 variant expressing nano luciferase (NLuc) in place of Nef. At 24 hpi, we observed a significant, approximately threefold increase in total viral RNA expression in the +DF2 cells when compared with Ctrl cells (Fig. 2B). Similarly, expression of the virally encoded NLuc was approximately fourfold higher in the +DF2 cells compared with Ctrl cells at both 24 and 48 hpi, demonstrating that this indicator virus was, as expected, able to effectively spread through the CD4 + 293T cell culture while maintaining the +DF2 associated higher level of NLuc expression seen at 24 hpi.
Figure 2.
YTHDF2 enhances HIV-1 replication, yet is not packaged into virions. (A–C) 293T cells, transfected with either empty vector (Ctrl) or a YTHDF2 expression plasmid (+DF2), were infected with NL-NLuc reporter virus and collected at 24 hpi for Western blot detection of overexpressed YTHDF2 (A) or qRT-PCR analysis of viral RNA expression (B). n = 8. (C) Quantification of virally encoded NLuc as a measure of viral replication at 24 hpi (n = 8) and 48 hpi (n = 4). (D) 293T cells were cotransfected with NL-NLuc along with expression vectors expressing FLAG-tagged YTHDF readers or agmA3G and treated with the HIV-1 protease inhibitor Indinavir. Lysates of the virus producer cells (top panel) and purified virions from the supernatant media (two bottom panels) were analyzed by Western blot for the FLAG-tagged YTHDF or agmA3G proteins and the HIV-1 Gag protein. Statistical analysis by two-tailed Student's t-test. (*) P < 0.05, (**) P < 0.01. . Error bars indicate SD.
As noted above, one group has reported that all three cytoplasmic YTHDF readers are incorporated into HIV-1 virions, where they inhibit reverse transcription, while a second group has reported that virion packaging is specific for YTHDF3, which is then degraded by the viral protease (Tirumuru et al. 2016; Jurczyszak et al. 2020). To determine whether the YTHDF proteins are indeed incorporated into HIV-1 virions, we cotransfected 293T cells with a molecular clone of HIV-1 along with vectors expressing either FLAG-tagged forms of the m6A readers YTHDF1, YTHDF2, or YTHDF3 or expressing FLAG-tagged African green monkey APOBEC3G (agmA3G), which is known to be packaged into virions (Bogerd et al. 2004; Schrofelbauer et al. 2004). While all three YTHDF proteins, along with agmA3G, were detected at comparable levels in virus-producing cells (Fig. 2D, top panel), only agmA3G was detected in highly purified virion particles isolated from the supernatant media by centrifugation through a sucrose cushion followed by banding on an iodixanol gradient (Fig. 2D, middle panel). The failure to detect virion-packaged m6A readers could not be explained by their HIV-1 protease-mediated degradation, as virions were prepared in the presence of the viral protease inhibitor indinavir, as confirmed by the fact that only full-length p55-Gag and p47-Gag were detected in these virions by Western, with no mature p21 CA protein observed (Fig. 2D, bottom panel). The p47-Gag product has previously been detected by others in virion particles produced in the presence of protease inhibitors (Davis et al. 2012) and is of uncertain origin.
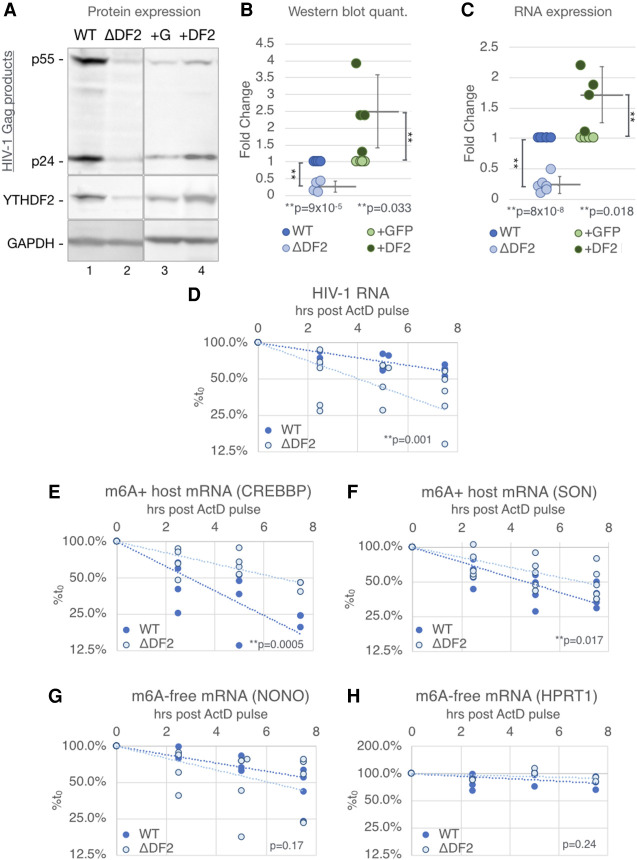
We next sought to define the mechanism by which YTHDF2 enhances HIV-1 gene expression using a more biologically relevant system; i.e., single-cycle infection by HIV-1 of the CD4+ T-cell line CEM-SS. YTHDF2 knockout (ΔDF2) CEM-SS cells, and YTHDF2-overexpressing (+DF2) or GFP-expressing (+GFP) CEM-SS cells, have been previously described (Kennedy et al. 2016). Wild-type (WT), ΔDF2, +GFP, and +DF2 CEM-SS cells were infected with HIV-1, RT-blocked at 16 hpi using nevirapine to ensure a single-cycle infection, and then harvested at 48 hpi. We observed not only reduced Gag protein expression in the ΔDF2 cells (Fig. 3A, lanes 1,2) but also increased Gag expression in the +DF2 cells (Fig. 3A, lanes 3,4), confirming that YTHDF2 indeed enhances viral gene expression in a single-cycle infection of T cells (Fig. 3B). The inhibition of Gag production seen upon loss of YTHDF2 expression was also observed when a second, independent ΔDF2 CEM-SS clone was analyzed (Supplemental Fig. S2). We also observed an ∼80% drop in viral RNA levels in the ΔDF2 cells and an ∼80% increase in viral RNA in the infected +DF2 CEM-SS cells, respectively, when compared with control cells (Fig. 3C), again suggesting that YTHDF2 up-regulates viral gene expression at the RNA level. YTHDF2 has been reported to be cytosolic (Wang et al. 2014), which we confirmed in CEM-SS T cells (Supplemental Fig. S3A), and we therefore hypothesized that YTHDF2 was likely regulating HIV-1 RNA stability rather than synthesis. To test this, we again performed a single-cycle infection of the WT and ΔDF2 CEM-SS cells but added the transcription inhibitor actinomycin D (ActD) at 48 hpi. HIV-1 RNA levels were then quantified by qRT-PCR at multiple time points to monitor the rate of RNA decay. We indeed observed a significantly (P < 0.002) faster decay (t1/2 = 4 h) of HIV-1 transcripts in the ΔDF2 cells, when compared with WT CEM-SS cells (estimated t1/2 = 8 h) (Fig. 3D). As YTHDF2 binding to m6A residues has previously been reported to result in the destabilization of many host transcripts (Wang et al. 2014; Park et al. 2019; Zaccara and Jaffrey 2020), we next asked whether the stabilization of HIV-1 mRNA by YTHDF2 was due to a different intracellular environment—for example, the use of T cells—or is specific to the m6A+ transcript being analyzed. We thus measured, in exactly the same RNA samples, the stability of the host cell transcripts CREBBP and SON, which are both known to be m6A+ and have been reported to be destabilized by YTHDF2 binding (Wang et al. 2014). Surprisingly, we found that both of these cellular m6A+ transcripts indeed decayed more slowly in the ΔDF2 cells (Fig. 3E,F), confirming this previous report. YTHDF2 expression therefore simultaneously exerts diametrically opposite effects on the stability of viral and cellular transcripts in the same culture. Furthermore, no effect on RNA stability was observed when two m6A-free host mRNAs, encoding NONO and HPRT1, were tested in WT or ΔDF2 T cells (Fig. 3G,H), thus arguing that the observed changes in RNA stability in the presence or absence of YTHDF2 are indeed m6-dependent (Dominissini et al. 2012; Wang et al. 2014).
Figure 3.
The cytosolic m6A reader YTHDF2 enhances HIV-1 RNA stability while destabilizing m6A+ host mRNAs in CD4+ T cells. YTHDF2 knockout CD4+ T cells (ΔDF2), WT control cells (WT), and cells transduced with a lentiviral YTHDF2 or GFP expression vector (+DF2 or +G) were used for the following single-cycle infection assays. (A) Viral Gag protein expression levels analyzed by Western blot. (B) Quantification of protein band intensities of Western blots as shown in A (n = 4). (C) Viral RNA expression assayed by qRT-PCR (WT & ΔYTHDF2 n = 6, +GFP & +YTHDF2 n = 5). (D–H) Stability of RNA transcripts assayed by treating infected WT or ΔDF2 cells at 2 dpi with actinomycin D (ActD), quantifying the transcript at the indicated time points by qRT-PCR, shown as percentage of the RNA level at time point 0. (D) Stability of HIV-1 transcripts. (E,F) Stability of host m6A+ transcripts known to be destabilized by YTHDF2, including CREBBP and SON. (G,H) Stability of m6A-free host transcripts NONO and HPRT1. Statistical analysis of data in B and C used the two-tailed Student's t-test. Error bars indicate SD. For D–H, slopes of regression lines were compared by ANCOVA, n = 4–5.
Both YTHDC1 and YTHDF2 bind to m6A sites on HIV-1 transcripts
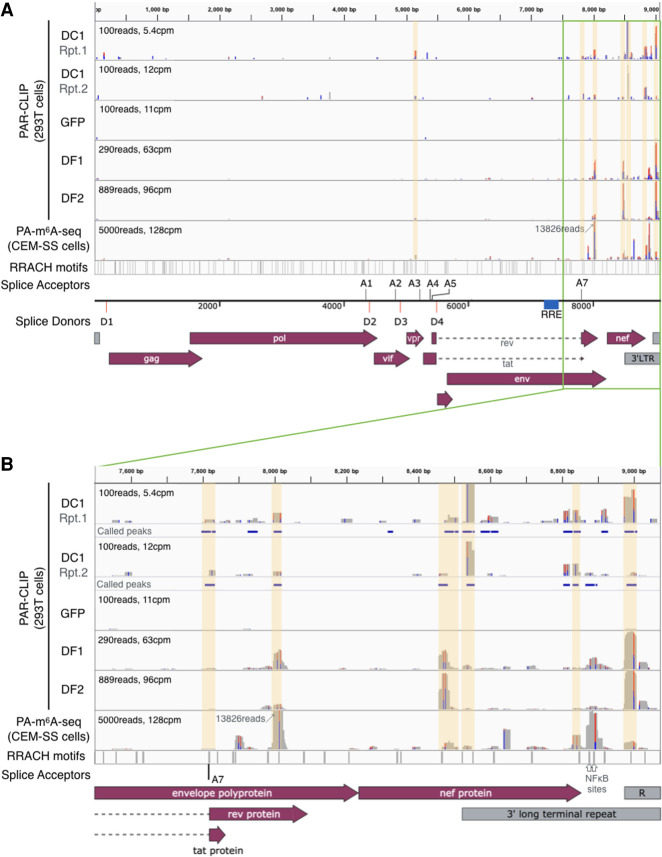
HIV-1 replication is tightly regulated by a complex pattern of alternative splicing that results in the ordered expression of up to 30 unspliced, partially spliced, and fully spliced viral mRNAs (Cullen and Greene 1990; Stoltzfus 2009). Both m6A and the nuclear m6A reader YTHDC1 have been reported to affect cellular mRNA splicing (Dominissini et al. 2012; Xiao et al. 2016), and we therefore sought to determine whether HIV-1 RNA splicing is also regulated by YTHDC1 binding. We used the photo-assisted cross linking and immunoprecipitation (PAR-CLIP) technique (Hafner et al. 2010) to map YTHDC1 binding sites on HIV-1 transcripts in infected cells and compared these with the previously mapped binding sites for YTHDF1 and YTHDF2 as well as sites of m6A addition mapped by antibody binding (PA-m6A-seq) (Fig. 4; Kennedy et al. 2016; Courtney et al. 2019b). We identified at least seven reproducible YTHDC1 binding sites across two independent PAR-CLIP experiments and found that these largely coincided with previously mapped sites of m6A addition (Courtney et al. 2019b) as well as with known YTHDF1 and YTHDF2 binding sites (Kennedy et al. 2016). Moreover, these sites all contained the m6A addition motif 5′-RRACH-3′ (Dominissini et al. 2012; Meyer et al. 2012). Due to splicing, the read depth in the first ∼7500 nt of the viral genome is lower than in the 3′ untranslated region of viral mRNAs in infected cells. Nevertheless, we were able to map a YTHDC1 binding site adjacent to HIV-1 splice acceptor A3 (Fig. 4A), as well as a binding site overlapping splice acceptor A7 (Fig. 4B), in addition to YTHDC1 binding sites coincident with the previously reported m6A sites in the env/rev overlap, the nef gene, and in the TAR hairpin (Kennedy et al. 2016). Conversely, the m6A sites at ∼8900 nt located in the NF-kB sites in the viral LTR U3 region are at best weakly bound by YTHDC1 (Fig. 4B). We observed no binding of YTHDC1, YTHDF1, YTHDF2, or the m6A-specific antibody to the proposed m6A site in the RRE (Lichinchi et al. 2016a), which agrees with m6A mapping data on HIV-1 RNA reported by others (Tirumuru et al. 2016).
Figure 4.
The nuclear m6A reader YTHDC1 binds HIV-1 RNA at previously mapped m6A sites. PAR-CLIP was performed on 293T cells transfected with FLAG-GFP or FLAG-YTHDC1, and infected with HIV-1. Sequencing reads were mapped to the HIV-1 genome, with two independent repeats of YTHDC1 PAR-CLIP (DC1 lanes) shown alongside previously published YTHDF1 and YTHDF2 PAR-CLIP (DF1 and DF2 lanes) and m6A mapping results (PA-m6A-seq lane) (Kennedy et al. 2016; Courtney et al. 2019b). A schematic of the HIV-1 genome is shown in A, with the 3′ end (3′ of 7500 bp) shown in B where most m6A sites are located. Locations of the m6A motif 5′-RRACH-3′ are shown in the bottom lane, with the location of viral splice donors and acceptors indicated. Significant YTHDC1 peaks called by PARalyzer are shown as blue bars below each DC1 lane in B. PARalyzer-called peaks that overlap with m6A sites and 5′-RRACH-3′ motifs are highlighted in yellow.
YTHDC1 differentially regulates HIV-1 transcript expression
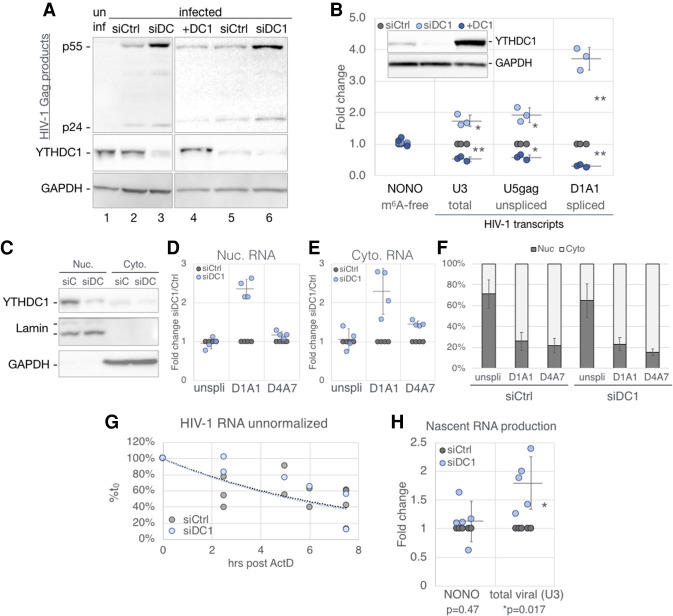
Having confirmed that YTHDC1 indeed binds m6A residues on HIV-1 transcripts in infected cells, we next asked whether YTHDC1 regulates HIV-1 RNA expression. Because we were unable to obtain YTHDC1 knockout cells, implying that YTHDC1 may be essential, we instead used RNAi to knockdown YTHDC1 expression. We transfected 293T cells with a plasmid encoding CD4 and also transfected the cells on two successive days with a mixture of three siRNAs targeting YTHDC1 mRNA (siDC1) or control siRNAs (siCtrl). As expected, we observed decreased YTHDC1 protein expression only in the siDC1 transfected cells, and this correlated with a surprising increase in HIV-1 Gag expression (Fig. 5A, cf. lanes 2 and 3). If the siDC1-induced increase in viral Gag expression is indeed a direct consequence of reduced YTHDC1 expression, then one might predict a decrease in viral Gag expression upon overexpression of YTHDC1 (+DC1) as compared to siCtrl cells expressing physiological levels of YTHDC1, and this was indeed observed (Fig. 5A, cf. lanes 4 and 5). A similar effect was seen at the RNA level, with quantification of total, unspliced, and spliced viral RNA (specifically, viral RNAs spliced from D1 to A1) using qRT-PCR revealing that knockdown of YTHDC1 using RNAi resulted in a significantly higher level of viral RNA expression, whereas YTHDC1 overexpression resulted in reduced viral RNA expression (Fig. 5B). This effect was not observed for the NONO transcript lacking m6A sites, was rather modest (1.7×) at the total viral RNA level, and was more prominent (∼3.7×) for spliced viral RNAs bearing the D1/A1 splice junction than for the unspliced viral RNA, thus suggesting that the nuclear YTHDC1 protein (Supplemental Fig. S3B) might regulate HIV-1 RNA expression post-transcriptionally.
Figure 5.
YTHDC1 regulates HIV-1 gene expression with no detectable effect on RNA nuclear export. 293T cells transfected with nontargeting (siCtrl, gray) or YTHDC1-targeting (siDC1, light blue) siRNAs or were transfected with a YTHDC1 expression vector (+DC1, dark blue) or empty vector. HIV-1 single-cycle infections were performed for the following analyses: (A) Viral Gag protein expression assayed by Western blot, costained with a YTHDC1 antibody. (B) Viral RNA levels assayed by qRT-PCR, n = 3, with the m6A-free host NONO mRNA as a control. A representive Western blot shown in the top left inset depicts the validation of YTHDC1 levels for samples used in this panel. (C–F) Subcellular fractionation assay of infected siCtrl and siDC1 cells. (C) Western blot validation of fractionation, stained for YTHDC1, nuclear Lamin A/C, and cytosolic GAPDH. (D–F) HIV-1 transcript alternatively spliced isoforms were quantified by qRT-PCR with primers targeting unspliced (unspli, U5-gag), and the D1/A1 and D4/A7 splice junctions, calculated as fold change of siDC1 over siCtrl in the nuclear (D), and cytosolic fraction (E). (F) The same RNA quantification as in D and E calculated as percent nuclear and percent cytoplasmic. (G) Stability of RNA transcripts in siCtrl and siDC1 cells assayed by treating infected cells at 2 dpi with ActD and the viral RNA levels from the indicated time points analyzed by qRT-PCR and shown as percentage of the RNA level at time point 0. (H) Production of nascent HIV-1 transcripts in infected siCtrl and siDC1 cells was measured by pulsing cells with the nucleoside analog 4SU for 1.5 h. The 4SU+ transcripts were then isolated and quantified by qRT-PCR. Statistical analysis used Student's t-test. Error bars indicate SD. (*) P < 0.05, (**) P < 0.01.
YTHDC1 does not regulate the nuclear export or stability of HIV-1 transcripts
YTHDC1 has been proposed to regulate both the alternative splicing and nuclear export of cellular m6A methylated mRNAs (Xiao et al. 2016; Roundtree et al. 2017), and we first asked whether YTHDC1 might regulate the nuclear export of HIV-1 transcripts. To determine whether knockdown of YTHDC1 expression had any effect on the nucleocytoplasmic localization of fully spliced or incompletely spliced HIV-1 RNAs, we isolated nuclear or cytoplasmic fractions from WT or YTHDC1 knockdown 293T cells. Analysis of these fractions by Western analysis for nuclear Lamin A/C or cytoplasmic GAPDH confirmed the integrity of the fractionation process and further confirmed not only the nuclear localization of YTHDC1 but also the effective knockdown of YTHDC1 protein expression by RNAi (Fig. 5C). Analysis of viral RNA expression in the isolated nuclear and cytoplasmic fractions confirmed that knockdown of YTHDC1 expression substantially enhanced the expression of HIV-1 transcripts bearing the D1/A1 splice junction (Fig. 5D,E), as previously noted in total cellular RNA (Fig. 5B). However, this analysis did not reveal any changes in the subcellular localization of either unspliced viral RNA, or fully spliced viral RNAs bearing the D4/A7 splice junction, when YTHDC1 expression was inhibited (Fig. 5D–F). As YTHDC1 may also regulate the stability of some m6A+ cellular transcripts (Liu et al. 2020), we analyzed the stability of HIV-1 transcripts in infected WT or YTHDC1 knockdown 293T cells, yet observed no evidence that YTHDC1 expression, unlike YTHDF2 expression, had any effect on HIV-1 RNA stability (Fig. 5G).
Our inability to observe an effect of YTHDC1 knockdown on HIV-1 RNA stability was puzzling given our observation, shown in Figure 5B, that YTHDC1 mRNA knockdown increased viral RNA expression while YTHDC1 overexpression decreased HIV-1 RNA expression. Whereas the differential effect of YTHDC1 on unspliced versus spliced HIV-1 RNA expression suggested an effect on splicing (see below), this seemed unlikely to account for the modest but significant, less than twofold effects seen when total HIV-1 RNA expression was measured. We note that it has recently been reported that YTHDC1 can epigenetically repress the expression of specific cellular genes, including especially retrotransposons, via binding to m6A+ nuclear noncoding RNAs (Liu et al. 2020, 2021), and we therefore wondered whether YTHDC1 might also inhibit HIV-1 transcription. In fact, we did see a significant, less than twofold increase in the production of nascent HIV-1 transcripts in infected 293T cells when YTHDC1 expression was inhibited (Fig. 5H). This effect was specific in that transcription of the host NONO mRNA, lacking any m6A sites, was unaffected by YTHDC1 knockdown. Whether the modest inhibitory effect of YTHDC1 on viral mRNA synthesis is indeed indirect, as we hypothesize, is not currently clear.
YTHDC1 regulates the alternative splicing of HIV-1 transcripts
The observation that knockdown of YTHDC1 expression using RNAi strongly enhanced the expression of HIV-1 transcripts bearing the D1/A1 splice junction, while only modestly affecting both unspliced and total viral RNA expression (Fig. 5), suggested that YTHDC1 might regulate the alternative splicing of HIV-1 transcripts, as has been proposed for cellular mRNAs (Xiao et al. 2016). To determine whether this is the case, we used Primer-ID-based deep sequencing of splice forms (Emery et al. 2017) to quantitatively measure the effect of YTHDC1 on HIV-1 alternative splicing. In this assay, a common forward primer that anneals 5′ of the gag gene can be paired with either random reverse primers, or splice form-specific 4- or 1.8-kb reverse primers, to selectively amplify viral transcripts (Fig. 6A). HIV-1 transcripts are divided by size into the ∼9-kb unspliced transcript, ∼4-kb incompletely spliced transcripts (which retain the D4/A7 env intron), and ∼1.8-kb fully spliced transcripts (lacking the D4/A7 intron). While the random reverse primer amplifies all viral transcripts, the 4-kb reverse primer, located within the D4/A7 intron, only amplifies incompletely spliced transcripts. The 1.8-kb reverse primer spans the D4/A7 splice junction and thus only amplifies fully spliced transcripts. Using the random reverse primer, we noted that ∼33% of viral transcripts are spliced in the siCtrl cells, while this increased to ∼42% in the siDC1 cells but decreased to ∼25% in the +DC1 cells (Fig. 6B). However, among spliced transcripts, a constant ∼85% of all spliced transcripts were fully spliced, that is, lacking the D4/A7 intron, regardless of YTHDC1 expression level (Fig. 6C). Thus, YTHDC1 has a stronger inhibitory effect on utilization of splice donor D1 than D4. Quantification of the level of splicing between donor D1 and the central splice acceptors A1–A5 (Fig. 6D), using the random reverse primer, revealed that YTHDC1 overexpression led to a significant bias toward utilization of the more 3′ acceptors A3, A4, and A5, at the expense of the more 5′ acceptors A1 and A2 (Fig. 6D), and, as expected, the opposite result was observed in cells in which YTHDC1 expression was knocked down; i.e., increased utilization of A1 and A2 and a concomitant reduction in the utilization of A3, A4, and A5. The class-specific 1.8- and 4-kb reverse primers confirmed this bias (Fig. 6E,F). YTHDC1 overexpression also led to a significant, ∼2.5× increase in the detection of the normally very rare D1 to A7 splice (Fig. 6G), consistent with the observed bias toward splicing D1 to more 3′ splice acceptors in YTHDC1-overexpressing cells. Finally, we noted that among the transcripts that used splice acceptor A2, YTHDC1 significantly decreased the proportion of viral RNAs that were additionally spliced using splice donor D3 (Fig. 6H), thus incorporating the small noncoding exon located between A2 and D3 (Fig. 6A).
Figure 6.
YTHDC1 regulates the alternative splicing of HIV-1 RNAs. Viral transcript spliced isoforms in infected siCtrl, siDC1, and +DC1 cells 48 hpi were analyzed by PrimerID RNA-seq. n = 3. (A) Schematic of HIV-1 splice donors and acceptors relative to the three major classes of spliced viral RNAs. Top arrows depict the primers used to amplify sequences for RNA-seq, including the common 5′ forward primer in blue, the reverse 4-kb primer in green, and the reverse 1.8-kb primer in red spanning the D4/A7 splice junction. A random reverse primer was also used but is not shown. (B) Spliced transcripts are given as a percentage of all transcripts (1.8-kb + 4-kb class read counts/total read counts). (C) Percent of fully spliced transcripts over all spliced transcripts (1.8-kb/[1.8-kb + 4-kb] read counts). (D–F) Splice acceptor usage assayed using the common forward primer in conjunction with the random reverse primer (D), 1.8-kb reverse primer (E), and 4-kb reverse primer (F). Use of acceptor A3 results in long transcripts that are biased against when amplifying using the 1.8- or 4-kb reverse primers, A3 usage is thus only shown with the random reverse primer. (G) Percent occurrence of D1/A7 splices. (H) Percent occurrence of A2-using spliced RNAs that subsequently splice from D3. Statistical analysis used Student's t-test. Error bars indicate SD. (*) P < 0.05, (**) P < 0.01.
Discussion
Previously, knockdown of the m6A writers METTL3 and METTL14 using RNAi was reported to inhibit HIV-1 gene expression, while knockdown of the m6A erasers FTO and ALKBH5 enhanced HIV-1 gene expression (Lichinchi et al. 2016a; Tirumuru et al. 2016). The resultant conclusion that m6A must facilitate one or more aspects of the HIV-1 replication cycle is consistent with the finding that HIV-1 genomic RNAs contain higher levels of m6A than the average cellular mRNA (Courtney et al. 2019b). More recently, m6A was shown to enhance the replication and pathogenicity of a wide range of DNA and RNA viruses (Tsai and Cullen 2020), thus implying that the underlying mechanism of action of m6A, though still unclear, can promote the expression and/or function of a range of viral RNA transcripts. These observations, however, seemed to contradict data from several groups showing that addition of m6A can result in the destabilization of cellular mRNAs (Wang et al. 2014; Ke et al. 2017; Park et al. 2019; Zaccara and Jaffrey 2020).
While m6A has the potential to affect RNA secondary structure, it is nevertheless clear that the phenotypic consequences of m6A addition are largely mediated by m6A readers, which include the nuclear m6A reader YTHDC1 and the cytoplasmic m6A readers YTHDF1, YTHDF2, and YTHDF3. While we have previously reported that all of the cytoplasmic readers, including especially YTHDF2, significantly boost HIV-1 RNA expression in infected cells (Kennedy et al. 2016), others have proposed that the YTHDF proteins are bound to m6A residues on the HIV-1 genomic RNA and then packaged into HIV-1 virions where they inhibit reverse transcription (Tirumuru et al. 2016). This latter result is clearly inconsistent with the finding, by this group and others (Kennedy et al. 2016; Lichinchi et al. 2016a; Tirumuru et al. 2016), that m6A promotes HIV-1 replication. Moreover, as m6A is added to RNAs at the consensus sequence 5′-RRACH-3′, mutants lacking this motif should be rapidly selected during passage of a rapidly replicating, error-prone RNA virus such as HIV-1 if m6A indeed exerted an inhibitory effect in cis. Recently, it was been reported that, if YTHDF proteins are indeed packaged into HIV-1 virions, then they are effectively inactivated due to degradation by the viral protease (Jurczyszak et al. 2020). However, we were unable to detect virion packaging of any YTHDF reader protein even when viral protease activity was inhibited (Fig. 2D).
In this report, we have sought to define the precise step(s) in the HIV-1 replication cycle that is regulated by m6A-bound reader proteins. In the case of YTHDF2, we show that this reader primarily acts to increase HIV-1 RNA expression, and not translation, and demonstrate that this effect is mediated by enhanced viral RNA stability (Figs. 2, 3). This was a surprising result given that several previous studies have reported that YTHDF2 binding to m6A residues on cellular mRNAs actually destabilizes that RNA (Wang et al. 2014; Park et al. 2019; Zaccara and Jaffrey 2020). We considered the possibility that the effect of YTHDF2 binding might vary depending on cellular origin, in this case, a CD4+ T-cell line, but in fact we found that the m6A+ cellular mRNAs encoding CREBBP and SON, previously reported to be destabilized by YTHDF2 binding, are also destabilized by YTHDF2 in T cells (Fig. 3E,F). In contrast, and as predicted, cellular mRNAs lacking m6A were unaffected by YTHDF2 expression (Fig. 3G,H). We note that m6A is known to promote the expression of mRNAs encoded by a wide range of viruses (Tsai and Cullen 2020), which is inconsistent with the idea that the recruitment of cytoplasmic readers such as YTHDF2 invariably inhibits mRNA expression. Moreover, several reports have suggested that m6A can stabilize specific cellular transcripts (Meyer and Jaffrey 2014), with one report demonstrating that m6A addition stabilizes key cellular mRNAs during hypoxia (Fry et al. 2017). Why YTHDF2, and potentially other cytoplasmic m6A readers, can stabilize m6A-containing viral RNAs yet destabilize m6A-containing cellular RNAs expressed in the same cells (Fig. 3) is currently unknown. However, this effect is unlikely to be due to viral factors acting in trans, as we have previously shown that the 3′ untranslated region (3′ UTR) of HIV-1 can enhance indicator gene expression in cis in uninfected cells, an activity that was lost when the m6A sites located in the HIV-1 3′ UTR were mutated (Kennedy et al. 2016). The functions exerted by readers of methylated nucleotides on mRNA have in fact previously been proposed to be context-dependent (Shi et al. 2019), which could also provide an explanation for the disparate effect of YTHDF2 binding on mRNA stability reported here.
While there has been little previous work looking at the effect of m6A on viral RNA splicing, with the exception of a recent paper showing that depletion of the m6A writer METTL3 reduces the efficiency of splicing of adenoviral late mRNAs (Price et al. 2020), it is known that m6A addition can regulate cellular mRNA splicing, acting predominantly via the nuclear m6A reader YTHDC1 (Dominissini et al. 2012; Alarcón et al. 2015; Xiao et al. 2016; Bartosovic et al. 2017). As HIV-1 transcripts are among the most extensively alternatively spliced RNAs ever identified (Purcell and Martin 1993; Stoltzfus 2009), we were interested in whether YTHDC1 regulated this process in infected cells. Using the PAR-CLIP technique, we mapped at least seven distinct YTHDC1 binding sites on HIV-1 transcripts that largely, but not entirely, coincided with previously mapped YTHDF1 and YTHDF2 binding sites as well as sites of m6A addition previously mapped using an antibody-dependent technique (Fig. 4; Kennedy et al. 2016; Courtney et al. 2019b). As we were unable to knock out YTHDC1 using CRISPR/Cas, we decided to knock down YTHDC1 expression using RNAi, and also overexpress YTHDC1 by transfection, to determine whether changing the level of YTHDC1 expression affected HIV-1 RNA expression and/or alternative splicing. As shown in Figure 5, A and B, we unexpectedly observed that knockdown of YTHDC1 actually increased HIV-1 Gag protein and RNA expression while overexpression of YTHDC1 inhibited HIV-1 gene expression. This effect, which is the opposite of what we saw when YTHDF2 was knocked out or overexpressed (Figs. 2, 3), was particularly significant for viral RNAs spliced from donor D1 to acceptor A1 (Fig. 5B), thus suggesting that YTHDC1 indeed regulated some aspect(s) of HIV-1 RNA metabolism. Despite previous reports indicating that YTHDC1 can regulate the nuclear export of cellular mRNAs (Roundtree et al. 2017), and our report that m6A regulates the nuclear export of SV40 transcripts (Tsai et al. 2018), we did not detect any effect of YTHDC1 knockdown on the nuclear export of either unspliced HIV-1 transcripts, which is dependent on the viral Rev protein, or of fully spliced viral transcripts bearing the D4/A7 splice junction, which is mediated by the canonical cellular nuclear mRNA export factor NXF1, as determined by their similar relative nuclear and cytoplasmic expression levels (Fig. 5D–F; Cullen 2003). However, using the previously described “primer ID-based deep sequencing of splice forms” technique (Emery et al. 2017), we were able to document clear effects of YTHDC1 knockdown or overexpression on the relative utilization of specific HIV-1 splice acceptors. Specifically, knockdown of YTHDC1 modestly increased viral RNA splicing (Fig. 6B) by increasing the utilization of splice acceptors A2 and particularly A1, while simultaneously reducing the utilization of splice acceptors A3, A4, and A5 (Fig. 6D–F), thus explaining the increased level of D1 to A1 spliced RNA detected by qRT-PCR in Figure 5. Overexpression of YTHDC1, in contrast, reduced utilization of A1 and A2 while simultaneously increasing the utilization of acceptors A3, A4, and A5, an effect that was most clearly seen when the random reverse primer splicing assay was used (Fig. 6D).
Previous work looking at the effect of YTHDC1 on splicing of cellular mRNAs largely looked at the effect of YTHDC1 on the inclusion or skipping of cassette exons and concluded that YTHDC1 promoted exon inclusion by recruiting the pre-mRNA splicing factor SRSF3 (Xiao et al. 2016). However, this report did not address whether the location of m6A addition sites relative to splice sites was important. As alternative splicing of HIV-1 RNAs is a much more complex process than simple exon inclusion or exclusion, it is hard to address whether our data are consistent with this earlier report. However, we note that none of the mapped YTHDC1 binding sites on the HIV-1 genome colocalize with either known viral splicing enhancer/repressor elements or with any intronic branch points (consensus motif 5′-YNYURAY-3′) (Tazi et al. 2010). A minor peak of YTHDC1 binding does precisely overlap with splice acceptor A7, with the only m6A motif present in this peak (5′-RRm6ACH-3′) positioning the splice junction exactly 5′ of the m6A (Fig. 4B). YTHDC1 overexpression also biases D1-originating splices away from acceptors A1 and A2 and toward A3, A4, A5, and A7 (Fig. 6), and we mapped a YTHDC1 binding site between A2 and A3 (Fig. 4A), which demarcates the border of this effect. However, silent mutagenesis of this latter m6A site did not detectably affect the utilization of A3 (data not shown). While we therefore strongly favor the hypothesis that YTHDC1 binding to m6A residues on HIV-1 transcripts is modulating HIV-1 RNA alternative splicing, potentially by regulating splicing factor recruitment as reported for cellular pre-mRNAs (Xiao et al. 2016), we cannot currently totally eliminate the possibility that this effect is indirect.
Materials and methods
Cell lines and virus clones
HEK293T cells (referred to as 293T) were cultured in Dulbecco's modified Eagle's medium (DMEM) with 6% fetal bovine serum (FBS) and 1% antibiotic–antimycotic (Gibco 15240062). The CD4+ T-cell lines CEM and CEM-SS were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium with 10% FBS and 1% antibiotic–antimycotic.
+ALKBH5 cells were produced by transducing CEM cells with a lentiviral vector expressing FLAG-ALKBH5 followed by puromycin selection and single-cell cloning. Three clones that strongly expressed a FLAG-ALKBH5 protein band slightly larger than endogenous ALKBH5 were selected. YTHDF2 knockout and -overexpressing CEM-SS cells were previously described (Kennedy et al. 2016). YTHDC1 mRNA knockdown in 293T cells was performed using a mixture of three siRNAs (Origene # SR314128) transfected using Lipofectamine RNAiMax (Invitrogen).
Recombinant virus clones used include the laboratory strain NL4-3 (Adachi et al. 1986) and the reporter virus NL-NLuc, in which the viral nef gene has been substituted with the NLuc indicator gene (Mefferd et al. 2018).
Antibodies
Antibodies used in this study were anti-FLAG (M2; Sigma F1804), anti-HIV-1 p24 gag #24-3 (ARP-6458), anti-GAPDH (Proteintech 60004-1-Ig), anti-ALKBH5 (Proteintech 16837-1-AP), anti-YTHDF2 (Proteintech 24744-1-AP), anti-YTHDC1 (Abcam ab122340), anti-mouse IgG (Sigma A9044), and anti-rabbit IgG (Sigma A6154).
Cloning of expression plasmids
The ALKBH5 and YTHDC1 ORFs were isolated from cDNA produced from CEM cells using PCR and ligated into the Not I and Xho I sites of the lentiviral vector pLEX-FLAG, which encodes double-FLAG tags 5′ of the Not I site (Kennedy et al. 2016). FLAG-YTHDC1 was subsequently PCR-cloned into the Hind III and Eco RI sites of pK to generate pK-FLAG-YTHDC1. See Supplemental Table S1 for primers used.
HIV-1 replication assays in 293T cells
Virus was packaged in 293T cells in 10-cm plates by transfection of 10 µg of pNL-NLuc using PEI (polyethylenimine). For infection target cells, 293T cells in six-well plates were cotransfected with 1 µg of empty vector or pEFtak-FLAG-YTHDF2 (Kennedy et al. 2016) and 0.5 µg of pCMV-CD4 using PEI, media were changed at 24 h post-transfection (hpt), and cells were passaged 48 hpt, infected with NL-NLuc 72 hpt, and harvested 24 or 48 hpi. Harvested cells were washed in PBS, lysed in passive lysis buffer (Promega E1941), and assayed using an NLuc assay kit (Promega N1120).
YTHDF protein virion packaging assay
Five micrograms of pNL-NLuc was cotransfected with 5 µg of pEFtak-FLAG-YTHDF1, pEFtak-FLAG-YTHDF2, pEFtak-FLAG-YTHDF3 (Kennedy et al. 2016), or pCDNA3-FLAG-agmA3G (Bogerd et al. 2004) into 293T cells using PEI and the cells immediately treated with 5 µM Indinavir. Three days after transfection, producer cells and supernatant media were collected, passed through a 0.45-µm filter, and virions purified by pelleting through a 20% sucrose cushion, followed by banding on a 6%–18% Iodixanol gradient (Sigma D1556) each by 90 min of ultracentrifugation at 38,000 rpm (Courtney et al. 2019b). Producer cells and purified virions were then lysed and analyzed by Western blot.
Infection of YTHDC1 knockdown and overexpression cells
Two million 293T cells were seeded per 10-cm plate and 0.4 million cells per six-well plate well. The next day (day 1), the 10-cm plate was transfected with 10 µg of pNL4-3 plasmid using PEI. Cells in six-well plates were transfected with nontargeting (siCtrl) or YTHDC1-targeted siRNAs (siDC1; three YTHDC1 siRNAs premixed at a 1:1:1 ratio; Origene SR314128) using Lipofectamine RNAiMax (Invitrogen), with the media changed the next day. The cells in six-well plates were subsequently cotransfected with 0.5 µg of pCMV-CD4, 1.5 µg of pBC-CXCR4, and 1 µg of empty pK vector or pK-FLAG-YTHDC1, using PEI. On day 2, the media of 10-cm and six-well plates were exchanged for fresh DMEM, and the six-well plates were subject to a second round of siRNA transfection. Transfections were coordinated for a control well transfected with pK vector + siCtrl, a knockdown well of pK vector + siDC1, and an overexpression well of pK-FLAG-YTHDC1 + siCtrl. On day 3, the media was changed again on the six-well plates to remove the transfection mix. On day 4, the cells in the six-well plates were trypsinized, counted, and seeded at 0.5 million per well in 12-well plates. The virus-containing supernatant media from the 10-cm plates was passed through a 0.45-µm filter and added to the newly seeded target cells at 1 mL/well.
Single-cycle replication assays in T cells
Infected cells were treated with 133 µM reverse transcriptase inhibitor Nevirapine (Sigma SML0097) at 16 hpi, then harvested at 48 hpi, as previously described (Tsai et al. 2020). Subsequent Western blots and qRT-PCRs were also done as described (Tsai et al. 2020). Western blot band quantifications were performed using Image J v1.51s (Schneider et al. 2012). qRT-PCR readouts were normalized to GAPDH levels using the ΔΔCt method. All PCR primers used are listed in Supplemental Table S1.
Subcellular fractionation assays
HIV-1 single-cycle infected cells were lysed and divided into nuclear and cytoplasmic fractions as previously described (Tsai et al. 2018).
RNA decay assay
RNA stability was measured using the transcription stop method as previously described (Tsai et al. 2020). Single-cycle infected cells were treated at 2 d postinfection with 5 µg/mL actinomycin D (Sigma A9415) and harvested 0, 2.5, 5, and 7.5 h later. Viral RNA levels at each time point were assayed by qRT-PCR using the D1/A1 splice junction primer set to avoid detection of residual DNA (Supplemental Table S1). See Supplemental Table S1 for primer sets used for host transcripts.
Nascent RNA transcription assay
Single-cycle infected cells were pulsed at 48 hpi with 150 µM 4SU for 1.5 h, the cells were then harvested and RNA was extracted with Trizol. Nascent RNA was isolated as previously described (Dolken et al. 2008; Duffy et al. 2015; Tsai et al. 2020) . Briefly, 4SU+ RNA was biotinylated with MTSEA-biotin-XX (Biotium 89139-636) in dimethyl formamide, purified with streptavidin magnetic beads, eluted with DTT, and quantified by qRT-PCR.
YTHDC1 PAR-CLIP
PAR-CLIP was performed as before using 4SU-pulsed, HIV-1-infected cells with minor modifications (Hafner et al. 2010; Kennedy et al. 2016). Briefly, cells were lysed with RIPA buffer (150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 25 mM Tris-HCl at pH 7.4) instead of PAR-CLIP lysis buffer to ensure sufficient release of nuclear proteins. The subsequent pulldown used FLAG antibody (M2; Sigma). Illumina sequencing library preparation was done using the Illumina TruSeq small RNA library preparation kit. Data analysis was done as previously described (Tsai et al. 2020). Index-removed reads <15 nt with a fastq quality score >Q33 were aligned to the human genome (hg19) using Bowtie, and the human nonaligning reads then aligned to the HIV-1 NL4-3 sequence with a single-copy LTR (U5 on the 5′ end, and U3-R on the 3′ end), and thus 551–9626 nt of GenBank AF324493.2, allowing one mismatch. An in-house Perl script was used to only retain reads containing the T > C conversions resulting from UV cross-linked 4SU. Peak calling was done using PARalyzer v1.1 (Corcoran et al. 2011).
Primer-ID RNA-seq splicing assay
YTHDC1 knockdown or -overexpressing 293T cells treated and infected as mentioned above were grown in six-well plates, two wells per sample were collected, and the RNA extracted with Trizol (Invitrogen). RNA samples were subsequently amplified and sequenced as previously described (Emery et al. 2017).
Quantification and statistical analysis
All statistical details are listed in the figure legends. All averaged data include error bars that denote standard deviation. All statistical analysis was done by Student's t-test comparing subjects to control, with the exception of RNA decay studies, where the slopes of regression lines were compared by analysis of covariance (ANCOVA) using R suite.
Data availability
Deep sequencing data for the YTHDC1 PAR-CLIP have been deposited at the NCBI GEO database under accession number GSE165473, while the splicing analysis data are under accession number GSE166237.
Supplementary Material
Acknowledgments
This research was funded in part by National Institutes of Health (NIH) grant R01-DA046111 to B.R.C., and a Duke University Center for AIDS Research (CFAR; P30-AI064518) pilot award to K.T. B.R.C and R.S. are both supported by the Center for HIV RNA Studies (CRNA; NIH award U54-AI15047). This research received infrastructure support from the Duke University CFAR, the University of North Carolina (UNC) CFAR (P30-AI50410), and the UNC Lineberger Comprehensive Cancer Center (P30-CA16068). The following reagents were obtained through the NIH AIDS Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, NIH: HIV-1 p24 Gag monoclonal (#24-3) from Michael Malim, and infectious HIV-1 clone pNL4-3 (#114) from Malcolm Martin. We thank Ananda Vasudevan for cloning assistance, Joanne Lin for guidance on statistical analysis, and the laboratories of Christopher Holley, Nicholas Heaton, Dennis Ko, and Qi-Jing Li for use of equipment.
Author contributions: K.T., H.P.B., E.M.K., and A.E. performed the experiments. K.T., H.P.B., E.M.K. A.E., R.S., and B.R.C. designed the experiments. R.S and B.R.C. obtained funding for this work. K.T. and B.R.C. wrote the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.348508.121.
Competing interest statement
The authors declare no competing interests.
References
- Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol 59: 284–291. 10.1128/JVI.59.2.284-291.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcón CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF. 2015. HNRNPA2B1 is a mediator of m6A-dependent nuclear RNA processing events. Cell 162: 1299–1308. 10.1016/j.cell.2015.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartosovic M, Molares HC, Gregorova P, Hrossova D, Kudla G, Vanacova S. 2017. N6-methyladenosine demethylase FTO targets pre-mRNAs and regulates alternative splicing and 3′-end processing. Nucleic Acids Res 45: 11356–11370. 10.1093/nar/gkx778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd HP, Doehle BP, Wiegand HL, Cullen BR. 2004. A single amino acid difference in the host APOBEC3G protein controls the primate species specificity of HIV type 1 virion infectivity factor. Proc Natl Acad Sci 101: 3770–3774. 10.1073/pnas.0307713101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokar JA, Shambaugh ME, Polayes D, Matera AG, Rottman FM. 1997. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 3: 1233–1247. [PMC free article] [PubMed] [Google Scholar]
- Chu CC, Liu B, Plangger R, Kreutz C, Al-Hashimi HM. 2019. m6A minimally impacts the structure, dynamics, and Rev ARM binding properties of HIV-1 RRE stem IIB. PLoS One 14: e0224850. 10.1371/journal.pone.0224850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran DL, Georgiev S, Mukherjee N, Gottwein E, Skalsky RL, Keene JD, Ohler U. 2011. PARalyzer: definition of RNA binding sites from PAR-CLIP short-read sequence data. Genome Biol 12: R79. 10.1186/gb-2011-12-8-r79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney DG, Chalem A, Bogerd HP, Law BA, Kennedy EM, Holley CL, Cullen BR. 2019a. Extensive epitranscriptomic methylation of A and C residues on murine leukemia virus transcripts enhances viral gene expression. MBio 10: e01209–e01219. 10.1128/mBio.01209-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney DG, Tsai K, Bogerd HP, Kennedy EM, Law BA, Emery A, Swanstrom R, Holley CL, Cullen BR. 2019b. Epitranscriptomic addition of m5C to HIV-1 transcripts regulates viral gene expression. Cell Host Microbe 26: 217–227.e6. 10.1016/j.chom.2019.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen BR. 2003. Nuclear mRNA export: insights from virology. Trends Biochem Sci 28: 419–424. 10.1016/S0968-0004(03)00142-7 [DOI] [PubMed] [Google Scholar]
- Cullen BR, Greene WC. 1990. Functions of the auxiliary gene products of the human immunodeficiency virus type 1. Virology 178: 1–5. 10.1016/0042-6822(90)90373-Y [DOI] [PubMed] [Google Scholar]
- Davis DA, Soule EE, Davidoff KS, Daniels SI, Naiman NE, Yarchoan R. 2012. Activity of human immunodeficiency virus type 1 protease inhibitors against the initial autocleavage in Gag-Pol polyprotein processing. Antimicrob Agents Chemother 56: 3620–3628. 10.1128/AAC.00055-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desrosiers R, Friderici K, Rottman F. 1974. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci 71: 3971–3975. 10.1073/pnas.71.10.3971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolken L, Ruzsics Z, Radle B, Friedel CC, Zimmer R, Mages J, Hoffmann R, Dickinson P, Forster T, Ghazal P, et al. 2008. High-resolution gene expression profiling for simultaneous kinetic parameter analysis of RNA synthesis and decay. RNA 14: 1959–1972. 10.1261/rna.1136108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. 2012. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485: 201–206. 10.1038/nature11112 [DOI] [PubMed] [Google Scholar]
- Duffy EE, Rutenberg-Schoenberg M, Stark CD, Kitchen RR, Gerstein MB, Simon MD. 2015. Tracking distinct RNA populations using efficient and reversible covalent chemistry. Mol Cell 59: 858–866. 10.1016/j.molcel.2015.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery A, Zhou S, Pollom E, Swanstrom R. 2017. Characterizing HIV-1 splicing by using next-generation sequencing. J Virol 91: e02515-16. 10.1128/JVI.02515-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry NJ, Law BA, Ilkayeva OR, Holley CL, Mansfield KD. 2017. N6-methyladenosine is required for the hypoxic stabilization of specific mRNAs. RNA 23: 1444–1455. 10.1261/rna.061044.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhale NS, McIntyre ABR, McFadden MJ, Roder AE, Kennedy EM, Gandara JA, Hopcraft SE, Quicke KM, Vazquez C, Willer J, et al. 2016. N6-methyladenosine in Flaviviridae viral RNA genomes regulates infection. Cell Host Microbe 20: 654–665. 10.1016/j.chom.2016.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jungkamp AC, Munschauer M, et al. 2010. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141: 129–141. 10.1016/j.cell.2010.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imam H, Khan M, Gokhale NS, McIntyre ABR, Kim GW, Jang JY, Kim SJ, Mason CE, Horner SM, Siddiqui A. 2018. N6-methyladenosine modification of hepatitis B virus RNA differentially regulates the viral life cycle. Proc Natl Acad Sci 115: 8829–8834. 10.1073/pnas.1808319115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, et al. 2011. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol 7: 885–887. 10.1038/nchembio.687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurczyszak D, Zhang W, Terry SN, Kehrer T, Bermúdez González MC, McGregor E, Mulder LCF, Eckwahl MJ, Pan T, Simon V. 2020. HIV protease cleaves the antiviral m6A reader protein YTHDF3 in the viral particle. PLoS Pathog 16: e1008305. 10.1371/journal.ppat.1008305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke S, Pandya-Jones A, Saito Y, Fak JJ, Vågbø CB, Geula S, Hanna JH, Black DL, Darnell JE Jr., Darnell RB. 2017. m6A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev 31: 990–1006. 10.1101/gad.301036.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy EM, Bogerd HP, Kornepati AV, Kang D, Ghoshal D, Marshall JB, Poling BC, Tsai K, Gokhale NS, Horner SM, et al. 2016. Posttranscriptional m6A editing of HIV-1 mRNAs enhances viral gene expression. Cell Host Microbe 19: 675–685. 10.1016/j.chom.2016.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasman L, Krupalnik V, Viukov S, Mor N, Aguilera-Castrejon A, Schneir D, Bayerl J, Mizrahi O, Peles S, Tawil S, et al. 2020. Context-dependent functional compensation between ythdf m6A reader proteins. Genes Dev 34: 1373–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichinchi G, Gao S, Saletore Y, Gonzalez GM, Bansal V, Wang Y, Mason CE, Rana TM. 2016a. Dynamics of the human and viral m6A RNA methylomes during HIV-1 infection of T cells. Nat Microbiol 1: 16011. 10.1038/nmicrobiol.2016.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichinchi G, Zhao BS, Wu Y, Lu Z, Qin Y, He C, Rana TM. 2016b. Dynamics of human and viral RNA methylation during Zika virus infection. Cell Host Microbe 20: 666–673. 10.1016/j.chom.2016.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, et al. 2014. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol 10: 93–95. 10.1038/nchembio.1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Dou X, Chen C, Chen C, Liu C, Xu MM, Zhao S, Shen B, Gao Y, Han D, et al. 2020. N6-methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription. Science 367: 580–586. 10.1126/science.aay6018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Gao M, He J, Wu K, Lin S, Jin L, Chen Y, Liu H, Shi J, Wang X, et al. 2021. The RNA m6A reader YTHDC1 silences retrotransposons and guards ES cell identity. Nature 591: 322–326. 10.1038/s41586-021-03313-9 [DOI] [PubMed] [Google Scholar]
- Lu M, Zhang Z, Xue M, Zhao BS, Harder O, Li A, Liang X, Gao TZ, Xu Y, Zhou J, et al. 2020. N6-methyladenosine modification enables viral RNA to escape recognition by RNA sensor RIG-I. Nat Microbiol 5: 584–598. 10.1038/s41564-019-0653-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mefferd AL, Bogerd HP, Irwan ID, Cullen BR. 2018. Insights into the mechanisms underlying the inactivation of HIV-1 proviruses by CRISPR/Cas. Virology 520: 116–126. 10.1016/j.virol.2018.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Jaffrey SR. 2014. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat Rev Mol Cell Biol 15: 313–326. 10.1038/nrm3785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. 2012. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149: 1635–1646. 10.1016/j.cell.2012.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park OH, Ha H, Lee Y, Boo SH, Kwon DH, Song HK, Kim YK. 2019. Endoribonucleolytic cleavage of m6A-containing RNAs by RNase P/MRP complex. Mol Cell 74: 494–507.e8. 10.1016/j.molcel.2019.02.034 [DOI] [PubMed] [Google Scholar]
- Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, et al. 2014. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res 24: 177–189. 10.1038/cr.2014.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AM, Hayer KE, McIntyre ABR, Gokhale NS, Abebe JS, Della Fera AN, Mason CE, Horner SM, Wilson AC, Depledge DP, et al. 2020. Direct RNA sequencing reveals m6A modifications on adenovirus RNA are necessary for efficient splicing. Nat Commun 11: 6016. 10.1038/s41467-020-19787-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell DF, Martin MA. 1993. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J Virol 67: 6365–6378. 10.1128/JVI.67.11.6365-6378.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roundtree IA, Luo GZ, Zhang Z, Wang X, Zhou T, Cui Y, Sha J, Huang X, Guerrero L, Xie P, et al. 2017. YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. Elife 6: e31311. 10.7554/eLife.31311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrofelbauer B, Chen D, Landau NR. 2004. A single amino acid of APOBEC3G controls its species-specific interaction with virion infectivity factor (Vif). Proc Natl Acad Sci 101: 3927–3932. 10.1073/pnas.0307132101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Wei J, He C. 2019. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol Cell 74: 640–650. 10.1016/j.molcel.2019.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltzfus CM. 2009. Chapter 1. Regulation of HIV-1 alternative RNA splicing and its role in virus replication. Adv Virus Res 74: 1–40. 10.1016/S0065-3527(09)74001-1 [DOI] [PubMed] [Google Scholar]
- Tazi J, Bakkour N, Marchand V, Ayadi L, Aboufirassi A, Branlant C. 2010. Alternative splicing: regulation of HIV-1 multiplication as a target for therapeutic action. FEBS J 277: 867–876. 10.1111/j.1742-4658.2009.07522.x [DOI] [PubMed] [Google Scholar]
- Tirumuru N, Zhao BS, Lu W, Lu Z, He C, Wu L. 2016. N6-methyladenosine of HIV-1 RNA regulates viral infection and HIV-1 Gag protein expression. Elife 5: e15528. 10.7554/eLife.15528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai K, Cullen BR. 2020. Epigenetic and epitranscriptomic regulation of viral replication. Nat Rev Microbiol 18: 559–570. 10.1038/s41579-020-0382-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai K, Courtney DG, Cullen BR. 2018. Addition of m6A to SV40 late mRNAs enhances viral structural gene expression and replication. PLoS Pathog 14: e1006919. 10.1371/journal.ppat.1006919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai K, Jaguva Vasudevan AA, Martinez Campos C, Emery A, Swanstrom R, Cullen BR. 2020. Acetylation of cytidine residues boosts HIV-1 gene expression by increasing viral RNA stability. Cell Host Microbe 28: 306–312.e6. 10.1016/j.chom.2020.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. 2014. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505: 117–120. 10.1038/nature12730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C. 2015. N6-methyladenosine modulates messenger RNA translation efficiency. Cell 161: 1388–1399. 10.1016/j.cell.2015.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, Sun HY, Li A, Ping XL, Lai WY, et al. 2016. Nuclear m6A reader YTHDC1 regulates mRNA splicing. Mol Cell 61: 507–519. 10.1016/j.molcel.2016.01.012 [DOI] [PubMed] [Google Scholar]
- Zaccara S, Jaffrey SR. 2020. A unified model for the function of YTHDF proteins in regulating m6A-modified mRNA. Cell 181: 1582–1595.e18. 10.1016/j.cell.2020.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vågbø CB, Shi Y, Wang WL, Song SH, et al. 2013. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell 49: 18–29. 10.1016/j.molcel.2012.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Deep sequencing data for the YTHDC1 PAR-CLIP have been deposited at the NCBI GEO database under accession number GSE165473, while the splicing analysis data are under accession number GSE166237.