Abstract
Coronaviruses (CoVs) were first discovered in the 1960s. Severe acute respiratory syndrome CoV-2 (SARS-CoV-2) has been identified as the cause of COVID-19, which spread throughout China and subsequently, across the world. As COVID-19 causes serious public health concerns across the world, investigating the characteristics of SARS-CoV-2 and its interaction with the host immune responses may provide a clearer picture of how the pathogen causes disease in some individuals. Interestingly, SARS-CoV-2 has 80% sequence homology with SARS-CoV-1 and 96–98% homology with CoVs isolated from bats. Therefore, the experience acquired in SARS and Middle East Respiratory Syndrome (MERS) epidemics may improve our understanding of the immune response and immunopathological changes in COVID-19 patients. In the present paper, we have reviewed the immune responses (including the innate and adaptive immunities) to SARS-CoV, MERS-CoV, and SARS-CoV-2, so as to improve our understanding of the concept of the COVID-19 disease, which will be helpful in developing vaccines and medications for treating the COVID-19 patients.
Keywords: Coronavirus disease 2019, MERS-CoV, SARS-CoV, Immune response, Vaccine
Introduction
In the past two decades, there have been two major coronavirus (CoV) outbreaks, including the severe acute respiratory syndrome CoV (SARS-CoV) in 2002, and the Middle East Respiratory Syndrome CoV (MERS-CoV) in 2012 [1, 2]. The recent CoV outbreak, which happened in the Wuhan city of China, is known as the 2019-nCoV outbreak and has been recently renamed as SARS-CoV-2 outbreak or COVID-19 [3].
The first case of SARS-CoV-2 infection was reported with presentation of the symptoms of atypical pneumonia. This case was further confirmed to be caused by the novel CoV, SARS-CoV-2 [4]. The most potential risk for the spread of COVID-19 worldwide is related to travel, which leads to the regional and global spread of the disease [5]. The origin of CoVs is primarily related to animals. The outbreaks occur when these viruses cross the species barrier and infect humans. SARS and COVID-19 share many similarities in terms of their transmission and pathogenicity. They both cause acute respiratory illnesses and follow human-to-human transmission. Although SARS-CoV-2, which is responsible for COVID-19 infection, has been successfully isolated and the viral infectivity and pathogenicity have been understood, further investigations are still required to understand the viral antigenic structure, mode of action, and pathogenicity of this pathogen [2].
SARS-CoV-2 is a novel emerging contagious agent that has found a way into human civilization. The prediction of Fan et al. [6] about the emergence of a future SARS or MERS-like CoVs epidemic in China with a probable bat source turned into reality when the first case of concentrated viral pneumonia was reported in Wuhan. Later on, the novel CoV, designated as SARS-CoV-2, was found to be responsible for the viral outbreak of pneumonia in Wuhan [7]. Generally, emerging and reemerging of viral infections belong to the RNA family of viruses since these viruses have high mutation rates that allow their eminent environmental adaptation with rapid evolution [8]. To date, little knowledge is available about SARS-CoV-2.
A recently published research suggests that SARS-CoV-2 shares 79% nucleotide identity to SARS-CoV and 51.8% identity to MERS-CoV [9], indicating a high genetic homology among SARS-CoV-2, MERS-CoV, and SARS-CoV. In SARS-CoV and MERS-CoV-infected animal models, marked inflammatory and immune responses may activate a “cytokine storm” and apoptosis of epithelial and endothelial cells. Subsequently, vascular leakage as well as abnormal T cell and macrophage responses ensue and induce acute lung injury/acute respiratory distress syndrome (ARDS) or even death [10].
However, the systemic landscape of the immune responses in patients with COVID-19 is unclear. Since there are some similarities among the clinical features and immunopathogenesis of SARS-CoV-2 and those of SARS-CoV and MERS-CoV [11], the knowledge learned from SARS-CoV and MERS-CoV has important implications for understanding this new CoV [12].
In order to contain the infection and develop effective management systems to handle viral infections in an outbreak scenario, we should understand the nature of infection and response of the immune system to the novel virus and evaluate the similarities and dissimilarities of the novel virus with the viruses that had caused outbreaks in the past. This review aims at exploring the immune system responses against the SARS-CoV-2, compared to the cases of other CoVs (SARS and MERS).
Clinical and Biochemical Indices
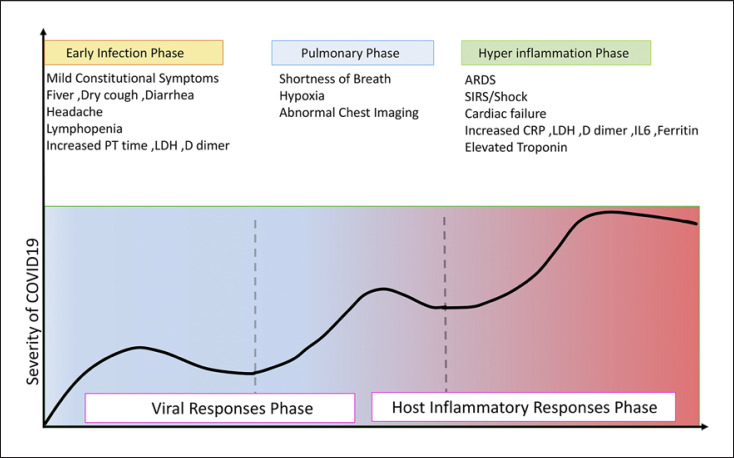
The most common laboratory abnormalities related to the new CoV include hypoalbuminemia, lymphopenia, decreased percentage of neutrophils, elevated C-reactive protein (CRP), and lactate dehydrogenase (LDH) levels, as well as decreased CD8 count. The viral load of SARS-CoV-2, detected through the patients' respiratory tracts, has been found to be positively linked to the lung disease severity. Albumin, lymphocytes, LDH, neutrophils, and CRP are highly correlated with acute lung injury. Age, viral load, lung injury score, and blood biochemistry indices, albumin, CRP, LDH, lymphocytes (%), and neutrophils (%), are possibly the predictors of disease severity [13]. In addition, nonsurvivors had higher levels of neutrophils, D-Dimer, blood urea nitrogen, and creatinine than survivors [14] (Fig. 1).
Fig. 1.
The illustration of escalating phases of COVID-19 disease progression, with associated signs and symptoms from the onset to recovery or death. Infection with SARS-CoV-2 (COVID-19) can be classified into three stages of increasing severity: early infection, pulmonary phase, and hyperinflammation phase. The first phase is related to the onset of the disease and is generally characterized by the development of influenza-like symptoms from mild to moderate. Some individuals recover and some progress to the second phase. In phase 2, it is possible to detect pneumonia-like symptoms evidenced as lung opacities. Phase 3 is characterized by hyperinflammation and sepsis of lungs and patient often requires ICU and most of them, unfortunately, cannot overcome the infection and eventually die. SARS-CoV-2, Severe acute respiratory syndrome coronavirus-2; ICU, intensive care unit; ARDS, acute respiratory distress syndrome; LDH, lactate dehydrogenase; CRP, C-reactive protein.
Innate Immune Responses to SARS-CoV-2
Based on the available accumulated data for previous CoV infections, the innate immune response plays a crucial role in the protective or destructive responses and may open a window for immune intervention. Active viral replication postpones the hyperproduction of interferon (IFN) type I and influx of neutrophils and macrophages, as the major sources of pro-inflammatory cytokines [15].
Cytokines and Chemokines
Cytokines and chemokines have been long thought to play an important role in immunity and immunopathology during virus infections. A rapid and well-coordinated innate immune response is the first line of defense against viral infections, but dysregulated and excessive immune responses may cause immunopathology [16]. Although there is no direct evidence for the involvement of pro-inflammatory cytokines and chemokines in lung pathology during SARS and MERS, correlative evidence from patients with severe disease suggests a role for hyper-inflammatory responses in the human CoV (hCoV) pathogenesis [10].
Analysis of serum cytokine levels and lymphocyte composition suggests that SARS-CoV-2 infection is associated with lymphopenia (particularly in CD4+ T cells and CD8+ T cells, but not in B cells), overproduction of cytokines such as interleukin (IL)-1, IL-6, IL-8, IL-2 receptor (IL-2R), IL-10, tumor necrosis factor-alpha (TNF-α), C-C motif chemokine 2 (CCL2), CCL3, CCL5, and decreased IFN-γ-expression in CD4+ T cells in severe COVID-19, being correlated with COVID-19 disease severity. Levels of IL-6, IL-2R, IL-10, and TNF-α were mildly elevated or within the normal range in moderate cases but markedly elevated in most severe cases. These cytokines are probably produced by highly inflammatory cells that have been implicated in a cytokine storm [17]. It is believed that dysregulated host immune response and cytokine storm are correlated with disease severity and poor prognosis during SARS-CoV and MERS-CoV infection [1, 18]. Unregulated levels of inflammatory cytokines, which may lead to activated T-helper-1 (Th1) cell responses, have been observed in COVID-19 patients [19]. However, in SARS-CoV-2 patients, excessive secretion of IL-4 and IL-10 has been reported, which may suppress inflammation via T-helper cell-2 (Th2) [15] (Fig. 2).
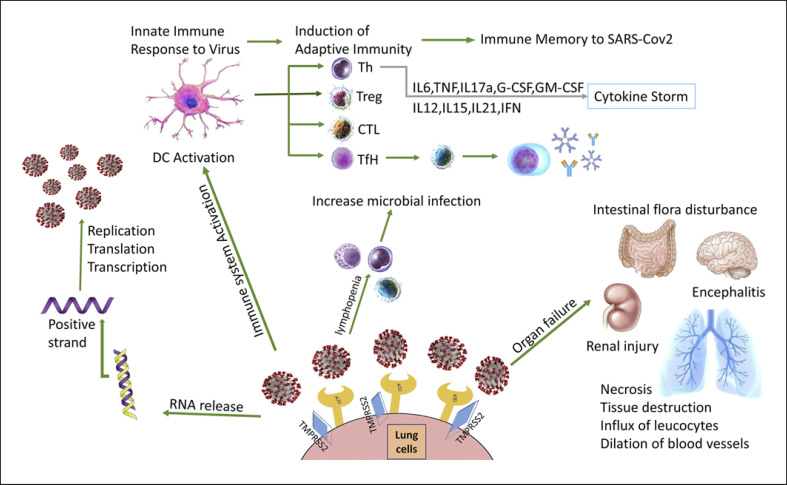
Fig. 2.
Manifestations of COVID-19 in body. Spike protein on the virion binds to ACE2, a cell-surface protein. TMPRSS2, an enzyme, helps the virion enter. The virion releases its RNA. Some RNA is translated into proteins by the cell's machinery, and some of these proteins form a replication complex to make more RNA. Then, RNAs are assembled into a new virion in the Golgi and released. Infection with SARS-CoV-2 leads to activation of innate immunity and DCs, which will drive the induction of virus-specific T-cell and B-cell responses. Hyperinflammation by innate and adaptive leads to cytokine storm through inflammatory cytokine secretion. COVID-19 manifestations including pulmonary involvement, ARDS, encephalitis, renal injury, intestinal flora disturbance, and pneumonia are well recognized. CTL, cytotoxic T lymphocyte; TFH, T follicular helper cell; TH, T-helper cell; Treg, regulatory T cell; DCs, dendritic cell; SARS-CoV-2, Severe acute respiratory syndrome coronavirus-2; ARDS, acute respiratory distress syndrome; ACE2, angiotensin-converting enzyme 2; IFN, interferon; GM-CSF, granulocyte-macrophage colony-stimulating factor; IL, interleukin; TNF, tumor necrosis factor.
While SARS-CoV productively infects airway and alveolar epithelial cells, infection of hematopoietic cells such as dendritic cells (DCs), monocyte-macrophages, and other peripheral blood mononuclear cells (PBMCs) is abortive. SARS-CoV infection of DCs induces low-level expression of antiviral IFN-α and IFN-β cytokines, moderate upregulation of pro-inflammatory cytokines TNF-α and IL-6, and a significant upregulation of inflammatory chemokines [20]. Similarly, SARS-CoV-infected macrophages show delays in the secretion of pro-inflammatory cytokines [20]. The delayed but excessive production of these cytokines and chemokines is thought to induce a dysregulated innate immune response to SARS-CoV infection. High serum levels of pro-inflammatory cytokines and chemokines have been found in SARS patients with severe disease, compared to individuals with uncomplicated SARS infection [21, 22]. These studies imply that dysregulated and/or exaggerated cytokine and chemokine responses by SARS-CoV-infected airway epithelial cells, DCs, and macrophages could play an important role in SARS pathogenesis.
Similar to the case of SARS, MERS-CoV infection of human airway epithelial cells induces significant but delayed IFN and pro-inflammatory cytokine (IL-1β, IL-6, and IL-8) responses [23]. Interestingly, there was a significant upregulation in the expression level of IL-17 in MERS-CoV-infected patients [24]. Th cells, especially Th17 cells, produce the pro-inflammatory cytokine IL-17 via the signal transducer and activator of transcription-3 (STAT3) and NF-κB signaling pathways [25]. MERS-CoV infection promotes the secretion of Th17 cytokines, which can recruit neutrophils and monocytes to the site of infection or inflammation and lead to the activation of other downstream cytokine and chemokine cascades, such as IL-1, IL-6, TNF-α, transforming growth factor-beta (TGF-β), IL-8, and monocyte chemoattractant protein-1 (MCP-1) [26].
It seems that cytokine storm can initiate viral sepsis and inflammatory-induced lung injury that leads to other complications such as pneumonitis, ARDS, respiratory failure, shock, organ failure, and even death [27]. It has been reported that patients in the intensive care unit have higher plasma levels of many innate cytokines, IFN-γ-inducible protein 10 (IP-10), MCP-1, macrophage inflammatory protein-1a, and TNF-α, and these clinical features have an association with disease progression and severity [24].
Interferons
The effective innate immune response against viral infections relies heavily on the IFN type I responses and its downstream cascade that culminates in controlling viral replication and induction of efficient adaptive immune response [28]. To counter innate antiviral cytokine responses, SARS-CoV and MERS-CoV encode several structural and nonstructural proteins (NSPs) that antagonize antiviral immune responses. SARS-CoV encodes nsp1, nsp3-macrodomain, nsp3 deubiquitinase, as well as ORF3b, ORF6, and ORF9b subvert antiviral responses via antagonizing IFN and interferon-stimulated gene responses [29]. Additionally, structural proteins such as the membrane (M) and nucleocapsid (N) proteins dampen IFN signaling by inhibiting TBK1/IKKe [30, 31]. Similarly, MERS-CoV structural proteins M and N and accessory proteins ORF3, ORF4a, and ORF4b antagonize IFN responses [32, 33]. Structural and NSP antagonisms of IFN responses further amplify inflammatory responses by promoting unrestrained virus replication, resulting in an increased viral pathogen-associated molecular pattern that further dampens IFN signaling. The lack of IFN signaling also leads to an excessive accumulation of Ly6Clow monocytes and neutrophils [10].
Early evidences have demonstrated that SARS-CoV-2 is sensitive to IFN-I/III pretreatment in vitro, perhaps to a greater degree than SARS-CoV-1 [34, 35]. Moreover, IFN induced transmembrane family proteins inhibit SARS-CoV-2 entry, as demonstrated for SARS-CoV [36] although their action in promoting infection has been also described for other CoVs [37].
The results of several studies on testing antiviral treatments against SARS-CoV replication indicate that administration of IFN type I inhibits SARS-CoV growth in cell culture as well as viral replication in cynomolgus macaques and mouse models [38, 39]. In response to viral infections, mononuclear phagocytes induce IFN-I and IFN-III production, resulting in inflammasome activation, induction of pathogenic Th1 and Th17 cell responses, recruitment of effector immune cells, and cytokine release syndrome pathology [40]. A study conducted by Chu et al. [41] has demonstrated that monocyte-derived DCs (Mo-DCs) infected with MERS-CoV exhibit no expression of IFN-β despite the marginally early expression of IFN-α. However, another recent study has failed to stimulate the pro-inflammatory innate response and production of IFN type I in vitro in cultured infected cells, including primary human airway epithelial cells, and Mo-DCs infected with MERS-CoV [42]. The mechanisms behind this response may be initially related to interference with the NF-κB signaling pathway, which is usually responsible for the induction of pro-inflammatory responses [43]. The effect of applying IFN-α in MERS-CoV-infected cells has been 50–100 folds greater than that in SARS-CoV-infected cells [44].
Upregulation of IFN type I and interferon-stimulated genes is not observed until 2 days after infection. It has been reported that IFN deficiency does not exacerbate SARS-CoV disease in animals, while treatment with IFN type I was helpful in controlling SARS-CoV replication [29].
Evasion Mechanisms by CoV
CoVs have developed several mechanisms to inhibit IFN-I induction [45]. In order to prevent IFN release, CoV proteins can inhibit several steps of the signal transduction pathway that bridges the IFN-α receptor 1 subunit (IFNAR1) and IFNAR2 to the STAT proteins that activate transcription. In the case of SARS-CoV-1, these mechanisms include IFNAR1 degradation by ORF3a [46], decreased STAT1 phosphorylation by NSP1 [47], and antagonism of STAT1 nuclear translocation by ORF6 [48]. However, SARS-CoV-2 ORF6 shares only 69% sequence homology with SARS-CoV-1, suggesting that this function may not be conserved. In support of this notion, SARS-CoV-2 infection fails to limit STAT1 phosphorylation, unlike what happens in SARS-CoV-1 infection [49].
SARS-CoV-2 can evade by inhibiting the production of type I/III IFNs by the infected cells [50]. In fact, patients with severe COVID-19 show remarkably impaired IFN-I signatures, compared to mild or moderate cases [51]. CoV-mediated antagonism of innate immunity begins with evasion of PRR sensing. CoVs can avoid PRR activation through inhibiting recognition and antagonizing PRR action [52, 53]. Viral RNA is guanosine-capped and methylated at the five ends by CoVs NSPs 10, 13, 14, and 16 [52], thereby resembling host messenger RNA (mRNA) to promote translation, prevent degradation, and evade RIG-I like receptor sensing [53]. SARS-CoV-1 and SARS-CoV-2 ORF9b indirectly suppress mitochondrial antiviral signaling protein via its association with mitochondrial membrane (Tom) 70 [54].
Macrophages and DCs
Mucosal immune responses to infectious agents are orchestrated and regulated by myeloid cells with specialized functions, including conventional DCs (cDCs), Mo-DCs, plasmacytoid DCs, and macrophages [55]. A growing body of evidence highlights dysregulated myeloid responses that potentially drive the COVID-19 hallmark syndromes, such as ARDS, cytokine release syndrome, and lymphopenia [56].
MERS-CoV infects and replicates inside macrophages and subsequently induces the expression of major histocompatibility complex class I molecules (MHC-I), MHC-II, and stimulation-related genes [57]. Due to homeostasis, macrophages and DCs act as vehicles and seem to disseminate viruses through the efferent lymphatic system. Meanwhile, activation of DC and macrophage by SARS-CoV leads to excessive pro-inflammatory cytokine responses [58].
The studies performed on pulmonary tissues of patients with severe COVID-19 disease have revealed an expansion of inflammatory monocytes and Ficolin-1+ monocyte-derived macrophages at the expense of tissue-resident reparative alveolar macrophages [59]. Additionally, alternative macrophages can increase airway hypersensitivity, thus exacerbating SARS-associated fibrosis [60]. Some studies ascertain the role of lung-resident and recruited granulocytes in SARS-CoV-2 control and pathogenesis [61, 62]. In contrast to their early protective role, neutrophil NETosis and macrophage crosstalk can trigger later-stage inflammatory cascades [63], underscoring the overall pathogenic nature of damage-sensing host responses. Existing evidences reveal that high levels of macrophage CXCL10/IP-10 and CCL2/MCP-1 and neutrophil chemoattractant CXCL2 and CXCL8 facilitate the migration of these immune cells to the site of infection, which is consistent with infiltration of mononuclear cell in lung tissues of COVID-19 patients [64].
Innate Lymphoid Cells
Innate lymphoid cells (ILCs) are innate immune effector cells that lack the expression of rearranged antigen receptors, namely T-cell and B-cell receptor. The ILC family is divided into two main groups: the cytotoxic natural killer (NK) cells and the noncytotoxic helper ILCs, which include ILC1, ILC2, and ILC3 [65]. Conventional NK cells include CD56brightCD16+ NK cells and CD56dimCD16+ cells that are specialized in cytokine production or cytotoxicity, respectively.
NK Cells
Multiple studies have reported reduced numbers of NK cells in the peripheral blood of COVID-19 patients, which is associated with the severity of the disease [66, 67]. Although lung NK cells are susceptible to infection with the influenza virus, they do not express angiotensin-converting enzyme 2 (ACE2) and, therefore, are unlikely to be directly infected by SARS-CoV-2 [68]. However, frequencies of NK cells expressing CD16 and/or KIRs are decreased in the blood following SARS-CoV-2 and SARS-CoV infection, respectively [69]. In vitro, CXCR3 ligands (CXCL9-11) are increased in SARS-CoV-2-infected human lung tissue [70], and CXCR3-ligand-producing monocytes are expanded in the lungs of COVID-19 patients [59]. This suggests that the CXCR3 pathway might facilitate NK cell recruitment from the peripheral blood to the lungs in the COVID-19 patients. Interaction with virus antigen causes both cytokine production by NK cells and lysis of infected cells through antibody-mediated cellular cytotoxicity [71]. These findings suggest that triggering NK cell activation may contribute not only to the resolution of infection but also to the cytokine storm in ARDS. Ex vivo NK cells from peripheral blood of COVID-19 patients have reduced intracellular expression of CD107a, granulysin, and granzyme B, suggesting the impaired cytotoxicity and production of cytokines [72].
Adaptive Immune Responses
Adaptive immune responses are the key players against viral infections. CD4+ T cells facilitate virus-specific antibody production through T-dependent activation of B cells. However, CD8+ T cells are cytotoxic and kill virus-infected cells [73]. As the immune system cannot effectively control the virus in the acute phase (pneumonia phase), the patient state will become severe or critical type. It seems that T cells and B cells are further reduced, while inflammatory cytokines and D-Dimer continue to increase in the severe type patients [74].
Cell-Mediated Responses
When a virus is inhaled and infects respiratory epithelial cells, DCs phagocytose the virus and present antigens to T cells. Effector T cells function through killing the infected epithelial cells, and cytotoxic CD8+ T cells produce and release pro-inflammatory cytokines which induce cell apoptosis [75]. Both activated CD8+ cells and anti-MERS-CoV antibodies have been reported to be crucial for the clearance of the initial infection and protection against a subsequent challenge with the virus, respectively. This finding implies that the response to MERS-CoV generally occurs through antibody-mediated immunity. Hence, the antiviral effects of the depleted cells may be important during later infection time points, leading to the persistence of viral infection and promotion of viral survival. SARS-CoV triggers and amplifies the immune response. The exacerbation of cytokine production, excessive recruitment of immune cells, and the uncontrollable epithelial damage generate a vicious circle for infection-related ARDS [76]. Both CD4+ and CD8+ T cells isolated from human peripheral blood, tonsils, spleens, and lymphoid organs could be infected with MERS-CoV but not with SARS-CoV. This infection pattern might be attributed to the low expression of the SARS-CoV receptor, namely ACE2, in T cells [77].
Evidence strongly indicates that Th1 type response is a key for successful control of SARS-CoV and MERS-CoV and this is probably true for SARS-CoV-2, as well. It has been shown that the patients infected with SARS-CoV-2 also had high levels of IL-1, IFN-γ, IP-10, and MCP-1, probably leading to activated Th1 cell responses [15]. On the other hand, SARS-CoV-2 infection also initiated increased secretion of Th2 cytokines (e.g., IL-4 and IL-10) that suppress inflammation, a finding that differs from the case of SARS-CoV infection [78].
Flow cytometric analyses of PBMCs obtained from symptomatic COVID-19 patients have shown a significant influx of granulocyte-macrophage colony-stimulating factor-producing, activated CD4+ T cells and CD14+ HLA-DRlo monocytes [79]. Another study reported a significantly increased PBMC frequency of polyclonal granulocyte-macrophage colony-stimulating factor+ CD4+ T cells capable of prodigious ex vivo IL-6 and IFN-γ production in patients with severe COVID-19 [80].
Xu et al. [81] showed that peripheral blood of a patient with severe COVID-19 had a strikingly high number of CCR6+ Th17 cells, further supporting the occurrence of a Th17 type cytokine storm in this disease. Elevated Th17 responses or enhanced IL-17-related pathways are also observed in MERS-CoV and SARS-CoV patients [82]. In MERS-CoV patients, higher IL-17 with lower IFN-γ and IFN-α levels have a worse outcome than the reversed phenotype [83].
Additionally, a study has reported reduced frequencies of regulatory T cells (Treg cells) in severe COVID-19 cases [84]. Since Treg cells have been shown to help resolving ARDS inflammation in mouse models [85], the loss of Treg cells might facilitate the development of COVID-19 lung immunopathology [86].
In severe COVID-19, T cells seem to be more activated and may exhibit a trend toward exhaustion based on the continuous expression of inhibitory markers such as programmed death 1 (PD-1) and T-cell immunoglobulin-3 as well as an overall reduced activity and cytotoxicity. Conversely, recovering patients were found to have an increase in the count of follicular helper CD4+ T cells (TFH) as well as decreasing levels of inhibitory markers along with enhanced levels of effector molecules such as granzyme and perforin [87].
Since most epitopes identified for both viruses concentrate on the viral structural proteins, it will be informative to map the epitopes identified with SARS-CoV/MERS-CoV with those related to SARS-CoV-2. In SARS-CoV, lymphocyte epitopes were extensively mapped for the structural proteins, that is, S, N, M, and E proteins [88]. Although all SARS-CoV surface proteins, including S, M, E, and N proteins, were involved in T cell responses, S protein contributed to most of the T-cell recognition epitopes. In patients recovering from mild COVID-19, robust T cell responses specific for viral N, M, and S proteins detected by IFN-γ ELISPOT, were weakly correlated with neutralizing antibody concentrations (like convalescent SARS-CoV-1 patients) [89]. Identification of overlapping epitopes among the three viruses can be useful for designing a cross-reactive vaccine that provides protection against all three types of human CoV in the future [27].
In SARS-CoV survivors, the magnitude and frequency of specific CD8+ memory T cells exceeded that of CD4+ memory T cells, and virus-specific T cells persisted for at least 6–11 years, suggesting that T cells may confer long-term immunity [90]. Both virus-specific CD4+ and CD8+ T cells were detected in all patients at average frequencies of 1.4 and 1.3%, respectively, and very limited frequency of CD4+ T-cell central memory or CD8+ T-cell effector memory and effector memory RA cells. This study is notable for the use of large complementary peptide pools comprising 1,095 SARS-CoV-2 epitopes [91].
In the acute phase of SARS-CoV infection, rapid reduction of lymphocytes in peripheral blood [92], mainly T lymphocytes, is observed, and both CD4+ and CD8+ T lymphocytes are decreased. However, CD4+ T cells are more susceptible to infection. Depletion of CD4+ T cells is associated with reduced pulmonary recruitment of lymphocytes and neutralizing antibody and cytokine production, resulting in a strong immune-mediated interstitial pneumonitis and delayed clearance of SARS-CoV from lungs [93]. The loss of lymphocytes precedes even the abnormal changes on the chest X-ray [94]. After a 1-year follow-up of SARS patients, CD3+, CD4+, and CD8+ T cells recovered rapidly during the disease recovery period, and CD8+ T lymphocytes returned to normal within 2–3 months after onset. The memory CD4+ T cells returned to normal 1 year after onset, whereas other cell counts including total T lymphocytes, CD3+ cells, CD4+ cells, and naive CD4+ T cells were still lower than healthy controls [95].
It seems that lymphopenia in SARS and COVID-19 patients is more likely caused by cytokines such as IFN-I and TNF-a may inhibit T-cell recirculation in blood by promoting retention in lymphoid organs and attachment to the endothelium [96] or endogenous or exogenous glucocorticoids which ultimately led to apoptosis of lymphocytes, rather than direct viral infection of these cells [97, 98].
Antibody-Mediated Responses
The antibody-mediated humoral response is crucial for preventing viral infections. A subset of these antibodies, which reduce viral infectivity by binding to the surface epitopes of viral particles and thereby blocking the entry of the virus to the infected cell, is defined as neutralizing antibodies [99].
Virus neutralizing antibodies induced by vaccines or infected viruses play vital roles in controlling viral infection [100]. They target S1-RBD, S1-NTD, or the S2 region, blocking the binding of RBDs to their respective receptors and interfering with S2-mediated membrane fusion or entry into the host cell, thus inhibiting viral infections [101]. Most of them target the RBD, while a few of them target regions in the S2 subunit or the S1/S2 proteolytic cleavage site [102].
It has been reported that neutralizing antibodies decline within 2–3 months in COVID-19 recovered patients. One mathematical model has also suggested shortly durable immunity [103]. The durability of neutralizing antibodies in other human CoV may be relevant for comparison. Among the seven pathogenic CoVs of human beings, HCoV-229E, HCoV-NL63, HCoVOC43, and HCoV-HKU1 cause mild disease (common cold), whereas SARS-CoV, MERS-CoV, and SARS-CoV-2 are highly pathogenic. Antibody titers lack longevity and wane substantially 1 year after infection in common cold CoV; 3 years in SARS-CoV; and persist for 2 years after recovery from severe MERS-CoV infection [104]. As SARS-CoV-2 infection has usually asymptomatic or mild clinical presentation, like common cold CoV, rapidly waning antibody responses following primary infection or immunization (compared to severe cases) may allow susceptibility to reinfection. The secretory IgA as protective neutralizing antibody against SARS-CoV-2 should also be explored because mucosal immunity provides protection through intranasal immunization against closely related SARS-CoV and MERS-CoV [105]. In a study on 175 COVID-19 recovered patients with mild symptoms, SARS-CoV-2-specific neutralizing antibodies were detected at the convalescent phase of infection from days 10 to 15 after the onset of the disease and remained thereafter. The titers of neutralizing antibodies were variable in different patients. Plasma neutralizing antibody titers in elderly and middle-aged patients were significantly higher. Neutralizing antibody titers were correlated positively with CRP levels and negatively with the lymphocyte counts of patients. It could be suggested that other immune responses, including T cells or cytokines, may contribute to the recovery of these patients [106].
Neutralizing IgGs against SARS-CoV reached a peak in serum during the convalescent phase and diminished after recovery [107]. A previous study has shown that antibodies from some recovered SARS-CoV-2 patients might cross-react or neutralize SARS-CoV from other patients [108]. Like SARS-CoV-1 infection [109], seroconversion occurs in most COVID-19 patients between 7 and 14 days after the onset of symptoms, and antibody titers persist in the weeks following virus clearance [110].
It seems that antibodies binding the SARS-CoV-2 internal N protein and the external S glycoprotein are commonly detected [111]. The S protein is highly immunogenic, and specific antibodies against the RBD can neutralize and block virus interactions with ACE2 as the host entry receptor [112]. SARS-CoV-2 S protein can bind to ACE2 with a higher affinity compared to SARS-CoV S [113]. The high affinity of the S protein for human ACE2 may lead to a great human-to-human transmission of SARS-CoV-2. Due to the key role of the S protein, it is the main target for antibody-mediated neutralization [12].
In a previous study, RBD-specific CD19+ IgG+ memory B cells were single-cell sorted from COVID-19 donors between days 9 and 28 after the onset of symptoms. From their antibody gene sequences, many SARS-CoV-2-specific monoclonal antibodies were produced. The monoclonal antibodies had a diverse repertoire, relatively low or no somatic mutations, and variable binding reactivity, with dissociation constants reaching 10−8 to 10−9, like antibodies isolated during acute infections. In addition, two potent neutralizing SARS-CoV-2 RBD-specific monoclonal antibodies were characterized that did not cross-react with the RBD of SARS-CoV-1 or MERS-CoV [114]. Together, these results demonstrate that antibody-mediated neutralization is virus-specific and likely driven by the binding of epitopes within the RBD.
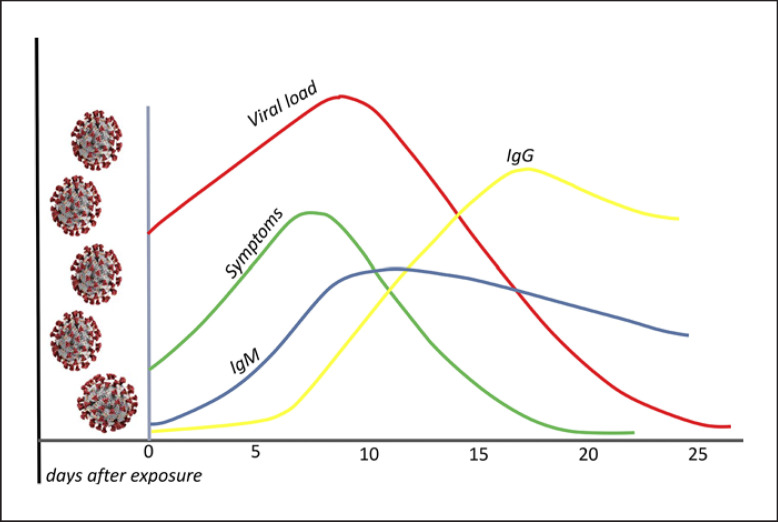
It was demonstrated that the IgM response to SARS-CoV-2 occurred and was maximized before the IgG antibody response. Furthermore, the IgM antibody response began to decline at week 3 of the illness (Fig. 3), while the IgG antibody response persisted and was maintained in patients with COVID-19. Severe cases of COVID-19 tended to have a more vigorous response in both IgG and IgM antibodies to COVID-19 illness. Importantly, the timing of IgM and IgG antibody occurrence in patients varies greatly, and this variation in timing may be associated with age as well as a comorbidity [115].
Fig. 3.
The time kinetics between viral load, symptoms, and host immunoglobulins (IgM, IgG) in COVID-19. The onset of symptoms is usually 5 days after infection. Seroconversion may usually be detectable between 5–7 and 14 days after the onset of symptoms. Viral RNA is inversely correlated with neutralizing antibody titers. Higher titers have been observed in critically ill patients. The humoral response in SARS-CoV-1 is relatively short lived, altogether, suggesting that immunity with SARS-CoV-2 may reduce 1–2 years after primary infection. SARS-CoV-2, Severe acute respiratory syndrome coronavirus-2.
Vaccine Candidates against SARS-CoV-2
Since the emergence of SARS-CoV-2, the scientific community has been working restlessly to find both short-term therapeutic approaches and a long-term vaccine solution to reduce spread and curb COVID-19 morbidity and mortality. The three main criteria which should be taken into consideration while developing a vaccine are speed, scale-up manufacturing, and global access. The astonishing efforts by researchers across the globe in terms of scale and speed of vaccine development have fastened the vaccine development journey from bench to bedside within a few months only. Along with speeding up the development process, it is equally important to evaluate the effectiveness and safety of vaccine at each step, and this has been the major hurdle for researchers in establishing the vaccine's efficacy so far.
Live-Attenuated Vaccines
The most common traditional method which involves manually weakened live pathogen which is no longer able to induce infection but able to induce immune response and, hence, mimic features of natural infection. It is capable of inducing both humoral and cellular immune responses. Live-attenuated vaccines on intranasal administration induce secretion of IgA and, hence, provide local mucosal immunity [116]. These vaccines are popular to induce strong lifelong immune responses within 2 doses. These are easy to produce for some viruses but challenging for complex pathogens. Codagenix Biotec Inc., collaboration with the Serum Institute of India Ltd., developing a live-attenuated SARS-CoV-2 vaccine in which the sequence of the target gene of interest has been changed by swapping its optimized codons with nonoptimized ones [117].
Nucleic Acid Vaccines
Plasmid-Based DNA Vaccines
DNA vaccine eliminates the need for using live viruses hence having a better safety profile. The manufacturing process of plasmid DNA is relatively straightforward, and the double-strand DNA molecules are more stable than virus, protein, and mRNA and can be freeze-dried for long-term storage. The main prohibitory factor for the plasmid DNA vaccine is the low transfection efficacy, requiring transfection modalities. For example, the Inovio's COVID-19 vaccine candidate, INO-4800, uses a handheld electroporation device, CELLECTRA [118]. The vaccine is injected intradermally along with electrodes, and then an electric pulse is applied to open the cell membrane, allowing the plasmid to enter the cells.
mRNA Vaccines
mRNA is an emerging, noninfectious, and a nonintegrating platform with almost no potential risk of insertional mutagenesis. The immunogenicity of the mRNA can be minimized, and alterations can be made to increase the stability of these vaccines. Furthermore, the anti-vector immunity is also avoided as the mRNA is the minimally immunogenic genetic vector, allowing repeated administration of the vaccine [119]. This platform has empowered the rapid vaccine development program due to its flexibility and ability to mimic the antigen structure and expression as seen in the course of a natural infection [120].
mRNA-1273 (Moderna TX, Inc.) is a vaccine composed of synthetic mRNA encapsulated in lipid nanoparticle (LNP) which codes for the full-length, pre-fusion stabilized S protein of SARS-CoV-2. It has the potential to elicit a highly S protein-specific antiviral response. Furthermore, it is considered to be relatively safe as it is neither made up of the inactivated pathogen nor the subunits of the live pathogen [121]. The vaccine has got a fast-track approval from FDA. BNT162b1 (BioNTech/FosunPharma/Pfizer) is another codon-optimized mRNA vaccine that encodes for the trimerized SARS-CoV-2 RBD, a critical target of the virus nAb. The vaccine portrays an increased immunogenicity due to the addition of T4 fibritin-derived fold on trimerization domain to the RBD antigen. The mRNA is encapsulated in 80 nm ionizable cationic LNPs, which ensures its efficient delivery [121]. Both mRNA vaccines use a lipid-based nanoparticle (LNP) carrier system, which also acts as an adjuvant. The LNPs are stabilized with polyethylene glycol, prolonging their lifespan. Scientists speculate that these allergic reactions might be related to either the lipid or the polyethylene glycol component of these vaccines [122].
Protein Subunit Vaccines
Subunit vaccines primarily induce CD4+ Th cell and antibody responses. Therefore, most of these vaccines contain full-length SARS-CoV-2 S protein that induces neutralizing antibodies, similarly to the majority of SARS and MERS vaccines, which had differing levels of efficacy [123, 124, 125]. Proteins or peptides alone are poorly immunogenic and generally require not only an adjuvant but also repeated administration, and they are poor activators of CD8+ T cell responses. Furthermore, this platform is generally unsuitable for respiratory mucosal vaccination [126]. In this regard, subunit COVID-19 vaccines being developed by GlaxoSmithKline and Novavax use AS03 and Matrix-M adjuvants, respectively [127].
Virus-Like Particles
These are protein multimers mimicking the structure of real virus but lacking genetic material and hence are noninfectious in nature. Virus-like particles (VLPs) act by stimulating antigen-presenting cells mediated activation of B- and T-cell immune responses. These are also involved in CD8+ cytotoxic T-cell mediated killing of pathogenic cells. The immune system recognizes VLPs in the same way as it recognizes original virus and thereby induces immune responses [128]. VLP formulations because of their poor immunogenicity require adjuvants in most of the cases. VLP-based vaccines are well-established platform for prophylactic use. These are less time taking and production cost depends upon the expression system used which is comparatively low for bacterial system than the mammalian expression system. The licensed vaccines based on this platform are currently in use for human papillomavirus [129]. Currently, there are two COVID-19 vaccine candidates developed as VLPs in clinical evaluation and COVID-19 vaccine candidates in preclinical evaluation stage developed [130].
Conclusion
The outbreak of COVID-19 caused by the novel virus SARS-CoV-2 started at the end of December 2019. In less than 2 months, it spread in many countries around the world. The rapid spread of SARS-CoV-2 and the unprecedented nature of COVID-19 have demanded urgency in basic science, clinical research, and vaccine strategies, and the scientific community has met that call with remarkable productivity. Within months, there has been a significant generation of scientific knowledge that has shed some light on the immunology of SARS-CoV-2 infections.
It is imperative that immune responses against SARS-CoV-2 and its immunopathological mechanisms are further elucidated to better define therapeutic strategies for COVID-19. Since SARS-CoV-2 is very similar to SARS-CoV and MERS-CoV and the symptoms are also similar between COVID-19, SARS, and MERS, the outbreak of COVID-19 has created a sense of SARS and MERS recurring. However, there are some remarkable differences between these CoVs, which are essential for containing the epidemic and treating the patients.
Conflict of Interest Statement
The authors have no conflicts of interest to declare.
Funding Sources
The authors did not receive any funding.
Author Contributions
Rasoul Baharlou participated in the study design and drafted the manuscript. Arefe Vafaeinezhad was responsible for writing the manuscript. Mohammad Reza Atashzar participated in the figure design of the paper.
References
- 1.de Wit E, van Doremalen N, Falzarano D, Munster VJ. SARS and MERS recent insights into emerging coronaviruses. Nat Rev Microbiol. 2016 Aug;14((8)):523–34. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019 Mar;17((3)):181–92. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang C, Horby PW, Hayden FG, Gao GF. A novel coronavirus outbreak of global health concern. Lancet. 2020 Feb 15;395((10223)):470–3. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health, Organization. Coronavirus disease 2019 (COVID-19) situation report 57. Geneva, Switzerland: WHO. 2020 [Google Scholar]
- 5.Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, et al. Early transmission dynamics in Wuhan of novel oronavirus-infected pneumonia. N Engl J Med. 2020 Mar 26;382((13)):1199–207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fan Y, Zhao K, Shi ZL, Zhou P. Bat coronaviruses in China. Viruses. 2019 Mar 2;11((3)):210. doi: 10.3390/v11030210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rehman SU, Shafique L, Ihsan A, Liu Q. Evolutionary trajectory for the emergence of novel coronavirus SARS-CoV-2. Pathogens. 2020 Mar 23;9((3)):240. doi: 10.3390/pathogens9030240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hui EK. Reasons for the increase in emerging and re-emerging viral infectious diseases. Microbes Infect. 2006 Mar;8((3)):905–16. doi: 10.1016/j.micinf.2005.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ren LL, Wang YM, Wu ZQ, Xiang ZC, Guo L, Xu T, et al. Identification of a novel coronavirus causing severe pneumonia in human a descriptive study. Chin Med J. 2020 May5;133((9)):1015–24. doi: 10.1097/CM9.0000000000000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Channappanavar R, Perlman S. Pathogenic human coronavirus infections causes and consequences of cytokine storm and immunopathology. Semin Immunopathol. 2017 Jul;39((5)):529–39. doi: 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu J, Zheng X, Tong Q, Li W, Wang B, Sutter K, et al. Overlapping and discrete aspects of the pathology and pathogenesis of the emerging human pathogenic coronaviruses SARS-CoV and 2019-nCoV. J Med Virol. 2020 May;92((5)):491–4. doi: 10.1002/jmv.25709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou G, Zhao Q. Perspectives on therapeutic neutralizing antibodies against the novel coronavirus SARS-CoV-2. Int J Biol Sci. 2020;16((10)):1718–23. doi: 10.7150/ijbs.45123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y, Yang Y, Zhang C, Huang F, Wang F, Yuan J, et al. Clinical and biochemical indexes from 2019-nCoV infected patients linked to viral loads and lung injury. Sci China Life Sci. 2020 Mar;63((3)):364–74. doi: 10.1007/s11427-020-1643-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan China. JAMA. 2020 Feb 7;323((11)):1061–9. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan China. Lancet. 2020 Feb 15;395((10223)):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Channappanavar R, Fehr AR, Vijay R, Mack M, Zhao J, Meyerholz DK, et al. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe. 2016 Feb10;19((2)):181–93. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Griffith JW, Sokol CL, Luster AD. Chemokines and chemokine receptors positioning cells for host defense and immunity. Annu Rev Immunol. 2014;32:659–702. doi: 10.1146/annurev-immunol-032713-120145. [DOI] [PubMed] [Google Scholar]
- 18.Fehr AR, Channappanavar R, Perlman S. Middle east respiratory syndrome emergence of a pathogenic human coronavirus. Annu Rev Med. 2017 Jan 14;68:387–99. doi: 10.1146/annurev-med-051215-031152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen L, Liu HG, Liu W, Liu J, Liu K, Shang J, et al. [Analysis of clinical features of 29 patients with 2019 novel coronavirus pneumonia]. Zhonghua Jie He He Hu Xi Za Zhi. 2020 Mar12;43((3)):203–8. doi: 10.3760/cma.j.issn.1001-0939.2020.03.013. [DOI] [PubMed] [Google Scholar]
- 20.Law HK, Cheung CY, Ng HY, Sia SF, Chan YO, Luk W, et al. Chemokine up-regulation in SARS-coronavirus-infected monocyte-derived human dendritic cells. Blood. 2005 Oct 1;106((7)):2366–74. doi: 10.1182/blood-2004-10-4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang CH, Liu CY, Wan YL, Chou CL, Huang KH, Lin HC, et al. Persistence of lung inflammation and lung cytokines with high-resolution CT abnormalities during recovery from SARS. Respir Res. 2005 May11;6((1)):42. doi: 10.1186/1465-9921-6-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chien JY, Hsueh PR, Cheng WC, Yu CJ, Yang PC. Temporal changes in cytokine/chemokine profiles and pulmonary involvement in severe acute respiratory syndrome. Respirology. 2006 Nov;11((6)):715–22. doi: 10.1111/j.1440-1843.2006.00942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lau SK, Lau CC, Chan KH, Li CP, Chen H, Jin DY, et al. Delayed induction of proinflammatory cytokines and suppression of innate antiviral response by the novel middle east respiratory syndrome coronavirus implications for pathogenesis and treatment. J Gen Virol. 2013 Dec;94((Pt 12)):2679–90. doi: 10.1099/vir.0.055533-0. [DOI] [PubMed] [Google Scholar]
- 24.Mahallawi WH, Khabour OF, Zhang Q, Makhdoum HM, Suliman BA. MERS-CoV infection in humans is associated with a pro-inflammatory Th1 and Th17 cytokine profile. Cytokine. 2018 Apr;104:8–13. doi: 10.1016/j.cyto.2018.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manni ML, Robinson KM, Alcorn JF. A tale of two cytokines IL-17 and IL-22 in asthma and infection. Expert Rev Respir Med. 2014 Feb;8((1)):25–42. doi: 10.1586/17476348.2014.854167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin W, Dong C. IL-17 cytokines in immunity and inflammation. Emerg Microbes Infect. 2013 Sep;2((9)):e60.. doi: 10.1038/emi.2013.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prompetchara E, Ketloy C, Palaga T. Immune responses in COVID-19 and potential vaccines lessons learned from SARS and MERS epidemic. Asian Pac J Allergy Immunol. 2020 Mar;38((1)):1–9. doi: 10.12932/AP-200220-0772. [DOI] [PubMed] [Google Scholar]
- 28.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020 Mar;579((7798)):270–3. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Totura AL, Baric RS. SARS coronavirus pathogenesis host innate immune responses and viral antagonism of interferon. Curr Opin Virol. 2012 Jun;2((3)):264–75. doi: 10.1016/j.coviro.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frieman M, Ratia K, Johnston RE, Mesecar AD, Baric RS. Severe acute respiratory syndrome coronavirus papain-like protease ubiquitin-like domain and catalytic domain regulate antagonism of IRF3 and NF-kappaB signaling. J Virol. 2009 Jul;83((13)):6689–705. doi: 10.1128/JVI.02220-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siu KL, Chan CP, Kok KH, Chiu-Yat Woo P, Jin DY. Suppression of innate antiviral response by severe acute respiratory syndrome coronavirus M protein is mediated through the first transmembrane domain. Cell Mol Immunol. 2014 Mar;11((2)):141–9. doi: 10.1038/cmi.2013.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Y, Zhang L, Geng H, Deng Y, Huang B, Guo Y, et al. The structural and accessory proteins M ORF 4a ORF 4b and ORF 5 of middle east respiratory syndrome coronavirus (MERS-CoV) are potent interferon antagonists. Protein Cell. 2013 Dec;4((12)):951–61. doi: 10.1007/s13238-013-3096-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lui PY, Wong LY, Fung CL, Siu KL, Yeung ML, Yuen KS, et al. Middle east respiratory syndrome coronavirus M protein suppresses type I interferon expression through the inhibition of TBK1-dependent phosphorylation of IRF3. Emerg Microbes Infect. 2016 Apr20;5((4)):e39. doi: 10.1038/emi.2016.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mantlo E, Bukreyeva N, Maruyama J, Paessler S, Huang C. Antiviral activities of type I interferons to SARS-CoV-2 infection. Antiviral Res. 2020 Jul 1;179:104811. doi: 10.1016/j.antiviral.2020.104811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stanifer ML, Kee C, Cortese M, Triana S, Mukenhirn M, Kraeusslich H-G, et al. Critical role of type III interferon in controlling SARS-CoV-2 infection, replication and spread in primary human intestinal epithelial cells. bioRxiv. 2020 doi: 10.1016/j.celrep.2020.107863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang IC, Bailey CC, Weyer JL, Radoshitzky , SR, Becker MM, Chiang JJ, et al. Distinct patterns of IFITM-mediated restriction of filoviruses SARS coronavirus and influenza A virus. PLoS Pathog. 2011 Jan 6;7((1)):e1001258. doi: 10.1371/journal.ppat.1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao X, Sehgal M, Hou Z, Cheng J, Shu S, Wu S, et al. Identification of residues controlling restriction versus enhancing activities of IFITM proteins on entry of human coronaviruses. J Virol. 2018;92((6)):e01535–17. doi: 10.1128/JVI.01535-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dahl H, Linde A, Strannegård O. In vitro inhibition of SARS virus replication by human interferons. Scand J Infect Dis. 2004;36((11–2)):829–31. doi: 10.1080/00365540410021144. [DOI] [PubMed] [Google Scholar]
- 39.Kumaki Y, Ennis J, Rahbar R, Turner JD, Wandersee MK, Smith AJ, et al. Single-dose intranasal administration with mDEF201 (adenovirus vectored mouse interferon-alpha) confers protection from mortality in a lethal SARS-CoV BALB/c mouse model. Antiviral Res. 2011 Jan;89((1)):75–82. doi: 10.1016/j.antiviral.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prokunina-Olsson L, Alphonse N, Dickenson RE, Durbin JE, Glenn JS, Hartmann R, et al. COVID-19 and emerging viral infections the case for interferon lambda. J Exp Med. 2020;217((5)):e20200653. doi: 10.1084/jem.20200653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chu H, Zhou J, Wong BH, Li C, Cheng ZS, Lin X, et al. Productive replication of middle east respiratory syndrome coronavirus in monocyte-derived dendritic cells modulates innate immune response. Virology. 2014 Apr;5:197–205. doi: 10.1016/j.virol.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zielecki F, Weber M, Eickmann M, Spiegelberg L, Zaki AM, Matrosovich M, et al. Human cell tropism and innate immune system interactions of human respiratory coronavirus EMC compared to those of severe acute respiratory syndrome coronavirus. J Virol. 2013 May;87((9)):5300–4. doi: 10.1128/JVI.03496-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Canton J, Fehr AR, Fernandez-Delgado R, Gutierrez-Alvarez FJ, Sanchez-Aparicio MT, García-Sastre A, et al. MERS-CoV 4b protein interferes with the NF-κB-dependent innate immune response during infection. PLoS Pathog. 2018 Jan;14((1)):e1006838. doi: 10.1371/journal.ppat.1006838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Wilde AH, Raj VS, Oudshoorn D, Bestebroer TM, van Nieuwkoop S, Limpens RW, et al. MERS-coronavirus replication induces severe in vitro cytopathology and is strongly inhibited by cyclosporin A or interferon-α treatment. J Gen Virol. 2013 Aug;94((Pt 8)):1749–60. doi: 10.1099/vir.0.052910-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cameron MJ, Kelvin AA, Leon AJ, Cameron CM, Ran L, Xu L, et al. Lack of innate interferon responses during SARS coronavirus infection in a vaccination and reinfection ferret model. PLoS One. 2012;7((9)):e45842. doi: 10.1371/journal.pone.0045842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Minakshi R, Padhan K, Rani M, Khan N, Ahmad F, Jameel S. The SARS coronavirus 3a protein causes endoplasmic reticulum stress and induces ligand-independent downregulation of the type 1 interferon receptor. PLoS One. 2009 Dec 17;4((12)):e8342. doi: 10.1371/journal.pone.0008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wathelet MG, Orr M, Frieman MB, Baric RS. Severe acute respiratory syndrome coronavirus evades antiviral signaling role of nsp1 and rational design of an attenuated strain. J Virol. 2007 Nov;81((21)):11620–33. doi: 10.1128/JVI.00702-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kopecky-Bromberg SA, Martínez-Sobrido L, Frieman M, Baric RA, Palese P. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6 and nucleocapsid. proteins function as interferon antagonists. J Virol. 2007 Jan;81((2)):548–57. doi: 10.1128/JVI.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lokugamage KG, Hage A, de Vries M, Valero-Jimenez AM, Schindewolf C, Dittmann M, et al. Type I interferon susceptibility distinguishes SARS-CoV-2 from SARS-CoV. bioRxiv. 2020 doi: 10.1128/JVI.01410-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blanco-Melo D, Nilsson-Payant BE, Liu WC, Uhl S, Hoagland D, Møller R, et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020 May28;181((5)):1036–45. doi: 10.1016/j.cell.2020.04.026. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hadjadj J, Yatim N, Barnabei L, Corneau A, Boussier J, Smith N, et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020 Aug7;369((6504)):718–24. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bouvet M, Debarnot C, Imbert I, Selisko B, Snijder EJ, Canard B, et al. In vitro reconstitution of SARS-coronavirus mRNA cap methylation. PLoS Pathog. 2010 Apr22;6((4)):e1000863. doi: 10.1371/journal.ppat.1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deng X, Hackbart M, Mettelman RC, O'Brien A, Mielech AM, Yi G, et al. Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc Natl Acad Sci U S A. 2017;114((21)):E4251–60. doi: 10.1073/pnas.1618310114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM, et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020 Jul1;583((7816)):459–68. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guilliams M, Lambrecht BN, Hammad H. Division of labor between lung dendritic cells and macrophages in the defense against pulmonary infections. Mucosal Immunol. 2013 May 1;6((3)):464–73. doi: 10.1038/mi.2013.14. [DOI] [PubMed] [Google Scholar]
- 56.Mehta P, McAuley DF, Brown M, Sanchez E, Tattersall RS, Manson JJ. COVID-19 consider cytokine storm syndromes and immunosuppression. Lancet. 2020 Mar 28;395((10229)):1033–4. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou J, Chu H, Li C, Wong BH, Cheng ZS, Poon VK, et al. Active replication of middle east respiratory syndrome coronavirus and aberrant induction of inflammatory cytokines and chemokines in human macrophages implications for pathogenesis. J Infect Dis. 2014 May1;209((9)):1331–42. doi: 10.1093/infdis/jit504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tseng CT, Perrone LA, Zhu H, Makino S, Peters CJ. Severe acute respiratory syndrome and the innate immune responses modulation of effector cell function without productive infection. J Immunol. 2005 Jun 15;174((12)):7977–85. doi: 10.4049/jimmunol.174.12.7977. [DOI] [PubMed] [Google Scholar]
- 59.Liao M, Liu Y, Yuan J, Wen Y, Xu G, Zhao J, et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med. 2020 Jun1;26((6)):842–4. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 60.Page C, Goicochea L, Matthews K, Zhang Y, Klover P, Holtzman MJ, et al. Induction of alternatively activated macrophages enhances pathogenesis during severe acute respiratory syndrome coronavirus infection. J Virol. 2012 Dec;86((24)):13334–49. doi: 10.1128/JVI.01689-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Camp JV, Jonsson CB. A role for neutrophils in viral respiratory disease. Front Immunol. 2017;8:550. doi: 10.3389/fimmu.2017.00550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Flores-Torres AS, Salinas-Carmona MC, Salinas E, Rosas-Taraco AG. Eosinophils and respiratory viruses. Viral Immunol. 2019;32((5)):198–207. doi: 10.1089/vim.2018.0150. [DOI] [PubMed] [Google Scholar]
- 63.Barnes BJ, Adrover JM, Baxter-Stoltzfus A, Borczuk A, Cools-Lartigue J, Crawford JM, et al. Targeting potential drivers of COVID-19 neutrophil extracellular traps. J Exp Med. 2020;217((6)):e20200652. doi: 10.1084/jem.20200652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu Z, Shi L, Wang Y, Zhang J, Huang L, Zhang C, et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8((4)):420–2. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vivier E, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, et al. Innate lymphoid cells 10 years on. Cell. 2018 Aug23;174((5)):1054–66. doi: 10.1016/j.cell.2018.07.017. [DOI] [PubMed] [Google Scholar]
- 66.Song C-Y, Xu J, He J-Q, Lu Y-Q. COVID-19 early warning score a multi-parameter screening tool to identify highly suspected patients. medRxiv. 2020 [Google Scholar]
- 67.Yu L, Tong Y, Shen G, Fu A, Lai Y, Zhou X, et al. Immunodepletion with hypoxemia a potential high risk subtype of coronavirus disease. medRxiv. 2019 [Google Scholar]
- 68.Travaglini KJ, Nabhan AN, Penland L, Sinha R, Gillich A, Sit RV, et al. A molecular cell atlas of the human lung from single cell RNA sequencing. bioRxiv. 2020 doi: 10.1038/s41586-020-2922-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang F, Nie J, Wang H, Zhao Q, Xiong Y, Deng L, et al. Characteristics of peripheral lymphocyte subset alteration in COVID-19 pneumonia. J Infect Dis. 2020;221((11)):1762–9. doi: 10.1093/infdis/jiaa150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chu H, Chan JF, Wang Y, Yuen TT, Chai Y, Hou Y, et al. Comparative replication and immune activation profiles of SARS-CoV-2 and SARS-CoV in human lungs an ex vivo study with implications for the pathogenesis of COVID-19. Clin Infect Dis. 2020;71((6)):1400–9. doi: 10.1093/cid/ciaa410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Von Holle TA, Moody MA. Influenza and antibody-dependent cellular cytotoxicity. Front Immunol. 2019;10:1457. doi: 10.3389/fimmu.2019.01457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wilk AJ, Rustagi A, Zhao NQ, Roque J, Martínez-Colón GJ, McKechnie JL, et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat Med. 2020 Jul;26((7)):1070–6. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xu X, Gao X. Immunological responses against SARS-coronavirus infection in humans. Cell Mol Immunol. 2004 Apr;1((2)):119–22. [PubMed] [Google Scholar]
- 74.Lin L, Lu L, Cao W, Li T. Hypothesis for potential pathogenesis of SARS-CoV-2 infection-a review of immune changes in patients with viral pneumonia. Emerg Microbes Infect. 2020 Dec;9((1)):727–32. doi: 10.1080/22221751.2020.1746199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rogers MC, Williams JV. Quis custodiet ipsos custodes? regulation of cell-mediated immune responses following viral lung infections. Annu Rev Virol. 2018 Sep 29;5((1)):363–83. doi: 10.1146/annurev-virology-092917-043515. [DOI] [PubMed] [Google Scholar]
- 76.Yang CY, Chen CS, Yiang GT, Cheng YL, Yong SB, Wu MY, et al. New insights into the immune molecular regulation of the pathogenesis of acute respiratory distress syndrome. Int J Mol Sci. 2018 Feb16;19((2)):588. doi: 10.3390/ijms19020588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ying T, Li W, Dimitrov DS. Discovery of T-cell infection and apoptosis by middle east respiratory syndrome coronavirus. J Infect Dis. 2016 Mar 15;213((6)):877–9. doi: 10.1093/infdis/jiv381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wong CK, Lam CW, Wu AK, Ip WK, Lee NL, Chan IH, et al. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin Exp Immunol. 2004 Apr;136((1)):95–103. doi: 10.1111/j.1365-2249.2004.02415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhou Y, Fu B, Zheng X, Wang D, Zhao C, Qi Y, et al. Aberrant pathogenic GM-CSF+ T cells and inflammatory CD14+CD16+ monocytes in severe pulmonary syndrome patients of a new coronavirus. bioRxiv. 2020 [Google Scholar]
- 80.Zhou Y, Fu B, Zheng X, Wang D, Zhao C, Qi Y, et al. Pathogenic T-cells and inflammatory monocytes incite inflammatory storms in severe COVID-19 patients. Natl Sci Rev. 2020;7((6)):998–1002. doi: 10.1093/nsr/nwaa041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xu Z, Shi L, Wang Y, Zhang J, Huang L, Zhang C, et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020 Apr;8((4)):420–2. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Josset L, Menachery VD, Gralinski LE, Agnihothram S, Sova P, Carter VS, et al. Cell host response to infection with novel human coronavirus EMC predicts potential antivirals and important differences with SARS coronavirus. mBio. 2013 Apr30;4((3)):e00165–e13. doi: 10.1128/mBio.00165-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Faure E, Poissy J, Goffard A, Fournier C, Kipnis E, Titecat M, et al. Distinct immune response in two MERS-CoV-infected patients can we go from bench to bedside? PLoS One. 2014;9((2)):e88716. doi: 10.1371/journal.pone.0088716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Qin C, Zhou L, Hu Z, Zhang S, Yang S, Tao Y, et al. Dysregulation of immune response in patients with coronavirus 2019 (COVID-19) in Wuhan China. Clin Infect Dis. 2020;71((15)):762–8. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Walter JM, Helmin KA, Abdala-Valencia H, Wunderink RG, Singer BD. Multidimensional assessment of alveolar T cells in critically ill patients. JCI Insight. 2018;3((17)):e123287. doi: 10.1172/jci.insight.123287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dong P, Ju X, Yan Y, Zhang S, Cai M, Wang H, et al. γδ T cells provide protective function in highly pathogenic avian H5N1 influenza A virus infection. Front Immunol. 2018;9:2812. doi: 10.3389/fimmu.2018.02812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Thevarajan I, Nguyen THO, Koutsakos M, Druce J, Caly L, van de Sandt CE, et al. Breadth of concomitant immune responses prior to patient recovery a case report of non-severe COVID-19. Nat Med. 2020 Apr1;26((4)):453–5. doi: 10.1038/s41591-020-0819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu WJ, Zhao M, Liu K, Xu K, Wong G, Tan W, et al. T-cell immunity of SARS-CoV implications for vaccine development against MERS-CoV. Antiviral Res. 2017 Jan;137:82–92. doi: 10.1016/j.antiviral.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li CK, Wu H, Yan H, Ma S, Wang L, Zhang M, et al. T cell responses to whole SARS coronavirus in humans. J Immunol. 2008 Oct15;181((8)):5490–500. doi: 10.4049/jimmunol.181.8.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tang N, Li D, Wang X, Sun Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J Thromb Haemost. 2020 Apr;18((4)):844–7. doi: 10.1111/jth.14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Weiskopf D, Schmitz KS, Raadsen MP, Grifoni A, Okba NMA, Endeman H, et al. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci Immunol. 2020 Jun26;5((48)):5. doi: 10.1126/sciimmunol.abd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li T, Qiu Z, Zhang L, Han Y, He W, Liu Z, et al. Significant changes of peripheral T lymphocyte subsets in patients with severe acute respiratory syndrome. J Infect Dis. 2004 Feb15;189((4)):648–51. doi: 10.1086/381535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen J, Lau YF, Lamirande EW, Paddock CD, Bartlett JH, Zaki , SR, et al. Cellular immune responses to severe acute respiratory syndrome coronavirus (SARS-CoV) infection in senescent BALB/c mice CD4+ T cells are important in control of SARS-CoV infection. J Virol. 2010;84((3)):1289–301. doi: 10.1128/JVI.01281-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li T, Qiu Z, Han Y, Wang Z, Fan H, Lu W, et al. Rapid loss of both CD4+ and CD8+ T lymphocyte subsets during the acute phase of severe acute respiratory syndrome. Chin Med J. 2003 Jul;116((7)):985–7. [PubMed] [Google Scholar]
- 95.Xie J, Fan HW, Li TS, Qiu ZF, Han Y. [Dynamic changes of T lymphocyte subsets in the long-term follow-up of severe acute respiratory syndrome patients]. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2006 Apr;28((2)):253–5. [PubMed] [Google Scholar]
- 96.Kamphuis E, Junt T, Waibler Z, Forster R, Kalinke U. Type I interferons directly regulate lymphocyte recirculation and cause transient blood lymphopenia. Blood. 2006;108((10)):3253–61. doi: 10.1182/blood-2006-06-027599. [DOI] [PubMed] [Google Scholar]
- 97.Panesar NS. Glucocorticoid treatment of patients with SARS implications for mechanisms of immunopathology. Nat Rev Immunol. 2006;6((4)):334. [Google Scholar]
- 98.Panesar NS. What caused lymphopenia in SARS and how reliable is the lymphokine status in glucocorticoid-treated patients? Med Hypotheses. 2008 Aug;71((2)):298–301. doi: 10.1016/j.mehy.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Klasse PJ. Neutralization of virus infectivity by antibodies old problems in new perspectives. Adv Biol. 2014;2014:157895. doi: 10.1155/2014/157895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhou Y, Yang Y, Huang J, Jiang S, Du L. Advances in MERS-CoV vaccines and therapeutics based on the receptor-binding domain. Viruses. 2019;11((1)):60. doi: 10.3390/v11010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jiang S, Du L, Shi Z. An emerging coronavirus causing pneumonia outbreak in Wuhan, China calling for developing therapeutic and prophylactic strategies. Emerg Microbes Infect. 2020 Jan1;9((1)):275–7. doi: 10.1080/22221751.2020.1723441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhu Z, Chakraborti S, He Y, Roberts A, Sheahan T, Xiao X, et al. Potent cross-reactive neutralization of SARS coronavirus isolates by human monoclonal antibodies. Proc Natl Acad Sci U S A. 2007;104((29)):12123–8. doi: 10.1073/pnas.0701000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Long QX, Tang XJ, Shi QL, Li Q, Deng HJ, Yuan J, et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med. 2020 Aug;26((8)):1200–4. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- 104.Sariol A, Perlman S. Lessons for COVID-19 immunity from other coronavirus infections. Immunity. 2020 Aug 18;53((2)):248–63. doi: 10.1016/j.immuni.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Priyanka, Choudhary OP, Singh I. Protective immunity against COVID-19 unravelling the evidences for humoral vs. cellular components. Travel Med Infect Dis. 2021 Jan-Feb;39:101911. doi: 10.1016/j.tmaid.2020.101911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wu F, Liu M, Wang A, Lu L, Wang Q, Gu C, et al. Evaluating the association of clinical characteristics with neutralizing antibody levels in patients who have recovered from mild COVID-19 in Shanghai China. JAMA Intern Med. 2020 Oct 1;180((10)):1356–62. doi: 10.1001/jamainternmed.2020.4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cao WC, Liu W, Zhang PH, Zhang F, Richardus JH. Disappearance of antibodies to SARS-associated coronavirus after recovery. N Engl J Med. 2007 Sep 13;357((11)):1162–3. doi: 10.1056/NEJMc070348. [DOI] [PubMed] [Google Scholar]
- 108.Du L, Ma C, Jiang S. Antibodies induced by receptor-binding domain in spike protein of SARS-CoV do not cross-neutralize the novel human coronavirus hCoV-EMC. J Infect. 2013 Oct;67((4)):348–50. doi: 10.1016/j.jinf.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hsueh PR, Huang LM, Chen PJ, Kao CL, Yang PC. Chronological evolution of IgM, IgG and . neutralisation antibodies after infection with SARS-associated coronavirus. Clin Microbiol Infect. 2004 Dec 1;10((12)):1062–6. doi: 10.1111/j.1469-0691.2004.01009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Okba NMA, Müller MA, Li W, Wang C, GeurtsvanKessel CH, Corman VM, et al. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease patients. Emerg Infect Dis. 2020 Jul;26((7)):1478–88. doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Amanat F, Stadlbauer D, Strohmeier S, Nguyen THO, Chromikova V, McMahon M, et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat Med. 2020 Jul1;26((7)):1033–6. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ju B, Zhang Q, Ge J, Wang R, Sun J, Ge X, et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020 Aug1;584((7819)):115–9. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- 113.Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh C-L, Abiona O, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020 Mar13;367((6483)):1260–3. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Huang AT, Garcia-Carreras B, Hitchings MDT, Yang B, Katzelnick LC, Rattigan SM, et al. A systematic review of antibody mediated immunity to coronaviruses kinetics, correlates of protection, and association with severity. Nat Commun. 2020 Sep 17;11((1)):4704. doi: 10.1038/s41467-020-18450-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.To KK, Tsang OT, Leung WS, Tam AR, Wu TC, Lung DC, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2 an observational cohort study. Lancet Infect Dis. 2020 May;20((5)):565–74. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Barría MI, Garrido JL, Stein C, Scher E, Ge Y, Engel SM, et al. Localized mucosal response to intranasal live attenuated influenza vaccine in adults. J Infect Dis. 2012;207((1)):115–24. doi: 10.1093/infdis/jis641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kaur SP, Gupta V. COVID-19 vaccine a comprehensive status report. Virus Res. 2020;288:198114. doi: 10.1016/j.virusres.2020.198114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Montel Mendoza G, Pasteris SE, Otero MC, Fatima Nader-Macías. ME. Survival and beneficial properties of lactic acid bacteria from raniculture subjected to freeze-drying and storage. J Appl Microbiol. 2014 Jan;116((1)):157–66. doi: 10.1111/jam.12359. [DOI] [PubMed] [Google Scholar]
- 119.Zhang C, Maruggi G, Shan H, Li J. Advances in mRNA vaccines for infectious diseases. Front Immunol. 2019;10:594. doi: 10.3389/fimmu.2019.00594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mulligan MJ, Lyke KE, Kitchin N, Absalon J, Gurtman A, Lockhart S, et al. Phase 1/2 study to describe the safety and immunogenicity of a COVID-19 RNA vaccine candidate (BNT162b1) in adults 18 to 55 years of age interim report. medRxiv. 2020 [Google Scholar]
- 121.Tu Y-F, Chien C-S, Yarmishyn AA, Lin Y-Y, Luo Y-H, Lin Y-T, et al. A Review of SARS-CoV-2 and the ongoing clinical trials. IJMS. 2020;21((7)):2657. doi: 10.3390/ijms21072657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Castells MC, Phillips EJ. Maintaining safety with SARS-CoV-2 vaccines. N Engl J Med. 2021 Feb 18;384((7)):643–9. doi: 10.1056/NEJMra2035343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Guo Y, Sun S, Wang K, Zhang S, Zhu W, Chen Z. Elicitation of immunity in mice after immunization with the S2 subunit of the severe acute respiratory syndrome coronavirus. DNA Cell Biol. 2005 Aug;24((8)):510–5. doi: 10.1089/dna.2005.24.510. [DOI] [PubMed] [Google Scholar]
- 124.Mou H, Raj VS, van Kuppeveld FJ, Rottier PJ, Haagmans BL, Bosch BJ. The receptor binding domain of the new middle east respiratory syndrome coronavirus maps to a 231-residue region in the spike protein that efficiently elicits neutralizing antibodies. J Virol. 2013;87((16)):9379–83. doi: 10.1128/JVI.01277-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhou Y, Jiang S, Du L. Prospects for a MERS-CoV spike vaccine. Expert Rev Vaccines. 2018;17((8)):677–86. doi: 10.1080/14760584.2018.1506702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Diamond MS, Pierson TC. The challenges of vaccine development against a new virus during a pandemic. Cell Host Microbe. 2020;27((5)):699–703. doi: 10.1016/j.chom.2020.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Jeyanathan M, Afkhami S, Smaill F, Miller MS, Lichty BD, Xing Z. Immunological considerations for COVID-19 vaccine strategies. Nat Rev Immunol. 2020 Oct;20((10)):615–32. doi: 10.1038/s41577-020-00434-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hemann EA, Kang SM, Legge KL. Protective CD8 T cell-mediated immunity against influenza A virus infection following influenza virus-like particle vaccination. J Immunol. 2013 Sep 1;191((5)):2486–94. doi: 10.4049/jimmunol.1300954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhao Q, Li S, Yu H, Xia N, Modis Y. Virus-like particle-based human vaccines quality assessment based on structural and functional properties. Trends Biotechnol. 2013 Nov;31((11)):654–63. doi: 10.1016/j.tibtech.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 130.Rawat K, Kumari P, Saha L. COVID-19 vaccine a recent update in pipeline vaccines, their design and development strategies. Eur J Pharmacol. 2021;892:173751. doi: 10.1016/j.ejphar.2020.173751. [DOI] [PMC free article] [PubMed] [Google Scholar]