Abstract
Since the reporting of the first cases of coronavirus in China and the publication of the first sequence of SARS-CoV-2 in December 2019, the virus has undergone numerous mutations. In Europe, the spring outbreak (March–April) was followed by a drop in the number of cases and deaths. The disease may have evolved into a milder form. The increase in PCR-positive cases in late summer 2020 did not lead to the expected increase in hospitalizations, ICU admissions, and deaths, based on the severity of the disease in the spring. This difference in disease severity could be due to factors independent of the virus or to the evolution of the virus. This review attempts to identify the mutations that have appeared since the beginning of the pandemic and their role in the temporal evolution of the pandemic. There are a cell and humoral type cross-reactivity in a large part of the population to common cold coronaviruses (HCoVs) and SARS-CoV-2. Evolutionarily important mutations and deletions have emerged in the SARS-CoV-2 genes encoding proteins that interact with the host immune system. In addition, one of the major mutations (in viral polymerase) is logically associated with a higher frequency of mutations throughout the genome. This frequency fluctuates over time and shows a peak at the time when the epidemic was most active. The rate of mutations in proteins involved in the relationship to the immune system continues to increase after the first outbreak. The cross-reactivity on the 1 hand and the viral mutations observed on the other hand could explain the evolution of the pandemic until the summer of 2020, partly due to the evolution of the virus in relation to the host immune system. The immunization campaign began in December 2020: concerns are emerging about a possible escape of the circulating variants vaccines in early 2021. These variants could also escape immunity acquired through infection with the 2020 strains.
Keywords: SARS-CoV-2, Evolution, Immunity, Cross-reactivity, Cross-immunity
Evolution of the Epidemic
The SARS-CoV-2 pandemic has been occurring in several phases since the emergence of the virus in China at the end of 2019. In Europe, the spring outbreak (March–April) was followed by a decrease in the number of cases and deaths. According to some physicians who treated coronavirus 2019 (COVID-19) patients during the consecutive phases of the outbreak, the virus is believed to have evolved into a milder phenotype from the end of May 2020. But several factors independent of the virus could have impacted the case-fatality rate (CFR) (number of deaths/case numbers). At the beginning of 2020, testing capabilities were poorly developed. Mask use was not widespread and its use became widespread late, which may have reduced the inoculum of infected persons and the severity of cases, and the clinical management of COVID-19 had to improve over time.
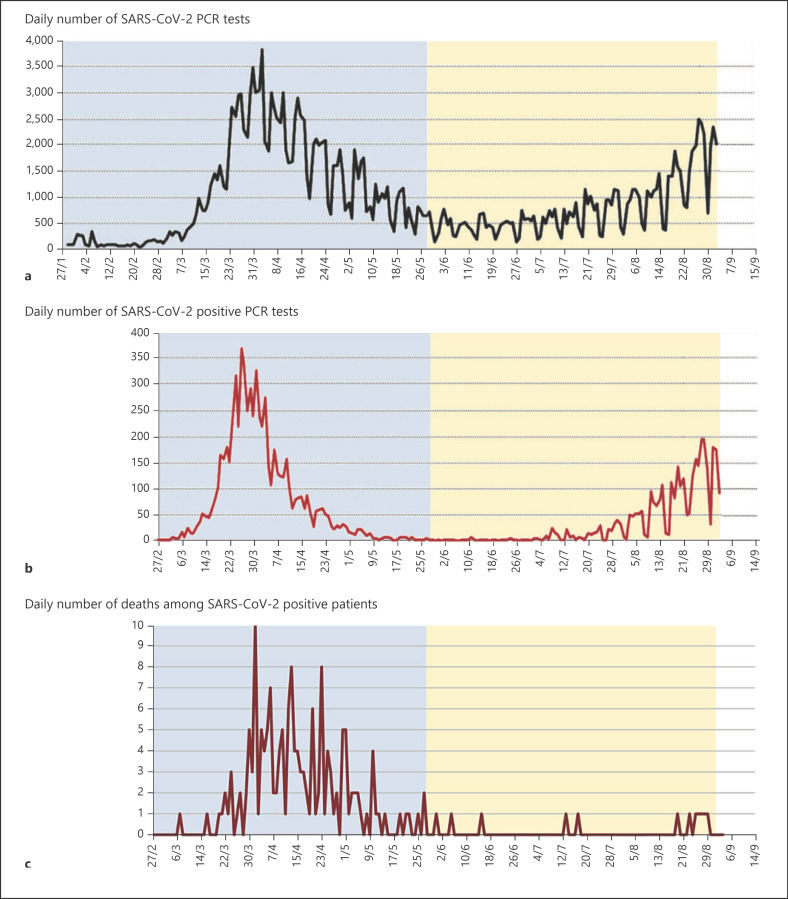
However, this is not the case everywhere: according to the IHU Marseille (France) [1], the mortality of patients hospitalized since mid-June is lower than that of the March–April phase. This cannot be due to a bias in comparison of the mortality rate between these 2 periods because the strategy of testing and clinical and therapeutic management has remained the same (Fig. 1).
Fig. 1.
From Colson et al. [1]: number of PCR tests, positive diagnoses, and deaths from February to September. Number of PCR tests performed at IHU Méditerranée Infection (a); number of PCR-positive patients performed at IHU Méditerranée Infection (b); number of deaths among SARS-CoV-2-positive patients in Marseille public hospitals (Assistance Publique-Hôpitaux de Marseille) (c).
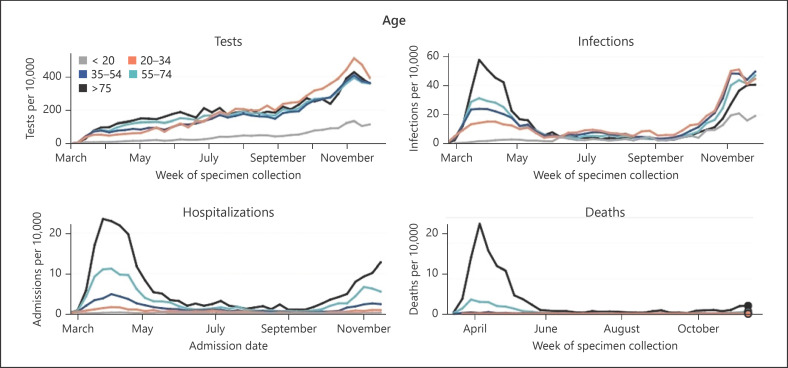
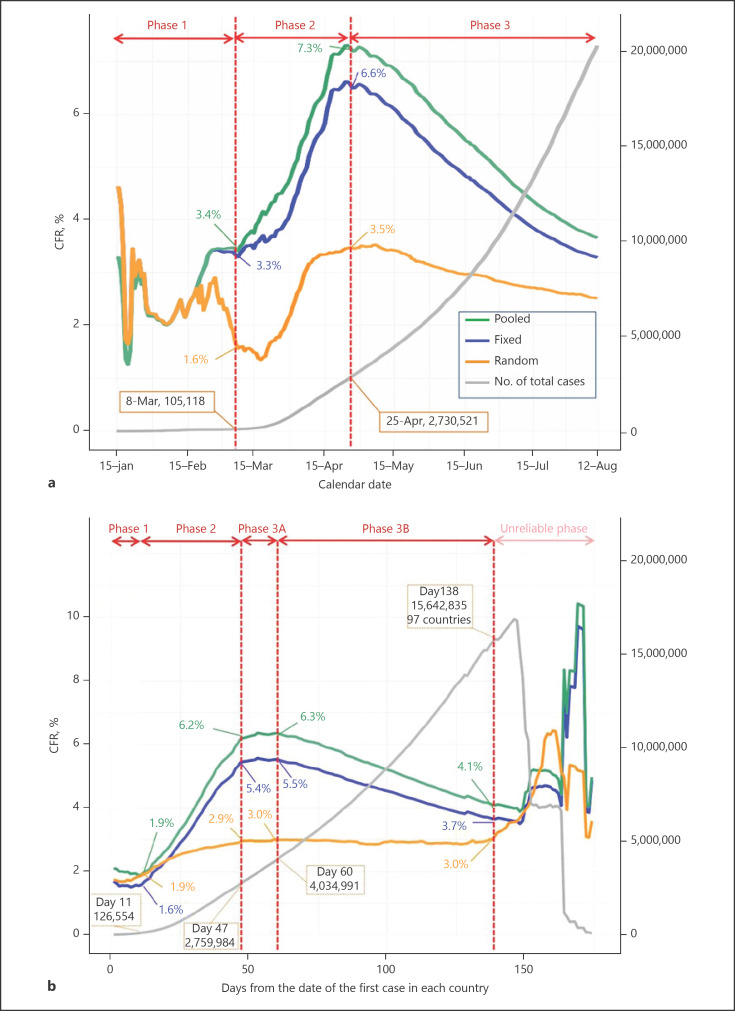
In Philadelphia (Pennsylvania), where the massive testing campaign started late, in contrast to Marseille (France), the curves of positive cases, hospitalizations and deaths also show this epidemic peak in March–April followed by a very clear decline. The increase in hospitalizations and deaths at the end of the summer is also not proportional to the number of infections (Fig. 2). According to Ghayda et al. [2], who evaluate the best way to estimate the CFR dynamically as the epidemic evolves, the CFR would appear to decrease regardless of the evaluation method (Fig. 3).
Fig. 2.
https://www.phila.gov/programs/coronavirus-disease-2019-covid-19/testing-and-data/#/. Number of tests performed, infections, hospitalizations, and deaths between March and November 2020, Philadelphia, Pennsylvania.
Fig. 3.
Timeline of CFR worldwide among countries with COVID-19 reports until August 12, 2020: according to date (a) and according to days (b) since the first confirmed case. COVID-19, coronavirus 2019; CFR, case-fatality rate; Fixed, fixed-effect model; Random, random-effect model; Pooled, calculated CFR based on incidence and mortality data; N, number. Gayda et al. [2].
Brewer et al. [3] have proposed that RNA viruses undergo natural genetic attenuation that contributes to the transient nature of pandemics caused by these viruses. Hygiene measures (such as the physical distancing provided by wearing masks) and host immune defenses (partial immunity that may result in the case of COVID-19 from cross-reactivity with HCoVs) would reduce inoculum levels and accelerate the mutation rate, which should result in greater bottlenecks and accelerated decline.
The emergence of another human coronavirus, HCoV-OC43 has been proposed to be linked to a host change around 1890, a time that coincides with a pandemic of respiratory disease in humans [4]. Could SARS-CoV-2, responsible for COVID-19, follow this evolutionary path as suggested by Benedetti et al. [5]?
Cross-Reactivity between SARS-CoV-2 and HCoVs
Cross-reactivity to common cold viruses and SARS-CoV-2 is now established [6]. This reactivity is logically directed against antigens common to all coronaviruses and not against SARS-CoV-2 specific antigens. These common antigens are found on the structural proteins N, M, and Spike and also on nonstructural proteins (NSPs, including viral RNA replication enzymes). Important differences in cellular reactivity to coronavirus epitopes are found between exposed and unexposed individuals and between severely affected and asymptomatic or poorly affected individuals. According to Grifoni et al. [7], the unexposed preferentially (compared to the exposed) reacts to ORF1 proteins (open reading frame = area of the genome that codes for many overlapping proteins) whereas the exposed recognize structural proteins; according to Mateus et al. [8], epitopes of M (the membrane protein) are not recognized by the unexposed, whereas they are recognized in a robust way in COVID (CD4+) cases. Li et al. [9] also did not find the same type of cellular response according to the severity of the disease in 2003 SARS-CoV.
NSPs are highly conserved among coronaviruses. According to Le Bert et al. [10] exposed but uninfected individuals would develop cellular reactivity toward NSPs involved in viral RNA replication, in particular NSP1 (encoded by the OrF1 region). Indeed, this protein is essential for virus replication and is, therefore, expressed first. Therefore, ORF1-specific T cells could hypothetically interrupt viral production by lysing SARS-CoV-2 infected cells before the formation of mature virions. NSP1 is involved in escape to the host immune system (it blocks innate immunity and interferon synthesis) [11].
While most of the scientific literature assumed preexisting T cells could be beneficial, there is also the possibility that preexisting immunity might actually be detrimental (Sette and Crotty [12]). According to Bacher et al. [13], cross-reactive T cells have a low avidity, negatively impacting the response quality against neoantigens such as SARS-CoV-2 and inappropriate immune reactions. But these authors suggest that preexisting memory T cells targeting HCoVs epitopes and having a low cross-reactivity with SARS-CoV-2 epitopes could prevent the activation of naïve T cells and the selection of high-affinity clones. This is inconsistent with the documented presence of cross-reactive T cells in all donors exposed or unexposed to COVID-19 and with the fact that the majority of individuals infected with SARS-CoV-2 have a mild or asymptomatic infection. If there is a harmful role for these low-affinity memory T cells, it appears to correlate with the immunological age of the patient. Moreover, according to Sagar et al. [14], a recent HCoV infection would protect against a severe COVID.
According to Ng et al. [15], antibodies directed against the S2 subunit of the spike protein of SARS-CoV-2 would preexist in many uninfected subjects, they would come from previous HCoV infections. In Africa, where COVID has had little effect, Tso et al. [16] find a predominance of cross-reactive antibodies directed against the N protein. The prevalence of these antibodies is 6–8 times higher than that found in the USA where the incidence and morbidity of COVID have been higher.
Evolution of the Virus
It seems that the emergence of the virus dates back to late summer 2019 in China and that the virus entered the West as early as October 2019 (or earlier?). This is suggested by Li et al. [17] based on mutation analysis. Van Dorp et al. [18] indicate that the disease spread worldwide probably from the beginning of the pandemic. Chaw et al. [19] also believe that SARS-CoV-2 circulated cryptically well before the late 2019 outbreak in China, Gambaro et al. [20] say the same for France.
A hypothesis on the origin of SARS-CoV-2, discussed by Sallard et al. [21], proposes that SARS-CoV-2 is derived from a virus that appeared in a mine in China in 2012, was collected in a laboratory, and may have escaped from it during manipulations in 2018 or 2019. This hypothesis could account for the circulation of the virus before the outbreak, during this early period the virus could have undergone undetected mutations. Thus, there would be a lack of archives from the early period of the pandemic and mutations that occurred during this period would have gone unnoticed. Moreover, only a few tens of thousands of sequences have been analyzed out of the 34 million suspected cases of COVID-19. Of the 295,000 published sequences (Gisaid https://www.gisaid.org) a minority were analyzed for their biological and evolutionary significance. It will, therefore, be necessary to follow in the coming months the publications on late-phase isolated sequences.
What are the main mutations found on SARS-CoV-2 compared to the first published Chinese sequence dated December 2019 (see Table 1)? This work will be mainly limited to the first 2 phases of the pandemic (from December 2019 to summer 2020).
Table 1.
Main mutations, role of the mutated regions
| Genome region | Protein | Mutation | Remark | Role or assumed role |
|---|---|---|---|---|
| Leader 5′UTR | Mutated nucleotide | |||
| C251T | G clade | Not transcribed | ||
| Mutated amino acid | ||||
| orflab | NSP1 | Excess of mutations | Immune evasion | |
| NSP2 | T265I | |||
| NSP3 | F106F | G clade | Inflammasome | |
| Silent |
Interaction | |||
| NSP6 | L37F | V clade | Innate immunity Interaction | |
| L3606F | ||||
| NSP12 | P323L | G clade | RNA dependent RNA polymerase | |
| NSP13 | P504L | Helicase | ||
| Y541C | ||||
| NSP14 | Exonuclease proofreading | |||
| Spike | S | D614G | Not in RBD | Increase infectivity |
| G clade | Increase viral load URT | |||
| N439K | In RBD | Increase infectivity | ||
| Deletions | ||||
| orf3a | Viroporin | Q57H | USA | Cellular release of virus |
| G215V | V clade (China) | Virulence Viral replication | ||
| orf8 | ORF8 | L84S | S clade | Host immune response |
| Deletions | Immunogenic protein | |||
| Nucleocapsid | N | R203K | GR clade | Increase infectivity |
| R202K | These 3 mutations are linked | and replication | ||
| G204R | ||||
| orf10 | Deletions | Noncoding region Not essential | ||
NSP, nonstructural proteins; ORF, open reading frame; RBD, receptor-binding domain.
Deletions
They are mainly found in regions coding for proteins that interact with the host response and in the spike protein. A hotspot of deletion in the NSP1 region has been found in several countries, suggesting that it is due to potential convergent evolution. Particularly, the Δ500–532 is correlated with lower viral load, nonsevere traits, and lower serum IFN-β, with possible implications for proper immune response against SARS-CoV-2 infection [22].
In April 2020 [23], a deletion is found in the ORF7 region from the original sequence. It concerns a region that could be important for the adaptation of the virus to humans (because it is close to the ORF8 region identified in 2003 for SARS-CoV-1) [24].
Between January and February 2020, a team from Singapore recovered a deletion of 382 nucleotides in the ORF8 region [25]. The authors suggest that the deletion could lead to an attenuated SARS-CoV-2 phenotype. This mutation then disappeared after March 2020. Hospitalized patients with this mutation had less severe COVID than those without it. The ORF8 region is believed to be involved in immune evasion.
Patients carrying this variant have better T cell responses and higher production of gamma interferon. The proteins produced by orf8 are highly immunogenic and induce early antibody synthesis during the disease. The absence of these proteins in patients carrying the mutation could explain the lesser inflammatory response.
This deletion could have been selected by the pressure of the host immune system: Muth et al. [24] had already proposed this for a deletion in ORF8 found on a strain of SARS-CoV-1.
McCarthy et al. [26] highlight the importance of deletions in immune evasion on the evolutionary trajectory of SARS-CoV-2 to an endemic virus. According to Parvez et al. [27] and Rahman et al. [28], deletions in ORF7, ORF8, and ORF10 found in Bangladesh were associated with reduced virulence. According to Liu et al. [29], isolates containing deletions in or near the furin polybasic site of the spike protein have been associated with mild or asymptomatic outcome. Variants with deletions may be underrepresented in the databases due to their low frequency and thus their elimination, Nagy et al. [30]. Moreover, very few complete sequences have been deposited at GISAID, Wang et al. [31].
Observed Mutations
The selected point mutations (SNPs) would be in epistatic interaction: they are the signatures of the observed clades which present distinct spatial and temporal dynamics [32]. According to Rice et al. [33], untranscribed regions are subject to selection (e.g., orf10). According to Nagy et al. [30], the absence or modification of a NSP and nontranscribed regions can influence the expression of the viral genome and have as much effect on pathogenicity as nonsynonymous mutations.
Below will be listed possible correlations between point mutations and disease severity and/or infectivity. But it should be kept in mind that SNPs have coevolved to form the clades that have become dominant and these associations of synonymous and nonsynonymous mutations and deletions could strongly influence fitness and virulence.
Mutations on the Spike Protein (D614G and Others)
The D614G mutation (located outside the receptor-binding domain [RBD]) resulted in the replacement of an asparagine by a glycine on the C-terminal part of the surface spike protein. This mutation never appears alone but is part of a haplotype of 4 mutations (including those that alter NSP12 [NSP], 5′ TRU, and silently NSP3), which constitute the clade G originating from China and established in Europe [34].
It is not certain that this mutation increases infectivity, but it is now accepted that it does not increase the severity of the disease [35]. However, in April 2020 (first phase of the pandemic), Becerra-Flores and Cardozo noted a correlation between the frequency of the D614G variant and the CFR [36] although this kind of analysis can be complicated by different availability of testing and care in different nations [35]. Long et al. [37] validate previous studies showing that patients infected with variant D614G have higher viral loads in the upper respiratory tract without worsening the disease. Plante et al. [38] propose that strains carrying D614G would be less adapted to the lower respiratory tract.
Other mutations appeared during the summer in the spike protein, in particular, N439K in RBD: according to Chen et al. [39], the most frequent mutations in the spike (including D614G, N439K, and S477N) increase its transmissibility. A strain with D614G associated with mutations at RBD is more infective and resistant to some neutralizing antibodies with obvious implication for the recovery of COVID-19 patients [40].
Mutations Related to the Host Immune System
Wang et al. [41] show by studying C > T mutations in the viral genome that about 65% of these are imposed by the host immune response: the APOBEC system (apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like) edits the viral genes and in return T > C mutations are caused by the virus' protection mechanism, T > C.
This C > T ratio increases with age (older people are more affected) and may also explain the differences in the populations' level of infection (Africans and Oceanians are less affected by this mutation and the epidemic in their geographical area has been less severe than elsewhere).
According to Rice et al. [33], this codon bias could be a route to attenuation of the virus through reduced immunogenicity. Jaroszewski et al. [42] showed an excess of mutations in proteins NSP1 and NSP2 and in ORF3a, ORF8b, and ORF14 (involved in virus-host interactions) compared to other sites. The spike, the membrane M protein and RdRp (RNA-dependent RNA polymerase virale = NSP12) show a lower rate of false-sense mutations than the others: this would be due to the purifying selection effect given the importance of RdRp for the biology of SARS-CoV-2.
The NSP1 protein allows immune evasion of the virus [11] a mutation in this location can make the virus vulnerable to immune clearance. NSP1 plays a role in inhibiting host RNA expression [43] and in IFN-ß response, as seen above [22]. Exposed and unexposed individuals have a different T cell response to the NSP1 protein [10].
NSP3 could be involved in the cytokine releasing inflammation observed in severe COVID; this protein would interact with the inflammasome (protein complex involved in inflammation and innate immunity) and in particular through its hypervariable part. The expression of the NSP3 protein in IFN-activated macrophages (IFN: interferon) would indirectly promote prolonged pro-inflammatory expression of IFN-stimulated genes. This would participate in the cytokine storm characteristic of severe cases of COVID-19 [44].
The synonymous mutation in NSP3 (F106F) has coevolved with the other signature mutations of clade G and could, although silent, affect the fitness of the virus [34]. According to Wang et al. [45], the NSP6 L37F mutation, frequently found at the beginning of the pandemic, mainly in Asia, is associated with a high frequency of asymptomatic cases. However, it disappeared in the later phases of the pandemic and belonged to the GISAID clade V [46]. NSP6 decreases the autophagic capacity of infected cells, which provides an innate defense against viral infections. This capacity also promotes cell death and morbidity.
The Q57H mutation is found on the ORF3a region, Wang et al. [47]. ORF3a proteins are involved in apoptosis and activate the inflammasome. The ORF8 protein is involved in immune system evasion: the temporal evolution of the L84S mutation, which mainly appeared in the USA and is associated with 2 other helicase mutations (NSP13, P504L, and Y541C), follows the evolution of deaths in the first phase of the epidemic (March–June 2020). The Y541C and P504L mutations would prevent the SARS-CoV-2 from interacting effectively with the host interferon signaling molecules and NSP13 from participating effectively in the replication/transcription process.
On the contrary, according to Nagy et al. [30], mutations associated with a lower result were localized in the surface (S) glycoprotein, in RdRp, in exonuclease 3′-5′, in ORF3a, NSP2, and N. Mutations associated with a low result were localized in ORF8, NSP6, ORF3a, NSP4 and in nucleocapsid phosphoprotein N. However, only isolates associated with clinical data were studied (10% of the total published sequences) and the geographical distribution of the sequences obtained is unbalanced (very few come from America and Africa, the majority from Asia). This could bias the results.
As Wang et al. [45], Banerjee et al. [48], and Pachetti et al. [49] find spatial-temporal variations as well as Zhao et al. [50] for whom the incidence of some of these sites decreased after reaching a (often local) peak. This could indicate a potential signal for positive selection in genes encoding ORF1ab and structural proteins. Mutations in NSP6, NSP13, ORF3a, and ORF8 show high-frequency peaks at the beginning of the epidemic and only in some regions but have subsequently declined sharply and are now low.
On the other hand, the frequency of mutations in NSP2, NSP12, ORF3a 57, N, and S has increased since their introduction into the viral genome. They are present in great abundance in the second half of March, either globally or in certain continents. Eskier et al. [51] also study the temporal evolution of the co-mutations of the G and GH clades, unlike Europe, North and South America, where RdRp-P323L became the dominant form with its co-mutations, RdRp-323, and its co-mutations remained the minor form in Asia, which may explain the epidemiological differences between these continents.
The results suggest that SARS-CoV-2 genomes with the RdRp-323 mutation are 1.5 times more likely to have high mutation rates in other parts of the genome in terms of time and location: a more error-prone mutant RdRp should increase viral genetic diversity and allow the virus to spread under different selective pressures in different populations. Mutations in NSP14, an error-correcting protein exonuclease [41] are most strongly associated with an increased mutation load throughout the genome.
Finally, there is a relationship between SARS-CoV-2 mutation densities and the dynamics of viral transmission in human populations, when the number of new daily cases started to reach a plateau. The increase in mutation density ended when the number of new daily cases began to reach a plateau. But synonymous and nonsynonymous mutations in the S and ORF1a genes continued to accumulate until the number of cases and deaths declined sharply. According to Alam et al. [52], the progression of the pandemic and the number of deaths are correlated over time with the frequency of G clades, and then GH and GR derived from it. The exponential increase in deaths during the first phase of the pandemic is correlated with the frequency of G clade except in the Western Pacific zone. The GR clade is significantly associated with a low death/case ratio. The progression of the disease and the death/case ratio lead to infer an adaptation of the virus (fitness would compromise the virulence of the virus).
Tomaszewski et al. [34] also studied the rate of mutations in the SARS-CoV-2 genome between January and May 2020. Between January and April, the mutation rate increased and then slowed for Spike and NSP12 and NSP13. Conversely, certain regions of the genome have a mutation rate that continues to increase after April: this concerns the N protein (particularly the R203K and G204R mutations) and viroporin 3a (encoded by ORF3a, Q57H, and G251V mutations). These viral molecules can subvert the immune response, in particular the response to interferon: protein N, protein 3a, and NSP6 are antagonists of interferon β which operate in coronavirus diseases.
Explanatory Assumptions
As demonstrated by Wang et al. [41], the majority of SARS-CoV-2 mutations are imposed by the APOBEC antiviral defense system and have a priori no adaptive value. The evolution of emerging viruses results from selective pressures (adaptation to the host immune system, increased transmissibility) but most mutations are selectively neutral or slightly deleterious.
According to Chen et al. [39], mutations on the spike protein all increase transmissibility.
As described above, no single SNP studied in isolation is clearly associated with a difference in disease severity. However, deletions on proteins interacting with the immune system could easily go unnoticed (on ORF7 and 8). Clades that have become dominant are characterized by associations of synonymous and nonsynonymous mutations (and possibly deletions): positive selection could have selected these associations. For example, the silent mutation (because on an untranscribed region) on the leader 5′UTR (C251T) is part of the signature of the clade G that has rapidly become dominant overall [52].
In addition to the pressure on transmissibility, the selective pressure that can act globally on the evolution of the virus is that of innate and preexisting immunity (perhaps by cross-reaction with other coronaviruses). Indeed, adaptive immunity plays a much less important role because it appears later, once the infected person has been able to eventually transmit the virus.
Severely ill people fought the virus ineffectively, but those exposed but not ill destroyed most of the infecting virus and were able to select the less virulent (less efficiently replicating) forms not affected by the innate immune system. Indeed, immunopathological phenomena seem to be responsible for the severity of the disease [6]. Virions that stimulate these phenomena less, by interacting less with innate immunity, would be selected, and the virus would evolve toward a benign phenotype. Most of the available sequences were isolated from severely ill patients [20].
In asymptomatic patients (the majority of infected individuals), few complete sequences are isolated, and therefore, little is known about the mutations responsible for this attenuation. But we can assume that these “less agressive” viruses are finally the ones that circulate the most in the general population now, to the point of completely supplanting the “more aggressive”?
The work of Eskier et al. [51] Wang et al. [45], and Tomaszewski et al. [34] would tend to show that mutations favoring the virus can also disadvantage it: RdRp makes more errors when it is faster. If these mutations are associated with mutations on the NSP14 that repair these errors, mutations in structural proteins may accumulate that could also explain the decline of the epidemic. The variation in the mutation rate of the dominant strains in Europe and the USA is correlated with the temporal evolution of the epidemic: this concerns mutations related to the immune system especially in the declining phase of the epidemic.
Conclusion
It is not impossible that the known common cold coronaviruses, when they jumped from animals to humans (they are all originally responsible for zoonoses), began their evolutionary course with a pandemic like COVID-19. But at the time, the means of investigation in virology and molecular biology did not exist and were not identified.
There is a cross-reactivity of cellular and humoral type in a large part of the population to common cold coronaviruses and SARS-CoV-2. Vijgen et al. [4] proposed that the common cold coronavirus HCoV-OC43 may have followed the same trajectory as the SARS-CoV-2: a species jump resulting in a pandemic, followed by an evolution to a more benign common cold coronavirus.
This evolution seems to be common to emerging viruses (viral attenuation). Evolutionarily important mutations have appeared in the SARS-CoV-2 genes encoding proteins that interact with the host immune system. One of the major mutations (in viral polymerase) is logically associated with a higher frequency of mutations throughout the genome. This frequency fluctuates over time and peaks at the epidemic peak in the spring of 2020.
Cross-reactivity on the one hand and the viral mutations observed on the other hand could explain the evolution toward a benign phenotype of SARS-CoV-2, in part by evolution of the virus against the host immune system, at least until the summer of 2020. Sustained attention will need to be paid to mutations that could reduce the efficacy of vaccines (directed against spike protein) [53] and PCRs depending on the probes used [54].
In December 2020, a new variant was identified in the UK (VUI 202012/01 or B.1.17), defined by multiple mutations in the spike protein (Δ69–70, Δ144, N501Y, A570D, D614G, P681H, T716I, S982A, and D1118H). This variant would not be associated with more severe infections, would not preferentially affect an age-group compared to previously circulating viruses, but being more contagious it resulted in January 2021 in the highest mortality rate in the UK since the start of the pandemic [55]. The B.1.1.7 variant is unlikely to escape recognition by antibodies generated by prior infection or the vaccines [56] although a recent report suggested that the full set of Spike mutations present in the B.1.1.7 variant may reduce the neutralizing activity of the Pfizer vaccine BNT162b2 [57].
In December 2020, the variant B.1.351 (501Y.V2) was first isolated in South Africa, it carries 8 characteristic mutations in the spike protein [58] and may have increased transmissibility, but no change in disease severity has been shown to date. It has been identified in several European countries [59].
Of note, recent statements by Johnson & Johnson [60]and Novavax [61] reported that vaccine efficacy may be reduced against the South Africa variant. Viral neutralization by sera induced by the ChAdOx1 nCoV-19 coronavirus vaccine against the B.1.351 coronavirus variant was substantially reduced when compared with the original strain of the coronavirus [62].
The P.1 variant has so far only been identified in Brazil, and in travelers from Brazil reported in Japan and South Korea. This variant includes 20 unique mutations, some of which could be responsible for an escape of the antibodies [63].
Several data are emerging regarding the effect of SARS-CoV-2 mutation on neutralizing antibodies in convalescent patients with possible implication for disease recurrence. Lineage 501Y.V2 exhibits complete escape from 3 classes of therapeutically relevant monoclonal antibodies [64], and serum antibodies [65]. In lineages carrying the E 484 mutation (present in South Africa and Brazil variants), neutralization by some sera is reduced >10 fold [66].
Conflict of Interest Statement
The author has no conflicts of interest to declare.
Funding Sources
The author did not receive any funding.
References
- 1.Colson P, Levasseur A, Delerce J, Chaudet H, Bossi V, Ben Khedher M, et al. Dramatic increase in the SARS-CoV-2 mutation rate and low mortality rate during the second epidemic in summer in Marseille. Preprint IHU. 2020 [Google Scholar]
- 2.Ghayda RA, Lee KH, Han YJ, Ryu S, Hong SH, Yoon S, et al. Estimation of global case fatality rate of coronavirus disease 2019 (COVID-19) using meta-analyses: comparison between calendar date and days since the outbreak of the first confirmed case. Int J Infect Dis. 2020 Nov;100:302–8. doi: 10.1016/j.ijid.2020.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brewer WH, Smith FD, Sanford JC. Information loss: potential for accelerating natural genetic attenuation of RNA viruses. Biol Inf. 2013 Jul;:369–84. [Google Scholar]
- 4.Vijgen L, Keyaerts E, Moës E, Thoelen I, Wollants E, Lemey P, et al. Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J Virol. 2005;79((3)):1595–604. doi: 10.1128/JVI.79.3.1595-1604.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benedetti F, Pachetti M, Marini B, Ippodrino R, Ciccozzi M, Zella D. SARS-CoV-2: march toward adaptation. J Med Virol. 2020 Jun 29;:10. doi: 10.1002/jmv.26233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banoun H. COVID19: cross-immunity with other coronaviruses, immunopathological phenomena. SSRN Electron J. 2020 [Google Scholar]
- 7.Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR, et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181((7)):1489–e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mateus J, Grifoni A, Tarke A, Sidney J, Ramirez SI, Dan JM, et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020 Oct 2;370((6512)):89–94. doi: 10.1126/science.abd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li CK, Wu H, Yan H, Ma S, Wang L, Zhang M, et al. T cell responses to whole SARS coronavirus in humans. J Immunol. 2008;181((8)):5490–500. doi: 10.4049/jimmunol.181.8.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le Bert N, Tan AT, Kunasegaran K, Tham CYL, Hafezi M, Chia A, et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584((7821)):457–62. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 11.Thoms M, Buschauer R, Ameismeier M, Koepke L, Denk T, Hirschenberger M, et al. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science. 2020 Sep 4;369((6508)):1249–55. doi: 10.1126/science.abc8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sette A, Crotty S. Pre-existing immunity to SARS-CoV-2: the knowns and unknowns. Nat Rev Immunol. 2020;20((8)):457–8. doi: 10.1038/s41577-020-0389-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bacher P, Rosati E, Esser D, Rios G, Saggau C, Schiminsky E, et al. Pre-existing T cell memory as a risk factor for severe 1 COVID-19 in the elderly. MedRxiv. 2020 Sep 18 [Google Scholar]
- 14.Sagar M, Reifler K, Rossi M, Miller NS, Sinha P, White L, et al. Recent endemic coronavirus infection is associated with less severe COVID-19. J Clin Invest. 2020 Sep 30;:143380. doi: 10.1172/JCI143380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ng KW, Faulkner N, Cornish GH, Rosa A, Harvey R, Hussain S, et al. Preexisting and de novo humoral immunity to SARS-CoV-2 in humans. Science. 2020;370((6522)):1339–43. doi: 10.1126/science.abe1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tso FY, Lidenge SJ, Peña PB, Clegg AA, Ngowi JR, Mwaiselage J, et al. High prevalence of pre-existing serological cross-reactivity against SARS-CoV-2 in sub-Sahara Africa. Int J Infect Dis. 2020 Nov 8;102:577–83. doi: 10.1016/j.ijid.2020.10.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y, Liu B, Wang Z, Cui J, Shen Y, Xu Y, et al. COVID-19 evolves in human hostss. SSRN Electron J. 2020 Mar [Google Scholar]
- 18.Van Dorp L, Acman M, Richard D, Shaw LP, Ford CE, Ormond L, et al. Emergence of genomic diversity and recurrent mutations in SARS-CoV-2. Infect Genet Evol. 2020 Sep;83:104351. doi: 10.1016/j.meegid.2020.104351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chaw SM, Tai JH, Chen SL, Hsieh CH, Chang SY, Yeh SH, et al. The origin and underlying driving forces of the SARS-CoV-2 outbreak. J Biomed Sci. 2020;27((1)):73. doi: 10.1186/s12929-020-00665-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gámbaro F, Behillil S, Baidaliuk A, Donati F, Albert M, Alexandru A, et al. Introductions and early spread of SARS-CoV-2 in France. Euro Surveill. 2020 Jul;25((26)):2001200. doi: 10.2807/1560-7917.ES.2020.25.26.2001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sallard E, Halloy J, Casane D, Decroly E, van Helden J. Tracing the origins of SARS-COV-2 in coronavirus phylogenies: a review. Med Sci. 2020 Aug–Sep;36((8–9)):783–96. doi: 10.1051/medsci/2020123. [DOI] [PubMed] [Google Scholar]
- 22.Lin JW, Tang C, Wei HC, Du B, Chen C, Wang M, et al. Genomic monitoring of SARS-CoV-2 uncovers an Nsp1 deletion variant that modulates type I interferon response. Cell Host Microbe. 2021 Jan 29;S1931–3128((21)):00045–7. doi: 10.1016/j.chom.2021.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holland LA, Kaelin EA, Maqsood R, Estifanos B, Wu LI, Varsani A, et al. An 81-nucleotide deletion in SARS-CoV-2 ORF7a Identified from sentinel surveillance in Arizona (January to March 2020) J Virol. Jul. 2020;94((14)):e00711–20. doi: 10.1128/JVI.00711-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muth D, Corman VM, Roth H, Binger T, Dijkman R, Gottula LT, et al. Attenuation of replication by a 29 nucleotide deletion in SARS-coronavirus acquired during the early stages of human-to-human transmission. Sci Rep. 2018 Oct 11;8((1)):15177. doi: 10.1038/s41598-018-33487-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Young BE, Fong SW, Chan YH, Mak TM, Ang LW, Anderson DE, et al. Effects of a major deletion in the SARS-CoV-2 genome on the severity of infection and the inflammatory response: an observational cohort study. Lancet. 2020 Aug 29;396((10251)):603–11. doi: 10.1016/S0140-6736(20)31757-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCarthy KR, Rennick LJ, Nambulli S, Robinson-McCarthy LR, Bain WG, Haidar G, et al. Natural deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape. BioRxiv. 2020 Nov 19; doi: 10.1126/science.abf6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parvez MSA, Rahman MM, Morshed MN, Rahman D, Anwar S, Hosen MJ. Genetic analysis of SARS-CoV-2 isolates collected from Bangladesh: insights into the origin, mutational spectrum and possible pathomechanism. Comput Biol Chem. 2021 Feb;90:107413. doi: 10.1016/j.compbiolchem.2020.107413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rahman M, Kader SB, Rizvi SMS. Molecular characterization of SARS-CoV-2 from Bangladesh: Implications in genetic diversity, possible origin of the virus, and functional significance of the mutations. BioRxiv. 2020 Oct 13; doi: 10.1016/j.heliyon.2021.e07866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Z, Zheng H, Lin H, Li M, Yuan R, Peng J, et al. Identification of common deletions in the spike protein of severe acute respiratory syndrome coronavirus 2. J Virol. Aug 2020;94((17)):e00790–20. doi: 10.1128/JVI.00790-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagy Á, Pongor S, Győrffy B. Different mutations in SARS-CoV-2 associate with severe and mild outcome. Int J Antimicrob Agents. 2021 Feb;57((2)):106272. doi: 10.1016/j.ijantimicag.2020.106272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang R, Hozumi Y, Yin C, Wei GW. Mutations on COVID-19 diagnostic targets. Genomics. 2020 Nov;112((6)):5204–13. doi: 10.1016/j.ygeno.2020.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rochman ND, Wolf YI, Faure G, Zhang F, Koonin EV. Ongoing adaptive evolution and globalization of Sars-Cov-2. BioRxiv. 2020 Oct 13 [Google Scholar]
- 33.Rice AM, Castillo Morales A, Ho AT, Mordstein C, Mühlhausen S, Watson S, et al. Evidence for strong mutation bias toward, and selection against, u content in SARS-CoV-2: implications for vaccine design. Mol Biol Evol. 2021 Jan 4;38((1)):67–83. doi: 10.1093/molbev/msaa188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomaszewski T, DeVries RS, Dong M, Bhatia G, Norsworthy MD, Zheng X, et al. New pathways of mutational change in SARS-CoV-2 proteomes involve regions of intrinsic disorder important for virus replication and release. Evol Bioinform Online. 2020 Oct 23;16:1176934320965149. doi: 10.1177/1176934320965149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020 Aug 20;182((4)):812–e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Becerra-Flores M, Cardozo T. SARS-CoV-2 viral spike G614 mutation exhibits higher case fatality rate. Int J Clin Pract. 2020 Aug;74((8)):e13525. doi: 10.1111/ijcp.13525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Long SW, Olsen RJ, Christensen PA, Bernard DW, Davis JJ, Shukla M, et al. Molecular architecture of early dissemination and massive second wave of the SARS-CoV-2 virus in a major metropolitan area. MedRxiv. 2020 Sep; doi: 10.1128/mBio.02707-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Plante JA, Liu Y, Liu J, Xia H, Johnson BA, Lokugamage KG, et al. Spike mutation D614G alters SARS-CoV-2 fitness and neutralization susceptibility. BioRxiv. 2020 Sep 02 [Google Scholar]
- 39.Chen J, Wang R, Wang M, Wei GW. Mutations strengthened SARS-CoV-2 infectivity. J Mol Biol. 2020 Sep 4;432((19)):5212–26. doi: 10.1016/j.jmb.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Q, Wu J, Nie J, Zhang L, Hao H, Liu S, et al. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell. 2020 Sep 3;182((5)):1284–e9. doi: 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang R, Hozumi Y, Zheng Y-H, Yin C, Wei G-W. Host immune response driving SARS-CoV-2 evolution. Viruses. 2020;12((10)):1095. doi: 10.3390/v12101095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jaroszewski L, Iyer M, Alisoltani A, Sedova M, Godzik A. The interplay of SARS-CoV-2 evolution and constraints imposed by the structure and functionality of its proteins. BioRxiv. 2020 Aug 10; doi: 10.1371/journal.pcbi.1009147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmidtk P. SARS-Cov-2 − part 3 − nsp1: a hopefully more detailed analysis of the cellular saboteur. 2020. Apr 6. Available from: https://www.discngine.com/blog/2020/3/29/sars-cov-2-3-nsp1-a-detailed-analysis.
- 44.Claverie J-M. A putative role of de-mono-adp-ribosylation of STAT1 by the SARS-CoV-2 Nsp3 protein in the cytokine storm syndrome of COVID-19. Viruses. 2020 Jun 15;12((6)):646. doi: 10.3390/v12060646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang R, Chen J, Hozumi Y, Yin C, Wei GW. Decoding asymptomatic COVID-19 infection and transmission. J Phys Chem Lett. 2020 Dec 3;11((23)):10007–15. doi: 10.1021/acs.jpclett.0c02765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mercatelli D, Giorgi FM. Geographic and genomic distribution of SARS-CoV-2 mutations. Front Microbiol. 2020;11:1800. doi: 10.3389/fmicb.2020.01800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang R, Chen J, Gao K, Hozumi Y, Yin C, Wei GW. Characterizing SARS-CoV-2 mutations in the United States. Res Sq. 2020 Aug 11 [Google Scholar]
- 48.Banerjee R, Basak K, Ghosh A, Rajachandran V, Sureka K, Ganguly D, et al. Spike protein mutational landscape in India: Could Muller's ratchet be a future game-changer for COVID-19? BioRxiv. 2020 Oct 7; doi: 10.1016/j.meegid.2021.104874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pachetti M, Marini B, Benedetti F, Giudici F, Mauro E, Storici P, et al. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J Transl Med. 2020;18((1)):179. doi: 10.1186/s12967-020-02344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao J, Zhai X, Zhou J. Snapshot of the evolution and mutation patterns of SARS-CoV-2. BioRxiv. 2020 Jul 05 [Google Scholar]
- 51.Eskier D, Suner A, Karakülah G, Oktay Y. Mutation density changes in SARS-CoV-2 are related to the pandemic stage but to a lesser extent in the dominant strain with mutations in spike and RdRp. PeerJ. 2020 Aug 19;8:e9703. doi: 10.7717/peerj.9703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alam ASM, Islam KO, Hasan S, Islam R, Mahmud S, AlEmran HM, et al. Evolving infection paradox of SARS-CoV-2: fitness costs virulence? MedRxiv. 2021 Feb 23 [Google Scholar]
- 53.Chen J, Gao K, Wang R, Wei G. Prediction and mitigation of mutation threats to COVID-19 vaccines and antibody therapies. MedRxiv. 2020 Oct13:06357v1. doi: 10.1039/d1sc01203g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang R, Hozumi Y, Yin C, Wei GW. Decoding SARS-CoV-2 transmission and evolution and ramifications for COVID-19 diagnosis, vaccine, and medicine. J Chem Inf Model. 2020 Jun 25;60((12)):5853–65. doi: 10.1021/acs.jcim.0c00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Public Health England Investigation of novel SARS-CoV-2 variant. 2020. Dec, Available from: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/948152/Technical_Briefing_VOC202012-2_Briefing_2_FINAL.pdf.
- 56.Muik A, Wallisch AK, Sänger B, Swanson KA, Mühl J, Chen W, et al. Neutralization of SARS-CoV-2 lineage B.1.1.7 pseudovirus by BNT162b2 vaccine-elicited human sera. Science. 2021 Mar 12;371((6534)):1152–3. doi: 10.1126/science.abg6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Collier D-A, De Marco A, Ferreira I-ATM, Meng B, Datir R, Walls AC, et al. SARS-CoV-2 B.1.1.7 escape from mRNA vaccine-elicited neutralizing antibodies. MedRxiv. 2021 Feb 02 [Google Scholar]
- 58.Tegally H, Wilkinson E, Giovanetti M, Iranzadeh A, Fonseca V, Giandhari J, et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. MedRxiv. 2020 Dec 22 [Google Scholar]
- 59.Priesemann V, Balling R, Brinkmann MM, Ciesek S, Czypionka T, Eckerle I, et al. An action plan for pan-European defence against new SARS-CoV-2 variants. Lancet. 2021 Feb 6;397((10273)):469–70. doi: 10.1016/S0140-6736(21)00150-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Novavax Novavax COVID-19 vaccine demonstrate 89.3% efficacy in UK phase 3 trial. 2021. Jan 28. Available from: https://ir.novavax.com/news-releases/news-release-details/novavax-covid-19-vaccine-demonstrates-893-efficacy-uk-phase-3.
- 61.Johnson & Johnson Johnson & Johnson announces single-shot janssen COVID-19 vaccine candidate met primary endpoints in interim analysis of its phase 3 ensemble trial. 2021. Jan, Available from: https://www.jnj.com/johnson-johnson-announces-single-shot-janssen-covid-19-vaccine-candidate-met-primary-endpoints-in-interim-analysis-of-its-phase-3-ensemble-trial.
- 62.University of the Witwatersrand, Johannesburg Oxford Covid-19 vaccine trial results. 2021. Feb, Available from: https://www.wits.ac.za/covid19/covid19-news/latest/oxford-covid-19-vaccine-trial-results.html.
- 63.COG-UK, Covid-19 Genomics UK Consortium COG-UK report on SARS-CoV-2 spike mutations of interest in the UK. 2021. Jan 15. Available from: https://www.cogconsortium.uk/wp-content/uploads/2021/01/Report-2_COG-UK_SARS-CoV-2-Mutations.pdf.
- 64.Wibmer CK, Ayres F, Hermanus T, Madzivhandila M, Kgagudi P, Lambson BE, et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. BioRxiv. 2021 Jan 19; doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- 65.Liu Z, VanBlargan LA, Bloyet LM, Rothlauf PW, Chen RE, Stumpf S, et al. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe. 2021 Jan 27;S1931–3128((21)):00044–5. doi: 10.1016/j.chom.2021.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Greaney AJ, Loes AN, Crawford KHD, Starr TN, Malone KD, Chu HY, et al. Comprehensive mapping of mutations to the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human serum antibodies. BioRxiv. 2021 Jan 4; doi: 10.1016/j.chom.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]