Abstract
BACKGROUND
Aedes aegypti is a remarkably effective mosquito vector of epidemiologically important arboviral diseases including dengue fever, yellow fever and Zika. The present spread of resistance against pyrethroids, the primary insecticides used for mosquito control, in global populations of this species is of great concern. The voltage‐gated sodium channel (VGSC) in the nervous system is the known target site of pyrethroids in insects. Past studies have revealed several amino‐acid substitutions in this channel that confer pyrethroid resistance, which are known as knockdown resistance (kdr) mutations.
RESULTS
This study investigated a laboratory colony of Ae. aegypti, MCNaeg, established from larvae collected in Rio de Janeiro, Brazil in 2016. The MCNaeg colony showed strong resistance against pyrethroids without laboratory selection. Of the two VGSC gene haplotypes present within this colony, one harbored three known kdr mutations, V410L, V1016I, and F1534C, and the other harbored only the known F1534C mutation. In latter haplotype, we also found novel amino‐acid substations including V253F. Previous molecular modeling and electrophysiological studies suggest that this residue serves a pyrethroid‐sensing site in the second receptor, PyR2. Our genetical analysis showed that the haplotype harboring V253F and F1534C is associated with equal or slightly stronger resistance than the other triple kdr haplotype to both Type I and Type II pyrethroids.
CONCLUSION
The novel substitution V253F is potentially involved in pyrethroid resistance in Ae. aegypti. Further studies are needed to elucidate the role of this substitution in the pyrethroid susceptibility of VGSC.
© 2021 The Authors. Pest Management Science published by John Wiley & Sons Ltd on behalf of Society of Chemical Industry.
Keywords: pyrethroids, knockdown resistance, Aedes aegypti, sodium channel
We found a novel amino‐acid substitution V253F in the voltage‐gated sodium channel in Aedes aegypti which is potentially involved in pyrethroid resistance of this vector mosquito
![]()
1. INTRODUCTION
Pyrethroids include a number of natural and synthetic chemicals used as the active ingredients of insecticides. Because of their rapid effect and highly selective toxicity to insects versus mammals, pyrethroids are essential for the control of medically important insect vectors. They are used in indoor residual spraying formulations and insecticide‐treated bed net. The insecticidal activity of pyrethroids results from their inhibition of voltage‐gated sodium channel (VGSC) in the nervous system. Pyrethroids bind to the insect VGSC and prolong the channel's open state, hereby prohibiting normal signal transduction and causing paralysis. 1 Several specific amino‐acid substitutions in the VGSC are known to decrease its sensitivity to pyrethroids. 2 , 3 These amino‐acid substitutions, which are known as genetic factor kdr (knockdown resistance), has been observed in many agricultural and medically important arthropod pests. 2 , 3
The alpha subunit of eukaryotic VGSC consists of a single polypeptide chain including four homologous repeat domains (I–IV), each having six transmembrane segments (S1–S6). Recent molecular modeling and electrophysiological studies 4 , 5 suggest the presence of two pyrethroid receptors, PyR1 and PyR2, within the VGSC. In the proposed model, each receptor includes the residues located in IIL45 (the loop between the transmembrane segments S4 and S5 in domain II), IIS5 (the transmembrane segment S5 in domain II), IIS6, and IIIS6 and in IL45, IS5, IS6, and IIS6, respectively, with rotationally quasi‐symmetric disposition. It is considered that simultaneous binding of pyrethroid molecules to each receptor is required for inhibition of VGSC. Many of the residues located in the two receptors correspond known kdr substitutions associated with pyrethroid resistance in nature. 5
Aedes aegypti, the yellow fever mosquito, is a remarkably effective vector for numerous important human arbovirus diseases, including dengue fever, yellow fever, Chikungunya, and Zika. In this species, the amino‐acid variation associated with pyrethroid resistance include V410L, G923V, L982W, S989P, A1007G, I1011V/M, V1016G/I, T1520I, F1534C/L, and D1763Y (amino‐acid positions corresponding to the Musca domestica VGSC model) (see references in Fan et al., 6 Du et al., 7 and Chen et al. 8 ). In Latin America, F1534C is the kdr substitution most frequently reported to date. V1016I (V1023I in some studies) is another kdr variant common in Latin America, along with the F1534C substitution. 9 A congenic strain that inherited the F1534C haplotype from Thailand in the susceptible strain (ROCK) genetic background exhibited seven‐fold and 16‐fold resistance against permethrin (a type I pyrethroid) and deltamethrin (a type II pyrethroid), respectively. 10 The contribution of F1534C on the deltamethrin resistance phenotype, however, is still debatable. Multiple electrophysiological studies on VGSC heterologously expressed in Xenopus oocytes have shown that F1534C alone confers slight resistance to permethrin and dichlorodiphenyltrichloroethane (DDT) but not to deltamethrin. 11 , 12 , 13 Interestingly, V1016I enhances the effect of F1534C on the permethrin resistance in the electrophysiological experiment; this V1016I + F1534C double mutant even confers resistance to deltamethrin even though neither of the two mutations alone confers notable deltamethrin resistance. 11 The V410L variant, which often associates with F1534C, was first found in Brazil. 13 An electrophysiological study 13 of encoded channel indicates that this mutation confers strong resistant to both permethrin and deltamethrin. The channel with the double mutation V410L + F1534C is more resistant to permethrin than channels with either of the mutation alone. Concerningly, several reports show that a haplotype harboring all three mutations (V410L + V1016I + F1534C) already exists in natural population in America 6 , 14 , 15 , 16 and more recently in Africa. 17
In this article, we report several novel substitutions found in the Ae. aegypti VGSC from a pyrethroid‐resistant colony collected in Brazil. Among those substitutions, V253F resided at the residue V1k11, which has been implicated as one of the pyrethroid‐sensing residues in PyR2 under the dual‐receptor model. 4 The results obtained in this study prompt further electrophysiological and population genetical characterizations of this mutation to evaluate potential impact for future control of Ae. aegypti.
2. MATERIALS AND METHODS
2.1. Insects
Twelve Aedes aegypti larvae were collected on March 13, 2016 at Maracanã, Rio de Janeiro, Brazil. The larvae were kept in the laboratory insectarium and finally emerged into five males and seven females. Random crossing of those adult mosquitoes founded the MCNaeg colony. The SMK strain, which was originally from the United States and has been maintained in the laboratory for at least 20 years without exposure to insecticides, 18 was used as susceptible control. Larvae were fed insect foods purchased from the Oriental Yeast Company (Tokyo, Japan) and adults were fed 2% sugar water. Female mosquitoes were fed blood from a live mouse (Slc:ICR). The environmental condition of the insectarium was set on 25 °C under 16 h light/8 h dark photoperiodic cycle. Animal care and protocols were approved by the Animal Ethics Committee of National Institute of Infectious Diseases, Japan (approval number 119051).
2.2. Chemicals
Two pyrethroids were used for testing insecticide susceptibility: Permethrin (91.2%) was obtained from Sumitomo Chemical (Tokyo, Japan) and deltamethrin (99.4%) was obtained from GL Sciences (Tokyo, Japan). Piperonyl butoxide (PBO, 98.0%) was obtained from FUJIFILM Wako Pure Chemical Co. (Osaka, Japan).
2.3. Adult bioassay
Bioassays were conducted to assess the pyrethroid resistance of adult mosquitoes (mated females at the second inbred generation) by topical application as described previously. 18 Mosquitoes unable to stand at the bottom of the cup 24 h after treatment were considered dead. For the original MCNaeg colony, four batches of 20 females (total 80 females) were treated with 5.87 and 58.7 ng of permethrin. To estimate the half lethal dose (LD50), at least five different doses of permethrin or deltamethrin were used to achieve mortality > 0% and < 100%. At least 40 mosquitoes were used per single dose. To estimate the contribution of cytochrome P450s to resistance, 2 μg PBO (in 0.22 μL acetone) was applied to the thoracic notum of the mosquitoes 2 h before pyrethroid application. LD50 values for each population were calculated by fitting numbers of dead and surviving mosquitoes to log‐probit model 19 using the glm function in R v3. 20
2.4. Association between kdr genotypes and permethrin susceptibility
For the polymerase chain reaction (PCR) template, genomic DNA (gDNA) from dead and surviving mosquitoes of the MCNaeg colony after the exposure to 58.7 ng permethrin was prepared from two hind legs using the alkaline lysis method 21 modified for mosquito legs. 22 Domains I, II, and III of the VGSC genes were partially amplified and directly sequenced as previously described. 23 For genotyping the domain I V410 residue, primers Ae410F1 (5′‐TTACGATCAGCTGGACCGTG‐3′) and Ae434R3 (5′‐CTTCTTCCTCGGCGGCCTC‐3′) were used for PCR amplification. The cycling conditions for PCR were as follows: initial denaturation at 95 °C for 2 min, followed by 35 cycles of 98 °C for 10 s, 55 °C for 30 s, 68 °C for 30 s, and a final extension step at 68 °C for 5 min. The 180 bp amplicon was directly sequenced with primer Ae410F2 (5′‐ATCAGCTGGACCGTGGCA‐3′) and genotyped from the electropherogram. Fisher's exact test was conducted for difference in allele frequency between survived and dead group using the fisher. test function in R v3.
2.5. Isolation of two sub‐colonies from the MCNaeg colony
Two distinct haplotypes were identified in the MCNaeg colony that harbored the V410 and L410 alleles. To separate these haplotypes into two sub‐colonies, pupae of the MCNaeg colony were isolated, and each emerged adult was genotyped by PCR and direct sequencing using a hind leg as described earlier. Individuals with a homozygous genotype for either allele were selected and crossed separately. Isolated sub‐colonies were designated as MCNaeg‐C (with V410) or MCNaeg‐LIC (with L410).
2.6. Targeted capture sequencing
Genomic DNA was extracted from four individual pupae in each sub‐colony using MagExtractor ‐Genome‐ (Toyobo, Japan) as described previously. 16 Illumina library construction and hybridization capture was conducted with the biotinylated oligo probe designed from the Ae. albopictus VGSC gene, whose exons show > 92.5% homologies to corresponding exons in Ae. agypti except one tiny optional exon 16.5. 16 The libraries were pooled along with libraries for other projects and sequenced for 150 bp at both ends in MiniSeq (Illumina, Inc., Foster City, CA, USA) with the 300‐cycle Mid‐Output Sequencing kit. A range of 188 481 to 240 793 read pairs (50–72 Mb) was obtained per individual. The row next generation sequencing (NGS) reads were deposited in the DNA Data Bank of Japan (DDBJ) Sequence Read Archive (DRA) under accession number DRR234414–DRR234421.
2.7. Bioinformatic analysis
The obtained NGS read data were used to genotype and functionally annotate variants with respect to VGSC gene coding sequence (CDS) using the automated pipeline MoNaS (https://github.com/ItokawaK/MoNaS), 16 which depends on BWA, 24 SAMTools, 25 BEDTools, 26 and FreeBayes. 27 SKESA v.2.3.0 28 with default parameters (minimal kmer length = 21, maximum noise‐to‐signal ratio = 0.1) was used to assemble the NGS reads obtained from the targeted capture sequencing. The contigs containing the VGSC exons and flanking introns for each sub‐colony were submitted to DDBJ under accession number LC557523–LC557562.
3. RESULTS
3.1. Pyrethroid susceptibility and two VGSC haplotypes in MCNaeg
After permethrin exposure of 5.87 and 58.7 ng per mosquito doses, the females of the MCNaeg colony exhibited mortality rates of only 10.0% (n = 80) and 28.8% (n = 80), respectively. Because these doses were expected to eliminate almost 100% susceptible Ae. aegypti, 18 and corresponded to 99% lethal dose (LD99) and its ten times equivalent, respectively, for the susceptible strain of Ae. albopictus (the HKM strain), 23 the MCNaeg colony was considered highly resistance against permethrin. All 80 females used for the 58.7 ng permethrin challenge (56 survivors and 24 dead) were genotyped by direct sequencing of three regions of the VGSC gene that include codons of known kdr variations (V410L, V1016G/I, and F1534C). All of these mosquitoes were homozygous for the C1534 allele, while segregations of genotypes at the residue 410 (V or L) and 1016 (V or I) were observed. Because there were only three combinations of genotypes at the two segregating sites among the 80 females (Table 1), we suspected the MCNaeg colony had only two VGSC haplotypes, V–V–C or L–I–C corresponding to the residues 410, 1016, and 1534. Frequency of the L–I–C haplotype among dead and surviving mosquitoes was 70.8% and 37.5%, respectively. Intriguingly, the V–V–C haplotype seemed to have a slightly higher chance of being selected over the L–I–C haplotype under this permethrin exposure (Table 1, P < 0.05), even though the V–V–C haplotype had only F1534C as a ‘known’ kdr mutation.
Table 1.
Association between voltage‐gated sodium channel (VGSC) genotypes and responses to 58.7 ng permethrin challenge in the MCNaeg colony
| Genotype | Dead | Survived | Mortality (95% confidence interval) | ||
|---|---|---|---|---|---|
| V410L | V1016I | F1534C | n = 24 | n = 56 | |
| V/V | V/V | C/C | 1 | 4 | 0.20 (0.0051–0.72) |
| V/L | V/I | C/C | 6 | 31 | 0.16 (0.062–0.32) |
| L/L | I/I | C/C | 17 | 21 | 0.45 (0.29–0.62) |
|
Frequency of the V‐V‐C allele a (95% confidence interval) |
0.17 (0.075–0.30) |
0.35 (0.26–0.44) |
|||
Significant difference in the allele frequency between dead and survived (P = 0.023) in Fisher's exact test.
3.2. Isolation of two sub‐colonies with fixed VGSC haplotypes
We isolated 144 unmated mosquitoes (96 females; 48 males) in the MCNaeg colony and genotyped them at V410L. Frequencies of each genotype (V/V, V/L, and L/L) were 9, 42, and 45, respectively, among female and 4, 22, and 22, respectively, among males. The L410 homozygous mosquitoes (45 females; 22 males) were separated from the V410 homozygous mosquitoes (nine females; four males) kept in separate cages, and mated. This selection by genotype resulted in the isolation of two sub‐colonies, MCNaeg‐LIC (fixed with respect to L410) and MCNaeg‐C (fixed with respect to V410) (Fig. 1(A)). As expected from the results of phenotype–genotype associations in the original MCNaeg colony (Table 1), the MCNaeg‐C sub‐colony had equal or slightly higher resistance for both permethrin and deltamethrin (resistance ratio, RR, compared to the SMK strain were 42‐ and 64‐fold, respectively) than did the MCNaeg‐LIC (24‐ and 43‐fold) (Fig. 1(B)). Treatment with PBO, an inhibiter of cytochrome P450 detoxification enzymes that may be involved in pyrethroid resistance 18 along with the kdr mechanism, did not dramatically reduce the resistance ratios both in MCNaeg‐LIC (22‐ and 44‐fold) and MCNaeg‐C (34‐ and 71‐fold) (Fig. 1(B)).
Figure 1.
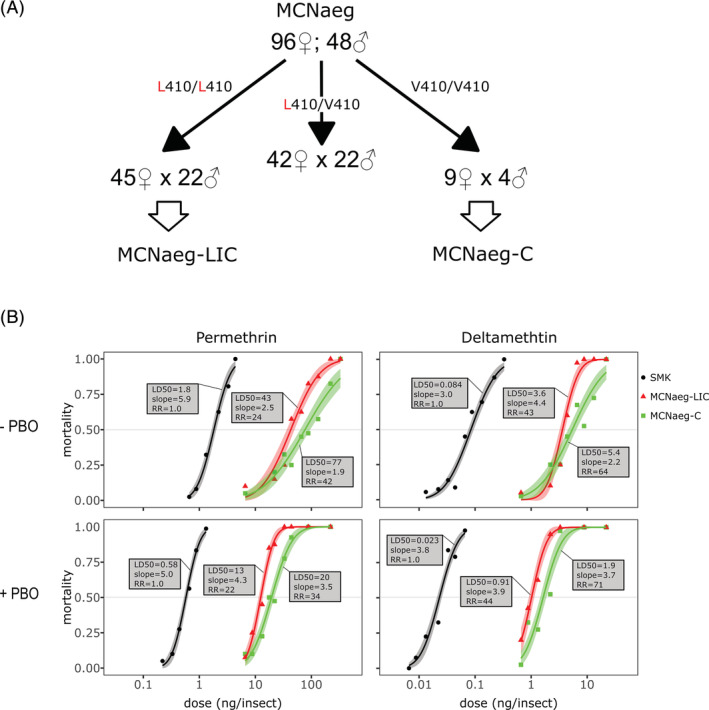
(A) Schematic diagram depicting the selection of two sub‐colonies MCNaeg‐LIC and MCNaeg‐C from the MCNaeg colony with the V410L genotype. (B) Dose‐response curve for permethrin and deltamethrin either with (+) or without (−) PBO for MCNaeg‐LIC, MCNaeg‐C and susceptible (SMK) adult females. Lines represent fitted curves for the log‐probit model, and bands with translucent colors are 95% confidential interval for morality. The LD50, slope and RR of each fitted curve are shown in the box.
3.3. Sequencing the VGSC coding sequence
The entire CDS of the VGSC gene was sequenced using targeted capture sequencing with hybridization probes designed from the Ae. albopictus VGSC gene. 16 Four individuals were sampled from each sub‐colony for sequencing analysis. In addition to the known variants V410L, V1016I, and F1534C, we identified four other amino‐acid substitutions: V253F (GTC > TTC at exon 7), M374I (ATG > ATA at exon 9), S723T (TCT > ACT at exon 15), and G923S (GGT > AGT at a mutually exclusive exon 19d) (Fig. 2(A)). The haplotype in the MCNaeg‐LIC sub‐colony appeared to harbor amino‐acid substitutions V410L, S723T, V1016I, and F1534C. The haplotype in the MCNaeg‐C sub‐colony, however, harbored amino‐acid substitutions V253F, M374I, G923S, and F1534C (Fig. 2(B, C)).
Figure 2.
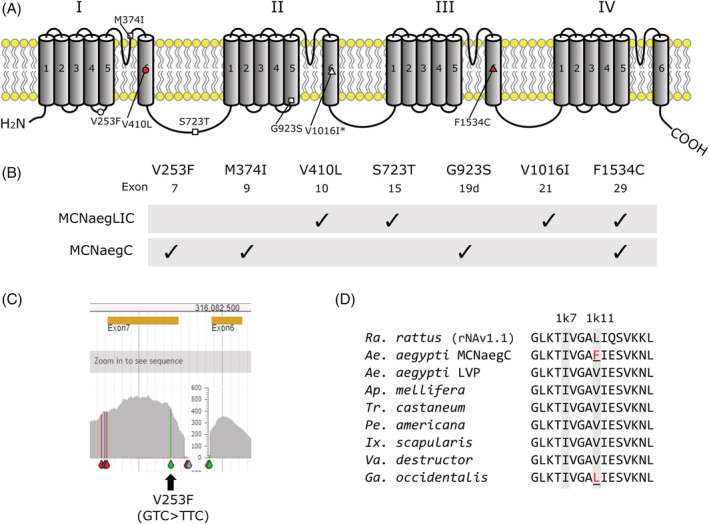
(A) Mapping of the amino‐acid substitutions discovered in haplotypes of the MCNaeg‐LIC and MCNaeg‐C sub‐colonies to the VGSC protein. Triangle and circles points indicate residues consisting of pyrethroid receptors 1 and 2, respectively, according to the dual‐receptor model by Du et al. 4 , 5 Rectangle points indicate residues with no known association with insecticide resistance. Filled points indicate known resistance substitutions with evidence from electrophysiological studies. V1016I annotated by an asterisk has been shown to enhance pyrethroid insensitivity mediated by F1534C. 11 (B) Amino‐acid substitutions in each VGSC haplotype. The exon in which each substituting mutation resides is indicated. (C) NGS read mapping view for a MCNaeg‐C individual in exon 7 of VGSC (chromosome III), including the V253F substation. (D) An alignment of the IL45 linker region of the sodium channel in the rat Rattus rattus rNAv1.1 (NP_110502), and Ae. aegypti Liverpool (LVP) and MCNaeg‐C (LC557527) strains, the bee Apis mellifera (XP_006561583.1), the beetle Tribolium castaneum (XP_015837360.1), the cockroach Periplaneta americana (ACX44801.1), the tick Ixodes scapuraris (EEC03677.1), the mite Varroa destructor (XP_022662766.1), and the mite Galendromus (Metaseiulus) occidentalis (XP_028966827.1). Two pyrethroid‐sensing residues, 1 k7 and 1 k11 (according to Du et al. 4 ), are highlighted.
No heterozygous genotype within the CDS was found in any of the individuals sampled from either sub‐colony, allowing us to reconstruct the full sequences for each VGSC haplotype by assembling the NGS reads. The complete CDSs of the VGSC gene were compared to that of three other haplotypes in two Ae. aegypti laboratory colonies; SP‐01 from a colony established from mosquitoes collected in Singapore 2009, and Mex‐03 and Mex‐06 from colony established from mosquitoes collected in Monterrey, Mexico 2008 16 (Fig. 3). A total of 26 synonymous and non‐synonymous polymorphic sites were present within the whole CDS (approximately 6 kb) compared to the reference genome assembly the Liverpool strain, AaegL5. 29 The two haplotypes MCNaeg‐LIC and Mex‐06, which shared identical amino‐acid substitutions (V410L + S723T + V1016I + F1534C) 16 had exactly the same set of nucleotide substitutions across the entire CDS.
Figure 3.
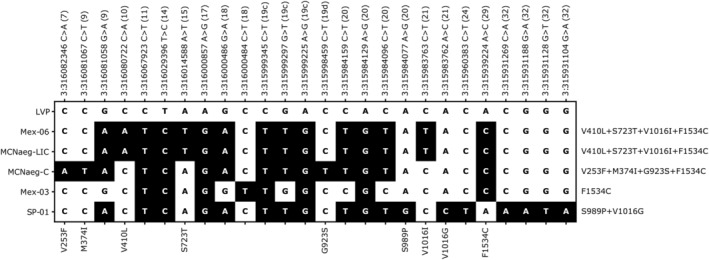
All synonymous and non‐synonymous nucleotide substitutions found in entire CDSs of VGSC genes in the Liverpool strain (genome reference, AaegL5), MCNaeg‐LIC and MCNaeg‐C sub‐colonies in this study, and specimens Mex‐03, Mex‐06, and SP‐01 from our previous study (SRA accession: DRR167899, DRR167902, and DRR167913, respectively), 16 as shown at the left side of the table. Genomic coordination of each substitution in the AaegL5 assembly are shown at the upper side of the table. Characters in parenthesis indicate VGSC gene exon numbers in which each nucleotide locus resides. Corresponding amino‐acid substitutions are shown at the bottom of the table if a substitution is non‐synonymous. Haplotypes with respect to amino‐acid substitutions are shown on the right side of the table.
4. DISCUSSION AND CONCLUSION
The Ae. aegypti colony, MCNaeg, which has been founded from mosquitoes collected in Rio de Janeiro Brazil, showed > 50% survivability on 58 ng/insect dose of permethrin, which was 38‐fold higher dose than LD50 of susceptible SMK strain (Fig. 1(B)), without laboratory selection. The colony included two different haplotypes of the VGSC gene harboring amino‐acid substitutions V410L + S723T + V1016I + F1534C (MCNaeg‐LIC) and V253F + M374I + G923S + F1534C (MCNaeg‐C) compared to the reference genome assembly (AaegL5). 29
Saavedra‐Rodriguez et al. 15 found VGSC haplotypes harboring the three kdr substitutions V410L, V1016I, and F1534C in specimens collected during 2002–2005. The S723T substitution located at the intracellular linker between domains I and II has been reported in at least three studies on Ae. aegypti collected in Mexico 16 , 30 and Puerto Rico. 31 In two of these studies, 16 , 31 the S723T substitution was associated with V410L, V1016I, and F1534C. Of note, the nucleotide sequence of the MCNaeg‐LIC VGSC haplotype is identical to that reported for the entire CDS region of the V410L + S723T + V1016I + F1534C haplotype found in mosquitoes collected in Mexico 2008. 16 This indicates that these haplotypes share the same evolutionally origin. Thus far, no evidence, yet, suggests that S723T is involved in VGSC to pyrethroids. Electrophysiological study on Ae. aegypti VGSC with the triple (V410L + V1016I + F1534C) and quadruple (V410L + S723T + V1016I + F1534C) substitutions are needed to correctly understand the selective advantage of these haplotypes in natural population.
The other haplotype (V253F + M374I + G923S + F1534C), isolated in the MCNaeg‐C sub‐colony, was genetically associated with at least comparable pyrethroid resistance to the V410L + S723T + V1016I + F1534C haplotype even though the F1534C is the only known kdr mutation implicated in pyrethroid resistance in the MCNaeg‐C thus far. Other three substitutions, V253F (at IL4), M374I (at IL5), and G923S (at IIS5) are novel in Ae. aegypti, and probably also in arthropods. Of these amino‐acid substitutions, we consider V253F to be particularly noteworthy. A valine in this residue is highly conserved among arthropods (Fig. 2(D)). Du et al. 4 , 5 state that this residue (also indexed as V1k11) constitutes the VGSC's second pyrethroid‐receptor, PyR2, in their dual‐receptor model. An electrophysiological study using Xenopus oocytes showed that substitution of this valine residue to alanine (V253A) in Ae. aegypti VGSC dramatically reduces the sensitivity of the channel to both deltamethrin and permethrin. 4 The effect of the V253F substitution on the electrophysiological characteristics of the channel is yet unknown. While G923S is a novel substitution, another substitution at the same amino‐acid site, G923V, has been reported in Ae. aegypti collected in three Latin American countries. 32 Although these colonies had pyrethroid resistance phenotypes, it is unclear whether G923V is responsible for the resistance because the I1011M kdr substitution was present in the same haplotype. 32
To the best of our knowledge, no amino‐acid substitution at the V1k11 residue has been reported in arthropods in nature. However, our search of the Genbank database (July 2020) identified the L253 VGSC allele in a predatory mite species Galendromus (Metaseiulus) occidentalis, which is used as a commercial biological insecticide, from entries of the genome assembled for a carbaryl‐organophosphate‐sulfur selected resistance inbred strain 33 (Fig. 2(D)). Intriguingly, leucine at this site is homologous to that of the mammalian VGSC alpha subunits. According to Du et al., 4 L1k11V conversion on one of the rat sodium channels (rNav1.4) significantly increased the channel sensitivity to deltamethrin, indicating that the differing residues at this site may explain the selective action of pyrethroids between mammalian and insect sodium channels. 4 Thus, the prospect that a V1k11L mutation possibly occurred in this mite VGSC (conversion to mammalian type) confers the pyrethroid resistance of this beneficial predator 34 is intriguing.
We observed the MCNaeg‐C sub‐colony fixed with the V253F + M374I + G923S + F1534C haplotype showed comparable or rather slightly higher resistance to the MCNaeg‐LIC sub‐colony with three kdr mutations (V410L, V1016I, and F1534C). At present, however, whether the V253F + M374I + G923S + F1534C haplotype actually confers the same level of resistance as V410L + S723T + V1016I + F1534C is unclear because genetic studies alone cannot exclude the presence of another resistance factor hitchhiking to the VGSC haplotype. Further studies, especially electrophysiological and genome editing analyses, 35 are required to fully understand the contribution of the V253F substitution to the resistance phenotype.
CONFLICT OF INTEREST
There is no interest to declare with this study.
ACKNOWLEDGEMENTS
This study was partially supported by the Research Program on Emerging and Re‐emerging Infectious Diseases from Japan Agency for Medical Research and Development (AMED, grant numbers: JP20wm0225007 and JP20fk0108067) and by Japan Initiative for Global Research Network on Infectious Diseases (J‐GRID) from Ministry of Education, Culture, Sports, Science and Technology in Japan and AMED (grant number: JP17fm0108019).
REFERENCES
- 1. Narahashi T, Neuroreceptors and ion channels as the basis for drug action: past, present, and future. J Pharmacol Exp Ther 294:1–26 (2000). [PubMed] [Google Scholar]
- 2. Rinkevich FD, Du Y and Dong K, Diversity and convergence of sodium channel mutations involved in resistance to pyrethroids. Pestic Biochem Physiol 106:93–100 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dong K, Du Y, Rinkevich F, Nomura Y, Xu P, Wang L et al., Molecular biology of insect sodium channels and pyrethroid resistance. Insect Biochem Mol Biol 50:1–17 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Du Y, Nomura Y, Satar G, Hu Z, Nauen R, He SY et al., Molecular evidence for dual pyrethroid‐receptor sites on a mosquito sodium channel. Proc Natl Acad Sci 110:11785–11790 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Du Y, Nomura Y, Zhorov BS and Dong K, Rotational symmetry of two pyrethroid receptor sites in the mosquito sodium channel. Mol Pharmacol 88:273–280 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fan Y, O'Grady P, Yoshimizu M, Ponlawat A, Kaufman PE and Scott JG, Evidence for both sequential mutations and recombination in the evolution of kdr alleles in Aedes aegypti , ed. by Sunil S. PLoS Negl Trop Dis 14:e0008154 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Du Y, Nomura Y, Zhorov BS and Dong K, Sodium channel mutations and pyrethroid resistance in Aedes aegypti . Insects 7:60 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen M, Du Y, Nomura Y, Zhorov BS and Dong K, Chronology of sodium channel mutations associated with pyrethroid resistance in Aedes aegypti . Arch Insect Biochem Physiol 104:e21686 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Saavedra‐Rodriguez K, Urdaneta‐Marquez L, Rajatileka S, Moulton M, Flores AE, Fernandez‐Salas I et al., A mutation in the voltage‐gated sodium channel gene associated with pyrethroid resistance in Latin American Aedes aegypti . Insect Mol Biol 16:785–798 (2007). [DOI] [PubMed] [Google Scholar]
- 10. Fan Y and Scott JG, The F1534C voltage‐sensitive sodium channel mutation confers 7‐ to 16‐fold resistance to pyrethroid insecticides in Aedes aegypti . Pest Manag Sci 76:2251–2259 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen M, Du Y, Wu S, Nomura Y, Zhu G, Zhorov BS et al., Molecular evidence of sequential evolution of DDT‐ and pyrethroid‐resistant sodium channel in Aedes aegypti , ed. by Lenhart A. PLoS Negl Trop Dis 13:e0007432 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hirata K, Komagata O, Itokawa K, Yamamoto A, Tomita T and Kasai S, A single crossing‐over event in voltage‐sensitive Na+ channel genes may cause critical failure of dengue mosquito control by insecticides. PLoS Negl Trop Dis 8:e3085 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Haddi K, Tomé HVV, Du Y, Valbon WR, Nomura Y, Martins GF et al., Detection of a new pyrethroid resistance mutation (V410L) in the sodium channel of Aedes aegypti: a potential challenge for mosquito control. Sci Rep 7:46549 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Granada Y, Mejía‐Jaramillo A, Strode C and Triana‐Chavez O, A point mutation V419L in the sodium channel gene from natural populations of Aedes aegypti is involved in resistance to λ‐cyhalothrin in Colombia. Insects 9:23 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Saavedra‐Rodriguez K, Maloof FV, Campbell CL, Garcia‐Rejon J, Lenhart A, Penilla P et al., Parallel evolution of vgsc mutations at domains IS6, IIS6 and IIIS6 in pyrethroid resistant Aedes aegypti from Mexico. Sci Rep 8:1–9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Itokawa K, Sekizuka T, Maekawa Y, Yatsu K, Komagata O, Sugiura M et al., High‐throughput genotyping of a full voltage‐gated sodium channel gene via genomic DNA using target capture sequencing and analytical pipeline MoNaS to discover novel insecticide resistance mutations, ed. by Asgari S. PLoS Negl Trop Dis 13:e0007818 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ayres CFJ, Seixas G, Borrego S, Marques C, Monteiro I, Marques CS et al., The V410L knockdown resistance mutation occurs in Island and continental populations of Aedes aegypti in west and Central Africa, Bonizzoni M. PLoS Negl Trop Dis 14:e0008216 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kasai S, Komagata O, Itokawa K, Shono T, Ng LC, Kobayashi M et al., Mechanisms of pyrethroid resistance in the dengue mosquito vector, Aedes aegypti: target site insensitivity, penetration, and metabolism. PLoS Negl Trop Dis 8:e2948 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Finney DJ, Probit Analysis. Cambridge University Press, Cambridge: (1977). [Google Scholar]
- 20. R_Development_Core_Team, R , A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna: (2014). [Google Scholar]
- 21. Rudbeck L and Dissing J, Rapid, simple alkaline extraction of human genomic DNA from whole blood, buccal epithelial cells, semen and forensic stains for PCR. Biotechniques 25:588–590, 592 (1998). [DOI] [PubMed] [Google Scholar]
- 22. Itokawa K, Komagata O, Kasai S, Ogawa K and Tomita T, Testing the causality between CYP9M10 and pyrethroid resistance using the TALEN and CRISPR/Cas9 technologies. Sci Rep 6:24652 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kasai S, Caputo B, Tsunoda T, Cuong TC, Maekawa Y, Lam‐Phua SG et al., First detection of a Vssc allele V1016G conferring a high level of insecticide resistance in Aedes albopictus collected from Europe (Italy) and Asia (Vietnam), 2016: a new emerging threat to controlling arboviral diseases. Eurosurveillance 24:1700847 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li H and Durbin R, Fast and accurate short read alignment with Burrows‐Wheeler transform. Bioinformatics 25:1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N et al., The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Quinlan AR and Hall IM, BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Garrison E and Marth G, Haplotype‐Based Variant Detection from Short‐Read Sequencing (2012).
- 28. Souvorov A, Agarwala R and Lipman DJ, SKESA: strategic k‐mer extension for scrupulous assemblies. Genome Biol 19:153 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Matthews BJ, Dudchenko O, Kingan SB, Koren S, Antoshechkin I, Crawford JE et al., Improved reference genome of Aedes aegypti informs arbovirus vector control. Nature 563:501–507 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Saavedra‐Rodriguez K, Campbell CL, Lenhart A, Penilla P, Lozano‐Fuentes S and Black WC, Exome‐wide association of deltamethrin resistance in Aedes aegypti from Mexico. Insect Mol Biol 28:591–604 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pinch M, Rodriguez SD, Mitra S, Kandel Y, Moore E and Hansen IA, Low levels of pyrethroid resistance in hybrid offspring of a highly resistant and a more susceptible mosquito strain. J Insect Sci 20:1 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Brengues C, Hawkes NJ, Chandre F, Mccarroll L, Duchon S, Guillet P et al., Pyrethroid and DDT cross‐resistance in Aedes aegypti is correlated with novel mutations in the voltage‐gated sodium channel gene. Med Vet Entomol 17:87–94 (2003). [DOI] [PubMed] [Google Scholar]
- 33. Hoy MA, Waterhouse RM, Wu K, Estep AS, Ioannidis P, Palmer WJ et al., Genome sequencing of the phytoseiid predatory mite metaseiulus occidentalis reveals completely atomized hox genes and superdynamic intron evolution. Genome Biol Evol 8:1762–1775 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. HOY MA and KNOP NF, Selection for and genetic analysis of permethrin resistance in metaseiulus occidentals: genetic improvement of a biological control agent. Entomol Exp Appl 30:10–18 (1981). [Google Scholar]
- 35. Samantsidis GR, O'Reilly AO, Douris V and Vontas J, Functional validation of target‐site resistance mutations against sodium channel blocker insecticides (SCBIs) via molecular modeling and genome engineering in drosophila. Insect Biochem Mol Biol 104:73–81 (2019). [DOI] [PubMed] [Google Scholar]


