Abstract
In amyotrophic lateral sclerosis (ALS), early diagnosis is essential for both current and potential treatments. To find a supportive approach for the diagnosis, we constructed an artificial intelligence‐based prediction model of ALS using induced pluripotent stem cells (iPSCs). Images of spinal motor neurons derived from healthy control subject and ALS patient iPSCs were analyzed by a convolutional neural network, and the algorithm achieved an area under the curve of 0.97 for classifying healthy control and ALS. This prediction model by deep learning algorithm with iPSC technology could support the diagnosis and may provide proactive treatment of ALS through future prospective research. ANN NEUROL 2021;89:1226–1233
Amyotrophic lateral sclerosis (ALS) is an intractable motor neuron disease with a survival period of 3 to 5 years after onset if a mechanical ventilator is not adopted. 1 The clinical diagnosis of ALS is based on neurological findings including muscle weakness and increased tendon reflexes, and these do not manifest themselves until the disease has progressed. 2 In ALS, early diagnosis is necessary even for the few approved therapies to treat symptoms 3 and for further effective disease‐modifying therapies to protect motor neurons that are expected to be discovered in the future. We hereby propose a prediction model of ALS using deep learning with images of motor neurons derived from patient induced pluripotent stem cells (iPSCs) to support ALS diagnosis.
Deep learning algorithms have recently been shown to perform well in classification in certain fields such as skin cancer diagnosis by pictures of skin legions, 4 lung cancer diagnosis by computed tomographic (CT) imaging, 5 pathological diagnosis of cancers for prediction of therapeutic efficacy, 6 and biological science. 7 Here, we classified images of motor neurons derived from iPSCs of healthy control subjects and ALS patients using deep learning algorithms. This study investigated the concept of predicting ALS by a combination of deep learning algorithms and iPSC technology.
Subjects and Methods
Ethics
The generation and use of human iPSCs was approved by the ethics committees at Kyoto University. All methods were performed in accordance with approved guidelines. Formal informed consent was obtained from all subjects.
Generation of iPSCs
iPSCs were generated from fibroblasts or peripheral blood mononuclear cells (PBMCs) of healthy control subjects and ALS patients using episomal vectors for OCT3/4, Sox2, Klf4, L‐Myc, Lin28, and dominant‐negative p53 or OCT3/4, Sox2, Klf4, L‐Myc, Lin28, and shRNA for p53, respectively, as previously reported. 8 They were cultured by feeder‐free and xeno‐free culture systems with StemFit (Ajinomoto, Tokyo, Japan) with penicillin/streptomycin.
Analysis for ALS‐Related Genes
Genomic DNA was isolated from fibroblasts or PBMCs from healthy controls or ALS patients using PureLink Genomic DNA Mini Kit (Thermo Fisher Scientific, Waltham, MA).
To investigate the presence of mutations in ALS‐related genes, including SOD1, TARDBP, FUS, OPTN, VCP, SQSTM1, SIGMAR1, UBQLN2, and TBK1, target sequencing was performed using the Ion Proton System (Life Technologies, Carlsbad, CA) according to the protocol for the Ion AmpliSeq Library Kit 2.0 (Life Technologies) and Ion PI Hi‐Q Chef Kit (Life Technologies) in the same manner as already reported. 9 Amplicon coverage was investigated after Ion Proton sequencing. Variants were identified using Torrent Suite software 5.10 and verified by standard polymerase chain reaction–based amplification followed by Sanger sequencing using an Applied Biosystems 3130 DNA sequencer (Life Technologies). Variants were evaluated by predictive algorithms (Sorting Intolerant from Tolerant [http://sift.jcvi.org/], PolyPhen‐2 [http://genetics.bwh.harvard.edu/pph2/], MutationTaster [http://www.mutationtaster.org/], and Combined Annotation Dependent Depletion [CADD; https://cadd.gs.washington.edu/]) that predicted whether amino acid substitutions would affect protein function. Furthermore, dbSNP137 (http://www.ncbi.nlm.nih.gov/SNP/), ToMMo (https://jmorp.megabank.tohoku.ac.jp/), 1000 Genomes (http://www.1000genomes.org/), and ExAC (https://gnomad.broadinstitute.org/) were also used. Variants were evaluated for the pathogenicity under the following conditions: that 2 or more prediction algorithms were positive (CADD score threshold was set as >20) and that the frequency of the variant was ≤0.005, and that the registration information in ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) was also used. C9ORF72 repeat expansion was evaluated according to the previously established protocol. 10
Motor Neuron Differentiation from iPSCs
Motor neurons were differentiated from iPSCs as previously described. Briefly, iPSCs were dissociated to single cells and quickly reaggregated in low cell adhesion U‐shaped 96‐well plates (Lipidule‐Coated Plate A‐U96; NOF Corporation, Tokyo, Japan). Aggregations were cultured in EB medium containing Dulbecco modified Eagle medium/Ham's F12 (Thermo Fisher Scientific), 5% KSR (Invitrogen, Waltham, MA), minimum essential medium–nonessential amino acids (Invitrogen), L‐glutamine (Sigma‐Aldrich, St Louis, MO), 2‐mercaptoethanol (Wako, Osaka, Japan), 2μM dorsomorphin (Sigma‐Aldrich), 10μM SB431542 (Cayman, Ann Arbor, MI), 3μM CHIR99021 (Cayman), and 12.5ng/ml fibroblast growth factor (Wako) in a neural inductive stage for 11 days. One hundred nanomolars retinoic acid (Sigma‐Aldrich) and 500nM smoothened ligand (Enzo Life Sciences, Farmingdale, NY) were added on day 4. After patterning with neurobasal medium (Thermo Fisher Scientific) supplemented with B27 Supplement (Thermo Fisher Scientific), 100nM retinoic acid, 500nM smoothened ligand, and 10μM DAPT (Selleck, Houston, TX), the aggregates were separated by Accumax (Innovative Cell Technologies, San Diego, CA), dissociated into single cells, and adhered to Matrigel (BD Biosciences, Franklin Lakes, NJ)‐coated dishes on day 16. Adhesive cells were cultured in neurobasal medium with 10ng/ml brain‐derived neurotrophic factor (R&D Systems, Minneapolis, MN), 10ng/ml glial cell line‐derived neurotrophic factor (R&D Systems), and 10ng/ml neurotrophin‐3 (R&D Systems). On day 21, cells were dissociated to single cells using Accumax and seeded to iMatrix‐coated 24‐well plates (Corning, Corning, NY) at 2 × 105 cells/well. On day 23, cells were fixed with 4% paraformaldehyde followed by immunostaining and imaging.
Immunostaining and Image Acquisition
Cells were fixed in 4% paraformaldehyde for 30 minutes at room temperature, washed twice with phosphate‐buffered saline (PBS), and permeabilized in PBS containing 0.1% Triton X‐100 for 10 minutes at room temperature, followed by blocking for 30 minutes with Block Ace (Yukijirushi, Tokyo, Japan). After incubation with primary antibody, anti–neurofilament H antibody (1:1,000, AB1989; Millipore, Billerica, MA), overnight at 4°C, cells were washed 3 times with PBS and then incubated with secondary antibody with Alexa 488 for 1 hour at room temperature. Cell images were acquired with IN Cell Analyzer 6000 (GE Healthcare, Chicago, IL). The number of cells, neurite length, and soma size were quantified with IN Cell Developer toolbox software v1.9 (GE Healthcare).
For the analysis of cleaved caspase‐3, cells were stained with anti–cleaved caspase‐3 antibody (1:500; Cell Signaling Technology, Danvers, MA) and anti–βIII tubulin antibody (1:1,000, Millipore), and the number of cleaved caspase‐3‐positive cells and βIII tubulin‐positive cells were quantified using IN Cell Developer toolbox software v1.9 (GE Healthcare) following image acquisition by IN Cell Analyzer 6000 (GE Healthcare).
Data Preparation and Training of Convolutional Neural Networks
Control and ALS motor neurons were assigned an encoded anonymous number for the samples with sample names encoded, and randomly cultured in the same 24‐well plates, considering that potential confounders such as plate location varied sufficiently between control and disease clones. Immunohistochemical staining was conducted in a blinded manner so that the clone name was not recognizable. After image acquisition, the sample names were matched to use them for deep learning. The motor neuron images stored as 16‐bit grayscale in tiff format were converted to 8‐bit RGB images. To correct low‐biased intensity distribution of the original images and reduce the variance of the intensity distribution range between samples, we normalized and rescaled the intensity so that the median value of each image aligned to 63. If the intensity exceeded 255, the values were clipped. We recruited a VGG‐16 network pretrained by the ImageNet dataset and substituted fully connected layers with new layers with regularization effect (Supplementary Table S1). The input images were rescaled to 256 × 256, and a mean of ImageNet images was subtracted. The images in training datasets were randomly rotated, shifted, zoomed, and flipped when input into the network. In the training, only weights of the substituted fully connected layers were updated. The weights were optimized by momentum stochastic gradient descent algorithm. Learning rate (10−4 ≤ learning rate ≤ 10−2) and momentum (0.7 ≤ momentum ≤ 1.0) were tuned by Hyperas (v0.4) module according to the accuracy for validation data. The cost function of the network was computed as the cross‐entropy errors between the distribution of the predicted class and that of the actual class. The size of the training batch was 32. Then, a network was trained for 64 epochs with tuned hyper parameters, and prediction performance was tested with test data. Accuracy and area under the curve (AUC) of receiver operating characteristic (ROC) curve were evaluated. To interpret where the convolutional neural network (CNN) focused so as to process an image, gradient‐weighted class activation mapping (Grad‐CAM) and guided Grad‐CAM algorithm were used. 11 All of the above processes were implemented in the Keras/TensorFlow framework (v1.13.1) 12 on GTX1080Ti by using CUDA 10.0.
Classification with Random Forest Classifiers
The total neurite length, number of cells, and average size of neuronal soma were obtained with IN Cell Analyzer 6000. Using the 3 features, random forest classifiers were trained to estimate whether the cell images were derived from healthy control subjects or ALS patients. AUCs of ROC curves were evaluated to compare the performance with CNNs. Grouping for training datasets and test datasets was exactly the same as that used in the classification by CNNs. The random forest classifiers were implemented with scikit‐learn (v0.23.2).
Results
A strategy for predicting ALS was constructed by deep CNNs using images of iPSC‐derived motor neurons (Fig 1). iPSCs from 15 healthy control subjects and 15 sporadic ALS patients without mutations in ALS‐related genes (Table 1, Supplementary Table S2) were differentiated to spinal motor neurons as previously described. 13 , 14 The samples on day 23 were immunostained with a neuronal marker, neurofilament H, followed by acquisition of images using confocal microscopy in a blinded manner. Representative motor neuron figures from healthy control subjects and ALS patients are presented in Figure 1D. Tensorflow/Keras 12 was used to construct a VGG‐16 network 15 pretrained with ImageNet 16 dataset and to conduct transfer learning for classifying the images of healthy control motor neurons and of ALS motor neurons. The fully connected layers of the network were substituted with new layers, but only with those that were trained by the cell images. A series of 225 images was collected by each clone, and a total of 4,500 images from 10 clones from healthy control subjects and 10 clones from ALS patients were used as a training dataset. A total of 1,350 images from 3 independent healthy control clones and 3 ALS clones were used as a validation dataset, and 900 images from 2 independent healthy control clones and 2 ALS clones were used as a test dataset. The clones used for training, validation, and test, respectively, did not overlap. For unbiased evaluation, we constructed a minimal set of analyses to ensure that all clones were included in both validation and test at least once. The data splits were chosen in an unbiased manner, beyond making sure that all different clones were used in different groups. We independently formed 55 sets composed of training/validation/test datasets and ran evaluation (Fig 2, Supplementary Table S3). The CNNs learned training datasets without obvious increase in validation loss during training, which suggests that the prediction models were trained without clear overfitting. 17 The average accuracy of the classification was 0.90 ± 0.10 (mean ± standard deviation [SD]), and the AUC of the ROC curve for classification of healthy control motor neurons and ALS motor neurons was 0.97 ± 0.04 (mean ± SD). To visualize and understand how the images were processed in the pretrained CNN, Grad‐CAM was applied. It presented the detection of neurites in the output from block 2 and soma in that from block 3, indicating that CNN captured the alteration of the morphology of motor neurons, especially neurites and soma, although the findings could reflect the differences in intensity or pattern that would not necessarily be indicative of a morphological change. It is also important to note the experimental batch effect. 18 We evaluated the images obtained from 3 independent differentiation experiments as test data, using the training and validation sets already obtained, and found a high degree of accuracy in these batches (Supplementary Table S4). Furthermore, there was no apparent association of classification accuracy with the number of iPSC passages (n = 30 included 15 control and 15 ALS iPSCs, Spearman coefficient r = 0.33, p = 0.231).
FIGURE 1.
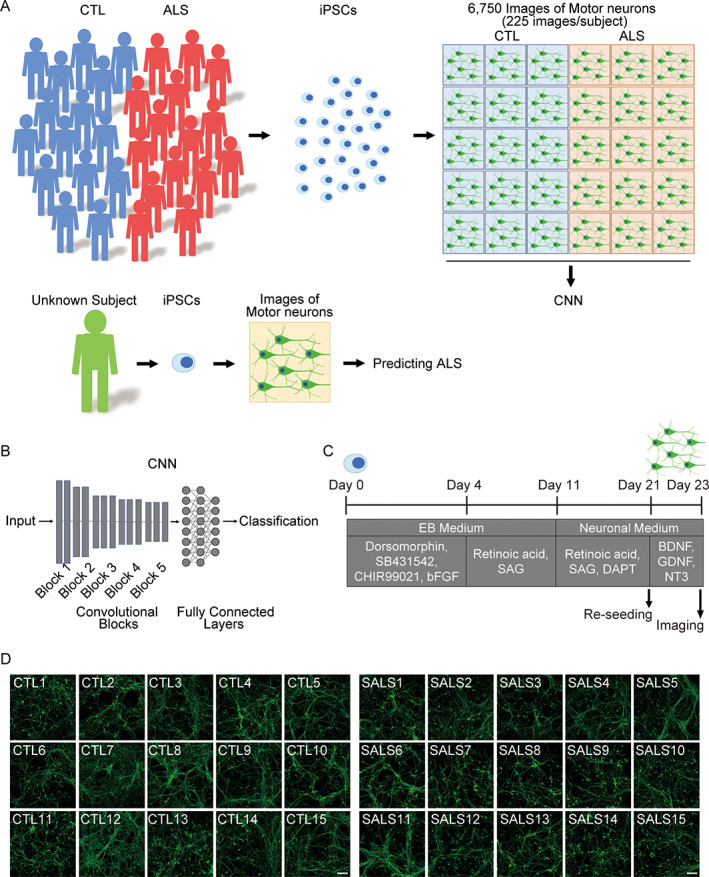
Amyotrophic lateral sclerosis (ALS) prediction model using induced pluripotent stem cell (iPSC)‐derived motor neurons. (A) Schematic illustration of the prediction model for ALS using iPSC‐derived motor neurons and convolutional neural network (CNN). (B) Schema of CNN. VGG‐16 network pretrained by ImageNet dataset and substitute fully connected layers were recruited. (C) Differentiation schedule of motor neurons from iPSCs for imaging. (D) Representative images of motor neurons derived from iPSCs of healthy control (CTL) subjects and sporadic ALS (SALS) patients. Scale bars = 100μm. BDNF = brain‐derived neurotrophic factor; bFGF = basic fibroblast growth factor; GDNF = glial cell line‐derived neurotrophic factor; NT‐3 = neurotrophin‐3; SAG = smoothened agonist.
TABLE 1.
Clinical Information for Subjects Included in Deep Learning
| Healthy Controls, n = 15 | ALS, n = 15 | |
|---|---|---|
| Gender, n (%) | ||
| F | 8 (53.3%) | 9 (60.0%) |
| M | 7 (47.7%) | 6 (40.0%) |
| Age at iPSC establishment, yr | 64.3 ± 13.5 | 56.8 ± 9.7 |
| Duration of illness, yr | NA | 3.9 ± 4.2 |
| Bulbar type, n (%) | NA | 2 (13.3%) |
Duration of illness refers to the period from the onset of the disease to the collection of somatic cells for iPSC generation.
ALS = amyotrophic lateral sclerosis; F = female; iPSC = induced pluripotent stem cell; M = male; NA, not applicable.
FIGURE 2.
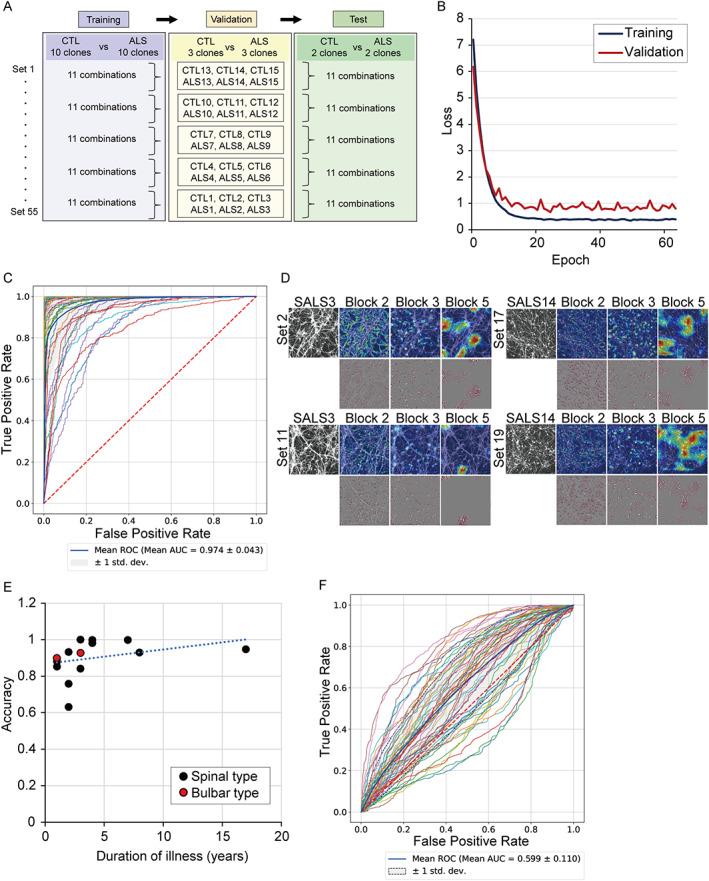
Classification performance in amyotrophic lateral sclerosis (ALS) motor neurons. (A) Strategy of model construction of learning, validation, and test. (B) Loss function of learning. There was no increase in loss even in the late stage of learning. (C) Receiver operating characteristic (ROC) curve of classification for healthy controls and ALS subjects from 55 test datasets. The average curve is shown by the thick blue line. (D) Saliency maps for ALS motor neurons by gradient‐weighted class activation mapping (Grad‐CAM) in upper row and guided Grad‐CAM in lower row. The saliency map was centered around neurites in block 2 and soma of motor neurons in block 3. (E) Correlation of the duration of illness and accuracy of the classification. The accuracy of each ALS sample averaged in each set is shown. The positive correlation between the duration of illness and accuracy is presented; n = 15, Spearman coefficient r = 0.61, p = 0.018. Duration of illness refers to the period from the onset of the disease to the collection of somatic cells for iPSC generation. Correlation of accuracy was not seen between the bulbar type and spinal type of ALS. (F) The random forest classifier resulted in low classification accuracy using these features. Using the same sample set as deep learning experiments, area under the curve (AUC) for ALS diagnosis by the random forest was approximately 0.60. CTL = control.
Next, we examined the relevance between clinical information and accuracy of the classification. There was a correlation between the duration of illness, that is, the duration from the onset of the disease to the sampling for iPSC generation, and accuracy, although the analysis was based on a limited number of samples (see Fig 2E). The correlation between clinical types of ALS and accuracy was not determined due to the small sample size.
We evaluated the performance when the same task was achieved by 10 well‐trained cell biologists, and found that the correct answer rate was 0.54 ± 0.08 (mean ± SD), showing that CNN presented a higher performance of classification than humans could achieve. The ratio of cell death detected by cleaved caspase‐3 was not different between control and ALS (control, n = 15; ALS, n = 15; mean ± SD, not significant by Student's t‐test). We also evaluated the original images used in deep learning by a conventional analytic method using high content analysis. ALS motor neurons showed a tendency to have smaller values for cell number, soma size, and neurite length compared to controls, but we could not find any significant differences in these cellular features between control motor neurons and ALS motor neurons (control, n = 15; ALS, n = 15; not significant by Student's t‐test). Furthermore, the random forest classifier, a conventional machine learning model, resulted in low classification accuracy using these features (see Fig 2F).
Discussion
We demonstrated a prediction model of ALS using deep learning algorithms and images of iPSC‐derived motor neurons. Research using iPSCs presents advantages such as utilizing cells with the same genetic background as the donor for various disease models and drug screenings. 19
The diagnosis of ALS depends on clinical observation and electrophysiological analysis, and reducing the diagnostic delay is crucial for the avoidance of treatment delay as well as for patient well‐being. 20 The disease staging is important, because ALS is a continuously evolving process and treatments might need to be administered at precise time points of the disease course to be beneficial. 21 Progression of the disease in patients is considered to be one of the causes of clinical trial failure, because motor neurons start to disappear, and the urgency for diagnosis is increasingly emphasized. Attempts to create new diagnostic criteria 22 and the development of biomarkers 23 , 24 are promising, but they have as yet not predicted ALS diagnosis. 25 Thus, by combining our model, which is expected to be a supportive tool for ALS diagnosis, and clinical biomarkers including blood/spinal fluid neurofilament levels, it may be possible to classify ALS with even greater accuracy. Furthermore, for ALS patients with progressed stage, there may be a possibility of applying this model to prognosis prediction and the stratification of patients for treatment selection in the future.
A previous report regarding the etiology of twins with ALS indicated that ALS has high heritability. 26 The genome‐wide association analysis identified risk single nucleotide polymorphisms in sporadic ALS, 24 and the shared polygenic risk and causal association in sporadic ALS has been reported. 27 Thus, ALS is closely related to the genetic background even in sporadic cases. iPSCs retain genetic information of the donor, although some epigenetic information is lost in the reprogramming procedure. 28 Several cellular phenotypes of motor neurons derived from sporadic ALS iPSCs have been reported previously. 29 , 30 , 31 The detection of motor neuron phenotypes may depend on the culture period and the differentiation protocol, and our relatively short culture period did not allow us to detect a significant cellular phenotype with high content analysis. Although each genetic alteration may present subtle cellular phenotypes by human eyes, deep learning algorithms succeeded in predicting the disease using imaging data.
Deep learning algorithms present high performance in the classification of images for classification in multiple medical situations, including skin cancer diagnosis by pictures of skin legions, 4 lung cancer diagnosis by CT imaging, 5 and pathological diagnosis for cancers. 6 They are expected to be applicable to medical examinations using materials from patients. On the other hand, our system, which is applied prior to the clinical stage, has 2 advantages: (1) the utility of motor neuron data not available from patients clinically and (2) the potential of predictive diagnosis of ALS. This shows advantages of the combination of deep learning algorithms and iPSC technology.
On the other hand, although we tried to address this, there are general issues regarding deep learning, for example, “black boxes” of the resulting network. 32 This study is based on a laboratory resource experiment, and we have not yet fully determined whether deep learning is looking at the actual features of the disease. Furthermore, the experimental batch effects related to the technical matter need to be considered carefully, 18 and our experiments showed high accuracy in different batches. However, the generalizability of an individual model to a large number of clones from multiple patients has not been demonstrated. Nonetheless, careful evaluation of the generalization ability of the prediction models is still needed to exclude the possibility of overfitting to laboratory‐specific technical artifacts. Furthermore, consideration of the alteration of epigenetic memory by passage of iPSCs is also needed. Although this study did not find any difference in accuracy because of differences in the number of passages of iPSCs, it has been reported that the epigenome is altered by the passage of iPSCs, 33 and this is an issue requiring further study.
We have demonstrated a prediction model of ALS by combining deep learning algorithms and iPSC technology as a proof‐of‐concept study. Further study with expanded information from increased datasets will be needed in the future.
Author Contributions
H.I. contributed to the conception and design of the study; all authors contributed to the acquisition and analysis of data; K.I., Y.Y., and H.I. contributed to drafting the text and preparing the figures.
Potential Conflicts of Interest
Nothing to report.
Supporting information
APPENDIX S1. Supporting Information
Acknowledgments
This research was funded in part by a grant for Core Center for iPS Cell Research of the Research Center Network for Realization of Regenerative Medicine from Japan Agency for Medical Research and Development (AMED) and the Uehara Memorial Foundation to H.I., and from KAKENHI (18 K18452) to H.I. and Y.Y.
We thank our coworkers and collaborators: Dr. A. Kinoshita, Dr. R. Matsumoto, Dr. N. Kohara, Dr. R. Matsumura, Dr. M. Oda, Dr. K. Ito, Dr. M. Moritani, Dr. K. Yasui, and Dr. T. Kiyozuka for cooperation with patient recruitment; and Mrs. M. Iijima, Mrs. N. Kawabata, Mrs. T. Urai, Mrs. M. Nagata, and Mrs. M. Yasui for their valuable administrative support.
References
- 1. Maurel C, Dangoumau A, Marouillat S, et al. Causative genes in amyotrophic lateral sclerosis and protein degradation pathways: a link to neurodegeneration. Mol Neurobiol 2018;55:6480–6499. [DOI] [PubMed] [Google Scholar]
- 2. Brooks BR, Miller RG, Swash M, Munsat TL. El Escorial revisited: revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph Lateral Scler Other Motor Neuron Disord 2000;1:293–299. [DOI] [PubMed] [Google Scholar]
- 3. Hardiman O, van den Berg LH, Kiernan MC. Clinical diagnosis and management of amyotrophic lateral sclerosis. Nat Rev Neurol 2011;7:639–649. [DOI] [PubMed] [Google Scholar]
- 4. Esteva A, Kuprel B, Novoa RA, et al. Dermatologist‐level classification of skin cancer with deep neural networks. Nature 2017;542:115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Causey JL, Zhang J, Ma S, et al. Highly accurate model for prediction of lung nodule malignancy with CT scans. Sci Rep 2018;8:9286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kather JN, Pearson AT, Halama N, et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat Med 2019;25:1054–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Christiansen EM, Yang SJ, Ando DM, et al. In Silico labeling: predicting fluorescent labels in unlabeled images. Cell 2018;173:792–803.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Okita K, Yamakawa T, Matsumura Y, et al. An efficient nonviral method to generate integration‐free human‐induced pluripotent stem cells from cord blood and peripheral blood cells. Stem Cells 2013;31:458–466. [DOI] [PubMed] [Google Scholar]
- 9. Tada Y, Kume K, Matsuda Y, et al. Genetic screening for potassium channel mutations in Japanese autosomal dominant spinocerebellar ataxia. J Hum Genet 2020;65:363–369. [DOI] [PubMed] [Google Scholar]
- 10. Renton AE, Majounie E, Waite A, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21‐linked ALS‐FTD. Neuron 2011;72:257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Philbrick KA, Yoshida K, Inoue D, et al. What does deep learning see? Insights from a classifier trained to predict contrast enhancement phase from CT images. AJR Am J Roentgenol 2018;211:1184–1193. [DOI] [PubMed] [Google Scholar]
- 12. Rampasek L, Goldenberg A. TensorFlow: biology's gateway to deep learning? Cell Syst 2016;2:12–14. [DOI] [PubMed] [Google Scholar]
- 13. Murakami N, Imamura K, Izumi Y, et al. Proteasome impairment in neural cells derived from HMSN‐P patient iPSCs. Mol Brain 2017;10:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Matsuzono K, Imamura K, Murakami N, et al. Antisense oligonucleotides reduce RNA foci in spinocerebellar ataxia 36 patient iPSCs. Mol Ther Nucleic Acids 2017;8:211–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Badrinarayanan V, Kendall A, Cipolla R. SegNet: a deep convolutional encoder‐decoder architecture for image segmentation. IEEE Trans Pattern Anal Mach Intell 2017;39:2481–2495. [DOI] [PubMed] [Google Scholar]
- 16. Hohman F, Park H, Robinson C, Chau DH. Summit: scaling deep learning interpretability by visualizing activation and attribution summarizations. IEEE Trans Vis Comput Graph 2020;26:1096–1106. [DOI] [PubMed] [Google Scholar]
- 17. Cao C, Liu F, Tan H, et al. Deep learning and its applications in biomedicine. Genomics Proteomics Bioinformatics 2018;16:17–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang SJ, Lipnick SL, Makhortova NR, et al. Applying deep neural network analysis to high‐content image‐based assays. SLAS Discov 2019;24:829–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Imamura K, Izumi Y, Watanabe A, et al. The Src/c‐Abl pathway is a potential therapeutic target in amyotrophic lateral sclerosis. Sci Transl Med 2017;9:eaaf3962. [DOI] [PubMed] [Google Scholar]
- 20. Schrooten M, Smetcoren C, Robberecht W, Van Damme P. Benefit of the Awaji diagnostic algorithm for amyotrophic lateral sclerosis: a prospective study. Ann Neurol 2011;70:79–83. [DOI] [PubMed] [Google Scholar]
- 21. Kiernan MC, Vucic S, Talbot K, et al. Improving clinical trial outcomes in amyotrophic lateral sclerosis. Nat Rev Neurol 2021;17:104–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Costa J, Swash M, de Carvalho M. Awaji criteria for the diagnosis of amyotrophic lateral sclerosis: a systematic review. Arch Neurol 2012;69:1410–1416. [DOI] [PubMed] [Google Scholar]
- 23. Gagliardi D, Meneri M, Saccomanno D, et al. Diagnostic and prognostic role of blood and cerebrospinal fluid and blood neurofilaments in amyotrophic lateral sclerosis: a review of the literature. Int J Mol Sci 2019;20:4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vijayakumar UG, Milla V, Cynthia Stafford MY, et al. A systematic review of suggested molecular strata, biomarkers and their tissue sources in ALS. Front Neurol 2019;10:400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Richards D, Morren JA, Pioro EP. Time to diagnosis and factors affecting diagnostic delay in amyotrophic lateral sclerosis. J Neurol Sci 2020;417:117054. [DOI] [PubMed] [Google Scholar]
- 26. Al‐Chalabi A, Fang F, Hanby MF, et al. An estimate of amyotrophic lateral sclerosis heritability using twin data. J Neurol Neurosurg Psychiatry 2010;81:1324–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bandres‐Ciga S, Noyce AJ, Hemani G, et al. Shared polygenic risk and causal inferences in amyotrophic lateral sclerosis. Ann Neurol 2019;85:470–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Polo JM, Anderssen E, Walsh RM, et al. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell 2012;151:1617–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hawrot J, Imhof S, Wainger BJ. Modeling cell‐autonomous motor neuron phenotypes in ALS using iPSCs. Neurobiol Dis 2020;134:104680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fujimori K, Ishikawa M, Otomo A, et al. Modeling sporadic ALS in iPSC‐derived motor neurons identifies a potential therapeutic agent. Nat Med 2018;24:1579–1589. [DOI] [PubMed] [Google Scholar]
- 31. Burkhardt MF, Martinez FJ, Wright S, et al. A cellular model for sporadic ALS using patient‐derived induced pluripotent stem cells. Mol Cell Neurosci 2013;56:355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gupta A, Harrison PJ, Wieslander H, et al. Deep learning in image cytometry: a review. Cytometry A 2019;95:366–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Horvath S. DNA methylation age of human tissues and cell types. Genome Biol 2013;14:R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
APPENDIX S1. Supporting Information


