Abstract
Site‐specific conditional inactivation technologies using Cre‐loxP or Flp‐FRT systems are becoming increasingly important for the elucidation of gene function and disease mechanism in vivo. A large number of gene knockout mouse models carrying complex conditional alleles have been generated by global community efforts and made available for biomedical researchers. The structures of conditional alleles in these mice are becoming increasingly complex and sophisticated, and so the validation of the genetic quality of these alleles is likewise becoming a laborious task for individual researchers. To ensure the reproducibility of conditional experiments, the researcher should confirm that loxP or FRT is integrated at the correct positions in the genome prior to start of the experiments. We report the successful design of universal PCR primers specific to loxP and FRT for the quick validation of conditional floxed and Flrted alleles. The primer set consists of forward and reverse primers complimentary to the loxP or FRT sequences with partial modifications at the 5ʹ end containing 6‐base restriction endonuclease recognition sites. The universal primer set was tested to detect genomic intervals between a pair of cis‐integrated loxP or FRT and was useful for quickly validating various floxed or Flrted alleles in conditional mice.
Keywords: conditional mice, Cre‐loxP, Flp‐FRT, genetic quality, reproducibility
The universal PCR primer set specific to loxP and FRT has been developed and can be used for quickly validating various floxed or Flrted alleles in conditional mice.
1. INTRODUCTION
Conditional knockout mouse models are essential tools for studying gene functions and disease mechanisms (Gossen & Bujard, 2002; Nagy, 2000). Comprehensive in vivo analyses of gene functions and developmental disorders via systemic inactivation of specific genes with null alleles in mice have revealed that approximately one‐third of protein‐coding genes are essential for embryonic and pre‐weaning life (Dickinson et al., 2016). To further annotate these embryonic lethal and sub‐viable genes in vivo, it is necessary to inactivate the gene in a site‐specific manner with conditional tools such as Cre‐loxP and Flp‐FRT systems (Nagy, 2000; Skarnes et al., 2011).
Originally discovered in bacteriophage P1, the Cre‐loxP system is the most widely used conditional tool and comprises a sequence‐specific Cre recombinase and its target loxP sequence (Nagy, 2000; Sauer & Henderson, 1988). Another tool, discovered in Saccharomyces cerevisiae and designated the Flp‐FRT system, uses Flp recombinase and its target FRT sequence and has also been reported for genomic modification (Buchholz et al., 1998; Sadowski, 1995). In conditional experiments, we use two genetic tools; one is a floxed strain that carries an allele containing a genomic region flanked by loxP (floxed allele), and the other is a Cre‐expressing strain containing a transgene that expresses Cre recombinase under the control of a tissue‐specific promoter for tissue‐specific gene modification. The floxed and Cre‐expressing strains are crossed to induce conditional gene activation or inactivation in specific tissues where the promoter is active. A Flrted strain carrying an allele that contains a genomic region flanked by FRT (Flrted allele) and a Flp‐expressing strain can also be used for the same purpose.
Recent advancements in genome editing together with DNA recombination technologies have resulted in the creation of thousands of conditional mouse strains carrying increasingly complex and sophisticated floxed and/or Flrted alleles for in vivo studies of gene functions and disease processes (Gurumurthy et al., 2019; Skarnes et al., 2011). To obtain reproducible and reliable experimental results using the floxed and/or Flrted mice, the genetic quality of the floxed and Flrted alleles, including their complete structure as originally designed using functional components, should be validated prior to experiments (Nakata et al., 2009). In large‐scale knockout mouse production facilities, the validation of conditional alleles is conducted by long‐range PCR with primers designed based on the information of gene cassette and genome sequences adjacent to the loxP followed by sequencing (Skarnes et al., 2011). To generate a specific primer set based on the genome information of every conditional line for validation of the genetic quality is a laborious task for individual researchers.
loxP and FRT have a unique structure in common that consists of an 8‐bp core sequence flanked by palindromic repeats of 13‐bp sequences (Gossen & Bujard, 2002; Nagy, 2000). If PCR primers specific to loxP and FRT sites are successfully designed, we can easily validate the genetic quality of various conditional mouse lines by examining each genomic interval flanked by loxP and/or FRT sites in a quick and cost‐effective manner. However, the creation of PCR primers specific to loxP and/or FRT sites seemed to be difficult due to the palindrome sequences which induce self‐annealing and formation of a hairpin structure.
We report here the successful creation of universal PCR primers specific to loxP and FRT sites enabling quick examination of floxed and/or Flrted alleles. The primer set consists of forward and reverse primers complimentary to the loxP or FRT sequences with partial modifications at the 5ʹ end containing 6‐base restriction endonuclease recognition sites.
2. RESULTS AND DISCUSSION
2.1. Primer design
The sequences of loxP (Figure 1a) and FRT (Figure 1b) are considered to form a hairpin structure due to palindrome sequences at both ends of the core sequence and not to be suitable for primer sequences as original. To prevent the complete hairpin formation, we modified the 5′ end of each primer with 8 to 11 bases containing restriction endonuclease recognition sites (Table 1; Figure 1a,b). Such modification of the 5′ end of PCR primer with restriction endonuclease recognition sites and a few spacer nucleotides was previously reported for cloning of PCR products (Zimmermann et al., 1998). The DNA melting temperature Tm (Schildkraut, 1965) for the 27–28 bp loxP‐ or FRT‐complementary portion of the primers was as follows: 47.5 and 47.0°C for loxP‐forward and reverse primers, 50.8 and 50.4°C for FRT‐forward and reverse primers, respectively. A set of the forward and reverse primers can be used to amplify a floxed or Flrted genomic fragment between two loxPs or FRTs as illustrated in Figure 1c. PCR products are expected to be generated with functional components of loxP or FRT and restriction sites protected by spacer nucleotides on both ends (Figure 1c).
FIGURE 1.
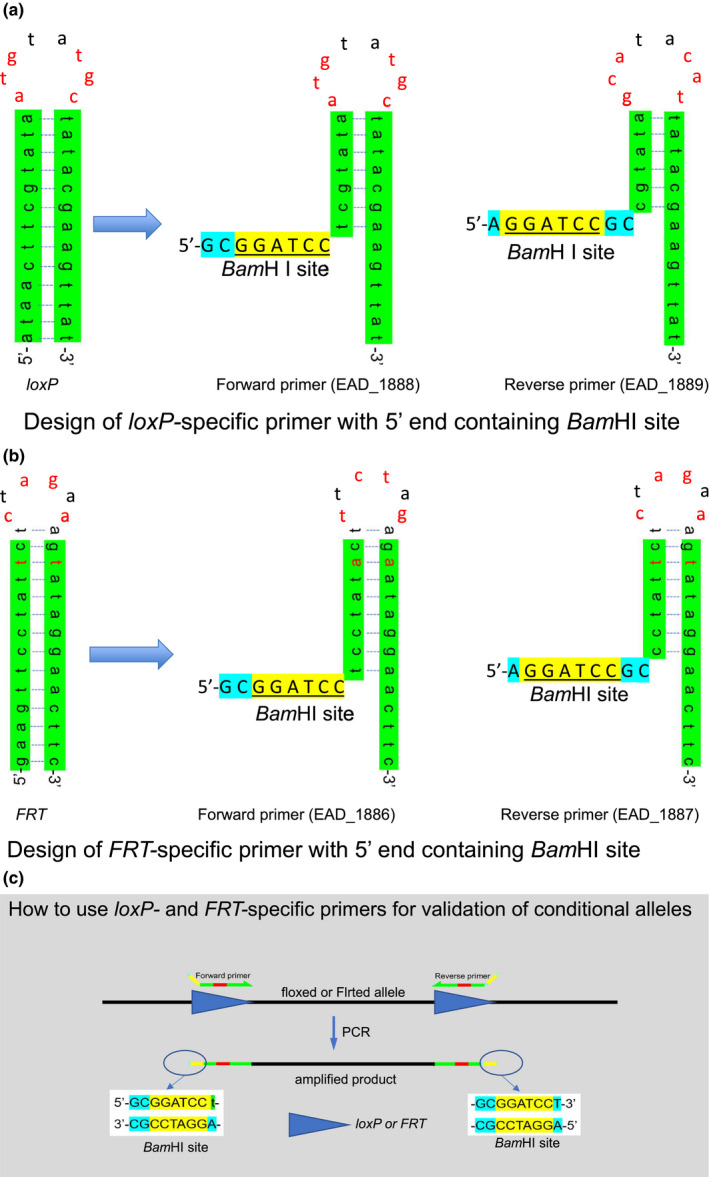
Universal PCR primers for loxP and FRT sites. (a) Design of loxP‐specific primer with modification at 5’ end containing BamHI site. (b) Design of FRT‐specific primer with modification at 5’ end containing BamHI site. (c) How to use loxP‐ and FRT‐specific primers for validation of conditional alleles. Green: palindrome right and left arms of loxP and FRT. Blue: spacer sequence to ensure the amplified fragment contains intact restriction site. Yellow: restriction endonuclease recognition site. Red nucleotide in (a) and (b): non complementary sequence. Red bar in (c): primer sequence complementary to the core sequence of loxP or FRT
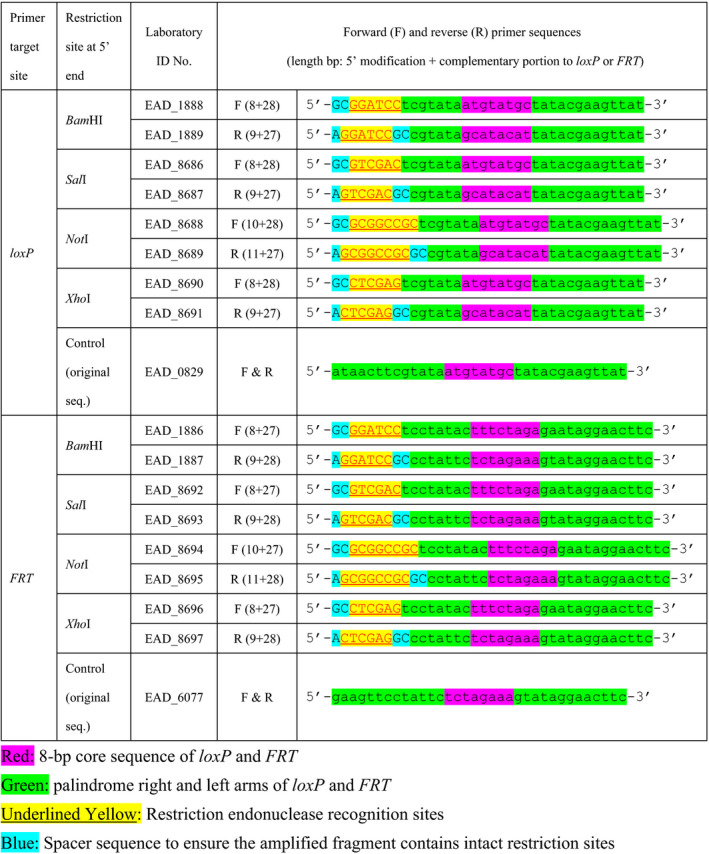
TABLE 1.
loxP‐ and FRT specific primers designed and tested in this study
2.2. PCR analysis of the floxed and Flrted alleles
We analyzed the mouse strain C57BL/6N‐Galtm1a(KOMP)Wtsi/G08 (RBRC05903) with a complex conditional allele designated as tm1a, which is widely available to the global biomedical community (Skarnes et al., 2011). The conditional allele contains three loxPs and two FRTs, all of which are cis‐integrated (Figure 2a). We tested loxP‐ and FRT‐specific PCR primer sets with 5’ end containing restriction sites as listed in Table 1. As results, all the primer sets could successfully amplify three floxed products of 0.7, 1.9, and 2.6 kb, respectively, by loxP‐test, and one Flrted product of 7.0 kb by FRT‐test, as predicted by in silico estimation (Figure 2b). However, the loxP‐primer set with NotI site seemed less efficient in amplification of 2.6 kb band. The original sequences of loxP and FRT for control did not work at all as PCR primers (Figure 2b). Further optimization of the primer sequences including spacer nucleotides is remained for the future study. The PCR primer sets with BamHI site were used in further experiments in this study.
FIGURE 2.
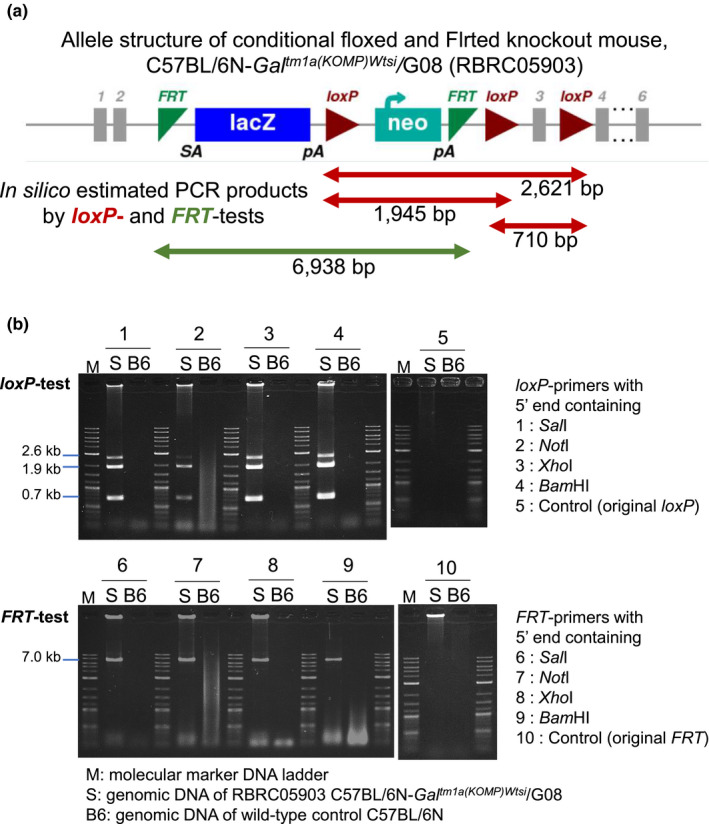
PCR analyses of conditional floxed and Flrted alleles. (a) The allele structure of the conditional floxed and Flrted knockout mouse C57BL/6N‐Galtm1a(KOMP)Wtsi/G08 (RBRC05903), posted on the IMPC website (https://www.mousephenotype.org/). Three floxed (red arrows) and two Flrted (green arrow) intervals expected as PCR products by loxP‐ and FRT‐PCR tests were estimated in silico. (b) Results of loxP‐ and FRT‐tests using primers with 5' end containing four different restriction sites and control. M: 1 kb Plus DNA Ladder (0.1–10.0 kb; New England BioLabs Japan). S: Genomic DNA of Galtm1a /+ heterozygous mice. B6: Genomic DNA of wild‐type B6 control
2.3. PCR analysis of various conditional strains
We further evaluated the efficacy of the primer sets by validating 6 different conditional strains carrying different numbers of loxPs and FRTs, as listed in Table 2. We detected PCR products of the sizes expected from in silico estimations for each floxed and/or Flrted allele. Furthermore, the conditional alleles of 215 mouse strains (including the above 7 strains in Figure 1; Table 2) publicly available at the RIKEN BioResource Research Center (BRC) were successfully validated using the developed universal primer sets with a high success rate at 96.7% (Table S1; Figure S1). In conclusion, our results indicated that the universal primer sets developed in this study were useful for quickly validating various conditional floxed and/or Flrted genomic intervals between cis‐integrated loxPs or FRTs.
TABLE 2.
Validation of various conditional floxed and Flrted alleles by universal PCR primers for loxP‐ and FRT‐tests
| RBRC no. | Strain name | No. of loxP | Product size (kb) by loxP‐test | No. of FRT | Product size (kb) by FRT‐test | Reference |
|---|---|---|---|---|---|---|
| 09446 | C57BL/6‐Islr tm1a(EUCOMM)Wtsi/Ea | 3 | 4.0, 2.0, 1.8 | 2 | 7.0 | Skarnes et al. (2011) |
| 04503 | B6.129P2‐Smad7 tm1Tchi | 2 | 3.5 | 2 | 1.1 | Tojo et al. (2012) |
| 05584 | C57BL/6N‐Ikzf5 tm1(KOMP)Wtsi/C06 | 2 | 1.8 | 2 | 7.0 | Skarnes et al. (2011) |
| 09534 | B6‐Pgk1 tm1(CAG‐eGFP‐NLS, PGK‐Neo)Koba | 2 | 6.0 | 2 | 1.5 | Kobayashi (2018) |
| 04608 | B6;129P2‐Runx1 tm1Homy | 2 | 3.6 | 1 | Not detected | Nagamachi et al. (2010) |
| 06221 | B6;129‐Slc39a10 tm1.1Tfk | 2 | 8.0 | 0 | Not detected | Miyai et al. (2014) |
2.4. Application of the developed universal primers
The genetic quality control (QC) of complex and sophisticated genetic tools such as conditional mouse models is essential for ensuring the reproducibility of experiments. PCR tests using the developed universal primers can be widely applied to the various conditional genetic models containing loxPs and FRTs currently available at the core facility of the National BioResource Project by the Japanese Ministry of Education, Culture, Sports, Science and Technology and the Agency for Medical Research and Development (Yoshiki et al., 2009). To establish a comprehensive genetic QC program, it is recommended that the loxP‐ and FRT‐PCR tests be conducted along with other genetic QC tests, such as marker gene detection (Nakata et al., 2009) and strain‐specific genotyping and sequencing. The developed universal primers should also play an important role in the quick validation of conditional mice with cis‐ versus trans‐integrated loxPs via genome editing knock‐in technology (Gurumurthy et al., 2019).
3. EXPERIMENTAL PROCEDURES
3.1. Mice
Conditional floxed and Flrted strains identified by Riken BRC numbers 09446, 04503, 09534, 04608, and 06221 have been deposited and maintained at the barrier facility for genetically modified mice at the RIKEN BRC. Conditional mice identified by Riken BRC numbers 05903, 05584, and 09446 were generated at the RIKEN BRC by using knockout mouse embryonic stem cell resources obtained from the International Mouse Phenotyping Consortium repositories (Skarnes et al., 2011). The C57BL/6JJcl mice were purchased from CLEA Japan and used as wild‐type controls. All experiments including genetically modified mice were approved by the Institutional Animal Care and Use Committee and the Genetic Recombination Experiments Committee of the RIKEN Tsukuba Branch.
3.2. PCR analysis for floxed and Flrted alleles
Mouse genomic DNA was extracted from tail tips as previously described using automated DNA isolator (GENE PREP STAR PI‐80X, Kurabo; Nakata et al., 2009; Figure S2). Primer sets with 5' end containing restriction sites for loxP and FRT as shown in Figure 1 and listed in Table 1 were used. PCR tests were performed in mixtures containing 100 ng of genomic DNA, 1 × PCR buffer, 0.4 mM dNTPs, 0.5 μM each of primer, and 0.4 U of KOD FX (Toyobo) in a volume of 20 µl (Figure S3). The reactions were carried out with an initial denaturation at 94°C for 120 s, followed by 38 cycles of denaturation at 98°C for 10 s, and annealing and extension at 68°C for 300 s. The PCR products were separated and detected on 1% agarose gels in 1X TAE buffer. The gel images were captured and recorded using an E‐Box CX5 gel documentation system (Vilber Lourmat Sté).
Supporting information
Fig S1
Fig S2
Fig S3
Table S1
ACKNOWLEDGMENTS
We thank Drs. T. Chiba, S. Kobayashi, H. Honda, and T. Fukada for kindly depositing invaluable conditional mouse strains RBRC 04503, 09534, 04608, and 06221, respectively. We also thank Y. Seki, Y. Shima, H. Honjo, A. Fukumoto, and the members of the Experimental Animal Division for technical assistance and advice; and Drs. Kuniya Abe, Toshihiko Shiroishi, and Yuichi Obata of the RIKEN BioResource Research Center for their kind advice and support. This study was conducted in cooperation with the National BioResource Project of the Japanese Ministry of Education, Culture, Sports, Science and Technology and the Agency for Medical Research and Development.
Nakata H, Hashimoto T, Yoshiki A. Quick validation of genetic quality for conditional alleles in mice. Genes Cells. 2021;26:240–245. 10.1111/gtc.12834
[The copyright line for this article was changed on 16 April 2021 after original online publication]
Communicated by: Yuji Kohara
REFERENCES
- Buchholz, F. , Angrand, P. O. , & Stewart, A. F. (1998). Improved properties of FLP recombinase evolved by cycling mutagenesis. Nature Biotechnology, 16, 657–662. 10.1038/nbt0798-657 [DOI] [PubMed] [Google Scholar]
- Dickinson, M. E. , Flenniken, A. M. , Ji, X. , Teboul, L. , Wong, M. D. , White, J. K. , Meehan, T. F. , Weninger, W. J. , Westerberg, H. , Adissu, H. , Baker, C. N. , Bower, L. , Brown, J. M. , Caddle, L. B. , Chiani, F. , Clary, D. , Cleak, J. , Daly, M. J. , Denegre, J. M. , … Murray, S. A. (2016). High‐throughput discovery of novel developmental phenotypes. Nature, 537(7621), 508–514. 10.1038/nature19356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen, M. , & Bujard, H. (2002). Studying gene function in eukaryotes by conditional gene inactivation. Annual Review of Genetics, 36, 153–173. 10.1146/annurev.genet.36.041002.120114 [DOI] [PubMed] [Google Scholar]
- Gurumurthy, C. B. , O’Brien, A. R. , Quadros, R. M. , Adams, J. , Alcaide, P. , Ayabe, S. , Ballard, J. , Batra, S. K. , Beauchamp, M.‐C. , Becker, K. A. , Bernas, G. , Brough, D. , Carrillo‐Salinas, F. , Chan, W. , Chen, H. , Dawson, R. , DeMambro, V. , D'Hont, J. , Dibb, K. M. , … Burgio, G. (2019). Reproducibility of CRISPR‐Cas9 methods for generation of conditional mouse alleles: A multi‐center evaluation. Genome Biology, 20(1), 171. 10.1186/s13059-019-1776-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, S. (2018). Live imaging of X‐chromosome inactivation and reactivation kinetics. Methods in Molecular Biology, 1861, 73–89. 10.1007/978-1-4939-8766-5_7 [DOI] [PubMed] [Google Scholar]
- Miyai, T. , Hojyo, S. , Ikawa, T. , Kawamura, M. , Irie, T. , Ogura, H. , Hijikata, A. , Bin, B.‐H. , Yasuda, T. , Kitamura, H. , Nakayama, M. , Ohara, O. , Yoshida, H. , Koseki, H. , Mishima, K. , & Fukada, T. (2014). Zinc transporter SLC39A10/ZIP10 facilitates antiapoptotic signaling during early B‐cell development. Proceedings of the National Academy of Sciences of the United States of America, 111(32), 11780–11785. 10.1073/pnas.1323549111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamachi, A. , Htun, P. W. , Ma, F. , Miyazaki, K. , Yamasaki, N. , Kanno, M. , Inaba, T. , Honda, Z. , Okuda, T. , Oda, H. , & Tsuji, K. , & Honda, H. (2010). A 5ʹ untranslated region containing the IRES element in the Runx1 gene is required for angiogenesis, hematopoiesis and leukemogenesis in a knock‐in mouse model. Developmental Biology, 345(2), 226–236. 10.1016/j.ydbio.2010.07.015 [DOI] [PubMed] [Google Scholar]
- Nagy, A. (2000). Cre recombinase: The universal reagent for genome tailoring. Genesis, 26(2), 99–109. [DOI] [PubMed] [Google Scholar]
- Nakata, H. , Hashimoto, T. , Seki, Y. , Mekada, K. , Obata, Y. , & Yoshiki, A. (2009). Simultaneous detection of multiple transgenes for genetically‐modified mouse strains. Experimental Animals, 58(4), 437–442. 10.1538/expanim.58.437 [DOI] [PubMed] [Google Scholar]
- Sadowski, P. D. (1995). The Flp recombinase of the 2‐microns plasmid of Saccharomyces cerevisiae. Progress in Nucleic Acid Research and Molecular Biology, 51, 53–91. [PubMed] [Google Scholar]
- Sauer, B. , & Henderson, N. (1988). Site‐specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proceedings of the National Academy of Sciences of the United States of America, 85(14), 5166–5170. 10.1073/pnas.85.14.5166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schildkraut, C. (1965). Dependence of the melting temperature of DNA on salt concentration. Biopolymers, 3(2), 195–208. 10.1002/bip.360030207 [DOI] [PubMed] [Google Scholar]
- Skarnes, W. C. , Rosen, B. , West, A. P. , Koutsourakis, M. , Bushell, W. , Iyer, V. , Mujica, A. O. , Thomas, M. , Harrow, J. , Cox, T. , Jackson, D. , Severin, J. , Biggs, P. , Fu, J. , Nefedov, M. , de Jong, P. J. , Stewart, A. F. , & Bradley, A. (2011). A conditional knockout resource for the genome‐wide study of mouse gene function. Nature, 474(7351), 337–342. 10.1038/nature10163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tojo, M. , Takebe, A. , Takahashi, S. , Tanaka, K. , Imamura, T. , Miyazono, K. , & Chiba, T. (2012). Smad7‐deficient mice show growth retardation with reduced viability. Journal of Biochemistry, 151(6), 621–631. 10.1093/jb/mvs022 [DOI] [PubMed] [Google Scholar]
- Yoshiki, A. , Ike, F. , Mekada, K. , Kitaura, Y. , Nakata, H. , Hiraiwa, N. , Mochida, K. , Ijuin, M. , Kadota, M. , Murakami, A. , Ogura, A. , Abe, K. , Moriwaki, K. , & Obata, Y. (2009). The mouse resources at the RIKEN BioResource center. Experimental Animals, 58(2), 85–96. 10.1538/expanim.58.85 [DOI] [PubMed] [Google Scholar]
- Zimmermann, K. , Schögl, D. , & Mannhalter, J. W. (1998). Digestion of terminal restriction endonuclease recognition sites on PCR products. BioTechniques, 24(4), 582–584. 10.2144/98244bm15 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Fig S2
Fig S3
Table S1


