Summary
Infections by more than one strain of a pathogen predominate under natural conditions. Mixed infections can have significant, though often unpredictable, consequences for overall virulence, pathogen transmission and evolution. However, effects of mixed infection on disease development in plants often remain unclear and the critical factors that determine the outcome of mixed infections remain unknown. The fungus Zymoseptoria tritici forms genetically diverse infections in wheat fields. Here, for a range of pathogen traits, we experimentally decompose the infection process to determine how the outcomes and consequences of mixed infections are mechanistically realized. Different strains of Z. tritici grow in close proximity and compete in the wheat apoplast, resulting in reductions in growth of individual strains and in pathogen reproduction. We observed different outcomes of competition at different stages of the infection. Overall, more virulent strains had higher competitive ability during host colonization, and less virulent strains had higher transmission potential. We showed that within‐host competition can have a major effect on infection dynamics and pathogen population structure in a pathogen and host genotype‐specific manner. Consequently, mixed infections likely have a major effect on the development of septoria tritici blotch epidemics and the evolution of virulence in Z. tritici.
Introduction
In nature, infectious diseases are typically caused by a genetic assortment of co‐infecting pathogen strains and/or species (van Baalen and Sabelis, 1995; Read and Taylor, 2001; Linde et al., 2002; López‐Villavicencio et al., 2011; Tollenaere et al., 2016). When co‐infecting microbes share the same space and resources or are exposed to the same immune responses, the fitness of the individual strains can be affected by their interactions both with the host and with co‐existing strains or species (Read and Taylor, 2001; Alizon and van Baalen, 2008; Tollenaere et al., 2016). Complex interactions between co‐infecting pathogens and their hosts can have a strong impact on the outcome of the infection, on the ecology of the pathosystem and on the evolution of the pathogen (Mosquera and Adler, 1998; Read and Taylor, 2001; Mideo, 2009). While numerous theoretical studies (Alizon, 2008; Alizon and van Baalen, 2008; Mideo, 2009) have investigated various aspects of mixed infections, more empirical investigations are needed to better understand realized outcomes across pathosystems.
The ecological and evolutionary consequences of mixed infections for both host and pathogen fitness are predicted to vary depending on the nature of the interactions among strains (Mosquera and Adler, 1998; Read and Taylor, 2001; Brown et al., 2002; Alizon and van Baalen, 2008; Mideo, 2009). For example, interactions involving individuals of the same species often involve competition for limited resources, including space and nutrients, within defined niches such as plant leaves (i.e., exploitation competition; Gold et al., 2009). In such scenarios, theoretical models often predict that mixed infections will result in higher virulence (defined here as damage to the host; Leggett et al., 2013) than expected based on single infections and lead to the evolution of increased virulence. This reflects the assumption that faster‐growing (and hence more virulent) strains will be favoured in mixed infections due to their greater ability to access shared resources, resulting in selection for higher overall virulence (Rochow and Ross, 1955; de Roode et al., 2005; Ben Ami et al., 2008; Mideo, 2009; Zinga et al., 2013). In other cases, mixed infections might result in a reduction in overall virulence (Hood, 2003; Schürch and Roy, 2004). For instance, pathogens might directly interfere with the progression of a competitor by the secretion of antimicrobials (interference competition) and this might result in lower overall virulence and pathogen fitness, particularly if the trait is expressed by more than one genotype (Riley and Gordon, 1999; Gardner et al., 2004; Massey et al., 2004; Mideo, 2009). Furthermore, hosts are not passive substrates for pathogen colonization. Differences in both the qualitative and quantitative outcomes of coinfection are possible depending on the makeup of the host and pathogen genotypes (Råberg et al., 2006; Susi et al., 2015). For example, in cases of apparent competition where a cross‐reaction of the immune response affects the co‐existing microorganism (Cui et al., 2005; Råberg et al., 2006; Spoel et al., 2007; Holt and Bonsall, 2017), mixed infections might also result in a reduction in overall virulence.
As a result of these types of complex interactions, mixed infections may strongly influence pathogen virulence and transmission, two traits critical to host‐pathogen interactions (Rochow and Ross, 1955; van Baalen and Sabelis, 1995; Read and Taylor, 2001; Gonzalez‐Jara et al., 2004; Gower and Webster, 2005; Rigaud et al., 2010; Susi et al., 2015). However, precise outcomes are likely context‐dependent and may be difficult to predict. We have little understanding about how coinfecting strains interact throughout an infection, or how outcomes of interactions between coinfecting strains are affected by variation in (i) the genetic composition of mixed infections; and (ii) host immunity.
Natural infections of the wheat fungal pathogen Zymoseptoria tritici are genetically diverse and several strains may coexist within lesions (Linde et al., 2002; Zhan et al., 2002a; Zhan et al., 2005). Earlier work showed that mixed infections of Z. tritici can reduce the production of reproductive structures called pycnidia (Zelikovitch and Eyal, 1991; Halperin et al., 1996; Schürch and Roy, 2004). Although mixed infections are now known to be the norm in natural epidemics, the great majority of research remains oriented around infections caused by single strains (O'Driscoll et al., 2014). In single infections, individual strains display different infection phenotypes, including variation in the latent period, the degree of leaf damage, and rates of formation of pycnidia (Stewart et al., 2018; Haueisen et al., 2019). Host genotype strongly influences pathogen fitness. For example, the wheat resistance gene Stb6 (for Septoria tritici blotch 6) is present in approximately 15% of the wheat cultivars grown in Europe (Brown et al., 2015; Saintenac et al., 2018) and confers qualitative resistance to Z. tritici strains harbouring the avirulence factor AvrStb6 (Zhong et al., 2017; Kema et al., 2018). However, the contribution of host‐specific resistance to disease control in mixed infections remains largely unexplored.
In this study, we investigate the effects of mixed infection on Z. tritici virulence and fitness over the full time‐course of the infection using two wheat genotypes. We thoroughly characterize the effects of mixed infection on pathogenicity (defined here as the ability of a pathogen to infect a host) by assessing multiple traits, including virulence (understood here as damage to the host), spore production and biomass. We then use this data to investigate our hypothesis that competition between strains is influenced by host genotype, pathogen infection strategy, and the phase of the infection cycle.
Results
The infection progress of Zymoseptoria tritici is strongly influenced by host genotype and infection treatment comprised of various strain combinations
We thoroughly characterized the infection dynamics of single and mixed infection treatments, comprising four strains of Z. tritici (3D1, 3D7, 1E4 and 1A5) in two wheat cultivars (Chinese Spring and Drifter). The four strains, collected on the same day from two wheat fields separated by 10 km (Zhan et al., 2002b), were shown to have different virulence phenotypes (Stewart et al., 2018; Fig. S1) in single infections. Chinese Spring carries the Stb6 resistance allele and 1E4 expresses the AvrStb6 avirulence allele; therefore, 1E4 is avirulent in this cultivar. We performed infections with spore suspensions of single strains and a mixture of two or four of the strains to evaluate the infection dynamics of single and mixed infections. In order to gain further insights into the infection process associated with the different infection treatments, we first estimated pathogen biomass (ng per cm2 of leaf), virulence (estimated as the percentage of leaf area covered by lesions, PLACL) and reproduction (pycnidia cm−2 of leaf) over time for all the investigated treatments.
Total biomass
We estimated the biomass of each strain in single and mixed infections throughout the infection cycle (10–19 days post infection, dpi; Figs S1 and S2) using qPCR with strain‐specific primers. We calculated total biomass in mixed infections by summing the biomass of co‐infecting strains. As expected, the incompatible interaction (1E4 infecting Chinese Spring) yielded very low levels of biomass. In all the compatible interactions, fungal growth was detected only after the appearance of the first symptoms (between 10 and 14 dpi; Figs S1 and S2). Overall, fungal growth ceased after the leaves were fully necrotized (Figs S1 and S2). At 19 dpi strains reached their maximum growth compared to the other analysed time points. We thus used the 19 dpi time point to evaluate growth differences between treatments as measured by pathogen biomass.
Linear modelling revealed significant effects of host, pathogen treatment, and their interaction on total pathogen biomass. There was a strong, consistent effect of host genotype across all pathogen treatments (Table 1, Fig. 1). For all treatments, total biomass was higher in the cultivar Drifter (Fig. 1A). The effects of fungal treatment on total biomass also varied among hosts. In Drifter, the 4‐strain treatment had significantly lower total biomass compared to many one and two strain treatments (3D1/3D7/1E4 + 3D1/1E4 + 3D7: p < 0.05; Table S1). For Chinese Spring, the biomass of the avirulent strain (1E4) treatment was significantly less (p < 0.05) than 3D7 alone (p < 0.05; Table S1). This data corroborates earlier experiments showing that 1E4 does not grow when it is recognized by the host (Kema et al., 2018).
Table 1.
Summary of results from generalized linear models of total fungal biomass (ng fungal DNA cm−2, 19 dpi), percentage of leaf area covered by lesions (PLACL, 14 dpi) and pycnidia density (pycnidia cm−2 leaf).
| Total fungal biomass | PLACL | Pycnidia | |||||||
|---|---|---|---|---|---|---|---|---|---|
| df | Deviance | p‐value | df | Deviance | p‐value | df | Deviance | p‐value | |
| Total | 65 | 96.31 | 131 | 84.84 | 130 | 307.20 | |||
| Host | 1 | 60.74 | <0.0001 | 1 | 0.229 | 0.35 a | 1 | 3.77 | 0.183 |
| Infection treatment | 10 | 15.13 | <0.0001 | 10 | 51.36 | <0.0001 | 10 | 126.63 | <0.0001 |
| Host × Treatment | 10 | 7.08 | 0.0096 | 10 | 6.16 | <0.0001 | 10 | 90.315 | <0.0001 |
| Residuals | 44 | 13.35 | 110 | 27.09 | 109 | 86.48 | |||
NS, non‐significant.
Fig 1.
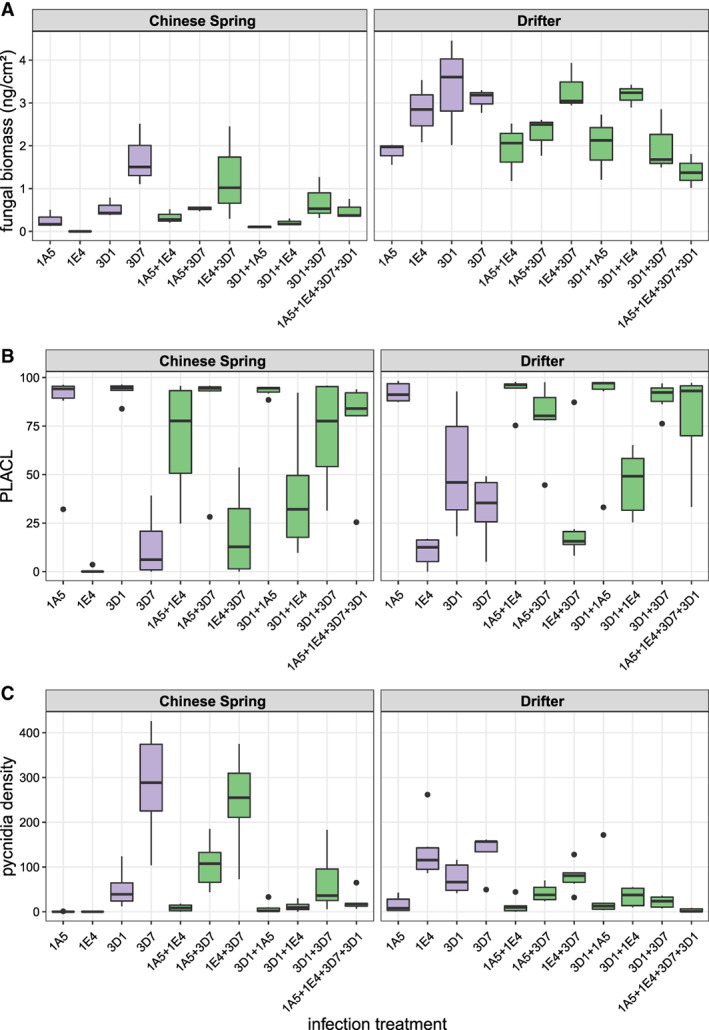
Single and mixed wheat infections affect Zymoseptoria tritici pathogenicity traits. Boxplots for total fungal biomass at 19 days post infection (dpi; ng cm−2 leaf); (A), percentage of leaf area covered by lesions (PLACL) at 14 dpi (B) and pycnidia density at 24 dpi (for Drifter) or 25 dpi (for Chinese Spring, pycnidia cm−2 leaf; (C) grouped according to infection treatment and host (Chinese Spring and Drifter). The boxes are bound by the lower (Q1) and upper quartile (Q3), within which the median is plotted as a solid line. The single and mixed infection treatments are shown in purple and green, respectively. [Color figure can be viewed at wileyonlinelibrary.com]
PLACL
Host damage was evaluated throughout the infection cycle by calculating PLACL at each time point using automated image analysis. To compare differences among infection treatments, the increase in lesion area was fitted with the logistic function [Eq. (1) in Note S1; Fig. S3] to infer the rate of increase in lesion area (r), the PLACL at the first time point 10 dpi (n 0), and the maximum host damage reached (K; Fig. S3, Note S1). We determined if these parameters differed among the treatments by using a model selection approach. Overall, n 0 and K did not vary between most of the treatments, while r varied between the different treatments (Note S1). The highest variation between treatments in PLACL occurred at 14 dpi (Table S2; Fig. S3) and at this time point, PLACL significantly correlated with the modelled rate of increase (r; p = 0.01; R 2 = 0.27). We therefore used PLACL at 14 dpi to examine the effects of the different treatments on host damage.
Linear modelling revealed significant effects of pathogen treatment and the interaction between host and treatment (Table 1) on PLACL at 14 dpi. Fungal treatment effects were highly variable and thus had large effects on symptom development, often consistently across the different hosts (Table 1, Fig. 1). For example, the 1A5 treatment and the four‐strain treatment were highly damaging to both hosts, whereas the 3D7 treatment was much less virulent (Fig. 1B; Fig. S3). There were also host‐specific differences in the development of symptoms. For example, 1E4 did not infect Chinese Spring due to the qualitative resistance in that host. 1E4 was more damaging on Drifter, although this strain maintained relatively low levels of virulence compared to others (Fig. 1B; Fig. S3).
Pycnidia density
We assessed levels of asexual reproduction of Z. tritici by analysing pycnidia density. Linear modelling indicated significant effects of pathogen treatment (p < 0.0001) and the interaction between host and treatment (p < 0.0001) on pycnidia density. There were strong interactions between host and pathogen treatment (Fig. 1C, Table 1), with relatively high pycnidia density in some treatments in Chinese Spring. No pycnidia were produced by the avirulent strain 1E4 on this resistant cultivar (Fig. 1C). In this host, the density of pycnidia in treatments comprising 3D7 was relatively high, with the exception of the 4‐strain treatment, where density was low (Fig. 1C).
Biomass was a significant predictor of pycnidia production in both hosts, with the density tending to increase with increasing biomass (Drifter: p < 0.001, R 2 = 0.41; Chinese Spring: p < 0.001, R 2 = 0.57). PLACL was a significant predictor of pycnidia production only in Drifter (p < 0.001, R 2 = 0.21), but in this case density tended to decline with increasing damage to the host. These results indicate that in the absence of the Stb6 resistance gene, 1E4 and 3D7 exploit the host more slowly but produce larger numbers of pycnidia, whereas 1A5 is a highly virulent strain that exploits the host more quickly while producing smaller numbers of pycnidia (Figs 1 and S1).
Virulent strains compete for limited resources and vary in competitive ability
We next evaluated the effect of mixed‐infections on the total fungal biomass by analysing the total biomass produced by single and mixed infections. There was a significant main effect of mixed infection (p = 0.01) on individual strain biomass, reflecting a general reduction in the biomass of individual strains in mixed infections. For example, in 2‐strain mixed infections, individual strains suffered a more than 2‐fold reduction in biomass, on average, in Chinese Spring (0.63 versus 0.26 ng cm−2) and Drifter (2.78 versus 1.23 ng), indicating that pathogen strains are likely competing. However, total pathogen biomass in mixed infections (i.e., the sum of all individual biomass estimates) was not significantly different (at p < 0.05) compared to the biomass produced by single infections (Table 2).
Table 2.
The effect of mixed infection and host genotype on the transmission potential of Zymoseptoria tritici. Results are from a linear mixed model analysis where mixed infection and host were treated as fixed effects and infection treatment as a random effect. The effects on total fungal biomass (19 days post inoculation, dpi), percentage of leaf area covered by lesions (PLACL, 14 dpi) and pycnidia density (pycnidia cm−2 leaf) are shown.
| Total fungal biomass | PLACL | Pycnidia | |||||
|---|---|---|---|---|---|---|---|
| df | MeanSq | Fvalue | MeanSq | Fvalue | MeanSq | Fvalue | |
| Coinfection | 1 | 0.0004 | 0.01 a | 0.46 | 1.13 a | 0.006 | 0.003 a |
| Host | 1 | 6.31 | 163.47*** | 1.68 | 4.09* | 15.83 | 7.53** |
| Coinfection:Host | 1 | 0.10 | 2.46 a | 0.20 | 0.50 a | 53.49 | 25.44*** |
p < 0.01;
p < 0.001;
p < 0.0001.
NS, non‐significant.
In order to investigate the competitive ability of individual strains during host colonization, we modelled biomass for each strain in the presence or absence of a competitor. As described in experimental procedures, for these analyses we adjusted biomass estimates in mixed infection treatments by multiplying biomass two‐fold (data from the four‐strain mixture was not included). The results indicate that strains vary in their response to competition, according to the identity of the competing strain and the host background (Table 3, Fig. 2), and in their effects on the growth of competing strains. For example, the highly virulent strain 1A5 generally hinders the growth of competitors (Table 3: 1A5 column), but itself is little influenced by the presence of competing strains (Table 3: 1A5 rows; Fig. 2), suggesting that 1A5 is a relatively strong competitor in terms of biomass accumulation. In contrast, 3D1 was inclined towards growth reductions in several mixed infection treatments (Fig. 2, Table 3). Despite this, the biomass of 3D1 in cultivar Drifter was relatively high in both mixed and single infections (Fig. 2), suggesting that 3D1 retains the potential to be highly competitive.
Table 3.
The competitive ability of each strain depends on the identity of the competing strain and the host background. Linear model coefficients show the individual fungal biomass response (19 dpi) of each Z. tritici isolate to a mixed infection with competing strains. Coefficients (response to a mixed infection) from individual analyses are shown across rows and represent how each strain responds to the presence of another strain in a mixed infection. A positive coefficient (in green) indicates that the fungal biomass increased relative to expectations based on the performance in single strain infections, while negative coefficients (in red) indicate a decrease in biomass relative to single strain infections. For each strain, rows show response coefficients for each target strain to the presence of the competitors shown in columns, whereas columns show the predicted effects of each strain on the growth of the other strains.
| Host | Target strain | 3D7 | 3D1 | 1A5 | 1E4 |
|---|---|---|---|---|---|
| Drifter | 3D7 → | – | −0.70** | −0.08 a | +0.40** |
| 3D1 → | −0.30 a | – | −0.46 a | +0.35 a | |
| 1A5 → | −0.06 a | +0.04 a | – | +0.35 a | |
| 1E4 → | −0.34* | −0.56** | −0.85*** | – | |
| Chinese Spring | 3D7 → | – | −0.66 a | −1.13 a | +0.37 a |
| 3D1 → | −0.02 a | – | −1.71** | −0.28 a | |
| 1A5 → | +0.67 a | −0.92 a | – | +0.90* | |
| 1E4 → (avirulent) | +3.58** | +2.72 a | +2.62 a | – |
p < 0.05;
p < 0.005;
p < 0.0005.
NS, non‐significant.
Negative values are in red and positive values are in green.
Fig 2.
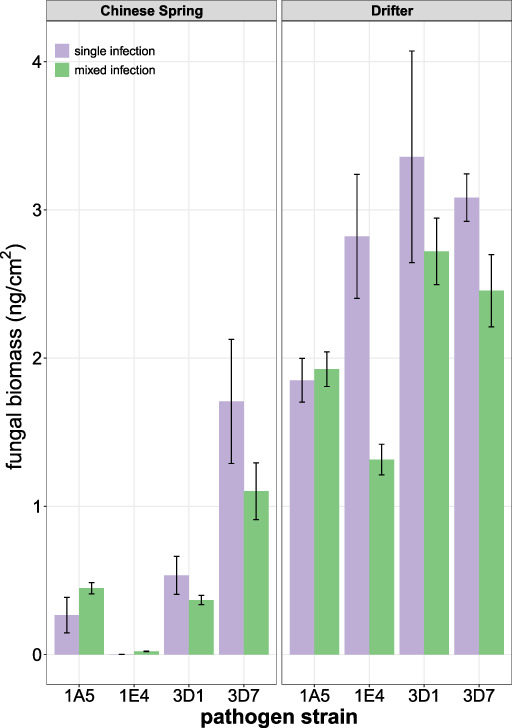
Zymoseptoria tritici strains vary in their competitive ability according to the host and the genotype of the coinfecting strains. For each leaf, fungal biomass (ng) per cm2 is shown for individual strains in mixed and single infections in wheat cultivars Chinese Spring and Drifter. Replicate biomass data for mixed infections was multiplied by the number of strains in the mixture prior to generating means and standard errors. [Color figure can be viewed at wileyonlinelibrary.com]
We also examined the relative growth of virulent strains in the presence of the avirulent strain 1E4 in the resistant cultivar Chinese Spring. In all coinfection treatments on Chinese Spring, the growth responses of 1E4 trend positively (Table 3: 1E4/Chinese Spring row), although the total increase was relatively small and the overall growth of 1E4 in Chinese Spring remained very low (Fig. 2). 1E4 did not significantly reduce the growth of any of the competing strains in Chinese Spring (Table 3: 1E4/CS column). In Drifter, 1E4 biomass was generally inhibited by the presence of competitors (Table 3: 1E4/Drifter row) while most strains tended to respond positively to the presence of 1E4 (Table 3: 1E4 column). In summary, our results showed that more virulent strains are better competitors than less virulent strains in terms of fungal biomass.
Overall patterns of disease progression are not altered in mixed infections
To test for general effects of mixed and single infections on pathogen virulence, pathogen treatments were assigned to either single infection or mixed infection categories. Overall, we did not observe significant differences in lesion area (PLACL at 14 dpi) between single and mixed infections (p > 0.05: Table 2). We also compared the observed PLACL at 14 dpi with the expected values calculated from single infections. Lesion area in mixed infections was, in all cases except one, higher than that predicted based on the mean of single infections of the corresponding strains. However, the confidence intervals around the mean frequently overlapped with predicted values (Fig. S4A). Likewise, compared to the maximum expected lesion area, observed values were mainly indistinguishable (Fig. S4B). Together, these results suggest that levels of host damage are not strongly affected by mixed infections.
We also evaluated how the expression of qualitative resistance affects co‐infecting strains. As reported, qualitative resistance encoded by Stb6 completely blocks the progression of infection for the avirulent strain 1E4 in a single infection (Fig. 1; Kema et al., 2018; Zhong et al., 2017). In the mixed infection with 3D1, 1E4 reduced the damage to the host (Fig. 1, Table S1). This result indicates that the qualitative resistance triggered by AvrStb6 recognition might reduce the damage caused to the host by some virulent strains.
Pathogen reproduction is reduced in mixed infections in a host‐dependent manner
The effect of mixed infections on transmission potential was evaluated by calculating pycnidia density in single and mixed infections. When pathogen treatments were assigned to either single‐infection or mixed‐infection categories there was a significant interaction between mixed infection and host genotype (Table 2). This result is largely driven by observations on the cultivar Drifter, with relatively low pycnidia density in mixed infections compared to single infections (Fig. 3). For example, 3D1 and 3D7 mixed infections in Drifter produced fewer pycnidia than the corresponding single infections (Fig. 1). While pycnidia density was also lower for mixed infections on Chinese Spring, the difference was not significant (Fig. 3).
Fig 3.
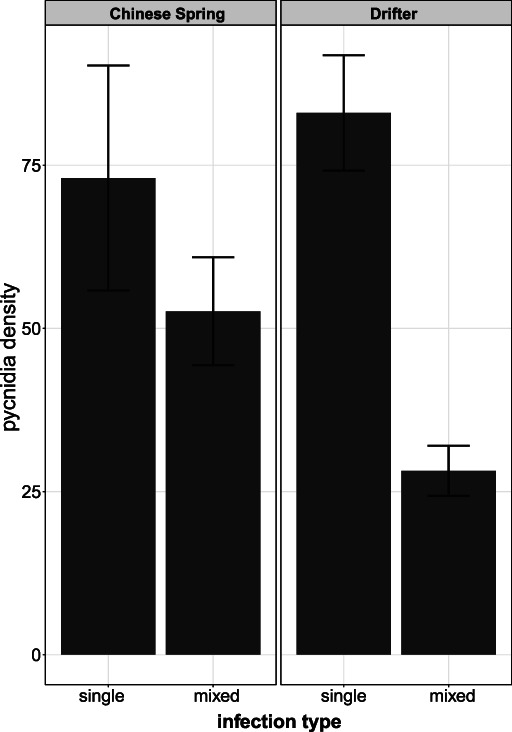
Effects of host cultivar and mixed infections on pathogen asexual reproduction. Mean and standard errors of Zymoseptoria tritici pycnidia density produced in mixed and single infections in the wheat cultivars Chinese Spring and Drifter at 24 days post infection.
We also compared the pycnidia density in mixture with the minimum, mean and maximum expected values calculated from single‐strain inoculations (Fig. 4). In comparisons using the mean and maximum expected values, we observed in most treatments a significantly lower pycnidia density in mixed infections than expected (Fig. 4A and B). For 5 of the 7 mixed infection treatments in Drifter, we also observed significantly lower pycnidia density compared to the minimum expected. Using the same comparison in Chinese Spring, many treatments were not significantly different from zero (Fig. 4C). Together, these results suggest that mixed infections have generally negative effects on pathogen reproduction, consistent with the predictions that competition among strains is antagonistic.
Fig 4.
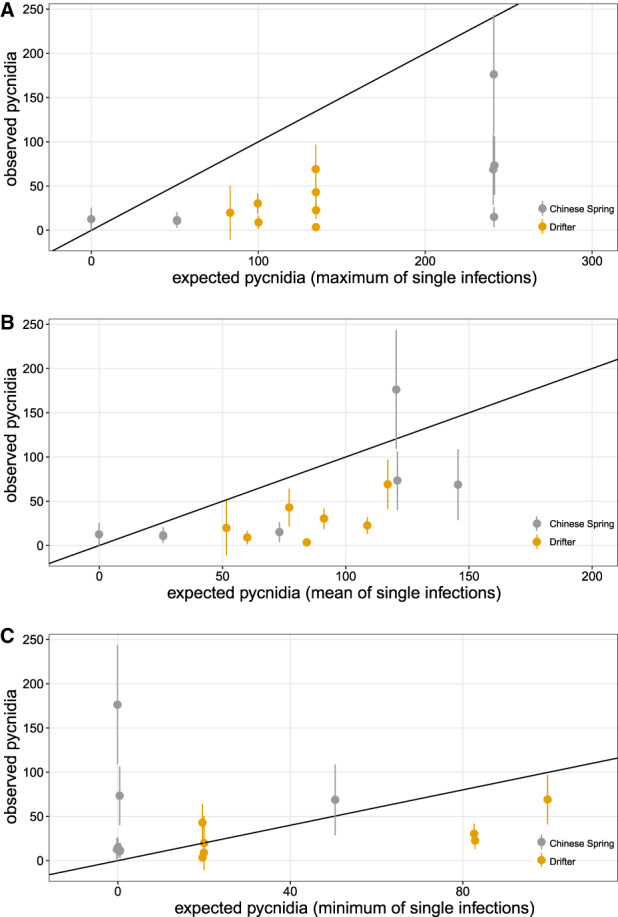
Mixed infections have generally negative effects on pathogen asexual reproduction. Observed values of Zymoseptoria tritici pycnidia density on the y‐axis are compared to expected values of pycnidia density on the x‐axis. Expected values are based on panel A, the strain producing the highest amount of pycnidia in a single infection; B, the mean pycnidia production of strains in a single infection; and C, the strain producing the lowest number of pycnidia in a single infection. Data for both wheat cultivars Chinese Spring and Drifter are shown. Errors bars show 95% confidence intervals around the means. [Color figure can be viewed at wileyonlinelibrary.com]
Skewed transmission in mixed infections
In order to evaluate the effect of mixed infections on the transmission potential of individual strains, we measured the relative frequencies of propagules for each strain in each treatment. With this aim, we collected the propagules formed on the leaf surfaces after 21 days of infection with a spore suspension of a mixture of either two or four strains in both cultivars Chinese Spring and Drifter and obtained the genotype of each propagule using PCR with strain‐specific primers. These data demonstrate that reproduction ratios in mixed infections varied depending on the mixture components, and that strains had significantly different competitive abilities (Fig. 5). In Drifter, 1E4 and 3D7 were the strongest competitors, dominating both 2‐ and 4‐strain mixtures. In the 1E4 + 3D7 treatment, 1E4 produced a significantly higher number of propagules (p < 0.001; Fig. 5) despite its lower biomass (Fig. 2; Fig. S2) and its lower virulence compared to the other strains in single infections (Fig. 1). 3D7 was the strongest competitor across all infection treatments in Chinese Spring, consistent with its high growth in mixed infections and despite not being the most virulent strain in single infections in this cultivar (Fig. 1). Interestingly, the avirulent strain 1E4 was able to produce propagules in mixed infections on Chinese Spring (Fig. 5), suggesting that coinfection with virulent strains may enable transmission of avirulent strains.
Fig 5.
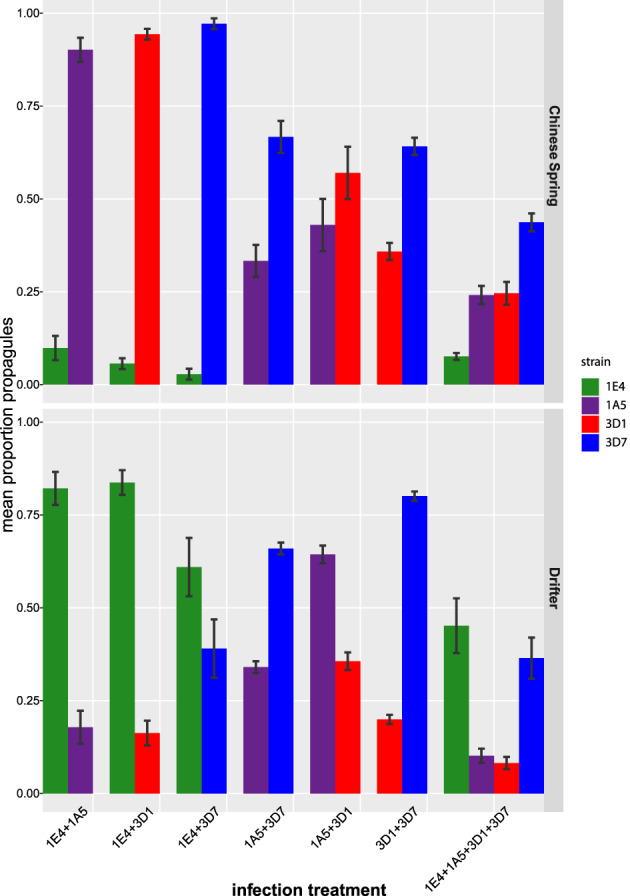
Evidence for unequal transmission potential in mixed infections. Proportion of Zymoseptoria tritici propagules produced by each strain in mixed infection for each strain combination and in each cultivar at 20 dpi. Means and standard errors are shown. [Color figure can be viewed at wileyonlinelibrary.com]
Co‐infecting strains occupy the same extracellular space
To follow the progression of co‐infecting strains in planta, we used transformed strains of 1E4 and 3D7 expressing either eGFP or mCherry (Fig. 6). In mixed infections with isogenic strains marked with different markers (3D7‐eGFP and 3D7‐mCherry) we observed frequent cases of hyphal fusion in the apoplastic space, resulting in an overlap of eGFP and mCherry signals that were detected in the same hyphae. The fused hyphae frequently colonized substomatal cavities (Fig. 6a). In contrast, when leaves were co‐infected with 1E4 and 3D7 strains labelled with different markers, no hyphal fusion was observed, most probably because of vegetative incompatibility. When co‐inoculated onto Drifter, the 1E4‐eGFP and 3D7‐mCherry strains formed two distinct networks of hyphae in the plant apoplastic space (Fig. 6C). Those hyphal networks grew into the same extracellular space and surrounded the same host cells, including the substomatal cavity (Fig. 6C and D). The reciprocal combination (3D7‐eGFP and 1E4‐mCherry) showed the same pattern of growth (Fig. 6B). These results show that co‐infecting strains do not mutually exclude each other and instead can coexist in the same extracellular space, enabling an intimate competition for the same host resources.
Fig 6.
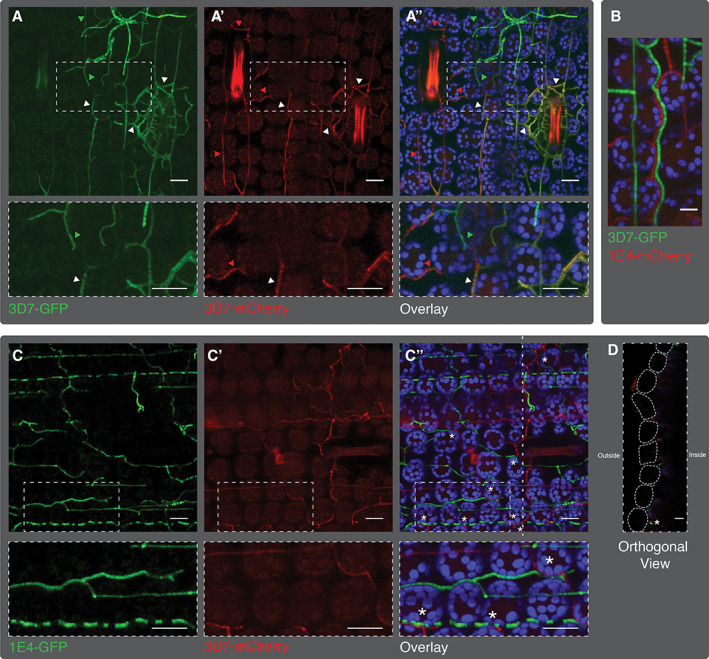
Co‐infecting hyphae of different strains co‐exist in cultivar Drifter. A. Plants infected with a mixture of 3D7 labelled with either cytosolic eGFP (green) or cytosolic mCherry (red). Hyphal fusions between strains were identified by finding hyphae containing both fluorescent proteins. Green arrows point to eGFP labelled hyphae, red arrows to mCherry labelled hyphae and white arrows to hyphae in which both fluorophores were detected. a, a′ represent, respectively, eGFP and mCherry channels and a″ the overlay. B. Image of 3D7 labelled with eGFP and 1E4 labelled with mCherry growing next to each other at 13 dpi. C. Confocal microscopy analysis at 9 dpi of cultivar Drifter infected with the 1E4 strain expressing cytosolic eGFP and the 3D7 strain expressing cytosolic mCherry. c and c′ represent eGFP and mCherry channels respectively, and c″ the overlay. D. Orthogonal view (yz) of the image shown in c″ at the position of the dotted line. Asterisks indicate 1E4 and 3D7 hyphae that grow next to each other. Lower panels in a–a″ and c–c″ represent a magnification of the upper panels indicated with the dotted squares. The blue signal in a″, b and c″ highlights the chloroplast autofluorescence. Bars in a and c represent 20 μm. Bars in panels B and D represent 10 μm. [Color figure can be viewed at wileyonlinelibrary.com]
Discussion
Infections are often genetically diverse with several pathogen strains co‐existing in the same host (Read and Taylor, 2001; Alizon, 2008; Tollenaere et al., 2016). Such mixed infections can lead to infection outcomes and epidemiological effects that differ from expectations developed from knowledge of single strain infections. We evaluated the effect of intra‐host competition during different stages of the infection cycle for the highly diverse plant pathogenic fungus Z. tritici. The results indicate that competition for resources and cross‐reaction of the immune system might lead to the observed reduction in transmission potential for the pathogen in mixed infections and that the effects of competition are host‐ and strain‐specific. We found evidence for dynamic changes in population composition during the infection cycle. Strains that suppressed the growth of competitors during host colonization (indicated by relative strain biomass) did not always produce more offspring (indicated by the relative proportions of spores) than the suppressed strains. We did not find an association between virulence (PLACL) in single infections and transmission potential (proportion of pycnidiospores produced) in mixed infections, suggesting that mixed infections in Z. tritici do not necessarily lead to selection of more virulent strains.
In two‐strain mixed infections, both strains formed separate mycelial networks that co‐existed during invasion of the same host apoplastic space, presumably drawing resources from the same plant cells. This intimate sharing of host space potentially leads to competition among strains, which likely explains the observed reduction in transmission potential during mixed infections. Hyphal fusion enables the formation of a functional unit in which growth, resource acquisition and nutritional exchange is coordinated across a mycelial network (Kasuga and Glass, 2008), increasing the fitness of mycelial colonies (Kasuga and Glass, 2008; Weichert and Fleißner, 2015). The absence of hyphal fusion between different strains might hinder the translocation of nutrients and/or signals needed to coordinate the formation of pycnidia. In addition, since pycnidia formation is locally confined to substomatal cavities and a mature pycnidium is formed by only one strain, competition for space in this constrained compartment potentially limits pycnidia production.
Our experimental design allowed us to investigate the impact of host genetic resistance on pathogen fitness in mixed infections. Host genotype had a significant effect on pathogen biomass, lesion area and fecundity. We consider it likely that these patterns reflect complex host‐pathogen‐pathogen interactions as well as cross‐reactions with the resistance response. Some wheat genotypes express strain‐specific resistance to Z. tritici, while remaining fully susceptible to other strains. Strain‐specific resistance was previously shown to contribute to the outcomes of mixed infections in other pathosystems (Råberg et al., 2006; Seifi et al., 2012; Susi et al., 2015; Woolhouse et al., 2015). Here we show that an avirulent strain of Z. tritici harbouring AvrStb6 hindered the development of symptoms in the presence of a virulent strain (3D1). Given that the avirulent strain only slightly increases in biomass in mixed infections and does not produce any symptoms when alone (Zhong et al., 2017; Kema et al., 2018), we consider it unlikely that this pattern reflects resource competition. Instead, cross‐host immune responses most probably hinder certain co‐existing virulent strains, presenting an example of apparent competition (Read and Taylor, 2001; Råberg et al., 2006). A previous experiment showed that the slight growth of the avirulent strain in mixed infections with virulent strains was sufficient to enable sexual reproduction between co‐infecting strains (Kema et al., 2018). Our experiments showed that avirulent strains can also produce asexual propagules in coinfections with virulent strains, illustrating that AvrStb6‐containing avirulent strains can possibly directly benefit from mixed infections and persist as distinct genotypes even when infecting a resistant host. This also illustrates the importance of evaluating the effectiveness and durability of genetic resistance under a scenario of mixed infections.
Numerous theoretical predictions suggest that mixed infections lead to an increase in overall virulence due to the higher competitive ability of the most virulent strains (Brown et al., 2002; Mideo, 2009). However, exceptions have been experimentally demonstrated in several pathosystems (Gower and Webster, 2005; Harrison et al., 2006). Our data shows that mixed infections of Z. tritici do not lead to synergistic increases in virulence, nor did they result in attenuated symptoms. Rather, our results indicate that in most cases the lesion area of mixed infections was very similar to that found in single infections based on the more virulent of the coinfecting strains. This pattern is in accordance with the capacity of the most virulent strains to inhibit competitors during host colonization.
Predicting the outcome of mixed infections requires knowledge of the within‐host interactions occurring between different strains, as different traits may be favoured in mixed compared to single infections (Levin and Pimentel, 1981; Brown et al., 2002; Gower and Webster, 2005; Mideo, 2009; Choisy and De Roode, 2010; Alizon et al., 2013). In our experiments, we observed a discrepancy between competitive dominance with respect to growth rate within the host (fungal biomass) and transmission potential (spore production). For example, the virulent strain 1A5 strongly suppressed the growth of co‐infecting strains, but in the absence of qualitative resistance, less virulent strains (3D7 and 1E4) produced more propagules than 1A5 in a mixed infection. This pattern agrees with theoretical models and empirical research on other pathosystems that have shown that the competitive ability of a pathogen can vary across different stages of infection, depending on the infection strategies of the coinfecting strains (Levin and Pimentel, 1981; Mosquera and Adler, 1998).
In the case of Z. tritici, faster growth of more virulent strains (such as 1A5 in cultivar Drifter) during the early infection may enable faster development of pycnidia (i.e., a shorter latent period). In contrast, less virulent strains (such as 3D7 and 1E4) that have reduced growth rates (i.e., a longer latent period) could predominate if they are more efficient at converting host resources into pathogen reproductive structures. Furthermore, precise outcomes are very likely context dependent. For example, it might be predicted that highly virulent, fast growing strains will have an advantage under conditions that enable nearly continuous pathogen transmission (e.g., rain every 3–4 days enabling nearly constant dispersal of the pycnidiospores). On the contrary, less virulent strains should have an advantage when rainfall is rare (e.g., rain every 20–25 days) because benefits associated with a shorter latent period likely disappear when opportunities for transmission are further separated in time.
Ultimately, the competitive success for pathogens depends on the frequency of successful transmission between hosts. Our experiments showed that spore production in mixed infections can be highly skewed, often favouring one strain over the others. Our data indicate that competitive success in mixed infections is determined at least in part by asexual spore production in single infections. For example, in Drifter the strains producing the highest amount of pycnidia in single infections (3D7 and 1E4) were also the most competitive in terms of propagule production. Likewise, the strain producing a higher amount of pycnidia in single infections on Chinese Spring (3D7) was also the most competitive in mixed infections. These data suggest that the fitness of a Z. tritici genotype in single infections may be a good predictor of its fitness in a mixed infection.
Within‐host interactions among competing strains are thought to be a major contributor to pathogen evolution (Mideo, 2009; Garbutt et al., 2011; Tollenaere et al., 2016). Our findings illustrate that mixed infections could favour the selection of less virulent pathogen strains, in contrast to what is frequently postulated. Due to the strong differential interaction between host and pathogen strain genotypes, the consequences of mixed infections for the evolution of virulence cannot be predicted in advance and need to be further investigated by performing between‐host transmission experiments (Alizon et al., 2013). Critically, the evolution of virulence in mixed infections is likely to depend on the biology of the pathosystem, suggesting that predicting the outcome of mixed infections in any pathosystem based on knowledge acquired in other pathosystems is likely to be a challenge.
Experimental procedure
Plant and fungal material
For the infection assays, the wheat (Triticum aestivum) cultivars Drifter (Delley Semences et Plants SA, DSP, Delley, Switzerland) and Chinese Spring Limagrain (Rosenthal, Germany) were used as hosts. We chose four Swiss strains of Z. tritici, ST99CH_3D1, ST99CH_3D7, ST99CH_1A5 and ST99CH_1E4 (3D1, 3D7, 1A5 and 1E4, respectively). 1A5 and 3D1 are mating type 2 whereas 1E4 and 3D7 are mating type 1. The strain 1E4 harbours AvrStb6 and is avirulent on the cultivar Chinese Spring, which harbours Stb6 (Zhong et al., 2017; Saintenac et al., 2018). The strains 1E4 and 3D7 used for confocal microscopy were transformed for this study by S. Kilaru and G. Steinberg with a codon optimized version of cytoplasmic enhanced green fluorescent protein (eGFP) and cytoplasmic monomeric Cherry (mCherry; Kilaru et al., 2015; Schuster et al., 2015). Fitness and virulence of the transformant lines were shown to be similar to that of the wild type strains by performing in vitro growth tests and virulence assays in cultivar Drifter (Fig. S5) as described previously (Meile et al., 2018). Strains were grown in 50 ml of yeast sucrose broth (YSB) medium (10 g l−1 yeast extract, 10 g l−1 sucrose) amended with 50 μg ml−1 kanamycin for 6 days at 18°C and 120 rpm. Spore cultures were filtered through cotton gauze, pelleted and resuspended in sterile water. Spore concentrations were estimated using KOVA® Glasstic slides (Hycor Biomedical, Inc., California).
Infection assays: evaluation of virulence, asexual reproduction and fungal biomass
The infections were performed with plants grown in the glasshouse, wherein wheat seedlings of each of the cultivars Drifter and Chinese Spring were infected with 11 different fungal treatments (described below), giving a total of 22 host x pathogen treatment combinations. Fifteen or 16 wheat seedlings of the cultivars Drifter and Chinese Spring were grown in peat soil (Tref, Netherlands) in square 11 × 11 × 12 cm plastic pots. Large trays, each containing a maximum of 15 pots, were placed in a glasshouse chamber with 18°C during the day and 15°C during the night, 16 h of light and 70% relative humidity. Seventeen‐day old plants were spray‐inoculated until run‐off (total volume of 35 ml) with a spore suspension containing 0.1% (v/v) of Tween 20 (Sigma Aldrich) either with one strain (1A5, 1E4, 3D1 and 3D7), or the six pairwise strain combinations (1A5 + 1E4, 1A5 + 3D7, 1E4 + 3D7, 3D1 + 1A5, 3D1 + 1E4 and 3D1 + 3D7) or with the four strains mixed together (1A5 + 1E4 + 3D7 + 3D1). The spore concentration for each strain in single and mixed infections was 106 spores ml−1. Therefore, the total concentration of the inoculum was 106 spores ml−1 multiplied by the number of strains used. To evaluate virulence, asexual reproduction and fungal growth, for each treatment, we spray‐inoculated a total of 60–64 individual plants growing in four pots. After inoculation, plants were kept at 100% humidity for 3 days by covering a small tray containing two pots with a plastic bag.
In order to minimize any potential influence of spatial position within trays or glasshouse benches on disease development, pots were randomized every 2–3 days by changing the location of the trays in the glasshouse chamber and the relative positions of the pots in the trays. Post infection, plants were sampled at 8 time points to capture the full range of disease progression. At each time point, the second leaf was destructively sampled from 6 replicate plants. These were randomly selected from among the four pots planted to each treatment combination (1–2 leaves were harvested per pot).
Damage to the host (a proxy for virulence), quantified as PLACL, was evaluated using automated image analysis (Stewart et al., 2018) at all the time points (10, 12, 14, 16, 17, 19 and 21 dpi). Since the phenotype of each strain was different (i.e., different necrotic lesions and pycnidia of different size and colour), we could not find optimal parameters to quantify pycnidia using automated image analysis. Therefore, pycnidia were counted manually using the multipoint selection tool in ImageJ (Schneider et al., 2012) at 24 dpi for Drifter and at 25 dpi for Chinese Spring and normalized to the leaf surface area to calculate pycnidia density (a proxy for asexual reproduction). Raw data is included in Tables S2 and S3.
Biomass quantification
Samples collected at 10–19 dpi were also analysed for fungal biomass. Immediately after scanning for image analysis, the top 2 cm of each leaf was discarded and the adjacent 8 cm section was used for DNA extraction. Two leaf segments were placed in a tube for each DNA extraction, three biological replicates were conducted for each extraction (six leaves total were extracted). Lyophilized infected leaf tissue was ground with the homogenizer Bio 101 FastPrep FP‐120 (Savant Instruments Inc.; Qbiogene). DNA was extracted using the DNeasy Plant kit (Qiagen). Quantitative PCR (qPCR) was performed on a LightCycler 480 (Roche Diagnostics Corp., Indianapolis, IN, USA) in 384‐well plates using specific primers designed for each strain (Table S4). The 10 μl reaction contained 1× HOT FIREPol EvaGreen qPCR Mix Plus master mix (Solis BioDyne, Tartu, Estonia), 0.1 μM of each primer pair and 1 μl of DNA. A standard curve was obtained using 0.01 ng, 0.1 ng, 1 ng, 10 ng and 20 ng of DNA of each strain grown in vitro, except for 3D1 for which 50 ng was used instead of 20 ng. All analysed samples gave values that were within the range of the standard curve. Amplification conditions were 15 min at 95°C, followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. Melting curve analysis was performed to confirm specificity. Absolute quantification was obtained using the second derivative method from the LightCycler 480 Software version 1.5 (Roche Diagnostics Corp., Indianapolis, IN, USA) and the total amount of fungal biomass normalized to the leaf area (ng of fungal DNA per cm2 of leaf) was calculated. Raw data is included in Table S5.
Measurement of propagule frequencies in mixed infections
To determine propagule frequency for each strain in mixed inoculations, 24 17‐day old plants grown under the same glasshouse conditions described above and in three pots (of 9 cm diameter and 7 cm height) were infected with 106 spores ml−1 of each of the mixed infection treatments described above. A spore suspension of 15 ml was used for each inoculation per cultivar. The spatial positions of the pots were randomized every 2–3 days. At 20 dpi, all infected sections of the second and third leaves in each pot were harvested, pooled and used to collect propagules. Each pot was considered a biological replicate and the experiment was performed twice independently. Leaves were kept overnight at 100% humidity in a glass Petri dish to facilitate extrusion of cirrhi from the pycnidia. Leaves were transferred to 50‐mL tubes containing 20–25 ml of sterile water and shaked vigorously to release the pycnidiospores. After filtering, spore concentrations were determined. Ten‐fold serial dilutions of the suspension in a final volume of 100 μl was placed on 20 yeast‐malt‐sucrose (YMS) agar plates (4 g l−1 yeast, 4 g l−1 malt, 4 g l−1 sucrose, 12 g l−1 agar) amended with kanamycin for obtaining well‐spaced single colonies. Ninety‐six colonies per replicate and treatment were transferred to 96 well plates with YSB. After 4–5 days of incubation at 18°C, direct PCR using the KAPA3G Plant PCR kit (KapaBiosystems, USA) was performed using strain‐specific primers (Table S4). Raw data is included in Table S6.
Confocal microscopy
An inverted Zeiss LSM 780 confocal microscope using both DPSS (561 nm) and Argon (488 nm) lasers as illumination sources was used for analysing the growth pattern of the strains in mixed infections. Signal detection for emissions were set as follows: eGFP (494.95–535.07 nm); mCherry (614.63–632.38 nm) and chloroplast autofluorescence (656.01–681.98 nm). Samples were prepared as follows: 13–15 day‐old Drifter plants were infected with eGFP and mCherry transformants, as described for the virulence assay; infected leaves were collected at either 9 or 13 days post infection; the top 3 cm of each leaf was discarded; the adjacent section of about 1.5 cm was collected and mounted in observation solution (milliQ water, 0.02% Tween20). Image processing was done using the Fiji package of ImageJ (http://fiji.sc; Schneider et al., 2012). Image processing included brightness, contrast adjustments and median filters (radius of 1 pixel). Orthogonal projections, cropping and scale bar additions were also made using Fiji. Tracing of the epidermal cells was done using Adobe Photoshop CC 2015. A total of six pictures were taken from three different plants. Two other independent datasets were obtained.
Statistical analyses
For all traits, data collected from second leaves sampled at single time points was used for linear modelling and biodiversity analyses. For pathogen biomass, data was collected at 19 dpi (when each treatment was at its maximum). To generate estimates of total fungal biomass in mixed infections, values recorded for individual strains were summed for each leaf. To quantify variation in asexual reproduction, pycnidia density at 24 dpi (for Drifter) or 25 dpi (for Chinese spring; the optimum time point for each treatment) was used for all analyses. For host damage (PLACL), analyses were performed using data collected at 14 dpi (reflecting the rate of increase in symptoms as modelled using the logistic function; see below). All analyses were conducted in R unless stated otherwise (R Core Team, 2020).
To investigate the main effects of host genotype (2 levels), pathogen treatment (11 levels), and their interactions, a linear modelling approach was used to test for differences among trait means. For total biomass data, we used a Generalized Linear Model (GLM) with a gaussian distribution and an identity link function. A GLM was also fitted to the pycnidia density data using a gamma distribution of errors and a log link function. For PLACL, we used a GLM with a quasibinomial distribution and a logit link function. Pairwise comparisons were subsequently calculated on model least square means to determine specific differences among treatment combinations using the package emmeans. GLM was also used to test for differences among counts of different strains collected in the final pycnidiospore assays, using a quasipoisson distribution of errors and a log link function. Experiment was included in the model as a fixed blocking effect.
To test for the effects of mixed and single infections on total biomass, PLACL and pycnidia density, pathogen treatments were assigned to either single‐infection or mixed‐infection categories (i.e., infection treatments comprising single strains were differentiated from those comprising two or four strains). Analyses were performed for each trait using a linear mixed model (LMM) where mixed infection (2 levels) and host (2 levels) were treated as fixed effects and infection treatment (11 levels) as a random effect. Data were transformed using cube‐root, log and rank‐based inverse normal for biomass, pycnidia density and PLACL, respectively. An additional LMM analysis was also performed using individual biomass as the response variable, where mixed infection and host were treated as fixed effects and infection treatment and strain were treated as random effects.
To assess the effects of strain identity on biomass in mixed infections, for each isolate/host combination, we used GLM to separately model individual strain biomass in response to the presence or absence of each competing strain. Data from the four‐strain treatment was excluded from this analysis as it was not possible to parse out relative effects of different strains. To facilitate a meaningful comparison of how the presence of competing strains influences biomass, estimates of biomass in two‐strain infections were doubled. This adjustment assumes that host resource availability does not change in the presence of multiple strains. Given this assumption, any differences in biomass among single and mixed infection treatments can be interpreted as the effect of the presence of a specific competing strain.
To further investigate how mixed infections influenced the infection outcome, we calculated the net biodiversity effect, calculated as the difference between trait means in mixed infections and expected means from single‐strain inoculations (Loreau and Hector, 2001). Expected values were calculated for each cultivar in three different ways: (i) based on the maximum of the single‐strain treatment means (maximum effect); (ii) based on the average of all component strains in single inoculation treatments (mean effect); and (iii) based on the minimum of the single‐strain treatment means (minimum effect). Observed values greater than the maximum indicate that mixed infection has positive synergistic effects on the infection progression; observed values less than the minimum indicate that mixed infection has negative synergistic effects. Observed values between the mean and the maximum may reflect either positive synergistic effects, or the competitive dominance of the more pathogenic strain; while values less than the mean indicate negative synergistic effects or the competitive dominance of the less pathogenic strain.
Changes in host damage (PLACL) over time were additionally modelled using the logistic function [Eq. (1) in Supporting Information: Note S1]. The shape of each curve is determined by three parameters: n 0 (PLACL at 10 dpi), r (rate of increase in PLACL) and K (stationary value of PLACL), which were used to estimate the virulence of each strain and the overall virulence of mixed infections. We determined which of these three parameters differed in different treatments by using the model selection approach (Johnson and Omland, 2004) based on the Bayesian information criterion (BIC; Schwarz, 1978) and nonlinear least squares fitting. BIC balances the goodness of fit with the number of parameters (or complexity) of the model. The analysis was conducted independently for each cultivar. Logistical modelling was conducted in the Python programming language (version 3.6.2, https://www.python.org) using the open‐source packages scipy (version 0.19.1), numpy (version 1.11.1) and matplotlib (version 1.5.3; Jones et al., 2001).
Supporting information
Appendix S1. Supporting information on materials and methods.
Fig. S1. Dynamics of single infections in the cultivars Chinese Spring (left panel) and Drifter (right panel). (A) Percentage of leaf area covered by lesions (PLACL) of wheat leaves infected with 3D1, 3D7, 1A5 and 1E4 in the cultivars Chinese Spring (left panel) and Drifter (middle panel) at different time points (10, 12, 14, 16, 17, 19, 21 dpi). Points represent the independent replicates and the curve represents the best model according to model selection based on the Bayesian information criterion (Note S1). Representative pictures of each treatment and for both cultivars are shown on the right panel. (B) The fungal biomass of each strain in single infections is shown for 1A5, 3D1, 3D7 and 1E4 based on the amount of DNA (ng) per cm2 of leaf at different time points (10, 12, 14, 16, 17 and 19 dpi).
Fig. S2. Fungal growth dynamics during host colonization in single infections or in mixed infections in the cultivars Drifter (A‐D) and Chinese Spring (E‐H). The fungal biomass of each strain in single and mixed infections is shown for 1A5 (A, E), 3D1 (B, F), 3D7 (C, G), and 1E4 (D, H) based on the amount of DNA (ng) per cm2 of leaf at different time points (10, 12, 14, 16, 17 and 19 dpi).
Fig. S3. The infection dynamics of mixed infections are altered compared to single infections and are both cultivar‐ and strain‐specific. Percentage of leaf area covered by lesions (PLACL) of wheat leaves infected with a single strain or with a combination of two strains or with all four strains in the cultivars Drifter (A–G) and Chinese Spring (H‐N) at different time points (10, 12, 14, 16, 17, 19, 21 dpi). PLACL for each treatment is shown in each graph (A, H) 3D1 + 1E4; (B, I) 3D7 + 1E4; (C, J) 1A5 + 1E4; (D, K) 3D1 + 3D7; (E, L) 3D7 + 1A5; (F, M) 3D1 + 1A5; (G, N) 3D1 + 3D7 + 1E4 + 1A5. Points represent the independent replicates and the curve represents the best model according to model selection based on the Bayesian information criterion (Note S1).
Fig. S4. Symptom development is not strongly influenced by mixed infections. The effect of mixed infections on PLACL (host damage). Observed values on the y‐axis should be compared to expected values on the x‐axis. Expected values are based on (A) mean PLACL of strains in monoculture; (B) maximum PLACL of strains in monoculture. Errors bars show 95% confidence intervals around the mean.
Fig. S5. Fluorescently labelled strains have phenotypes similar to the wildtype strains. (A) Percentage of leaf area covered by lesions and pycnidia formation produced by the strains 3D7, 1E4 and the corresponding strains expressing either eGFP or mCherry. No significant differences were detected (α = 0.1, KS test) between the wildtype and the mutant strains. (B) Growth of the labelled strains in rich media and in rich media appended with NaCl (0.5 M) and H2O2 (1 mM). Spores used were at a concentration of 106 and 105 spores ml−1. Pictures were taken after 5 days of growth at 18°C.
Table S1. Compact letter display showing pairwise comparisons of marginal means estimated from GLM analysis of total fungal biomass, pycnidia density and PLACL. Confidence levels for compact letter displays are set at 0.95.
Table S2. Host damage (PLACL) produced in single and mixed infections in the cultivars Chinese Spring and Drifter during infection.
Table S3. Pycnidia density and number produced in single and mixed infections in the cultivars Chinese Spring (25 dpi) and Drifter (24 dpi).
Table S4. Primers used to quantify the fungal biomass of individual Zymoseptoria tritici strains in infected leaves (qPCR) and to determine the composition of the propagules produced in mixed infections.
Table S5. Fungal biomass produced (ng cm−2 leaf) by each strain during host colonization in single and mixed infections in the cultivars Chinese Spring and Drifter. Crossing point (Cp), total DNA, leaf surface and biomass per leaf surface produced by each strain in single and mixed infections.
Table S6. Proportion of propagules produced in Chinese Spring and Drifter by each competing strain.
Acknowledgements
This project was supported by the ETH research grant ETH‐23 15‐2 to ASV and BAM. AM gratefully acknowledges financial support from the Swiss National Science Foundation through the Ambizione grant PZOOP3161453. Lukas Meile provided valuable comments to the manuscript. We thank Sreedhar Kilaru and Gero Steinberg for kindly providing the fluorescent Z. tritici strains (3D7‐eGFP, 3D7‐mCherry, 1E4‐eGFP and 1E4‐mCherry) and Petteri Karisto for designing the primers 1A5_5_F/R and 3D7_2_F/R. Confocal microscopy and qPCRs were performed, respectively, in the Scientific Center for Optical and Electron Microscopy (ScopeM) and in the Genetic Diversity Center (GDC) at ETH Zürich.
References
- Alizon, S. (2008) Decreased overall virulence in coinfected hosts leads to the persistence of virulent parasites. Am Nat 172: E67–E79. [DOI] [PubMed] [Google Scholar]
- Alizon, S. , and van Baalen, M. (2008) Multiple infections, immune dynamics, and the evolution of virulence. Am Nat 172: E150–E168. [DOI] [PubMed] [Google Scholar]
- Alizon, S. , de Roode, J.C. , and Michalakis, Y. (2013) Multiple infections and the evolution of virulence. Ecol Lett 16: 556–567. [DOI] [PubMed] [Google Scholar]
- Ben Ami, F. , Mouton, L. , and Ebert, D. (2008) The effects of multiple infections on the expression and evolution of virulence in a daphnia‐endoparasite system. Evolution (N Y) 62: 1700–1711. [DOI] [PubMed] [Google Scholar]
- van Baalen, M. , and Sabelis, M.W. (1995) The dynamics of multiple infection and the evolution of virulence. Am Nat 146: 881–910. [Google Scholar]
- Brown, J.K.M. , Chartrain, L. , Lasserre‐Zuber, P. , and Saintenac, C. (2015) Genetics of resistance to Zymoseptoria tritici and applications to wheat breeding. Fungal Genet Biol 79: 33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, S.P. , Hochberg, M.E. , and Grenfell, B.T. (2002) Does multiple infection select for raised virulence? Trends Microbiol 10: 401–405. [DOI] [PubMed] [Google Scholar]
- Choisy, M. , and De Roode, J.C. (2010) Mixed infections and the evolution of virulence: effects of resource competition, parasite plasticity, and impaired host immunity. Am Nat 175: E105–E118. [DOI] [PubMed] [Google Scholar]
- Cui, J. , Bahrami, A.K. , Pringle, E.G. , Hernandez‐Guzman, G. , Bender, C.L. , Pierce, N.E. , and Ausubel, F.M. (2005) Pseudomonas syringae manipulates systemic plant defenses against pathogens and herbivores. Proc Natl Acad Sci U S A 102: 1791–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbutt, J. , Bonsall, M.B. , Wright, D.J. , and Raymond, B. (2011) Antagonistic competition moderates virulence in Bacillus thuringiensis . Ecol Lett 14: 765–772. [DOI] [PubMed] [Google Scholar]
- Gardner, A. , West, S.A. , and Buckling, A. (2004) Bacteriocins, spite and virulence. Proc R Soc London Ser B Biol Sci 271: 1529–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold, A. , Giraud, T. , and Hood, M.E. (2009) Within‐host competitive exclusion among species of the anther smut pathogen. BMC Ecol 9: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez‐Jara, P. , Tenllado, F. , Martinez‐Garcia, B. , Atencio, F.A. , Barajas, D. , Vargas, M. , et al. (2004) Host‐dependent differences during synergistic infection by Potyviruses with potato virus X. Mol Plant Pathol 5: 29–35. [DOI] [PubMed] [Google Scholar]
- Gower, C.M. , and Webster, J.P. (2005) Intraspecific competition and the evolution of virulence in a parasitic trematode. Evolution (N Y) 59: 544–553. [PubMed] [Google Scholar]
- Halperin, T. , Schuster, S. , Pnini‐Cohen, S. , Zilberstein, A. , and Eyal, Z. (1996) The suppression of pycnidial production on wheat seedlings following sequential inoculation by isolates of Septoria tritici . Phytopathology 86: 728–732. [Google Scholar]
- Harrison, F. , Browning, L.E. , Vos, M. , and Buckling, A. (2006) Cooperation and virulence in acute Pseudomonas aeruginosa infections. BMC Biol 4: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haueisen, J. , Möller, M. , Eschenbrenner, C.J. , Grandaubert, J. , Seybold, H. , Adamiak, H. , and Stukenbrock, E.H. (2019) Highly flexible infection programs in a specialized wheat pathogen. Ecol Evol 9: 275–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt, R.D. , and Bonsall, M.B. (2017) Apparent competition. Annu Rev Ecol Evol Syst 48: 447–471. [Google Scholar]
- Hood, M.E. (2003) Dynamics of multiple infection and within‐host competition by the anther‐smut pathogen. Am Nat 162: 122–133. [DOI] [PubMed] [Google Scholar]
- Johnson, J.B. , and Omland, K.S. (2004) Model selection in ecology and evolution. Trends Ecol Evol 19: 101–108. [DOI] [PubMed] [Google Scholar]
- Jones, E. , Oliphant, T. , and Peterson, P. (2001). SciPy: open source scientific tools. Available at http://www.scipy.org/. [accessed 30 May 2018].
- Kasuga, T. , and Glass, N.L. (2008) Dissecting colony development of Neurospora crassa using mRNA profiling and comparative genomics approache. Eukaryot Cell 7: 1549–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kema, G.H.J. , Gohari, A.M. , Aouini, L. , Gibriel, H.A.Y. , Ware, S.B. , van Den Bosch, F. , et al. (2018) Stress and sexual reproduction affect the dynamics of the wheat pathogen effector AvrStb6 and strobilurin resistance. Nat Genet 50: 375–380. [DOI] [PubMed] [Google Scholar]
- Kilaru, S. , Schuster, M. , Studholme, D. , Soanes, D. , Lin, C. , Talbot, N.J. , and Steinberg, G. (2015) A codon‐optimized green fluorescent protein for live cell imaging in Zymoseptoria tritici . Fungal Genet Biol 79: 125–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leggett, H.C. , Buckling, A. , Long, G.H. , and Boots, M. (2013) Generalism and the evolution of parasite virulence. Trends Ecol Evol 28: 592–596. [DOI] [PubMed] [Google Scholar]
- Levin, S. , and Pimentel, D. (1981) Selection of intermediate rates of increase in parasite‐host systems. Am Nat 117: 308–315. [Google Scholar]
- Linde, C.C. , Zhan, J. , and McDonald, B.A. (2002) Population structure of Mycosphaerella graminicola: from lesions to continents. Phytopathology 92: 946–955. [DOI] [PubMed] [Google Scholar]
- López‐Villavicencio, M. , Courjol, F. , Gibson, A.K. , Hood, M.E. , Jonot, O. , Shykoff, J.A. , and Giraud, T. (2011) Competition, cooperation among kin, and virulence in multiple infections. Evolution (N Y) 65: 1357–1366. [DOI] [PubMed] [Google Scholar]
- Loreau, M. , and Hector, A. (2001) Partitioning selection and complementarity in biodiversity experiments. Nature 412: 72–76. [DOI] [PubMed] [Google Scholar]
- Massey, R.C. , Buckling, A. , and ffrench‐Constant, R. (2004) Interference competition and parasite virulence. Proc R Soc London Ser B Biol Sci 271: 785–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meile, L. , Croll, D. , Brunner, P.C. , Plissonneau, C. , Hartmann, F.E. , McDonald, B.A. , and Sánchez‐Vallet, A. (2018) A fungal avirulence factor encoded in a highly plastic genomic region triggers partial resistance to septoria tritici blotch. New Phytol 219: 1048–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mideo, N. (2009) Parasite adaptations to within‐host competition. Trends Parasitol 25: 261–268. [DOI] [PubMed] [Google Scholar]
- Mosquera, J. , and Adler, F.R. (1998) Evolution of virulence: a unified framework for coinfection and superinfection. J Theor Biol 195: 293–313. [DOI] [PubMed] [Google Scholar]
- O'Driscoll, A. , Kildea, S. , Doohan, F. , Spink, J. , and Mullins, E. (2014) The wheat‐Septoria conflict: a new front opening up? Trends Plant Sci 19: 602–610. [DOI] [PubMed] [Google Scholar]
- R Core Team . (2020) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: https://www.R-project.org/. [Google Scholar]
- Råberg, L. , de Roode, J.C. , Bell, A.S. , Stamou, P. , Gray, D. , and Read, A.F. (2006) The role of immune‐mediated apparent competition in genetically diverse malaria infections. Am Nat 168: 41–53. [DOI] [PubMed] [Google Scholar]
- Read, A.F. , and Taylor, L.H. (2001) The ecology of genetically diverse infections. Science 292: 1099–1102. [DOI] [PubMed] [Google Scholar]
- Rigaud, T. , Perrot‐Minnot, M.‐J. , and Brown, M.J.F. (2010) Parasite and host assemblages: embracing the reality will improve our knowledge of parasite transmission and virulence. Proc Biol Sci 277: 3693–3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley, M.A. , and Gordon, D.M. (1999) The ecological role of bacteriocins in bacterial competition. Trends Microbiol 7: 129–133. [DOI] [PubMed] [Google Scholar]
- Rochow, W.F. , and Ross, A.F. (1955) Virus multiplication in plants doubly infected by potato viruses X and Y. Virology 1: 10–27. [DOI] [PubMed] [Google Scholar]
- de Roode, J.C. , Pansini, R. , Cheesman, S.J. , Helinski, M.E.H. , Huijben, S. , Wargo, A.R. , et al. (2005) Virulence and competitive ability in genetically diverse malaria infections. Proc Natl Acad Sci U S A 102: 7624–7628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saintenac, C. , Lee, W.‐S. , Cambon, F. , Rudd, J.J. , King, R.C. , Marande, W. , et al. (2018) Wheat receptor‐kinase‐like protein Stb6 controls gene‐for‐gene resistance to fungal pathogen Zymoseptoria tritici . Nat Genet 50: 368–374. [DOI] [PubMed] [Google Scholar]
- Schneider, C.A. , Rasband, W.S. , and Eliceiri, K.W. (2012) NIH image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schürch, S. , and Roy, B.A. (2004) Comparing single‐vs. mixed‐genotype infections of Mycosphaerella graminicola on wheat: effects on pathogen virulence and host tolerance. Evol Ecol 18: 1–14. [Google Scholar]
- Schuster, M. , Kilaru, S. , Guo, M. , Sommerauer, M. , Lin, C. , and Steinberg, G. (2015) Red fluorescent proteins for imaging Zymoseptoria tritici during invasion of wheat. Fungal Genet Biol 79: 132–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz, G. (1978) Estimating the dimension of a model. Ann Stat 6: 461–464. [Google Scholar]
- Seifi, A. , Nonomura, T. , Matsuda, Y. , Toyoda, H. , and Bai, Y. (2012) An avirulent tomato powdery mildew isolate induces localized acquired resistance to a virulent isolate in a spatiotemporal manner. Mol Plant Microbe Interact 25: 372–378. [DOI] [PubMed] [Google Scholar]
- Spoel, S.H. , Johnson, J.S. , and Dong, X. (2007) Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proc Natl Acad Sci U S A 104: 18842–18847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, E.L. , Croll, D. , Lendenmann, M.H. , Sanchez‐Vallet, A. , Hartmann, F.E. , Palma‐Guerrero, J. , et al. (2018) Quantitative trait locus mapping reveals complex genetic architecture of quantitative virulence in the wheat pathogen Zymoseptoria tritici . Mol Plant Pathol 19: 201–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susi, H. , Barrès, B. , Vale, P.F. , and Laine, A.‐L. (2015) Co‐infection alters population dynamics of infectious disease. Nat Commun 6: 5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollenaere, C. , Susi, H. , and Laine, A.‐L. (2016) Evolutionary and epidemiological implications of multiple infection in plants. Trends Plant Sci 21: 80–90. [DOI] [PubMed] [Google Scholar]
- Weichert, M. , and Fleißner, A. (2015) Anastomosis and heterokaryon formation. In Genetic Transformation Systems in Fungi, Volume 2, van den Berg, M.A. , and Maruthachalam, K. (eds). Cham: Springer International Publishing, pp. 3–21. [Google Scholar]
- Woolhouse, M.E.J. , Thumbi, S.M. , Jennings, A. , Chase‐Topping, M. , Callaby, R. , Kiara, H. , et al. (2015) Co‐infections determine patterns of mortality in a population exposed to parasite infection. Sci Adv 1: e1400026‐844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelikovitch, N. , and Eyal, Z. (1991) Reduction in pycnidial coverage after inoculation of wheat with mixtures of isolates of Septoria tritici . Plant Dis 75: 907–910. [Google Scholar]
- Zhan, J. , Kema, G.H.J. , Waalwijk, C. , and McDonald, B.A. (2002a) Distribution of mating type alleles in the wheat pathogen Mycosphaerella graminicola over spatial scales from lesions to continents. Septoria tritici blotch Dis wheat Tools Tech to study Pathog Zymoseptoria tritici 36: 128–136. [DOI] [PubMed] [Google Scholar]
- Zhan, J. , Linde, C.C. , Jurgens, T. , Merz, U. , Steinebrunner, F. , and McDonald, B.A. (2005) Variation for neutral markers is correlated with variation for quantitative traits in the plant pathogenic fungus Mycosphaerella graminicola . Mol Ecol 14: 2683–2693. [DOI] [PubMed] [Google Scholar]
- Zhan, J. , Mundt, C.C. , Hoffer, M.E. , and McDonald, B.A. (2002b) Local adaptation and effect of host genotype on the rate of pathogen evolution: an experimental test in a plant pathosystem. J Evol Biol 15: 634–647. [Google Scholar]
- Zhong, Z. , Marcel, T.C. , Hartmann, F.E. , Ma, X. , Plissonneau, C. , Zala, M. , et al. (2017) A small secreted protein in Zymoseptoria tritici is responsible for avirulence on wheat cultivars carrying the Stb6 resistance gene. New Phytol 214: 619–631. [DOI] [PubMed] [Google Scholar]
- Zinga, I. , Chiroleu, F. , Legg, J. , Lefeuvre, P. , Komba, E.K. , Semballa, S. , et al. (2013) Epidemiological assessment of cassava mosaic disease in Central African Republic reveals the importance of mixed viral infection and poor health of plant cuttings. Crop Prot 44: 6–12. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1. Supporting information on materials and methods.
Fig. S1. Dynamics of single infections in the cultivars Chinese Spring (left panel) and Drifter (right panel). (A) Percentage of leaf area covered by lesions (PLACL) of wheat leaves infected with 3D1, 3D7, 1A5 and 1E4 in the cultivars Chinese Spring (left panel) and Drifter (middle panel) at different time points (10, 12, 14, 16, 17, 19, 21 dpi). Points represent the independent replicates and the curve represents the best model according to model selection based on the Bayesian information criterion (Note S1). Representative pictures of each treatment and for both cultivars are shown on the right panel. (B) The fungal biomass of each strain in single infections is shown for 1A5, 3D1, 3D7 and 1E4 based on the amount of DNA (ng) per cm2 of leaf at different time points (10, 12, 14, 16, 17 and 19 dpi).
Fig. S2. Fungal growth dynamics during host colonization in single infections or in mixed infections in the cultivars Drifter (A‐D) and Chinese Spring (E‐H). The fungal biomass of each strain in single and mixed infections is shown for 1A5 (A, E), 3D1 (B, F), 3D7 (C, G), and 1E4 (D, H) based on the amount of DNA (ng) per cm2 of leaf at different time points (10, 12, 14, 16, 17 and 19 dpi).
Fig. S3. The infection dynamics of mixed infections are altered compared to single infections and are both cultivar‐ and strain‐specific. Percentage of leaf area covered by lesions (PLACL) of wheat leaves infected with a single strain or with a combination of two strains or with all four strains in the cultivars Drifter (A–G) and Chinese Spring (H‐N) at different time points (10, 12, 14, 16, 17, 19, 21 dpi). PLACL for each treatment is shown in each graph (A, H) 3D1 + 1E4; (B, I) 3D7 + 1E4; (C, J) 1A5 + 1E4; (D, K) 3D1 + 3D7; (E, L) 3D7 + 1A5; (F, M) 3D1 + 1A5; (G, N) 3D1 + 3D7 + 1E4 + 1A5. Points represent the independent replicates and the curve represents the best model according to model selection based on the Bayesian information criterion (Note S1).
Fig. S4. Symptom development is not strongly influenced by mixed infections. The effect of mixed infections on PLACL (host damage). Observed values on the y‐axis should be compared to expected values on the x‐axis. Expected values are based on (A) mean PLACL of strains in monoculture; (B) maximum PLACL of strains in monoculture. Errors bars show 95% confidence intervals around the mean.
Fig. S5. Fluorescently labelled strains have phenotypes similar to the wildtype strains. (A) Percentage of leaf area covered by lesions and pycnidia formation produced by the strains 3D7, 1E4 and the corresponding strains expressing either eGFP or mCherry. No significant differences were detected (α = 0.1, KS test) between the wildtype and the mutant strains. (B) Growth of the labelled strains in rich media and in rich media appended with NaCl (0.5 M) and H2O2 (1 mM). Spores used were at a concentration of 106 and 105 spores ml−1. Pictures were taken after 5 days of growth at 18°C.
Table S1. Compact letter display showing pairwise comparisons of marginal means estimated from GLM analysis of total fungal biomass, pycnidia density and PLACL. Confidence levels for compact letter displays are set at 0.95.
Table S2. Host damage (PLACL) produced in single and mixed infections in the cultivars Chinese Spring and Drifter during infection.
Table S3. Pycnidia density and number produced in single and mixed infections in the cultivars Chinese Spring (25 dpi) and Drifter (24 dpi).
Table S4. Primers used to quantify the fungal biomass of individual Zymoseptoria tritici strains in infected leaves (qPCR) and to determine the composition of the propagules produced in mixed infections.
Table S5. Fungal biomass produced (ng cm−2 leaf) by each strain during host colonization in single and mixed infections in the cultivars Chinese Spring and Drifter. Crossing point (Cp), total DNA, leaf surface and biomass per leaf surface produced by each strain in single and mixed infections.
Table S6. Proportion of propagules produced in Chinese Spring and Drifter by each competing strain.


