Summary
Symbiotic association is universal in nature, and an array of symbionts play a crucial part in host life history. Aphids and their diverse symbionts have become a good model system to study insect‐symbiont interactions. Previous symbiotic diversity surveys have mainly focused on a few aphid clades, and the relative importance of different factors regulating microbial community structure is not well understood. In this study, we collected 65 colonies representing eight species of the aphid genus Mollitrichosiphum from different regions and plants in southern China and Nepal and characterized their microbial compositions using Illumina sequencing of the V3 − V4 hypervariable region of the 16S rRNA gene. We evaluated how microbiota varied across aphid species, geography and host plants and the correlation between microbial community structure and host aphid phylogeny. Heritable symbionts dominated the microbiota associated with Mollitrichosiphum, and multiple infections of secondary symbionts were prevalent. Ordination analyses and statistical tests highlighted the contribution of aphid species in shaping the structures of bacterial, symbiont and secondary symbiont communities. Moreover, we observed a significant correlation between Mollitrichosiphum aphid phylogeny and microbial community composition, providing evidence for a pattern of phylosymbiosis between natural aphid populations and their microbial associates.
Introduction
Eukaryotes engage in associations with a variety of microorganisms. Bacterial symbionts of sap‐feeding insects have been documented in numerous studies (Buchner, 1965; Baumann, 2005; Sudakaran et al., 2017). Phloem‐feeding aphids and their bacterial symbionts represent a good model system to study host‐symbiont interactions. Aphids rely on the primary endosymbiont Buchnera aphidicola, which is located in specialized bacteriocytes, to supply essential nutrition lacking in their diet (Buchner, 1965; Douglas, 1998). Buchnera persists in almost all aphid species (Baumann et al., 1995), is maintained within aphid populations by direct maternal transmission (Koga et al., 2012) and has diversified in parallel with host lineages (Munson et al., 1991; Clark et al., 2000; Liu et al., 2013; Xu et al., 2018). Furthermore, Buchnera has undergone extreme gene loss and degradation of functions due to long‐term living restricted to bacteriocytes (Rispe and Moran, 2000; Wernegreen, 2002). In some aphid species, other bacterial partners are involved in co‐obligate associations to compensate for the essential nutrient biosynthesis not ensured by Buchnera (e.g., the co‐obligate symbiont Serratia symbiotica in Cinara cedri) (Lamelas et al., 2011; Mccutcheon and Moran, 2012; Bennett and Moran, 2015).
In addition to the obligate heritable Buchnera, aphids harbour multiple heritable facultative symbionts that can provide diverse ecological benefits, such as conferring parasitoid and fungal resistance (Oliver et al., 2005; Scarborough et al., 2005; Łukasik et al., 2013; Heyworth and Ferrari, 2015), increasing tolerance to heat shock (Chen et al., 2000; Russell and Moran, 2006; Guay et al., 2009) and broadening host plant range (Tsuchida et al., 2004; Tsuchida et al., 2011; Wagner et al., 2015). Facultative symbionts inhabit various tissues of their aphid hosts (Oliver et al., 2010) and spread via vertical transmission and occasional horizontal transmission (Russell et al., 2003; Russell and Moran, 2005; Michalik et al., 2014; Pons et al., 2019). Nine facultative symbionts in aphids have been extensively reported, including Serratia symbiotica (Unterman et al., 1989), Rickettsia (Chen et al., 1996), Hamiltonella defensa (Darby et al., 2001), Regiella insecticola (Sandström et al., 2001), Spiroplasma (Fukatsu et al., 2001), Arsenophonus (Russell et al., 2003), Fukatsuia symbiotica (Guay et al., 2009), Rickettsiella viridis (Tsuchida et al., 2010) and Wolbachia (Augustinos et al., 2011). In aphids, multiple infections of secondary symbionts (i.e., infections of more than one symbiont in a host individual) occur moderately because of the cost of harbouring diverse assemblages of secondary symbionts (Oliver et al., 2014). For example, coinfection of H. defensa and R. viridis in the pea aphid Acyrthosiphon pisum caused a reduction in aphid survival and fecundity (Leclair et al., 2017).
To date, most studies on aphid symbiont diversity have focused on the pea aphid and species of subfamilies Aphidinae and Lachninae (Zytynska and Weisser, 2016). The occurrence of particular secondary symbionts within one aphid species has been reported to be mainly related to the host plant (Simon et al., 2003; Ferrari et al., 2012; Brady and White, 2013; Gauthier et al., 2015; Xu et al., 2020a) and geographic distribution (Tsuchida et al., 2002). Infection patterns can also vary from native to invasive regions (Bansal et al., 2014) and during seasonal shifts (Smith et al., 2015; Liu et al., 2019). At taxonomic levels higher than species, the patterns of secondary symbiont infections have been found to be associated with aphid species, characteristics of aphids and ecological conditions (Henry et al., 2015; Xu et al., 2020b, 2021). However, the factors influencing the symbiont community structure of aphids have rarely been explored and assessed across both ecological and aphid phylogenetic contexts. In addition, more studies on different aphid lineages are needed for a comprehensive understanding of the symbiont diversity landscape.
Mollitrichosiphum is a monophyletic aphid genus of the subfamily Greenideinae (Insecta: Hemiptera: Aphididae) that comprises 18 extant species worldwide (11 species recorded in China) (Favret, 2020) and is mainly distributed in eastern and southern Asia (Blackman and Eastop, 2020). Mollitrichosiphum species are monoecious with a holocyclic or anholocyclic life cycle. Some species are monophagous or oligophagous, feeding on young leaves or shoots of Fagaceae or Meliosma (Sabiaceae); some species colonize plants from different families, including Fagaceae, Betulaceae, Sabiaceae, Proteaceae and so on (Ghosh and Agarwala, 1993; Zhang and Qiao, 2010; Blackman and Eastop, 2020). Previous research has confirmed parallel evolution between Mollitrichosiphum aphids and Buchnera (Liu et al., 2013). In a survey study of Wolbachia infection in Chinese aphids (Wang et al., 2014), Wolbachia was detected in all sampled Mollitrichosiphum species. However, little is known about the bacterial flora of this genus.
In the present study, we used Illumina sequencing of the 16S rRNA gene to characterize the microbial communities of eight Mollitrichosiphum species collected from different plants and regions across southern China and Nepal. We fully assessed the variation in bacterial, symbiont (incl. Buchnera and secondary symbionts) and secondary symbiont communities according to different factors, including aphid species, geography and host plant, and revealed the microbial community determinant in Mollitrichosiphum aphids. Finally, we estimated the correlation between microbial community dissimilarity and aphid relatedness to further understand the eco‐evolutionary pattern of aphid‐symbiont interactions.
Results
Taxonomic composition of the microbial community associated with Mollitrichosiphum aphids
After all filtering steps, a total of 3,367,211 reads (51,803 reads per sample) were obtained. Ninety‐nine operational taxonomic units (OTUs) were clustered and assigned to 33 genera, 22 families, 15 orders, 13 classes and 6 phyla of bacteria. Bacterial communities of Mollitrichosiphum aphids were dominated by the phylum Proteobacteria (average relative abundance across all samples: 99.73%). The most highly dominant class and order were Gammaproteobacteria (93.58%) and Enterobacteriales (93.48%), respectively. Enterobacteriaceae (92.81%) was the most abundant family, followed by Rickettsiaceae (4.10%) (Table S1).
At the genus level, the primary endosymbiont Buchnera was detected in all samples and predominated in most, with an average relative abundance of 72.09%. Six secondary symbionts were detected: four showed relative abundances greater than 1% (Serratia symbiotica: 8.91%; Rickettsia: 4.10%; Arsenophonus: 3.10%; Wolbachia: 1.94%), with two having abundances lower than 1% (Fukatsuia symbiotica: 0.67%; Hamiltonella defensa: 0.40%) (Table S1). S. symbiotica was the most abundant secondary symbiont, and its relative abundance was even higher than that of Buchnera in several samples of M. nigrofasciatum (Fig. 1). Additionally, a high frequency of multiple infections of secondary symbionts was observed in Mollitrichosiphum. Each aphid sample simultaneously harboured 4–6 secondary symbionts. All samples were infected with S. symbiotica, Rickettsia, Arsenophonus and Wolbachia. F. symbiotica was detected in all Mollitrichosiphum species except M. nigrum (prevalence across all samples: 40/65). The prevalence of H. defensa was variable among different aphid species (28/65). H. defensa was represented by only one OTU, whereas each of the other secondary symbionts harboured 2–4 OTUs (Fig. 2). The dominant secondary symbiont OTUs generally differed among Mollitrichosiphum species, though most OTUs were widely distributed.
Fig. 1.
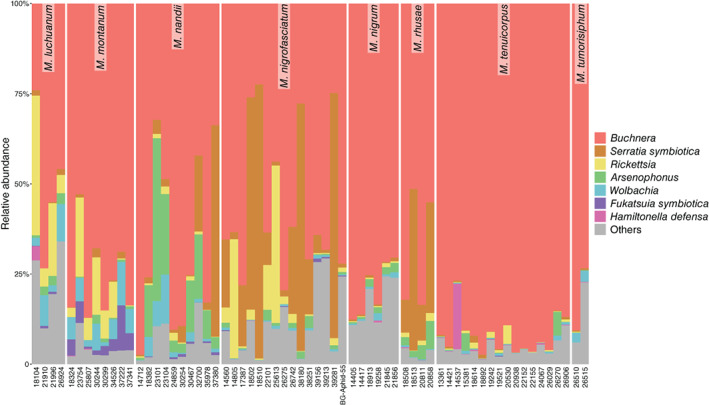
Microbial community composition associated with Mollitrichosiphum aphids.
Fig. 2.
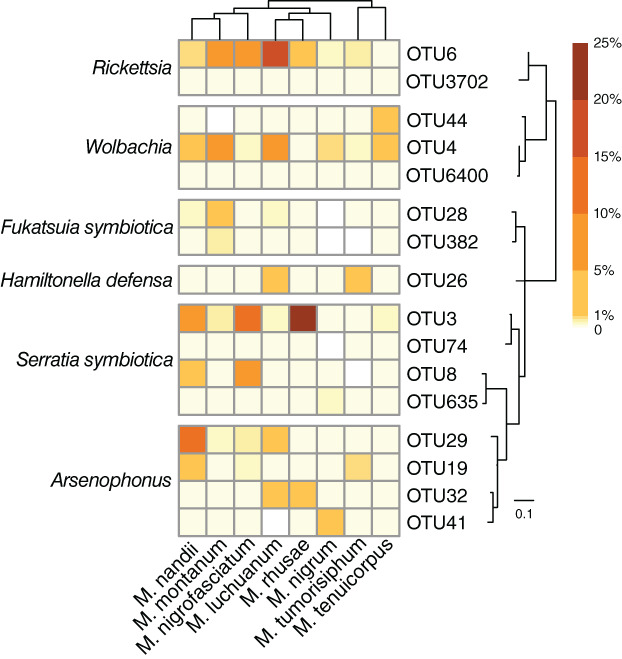
Heatmap representing the distribution and relative abundances of secondary symbiont OTUs among Mollitrichosiphum species. The maximum‐likelihood tree of secondary symbiont OTUs and a simplified cladogram displaying the phylogenetic relationships of Mollitrichosiphum species are presented.
Factors structuring Mollitrichosiphum microbial community diversity
The microbiota of Mollitrichosiphum displayed a pattern structured by aphid species. Kruskal–Wallis tests of alpha diversity indices of the bacterial, symbiont and secondary symbiont communities revealed significant differences among aphid species, which indicated greater interspecific microbiota variation than intraspecific variation (p < 0.05 for both Shannon and Simpson indices). Conversely, the microbial communities did not differ significantly among geographic region (Shannon, p = 0.081–0.901; Simpson, p = 0.060–0.922) or host plants (Shannon, p = 0.435–0.867; Simpson, p = 0.497–0.949). The results of three‐way ANOVA for alpha diversity indices also showed a significant impact of aphid species on the bacterial and symbiont communities (n ≥ 1, F (7,15) = 4.750–9.167, p ≤ 0.004; n ≥ 3, F (4,12) = 7.182–10.667, p ≤ 0.001) (Table S2). The community compositions of bacteria and symbionts were not significantly different among geographic regions or host plants (p > 0.05), and the secondary symbiont community was structured by none of these three factors (p > 0.05) (Table S2).
Regarding beta diversity, constrained PCoA (cPCoA) plots of Bray–Curtis distances displayed a separation tendency of microbial communities according to aphid species (p = 0.001) (Fig. 3A–C and Fig. S1A–C). The structures of the microbial community among geographic regions (p = 0.001–0.022) (Fig. 3D and F and Fig. S1D–F) were also significant, except for the symbiont community with a sample size ≥3 (p = 0.051) (Fig. 3E). However, aphid species usually explained more overall variance in the data (28.6%–39% of variance) than did geographic region (21%–38.7% of variance). Moreover, cPCoA analyses did not indicate a distinct structure constrained by host plant (4.66%–13.6% of variance, p = 0.12–0.68) (Fig. 3G–I and Fig. S1G–I). Unconstrained NMDS plots failed to uncover meaningful patterns structured by these three factors using either Bray–Curtis or unweighted UniFrac distances (Figs S2–S4).
Fig. 3.
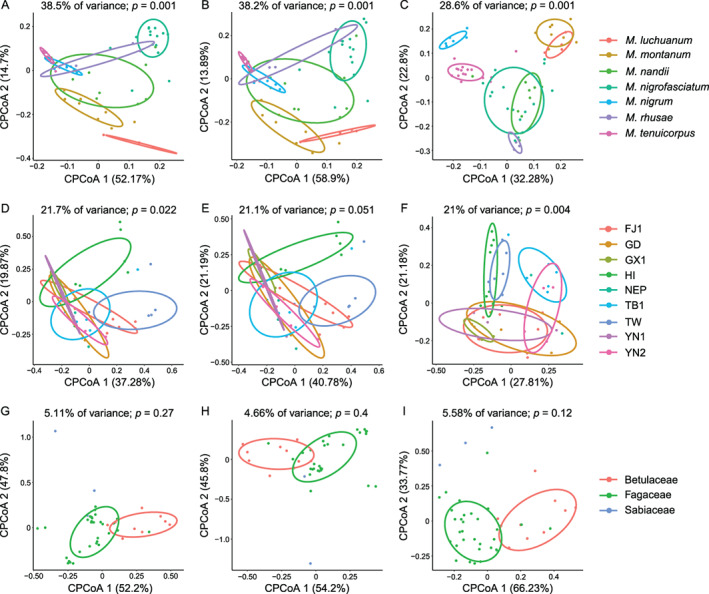
Constrained principal coordinate analysis (cPCoA) plots of Bray–Curtis distances of bacterial (A, D, G), symbiont (B, E, H) and secondary symbiont (C, F, I) communities (n ≥ 3). Plots are structured by aphid species (A–C), geographic region (D–F) and host plant (G–I). The overall variation explained by the constrained factor is displayed at the top of each plot. The percent variation shown on each axis refers to the fraction of the total variance explained by the projection. The abbreviations are given in Table S5.
ANOSIM corroborated that aphid species had the greatest effect on the microbial community structure of Mollitrichosiphum aphids (Table 1). Significant differences were observed among aphid species using all types of beta diversity data (R = 0.228–0.446; p < 0.001). The effects of host plant (R = −0.093–0.084; p = 0.117–0.877) and geographic region (R = −0.013 − 0.114; p = 0.055–0.553) were not statistically significant, except for a significant impact of geographic region on the secondary symbiont community (n ≥ 1, Bray–Curtis) (p = 0.016). Nonetheless, R values for this dataset suggested greater dissimilarity between samples from different aphid species (R = 0.397) than from different geographic regions (R = 0.137). The importance of aphid species in shaping microbiota composition was further confirmed by PERMANOVA, in which highly significant R 2 values of aphid species were obtained (R 2 = 0.292–0.465; p ≤ 0.007) (Table 1). Significant impacts of geographic region were found only in the analyses of Bray–Curtis distances (p = 0.002–0.017), with a minor R 2 value in most cases (R 2 = 0.229–0.410). The effect of host plant was not significant (R 2 = 0.041–0.144; p = 0.081–0.653), except for the bacterial community (unweighted UniFrac) (p = 0.004–0.008). But its contribution was limited (R 2 = 0.129–0.202) compared to aphid species (R 2 = 0.416–0.421).
Table 1.
Results of ANOSIM and PERMANOVA based on Bray–Curtis and unweighted UniFrac distances.
| Beta diversity distance | Microbial community | Sample size | Aphid species | Geographic region | Host plant | |||
|---|---|---|---|---|---|---|---|---|
| ANOSIM (R, p) | PERMANOVA (R 2, p) | ANOSIM (R, p) | PERMANOVA (R 2, p) | ANOSIM (R, p) | PERMANOVA (R 2, p) | |||
| Bray–Curtis | Bacteria | n ≥ 1 | 0.438, <0.001 | 0.465, <0.001 | 0.063, 0.176 | 0.410, 0.005 | −0.050, 0.667 | 0.095, 0.653 |
| n ≥ 3 | 0.446, <0.001 | 0.461, <0.001 | −0.013, 0.553 | 0.223, 0.054 | −0.016, 0.520 | 0.050, 0.310 | ||
| Symbionts | n ≥ 1 | 0.385, <0.001 | 0.432, <0.001 | 0.114, 0.055 | 0.465, 0.002 | 0.010, 0.430 | 0.109, 0.482 | |
| n ≥ 3 | 0.399, <0.001 | 0.428, <0.001 | −0.004, 0.495 | 0.204, 0.159 | 0.036, 0.341 | 0.054, 0.265 | ||
| Secondary symbionts | n ≥ 1 | 0.397, <0.001 | 0.444, <0.001 | 0.137, 0.016 | 0.375, 0.002 | 0.007, 0.463 | 0.134, 0.363 | |
| n ≥ 3 | 0.385, <0.001 | 0.438, <0.001 | 0.081, 0.078 | 0.229, 0.017 | 0.001, 0.485 | 0.050, 0.354 | ||
| Unweighted Unifrac | Bacteria | n ≥ 1 | 0.429, <0.001 | 0.421, <0.001 | 0.045, 0.205 | 0.328, 0.116 | 0.071, 0.135 | 0.202, 0.008 |
| n ≥ 3 | 0.425, <0.001 | 0.416, <0.001 | 0.057, 0.141 | 0.218, 0.085 | 0.084, 0.117 | 0.129, 0.004 | ||
| Symbionts | n ≥ 1 | 0.305, <0.001 | 0.405, <0.001 | 0.077, 0.105 | 0.317, 0.273 | 0.009, 0.429 | 0.144, 0.239 | |
| n ≥ 3 | 0.274, <0.001 | 0.391, <0.001 | 0.093, 0.057 | 0.230, 0.087 | 0.019, 0.381 | 0.098, 0.081 | ||
| Secondary symbionts | n ≥ 1 | 0.253, <0.001 | 0.341, 0.003 | 0.030, 0.305 | 0.302, 0.347 | −0.073, 0.808 | 0.080, 0.404 | |
| n ≥ 3 | 0.228, <0.001 | 0.292, 0.007 | 0.020, 0.332 | 0.161, 0.451 | −0.093, 0.877 | 0.041, 0.347 | ||
Statistically significant p values (p < 0.05) are highlighted in italics.
Correlation between microbial community composition and aphid relatedness
The correlation between microbial community composition and host aphid phylogeny was examined to further understand the pattern of aphid‐microbe associations. The divergence times of Mollitrichosiphum are depicted in Fig. S5. Mantel tests performed on unweighted UniFrac distances and aphid divergence times showed a significant positive correlation between microbial community structure and aphid phylogeny (p < 0.001; bacteria: r = 0.413; symbionts: r = 0.422; secondary symbionts: r = 0.396) (Fig. 4D–F). When analysed with Bray–Curtis distances, significant correlations were also observed for the bacterial (r = 0.087, p = 0.046) (Fig. 4A) and secondary symbiont communities (r = 0.137, p = 0.002) (Fig. 4C). Procrustes analyses revealed the same pattern, in which microbiota structure was related to aphid phylogeny (Procrustes M 2 = 0.768–0.856, p = 0.001) (Fig. 5A, B, D–F), except for the secondary symbiont community (Bray–Curtis) (M 2 = 0.998, p = 0.984) (Fig. 5C).
Fig. 4.
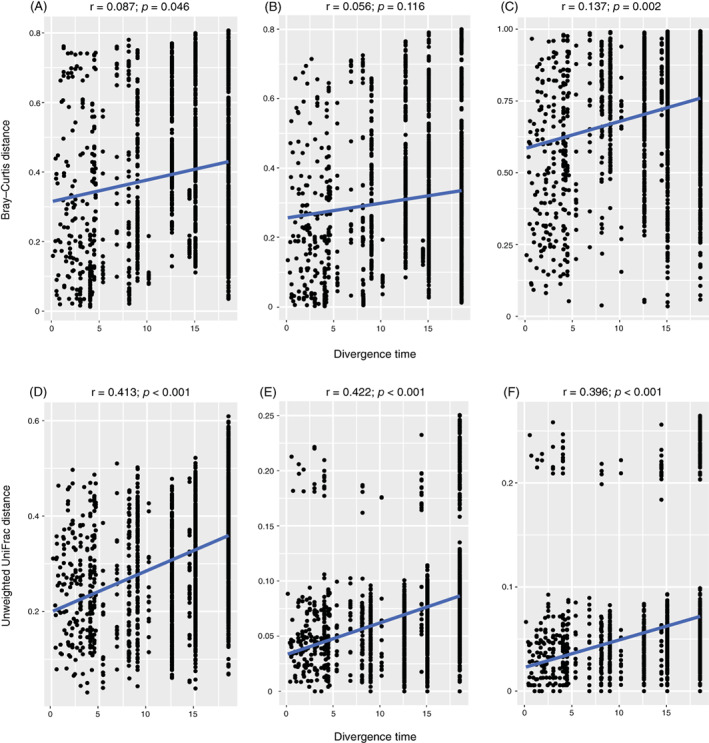
Correlations between microbiota dissimilarities and aphid divergence times estimated by Mantel tests in bacterial (A, D), symbiont (B, E) and secondary symbiont (C, F) communities. Microbiota dissimilarities were assessed by Bray–Curtis (A–C) and unweighted UniFrac distances (D–F).
Fig. 5.
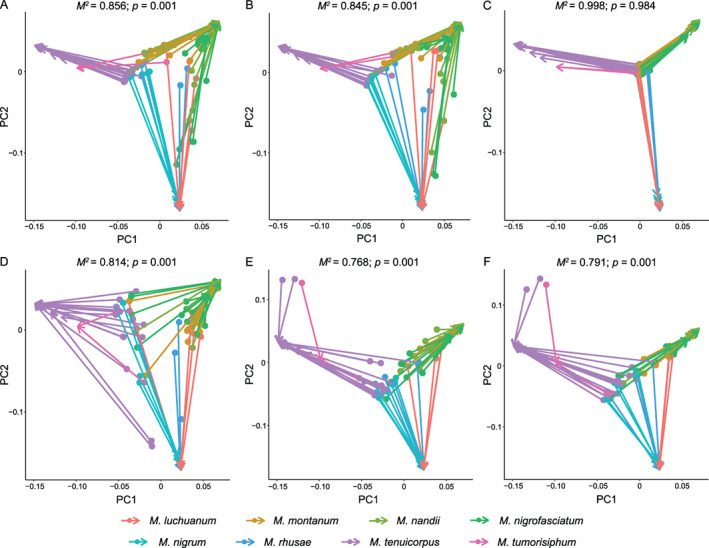
Procrustean superimpositions for PCA‐scaled aphid divergence times vs. variations in bacterial (A, D), symbiont (B, E) and secondary symbiont (C, F) communities. Bray–Curtis (A–C) and unweighted UniFrac distances (D–F) were used to estimate the microbiota variations. The Procrustes statistic, M 2, measures the degree of correspondence between two matrices after rotation.
Discussion
Symbiont composition of Mollitrichosiphum aphids
All of the top seven abundant genera associated with Mollitrichosiphum aphids were symbiotic bacteria (Table S1), which confirmed that the microbial communities of aphids are dominated by symbionts (Jousselin et al., 2016; Guyomar et al., 2018; Xu et al., 2020a,2021). High‐throughput 16S rRNA gene sequencing revealed a high symbiont diversity of Mollitrichosiphum. Buchnera and six secondary symbionts were detected, ranging from five to seven types of symbionts per sample. Buchnera was found in all samples with high relative abundance, which substantiated its obligate nutrient‐providing role in aphids (Douglas, 1998; Baumann, 2005; Wilson et al., 2010) and long‐term cospeciation history with host aphids (Munson et al., 1991; Liu et al., 2013, 2014; Xu et al., 2018).
Four types of secondary symbionts, including S. symbiotica, Rickettsia, Arsenophonus and Wolbachia, were detected in all samples. The resistance to heat shock conferred by S. symbiotica has been documented in a series of studies (Chen et al., 2000; Montllor et al., 2002; Russell and Moran, 2006), and Mollitrichosiphum aphids are mainly distributed in eastern and southern Asia, where the temperatures are relatively high (Blackman and Eastop, 2020). Considering the highest abundance and prevalence of S. symbiotica among the secondary symbiont flora of the examined samples, we infer that S. symbiotica may protect Mollitrichosiphum aphids from thermal stress. In addition, the high infection frequencies of Arsenophonus and Wolbachia in Mollitrichosiphum confirm their widespread distribution in aphids (Jousselin et al., 2013; Wang et al., 2014; Xu et al., 2020a). The majority of Mollitrichosiphum species also hosted F. symbiotica, which has been detected in A. pisum (Ferrari et al., 2012; Gauthier et al., 2015; Rock et al., 2018) and some Lachninae species (Manzano‐Marín et al., 2017; Meseguer et al., 2017). The frequent occurrence of F. symbiotica may be related to the mobile genetic elements in its genome, which encode toxins and pathogenicity factors that can facilitate heritable maintenance in hosts (Patel et al., 2019).
In previous studies, the defensive symbiont H. defensa has frequently been detected in field‐collected aphids (Ferrari et al., 2012; Brady et al., 2014; Henry et al., 2015; Zhao et al., 2016; Guo et al., 2019). However, the prevalence of H. defensa in Mollitrichosiphum aphids was not as high as other secondary symbionts, and its relative abundance and OTU diversity were quite low in this study. Mollitrichosiphum aphids move rapidly and their long siphunculi enable them to efficiently release alarm pheromones (Mondor et al., 2002) to escape from natural enemies. Henry et al. (2015) reported that the aphids with attendant ants that protected them from natural enemies tended not to harbour defensive symbionts. In general, ecological habits that confer defensive benefits may have resulted in the low infection frequency and abundance of H. defensa in Mollitrichosiphum.
Multiple infections of secondary symbionts
Previous studies found that multiple infections of many types of secondary symbionts within one aphid host were not frequent in natural populations (Sandström et al., 2001; Tsuchida et al., 2002; Haynes et al., 2003). Hughes et al. (2014) proposed that competitive interactions among microbes within the same host might give rise to the exclusion of less competitive microbes. The balance between physiological costs to hosts and mutualistic benefits (Oliver et al., 2006; Oliver et al., 2014; Leybourne et al., 2020) may also account for such coinfection patterns. Regardless, in this study, multiple infections were very common in Mollitrichosiphum aphids, and all samples examined harboured at least four secondary symbionts simultaneously.
Many studies have demonstrated that cohabitation of secondary symbionts may provide additional beneficial services for aphids. For example, pea aphids harbouring both S. symbiotica and Rickettsia were found to produce more winged morphs (Chen et al., 2000). Oliver et al. (2006) reported that pea aphids coinfected with S. symbiotica and H. defensa were more resistant to parasitism than were singly infected lines. Moreover, multiple infections provide opportunities for horizontal gene transfer among coharbouring symbionts, which may enhance microbial functions. Manzano‐Marín et al. (2020) confirmed that in some Cinara aphids, the symbiont Erwinia haradaeae acquired vitamin‐biosynthetic genes horizontally transferred from a Sodalis‐related bacterium and thereby gained a novel nutritional function. Finally, it is worth considering that some coinfecting secondary symbionts may contribute no benefit but only persist in aphid populations by hitchhiking alongside other beneficial symbionts (Smith et al., 2015; Doremus and Oliver, 2017). Further work should be performed to address the effects of such highly frequent multiple infections within Mollitrichosiphum aphids.
Host species‐specific and phylosymbiotic microbiota
The results of both alpha and beta diversity analyses highlighted that the aphid species had the strongest impact on the microbial communities associated with Mollitrichosiphum. Microbiota exhibited greater interspecific variation than intraspecific variation. At the OTU level, the secondary symbiont profiles were also different among aphid species (Fig. 2). Contributions of geography were found in several analyses but were generally weaker than those of aphid species. Geography has been found to be an important factor influencing the distribution of secondary symbionts in aphids (Tsuchida et al., 2002; Sepúlveda et al., 2017; Guo et al., 2019). Here, spatial variation may result from distinctive abiotic features such as temperatures and precipitation in different geographic regions (Sepúlveda et al., 2017).
Mantel tests and Procrustes analyses identified significant correlations between microbial community structure and host aphid phylogeny, which is referred to as ‘phylosymbiosis’ (Brucker and Bordenstein, 2013; Lim and Bordenstein, 2020). The microbiota were similar in closely related Mollitrichosiphum aphids and their dissimilarities increased along with the accumulation of host genetic variation. Such microbiota signature in Mollitrichosiphum is in line with previous study of McLean et al. (2019), and the pattern of phylosymbiosis has been reported in some insect, bird, fish and mammal groups (Sanders et al., 2014; Brooks et al., 2016; Groussin et al., 2017; Chiarello et al., 2018; Nishida and Ochman, 2018; Laviad‐Shitrit et al., 2019). Two alternative scenarios may account for this phylogenetic correlation: host‐microbe codiversification or ecological filtering by phylogenetically correlated factors (Sanders et al., 2014; Moran and Sloan, 2015; Lim and Bordenstein, 2020).
Buchnera is strictly maternally inherited, and parallel evolution between Buchnera and its corresponding Mollitrichosiphum hosts has been corroborated by Liu et al. (2013). Secondary symbionts primarily rely on maternal passage to persist in aphid generations. Chen and Purcell (1997) reported that S. symbiotica and Rickettsia could be transmitted from mother to offspring at a high rate under lab conditions. Theoretically, heritable secondary symbionts should also have codiversified with aphid hosts if they are strictly vertically transmitted. However, the fidelity of aphid‐secondary symbiont associations has been eroded over time due to occasional inheritance failures (Rock et al., 2018) and horizontal transmissions. This may explain why phylogenetic correlation was lacking within the microbiota of the ancient and typical heteroecious holocyclic aphid lineage Eriosomatinae (Xu et al., 2020b), in which repeated losses and horizontal gains of secondary symbionts might have occurred and consequently weakened or even erased phylosymbiosis signals during the long evolutionary period. In contrast, Mollitrichosiphum is a young clade (18.00–19.09 Mya, Fig. S5), and its monoecious life cycle may have greatly reduced interspecific horizontal transfer of secondary symbionts. In this study, S. symbiotica, Rickettsia, Arsenophonus and Wolbachia were observed in all examined Mollitrichosiphum samples. It has been found that some specific keystone or hub microbes may affect the colonization of other bacteria and determine the composition of the entire microbial community (Fisher and Mehta, 2014; Agler et al., 2016). Buchnera and these prevalent and abundant secondary symbionts may have served as keystones or hubs and are responsible for the phylosymbiosis of Mollitrichosiphum microbiota. Therefore, a shared diversification history between Mollitrichosiphum and its microbial associates at short time scales was uncovered. Similarly, mammalian gut microbiota display stronger phylosymbiosis signals in recently diverged host lineages (Groussin et al., 2017).
Nevertheless, another possible mechanism underlying the correlation between microbial community dissimilarities and Mollitrichosiphum phylogeny cannot be ruled out, namely, the filtering by environmental factors or host traits that have phylogenetic signals (Mazel et al., 2018). Closely related hosts generally possess similar physiologies or immune mechanisms, and they are more likely to select similar microbes from the environment. Chiarello et al. (2018) highlighted the role of diet in shaping the phylosymbiotic skin microbiome of coral reef fishes. In mammals and humans, diet is a strong selective filter for gut microbiota assemblage (Muegge et al., 2011; Wu et al., 2011). If such ecological filters themselves are phylogenetically non‐independent, the pattern of phylosymbiosis might be generated even in the absence of host‐microbe codiversification.
Conclusions
We provided the first systematic landscape of heritable symbionts associated with Mollitrichosiphum aphids in the present study, paving the way for further investigations of aphid‐bacterial symbiosis. The major role of aphid species in constraining microbiota was also confirmed. Finally, we detected a pattern of phylosymbiosis in Mollitrichosiphum, in which microbial community composition varied in accordance with host aphid relatedness. To elucidate how evolutionary and/or ecological driving forces have shaped phylosymbiotic Mollitrichosiphum‐microbe interactions, phylogenetic concordance between specific subsets of microbiota, especially keystone symbionts, and aphid hosts should be assessed, and candidate filtering factors should be identified and estimated quantitatively in the future.
Experimental procedures
Sample collection and identification
A total of 65 colonies of eight Mollitrichosiphum species were collected from seven families of plants and 19 geographic regions of southern China and Nepal. Detailed collection information is listed in Table S3. All samples were stored in 75% and 95% ethanol for slide mounting and molecular experiments, respectively, and frozen at −20°C. The aphids were identified by morphological examination and DNA barcoding. All samples and voucher specimens were deposited in the National Zoological Museum of China (NZMC), Institute of Zoology, Chinese Academy of Sciences, Beijing, China.
DNA extraction
A single adult viviparous female individual per colony was used for DNA extraction. The aphid was first surface sterilized with 70% ethanol for 5 min and five additional washes of sterile water. Total genomic DNA was extracted from the whole body of each individual using DNeasy Blood and Tissue Kit (QIAGEN, Hilden, Germany) following the manufacturer's protocol. Sterile ultrapure water was processed in the same way to serve as a negative control for DNA extraction. To identify aphid species and remove samples contaminated by parasitoid wasps, we quantified DNA extracts by PCR amplification of the cytochrome c oxidase subunit I (COI) gene with the primers LCO1490 (5′‐GGTCAACAAATCATAAAGATATTGG‐3′) and HCO2198 (5′‐TAAACTTCAGGGTGACCAAAAAATCA‐3′) (Folmer et al., 1994). The DNA samples were stored at −20°C.
High‐throughput 16S rRNA gene sequencing and sequence analyses
After extraction, the DNA was diluted to 1 ng μl−1 for use as a PCR template. Amplification of the V3 − V4 hypervariable region of the 16S rRNA gene was performed with the primers 341F (5′‐CCTAYGGGRBGCASCAG‐3′) and 806R (5′‐GGACTACNNGGGTATCTAAT‐3′) (Yu et al., 2005). A 30‐μl PCR mixture containing 15 μl Phusion High‐Fidelity PCR Master Mix (New England Biolabs, Ipswich, MA, USA), 3 μl primers and 10 μl PCR template was used. Triplicate reactions were performed under the following conditions: 98°C for 1 min; 30 cycles of 98°C for 10 s, 50°C for 30 s and 72°C for 30 s; and 72°C for 5 min. Negative controls for DNA extraction and amplification were included in PCR reactions. The PCR products were detected on a 2% agarose gel, and the positive samples with a bright band between 400–450 bp were chosen for purification with GeneJET Gel Extraction Kit (Thermo Scientific, Wilmington, DE, USA). The library was prepared using NEBNext Ultra DNA Library Prep Kit (New England Biolabs). Library quality was examined using a Qubit 2.0 Fluorometer (Thermo Scientific) and an Agilent Bioanalyzer 2100 system. Finally, the library pool was sequenced using the Illumina HiSeq 2500 PE250 platform (Illumina, San Diego, CA, USA).
Paired‐end reads were merged using FLASH v1.2.7 (Magoč and Salzberg, 2011) with a minimum overlap size of 10 bp and an error rate of 10%, and demultiplexed on basis of the unique barcodes. Merged sequences with quality score below 20 and length shorter than 300 bp were filtered by QIIME v1.9.1 (Caporaso et al., 2010). After removing chimeras with UCHIME v4.2.40 (Edgar et al., 2011), the remaining sequences were clustered into OTUs with a minimum identity of 97% using the UCLUST module (Edgar, 2010) in QIIME. The most abundant sequence in each OTU cluster was selected as the representative sequence. Classification of each OTU was performed using the RDP classifier (Wang et al., 2007) with a 0.80 confidence threshold based on the SILVA 128 reference database (Quast et al., 2013). Taxonomic assignments were then manually checked by BLAST against GenBank. For each OTU, the average number of sequences across three PCR replicates per sample was obtained for further analyses. Each sample was rarefied to the same sequencing depth in USEARCH v10.0 using the ‘otutab_norm’ function (Edgar, 2010). OTUs of which the sequences were less than 0.005% of the total sequences were discarded for quality filtering (Bokulich et al., 2013). Finally, an OTU table containing taxonomic definitions of bacterial taxa and sequence number per sample was generated (Table S4a).
Microbial community analyses
To better investigate the microbial diversity within Mollitrichosiphum aphids, two reduced OTU tables containing OTUs classified as known symbionts (incl. Buchnera and secondary symbionts) (Table S4b) and secondary symbionts of aphids (Table S4c) were produced. The relative abundance of each OTU was assessed by dividing the number of sequences assigned to each OTU by the sum of sequences in a given sample using the ‘decostand’ function and ‘total’ method of the package ‘vegan’ (Oksanen et al., 2010) in the R v3.5.1 programming environment (R Core Team, 2018). To visualize the relative abundance of secondary symbiont OTUs across aphid species, a heatmap was created using the ‘pheatmap’ function of the R package ‘pheatmap’ (Kolde and Kolde, 2015). The maximum‐likelihood tree showing the relatedness of these OTUs was generated in RAxML v8.2.7 (Stamatakis, 2014), and a simplified cladogram from the aphid divergence time estimation was presented to show the phylogeny of Mollitrichosiphum aphids (detailed dating methods are provided in the Supporting Information).
All statistical analyses were performed with bacterial, symbiont and secondary symbiont data. All samples were grouped by aphid species, geographic distribution and host plant. The detail grouping information is shown in Table S5. Downstream statistical analyses of microbial community variation (i.e., all the following analyses except Mantel test and Procrustes analysis) were performed on all groups and groups with a sample size ≥3. Samples with ambiguous host plant information were excluded from analyses.
Alpha diversity (Shannon and Simpson indices) measuring the community diversity within each aphid sample was calculated based on the OTU tables using the ‘diversity’ function in ‘vegan’. We investigated the variation in alpha diversity with respect to aphid species, geography and host plant. Nonparametric Kruskal–Wallis tests were performed because of the non‐normal distribution (Shapiro–Wilk test, p < 0.05) and variance heterogeneity of the alpha diversity data (Bartlett test, p < 0.05). We then used three‐way analysis of variance (ANOVA) to simultaneously evaluate the effect of each factor on the microbial alpha diversity. This method is useful in summarizing separate contributions of categorical variables by estimating the statistical significance of each variable (Vaughan and Corballis, 1969). Three‐way ANOVA was conducted using the ‘avop’ function in the R package ‘lmPerm’ (Wheeler and Torchiano, 2010).
Microbial community variation between aphid samples was also assessed. We used Bray–Curtis and unweighted UniFrac distances to quantify beta diversity. The Bray–Curtis distance considers the presence/absence and relative abundance of OTUs and the unweighted UniFrac distance uses phylogenetic information of OTUs to calculate community dissimilarity (Lozupone et al., 2011). The latter is more powerful because it provides insight into the complexity of phylogenetic compositions of microbial communities (Martin, 2002). The Bray–Curtis distance was assessed with the ‘vegdist’ function of ‘vegan’ and the unweighted UniFrac distance was calculated using the ‘GUniFrac’ function in ‘GUniFrac’ (Chen and Chen, 2018).
Based on both the Bray–Curtis and unweighted UniFrac distance matrices, we used ordination methods and statistical tests to assess the microbial community variation with respect to different factors. First, dissimilarity among samples was visualized using unconstrained nonmetric multidimensional scaling (NMDS) (‘metaMDS’ function in ‘vegan’; stress values <0.05 were regarded as indicative of excellent representations) and constrained principal coordinate analysis (cPCoA) (‘capscale’ and ‘anova.cca’ functions in ‘vegan’). NMDS is a robust unsupervised means to extract interpretable patterns from community dissimilarity data (Minchin, 1987), and the constrained ordination technique cPCoA can display community structures that may be masked in an unconstrained method (Anderson and Willis, 2003). CPCoA was performed based only on Bray–Curtis distances, as unweighted UniFrac distances are not suitable for this analysis.
Next, analysis of similarities (ANOSIM) and permutational multivariate analysis of variance (PERMANOVA) were applied based on the Bray–Curtis and unweighted UniFrac distance matrices to estimate statistically significant differences between groups. ANOSIM and PERMANOVA were performed using the ‘anosim’ and ‘adonis’ functions, respectively, with 10,000 permutations in ‘vegan’. The R value of ANOSIM is scaled to lie between −1 and + 1, and the values between 0 and + 1 indicate greater dissimilarity among samples between groups than occurs within groups (Anderson and Walsh, 2013). For PERMANOVA, a factor with a larger R 2 value is regarded as a more important component contributing to the overall variation (Anderson, 2017).
Finally, to explore the impact of aphid phylogeny on microbiota dissimilarity, the Mantel test and Procrustes analysis were conducted on all samples using matrices of aphid divergence times and beta diversity (Bray–Curtis and unweighted UniFrac distances). Aphid divergence times were estimated with BEAST v2.5.2 (Bouckaert et al., 2019) (detailed analysis methods are provided in the Supporting Information). The Mantel test is frequently employed to evaluate the statistical significance of the correlation between two dissimilarity matrices (Anderson and Walsh, 2013), and analyses were performed using Spearman's rank correlation method and the ‘mantel’ function in ‘vegan’ with 10,000 permutations. Procrustes analysis, which is more powerful for testing the concordance between matrices (Peres‐Neto and Jackson, 2001), was carried out with the ‘procrustes’ and ‘protest’ functions in ‘vegan’. We used the aphid divergence time matrix as the target matrix and the beta diversity matrix as the rotated matrix. These two matrices were first scaled using principal component analysis (PCA) and then rotated to find the optimal superimposition that maximized their fit. The fit of superimposition is represented as the M 2 value. The significance of Procrustes statistics was calculated using a Procrustean randomization test in which 999 permutations were performed (Jackson, 1995).
Conflict of Interest
The authors declare no conflict of interest.
Supporting information
Fig. S1 Structural segregation using constrained principal coordinate analyses (cPCoA) of Bray–Curtis distances of bacterial (A, D, G), symbiont (B, E, H) and secondary symbiont (C, F, I) communities (n ≥ 1). Plots are structured by aphid species (A–C), geographic region (D–F) and host plant (G–I). The overall variation explained by the constrained factor is displayed at the top of each plot. The percent variation shown on each axis refers to the fraction of the total variance explained by the projection. The abbreviations are given in Table S5.
Fig. S2. Nonmetric multidimensional scaling (NMDS) plots based on Bray–Curtis distances of bacterial (A, D, G), symbiont (B, E, H) and secondary symbiont (C, F, I) communities (n ≥ 1). Samples are coloured by aphid species (A–C), geographic region (D–F) and host plant (G–I). The stress value indicates the goodness of fit between the NMDS representation and the data. The abbreviations are given in Table S5.
Fig. S3. Nonmetric multidimensional scaling (NMDS) plots based on Bray–Curtis distances of bacterial (A, D, G), symbiont (B, E, H) and secondary symbiont (C, F, I) communities (n ≥ 3). Samples are coloured by aphid species (A–C), geographic region (D–F) and host plant (G–I). The stress value indicates the goodness of fit between the NMDS representation and the data. The abbreviations are given in Table S5.
Fig. S4. Nonmetric multidimensional scaling (NMDS) plots based on unweighted UniFrac distances of bacterial communities (n ≥ 1, A, C, E; n ≥ 3, B, D, F). The distance data of symbiont and secondary symbiont communities were insufficient for NMDS. Samples are coloured by aphid species (A, B), geographic region (C, D) and host plant (E, F). The stress value indicates the goodness of fit between the NMDS representation and the data. The abbreviations are given in Table S5.
Fig. S5. Time‐calibrated phylogenetic tree of Mollitrichosiphum. The red circle at the node shows the calibration point. Horizontal bars display the 95% highest posterior density intervals of the estimated node ages. The mean ages of nodes are presented above the bars.
Table S1. Relative abundance of the top 10 bacterial phyla, classes, orders, families and genera in Mollitrichosiphum.
Table S2. Results of three‐way ANOVA based on alpha diversity indices in bacterial, symbiont and secondary symbiont communities.
Table S3. Voucher information and GenBank accession numbers of aphid samples used in this study.
Table S5. Grouping information of Mollitrichosiphum aphid samples used in this study.
Table S4 The OTU tables of bacterial (a), symbiont (b) and secondary symbiont communities (c).
Acknowledgements
We would like to thank all sample collectors for their collecting in the field, Xue Yang for her help in molecular experiments and Fendi Yang for making the slide‐mounted specimens. This study was supported by the National Natural Science Foundation of China (Grant Nos. 31772492 and 31620103916), the Youth Innovation Promotion Association of the Chinese Academy of Sciences (Grant No. 2020087), and the Second Tibetan Plateau Scientific Expedition and Research (STEP) program (Grant No. 2019QZKK05010601).
Data Availability Statement
COI, Cytb and EF‐1α sequences obtained in this study were deposited in GenBank under accession numbers MT556450–MT556472 and MT563127–MT563158. Raw 16S rRNA gene amplicon reads were deposited in the NCBI Sequence Read Archive under BioProject accession number PRJNA637573.
References
- Agler, M.T. , Ruhe, J. , Kroll, S. , Morhenn, C. , Kim, S. , Weigel, D. , and Kemen, E. (2016) Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLoS Biol 14: e1002352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, M.J. (2017) Permutational multivariate analysis of variance (PERMANOVA). In Wiley StatsRef: Statistics Reference Online, Balakrishnan, N. , Colton, T. , Everitt, B. , Piegorsch, W. , Ruggeri, F. , and Teugels, J.L. (eds). Chichester: John Wiley and Sons, pp. 1–15. [Google Scholar]
- Anderson, M.J. , and Walsh, D.C. (2013) PERMANOVA, ANOSIM, and the mantel test in the face of heterogeneous dispersions: what null hypothesis are you testing? Ecol Monogr 83: 557–574. [Google Scholar]
- Anderson, M.J. , and Willis, T.J. (2003) Canonical analysis of principal coordinates: a useful method of constrained ordination for ecology. Ecology 84: 511–525. [Google Scholar]
- Augustinos, A.A. , Santos‐Garcia, D. , Dionyssopoulou, E. , Moreira, M. , Papapanagiotou, A. , Scarvelakis, M. , et al. (2011) Detection and characterization of Wolbachia infections in natural populations of aphids: is the hidden diversity fully unraveled? PLoS One 6: e28695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann, P. , Baumann, L. , Lai, C.Y. , Rouhbakhsh, D. , Moran, N.A. , and Clark, M.A. (1995) Genetics, physiology, and evolutionary relationships of the genus Buchnera: intracellular symbionts of aphids. Annu Rev Microbiol 49: 55–94. [DOI] [PubMed] [Google Scholar]
- Baumann, P. (2005) Biology bacteriocyte‐associated endosymbionts of plant sap‐sucking insects. Annu Rev Microbiol 59: 155–189. [DOI] [PubMed] [Google Scholar]
- Bansal, R. , Mian, M.R. , and Michel, A.P. (2014) Microbiome diversity of Aphis glycines with extensive superinfection in native and invasive populations. Environ Microbiol Rep 6: 57–69. [DOI] [PubMed] [Google Scholar]
- Bennett, G.M. , and Moran, N.A. (2015) Heritable symbiosis: the advantages and perils of an evolutionary rabbit hole. Proc Natl Acad Sci U S A 112: 10169–10176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackman, R.L. , and Eastop, V.F. (2020) Aphids on the world's plants: An online identification and information guide. http://www.aphidsonworldsplants.info. Accessed date 1 March 2020.
- Bokulich, N.A. , Subramanian, S. , Faith, J.J. , Gevers, D. , Gordon, J.I. , Knight, R. , et al. (2013) Quality‐filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods 10: 57–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert, R. , Vaughan, T.G. , Barido‐Sottani, J. , Duchêne, S. , Fourment, M. , Gavryushkina, A. , et al. (2019) BEAST 2.5: an advanced software platform for Bayesian evolutionary analysis. PLoS Comput Biol 15: e1006650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady, C.M. , Asplen, M.K. , Desneux, N. , Heimpel, G.E. , Hopper, K.R. , Linnen, C.R. , et al. (2014) Worldwide populations of the aphid Aphis craccivora are infected with diverse facultative bacterial symbionts. Microb Ecol 67: 195–204. [DOI] [PubMed] [Google Scholar]
- Brady, C.M. , and White, J.A. (2013) Cowpea aphid (Aphis craccivora) associated with different host plants has different facultative endosymbionts. Ecol Entomol 38: 433–437. [Google Scholar]
- Brucker, R.M. , and Bordenstein, S.R. (2013) The hologenomic basis of speciation: gut bacteria cause hybrid lethality in the genus Nasonia . Science 341: 667–669. [DOI] [PubMed] [Google Scholar]
- Brooks, A.W. , Kohl, K.D. , Brucker, R.M. , van Opstal, E.J. , and Bordenstein, S.R. (2016) Phylosymbiosis: relationships and functional effects of microbial communities across host evolutionary history. PLoS Biol 14: e2000225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchner, P. (1965) Endosymbiosis of Animals with Plant Microorganisms. New York: Interscience Publishers. [Google Scholar]
- Caporaso, J.G. , Kuczynski, J. , Stombaugh, J. , Bittinger, K. , Bushman, F.D. , Costello, E.K. , et al. (2010) QIIME allows analysis of high‐throughput community sequencing data. Nat Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, D.Q. , Campbell, B.C. , and Purcell, A.H. (1996) A new Rickettsia from a herbivorous insect, the pea aphid Acyrthosiphon pisum (Harris). Curr Microbiol 33: 123–128. [DOI] [PubMed] [Google Scholar]
- Chen, D.Q. , Montllor, C.B. , and Purcell, A.H. (2000) Fitness effects of two facultative endosymbiotic bacteria on the pea aphid, Acyrthosiphon pisum, and the blue alfalfa aphid, A. kondoi . Entomol Exp Appl 95: 315–323. [Google Scholar]
- Chen, D.Q. , and Purcell, A.H. (1997) Occurrence and transmission of facultative endosymbionts in aphids. Curr Microbiol 34: 220–225. [DOI] [PubMed] [Google Scholar]
- Chen, J. , and Chen, M.J. (2018) Package ‘GUniFrac’. R Package Version 1.1. https://cran.r-project.org/web/packages/GUniFrac.
- Chiarello, M. , Auguet, J. , Bettarel, Y. , Bouvier, C. , Claverie, T. , Graham, N.A. , et al. (2018) Skin microbiome of coral reef fish is highly variable and driven by host phylogeny and diet. Microbiome 6: 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, M.A. , Moran, N.A. , Baumann, P. , and Wernegreen, J.J. (2000) Cospeciation between bacterial endosymbionts (Buchnera) and a recent radiation of aphids (Uroleucon) and pitfalls of testing for phylogenetic congruence. Evolution 54: 517–525. [DOI] [PubMed] [Google Scholar]
- Darby, A.C. , Birkle, L.M. , Turner, S.L. , and Douglas, A.E. (2001) An aphid‐borne bacterium allied to the secondary symbionts of whitefly. FEMS Microbiol Ecol 36: 43–50. [DOI] [PubMed] [Google Scholar]
- Douglas, A.E. (1998) Nutritional interactions in insect‐microbial symbioses: aphids and their symbiotic bacteria Buchnera . Annu Rev Entomol 43: 17–37. [DOI] [PubMed] [Google Scholar]
- Doremus, M.R. , and Oliver, K.M. (2017) Aphid heritable symbiont exploits defensive mutualism. Appl Environ Microbiol 83: e03276–e03216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, R.C. (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461. [DOI] [PubMed] [Google Scholar]
- Edgar, R.C. , Haas, B.J. , Clemente, J.C. , Quince, C. , and Knight, R. (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27: 2194–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favret, C . (2020) Aphid Species File. http://Aphid.SpeciesFile.org. Accessed date 1 March 2020.
- Ferrari, J. , West, J.A. , Via, S. , and Godfray, H.C.J. (2012) Population genetic structure and secondary symbionts in host‐associated populations of the pea aphid complex. Evolution 66: 375–390. [DOI] [PubMed] [Google Scholar]
- Fisher, C.K. , and Mehta, P. (2014) Identifying keystone species in the human gut microbiome from metagenomic timeseries using sparse linear regression. PLoS One 9: e102451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukatsu, T. , Tsuchida, T. , Nikoh, N. , and Koga, R. (2001) Spiroplasma symbiont of the pea aphid, Acyrthosiphon pisum (Insecta: Homoptera). Appl Environ Microbiol 67: 1284–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folmer, O. , Black, M. , Hoeh, W. , Lutz, R. , and Vrijenhoek, R. (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol 3: 294–299. [PubMed] [Google Scholar]
- Gauthier, J.P. , Outreman, Y. , Mieuzet, L. , and Simon, J.C. (2015) Bacterial communities associated with host‐adapted populations of pea aphids revealed by deep sequencing of 16S ribosomal DNA. PLoS One 10: e0120664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh, A.K. , and Agarwala, B.K. (1993) Homoptera, Aphidoidea, Part 6. Subfamily: Greenideinae. The Fauna of India and the Adjacent Countries. Calcutta: Zoological Survey of India. [Google Scholar]
- Guay, J.F. , Boudreault, S. , Michaud, D. , and Cloutier, C. (2009) Impact of environmental stress on aphid clonal resistance to parasitoids: role of Hamiltonella defensa bacterial symbiosis in association with a new facultative symbiont of the pea aphid. J Insect Physiol 55: 919–926. [DOI] [PubMed] [Google Scholar]
- Guo, J. , Liu, X. , Poncelet, N. , He, K. , Francis, F. , and Wang, Z. (2019) Detection and geographic distribution of seven facultative endosymbionts in two Rhopalosiphum aphid species. MicrobiologyOpen 8: e00817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyomar, C. , Legeai, F. , Jousselin, E. , Mougel, C. , Lemaitre, C. , and Simon, J. (2018) Multi‐scale characterization of symbiont diversity in the pea aphid complex through metagenomic approaches. Microbiome 6: 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groussin, M. , Mazel, F. , Sanders, J.G. , Smillie, C.S. , Lavergne, S. , Thuiller, W. , and Alm, E.J. (2017) Unraveling the processes shaping mammalian gut microbiomes over evolutionary time. Nat Commun 8: 14319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes, S. , Darby, A.C. , Daniell, T.J. , Webster, G. , Van Veen, F.J.F. , Godfray, H.C.J. , et al. (2003) Diversity of bacteria associated with natural aphid populations. Appl Environ Microbiol 69: 7216–7223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry, L.M. , Maiden, M.C. , Ferrari, J. , and Godfray, H.C.J. (2015) Insect life history and the evolution of bacterial mutualism. Ecol Lett 18: 516–525. [DOI] [PubMed] [Google Scholar]
- Heyworth, E.R. , and Ferrari, J. (2015) A facultative endosymbiont in aphids can provide diverse ecological benefits. J Evol Biol 28: 1753–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, G.L. , Dodson, B.L. , Johnson, R.M. , Murdock, C.C. , Tsujimoto, H. , Suzuki, Y. , et al. (2014) Native microbiome impedes vertical transmission of Wolbachia in Anopheles mosquitoes. Proc Natl Acad Sci U S A 111: 12498–12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, D.A. (1995) PROTEST: a procrustean randomization test of community environment concordance. Écoscience 2: 297–303. [Google Scholar]
- Jousselin, E. , Coeur d'acier, A. , Vanlerberghe‐Masutti, F. , and Duron, O. (2013) Evolution and diversity of Arsenophonus endosymbionts in aphids. Mol Ecol 22: 260–270. [DOI] [PubMed] [Google Scholar]
- Jousselin, E. , Clamens, A. , Galan, M. , Bernard, M. , Maman, S. , Gschloessl, B. , et al. (2016) Assessment of a 16S rRNA amplicon Illumina sequencing procedure for studying the microbiome of a symbiont‐rich aphid genus. Mol Ecol Resour 16: 628–640. [DOI] [PubMed] [Google Scholar]
- Koga, R. , Meng, X. , Tsuchida, T. , and Fukatsu, T. (2012) Cellular mechanism for selective vertical transmission of an obligate insect symbiont at the bacteriocyte‐embryo interface. Proc Natl Acad Sci U S A 109: E1230–E1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolde, R. , and Kolde, M.R. (2015) Package “pheatmap” . https://cran.r-project.org/web/packages/pheatmap.
- Lamelas, A. , Gosalbes, M.J. , Manzano‐Marín, A. , Peretó, J. , Moya, A. , and Latorre, A. (2011) Serratia symbiotica from the aphid Cinara cedri: a missing link from facultative to obligate insect endosymbiont. PLoS Genet 7: e1002357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laviad‐Shitrit, S. , Izhaki, I. , Lalzar, M. , and Halpern, M. (2019) Comparative analysis of intestine microbiota of four wild waterbird species. Front Microbiol 10: 1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclair, M. , Polin, S. , Jousseaume, T. , Simon, J.C. , Sugio, A. , Morlière, S. , et al. (2017) Consequences of coinfection with protective symbionts on the host phenotype and symbiont titres in the pea aphid system. Insect Sci 24: 798–808. [DOI] [PubMed] [Google Scholar]
- Leybourne, D.J. , Bos, J.I. , Valentine, T.A. , and Karley, A.J. (2020) The price of protection: a defensive endosymbiont impairs nymph growth in the bird cherry‐oat aphid, Rhopalosiphum padi . Insect Sci 27: 69–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, S.J. , and Bordenstein, S.R. (2020) An introduction to phylosymbiosis. Proc R Soc B Biol Sci 287: 20192900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, L. , Huang, X. , Zhang, R. , Jiang, L. , and Qiao, G. (2013) Phylogenetic congruence between Mollitrichosiphum (Aphididae: Greenideinae) and Buchnera indicates insect‐bacteria parallel evolution. Syst Entomol 38: 81–92. [Google Scholar]
- Liu, L. , Li, X. , Huang, X. , and Qiao, G. (2014) Evolutionary relationships of Pemphigus and allied genera (Hemiptera: Aphididae: Eriosomatinae) and their primary endosymbiont, Buchnera aphidicola . Insect Sci 21: 301–312. [DOI] [PubMed] [Google Scholar]
- Liu, X. , Lei, H. , and Chen, F. (2019) Infection pattern and negative effects of a facultative endosymbiont on its insect host are environment‐dependent. Sci Rep 9: 4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone, C. , Lladser, M.E. , Knights, D. , Stombaugh, J. , and Knight, R. (2011) UniFrac: an effective distance metric for microbial community comparison. ISME J 5: 169–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Łukasik, P. , Van Asch, M. , Guo, H. , Ferrari, J. , and Godfray, H.C. (2013) Unrelated facultative endosymbionts protect aphids against a fungal pathogen. Ecol Lett 16: 214–218. [DOI] [PubMed] [Google Scholar]
- Magoč, T. , and Salzberg, S.L. (2011) FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27: 2957–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzano‐Marín, A. , Coeur d'acier, A. , Clamens, A.L. , Orvain, C. , Cruaud, C. , Barbe, V. , and Jousselin, E. (2020) Serial horizontal transfer of vitamin‐biosynthetic genes enables the establishment of new nutritional symbionts in aphids' di‐symbiotic systems. ISME J 14: 259–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzano‐Marín, A. , Szabó, G. , Simon, J.C. , Horn, M. , and Latorre, A. (2017) Happens in the best of subfamilies: establishment and repeated replacements of co‐obligate secondary endosymbionts within Lachninae aphids. Environ Microbiol 19: 393–408. [DOI] [PubMed] [Google Scholar]
- Martin, A.P. (2002) Phylogenetic approaches for describing and comparing the diversity of microbial communities. Appl Environ Microbiol 68: 3673–3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazel, F. , Davis, K.M. , Loudon, A. , Kwong, W.K. , Groussin, M. , and Parfrey, L.W. (2018) Is host filtering the main driver of phylosymbiosis across the tree of life? mSystems 3: e00097–e00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mccutcheon, J.P. , and Moran, N.A. (2012) Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol 10: 13–26. [DOI] [PubMed] [Google Scholar]
- McLean, A.H.C. , Godfray, H.C.J. , Ellers, J. , and Henry, L.M. (2019) Host relatedness influences the composition of aphid microbiomes. Environ Microbiol Rep 11: 808–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meseguer, A.S. , Manzano‐Marín, A. , Coeur d'Acier, A. , Clamens, A.L. , Godefroid, M. , and Jousselin, E. (2017) Buchnera has changed flatmate but the repeated replacement of co‐obligate symbionts is not associated with the ecological expansions of their aphid hosts. Mol Ecol 26: 2363–2378. [DOI] [PubMed] [Google Scholar]
- Michalik, A. , Szklarzewicz, T. , Jankowska, W. , and Wieczorek, K. (2014) Endosymbiotic microorganisms of aphids (Hemiptera: Sternorrhyncha: Aphidoidea): ultrastructure, distribution and transovarial transmission. Eur J Entomol 111: 91–104. [Google Scholar]
- Mondor, E.B. , Roitberg, B.D. , and Stadler, B. (2002) Cornicle length in macrosiphini aphids: a comparison of ecological traits. Ecol Entomol 27: 758–762. [Google Scholar]
- Montllor, C.B. , Maxmen, A. , and Purcell, A.H. (2002) Facultative bacterial endosymbionts benefit pea aphids Acyrthosiphon pisum under heat stress. Ecol Entomol 27: 189–195. [Google Scholar]
- Moran, N.A. , and Sloan, D.B. (2015) The hologenome concept: helpful or hollow? PLoS Biol 13: e1002311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muegge, B.D. , Kuczynski, J. , Knights, D. , Clemente, J.C. , Gonzalez, A. , Fontana, L. , et al. (2011) Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 332: 970–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munson, M.A. , Baumann, P. , Clark, M.A. , Baumann, L. , Moran, N.A. , Voegtlin, D.J. , and Campbell, B.C. (1991) Evidence for the establishment of aphid‐eubacterium endosymbiosis in an ancestor of four aphid families. J Bacteriol 173: 6321–6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minchin, P.R. (1987) An evaluation of relative robustness of techniques for ecological ordinations. Vegetatio 69: 89–107. [Google Scholar]
- Nishida, A.H. , and Ochman, H. (2018) Rates of gut microbiome divergence in mammals. Mol Ecol 27: 1884–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen, J. , Blanchet, F.G. , Kindt, R. , Legendre, P. , O'hara, R.B. , Simpson, G.L. , et al. (2010) Vegan: community ecology package. R package version 2.5‐2. https://cran.r-project.org/web/packages/vegan.
- Oliver, K.M. , Degnan, P.H. , Burke, G.R. , and Moran, N.A. (2010) Facultative symbionts in aphids and the horizontal transfer of ecologically important traits. Annu Rev Entomol 55: 247–266. [DOI] [PubMed] [Google Scholar]
- Oliver, K.M. , Moran, N.A. , and Hunter, M.S. (2005) Variation in resistance to parasitism in aphids is due to symbionts not host genotype. Proc Natl Acad Sci U S A 102: 12795–12800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver, K.M. , Moran, N.A. , and Hunter, M.S. (2006) Costs and benefits of a superinfection of facultative symbionts in aphids. Proc R Soc B‐Biol Sci 273: 1273–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver, K.M. , Smith, A.H. , and Russell, J.A. (2014) Defensive symbiosis in the real world – advancing ecological studies of heritable, protective bacteria in aphids and beyond. Funct Ecol 28: 341–355. [Google Scholar]
- Peres‐Neto, P.R. , and Jackson, D.A. (2001) How well do multivariate data sets match? The advantages of a procrustean superimposition approach over the mantel test. Oecologia 129: 169–178. [DOI] [PubMed] [Google Scholar]
- Pons, I. , Renoz, F. , Noël, C. , and Hance, T. (2019) Circulation of the cultivable symbiont serratia symbiotica in aphids is mediated by plants. Front Microbiol 10: 764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast, C. , Pruesse, E. , Yilmaz, P. , Gerken, J. , Schweer, T. , Yarza, P. , et al. (2013) The SILVA ribosomal RNA gene database project: improved data processing and web‐based tools. Nucleic Acids Res 41: D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . (2018) R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. https://cran.r-project.org/. [Google Scholar]
- Rispe, C. , and Moran, N.A. (2000) Accumulation of deleterious mutations in endosymbionts: Muller's ratchet with two levels of selection. Am Nat 156: 425–441. [DOI] [PubMed] [Google Scholar]
- Rock, D.I. , Smith, A.H. , Joffe, J. , Albertus, A. , Wong, N. , O'Connor, M. , et al. (2018) Context‐dependent vertical transmission shapes strong endosymbiont community structure in the pea aphid, Acyrthosiphon pisum . Mol Ecol 27: 2039–2056. [DOI] [PubMed] [Google Scholar]
- Russell, J.A. , Latorre, A. , Sabater‐Muñoz, B. , Moya, A. , and Moran, N.A. (2003) Side‐stepping secondary symbionts: widespread horizontal transfer across and beyond the Aphidoidea. Mol Ecol 12: 1061–1075. [DOI] [PubMed] [Google Scholar]
- Russell, J.A. , and Moran, N.A. (2005) Horizontal transfer of bacterial symbionts: heritability and fitness effects in a novel aphid host. Appl Environ Microbiol 71: 7987–7994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, J.A. , and Moran, N.A. (2006) Costs and benefits of symbiont infection in aphids: variation among symbionts and across temperatures. Proc R Soc B‐Biol Sci 273: 603–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders, J.G. , Powell, S. , Kronauer, D.J. , Vasconcelos, H.L. , Frederickson, M.E. , and Pierce, N.E. (2014) Stability and phylogenetic correlation in gut microbiota: lessons from ants and apes. Mol Ecol 23: 1268–1283. [DOI] [PubMed] [Google Scholar]
- Sandström, J.P. , Russell, J.A. , White, J.P. , and Moran, N.A. (2001) Independent origins and horizontal transfer of bacterial symbionts of aphids. Mol Ecol 10: 217–228. [DOI] [PubMed] [Google Scholar]
- Scarborough, C.L. , Ferrari, J. , and Godfray, H.C. (2005) Aphid protected from pathogen by endosymbiont. Science 310: 1781. [DOI] [PubMed] [Google Scholar]
- Sepúlveda, D.A. , Zepeda‐Paulo, F. , Ramírez, C.C. , Lavandero, B. , and Figueroa, C.C. (2017) Diversity, frequency, and geographic distribution of facultative bacterial endosymbionts in introduced aphid pests. Insect Sci 24: 511–521. [DOI] [PubMed] [Google Scholar]
- Simon, J.C. , Carre, S. , Boutin, M. , Prunier‐Leterme, N. , Sabater‐Muñoz, B. , Latorre, A. , and Bournoville, R. (2003) Host‐based divergence in populations of the pea aphid: insights from nuclear markers and the prevalence of facultative symbionts. Proc R Soc B Biol Sci 270: 1703–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, A.H. , Łukasik, P. , O'Connor, M.P. , Lee, A. , Mayo, G. , Drott, M.T. , et al. (2015) Patterns, causes and consequences of defensive microbiome dynamics across multiple scales. Mol Ecol 24: 1135–1149. [DOI] [PubMed] [Google Scholar]
- Stamatakis, A. (2014) RAxML version 8: a tool for phylogenetic analysis and post‐analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakaran, S. , Kost, C. , and Kaltenpoth, M. (2017) Symbiont acquisition and replacement as a source of ecological innovation. Trends Microbiol 25: 375–390. [DOI] [PubMed] [Google Scholar]
- Tsuchida, T. , Koga, R. , and Fukatsu, T. (2004) Host plant specialization governed by facultative symbiont. Science 303: 1989. [DOI] [PubMed] [Google Scholar]
- Tsuchida, T. , Koga, R. , Horikawa, M. , Tsunoda, T. , Maoka, T. , Matsumoto, S. , et al. (2010) Symbiotic bacterium modifies aphid body color. Science 330: 1102–1104. [DOI] [PubMed] [Google Scholar]
- Tsuchida, T. , Koga, R. , Matsumoto, S. , and Fukatsu, T. (2011) Interspecific symbiont transfection confers a novel ecological trait to the recipient insect. Biol Lett 7: 245–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida, T. , Koga, R. , Shibao, H. , Matsumoto, T. , and Fukatsu, T. (2002) Diversity and geographic distribution of secondary endosymbiotic bacteria in natural populations of the pea aphid, Acyrthosiphon pisum . Mol Ecol 11: 2123–2135. [DOI] [PubMed] [Google Scholar]
- Patel, V. , Chevignon, G. , Manzano‐Marin, A. , Brandt, J.W. , Strand, M.R. , Russell, J.A. , and Oliver, K.M. (2019) Cultivation‐assisted genome of Candidatus Fukatsuia symbiotica; the enigmatic “X‐type” symbiont of aphids. Genome Biol Evol 11: 3510–3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterman, B.M. , Baumann, P. , and McLean, D.L. (1989) Pea aphid symbiont relationships established by analysis of 16S rRNAs. J Bacteriol 171: 2970–2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan, G.M. , and Corballis, M.C. (1969) Beyond tests of significance: estimating strength of effects in selected ANOVA designs. Psychol Bull 72: 204–213. [Google Scholar]
- Wagner, S.M. , Martinez, A.J. , Ruan, Y. , Kim, K.L. , Lenhart, P.A. , Dehnel, A.C. , et al. (2015) Facultative endosymbionts mediate dietary breadth in a polyphagous herbivore. Funct Ecol 29: 1402–1410. [Google Scholar]
- Wang, Q. , Garrity, G.M. , Tiedje, J.M. , and Cole, J.R. (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73: 5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z. , Su, X. , Wen, J. , Jiang, L. , and Qiao, G. (2014) Widespread infection and diverse infection patterns of Wolbachia in Chinese aphids. Insect Sci 21: 313–325. [DOI] [PubMed] [Google Scholar]
- Wernegreen, J.J. (2002) Genome evolution in bacterial endosymbionts of insects. Nat Rev Genet 3: 850–861. [DOI] [PubMed] [Google Scholar]
- Wheeler, B. , and Torchiano, M. (2010) lmPerm: Permutation tests for linear models. R package version 1.1‐2. https://github.com/mtorchiano/lmPerm.
- Wilson, A.C.C. , Ashton, P.D. , Calevro, F. , Charles, H. , Colella, S. , Febvay, G. , et al. (2010) Genomic insight into the amino acid relations of the pea aphid, Acyrthosiphon pisum, with its symbiotic bacterium Buchnera aphidicola . Insect Mol Biol 19: 249–258. [DOI] [PubMed] [Google Scholar]
- Wu, G.D. , Chen, J. , Hoffmann, C. , Bittinger, K. , Chen, Y. , Keilbaugh, S.A. , et al. (2011) Linking long‐term dietary patterns with gut microbial enterotypes. Science 334: 105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, S. , Jiang, L. , Qiao, G. , and Chen, J. (2020a) The bacterial flora associated with the polyphagous aphid Aphis gossypii Glover (Hemiptera: Aphididae) is strongly affected by host plants. Microb Ecol 79: 971–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, T. , Chen, J. , Jiang, L. , and Qiao, G. (2018) Historical and cospeciating associations between Cerataphidini aphids (Hemiptera: Aphididae: Hormaphidinae) and their primary endosymbiont Buchnera aphidicola . Zool J Linn Soc 182: 604–613. [Google Scholar]
- Xu, T. , Chen, J. , Jiang, L. , and Qiao, G. (2021) Diversity of bacteria associated with Hormaphidinae aphids (Hemiptera: Aphididae). Insect Sci 28: 165–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, T. , Jiang, L. , Chen, J. , and Qiao, G. (2020b) Host plants influence the symbiont diversity of Eriosomatinae (Hemiptera: Aphididae). Insects 11: 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Y. , Lee, C. , Kim, J. , and Hwang, S. (2005) Group‐specific primer and probe sets to detect methanogenic communities using quantitative real‐time polymerase chain reaction. Biotechnol Bioeng 89: 670–679. [DOI] [PubMed] [Google Scholar]
- Zhang, D. , and Qiao, G. (2010) Mollitrichosiphum Suenaga from China (Hemiptera: Aphididae), with the description of one new species. Zootaxa 2608: 1–24. [Google Scholar]
- Zhao, Y. , Zhang, S. , Luo, J. , Wang, C. , Lv, L. , and Cui, J. (2016) Bacterial communities of the cotton aphid Aphis gossypii associated with Bt cotton in northern China. Sci Rep 6: 22958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zytynska, S.E. , and Weisser, W.W. (2016) The natural occurrence of secondary bacterial symbionts in aphids. Ecol Entomol 41: 13–26. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Structural segregation using constrained principal coordinate analyses (cPCoA) of Bray–Curtis distances of bacterial (A, D, G), symbiont (B, E, H) and secondary symbiont (C, F, I) communities (n ≥ 1). Plots are structured by aphid species (A–C), geographic region (D–F) and host plant (G–I). The overall variation explained by the constrained factor is displayed at the top of each plot. The percent variation shown on each axis refers to the fraction of the total variance explained by the projection. The abbreviations are given in Table S5.
Fig. S2. Nonmetric multidimensional scaling (NMDS) plots based on Bray–Curtis distances of bacterial (A, D, G), symbiont (B, E, H) and secondary symbiont (C, F, I) communities (n ≥ 1). Samples are coloured by aphid species (A–C), geographic region (D–F) and host plant (G–I). The stress value indicates the goodness of fit between the NMDS representation and the data. The abbreviations are given in Table S5.
Fig. S3. Nonmetric multidimensional scaling (NMDS) plots based on Bray–Curtis distances of bacterial (A, D, G), symbiont (B, E, H) and secondary symbiont (C, F, I) communities (n ≥ 3). Samples are coloured by aphid species (A–C), geographic region (D–F) and host plant (G–I). The stress value indicates the goodness of fit between the NMDS representation and the data. The abbreviations are given in Table S5.
Fig. S4. Nonmetric multidimensional scaling (NMDS) plots based on unweighted UniFrac distances of bacterial communities (n ≥ 1, A, C, E; n ≥ 3, B, D, F). The distance data of symbiont and secondary symbiont communities were insufficient for NMDS. Samples are coloured by aphid species (A, B), geographic region (C, D) and host plant (E, F). The stress value indicates the goodness of fit between the NMDS representation and the data. The abbreviations are given in Table S5.
Fig. S5. Time‐calibrated phylogenetic tree of Mollitrichosiphum. The red circle at the node shows the calibration point. Horizontal bars display the 95% highest posterior density intervals of the estimated node ages. The mean ages of nodes are presented above the bars.
Table S1. Relative abundance of the top 10 bacterial phyla, classes, orders, families and genera in Mollitrichosiphum.
Table S2. Results of three‐way ANOVA based on alpha diversity indices in bacterial, symbiont and secondary symbiont communities.
Table S3. Voucher information and GenBank accession numbers of aphid samples used in this study.
Table S5. Grouping information of Mollitrichosiphum aphid samples used in this study.
Table S4 The OTU tables of bacterial (a), symbiont (b) and secondary symbiont communities (c).
Data Availability Statement
COI, Cytb and EF‐1α sequences obtained in this study were deposited in GenBank under accession numbers MT556450–MT556472 and MT563127–MT563158. Raw 16S rRNA gene amplicon reads were deposited in the NCBI Sequence Read Archive under BioProject accession number PRJNA637573.


