Abstract
Background
New noninvasive and affordable molecular approaches that will complement current practices and increase the accuracy of Parkinson's disease (PD) diagnosis are urgently needed. Circular RNAs (circRNAs) are stable noncoding RNAs that accumulate with aging in neurons and are increasingly shown to regulate all aspects of neuronal development and function.
Objectives
Τhe aims of this study were to identify differentially expressed circRNAs in blood mononuclear cells of patients with idiopathic PD and explore the competing endogenous RNA networks affected.
Methods
Eighty‐seven circRNAs were initially selected based on relatively high gene expression in the human brain. More than half of these were readily detectable in blood mononuclear cells using real‐time reverse transcription‐polymerase chain reaction. Comparative expression analysis was then performed in blood mononuclear cells from 60 control subjects and 60 idiopathic subjects with PD.
Results
Six circRNAs were significantly down‐regulated in patients with PD. The classifier that best distinguished PD consisted of four circRNAs with an area under the curve of 0.84. Cross‐linking immunoprecipitation‐sequencing data revealed that the RNA‐binding proteins bound by most of the deregulated circRNAs include the neurodegeneration‐associated FUS, TDP43, FMR1, and ATXN2. MicroRNAs predicted to be sequestered by most deregulated circRNAs have the Gene Ontology categories “protein modification” and “transcription factor activity” mostly enriched.
Conclusions
This is the first study that identifies specific circRNAs that may serve as diagnostic biomarkers for PD. Because they are highly expressed in the brain and are derived from genes with essential brain functions, they may also hint on the PD pathways affected. © 2021 Biomedical Research Foundation, Academy of Athens. Movement Disorders published by Wiley Periodicals LLC on behalf of International Parkinson and Movement Disorder Society.
Keywords: circRNAs, biomarkers, Parkinson's disease, PBMCs, blood
The diagnosis of Parkinson's disease (PD) is currently based on clinical diagnostic criteria and neuroimaging and is monitored by rating scales related to motor and nonmotor features. 1 Rating scales are frequently subjective and influenced by periodic fluctuations in symptoms and effective symptomatic therapies, while neuroimaging techniques, such as dopamine transporter‐ single photon emission CT, offer a quantifiable measure of disease progression but are limited by practicality and costs. 2 In addition, protein biomarkers, such as those based on alpha‐synuclein (SNCA) and dopamine metabolic products, have yielded mixed results, do not reflect disease progression, and require an invasive lumbar puncture. 3
Circular RNAs (circRNAs) are a newly recognized class of single‐stranded regulatory RNAs that are formed by head‐to‐tail splicing in which a downstream 5′ splice site is covalently connected to an upstream 3′ splice site of an RNA molecule. The result is an enclosed nonpolyadenylated circular transcript. 4 , 5 , 6 Due to the lack of free ends, which are normally targeted by 3′ and 5′ exoribonucleases, circRNAs are extremely stable with a half‐life of more than 48 hours compared with approximately 6 hours for linear transcripts. 7 , 8 There are different subtypes of circRNAs, including exonic, intronic, and exo‐intronic. Exonic circRNAs are mostly localized in the cytoplasm, where they act as sponges for microRNAs (miRNAs) and RNA‐binding proteins (RBPs), thus inhibiting their interaction with mRNA targets. 4 , 9 , 10 , 11 In contrast, intronic or exo‐intronic circRNAs are mostly localized in the nucleus and have few or no binding sites for miRNAs; instead, they function to control transcription. 12 , 13 Interestingly, the cotranscriptional biogenesis of circRNAs has also been shown to reduce linear host mRNA levels and change downstream splice‐site choice in some mRNAs. 11 , 14 , 15
circRNAs are widely conserved and more abundant in the brain than in any other tissue, 16 with many being expressed in an organ‐specific manner, along with their host genes, which are enriched with tissue‐specific biological functions. 17 For instance, brain circRNA host genes are enriched in neurotransmitter secretion, synaptic activities, and neuron maturation. 17 Importantly, however, they are regulated independently from their linear counterparts, 16 , 18 with 60% of central nervous system circRNAs being up‐regulated throughout development, especially during synaptogenesis, whereas only 2% of their linear isoforms show this tendency. 17
Recent studies revealed the deregulation of circRNAs in neurodegenerative diseases and neuropsychiatric disorders (reviewed in Mehta et al. 19 ). Furthermore, several brain‐enriched circRNAs have been associated with pathogenetic processes of neurodegeneration. For instance, CDR1as (ciRS‐7), a highly abundant circRNA in the brain, is down‐regulated in the brain of patients with Alzheimer's disease (AD). 20 This circRNA contains 63 binding sites for miR‐7; therefore, it is acting as an efficient sponge for it. 4 Importantly, critical proteins for the neurodegeneration processes, such as the ubiquitin protein ligase A (UBE2A), which catalyzes the proteolytic clearing of toxic amyloid peptides in AD, and SNCA, which accumulates in PD/AD, are both targets of miR‐7. 21 , 22 More recently, another circRNA, circSLC8A1, was found to increase in the substantia nigra of individuals with PD and in cultured cells exposed to the oxidative stress‐inducing agent paraquat. 23 Importantly, circSLC8A1 carries seven binding sites for miR‐128, an abundant and brain‐restricted miRNA that governs neuronal excitability and motor behavior. 24 , 25 , 26 , 27
Peripheral blood mononuclear cells (PBMCs) inherit the same genetic information as brain cells and are armed with abundant signaling pathways that respond to pathological changes. Multiple studies have shown that genome‐wide transcriptional and alternative splicing profiles in peripheral blood parallel changes in gene expression in the brain, reflecting broad molecular and cellular impairments. 28 , 29 , 30 , 31 , 32 , 33 Therefore, PBMCs provide a powerful and minimally invasive tool for the identification of novel targets for neurodegeneration research. Considering that circRNAs: (1) are abundant in the brain modulating gene expression en masse, (2) are stable, (3) do not get modified like proteins and hence levels directly correlate with activity; and (4) can be accurately quantified by routine and fast laboratory methods, such as real‐time reverse transcription‐polymerase chain reaction (RT‐qPCR), suggests that they not only represent important constituents of the pathophysiological processes implicated in neurological diseases but also excellent candidate biomarkers. The purpose of this study was to identify differentially expressed brain‐enriched circRNAs in PBMCs from patients with idiopathic PD (iPD) and pinpoint competing endogenous RNA networks.
Subjects and Methods
Figure 1 provides a schematic representation of the workflow.
FIG. 1.
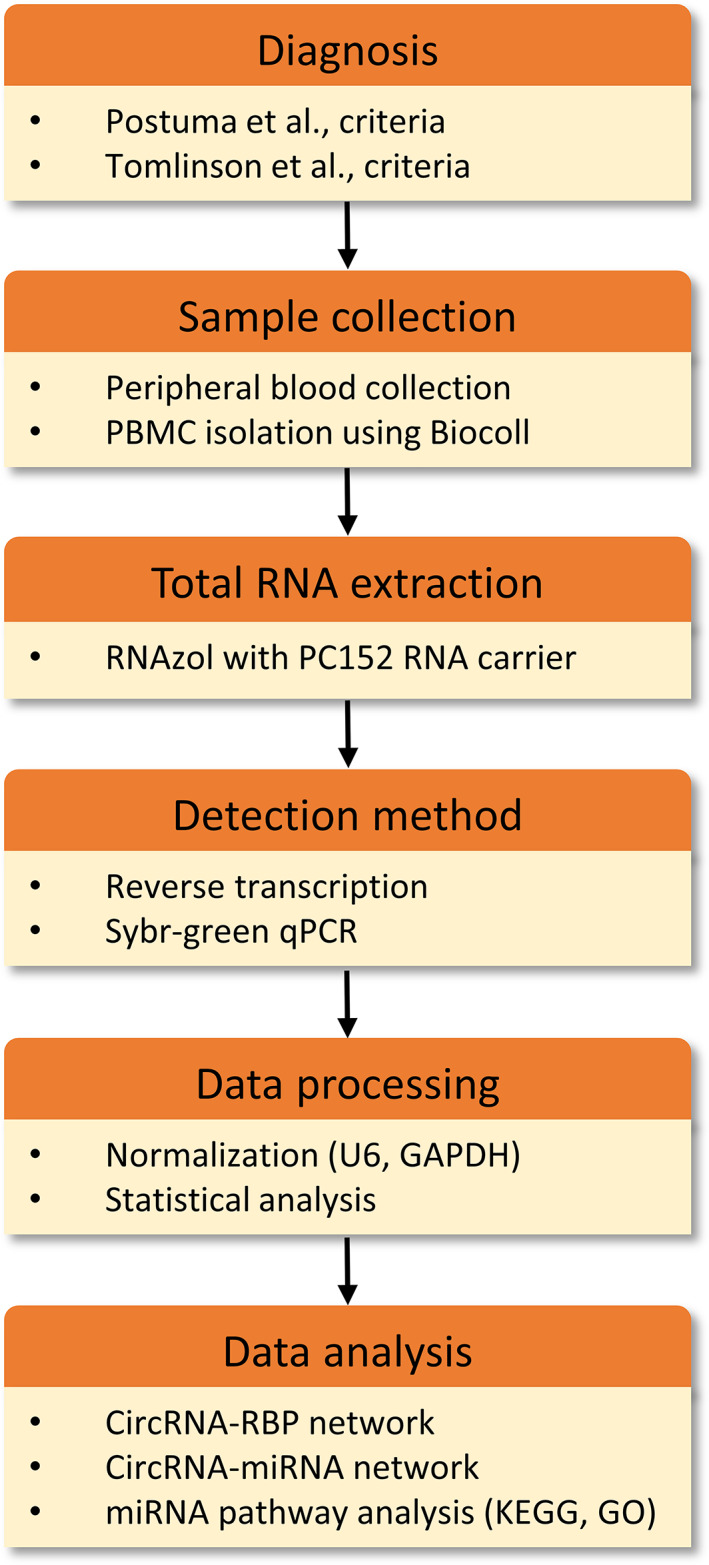
Schematic representation of the workflow. circRNA, circular RNA; GAPDH, glyceraldehyde 3‐phosphate dehydrogenase; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; miRNA, microRNA; PBMC, peripheral blood mononuclear cell; qPCR, quantitative polymerase chain reaction; RBP, RNA‐binding protein. [Color figure can be viewed at wileyonlinelibrary.com]
Study Population
This study included 60 patients with iPD and 60 healthy individuals in two separate cohorts. Patients were assessed with brain MRI or CT, and no relevant brain vascular lesions explaining the clinical phenotype were detected. The control group included spouses or unrelated companions of patients who had no known neurological disease, comorbidities, or PD family history. Individuals with concurrent malignant tumors, psychiatric disorders, collagen diseases, endocrine and cardiovascular diseases, or infections were excluded from this study, because these conditions are expected to alter the expression profile of transcripts. Patients affected by atypical parkinsonism were also excluded. All patients and control subjects were recruited from the National and Kapodistrian University of Athens' First Department of Neurology at Eginition Hospital. PD was diagnosed by two neurologists according to the criteria of Postuma et al. 1 In all cases, essential demographic and clinical information, including the study questionnaire for motor and nonmotor manifestations of the disease, and rating scales [Hoehn & Yahr stage, Mini‐Mental State Examination (MMSE) cognitive impairment score < 26, 34 Unified Parkinson's Disease Rating Scale part III (UPDRS III) in the on or off state] were collected and documented. The demographic and clinical features of patients and control subjects are summarized in Table 1. Levodopa equivalent daily dose (LEDD) was calculated for the patient group according to the criteria of Tomlinson et al. 35 The Eginition Hospital and Biomedical Research Foundation of the Academy of Athens ethics committees approved the study, and all participants provided written consent.
TABLE 1.
Demographic and clinical profiles of healthy control subjects and patients with Parkinon's disease
| Variables | Healthy Control Subjects | iPD | P Value |
|---|---|---|---|
| Subjects, n | 60 | 60 | Ν/Α |
| Age (years), ±SD | 64.38 ± 1.335 | 64.73 ± 1.33 | 0.84 |
| Sex (M/F) | 29/31 | 29/31 | 1 |
| Age of onset (years), ±SD | Ν/Α | 59.95 ± 10.97 | Ν/Α |
| Disease duration (years), ±SD | Ν/Α | 4.78 ± 0.55 | Ν/Α |
| Unified Parkinson's Disease Rating Scale part III, ±SD (on/off state) | Ν/Α | 25.77 ± 1.92 (42/18) | Ν/Α |
| Mini‐Mental State Examination, ±SD | Ν/Α | 27.53 ± 0.56 | Ν/Α |
| Hoehn & Yahr, ±SD | Ν/Α | 1.82 ± 0.10 | Ν/Α |
| Levodopa equivalent daily dose, ±SD | Ν/Α | 496.9 ± 59.71 | Ν/Α |
iPD, idiopathic Parkinson's disease; SD, standard deviation; N/A, not applicable
Isolation of PBMCs
PBMCs were isolated from whole blood by using density‐gradient centrifugation using the Biocoll Separating Solution according to the manufacturer's instructions (Biochrom, Cambridge, United Kingdom).
Total RNA Extraction and RT‐qPCR Analysis
Total RNA extraction was performed using the RNAzol‐RT reagent according to manufacturer's instructions (Molecular Research Center, Cincinnati, OH). To improve the yield of the small RNA fraction, we added a polyacryl carrier (PC152; Molecular Research Center) during the extraction method. Reverse transcription reactions were performed in triplicate for every sample. Similarly, qPCR was performed in triplicate on the Roche Lightcycler 96 using the SYBR FAST Universal 2X qPCR Master Mix from Roche Sequencing and Life Science Kapa Biosystems (Wilmington, MA). For the differential expression analysis, we selected only those circRNAs that were detected in PBMCs with a crossing threshold (Ct) value below 30, for improved detection accuracy. All primers span the splice junction. Noncoding U6 small nuclear 1 (RNU6‐1) and glyceraldehyde 3‐phosphate dehydrogenase (GAPDH) were used as reference genes. The relative expression level of circRNAs was calculated using the 2−ΔΔCt method between age‐ and sex‐matched counterparts. Primer sequences can be found in Supporting Information Table S1.
circRNA Selection Process
Eighty‐seven circRNAs were carefully selected by cross‐examining the data from three genome‐wide surveys. 4 , 18 , 36 We chose circRNAs that had high expression in the brain (Rybak–Wolf score > ~1,000) and low or no expression in other tissues. The host gene expression was also taken into account in the selection process. Initially, based on genotype‐tissue expression (GTEx) portal data, circRNAs for which host transcripts were specifically expressed in the brain were selected. However, we found that many circRNAs derived from these transcripts were not readily detectable in PBMCs. We therefore widened the analysis to host transcripts that are brain‐ or at least cerebellum‐enriched (ie, not exclusively expressed in the brain). Last, we included six brain‐abundant circRNAs deriving from host transcripts with low expression in the brain (UBXN7_circ_0001380, TMEM138_circ_0002058, ZNF292_circ_0004058, HAT1_circ_0008032, ZFAND6_circ_0000643, UIMC1_circ_0001558) and eight circRNAs that are hosted by brain‐relevant transcripts that have been found to be deregulated in AD (CORO1C_circ_0000437, WDR78_circ_0006677, PHC3_circ_0001359, SLAIN2_circ_0126525) 37 and autism (FAM120A_circ_0001875, CSNK1G3_circ_0001522, VMP1_circ_0006508, SMARCA5_circ_0001445). 38 For the list of circRNAs analyzed, see Supporting Information Tables S1 and S2.
circRNA Target Network
The interactions of the differentially expressed circRNAs with miRNAs and RBPs were identified by obtaining data from the Circular RNA Interactome (CircInteractome) and Interactional Database of Cancer‐Specific CircRNAs (IDCSC) databases, respectively. 39 , 40 CircInteractome uses the TargetScan algorithm to predict miRNA response elements (ie, miRNA‐binding sites), while IDCSC hosts circRNA cross‐linking immunoprecipitation sequencing (CLIP‐seq) data for the different RBPs extracted from starBase database. 41 The circRNA–miRNA and circRNA–RBP interactomes were then manually curated using the Cytoscape v.3.8.0 platform.
miRNA Pathway Analysis
The DIANA mirPath v.3 software suite was used to identify miRNA‐regulated pathways. This software renders possible the functional annotation of miRNAs using standard hypergeometric distributions, unbiased empirical distributions, and meta‐analysis statistics. 42 Here, predicted targets from the DIANA microT‐CDS algorithm with high‐quality experimentally supported interactions were used to identify Kyoto Encyclopedia of Genes and Genomes (KEGG) molecular pathways, as well as Gene Ontology (GO) terms targeted by each miRNA. The combinatorial effect of deregulated miRNAs was identified by simultaneously selecting multiple miRNAs in the software. The default values (P value threshold, 0.05; microT‐CDS threshold, 0.8; false discovery rate correction option ticked) were used for the analysis.
Statistical Analysis
Statistical analysis was performed using GraphPad PRISM v5.0 and R v3.5.3. All data underwent a normality test (Shapiro–Wilk) and were found to be nonnormally distributed. As a result, all circRNA data underwent a logarithmic transformation (with base 2), to better approximate the normal distribution. The parametric t test was used to observe differences between healthy control subjects and patients with PD. We applied the Benjamini–Hochberg false discovery rate correction in the resulting P values to account for the multiple numbers of tests. Spearman method with Bonferroni correction for multiple comparisons was used to correlate circRNA expression levels with participants' demographic and clinical characteristics (P value threshold, 0.0005).
To assess the possibility that sex is a confounding factor, we applied the two‐way ANOVA model to the log‐transformed data (normally distributed) with sex as an additional factor. No difference in the circRNAs that were statistically significant was found.
Receiver operating characteristic (ROC) curves were constructed, and the area under the curve (AUC) was calculated to evaluate the predictive sensitivity and specificity of PBMC circRNAs for PD diagnosis. The cutoff value for the ROC analysis was determined using the Youden Index. Data are presented as means ± standard error of the mean. circRNA selection was based on the stepwise removal approach. A logistic regression statistical model containing all available circRNAs as independent variables and PD status as the dependent variable was built. Then the circRNAs with the least contribution in the model (as determined by an F test) were removed. This process continued until no further removals were possible.
Data Availability
The datasets analyzed during this study are all available from the corresponding author on request.
Results
circRNAs Are Differentially Expressed in PBMCs of Patients With iPD
The demographic and clinical characteristics of 60 healthy control subjects and 60 patients with iPD are summarized in Table 1. The mean age of 64.5 years and the sex ratio were the same for both groups. The disease duration for the PD group was 4.8 ± 0.55 years, and the MMSE score 27.5 ± 0.56. Initially, RT‐qPCR was used to detect plasma levels of 32 circRNAs that are highly expressed by brain cells. It was anticipated that a sufficient quantity of brain‐derived circRNAs would find its way into the plasma. However, only two circRNAs derived from RMST (at Ct 29) and PSD3 (at Ct 28) genes were detected. Using the same amount of RNA, this time extracted from human brain tissue, it was revealed that all circRNAs were readily detectable with an average Ct value of 26, demonstrating that all primer pairs were functional (data not shown). This indicated that brain‐enriched circRNAs are not as abundant as brain‐enriched miRNAs (the average Ct value for 21 brain‐enriched miRNAs was 17.5 in the same human brain total RNA) and are not circulating in appreciable amounts in the blood (the average Ct value for the corresponding miRNAs in the plasma was 25). 43
Based on previous studies showing that genome‐wide transcriptional and alternative splicing profiles in peripheral blood cells parallel changes in gene expression in the brain, the levels of brain‐enriched circRNAs were next assessed in PBMCs. We increased the number of primer sets to 87 and found that 48 were detected with a Ct value <30, safeguarding accurate and reproducible detection. These circRNAs were then analyzed for differential expression in healthy control and iPD patient samples (Supporting Information Table S3).
After multiple comparison adjustment, six circRNAs were significantly altered in the PBMCs obtained from patients with PD compared with healthy control subjects. MAPK9_circ_0001566, HOMER1_circ_0006916, SLAIN1_circ_0000497, DOP1B_circ_0001187, RESP1_circ_0004368, and PSEN1_circ_0003848 were all down‐regulated in the PD cohort (Fig. 2 and Supporting Information Table S3). The swarm plots for the 42 circRNAs whose relative expression was not significantly altered in the PBMCs of patients with iPD are shown in Supporting Information Figure S1.
FIG. 2.
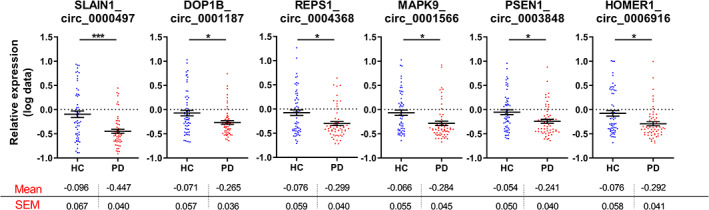
Swarm plots of deregulated circular RNAs (circRNAs) relative expression in the peripheral blood mononuclear cells (PBMCs) of control and idiopathic Parkinson's disease (iPD) cohorts. Mean levels ± standard error of the mean are included below each graph. Graphs demonstrate relative expression of log‐transformed data. Unpaired t test was used to determine the significance of differences between the two groups. *P < 0.05, **P < 0.01, ***P < 0.001. HC, healthy control subjects. [Color figure can be viewed at wileyonlinelibrary.com]
Association Between circRNA Levels and Clinical Features, Age or Sex
Spearman correlation test was used to relate circRNA levels to iPD patients' clinical features. We found no correlation between age at onset, disease duration, UPDRS III, MMSE, LEDD, Hoehn & Yahr, or patients' on/off state and circRNA levels (Supporting Information Table S4; data not shown). Finally, correcting clinical scores with LEDD did not reveal any more associations (data not shown). In addition, there was no significant correlation between circRNA expression and age or sex in either healthy control subjects or patients with PD.
Discriminant Analysis
To evaluate the utility of PBMC circRNA levels in discriminating subjects with iPD from healthy control subjects, we performed ROC curve analysis. The diagnostic sensitivity and specificity of a four‐circRNA panel (SLAIN1_circ_0000497, SLAIN2_circ_0126525, ANKRD12_circ_0000826, and PSEN1_circ_0003848) were 75.3% (62.1%–85.2%) and 78% (65.8%–88%), respectively, and the AUC was 0.84 (Fig. 3).
FIG. 3.
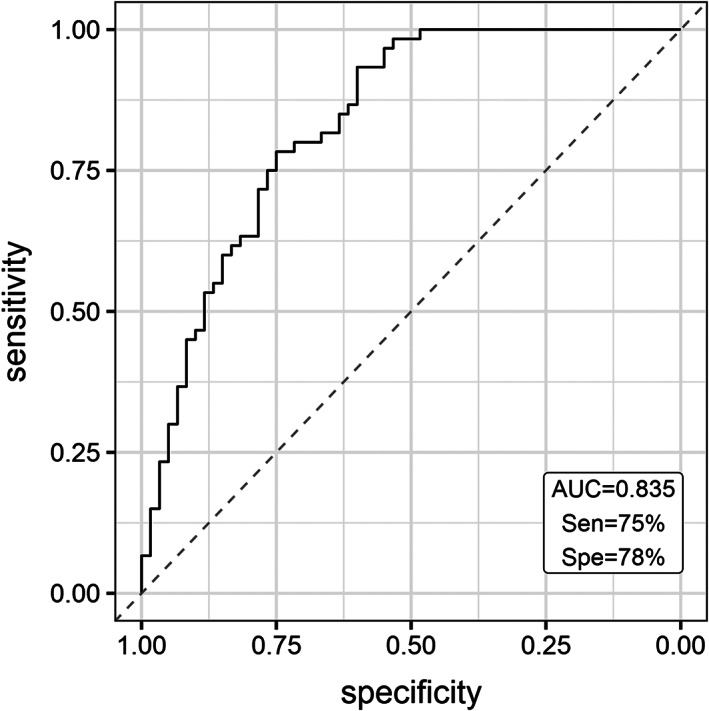
The receiver operating characteristic (ROC) curve analysis for discriminating idiopathic Parikinson's disease (iPD) from healthy control subjects. ROC curve of four circular RNAs (MAPK9_circ_0001566, SLAIN1_circ_0000497, SLAIN2_circ_0126525, and PSEN1_circ_0003848) differentiate iPD cases from healthy control subjects. AUC, area under the curve; Sen, sensitivity; Spe, specificity.
Competing Endogenous RNA Networks
circRNAs can act as miRNA and RBP sponges for regulating gene expression. To explore the functional role of the deregulated circRNAs, we identified all of their miRNA and RBP targets. Multiple miRNA binding sites are predicted for each circRNA, with SLAIN1_circ_0000497 and MAPK9_circ_0001566 having the most of the miRNA response elements (38 and 24, respectively) (Fig. 4A). Interestingly, five miRNAs were predicted to be sponged by half or more of the deregulated PD circRNAs. miR‐526b and miR‐659 are the top targets, sequestered by four deregulated PD circRNAs (Fig. 4A). Figure 4B shows the deregulated circRNA–RBP network. Like for miRNAs, CLIP‐seq data obtained from starBase database revealed that the deregulated circRNAs have multiple RBP binding sites. MAPK9_circ_0001566 and HOMER1_circ_0006916 host the most of these sites with 60 and 49 sites, respectively. Interestingly, 29 RBPs were sequestered by four or more deregulated circRNAs.
FIG. 4.
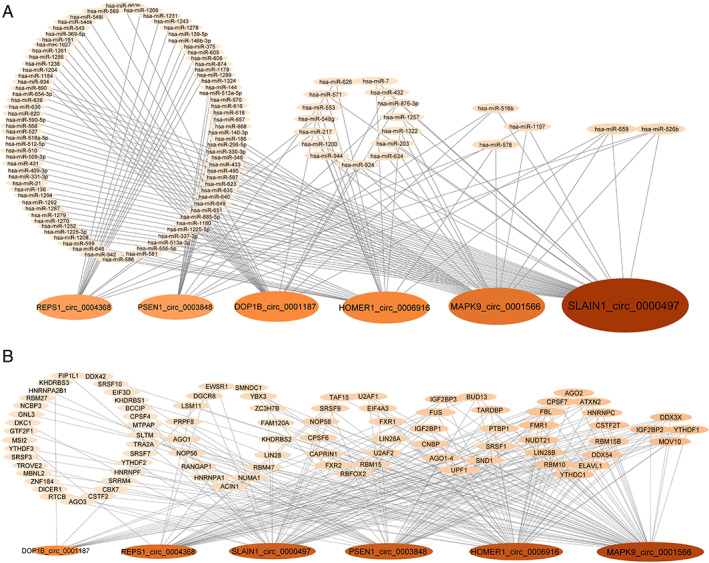
Circular RNA (circRNA) target networks. Diagrams show (A) the predicted microRNAs and (B) the cross‐linking immunoprecipitation (CLIP) sequencing that identified RNA‐binding proteins that bind to differentially expressed circRNAs. [Color figure can be viewed at wileyonlinelibrary.com]
circRNA–miRNA Pathway Analysis
To explore the biological pathways affected by the five miRNAs (miR‐516b‐5p, miR‐526b‐5p, miR‐578, miR‐659‐3p, and miR‐1197) sequestered by three or more of the deregulated circRNAs, we used the DIANA mirPath v3 tool to align miRNA predicted targets with KEGG pathways and GOslim categories. A priori gene union analysis of deregulated miRNA targets revealed 14 KEGG categories as significantly enriched; they included “thyroid hormone signaling pathway” (P < 0.0015, 24 genes), “regulation of actin cytoskeleton” (P < 0.015, 42 genes), “phosphatidylinositol signaling pathway” (P < 0.016, 16 genes), “MAPK signaling pathway” (P < 0.016, 46 genes), and “FoxO signaling pathway” (P < 0.016, 26 genes) (Supporting Information Table S5A). Similar findings were obtained using a posteriori analysis (Supporting Information Fig. S2A). Thirty‐nine GOslim categories that are controlled by the gene union of the deregulated miRNA targets were enriched following a priori analysis; these included “cellular protein modification” (P < 3.93E−19, 283 genes), “nucleic acid binding transcription factor activity” (P < 7.74E−7, 112 genes), “cytoskeletal protein binding” (P < 3.91E−9, 102 genes), “cell death” (P < 7.91E−8, 112 genes), “RNA binding” (P < 6.93E−7, 207 genes), and “response to stress” (P < 3.72E−5, 225 genes) (Supporting Information Table S5B). Similar findings were obtained using a posteriori analysis (Supporting Information Fig. S2B).
Discussion
We profiled brain‐enriched circRNAs in peripheral blood from control subjects and patients with PD using a RT‐qPCR‐based approach for three reasons. First, primers could be designed to span the splicing junction, which guarantees that only a message from the circRNA is amplified. Second, RT‐qPCR is the most sensitive method to accurately determine expression changes between cohorts; the alternative microarray approach is prone to errors at multiple levels and nearly always requires a follow‐up RT‐qPCR‐based analysis to validate findings. Third, we probed circRNAs that are abundantly expressed in the brain; in this way, we could identify differentially expressed or spliced circRNAs that are more likely associated with the neurological processes in PD.
Insights Into the Differentially Expressed circRNA Genes
We initiated our study with 87 brain‐enriched circRNAs from which more than half were confidently detected in PBMCs. From these circRNAs, six were differentially expressed in PD with a 17% decrease on average from healthy control subjects levels. These changes may appear subtle, but depending on the circRNA baseline expression levels and considering the relative importance of their multiple targets (transcription factors, RBPs, and miRNAs), as well as the added‐up deregulation of the common targets, the biological outcome is expected to be significant.
It has been observed that the biological role of host transcripts reflects on the function of the circRNAs. 17 We found that the host transcripts of the differentially expressed circRNAs are not exclusive to brain pathways; rather, they are housekeeping genes, whose functions are best characterized in the central nervous system because they are essential for neuronal homeostasis. A brief bibliographical overview of their properties follows.
Hsa_circ_0001566 is hosted by the mitogen‐activated protein kinase 9 (MAPK9) gene. MAPK9/JNK2 is a member of the c‐Jun n‐terminal kinase 1–3 family robustly activated by environmental stresses, including the PD‐related neurotoxins lipopolysaccharides, 1‐methyl‐4‐phenyl‐1,2,3,6‐tetrahydropyridine, and 6‐hydroxydopamine, to mediate neuronal degeneration. 44 It is indispensable during brain development for neuronal migration, axonal sprouting, and guidance, as well as neuronal survival. 45 Hsa_circ_0006916 is hosted by the homer scaffold protein 1 (HOMER1) gene. HOMER1 is a member of Homer 1–3 family constituting important scaffold proteins at the postsynaptic density that associate with a large number of Ca2+‐handling proteins, including channels, receptors, and shank scaffolding proteins to regulate intracellular Ca2+ homeostasis. 46 A single‐nucleotide polymorphism in the promoter of HOMER1 has been associated with psychotic symptoms in PD. 47 Further, circHomer1a is reduced in the prefrontal cortex of patients with schizophrenia and bipolar disorder, where it modulates the alternative splicing of mRNA transcripts involved in synaptic plasticity and psychiatric disease. 11 Hsa_circ_0000497 is hosted by the SLAIN motif family member 1 (SLAIN1) gene. SLAIN1 and SLAIN2 are microtubule‐associated proteins that promote persistent microtubule growth by recruiting the microtubule polymerase cytoskeleton‐associated protein 5 (CKAP5/ch‐TOG) to microtubule plus‐ends, and thus they are important for axon elongation in developing neurons. 48 Recently, SLAIN1 was identified as a candidate gene for intellectual disability. 49 Hsa_circ_0001187 is hosted by the DOP1 leucine zipper‐like protein B (DOP1B) gene. DOP1B/DOPEY2/C21orf5 and its ortholog DOP1A interact with partner MON2 to retrograde transport endosomes from the trans‐Golgi network to the Golgi. 50 , 51 DOP1B is a candidate gene for mental retardation in Down syndrome, 52 , 53 and copy number variations have been observed in AD. 54 , 55 Hsa_circ_0004368 is hosted by the RALBP1‐associated eps domain containing 1 (REPS1) gene. REPS1 is a signaling and endocytosis adaptor that interacts with adaptor Intersectin 1 (ITSN1) in clathrin‐coated pits and Amphiphysin 1 (AMPH) at the surface of synaptic vesicles. 56 Mutations in REPS1 are associated with neurodegeneration with brain iron accumulation in the basal ganglia. 57 Hsa_circ_0003848 is hosted by the presenilin 1 (PSEN1) gene. PSEN1 and its paralog PSEN2 are the endoprotease subunits of the gamma‐secretase complex that catalyzes the intramembrane cleavage of integral membrane proteins, such as Notch receptors and amyloid‐beta precursor protein. Mutations in either gene cause early‐onset AD 58 and SNCA accumulation in Lewy bodies (LBs) in these patients. 59 Besides their established role in mediating the formation of Aβ peptide, more recently mutant PS1 has been shown to impair numerous cellular functions, such as calcium flux, organization of proteins in different compartments, and protein turnover via vacuolar metabolism. 60 Interestingly, a novel PSEN1 mutation was recently identified as the likely cause for early‐onset parkinsonism. 61
Correlation Between circRNA Levels and Demographics
There was no significant correlation between the differential expression of a particular circRNA and clinical or demographic measures. Combined interactions with age and sex did not also appear to affect circRNA levels. These findings reinforce current knowledge that the etiology of PD is complex, involving a mix of genetic and environmental influences on aging brain. Similar findings have been observed in miRNA studies. 62 Further, a pool of four circRNAs discriminated patients with PD from control subjects with an AUC of 0.84.
circRNA–RBP and circRNA–miR Interactions
In silico approaches were used to identify potential biological roles for the deregulated circRNAs by identifying the RBPs and miRNAs that are sequestered preferentially by them. Because the circRNAs were all down‐regulated, it indicates that target RBP and miRNA functions will be enhanced in PD. The circRNA–RBP network, which is based on experimental CLIPS‐seq data, revealed that 29 RBPs were bound by four or more deregulated circRNAs. Importantly, several of these RBPs are implicated in familial neurodegeneration, including Fragile X Mental Retardation Protein 1 (FMR1, fragile X syndrome and associated disorders), Ataxin 2 (ATXN2, spinocerebellar ataxia 2, late‐onset PD), Fused in Sarcoma (FUS), and TAR DNA binding protein (TARDBP/TDP43) (amyotrophic lateral sclerosis, frontotemporal dementia). 63 , 64 , 65 , 66 , 67 , 68 , 69 , 70 , 71 , 72
The circRNA–miRNA network revealed five miRNAs that were predicted to be sponged by at least three down‐regulated PD circRNAs. miR‐659‐3p is of particular interest because it targets progranulin, a neuroprotective and anti‐inflammatory protein implicated in frontotemporal dementia. 73 , 74 , 75 , 76 , 77 To explore the molecular pathways controlled by the five miRNAs, we performed in silico analysis of KEGG pathways and GOslim terms. KEGG categories revealed multiple signaling pathways (thyroid hormone, phosphatidylinositol, MAPK, FoxO) implicated in neuronal survival and plasticity and “Regulation of actin cytoskeleton,” which is central to presynaptic and postsynaptic assembly as overrepresented. 78 , 79 , 80 , 81 GOslim analysis revealed “cellular protein modification,” “nucleic acid binding transcription factor activity,” “cytoskeletal protein binding,” “cell death,” and “response to stress” as overrepresented among the biological processes affected. “Cellular protein modifications,” such as phosphorylation, ubiquitination, truncation, acetylation, nitration, and sumoylation of PD‐linked proteins, have emerged as important modulators of pathogenic mechanisms in PD. 82 , 83 “Transcription factor” changes indicate that there is not only misexpression at the mRNA translation level by miRNA deregulation but also that there exists a second wave of en masse deregulation involving transcription‐mediated changes. Finally, deregulation of fine cytoskeletal dynamics is expected to impair trafficking and intracellular signaling pathways and has been recognized as a key insult in the pathogenesis of multiple neurodegenerative diseases, including PD. 84 , 85
Conclusions
We performed an RT‐qPCR‐based analysis on RNA extracted from PBMCs from a cohort of patients with PD and matched control subjects to identify deregulated circRNAs. The circRNAs investigated are highly expressed in the human brain. This is the first study of its kind in PD. The measurement of four out of six down‐regulated circRNAs provided reasonable sensitivity and specificity for PD in this discovery cohort. The deregulated circRNAs form a robust set of brain‐associated circRNAs that can now be further evaluated, along with other measures, as diagnostic and possible therapeutic targets for PD. In silico analysis provided a comprehensive guide of the pathways and processes they control, shedding light on their potential biological role. The impact of these findings will now await further exploration.
Author Roles
Conceived the study: E.D. Neurologically examined patients: A.B., N.P., and L.S. Peripheral blood processing: M.M. Differential expression analysis: S.R., D.K., and E.D. Analyzed data: S.R., N.P., and E.D. Bioinformatics analyses: E.D. Wrote the manuscript: E.D. All authors read, edited, and approved the final manuscript.
Financial Disclosures
S.R. is a postdoctoral researcher at Biomedical Research Foundation of the Academy of Athens (BRFAA). S.R. has no financial disclosures. A.B. is a neurology resident at Eginition Hospital. A.B. has no financial disclosures. D.K. is a former MSc student at BRFAA. D.K. has no financial disclosures. N.P. is a neurology resident at Eginition Hospital. N.P. has no financial disclosures. M.M. is a technician at BRFAA. M.M. has no financial disclosures. L.S. is employed by the National and Kapodistrian University of Athens and the Biomedical Research Foundation of the Academy of Athens. He has the following active grants: Fondation Sante research grant, a Michael J. Fox Foundation (MJFF) grant as a collaborator, and an ELIDEK grant. He has served on an Advisory Board for Abbvie, Novartis, and Roche and has received honoraria from Abbvie and Sanofi. E.D. is employed by BRFAA. He has one active MJFF research grant.
Supporting information
Figure S1. Swarm plots for the 42 circRNAs whose relative expression is not significantly altered in the PBMCs of idiopathic PD patients. Mean levels +/− SEM are included below each graph. Graphs demonstrate relative expression of log‐transformed data. Unpaired t‐test was used to determine the significance of differences between the two groups. *P < 0.05, **P < 0.01, ***P < 0.001.
Figure S2. KEGG and GOslim classifications of the circRNA‐miRNA target genes. (A) KEGG and (B) GOslim categories union of the RNA targets of the five miRNAs sequestered by three or more of the deregulated PD circRNAs (miR‐516b‐5p, miR‐526b‐5p, miR‐578, miR‐659‐3p, miR‐1197). They were prepared using the DIANA‐miRPath v3.0 interface using default values (P‐value threshold 0.05, microT‐CDS threshold 0.8).
Table S1. Primer sequences used for real‐time PCR.
Table S2. List of 39 circRNAs not detected in PBMCs.
Table S3. Basic characteristics and comparative PBMC circRNA expression in idiopathic PD patients and healthy controls.
CircRNA size and expression in the brain as well as host transcript expression in the body. Means with their respective standard deviation (std) for both groups are shown. Statistically significant differences in comparison to healthy controls (unpaired t‐test) are highlighted in grey. Multiple comparison analysis (adjusted P‐values) calculated according to Benjamini‐Hochberg false discovery rate method. *According to Rybak et al. ** According to genotype‐tissue expression portal.
Table S4. Correlation between relative circRNA expression and sex, age, age‐at‐onset, PD duration, HY, UPDRS and MMSE scores and LEDD of PD patients.
Significance level is set at P < 0.0005 after Bonferroni correction.
Table S5. KEGG and GOslim categories that are mostly deregulated in idiopathic PD. Gene union of the targets of the five miRNAs sequestered by three or more of the deregulated PD circRNAs (miR‐516b‐5p, miR‐526b‐5p, miR‐578, miR‐659‐3p, miR‐1197) in PD versus (A) KEGG and (B) GOslim categories created by the DIANA‐miRPath v3.0 interface using default values (P‐value threshold 0.05, microT‐CDS threshold 0.8).
Acknowledgments
The authors are grateful to patients, relatives, and volunteer healthy control subjects for their participation in this study. This research was financed by Greece and European Union (European Social Fund‐ESF) through the Operational Program «Human Resources Development, Education and Lifelong Learning 2014‐2020» in the context of the project “Development of diagnostic biomarker tests for Parkinson's disease” (MIS 5049385). It was also cofinanced by the action “Precision medicine Hellenic network in genetic neurodegenerative diseases” (2018∑Ε01300001) of the Hellenic public investments program of General Secretariat for Research and Technology and the Michael J. Fox Foundation for Parkinson's Research (Grant ID 13353).
Relevant conflicts of interest/financial disclosures: Nothing to report.
Funding agencies: This research was financed by Greece and European Union (European Social Fund‐ESF) through the Operational Program ‘Human Resources Development, Education and Lifelong Learning 2014‐2020’ in the context of the project “Development of diagnostic biomarker tests for Parkinson's disease” (MIS 5049385). It was also cofinanced by the action “Precision medicine Hellenic network in genetic neurodegenerative diseases” (2018∑Ε01300001) of the Hellenic public investments program of General Secretariat for Research and Technology and the Michael J. Fox Foundation for Parkinson's Research (Grant ID 13353).
References
- 1. Postuma RB, Berg D, Stern M, et al. MDS clinical diagnostic criteria for Parkinson's disease. Mov Disord 2015;30(12):1591–1601. [DOI] [PubMed] [Google Scholar]
- 2. Bhidayasiri R, Martinez‐Martin P. Clinical assessments in Parkinson's disease: scales and monitoring. Int Rev Neurobiol 2017;132:129–182. [DOI] [PubMed] [Google Scholar]
- 3. Miller DB, O'Callaghan JP. Biomarkers of Parkinson's disease: present and future. Metabolism 2015;64(3 Suppl 1):S40–S46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495(7441):333–338. [DOI] [PubMed] [Google Scholar]
- 5. Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 2012;7(2):e30733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zaphiropoulos PG. Circular RNAs from transcripts of the rat cytochrome P450 2C24 gene: correlation with exon skipping. Proc Natl Acad Sci U S A 1996;93(13):6536–6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013;19(2):141–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schwanhausser B, Busse D, Li N, et al. Global quantification of mammalian gene expression control. Nature 2011;473(7347):337–342. [DOI] [PubMed] [Google Scholar]
- 9. Abdelmohsen K, Panda AC, Munk R, et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol 2017;14(3):361–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495(7441):384–388. [DOI] [PubMed] [Google Scholar]
- 11. Zimmerman AJ, Hafez AK, Amoah SK, et al. A psychiatric disease‐related circular RNA controls synaptic gene expression and cognition. Mol Psychiatry 2020;25(11):2712–2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li Z, Huang C, Bao C, et al. Exon‐intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 2015;22(3):256–264. [DOI] [PubMed] [Google Scholar]
- 13. Zhang Y, Zhang XO, Chen T, et al. Circular intronic long noncoding RNAs. Mol Cell 2013;51(6):792–806. [DOI] [PubMed] [Google Scholar]
- 14. Ashwal‐Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with pre‐mRNA splicing. Mol Cell 2014;56(1):55–66. [DOI] [PubMed] [Google Scholar]
- 15. Koh W, Gonzalez V, Natarajan S, Carter R, Brown PO, Gawad C. Dynamic ASXL1 exon skipping and alternative circular splicing in single human cells. PLoS One 2016;11(10):e0164085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. You X, Vlatkovic I, Babic A, et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci 2015;18(4):603–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mahmoudi E, Cairns MJ. Circular RNAs are temporospatially regulated throughout development and ageing in the rat. Sci Rep 2019;9(1):2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rybak‐Wolf A, Stottmeister C, Glazar P, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell 2015;58(5):870–885. [DOI] [PubMed] [Google Scholar]
- 19. Mehta SL, Dempsey RJ, Vemuganti R. Role of circular RNAs in brain development and CNS diseases. Prog Neurobiol 2020;186:101746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lukiw WJ. Circular RNA (circRNA) in Alzheimer's disease (AD). Front Genet 2013;4:307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Doxakis E. Post‐transcriptional regulation of alpha‐synuclein expression by mir‐7 and mir‐153. J Biol Chem 2010;285(17):12726–12734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhao Y, Alexandrov PN, Jaber V, Lukiw WJ. Deficiency in the ubiquitin conjugating enzyme UBE2A in Alzheimer's disease (AD) is linked to deficits in a natural circular miRNA‐7 sponge (circRNA; ciRS‐7). Genes (Basel) 2016;7(12):116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hanan M, Simchovitz A, Yayon N, et al. A Parkinson's disease CircRNAs resource reveals a link between circSLC8A1 and oxidative stress. EMBO Mol Med 2020;12(9):e11942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Paschou M, Doxakis E. Neurofibromin 1 is a miRNA target in neurons. PLoS One 2012;7(10):e46773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Paschou M, Maier L, Papazafiri P, et al. Neuronal microRNAs modulate TREK two‐pore domain K(+) channel expression and current density. RNA Biol 2020;17(5):651–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tan CL, Plotkin JL, Veno MT, et al. MicroRNA‐128 governs neuronal excitability and motor behavior in mice. Science 2013;342(6163):1254–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang W, Kim PJ, Chen Z, et al. MiRNA‐128 regulates the proliferation and neurogenesis of neural precursors by targeting PCM1 in the developing cortex. eLife 2016;5(e11324). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Iturria‐Medina Y, Khan AF, Adewale Q, Shirazi AH, Alzheimer's Disease Neuroimaging I . Blood and brain gene expression trajectories mirror neuropathology and clinical deterioration in neurodegeneration. Brain 2020;143(2):661–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Naughton BJ, Duncan FJ, Murrey DA, et al. Blood genome‐wide transcriptional profiles reflect broad molecular impairments and strong blood‐brain links in Alzheimer's disease. J Alzheimers Dis 2015;43(1):93–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pinho R, Guedes LC, Soreq L, et al. Gene expression differences in peripheral blood of Parkinson's disease patients with distinct progression profiles. PLoS One 2016;11(6):e0157852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Soreq L, Bergman H, Israel Z, Soreq H. Exon arrays reveal alternative splicing aberrations in Parkinson's disease leukocytes. Neurodegener Dis 2012;10(1–4):203–206. [DOI] [PubMed] [Google Scholar]
- 32. van Heerden JH, Conesa A, Stein DJ, Montaner D, Russell V, Illing N. Parallel changes in gene expression in peripheral blood mononuclear cells and the brain after maternal separation in the mouse. BMC Res Notes 2009;2:195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Soreq L, Salomonis N, Bronstein M, et al. Small RNA sequencing‐microarray analyses in Parkinson leukocytes reveal deep brain stimulation‐induced splicing changes that classify brain region transcriptomes. Front Mol Neurosci 2013;6:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dubois B, Burn D, Goetz C, et al. Diagnostic procedures for Parkinson's disease dementia: recommendations from the movement disorder society task force. Mov Disord 2007;22(16):2314–2324. [DOI] [PubMed] [Google Scholar]
- 35. Tomlinson CL, Stowe R, Patel S, Rick C, Gray R, Clarke CE. Systematic review of levodopa dose equivalency reporting in Parkinson's disease. Mov Disord 2010;25(15):2649–2653. [DOI] [PubMed] [Google Scholar]
- 36. Maass PG, Glazar P, Memczak S, et al. A map of human circular RNAs in clinically relevant tissues. J Mol Med 2017;95(11):1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dube U, Del‐Aguila JL, Li Z, et al. An atlas of cortical circular RNA expression in Alzheimer disease brains demonstrates clinical and pathological associations. Nat Neurosci 2019;22(11):1903–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gokool A, Anwar F, Voineagu I. The landscape of circular RNA expression in the human brain. Biol Psychiatry 2020;87(3):294–304. [DOI] [PubMed] [Google Scholar]
- 39. Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol 2016;13(1):34–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xia S, Feng J, Chen K, et al. CSCD: a database for cancer‐specific circular RNAs. Nucleic Acids Res 2018;46(D1):D925–D929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA‐ceRNA, miRNA‐ncRNA and protein‐RNA interaction networks from large‐scale CLIP‐Seq data. Nucleic Acids Res 2014;42(Database issue):D92–D97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vlachos IS, Zagganas K, Paraskevopoulou MD, et al. DIANA‐miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res 2015;43(W1):W460–W466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ravanidis S, Bougea A, Papagiannakis N, et al. Circulating brain‐enriched MicroRNAs for detection and discrimination of idiopathic and genetic Parkinson's disease. Mov Disord 2020;35(3):457–467. [DOI] [PubMed] [Google Scholar]
- 44. Dzamko N, Zhou J, Huang Y, Halliday GM. Parkinson's disease‐implicated kinases in the brain; insights into disease pathogenesis. Front Mol Neurosci 2014;7:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Waetzig V, Zhao Y, Herdegen T. The bright side of JNKs‐multitalented mediators in neuronal sprouting, brain development and nerve fiber regeneration. Prog Neurobiol 2006;80(2):84–97. [DOI] [PubMed] [Google Scholar]
- 46. Jardin I, Lopez JJ, Berna‐Erro A, Salido GM, Rosado JA. Homer proteins in Ca(2)(+) entry. IUBMB Life 2013;65(6):497–504. [DOI] [PubMed] [Google Scholar]
- 47. De Luca V, Annesi G, De Marco EV, et al. HOMER1 promoter analysis in Parkinson's disease: association study with psychotic symptoms. Neuropsychobiology 2009;59(4):239–245. [DOI] [PubMed] [Google Scholar]
- 48. van der Vaart B, Franker MA, Kuijpers M, et al. Microtubule plus‐end tracking proteins SLAIN1/2 and ch‐TOG promote axonal development. J Neurosci 2012;32(42):14722–14728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Harripaul R, Vasli N, Mikhailov A, et al. Mapping autosomal recessive intellectual disability: combined microarray and exome sequencing identifies 26 novel candidate genes in 192 consanguineous families. Mol Psychiatry 2018;23(4):973–984. [DOI] [PubMed] [Google Scholar]
- 50. McGough IJ, de Groot REA, Jellett AP, et al. SNX3‐retromer requires an evolutionary conserved MON2:DOPEY2:ATP9A complex to mediate Wntless sorting and Wnt secretion. Nat Commun 2018;9(1):3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhao SB, Dean N, Gao XD, Fujita M. MON2 guides Wntless transport to the Golgi through recycling endosomes. Cell Struct Funct 2020;45(1):77–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lopes C, Chettouh Z, Delabar JM, Rachidi M. The differentially expressed C21orf5 gene in the medial temporal‐lobe system could play a role in mental retardation in down syndrome and transgenic mice. Biochem Biophys Res Commun 2003;305(4):915–924. [DOI] [PubMed] [Google Scholar]
- 53. Rachidi M, Delezoide AL, Delabar JM, Lopes C. A quantitative assessment of gene expression (QAGE) reveals differential overexpression of DOPEY2, a candidate gene for mental retardation, in down syndrome brain regions. Int J Dev Neurosci 2009;27(4):393–398. [DOI] [PubMed] [Google Scholar]
- 54. Swaminathan S, Huentelman MJ, Corneveaux JJ, et al. Analysis of copy number variation in Alzheimer's disease in a cohort of clinically characterized and neuropathologically verified individuals. PLoS One 2012;7(12):e50640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Swaminathan S, Shen L, Kim S, et al. Analysis of copy number variation in Alzheimer's disease: the NIALOAD/ NCRAD family study. Curr Alzheimer Res 2012;9(7):801–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Dergai O, Novokhatska O, Dergai M, et al. Intersectin 1 forms complexes with SGIP1 and Reps1 in clathrin‐coated pits. Biochem Biophys Res Commun 2010;402(2):408–413. [DOI] [PubMed] [Google Scholar]
- 57. Drecourt A, Babdor J, Dussiot M, et al. Impaired transferrin receptor palmitoylation and recycling in neurodegeneration with brain iron accumulation. Am J Hum Genet 2018;102(2):266–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bagyinszky E, Youn YC, An SS, Kim S. The genetics of Alzheimer's disease. Clin Interv Aging 2014;9:535–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lippa CF, Fujiwara H, Mann DM, et al. Lewy bodies contain altered alpha‐synuclein in brains of many familial Alzheimer's disease patients with mutations in presenilin and amyloid precursor protein genes. Am J Pathol 1998;153(5):1365–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Deaton CA, Johnson GVW. Presenilin 1 regulates membrane homeostatic pathways that are dysregulated in Alzheimer's disease. J Alzheimers Dis 2020;77(3):961–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gatto EM, Rojas GJ, Nemirovsky SI, et al. A novel mutation in PSEN1 (p.Arg41Ser) in an Argentinian woman with early onset Parkinsonism. Parkinsonism Relat Disord 2020;77:21–25. [DOI] [PubMed] [Google Scholar]
- 62. Ravanidis S, Bougea A, Papagiannakis N, et al. Validation of differentially expressed brain‐enriched microRNAs in the plasma of PD patients. Ann Clin Transl Neurol 2020;7(9):1594–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Abramzon YA, Fratta P, Traynor BJ, Chia R. The overlapping genetics of amyotrophic lateral sclerosis and frontotemporal dementia. Front Neurosci 2020;14:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Doxakis E. RNA binding proteins: a common denominator of neuronal function and dysfunction. Neurosci Bull 2014;30(4):610–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Egorova PA, Bezprozvanny IB. Molecular mechanisms and therapeutics for spinocerebellar ataxia type 2. Neurotherapeutics 2019;16(4):1050–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ravanidis S, Doxakis E. RNA‐binding proteins implicated in mitochondrial damage and mitophagy. Front Cell Dev Biol 2020;8:372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ravanidis S, Kattan FG, Doxakis E. Unraveling the pathways to neuronal homeostasis and disease: mechanistic insights into the role of RNA‐binding proteins and associated factors. Int J Mol Sci 2018;19(8):2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Salcedo‐Arellano MJ, Dufour B, McLennan Y, Martinez‐Cerdeno V, Hagerman R. Fragile X syndrome and associated disorders: clinical aspects and pathology. Neurobiol Dis 2020;136:104740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kim JM, Hong S, Kim GP, et al. Importance of low‐range CAG expansion and CAA interruption in SCA2 Parkinsonism. Arch Neurol 2007;64(10):1510–1518. [DOI] [PubMed] [Google Scholar]
- 70. Wang C, Xu Y, Feng X, et al. Linkage analysis and whole‐exome sequencing exclude extra mutations responsible for the parkinsonian phenotype of spinocerebellar ataxia‐2. Neurobiol Aging 2015;36(1):545 e541–545 e547. [DOI] [PubMed] [Google Scholar]
- 71. Gwinn‐Hardy K, Chen JY, Liu HC, et al. Spinocerebellar ataxia type 2 with parkinsonism in ethnic Chinese. Neurology 2000;55(6):800–805. [DOI] [PubMed] [Google Scholar]
- 72. Furtado S, Payami H, Lockhart PJ, et al. Profile of families with parkinsonism‐predominant spinocerebellar ataxia type 2 (SCA2). Mov Disord 2004;19(6):622–629. [DOI] [PubMed] [Google Scholar]
- 73. Martens LH, Zhang J, Barmada SJ, et al. Progranulin deficiency promotes neuroinflammation and neuron loss following toxin‐induced injury. J Clin Invest 2012;122(11):3955–3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Piscopo P, Grasso M, Fontana F, et al. Reduced miR‐659‐3p levels correlate with progranulin increase in hypoxic conditions: implications for frontotemporal dementia. Front Mol Neurosci 2016;9:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Rademakers R, Eriksen JL, Baker M, et al. Common variation in the miR‐659 binding‐site of GRN is a major risk factor for TDP43‐positive frontotemporal dementia. Hum Mol Genet 2008;17(23):3631–3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Xu J, Xilouri M, Bruban J, et al. Extracellular progranulin protects cortical neurons from toxic insults by activating survival signaling. Neurobiol Aging 2011;32(12):2326.e5–2326.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Yin F, Banerjee R, Thomas B, et al. Exaggerated inflammation, impaired host defense, and neuropathology in progranulin‐deficient mice. J Exp Med 2010;207(1):117–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Bucher M, Fanutza T, Mikhaylova M. Cytoskeletal makeup of the synapse: shaft versus spine. Cytoskeleton 2020;77(3–4):55–64. [DOI] [PubMed] [Google Scholar]
- 79. Pinho J, Marcut C, Fonseca R. Actin remodeling, the synaptic tag and the maintenance of synaptic plasticity. IUBMB Life 2020;72(4):577–589. [DOI] [PubMed] [Google Scholar]
- 80. Rai SN, Dilnashin H, Birla H, et al. The role of PI3K/Akt and ERK in neurodegenerative disorders. Neurotox Res 2019;35(3):775–795. [DOI] [PubMed] [Google Scholar]
- 81. Santo EE, Paik J. FOXO in neural cells and diseases of the nervous system. Curr Top Dev Biol 2018;127:105–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Junqueira SC, Centeno EGZ, Wilkinson KA, Cimarosti H. Post‐translational modifications of Parkinson's disease‐related proteins: phosphorylation, SUMOylation and ubiquitination. Biochim Biophys Acta Mol basis Dis 2019;1865(8):2001–2007. [DOI] [PubMed] [Google Scholar]
- 83. Pajarillo E, Rizor A, Lee J, Aschner M, Lee E. The role of posttranslational modifications of alpha‐synuclein and LRRK2 in Parkinson's disease: potential contributions of environmental factors. Biochim Biophys Acta Mol basis Dis 2019;1865(8):1992–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Calogero AM, Mazzetti S, Pezzoli G, Cappelletti G. Neuronal microtubules and proteins linked to Parkinson's disease: a relevant interaction? Biol Chem 2019;400(9):1099–1112. [DOI] [PubMed] [Google Scholar]
- 85. Pellegrini L, Wetzel A, Granno S, Heaton G, Harvey K. Back to the tubule: microtubule dynamics in Parkinson's disease. Cell Mol Life Sci 2017;74(3):409–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Swarm plots for the 42 circRNAs whose relative expression is not significantly altered in the PBMCs of idiopathic PD patients. Mean levels +/− SEM are included below each graph. Graphs demonstrate relative expression of log‐transformed data. Unpaired t‐test was used to determine the significance of differences between the two groups. *P < 0.05, **P < 0.01, ***P < 0.001.
Figure S2. KEGG and GOslim classifications of the circRNA‐miRNA target genes. (A) KEGG and (B) GOslim categories union of the RNA targets of the five miRNAs sequestered by three or more of the deregulated PD circRNAs (miR‐516b‐5p, miR‐526b‐5p, miR‐578, miR‐659‐3p, miR‐1197). They were prepared using the DIANA‐miRPath v3.0 interface using default values (P‐value threshold 0.05, microT‐CDS threshold 0.8).
Table S1. Primer sequences used for real‐time PCR.
Table S2. List of 39 circRNAs not detected in PBMCs.
Table S3. Basic characteristics and comparative PBMC circRNA expression in idiopathic PD patients and healthy controls.
CircRNA size and expression in the brain as well as host transcript expression in the body. Means with their respective standard deviation (std) for both groups are shown. Statistically significant differences in comparison to healthy controls (unpaired t‐test) are highlighted in grey. Multiple comparison analysis (adjusted P‐values) calculated according to Benjamini‐Hochberg false discovery rate method. *According to Rybak et al. ** According to genotype‐tissue expression portal.
Table S4. Correlation between relative circRNA expression and sex, age, age‐at‐onset, PD duration, HY, UPDRS and MMSE scores and LEDD of PD patients.
Significance level is set at P < 0.0005 after Bonferroni correction.
Table S5. KEGG and GOslim categories that are mostly deregulated in idiopathic PD. Gene union of the targets of the five miRNAs sequestered by three or more of the deregulated PD circRNAs (miR‐516b‐5p, miR‐526b‐5p, miR‐578, miR‐659‐3p, miR‐1197) in PD versus (A) KEGG and (B) GOslim categories created by the DIANA‐miRPath v3.0 interface using default values (P‐value threshold 0.05, microT‐CDS threshold 0.8).
Data Availability Statement
The datasets analyzed during this study are all available from the corresponding author on request.


