Summary
Most alpha‐gliadin genes of the Gli‐D2 locus on the D genome of hexaploid bread wheat (Triticum aestivum) encode for proteins with epitopes that can trigger coeliac disease (CD), and several contain a 33‐mer peptide with six partly overlapping copies of three epitopes, which is regarded as a remarkably potent T‐cell stimulator. To increase genetic diversity in the D genome, synthetic hexaploid wheat lines are being made by hybridising accessions of Triticum turgidum (AB genome) and Aegilops tauschii (the progenitor of the D genome). The diversity of alpha‐gliadins in A. tauschii has not been studied extensively. We analysed the alpha‐gliadin transcriptome of 51 A. tauschii accessions representative of the diversity in A. tauschii. We extracted RNA from developing seeds and performed 454 amplicon sequencing of the first part of the alpha‐gliadin genes. The expression profile of allelic variants of the alpha‐gliadins was different between accessions, and also between accessions of the Western and Eastern clades of A. tauschii. Generally, both clades expressed many allelic variants not found in bread wheat. In contrast to earlier studies, we detected the 33‐mer peptide in some A. tauschii accessions, indicating that it was introduced along with the D genome into bread wheat. In these accessions, transcripts with the 33‐mer peptide were present at lower frequencies than in bread wheat varieties. In most A. tauschii accessions, however, the alpha‐gliadins do not contain the epitope, and this may be exploited, through synthetic hexaploid wheats, to breed bread wheat varieties with fewer or no coeliac disease epitopes.
Keywords: D genome, gluten, alpha‐gliadin, re‐synthesised bread wheat, synthetic hexaploid wheat, SHW, T‐cell epitope, coeliac disease, Aegilops tauschii, Triticum aestivum
Significance Statement
We sequenced the expressed alpha‐gliadins in grains of Aegilops tauschii across its range. Some accessions contained the 33‐mer peptide with six overlapping coeliac disease epitopes, indicating that this was introduced into bread wheat from A. tauschii. We also found a large variation in the occurrence of coeliac disease epitopes, including accessions with small numbers of epitopes, suitable as basis for breeding bread wheat varieties with fewer or no coeliac disease epitopes.
INTRODUCTION
The genome of allohexaploid (2n = 6x = 42) bread wheat (Triticum aestivum) is composed of three subgenomes (A, B and D). It originated from hybridisation between allotetraploid Triticum turgidum (AB) and the diploid species Aegilops tauschii (D) around 8000 years ago (Nesbitt and Samuel, 1996). This hybridisation probably took place in agricultural fields, as bread wheat does not exist as a wild species. The genetic variation in the D genome of bread wheat is much lower than that present in the A and B genomes (Dubcovsky and Dvorak, 2007). This suggests that the hybridisation event involved only a small subset of A. tauschii genotypes, resulting in a strong genetic bottleneck (Dvorak et al., 1998). The notion of a hybridisation bottleneck is supported by four recent population genetics studies that show high levels of genetic diversity among genebank accessions of wild A. tauschii accessions sampled across the species range, based on the 10K Infinium single nucleotide polymorphism (SNP) array (Wang et al., 2013), on 15 D genome‐specific microsatellite markers (Jones et al., 2013), on an ultra‐high‐density 817K Affymetrix array (Winfield et al., 2016) and on 13 135 SNPs from genotyping‐by‐sequencing data (Singh et al., 2019). These studies confirmed the main division between Western (Iran, Turkey, Caucasus) and Eastern (Central Asia) A. tauschii accessions based on nuclear DNA (e.g. Lubbers et al., 1991; Dvorak et al., 1998; Pestsova et al., 2000), on chloroplast data (Dudnikov, 2012) or both (Mizuno et al., 2010). In addition, Mizuno et al. (2010), Wang et al. (2013), Jones et al. (2013) and Matsuoka et al. (2015) further distinguished several subgroups within the two regions. Interestingly, previous studies pinpoint a specific A. tauschii subgroup located south and southwest of the Caspian Sea in northern Iran as the main source of the bread wheat D genome. This subgroup was coded as 2E by Wang et al. (2013) (and S‐2 in Gill’s (2013) commentary on that paper) and as IIID/IIIE by Jones et al. (2013).
Wheat consumption may cause allergies and intolerances in some people. The prevalence of immunoglobulin E‐mediated allergy to wheat (and to cereals in general) is low (Gilissen et al., 2014), but 1–2% of the people can become intolerant to gluten proteins from wheat, rye and barley and may develop coeliac disease (CD), a chronic inflammation of the small intestine. This inflammation leads to a variety of symptoms and therefore most patients remain undiagnosed (Scherf et al., 2020). Coeliac disease is one of the best understood food intolerances with regard to human immunology and T‐cell specificity (Tye‐Din et al., 2010; Petersen et al., 2014, 2016; Jabri and Sollid, 2017; Sollid, 2017; Dahal‐Koirala et al., 2019; Scherf et al., 2020; Sollid et al., 2020). However, no treatment exists and the only way to prevent CD symptoms is to follow a gluten‐free (GF) diet, requiring complete exclusion of wheat, barley and rye. This is very difficult to adhere to, as gluten (especially from wheat) is added to a broad range of food products (Gallagher et al., 2004; Jouanin et al., 2018) due to its viscoelastic and binding properties (Atchison et al., 2010; Shewry, 2019).
Alpha‐gliadins, along with omega‐gliadins and gamma‐gliadins, are the most important source of immunogenic peptides triggering the T‐cell reaction in CD patients. Alpha‐gliadins are a multigene family, encoded by the Gli‐2 locus on the short arm of the group 6 chromosomes of bread wheat (Anderson et al., 2006), which includes intact as well as pseudogenes (van Herpen et al., 2006; Huo et al., 2019). Zhang et al. (2015) cloned and sequenced 23 alpha‐gliadins in Triticum urartu (A genome), of which 12 were intact genes. Huo et al. (2017) found 12 alpha‐gliadin genes clustered within a 550‐kb region, of which five were pseudogenes. Noma et al. (2016) sequenced alpha‐gliadins from the hexaploid variety Chinese Spring (CS), and found 90 genes, of which 50 were intact (16 on the A genome, 16 on the B genome and 18 on the D genome). Huo et al. (2018) re‐examined the CS set of alpha‐gliadin genes in the genome sequence of this variety (The International Wheat Genome Sequencing Consortium [IWGSC], 2018) using additional long‐read sequences, and annotated 47 alpha‐gliadin genes (26 on the A genome, 11 on the B genome, 10 on the D genome), of which 28 were expressed. Noma et al. (2019) used gene‐specific primers and found that 26 alpha‐gliadins were expressed in the developing endosperm of CS. Note that the number of genes varies between wheat varieties, and that expression levels vary among genes and between varieties (Shewry and Lookhart, 2003; Salentijn et al., 2009; Noma et al., 2019; Jouanin et al., 2020a).
A large amount of genetic variation exists for the presence of T‐cell‐stimulatory sequences among wheat species and accessions (Spaenij‐Dekking et al., 2005; Molberg et al., 2005; Van den Broeck et al., 2010a; Shewry and Tatham, 2016). Some alpha‐gliadin genes encode for proteins which contain more CD epitopes than others. The observed variation among the genes is genome specific, and the alpha‐gliadins from the D genome have the highest CD‐immunogenic potential (van Herpen et al., 2006; Salentijn et al., 2009; Mitea et al., 2010; Jouanin et al., 2019) as most of these genes contain at least one copy of each of three different DQ2 epitopes, of which glia‐α1 is the one to which most CD patients react. Several Gli‐D2 gliadins also contain a 33‐mer peptide with six partly overlapping copies of the T‐cell epitopes DQ2.5‐Glia‐α1a, DQ2.5‐Glia‐α1b and DQ2.5‐Glia‐α2; this is regarded as a remarkably potent T‐cell stimulator (Qiao et al., 2004). In addition, many of the alpha‐gliadins contain the p31‐43 epitope, which is involved in inducing the innate immune response that initiates the development of CD (Maiuri et al., 1996, 2003). This epitope is present in alpha‐gliadins from all three subgenomes. Screening studies suggest that there are no modern bread wheats that do not contain several alpha‐gliadin proteins encoded by the D genome locus with several CD epitopes each (Van den Broeck et al., 2010a,b; Jouanin et al., 2018).
As a means to introduce new genetic variation into the D genome of bread wheat, novel hexaploid wheats are being synthesised by hybridising T. turgidum subsp. durum with A. tauschii accessions, followed by chemical chromosome doubling. These so‐called re‐synthesised bread wheats or synthetic hexaploid wheats (SHWs) are made with the goal of introducing new functional trait diversity into modern germplasm (Kishii, 2019), notably disease resistance genes (Mujeeb‐Kazi et al., 1996; Das et al., 2016; Szabo‐Hever et al., 2018; Kishii et al., 2019; Mohler et al., 2020) but also, for example, a higher yield (Hao et al., 2019). The International Maize and Wheat Improvement Center (CIMMYT), Texcoco, Mexico has been developing many synthetic hexaploid wheats since the 1960s (Gordon et al., 2019), but several other programmes exist, for example at NIAB, Cambridge, UK. If these re‐synthesised bread wheats are made using A. tauschii accessions with a Gli‐D2 locus containing low‐immunogenic alpha‐gliadins, the resulting hexaploid would be safer for CD patients (Smulders et al., 2015). To this end, we need to identify A. tauschii accessions that contain gliadins with fewer CD epitopes.
Here we have screened the CD‐immunogenic potential of a diverse panel of A. tauschii accessions by deep sequencing of the N‐terminal region of alpha‐gliadin transcripts, which contains the repetitive domain with CD epitopes as described by Salentijn et al. (2013). Using the frequency of reads as a proxy for the level of gene expression, we estimated the occurrence of CD epitopes and the relative toxicity of A. tauschii accessions for CD patients. We also determined the occurrence of specific variants, including the 33‐mer, and whether safe gene variants exist. These results were related to the geographical region and genetic cluster to which the accessions belonged to determine the relative CD immunogenicity of subgroups of A. tauschii accessions and their potential for breeding safer wheat cultivars.
RESULTS
454 RNA‐amplicon sequencing
Sequencing of amplified transcript fragments, representing variants of the first variable domain of alpha‐gliadin that were expressed in the wheat grain endosperm, was performed from the 3′‐end, directly entering the first repetitive domain, instead of the 5′‐end (i.e. entering the signal peptide first) as done in Salentijn et al. (2013) in tetraploid durum wheat. This resulted, at similar sequence depth, in double the number of usable reads for corresponding samples than obtained by Salentijn et al. (2013), with qualitatively similar cDNA sequences (Table S1 in the online Supporting Information).
Diversity of unique alpha‐gliadin peptide variants
The reads were organised into ‘unique sequence clusters’ containing the cDNA sequences that were 100% similar and with a sequence depth of >20 reads. By translating the nucleotide sequences of these unique sequence clusters, ‘unique peptide variants’ (UPVs) were deduced (Table S2). Most of these peptide variants were represented by several different unique nucleotide sequences, so they may originate from different genes. As our analysis focused on the peptide variants and their characteristics, our results may therefore underestimate the number of different genes present in the genome and being expressed. Moreover, alpha‐gliadin genes that have an identical coding sequence may have different promoter regions, as found by Noma et al. (2016) who cloned all alpha‐gliadin genes from the hexaploid cultivar CS.
In developing grains of each accession, several alpha‐gliadin peptide variants were expressed (Table S3). Using a lower threshold of 0.1% of the reads for all peptide variants in a sample, an average of 66 variants were expressed in hexaploid bread wheat lines compared with 36 in the grains of diploid A. tauschii; however, the average number of abundant peptide variants (>5% of the total number of reads) was almost the same at six (ranging from five to seven) and five (ranging from three to seven). The number of peptide variants in the tetraploid T. turgidum accession Primadur was remarkably low at 14 variants (>0.1%), with four being abundant (>5%). All data are in Table S4.
In this study a total of 349 new alpha‐gliadin peptide variants were found, while 38 had been described before by Salentijn et al. (2013) in tetraploids (Table S3). Of these 387 peptide variants, 27 were present in both T. aestivum and A. tauschii, while 211 and 149 were unique to T. aestivum and A. tauschii, respectively. Salentijn et al. (2013) reported 171 unique peptide variants from 61 different T. turgidum accessions. Because of the sequencing technology used, these novel peptide variants included 62 short variants of only 50 amino acids in length. These were devoid of the immunodominant DQ2.5‐Glia‐α1a, DQ2.5‐Glia‐α1b, DQ2.5‐Glia‐α2 T‐cell epitopes, but some of them did contain the DQ2.5‐Glia‐α3 T‐cell epitope and/or the innate peptide p31‐43.
Patterns of expressed gene variants
Hierarchical cluster analysis of the abundance of alpha‐gliadin peptide variants across the T. aestivum and A. tauschii accessions identified several groups of accessions with a similar peptide variant expression pattern, although some A. tauschii accessions had a unique expression pattern (Figure 1). The main difference in the pattern of gene expression was between the Eastern (group II) and Western (group III) A. tauschii accessions. The pattern of gene expression was quite similar for the group II accessions, which is consistent with this group having a lower level of genetic diversity (Jones et al., 2013; Wang et al., 2013).
Figure 1.
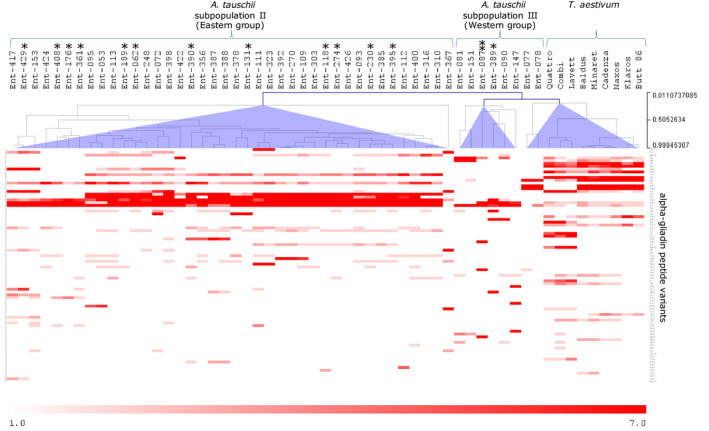
Alpha‐gliadin peptide variant expression profiles in grains of Aegilops tauschii (Ent lines) and Triticum aestivum accessions.
The peptide variants were differentially abundant in the accessions. The accessions were clustered based on their peptide variant expression profile using hierarchical clustering with Spearman rank correlation with average linking. Purple triangles highlight the main clusters. Ent lines marked with * and ** contain peptide variants with the 33‐mer. *Variant J234. **Variant J2 and J29. Variant J2 was also found in all of the nine bread wheat accessions.
The pattern of peptide variant expression in A. tauschii lines from subpopulations IIID and IIIE, and especially of the subpopulation IIID lines Ent‐077 and Ent‐078, was most similar to that observed in the bread wheat lines. This is in accordance with the suggestion of Jones et al. (2013) that, based on microsatellite data, A. tauschii from subpopulation III and, more specifically, subpopulations D and E, has the closest relationship to bread wheat.
Epitopes present in the alpha‐gliadin peptides
The alpha‐gliadin peptide variants cover the region of alpha‐gliadins with the p31‐43 epitope responsible for the innate immune response, and the region with immunodominant DQ2.5‐Glia‐α1a, DQ2.5‐Glia‐α1b, DQ2.5‐Glia‐α2 and DQ2.5‐Glia‐α3 T cell epitopes. The peptide variants differ in epitope makeup and therefore in putative toxicity. All A. tauschii accessions contain several alpha‐gliadin peptide variants with multiple sets of DQ2.5‐Glia‐α T‐cell epitopes, but some accessions [Ent‐095 (group IIB), Ent‐422 (group IIC), Ent‐151 (group IIID), Ent‐081 (group IIID)] also contain a significant number (8–10%) of peptide variants that lack any of the canonical epitopes (peptide variant C4 and J9) (Table S5). Furthermore, several accessions contain peptide variants with only the DQ2.5‐Glia‐α3 T‐cell epitope, which is regarded as the least toxic of the three (Anderson et al., 2006). In one such accession, Ent‐389 (group IIB), 40% of the peptide variants only contain the DQ2.5‐Glia‐α3 T‐cell epitope (variant J13, J17 and J28 with relative abundances of 17%, 15% and 8%, respectively).
Occurrence of the 33‐mer
The 33‐mer peptide with six overlapping CD epitopes, which is a unique feature of D genome alpha‐gliadins in bread wheat, was not found in A. tauschii in earlier studies (Ozuna et al., 2015; Huo et al., 2018) and these authors presumed that it may have developed after the hybridisation event. In our study, we ensured that we included A. tauschii accessions covering the whole distribution area of the species, i.e. the Eastern as well as the Western areas, as well as all subtypes within those regions. In our data we did find the 33‐mer in some of the A. tauschii accessions sequenced in this study, always as a small fraction of the transcripts. The 33‐mer was present in intact form in three alpha‐gliadin peptide variants: J2, J29 and J234. Variant J234 was found in 13 different A. tauschii accessions (12 of subpopulation II and one of subpopulation III; marked with * in Figure 1), always at low frequency (<0.5%). Variants J2 and J29 were only found in A. tauschii accession Ent‐087 (marked with ** in Figure 1) (subpopulation IIID) and were expressed at 0.05% and 6.8% of all transcripts in this accession, respectively. J2 was the only one that was found in both A. tauschii accession Ent‐087 and in all of the nine bread wheat accessions, in which it was more abundantly expressed (5–16% of all transcripts). In these nine hexaploids, ten other peptide variants (J60, J76, J89, J99, J208, J233, J234, J252, J273, J282) also contained the 33‐mer, but these were all present at low frequencies (maximum 0.7%). We have not detected these variants in our set of A. tauschii germplasm, which may be due to the limited number of A. tauschii accessions included that are from IIID.
Estimated CD epitope load
The alpha‐gliadin peptide variants differ in CD epitope composition (Table S3). The frequency of DQ2.5‐Glia‐α1a, DQ2.5‐Glia‐α1b, DQ2.5‐Glia‐α2 and DQ2.5‐Glia‐α3 epitope sequences in the total transcripts of different accessions was estimated by (i) determining the epitope composition for each individual alpha‐gliadin peptide variant and (ii) taking the frequency of the individual peptide variants into account. Epitope variants of DQ2.5‐Glia‐α1a (PFPQLQLPF and PFPHLQLPY) and DQ2.5‐Glia‐α2 (FLPQLPYPQ) for which toxicity has been demonstrated (Mitea et al., 2010) were included in the estimation.
In the alpha‐gliadin peptide variants of all A. tauschii and T. aestivum accessions a significant frequency of DQ2.5‐Glia‐α1a and DQ2.5‐Glia‐α3 epitopes was found, but all T. aestivum accessions had a lower frequency of these epitopes than most of the A. tauschii accessions (Figures 2 and S1).
Figure 2.
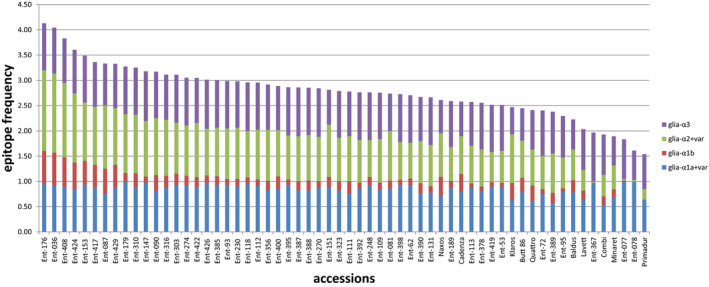
Accumulated frequency of different DQ2.5‐Glia‐alpha epitopes.
Accumulated frequency of different DQ2.5‐Glia‐alpha epitopes in transcripts of different accessions: glia‐ α1a+var, DQ2.5‐Glia‐α1a (PFPQPQLPY) and its toxic variants DQ2.5‐Glia‐α1‐varT1 and‐varT2 (PFPQPQLPF and PFPHPQLPY); glia‐ α1b, DQ2.5‐Glia‐α1b (PYPQPQLPY); glia‐α2+var, DQ2.5‐Glia‐α2 (PQPQLPYPQ) and its toxic variant DQ2.5‐Glia‐α2‐varT1 (FLPQLPYPQ); glia‐α3, DQ2.5‐Glia‐α3 (FRPQQPYPQ).
Epitope DQ2.5‐Glia‐α1b was far less abundant and showed more variation in frequency among all accessions and was (almost) absent in a few A. tauschii accessions (Figure S1). DQ2.5‐Glia‐α2 frequencies were most variable in the alpha‐gliadin peptides and were high in some A. tauschii accessions and very low in some others.
The lowest total frequency of DQ2.5‐Glia‐α epitopes was found in A. tauschii accessions Ent‐077, Ent‐078 and Ent‐367 (Figure S1).
Occurrence of the p31‐43 peptide
The gliadin peptide p31‐43, which induces an innate immunity response in CD patients, is present in 32% of the peptide variants and its presence shows no correlation (r = 0.42) with the presence of DQ2.5‐Glia‐α epitopes. The A. tauschii accession Ent‐367 appears to be completely devoid of gliadin peptide p31‐43, whereas in Ent‐77 and Ent‐78 only a few peptide fragments contain it (Figure 3). These accessions also had a low frequency of DQ2.5‐Glia‐α epitopes.
Figure 3.
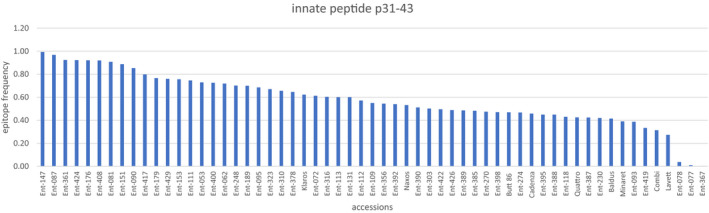
Relative frequencies of alpha‐gliadin peptide variants that contain the p31‐43 peptide in Aegilops tauschii and Triticum aestivum accessions.
DISCUSSION
Deep amplicon sequencing of the first repetitive domain of alpha‐gliadin gene transcripts enables prediction of the (relative) CD toxicity of the alpha‐gliadin protein fraction in wheat. Based on the results reported here we analysed the translated protein expression patterns of A. tauschii. This indicated that the expression profile of the alpha‐gliadins was different between accessions from Western and Eastern clades of A. tauschii. Within each of these groups the expression profile varied among accessions. Both clades expressed many allelic variants that are not found in bread wheat, confirming and extending the results of Yan et al. (2003). In particular, the alpha‐gliadin peptide variants J11, C1 and C2 are abundantly present in several A. tauschii lines (up to 35%, 17% and 18%, respectively) but are not found in bread wheat.
Alpha‐gliadins containing the 33‐mer peptide are regarded as the most CD‐toxic variants in bread wheat. This αGlia‐33‐mer fragment is naturally formed in the human gastrointestinal tract by digestion with gastric and pancreatic enzymes, it binds well to DQ2 after deamidation by tissue transglutaminase (TG2), and it is recognised much more effectively by intestinal T‐cell lines than shorter peptides covering only the DQ2‐α‐I, ‐α‐II or ‐α‐III epitopes (Shan et al., 2002). The 33‐mer is present in the alpha‐gliadins of the D genome of bread wheat (chromosome 6D, locus Gli‐2D) (Molberg et al., 2005; Spaenij‐Dekking et al., 2005; Van Herpen et al., 2006; Mitea et al., 2010; Salentijn et al., 2013). Ozuna et al. (2015) and we therefore expected it to be present in A. tauschii accessions, but we did not find it in 22 alpha‐gliadin genes from three A. tauschii accessions and concluded that the 33‐mer may have evolved in the hexaploid. We screened a wide set of accessions covering the complete distribution area of the species, and found the 33‐mer in only a few of the A. tauschii accessions. Just one of the A. tauschii accessions included in our study (Ent‐087) expressed the 33‐mer at a significant frequency. This accession was also the only A. tauschii accession in our study that shared an identical peptide variant containing a 33‐mer with the hexaploid bread wheat accessions in this study. Accession Ent‐087 is from the IIID subpopulation, which is from the region close to the Caspian Sea that is considered to be where the species hybridisation that led to bread wheat took place (Dvorak et al., 1998). This is consistent with the notion that the D genome in bread wheat has a narrow genetic basis.
Overall, sufficient genetic variation is present in the alpha‐gliadins in A. tauschii to try to identify A. tauschii accessions with a reduced CD epitope load. When we screened our sequences for variation in CD epitopes, no A. tauschii accessions were found without the DQ2.5‐Glia‐α T cell epitope but some accessions do express alpha‐gliadin proteins that have no or a few epitopes. Almost all accessions express alpha‐gliadin peptide variants that contain only the DQ2.5‐Glia‐α3 T cell epitope, which is regarded as least toxic of the three DQ2.5 alpha‐gliadin epitopes. This T‐cell epitope has been shown to make only a minor contribution to the gluten‐induced T‐cell response in HLA‐DQ2+ CD (Anderson et al., 2006). These accessions can be used to generate less toxic synthetic hexaploid wheat (SHW) lines, by hybridising these accessions with tetraploid wheat accessions, which would form a good basis for of a breeding programme to generate hypoimmunogenic or even CD‐safe bread wheat (Jouanin et al., 2018, 2020b). The tetraploid wheat accessions may be selected based on genetic characteristics or agronomic performance.
We have screened A. tauschii accessions as we believe that finding differences among them is more efficient than screening SHWs. First, some A. tauschii accessions tend to be used frequently for SHWs and others not, because they have interesting phenotypes unrelated to gluten, such as disease resistance. Second, another potential problem is that the expression of gliadins (particularly novel variants) may be masked or changed in the hexaploid background, and this may depend on the genotype with which it is hybridised, so that a broad screening becomes more complicated when using SHW lines. This is why recent gene identification from wild relatives has focused directly on the progenitor species [e.g. in the cloning of resistance genes using R gene enrichment sequencing (AgRenSeq) in A. tauschii by Arora et al. (2019)].
Several of the studied A. tauschii accessions have already been used at NIAB for production of synthetic hexaploid wheat lines (Table S6). In such a programme, targeted gene editing with CRISPR/Cas9 could be included later on to edit the remaining epitopes or remove some alpha‐gliadins (Smulders et al., 2015; Jouanin et al., 2019), as A. tauschii cannot easily be transformed or regenerated. Alternatively, one could apply Tilling to improve a specific A. tauschii line (Rawat et al., 2018).
Whether gene expression of alpha‐gliadins is affected by polyploidisation cannot be deduced from modern bread wheat varieties, as they are the result of 10 000 years of selection and breeding after the allopolyploidisation event and the original D genome donor is no longer available. It can be studied, though, by comparing the gene expression of developing grains of particular A. tauschii accessions with those of the grains of the synthetic hexaploids produced by hybridising these accessions to a few tetraploid lines, when grown side by side. For the use of specific A. tauschii accessions for introgression of less toxic variants it will be imperative to compare their gene expression with those at the allohexaploid level. Such synthetic hexaploids are currently being made.
EXPERIMENTAL PROCEDURES
Plant material
A subset of 51 different A. tauschii accessions used by Jones et al. (2013) in their diversity study and selected to be representative of the D genome diversity in A. tauschii, was sampled at NIAB. We adapted the subgroup codes of Jones et al. (2013), who used group II for the Eastern clade of A. tauschii accessions [1E and 1W in Wang et al. (2013), L1 in Singh et al. (2019)] and group III for the Western clade [2E and 2W in Wang et al. (2013), L2 in Singh et al. (2019)]. Furthermore, a panel of nine different bread wheat (T. aestivum) varieties and one durum wheat (T. turgidum) variety was included (Table S7). For transcript sequencing, immature spikes were harvested, frozen in liquid nitrogen and stored at −80°C until RNA extraction.
Extraction of RNA and cDNA synthesis
For each genotype, total RNA was extracted from wheat endosperm of a pool of five immature grains from a single spike at the milk to soft dough ripening stage. For some samples dry, mature grains were also used for total RNA isolation. The RNA was extracted according to Salentijn et al. (2013) but with a simple improvement to the RNA extraction protocol, consisting of removal of starch and polysaccharides prior to TRIzol extraction (Li and Trick, 2005). For this, 50–100 mg of fine‐ground grains was suspended in RNA extraction buffer (Li and Trick, 2005) and subjected to a chloroform/phenol extraction. The supernatant was then mixed with TRIzol and chloroform, and after TRIzol extraction the supernatant was further purified using the Qiagen RNeasy Plant Mini Kit (http://www.qiagen.com/). Total RNA was finally eluted from the RNeasy spin column using 50 µl of water. The RNA quality was checked using agarose gel electrophoresis and total RNA quantity was determined spectrophotometrically (NanoDrop ND1000, NanoDrop Products, http://www.thermofisher.com/). Prior to reverse transcription, traces of genomic DNA were removed by a DNaseI treatment using the Invitrogen DNase kit (http://www.thermofisher.com/). Subsequent cDNA synthesis and PCR amplification of alpha‐gliadin transcript fragments and pooling of PCR samples was performed exactly as described by Salentijn et al. (2013) using the same gene‐specific primers but with the unique 10 bp ID sequences included in the reverse primers.
454 sequencing and data analysis
Roche/454 amplicon sequencing and data analysis were performed essentially as described before (Salentijn et al., 2013). For this the signal peptide and the repetitive domain of alpha‐gliadin transcripts were amplified and sequenced. Sequencing was now started from the 3′‐end, directly entering the repetitive domain, instead of starting from the 5′‐end (signal peptide) as performed by Salentijn et al. (2013). On average 2184 reads per sample were obtained (minimum 199 reads, median 1632 reads, maximum 9409 reads). For some samples that yielded only a few hundred sequence reads this may give a bias towards the most abundant variants only. This was also found when we compared the abundance of gliadin transcripts in samples originating from dry grains with that of immature grains from the same genotype. Because RNA isolated from dry seed was of poorer quality, lower‐quality reads were obtained which represented the most abundant sequences that were found in transcripts from immature seeds (Table S8). For some samples replicate RNA extractions and sequencing from seeds of the same genotype were performed to show the reproducibility of the method (Table S8). After pre‐processing (as described in Salentijn et al., 2013) the transcript sequences were clustered using USEARCH v.4.0. First, all sequences were clustered at 100% homology, clusters were sorted according to the number of reads and for all clusters with more than 20 reads (a total of 280 571 sequences) the reads were then clustered at 99.5% homology in order to combine sequences from a single gene while allowing for typical 454 sequencing errors. The output of the pipeline consisted of the consensus cDNA sequences of these clusters (572 clusters in total), the deduced amino acid sequences (unique peptide fragments), the number of 454 reads per cluster per sample and the number of DQ2.5 CD epitopes in their non‐deamidated forms [DQ2.5‐Glia‐α1a (PFPQPQLPY) and its toxic variants DQ2.5‐Glia‐α1‐varT1 and‐varT2 (PFPQPQLPF and PFPHPQLPY), DQ2.5‐Glia‐α1b (PYPQPQLPY), DQ2.5‐Glia‐α2 (PQPQLPYPQ) and its toxic variant DQ2.5‐Glia‐α2‐varT1 (FLPQLPYPQ) and DQ2.5‐Glia‐α3 (FRPQQPYPQ)] and presence/absence of the innate peptide p31‐43 and the 33‐mer peptide. Several transcripts contain internal stop codons and can be regarded as pseudogenes. They were always present at low transcript abundance. If the stop codon was downstream of the DQ2.5 epitopes, these pseudogene transcripts were included in the epitope calling. Peptides in a sample were called based on the percentage of reads for that peptide relative to the total number of reads for all peptide variants in a sample. The call thresholds were 0.1% for rare variants and 5% for abundant transcripts.
Author contributions
NG, LJWJG and MJMS initiated the research. NG and ARB provided the material. EMJS, JGS and SVG extracted the RNA and produced the libraries. JGS, EMJS, CC and DGE analysed the sequence data. CC performed the epitope predictions. JGS and MJMS wrote the manuscript, with revisions from the others. All authors read and approved the final version.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
Table S1. Old versus new 454 approach.
Table S2. Clustered transcript sequence data and processing. Clustered transcript nucleotide sequences were translated into proteins and unique peptide variants were grouped.
Table S3. Unique peptide variants with epitope load.
Table S4. Unique peptide variants variant frequencies for each accession.
Table S5. Epitope frequency for each accession.
Table S6. List of Aegilops tauschii accessions captured in re‐synthesised hexaploid wheat (SHW). All SHW are available via the NIAB Breeder’s Toolkit (http://www.niab.com/pages/id/419/Breeders__Toolkit).
Table S7. Wheat accessions used. A list of the Aegilops tauschii lines studied (annotation from Jones et al., 2013) and the bread wheat and durum wheat lines included for comparison.
Table S8. Reproducibility of 454 sequencing.
Figure S1. Frequency of different DQ2.5‐Glia‐alpha epitopes in transcripts of different accessions.
ACKNOWLEDGEMENTS
This research was supported in part by the Coeliac Disease Consortium, an Innovative Cluster approved by the Netherlands Genomics Initiative and partially funded by the Dutch Government (BSIK03009), by the Ministry of Economic Affairs (KB15‐001‐007) and by the EFRO project ‘Nieuwe detectiemethoden voor coeliakie en coeliakie‐inducerende gluten in voeding’ (2011‐018974). The BBSRC ‘Designing Future Wheat’ (BB/P016855/1) supports ARB in the creation of synthetic wheat from diverse A. tauschii accessions.
Data Availability Statement
All relevant data on the peptides and epitopes can be found within the manuscript and its supporting materials.
REFERENCES
- Anderson, R.P. , van Heel, D.A. , Tye‐Din, J.A. , Jewell, D.P. and Hill, A.V. (2006) Antagonists and non‐toxic variants of the dominant wheat gliadin T cell epitope in coeliac disease. Gut, 55, 485–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora, S. , Steuernagel, B. , Gaurav, K. et al. (2019) Resistance gene cloning from a wild crop relative by sequence capture and association genetics. Nat. Biotechnol. 37, 139–143. [DOI] [PubMed] [Google Scholar]
- Dahal‐Koirala, S. , Ciacchi, L. , Petersen, J. et al. (2019) Discriminative T‐cell receptor recognition of highly homologous HLA‐DQ2–bound gluten epitopes. J. Biol. Chem. 294, 941–952. 10.1074/jbc.RA118.005736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, M.K. , Bai, G. , Mujeeb‐Kazi, A. and Rajaram, S. (2016) Genetic diversity among synthetic hexaploid wheat accessions (Triticum aestivum) with resistance to several fungal diseases. Genet. Resour. Crop Evol. 63, 1285–1296. [Google Scholar]
- Dubcovsky, J. and Dvorak, J. (2007) Genome plasticity a key factor in the success of polyploid wheat under domestication. Science, 316, 1862–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudnikov, A.J. (2012) Chloroplast DNA non‐coding sequences variation in Aegilops tauschii Coss.: evolutionary history of the species. Genet. Resour. Crop Evol. 59, 683–699. [Google Scholar]
- Dvorak, J. , Luo, M.C. , Yang, Z.L. and Zhang, H.B. (1998) The structure of the Aegilops tauschii genepool and the evolution of hexaploid wheat. Theor. Appl. Genet. 97, 657–670. [Google Scholar]
- Gallagher, E. , Gormley, T.R. and Arendt, E.K. (2004) Recent advances in the formulation of gluten‐free cereal‐based products. Trends Food Sci. Tech. 15, 143–152. [Google Scholar]
- Gill, B.S. (2013) SNPing Aegilops tauschii genetic diversity and the birthplace of bread wheat. New Phytol. 198, 641–642. [DOI] [PubMed] [Google Scholar]
- Gilissen, L.J.W.J. , van der Meer, I.M. and Smulders, M.J.M. (2014) Reducing the incidence of allergy and intolerance to cereals. J. Cereal Sci. 59, 337–353. [Google Scholar]
- Gordon, E. , Kaviani, M. , Kagale, S. , Payne, T. and Navabi, A. (2019) Genetic diversity and population structure of synthetic hexaploid‐derived wheat (Triticum aestivum L.) accessions. Genet. Resour. Crop Evol. 66, 335–348. [Google Scholar]
- Hao, M. , Zhang, L. , Zhao, L. et al. (2019) A breeding strategy targeting the secondary gene pool of bread wheat: introgression from a synthetic hexaploid wheat. Theor. Appl. Genet. 132, 2285–2294. [DOI] [PubMed] [Google Scholar]
- Huo, N. , Dong, L. , Zhang, S. et al. (2017) New insights into structural organization and gene duplication in a 1.75‐Mb genomic region harboring the α‐gliadin gene family in Aegilops tauschii, the source of wheat D genome. Plant J. 92, 571–583. [DOI] [PubMed] [Google Scholar]
- Huo, N. , Zhu, T. , Altenbach, S. , Dong, L. , Wang, Y. , Mohr, T. , Liu, Z. , Dvorak, J. , Luo, M.C. and Gu, Y.Q. (2018) Dynamic evolution of alpha‐gliadin prolamin gene family in homeologous genomes of hexaploid wheat. Sci. Rep. 8, 5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo, N. , Zhu, T. , Zhang, S. et al. (2019) Rapid evolution of α‐gliadin gene family revealed by analyzing Gli‐2 locus regions of wild emmer wheat. Funct. Integr. Genomics, 19, 993–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabri, B. and Sollid, L.M. (2017) T cells in celiac disease. J. Immunol. 198, 3005–3014. 10.4049/jimmunol.1601693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, H. , Gosman, N. , Horsnell, R. et al. (2013) Strategy for exploiting exotic germplasm using genetic, morphological, and environmental diversity: the Aegilops tauschii Coss. example. Theor. Appl. Genet. 126, 1793–1808. [DOI] [PubMed] [Google Scholar]
- Jouanin, A. , Gilissen, L.J.W.J. , Boyd, L.A. et al. (2018) Food processing and breeding strategies for coeliac‐safe and healthy wheat products. Food Res. Int. 110, 11–21. [DOI] [PubMed] [Google Scholar]
- Jouanin, A. , Schaart, J.G. , Boyd, L.A. , Cockram, J. , Leigh, F.J. , Bates, R. , Wallington, E.J. , Visser, R.G.F. and Smulders, M.J.M. (2019) Outlook for coeliac disease patients: towards bread wheat with hypoimmunogenic gluten by gene editing of α‐ and γ‐gliadin gene families. BMC Plant Biol. 19, 333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouanin, A. , Tenorio‐Berrio, R. , Schaart, J.G. , Leigh, F. , Visser, R.G.F. and Smulders, M.J.M. (2020a) Optimization of droplet digital PCR for determining copy number variation of α‐gliadin genes in mutant and gene‐edited polyploid bread wheat. J. Cereal Sci. 92, 102903. [Google Scholar]
- Jouanin, A. , Gilissen, L.J.W.J. , Schaart, J.G. et al. (2020b) CRISPR/Cas9 gene editing of gluten in wheat to reduce gluten content and exposure – reviewing methods to screen for coeliac safety. Front. Nutr. 7, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishii, M. (2019) An update of recent use of Aegilops species in wheat breeding. Front. Plant Sci. 10, 585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishii, M. , Huerta, J. , Tsujimoto, H. et al. (2019) Stripe rust resistance in wild wheat Aegilops tauschii Coss.: genetic structure and inheritance in synthetic allohexaploid Triticumwheat lines. Genet. Resour. Crop Evol. 66, 909–920. [Google Scholar]
- Li, Z. and Trick, H.N. (2005) Rapid method for high‐quality RNA isolation from seed endosperm containing high levels of starch. Biotechniques, 38(6), 872–876. [DOI] [PubMed] [Google Scholar]
- Lubbers, E.L. , Gill, K.S. , Cox, T.S. and Gill, B.S. (1991) Variation of molecular markers among geographically diverse accessions of Triticum tauschii . Genome, 34, 354–361. [Google Scholar]
- Maiuri, L. , Ciacci, C. , Ricciardelli, I. , Vacca, L. , Raia, V. , Auricchio, S. , Picard, J. , Osman, M. , Quaratino, S. and Londei, M. (2003) Association between innate response to gliadin and activation of pathogenic T cells in coeliac disease. Lancet, 362(9377), 30–37. [DOI] [PubMed] [Google Scholar]
- Maiuri, L. , Troncone, R. , Mayer, M. , Coletta, S. , Picarelli, A. , De Vincenzi, M. , Pavone, V. and Auricchio, S. (1996) In vitro activities of A‐gliadin‐related synthetic peptides: damaging effect on the atrophic coeliac mucosa and activation of mucosal immune response in the treated coeliac mucosa. Scand. J. Gastroenterol. 31, 247–253. [DOI] [PubMed] [Google Scholar]
- Matsuoka, Y. , Takumi, S. and Kawahara, T. (2015) Intraspecific lineage divergence and its association with reproductive trait change during species range expansion in central Eurasian wild wheat Aegilops tauschii Coss. (Poaceae). BMC Evol. Biol. 15, 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitea, C. , Salentijn, E.M. , van Veelen, P. et al. (2010) A universal approach to eliminate antigenic properties of alpha‐gliadin peptides in celiac disease. PLoS One, 5, e15637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno, N. , Yamasaki, M. , Matsuoka, Y. , Kawahara, T. and Takumi, S. (2010) Population structure of wild wheat D‐genome progenitor Aegilops tauschii Coss.: implications for intraspecific lineage diversification and evolution of common wheat. Mol. Ecol. 19, 999–1013. [DOI] [PubMed] [Google Scholar]
- Mohler, V. , Schmolke, M. , Zeller, F.J. et al. (2020) Genetic analysis of Aegilops tauschii‐derived seedling resistance to leaf rust in synthetic hexaploid wheat. J. Appl. Genet. 61, 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molberg, O. , Uhlen, A.K. , Jensen, T. , Flaete, N.S. , Fleckenstein, B. , Arentz‐Hansen, H. , Raki, M. , Lundin, K.E. and Sollid, L.M. (2005) Mapping of gluten T‐cell epitopes in the bread wheat ancestors: implications for celiac disease. Gastroenterology, 128, 393–401. [DOI] [PubMed] [Google Scholar]
- Mujeeb‐Kazi, A. , Rosas, V. and Roldan, S. (1996) Conservation of the genetic variation of Triticum tauschii (Coss.) Schmalh. (Aegilops squarrosa auct. non L.) in synthetic hexaploid wheats (T. turgidum L. s.lat. × T. tauschii; 2n = 6x = 42, AABBDD) and its potential utilization for wheat improvement. Genet. Resour. Crop Evol. 43, 129–134. [Google Scholar]
- Nesbitt, M. and Samuel, D. (1996) From staple crop to extinction? The archaeology and history of hulled wheats, pp. 41–100 in Proceedings of the First International Workshop on Hulled Wheats edited by S. Padulosi et al. International Plant Genetic Resources Institute. Rome.
- Noma, S. , Hayakawa, K. , Abe, C. , Suzuki, S. and Kawaura, K. (2019) Contribution of α‐gliadin alleles to the extensibility of flour dough in Japanese wheat cultivars. J. Cereal Sci. 86, 15–21. 10.1016/j.jcs.2018.12.017 [DOI] [Google Scholar]
- Noma, S. , Kawaura, K. , Hayakawa, K. , Abe, C. , Tsuge, N. and Ogihara, Y. (2016) Comprehensive molecular characterization of the alpha/beta‐gliadin multigene family in hexaploid wheat. Mol. Genet. Genomics 291, 65–77. [DOI] [PubMed] [Google Scholar]
- Ozuna, C.V. , Iehisa, J.C. , Gimenez, M.J. , Alvarez, J.B. , Sousa, C. and Barro, F. (2015) Diversification of the celiac disease alpha‐gliadin complex in wheat: a 33‐mer peptide with six overlapping epitopes, evolved following polyploidization. Plant J. 82, 794–805. [DOI] [PubMed] [Google Scholar]
- Pestsova, E. , Korzun, V. , Goncharov, N.P. , Hammer, K. , Ganal, M.W. and Roder, M.S. (2000) Microsatellite analysis of Aegilops tauschii germplasm. Theor. Appl. Genet. 101, 100–106. [Google Scholar]
- Petersen, J. , Montserrat, V. , Mujico, J.R. et al. (2014) T‐cell receptor recognition of HLA‐DQ2–gliadin complexes associated with celiac disease. Nat. Struct. Mol. Biol. 21, 480–488. 10.1038/nsmb.2817 [DOI] [PubMed] [Google Scholar]
- Petersen, J. , Kooy‐Winkelaar, Y. , Loh, K.L. , Tran, M. , Van Bergen, J. , Koning, F. , Rossjohn, J. and Reid, H.H. (2016) Diverse T cell receptor gene usage in HLA‐DQ8‐associated celiac disease converges into a consensus binding solution. Structure, 24, 1643–1657. 10.1016/j.str.2016.07.010 [DOI] [PubMed] [Google Scholar]
- Rawat, N. , Schoen, A. , Singh, L. et al. (2018) TILL‐D: an Aegilops tauschii TILLING resource for wheat improvement. Front. Plant Sci. 9, 1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao, S.W. , Bergseng, E. , Molberg, O. , Xia, J. , Fleckenstein, B. , Khosla, C. and Sollid, L.M. (2004) Antigen presentation to celiac lesion‐derived T cells of a 33‐mer gliadin peptide naturally formed by gastrointestinal digestion. J. Immunol. 173, 1757–1762. [DOI] [PubMed] [Google Scholar]
- Salentijn, E.M. , Esselink, D.G. , Goryunova, S.V. , van der Meer, I.M. , Gilissen, L.J.W.J. and Smulders, M.J.M. (2013) Quantitative and qualitative differences in celiac disease epitopes among durum wheat varieties identified through deep RNA‐amplicon sequencing. BMC Genomics, 14, 905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salentijn, E.M. , Goryunova, S.V. , Bas, N. , van der Meer, I.M. , van den Broeck, H.C. , Bastien, T. , Gilissen, L.J.W.J. and Smulders, M.J.M. (2009) Tetraploid and hexaploid wheat varieties reveal large differences in expression of alpha‐gliadins from homoeologous Gli‐2 loci. BMC Genomics, 10, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherf, K.A. , Catassi, C. , Chirdo, F.G. et al. (2020) Recent progress and recommendations on celiac disease from the Working Group on Prolamin Analysis and Toxicity. Front. Nutr. 7, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan, L. , Molberg, O. , Parrot, I. , Hausch, F. , Filiz, F. , Gray, G.M. , Sollid, L.M. and Khosla, C. (2002) Structural basis for gluten intolerance in celiac sprue. Science, 297, 2275–2279. [DOI] [PubMed] [Google Scholar]
- Shewry, P.R. (2019) What is gluten – why is it special? Front. Nutr. 6, 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewry, P.R. and Lookhart, G.L. (2003) Wheat gluten protein analysis. American Association of Cereal Chemists (AACC), St Paul, MN. [Google Scholar]
- Shewry, P.R. and Tatham, A.S. (2016) Improving wheat to remove coeliac epitopes but retain functionality. J. Cereal Sci. 67, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, N. , Wu, S. , Tiwari, V. , Sehgal, S. , Raupp, J. , Wilson, D. , Abbasov, M. , Gill, B. and Poland, J. (2019) Genomic analysis confirms population structure and identifies inter‐lineage hybrids in Aegilops tauschii . Front. Plant Sci. 10, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smulders, M.J.M. , Jouanin, A. , Schaart, J. et al. (2015) Development of wheat varieties with reduced contents of celiac‐immunogenic epitopes through conventional and GM strategies. Proceedings of the 28th meeting of the Working Group on Prolamin Analysis and Toxicity (P. Koehler, ed.), 25–27 September 2014, Nantes, France. pp. 47–56. https://www.wgpat.com/proceeding_28th.html [Google Scholar]
- Sollid, L.M. (2017) The roles of MHC class II genes and post‐translational modification in celiac disease. Immunogenetics, 69, 605–616. [DOI] [PubMed] [Google Scholar]
- Sollid, L.M. , Tye‐Din, J.A. , Qiao, S.W. , Anderson, R.P. , Gianfrani, C. and Koning, F. (2020) Update 2020: nomenclature and listing of celiac disease – relevant gluten epitopes recognized by CD4+ T cells. Immunogenetics, 72, 85–88. [DOI] [PubMed] [Google Scholar]
- Spaenij‐Dekking, L. , Kooy‐Winkelaar, Y. , van Veelen, P. , Drijfhout, J.W. , Jonker, H. , van Soest, L. , Smulders, M.J.M. , Bosch, D. , Gilissen, L.J.W.J. and Koning, F. (2005) Natural variation in toxicity of wheat: potential for selection of nontoxic varieties for celiac disease patients. Gastroenterology, 129, 797–806. [DOI] [PubMed] [Google Scholar]
- Szabo‐Hever, A. , Zhang, Q. , Friesen, T.L. et al. (2018) Genetic diversity and resistance to fusarium head blight in synthetic hexaploid wheat derived from Aegilops tauschii and diverse Triticum turgidum subspecies. Front. Plant Sci. 9, 1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International Wheat Genome Sequencing Consortium (IWGSC) , Appels, R. , Eversole, K. et al. (2018) Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science, 361, eaar7191. 10.1126/science.aar7191 [DOI] [PubMed] [Google Scholar]
- Tye‐Din, J.A. , Stewart, J.A. , Dromey, J.A. et al. (2010) Comprehensive, quantitative mapping of T cell epitopes in gluten in celiac disease. Science Transl. Med. 2(41), 41ra51. 10.1126/scitranslmed.3001012 [DOI] [PubMed] [Google Scholar]
- Van den Broeck, H.C. , De Jong, H.C. , Salentijn, E.M.J. et al. (2010a) Presence of celiac disease epitopes in modern and old hexaploid wheat varieties. Wheat breeding may have contributed to increased prevalence of celiac disease. Theor. Appl. Genet. 121, 1527–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Broeck, H.C. , Hongbing, C. , Lacaze, X. , Dusautoir, J.C. , Gilissen, L.J.W.J. , Smulders, M.J.M. and van der Meer, I.M. (2010b) In search of tetraploid wheat accessions reduced in celiac disease‐related gluten epitopes. Mol. BioSystems, 6, 2206–2213. [DOI] [PubMed] [Google Scholar]
- Van Herpen, T.W. , Goryunova, S.V. , van der Schoot, J. et al. (2006) Alpha‐gliadin genes from the A, B, and D genomes of wheat contain different sets of celiac disease epitopes. BMC Genomics, 7, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J.R. , Luo, M.C. , Chen, Z.X. , You, F.M. , Wei, Y.M. , Zheng, Y.L. and Dvorak, J. (2013) Aegilops tauschii single nucleotide polymorphisms shed light on the origins of wheat D‐genome genetic diversity and pinpoint the geographic origin of hexaploid wheat. New Phytol. 198, 925–937. [DOI] [PubMed] [Google Scholar]
- Winfield, M.O. , Allen, A.M. , Burridge, A.J. et al. (2016) High‐density SNP genotyping array for hexaploid wheat and its secondary and tertiary gene pool. Plant Biotechnol. J. 14, 1195–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, Y.M. , Hsam, S.L.K. , Yu, J.Z. , Jiang, Y. and Zeller, F.J. (2003) Genetic polymorphisms at Gli‐Dt gliadin loci in Aegilops tauschii as revealed by acid polyacrylamide gel and capillary electrophoresis. Plant Breed. 122, 120–124. [Google Scholar]
- Zhang, Y. , Luo, G. , Liu, D. , Wang, D. , Yang, W. , Sun, J. , Zhang, A. and Zhan, K. (2015) Genome‐, transcriptome‐ and proteome‐wide analyses of the gliadin gene families in Triticum urartu . PLoS One, 10, e0131559. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Old versus new 454 approach.
Table S2. Clustered transcript sequence data and processing. Clustered transcript nucleotide sequences were translated into proteins and unique peptide variants were grouped.
Table S3. Unique peptide variants with epitope load.
Table S4. Unique peptide variants variant frequencies for each accession.
Table S5. Epitope frequency for each accession.
Table S6. List of Aegilops tauschii accessions captured in re‐synthesised hexaploid wheat (SHW). All SHW are available via the NIAB Breeder’s Toolkit (http://www.niab.com/pages/id/419/Breeders__Toolkit).
Table S7. Wheat accessions used. A list of the Aegilops tauschii lines studied (annotation from Jones et al., 2013) and the bread wheat and durum wheat lines included for comparison.
Table S8. Reproducibility of 454 sequencing.
Figure S1. Frequency of different DQ2.5‐Glia‐alpha epitopes in transcripts of different accessions.
Data Availability Statement
All relevant data on the peptides and epitopes can be found within the manuscript and its supporting materials.


