Abstract
Intron retention (IR) has been implicated in the pathogenesis of complex diseases such as cancers; its association with Alzheimer's disease (AD) remains unexplored. We performed genome‐wide analysis of IR through integrating genetic, transcriptomic, and proteomic data of AD subjects and mouse models from the Accelerating Medicines Partnership‐Alzheimer's Disease project. We identified 4535 and 4086 IR events in 2173 human and 1736 mouse genes, respectively. Quantitation of IR enabled the identification of differentially expressed genes that conventional exon‐level approaches did not reveal. There were significant correlations of intron expression within innate immune genes, like HMBOX1, with AD in humans. Peptides with a high probability of translation from intron‐retained mRNAs were identified using mass spectrometry. Further, we established AD‐specific intron expression Quantitative Trait Loci, and identified splicing‐related genes that may regulate IR. Our analysis provides a novel resource for the search for new AD biomarkers and pathological mechanisms.
Keywords: alternative splicing, Alzheimer's disease, gene expression, integrative analysis, intron retention
1. NARRATIVE
1.1. Contextual background
Compared to exon skipping, exon mutual exclusion, and alternative donor/acceptor site, intron retention (IR) is probably the least understood mode of alternative splicing mechanisms. 1 , 2 , 3 Historically thought of as the consequence of mis‐splicing, IR has recently gained recognition for its role in regulating gene expression. 4 , 5 , 6 , 7 , 8 , 9 Most retained introns contain premature termination codons (PTCs), which often trigger the nonsense‐mediated decay (NMD) pathway to target intron‐retained mRNAs to be degraded. 4 , 8 Consequently, many intron‐retained mRNAs are highly unlikely to be translated into proteins. However, in some cases they are able to bypass NMD and produce proteins with modified functions, as seen for such genes as P element transposase, 10 Id3 (Inhibitor of DNA Binding 3), 11 SCN1B (Sodium Channel, Voltage Gated, Type I Beta Subunit), 12 PRX (Periaxin), 13 and CCND1 (cyclin D1). 14 In macrophages IR has recently been shown to play a key role in the retention of the mRNA in the nucleus, where it can be rapidly spliced, exported and translated in response to stimuli. 15 Recent studies show that IR is widespread in mammals, 8 and is implicated in cancers. 6 , 16 Specifically, IR was shown to diversify the transcriptomes in 16 cancer types, including acute myeloid leukemia and breast cancers. 7 These studies suggest that the largely understudied area of IR deserves more attention as a potential contributing factor in complex diseases.
Systematic Review: We performed genome‐wide detection of intron retention (IR) in human and mouse brain and analyzed its features and association with Alzheimer's disease (AD) by integrating genetic, transcriptomic, and proteomic data generated from the Accelerating Medicines Partnership‐Alzheimer's Disease project.
Interpretation: To the best of our knowledge, this is the first genome‐wide and multi‐omics integrative analysis of IR in AD. We found that IR was a widespread phenomenon in human and mouse brain. Our integrative analysis implied the functional association of IR with AD. We also identified genes in the splicing pathway that potentially regulated IR.
Future Direction: The identified association between IR with AD needs to be validated in independent samples. It would be valuable to identify genetic determinants of IR.
Alzheimer's disease (AD) is a complex, heterogeneous neurodegenerative brain disorder, and the most common form of dementia estimated to affect over 40 million people worldwide. 17 To understand disease mechanisms and eventually develop therapeutic drugs, extensive research has been focused on AD through large collaborative projects such as the International Genomics of Alzheimer's Project (IGAP), 18 the Alzheimer's Disease Sequencing Project (ADSP) and the Accelerating Medicines Partnership‐Alzheimer's Disease (AMP‐AD). Specifically, within the AMP‐AD consortium, we have generated a rich dataset from humans and model systems 19 , 20 , 21 at multiple molecular levels including genome, transcriptome, and proteome data, providing new opportunities to investigate AD, for example, by searching for perturbed regulatory networks. Alternative splicing such as exon skipping has previously been studied in AD. 22 , 23 , 24 However, the association between IR and AD remains unexplored.
Here we report the first genome‐wide analysis of IR in autopsied AD samples, 19 as well as in two transgenic mouse models of Aβ amyloidosis, namely, CRND8 25 and APPPS1, 26 using an integrated proteogenomic approach. We identified IR events in humans and mice, respectively, and characterized sequence features of retained introns. The IR events were validated with alternative approaches. We explored whether retained introns could be translated into proteins by analyzing mass spectrometry‐based proteomic data, and assessed the functional association of IR with AD. We computed intron expression quantitative trait loci (ieQTLs) for AD and control samples, respectively, aiming to identify potential genetic variants that determine intron expression and to identify ieQTLs that might be relevant to AD. As the splicing pathway is a key regulatory layer for IR, we constructed a Splicing Pathway‐based Intron RegulatiOn Network (SPIRON) to investigate how IR was correlated with splicing factors. We showed how the SPIRONs were differential between AD and control samples and studied the association of modules in SPIRONs with AD.
1.2. Study conclusions and disease associations
We performed a systematic analysis of IR in humans and mice with AD by integrating genomic, transcriptomic, and proteomic data, aiming to identify functional relevance of IR in the context of AD. For the sake of specificity, only independent introns that do not overlap with any exons of other isoforms/genes were considered. Of note, this work is only to investigate the association between IR and AD. Whether IR causes AD or is the result of AD remains to be explored.
The main findings of this paper are as follows: (1) IR was widespread in human and mouse brain, mainly in protein‐coding genes; (2) we found suggestive evidence for the translation of some retained introns; (3) IR provided additional power to identify dysregulated genes compared to conventional differential gene expression analyses that are based only on exonic reads, and most of the differentially expressed introns were upregulated in AD; (4) IR was associated with reduced level of protein expression based on our analysis of matched RNA‐seq and proteomic data; (5) we identified QTLs for intron expression, of which some were specific to AD or control samples; and (6) intron expression was correlated with splicing factors, and some modules in the SPIRON networks were correlated with severity of AD neuropathology as measured with Braak scores. We found that most introns, though regulated by the orchestrated network of multiple splicing factors, appear to be strongly regulated by a major splicing factor.
The numerous IR events in both human and mouse brain were mainly observed in protein‐coding genes, but their roles in noncoding RNAs should be of interest for future studies. We also found a progressive age‐related expression pattern of retained introns in mice, which might be relevant to understanding AD progression and normal brain aging.
We explored whether retained introns might be translated into peptides. In mice, we discovered possible novel intron‐retained protein isoforms for six genes (Farp1, Slc4a4, Rcbtb1, Rad23a, Plin4 and Dos), which were collectively supported by 14 intron‐specific non‐nested unique peptides. The novel intron‐retained peptides for all but two of the genes (Rcbtb1 and Rad23a) could be independently identified, when a different method, X!Tandem, 27 was used to search the mass spectra. The strongest proteomic evidence we found was for a lengthy intron of Farp1 gene (800 AAs), which was detected with nine unique peptides. The retention of this intron may result in a novel protein isoform of Farp1 in mice. Interestingly, Farp1 functions in promoting dendritic growth and synapse formation, processes known to be impaired in AD pathology. 28 , 29 We postulate that the novel protein may disrupt synapse formation due to loss‐of‐function likely resulting from missing critical domains. A gain‐of‐function scenario is also possible with novel protein domains from the intronic region altering Farp1 function and its associated networks. Of future interest is characterizing the Farp1 protein isoforms in a context relevant to synaptic formation and predicting the functional networks of each intron‐retained isoform. 30 For Farp1, we further looked into the annotation of its human counterpart FARP1, and found that the intron sequence retained in mouse was annotated to be an exon of the human transcript ENST00000319526 (Ensembl v77). The annotation evidence of this intron in mouse (Ensembl v75) is weak with a Transcript Support Level 5 (TSL5), which means being not supported at all by any mRNA or expressed sequence tag (EST). Therefore, our finding about the retention and translation of the Farp1 intron provides rationale for improving the current annotation of mouse transcriptomes, which is critical in identifying and comparing IR events between species.
In humans, peptides unique to intron‐retained isoforms of EIF2D (58AAs) and PLEC (174AAs) were identified. The retained intron of EIF2D has no homologous sequence in its mouse counterpart in Ensembl v75, and the retained intron of PLEC is not annotated to be exonic in its mouse homologue. PLEC is involved in interlinking cytoskeleton molecules, and EIF2D functions as a translation initiation factor. The peptides for the retained introns of these two genes were not detected independently by another method, JUMP, 31 making the evidence for translation of the introns putative. Further evidence is needed to confirm the translation of the retained introns identified in mice and humans. In addition, the full‐length sequence of the IR‐derived protein isoforms remains to be discovered. The structure of such isoforms and their detection could become very complicated when multiple introns are retained in the same transcript. Although determining the amino acid sequence of the full‐length protein isoforms is challenging and beyond the scope of this work, in the future we plan to focus on these retained introns and perform experiments toward the determination of their amino acid sequence. Specifically, one may focus on candidate transcripts showing retained introns in the 3′ untranslated region (UTR) as these are unlikely to undergo NMD or nuclear degradation. However, it should be noted that this would only provide a narrow validation encompassing a small subset of genes with IR in the 3′UTR. We also plan to experimentally test the relevance of IR proteins to AD and their functional consequences.
The proteomic results point to the existence of mechanisms through which intron‐retained transcripts can escape NMD. Such mechanisms may be a multifactorial orchestration of transcription and splice factor availability. Differences in IR events across a population are likely a function of genomic variants at splicing sites and RNA‐binding proteins in spliceosomes as well as differences in expression availability of splicing genes. We have noted that levels of retained introns are differentially expressed in AD versus controls, even when there is no differential expression of combined transcripts of that gene based on exonic reads. This suggests that differentially expressed isoforms resulting from IR events may be masked by other isoforms that may be more abundant or have the opposite direction of regulation. IR identifies evidence of differentially expressed transcripts and pathways which cannot be captured using conventional approaches that only consider exonic expression. We found that most of the retained differentially expressed introns (DEIs) were upregulated in AD samples, which was consistent with the previous finding that the expression level of intron‐retained transcripts of 12 AD‐associated genes were increased due to the deficiency of splicing machinery in AD cases. 23 Of interest, for three of the 12 genes, namely BACE1, BIN1 and PICALM, the IR level was also increased in AD samples in our data, suggesting that splicing defects might be associated with AD. The DEI that best correlated with Braak score was in HMBOX1 (positive correlation), a transcription factor involved in the innate immune system. This suggests potential roles of IR in regulating innate immune functions. This DEI could also be the result of different proportions or levels of immune cells found in the AD samples compared to controls or due to increased microglial activity. Immune processes are currently under investigation for their importance in AD. 32 , 33 , 34 By integrative analysis of the human RNA‐seq data and the matched proteomic data, we found that an increased level of IR was associated with reduced expression of proteins, an observation that could be explained either by the NMD or nuclear retention. 34
As IR may be regulated by genetic variants, we carried out an ieQTL analysis to discover loci that reside within 100 kb of genes (cis) and that associate with their intron levels (cis ieQTLs). We identified 2102 and 1583 cis ieQTLs for the AD and control samples, respectively, strongly supporting the existence of genetic determinants of intron expression. Using independent eQTL data from the ExSNP and GTEx databases, these ieQTLs were validated to be accurate. The ieQTL approach can help identify genetic variants associated with retained intron expression. However, the mechanism that mediates the association between single nucleotide polymorphisms (SNPs) and retained intron expression remains unclear. As the retained intron expression is the consequence of both transcription and splicing, a potential way to investigate the mechanism is to test whether SNPs are located in the transcription factor binding site and/or the RNA motif bound by the related proteins during splicing. Furthermore, it is of interest to experimentally investigate the relevance of IR‐associated SNPs to AD pathogenesis, which requires model organisms or cell‐based studies. In future studies, we plan to experimentally study the mechanism underlying SNP‐intron expression associations and the relevance of IR‐associated SNPs to AD pathogenesis, as they are beyond the scope of this work.
As a major mode of alternative splicing, IR appears to be most directly related to the splicing pathway that involves the formation of U1‐type spliceosome for the removal of introns, 4 which motivated us to build a SPIRON to map how IR might be affected by splicing factors. SPIRON builds a co‐expression network of introns and splicing factors to determine which splicing factors are most likely to influence retention of introns. On the same human dataset used for IR identification, we found that although multiple genes correlated with their expression, most introns were highly correlated with levels of a single splicing gene. We identified modules that may suggest the existence of sub‐pathways of alternative splicing. This splicing modularity was also observed in the mouse SPIRON. For example, the modules of several major splicing factors such as ACIN1, RNPC3, SNRNP70, PRPF40B, PUF60, FUS, and SRRM1 appeared in both the human and mouse SPIRONs. This observation suggests the conservation of the IR pathways between species. In future work, we plan to experimentally investigate the influence of the conserved splicing factors on IR. For example, we can knockdown the splicing factors and analyze how IR would change compared to the controls without knockdown. In addition to those introns that are mainly regulated by a single gene, there was also a proportion of retained introns that were linked to more than one major splicing factor. Such introns are presumably subject to co‐regulation by the above‐mentioned sub‐pathways.
Another finding from SPIRON was that the regulatory direction (to increase or decrease intron expression) of most splicing factors was robust in the cases studied, being either positive or negative. Positive correlation may reflect up‐regulation of splicing factor transcripts to compensate for loss of function at the protein level, and vice versa. For example, ACIN1 in the human AD‐specific SPIRON was mainly positively correlated with retained introns, while the correlation of PUF60 with introns was mainly negative. Only a few splicing factors were seen to be bi‐directional, presumably from a context‐dependent regulatory pattern. Overall, the regulatory patterns of splicing factors appeared to be conserved between humans and mice, but intron regulation by splicing factors also showed species specificity.
In conclusion, we systematically identified IR as a widespread phenomenon in both human and mouse brains, and explored its functional association with AD through integrating genomic, transcriptomic, and proteomic data. We identified intron‐retained genes that were associated with AD. There are also limitations in our study. For example, our identified IR events were specific to temporal cortex (human) and forebrain (mouse), which may not generalize to other brain regions or cell types because of the high heterogeneity of brain tissues. Indeed, the analysis of RNA‐seq data has shown differences in cell type composition between cases and controls. 20 , 35 Building a high resolution map of IR in different cell types and brain regions would be very useful for understanding specific regulatory mechanisms of IR as well as revealing other brain regions in which IR might be related to AD. For example, mapping hippocampus‐specific IR events could be of interest since it is particularly vulnerable in AD. 36 , 37 Another limitation is that mouse models only partially re‐capitulate features of AD, in our case representing models of amyloidosis. Since the type of introns that completely overlap with exons of other isoforms/genes was not considered, their possible retention could not be revealed. IR events were not distinguishable across splice isoforms, so further efforts are needed to achieve isoform‐level resolution of IR. 30 Despite these limitations, our work presents an initial attempt to exploit the association of IR with AD and opens up new ways for identifying biomarkers and therapeutic targets for AD. Our studies also have implications for single cell RNA‐seq studies in both humans and mice. Given the abundance of IR events we have noted, we believe that single cell studies could help understand the IR pattern in different cell types and how the pattern is related to AD.
2. CONSOLIDATED RESULTS AND STUDY DESIGN
2.1. Widespread IR in human and mouse AD brains
The overview of our work is depicted in Figure 1. A total of 164 human brain samples (AD: n = 84, Control: n = 80), as described in, 19 were used in this study. We identified 4535 IR events (originating from 2173 unique genes) (Figure 2). Most of these genes with IR are protein‐coding based on neXtProt 38 (Figure 3). Compared with non‐retained introns, retained introns were shorter (P < 1.0 × 10−6) and of higher guanine‐cytosine (GC) content (P < 1.0 × 10−6) (Figure 3), in line with a previous report. 8
FIGURE 1.
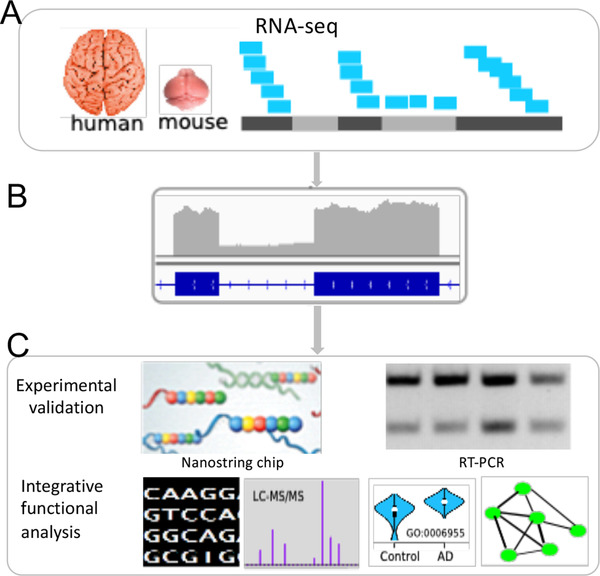
Overview of the identification and characterization of intron retention (IR) in Alzheimer's disease (AD). (A) AD cases and control brain samples were collected for both human samples (temporal cortex) and mouse models of amyloidosis, CRND8 and APPPS1 (forebrain) and their transcriptomes were sequenced. (B) IR detection from the RNA‐seq data. (C) Analysis and characterization of IR in AD. Sequence features such as guanine‐cytosine (GC) content were analyzed for both retained and non‐retained introns. Differentially expressed IRs were identified and Gene Ontology enrichment of the intron‐retained gene set was performed. Selected IR events were validated using RT‐PCR and customized Nanostring chips. Protein‐level expression of all identified IR events was examined using mass spectrometry‐based proteomic data. To explore how IR is regulated by the splicing pathway, we modeled intron expression as a function of expression levels of splicing genes and constructed a Splicing Pathway‐based Intron RegulatiOn Network (SPIRON), a unique and rich resource for understanding splicing‐level regulatory networks of IRs
FIGURE 2.
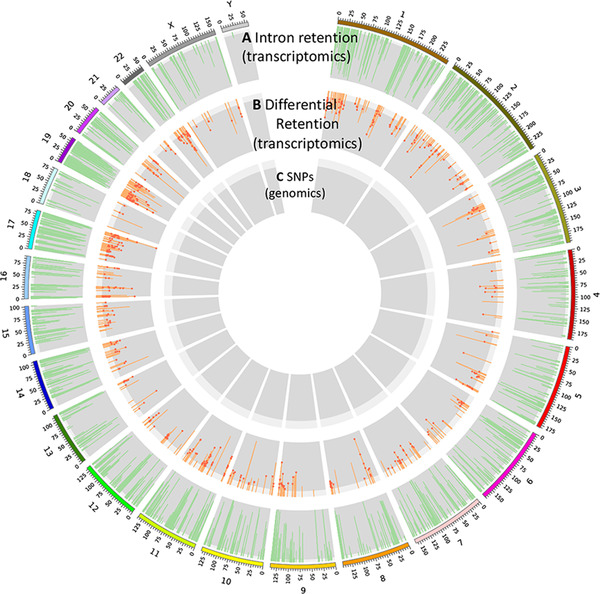
Genome‐wide intron retention (IR) in human brains with Alzheimer's disease (AD) and controls. (A) The distribution of IR events across chromosomes. Only introns with retention frequency >5% are displayed. (B) The Manhattan plot for retained introns with the false discovery rate (FDR)‐corrected P‐value calculated for differential expression between control and AD samples. The dot indicates a differentially‐expressed intron whose host gene is not differentially expressed based only on exonic reads. (C) The single nucleotide polymorphisms (SNPs) from genome sequencing of our samples were used to analyze intron expression quantitative trait locis in human AD and control samples, respectively
FIGURE 3.
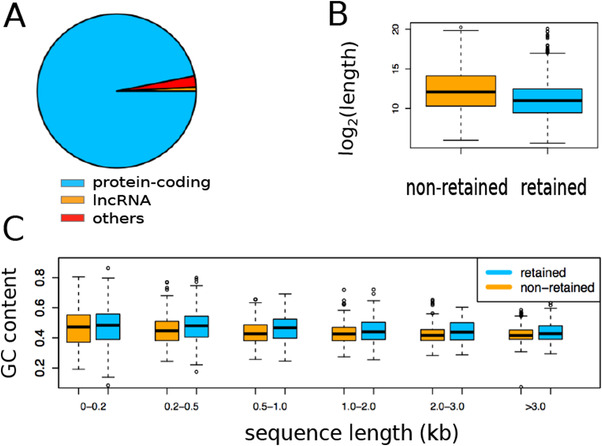
Intron retentions (IR) in temporal cortex of 164 human brains. (A) The biotype distribution of intron‐retained genes. (B) The length comparison between retained and non‐retained introns. (C) Guanine‐cytosine (GC) content comparison between retained and non‐retained introns binned by length
We also detected IR from mouse brain samples (n = 128) of two models of amyloidosis: CRND8 and APPPS1. We detected 4086 IR events (from 1736 genes). We observed that both the number and expression levels of retained introns vary across ages for both mouse models, implying development‐specific regulation of IR.
We tested the cell type specificity for intron‐retained genes. Using the brain cell type specific genes and the method described in, 39 we identified 111 human IR genes and 70 mouse IR genes that showed specific expression in one of the five brain cell types, namely, astrocyte, endothelial cell, microglia, neuron, and oligodendrocytes.
We investigated the expression correlation between retained introns and their parental genes. For both humans and mice, we found that the correlation could be either positive or negative, consistent with a previous report. 40 Overall, the correlation was weak in both humans and mice.
We validated the IR using two alternative methods: Nanostring chip and RT‐PCR (Figure 4). We tested a subset of retained introns in both humans and mice. In both species, we found that 90% of the selected retained introns could be validated experimentally, demonstrating the reliability of our identified IR events.
FIGURE 4.
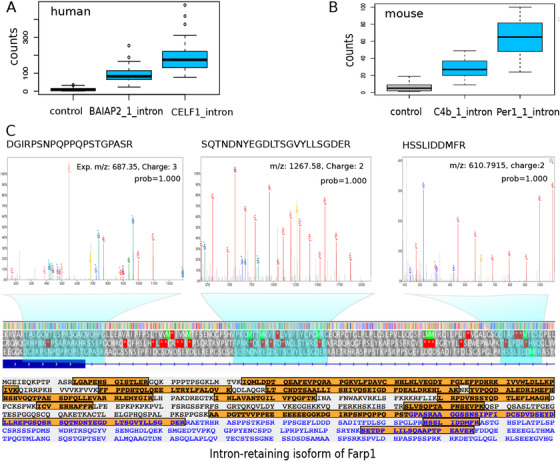
Transcriptomic and proteomic‐level validation of intron retention (IR). (A) Validation of IR using our custom‐designed Nanostring chip for BAIAP2 and CELF1 in humans. (B) Nanostring based validation of IR in C4b and Per1 in mice. (C) Mass spectrometry‐based protein expression validation of retained introns of the mouse protein Farp1 as an example. Shown in the upper part are tandem mass spectra of three peptides that are translated from the intronic region of the Farp1 gene. The probabilities of peptide‐spectrum‐match of these spectra are 1.00 with false discovery rate (FDR) < 0.001, calculated using PeptideProphet in the TPP (trans‐proteomics pipeline) software (v4.8.0). The lower part displays all peptides (orange) detected in the intron‐retained isoform of Farp1, with 45.3% (363/800 residues) of the amino acid sequence covered. Blue color indicates amino acid residues in introns
Splice isoforms with retained introns are rarely translated into proteins because they are often degraded by the NMD pathway. 4 We explored the possibility that our identified retained introns may be translated by interrogating mass spectrometry–based proteomic data (Figure 4). In mice, we identified likely translated introns for four genes (Farp1, Slc4a4, Plin4 and Dos). In humans, we obtained weak evidence for intron translation for PLEC and EIF2D.
2.2. Functional association of IR with AD
First, we tested retained introns for their association with AD through differential expression (Figure 5). We identified 2598 DEI retention events (false discovery rate [FDR] < 0.05). Most DEIs were up‐regulated. The parental genes of the DEIs were enriched in AD‐related functions such as neurodevelopment and AD pathology. A total of 63% of the parental genes of DEIs were not differentially expressed based on exonic reads, suggesting that IR provides additional discriminant information for the AD transcriptome compared to exonic expression.
FIGURE 5.
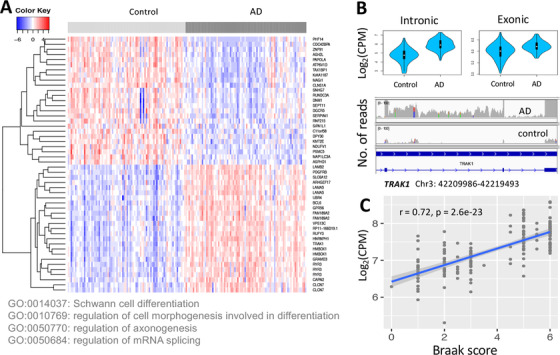
Association of intron retention (IR) with Alzheimers’ disease (AD). (A) Heat map based on the 50 most differentially expressed retained introns (DEIs) (the top 25 up‐regulated and the top 25 down‐regulated) and the Gene Ontology biological process terms enriched in all differentially expressed introns (DEIs). (B) An example of DEI (chr4:42209986‐42219493) whose parental gene ENSG00000182606 (TRAK1) was not differentially expressed based on exonic reads (CPM: counts per million). The read coverage of this intron in an AD and a control sample is shown using Integrative Genomics Viewer. (C) The correlation of the intron (chr8:29056685‐29063881) of ENSG00000147421 (HMBOX1) with Braak score, a measurement of tau pathology severity of AD
Second, to assess the association of IR with AD severity, we correlated intron expression with Braak scores for tau pathology severity (Figure 5). Interestingly, we found that 73% of DEIs were correlated with severity of AD tau neuropathology (FDR < 0.01), supporting their association with AD.
Third, by comparing the human RNA‐seq and the matched proteomic data, 41 we observed that genes with higher intron expression tended to have lower levels of protein expression, in support of an NMD mechanism (Figure 6). More importantly, we found that 70 proteins translated from intron‐retaining genes were differentially expressed (FDR <0.05), suggesting an association of IR with AD.
FIGURE 6.
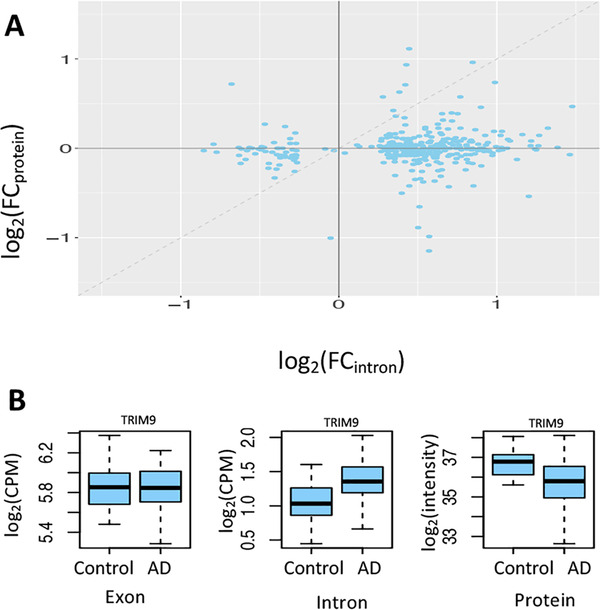
Influence of intron retention (IR) on protein production. (A) Increased intron expression is associated with reduced level of protein expression (note that the dashed line only indicates the diagonal and is not obtained by fitting the points with a linear regression model). Each point corresponds to a gene/protein. The x‐axis is the log2 transformed fold change (FC) of retained intron expression between AD and controls, denoted by log2(FCintron). The y‐axis is the log2 transformed fold change (FC) of protein expression between the same set of Alzheimer's disease (AD) and controls, denoted by log2(FCprotein). When intron expression is higher in AD (ie, log2[FCintron]) > 0), the value of log2(FCprotein) is mostly smaller than log2(FCintron) (ie, below the diagonal line). When intron expression is lower in AD (ie, log2[FCintron] < 0), the value of log2(FCprotein) is mostly higher than log2(FCintron) (ie, above the diagonal line). This observation suggests that the protein expression for the gene with higher retained intron expression tends to decrease more than that for the same gene with lower retained intron expression, likely secondary to nonsense‐mediated decay (NMD). (B) An example suggesting NMD. For the TRIM9 gene, its mRNA expression level is similar between AD and control samples. However, the intron expression level in AD is significantly higher than that in control samples, with reduced level of protein expression likely due to increased activity of NMD
2.3. Intron expression QTL in AD and control brains
To identify potential genetic determinants for intron expression, we performed a genome‐wide retained intron expression QTL analysis (ieQTL). We ran eQTL analysis on the adjusted intron expression data and for AD and control samples separately. We identified QTLs for 277 and 199 introns in AD and control samples, respectively (Figure 7). Further, we identified AD or control‐specific ieQTLs, suggesting that genetic regulation of intron expression may be differential in brains with AD and thus providing a new window into the molecular etiology of AD. We showed that the identified ieQTLs were reliable by indirectly validating them against gene‐level QTLs in two public databases: exSNP 42 and GTEx. 43
FIGURE 7.
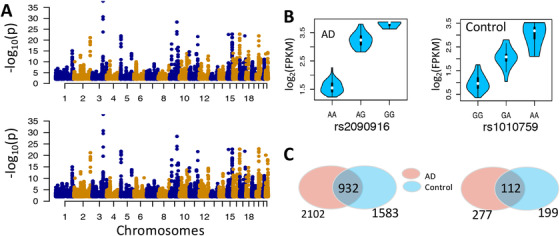
Intron expression quantitative trait loci (ieQTL) analysis. The ieQTLs were identified using MatrixEQTL at the threshold of false discovery rate (FDR) <0.05. (A) Manhattan plot of eQTLs for AD (upper panel) and control (lower panel) samples. (B) Example associations between intron expression level and genotypes. The retained introns shown the left and right are chr3:150584242‐150585393 and chr20:35545404‐35547261, respectively. (C) The sharing of ieQTL (left panel) and retained intron (right panel) between AD and control group
2.4. Splicing pathway‐based IR regulatory networks and their association with AD
Motivated by reports that intron expression is regulated partly by the regulatory network consisting of splicing factors in the splicing pathway, 40 , 44 , 45 , 46 we built the SPIRON to systematically explore the regulation of intron expression by splicing factors. In this network, an edge connects an intron to its corresponding splicing factor; the weight of the edge indicates the absolute correlation between introns and splicing factors.
For humans, both the AD and control‐specific SPIRONs showed patterns of highly structured modules, each containing a set of co‐regulated introns and centered on a major splicing factor (Figure 8). We observed that most introns appear to be dominantly regulated by one major splicing factor. We also observed the highly structured pattern in the mouse SPIRONs.
FIGURE 8.
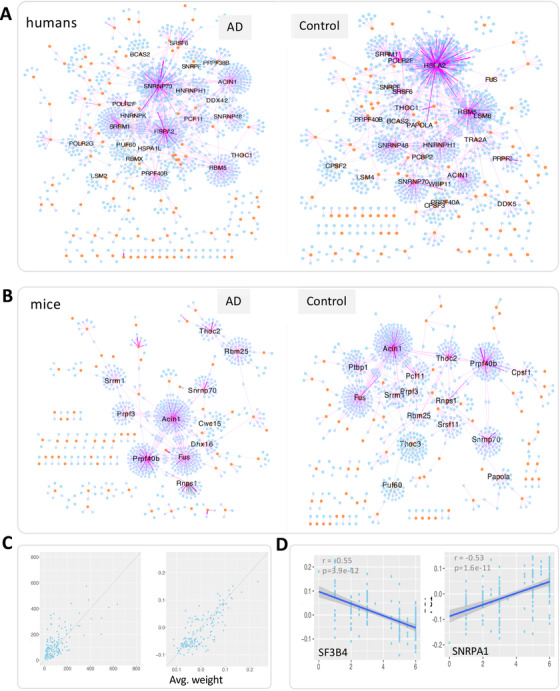
SPIRONs and the association of network modules with Braak score. (A) The human SPIRONs built with Alzheimer's disease (AD) and control samples, respectively. (B) The mouse SPIRONs built with transgenic and wild‐type control samples, respectively. In all these networks, lines represent edges that connect an intron to its splicing factor. Only edges with weight >0.2, indicating absolute correlation between an intron and its splicing factor are shown for visualization; yellow circles and blue hexagons represent splicing factors and introns, respectively. Purple and gray edges indicate positive and negative regression coefficients in the LASSO model, respectively. Genes with more than 15 first‐degree neighbors are labeled. (C) Comparison of the degree (left panel) and average weight (right panel) of each splicing factor between the control and AD‐specific SPIRONs. (D) The correlation of the eigengene (PC1 stands for the first principal component) of the SF3B4 and SNRPA1 module with Braak scores
Motivated by the finding that some splicing factors such as Snrnp70 and Prpf40b were major splicing factors in both the human and mouse SPIRONs, we tested the conservation of SPIRONs between the two species. We found that the regulatory patterns of more than half of the homologous splicing factors were conserved (FDR <0.05) (Figure 9).
FIGURE 9.
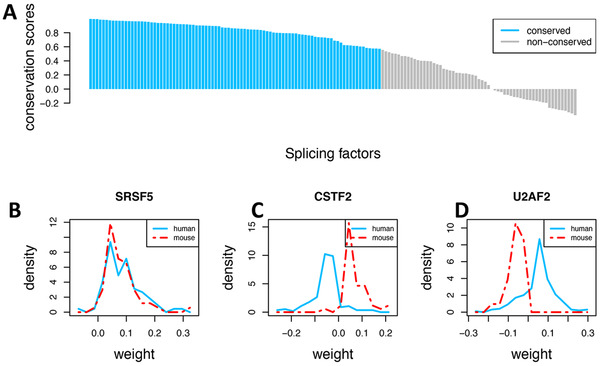
Comparison of the regulatory patterns of splicing factors between human and mice in the Alzheimer's disease (AD) samples. (A) For each splicing factor, we computed a conservation score (ranging from ‐1 to 1) between the human and mouse Splicing Pathway‐based Intron RegulatiOn Network (SPIRON; see Section 3.1, Methods). Most splicing factors show conserved regulatory patterns (false discovery rate [FDR] <0.01). (B) Illustration of conserved regulatory patterns with SRSF5. (C) Illustration of opposite regulatory directions of splicing factors between human and mouse with CSTF2, which negatively regulates retained introns in human but positively in mouse. (D) In contrast, U2AF2 positively correlates retained introns in human but negatively in mouse. Note: Weight is the edge weight between splicing factors and introns in the SPIRON network
Further, we investigated the network alteration between the AD and control‐specific SPIRONs. In humans, we found that some splicing factors showed large differences in their topological properties including both degree and the average weight (Figure 8). This finding held for the transgenic versus non‐transgenic mouse SPIRONs.
Next we tested whether the module in the SPIRON was associated with human AD traits using the method described in the weighted gene co‐expression network analysis (WGCNA). 47 We correlated the eigengene expression with Braak score and identified modules that were significantly correlated with Braak score (FDR <0.01) and had appreciable correlation (|r | >0.5) (Figure 8).
3. DETAILED METHODS AND RESULTS
3.1. Methods
3.1.1. Brain RNA‐seq and proteomic data
For human studies, 164 postmortem brain samples (84 with AD and 80 elderly controls without neurodegenerative disease) were collected. All AD samples were collected from the Mayo Clinic Brain Bank. The control samples were collected from two sources: the Mayo Clinic Brain Bank and the Banner Sun Health Institute. All brain samples were sequenced in the same place, namely, Mayo Clinic Genome Analysis Core. To ensure balance with respect to sex, age, RNA integrity number (RIN), Braak stages and diagnosis, the samples were randomized across flow cells. RIN for all samples were selected to be higher than 5.0. Total RNA was extracted from temporal cortex using Trizol reagent with RIN measured using an Agilent Technologies 2100 Bioanalyzer. A cDNA library was prepared using the TruSeq RNA Sample Prep Kit (Illumina, San Diego, CA), followed by 101 base pair paired‐end sequencing on Illumina HiSeq 2000 sequencers. The sample is sequenced with an average of 100 million reads, translating to a sequencing depth of approximately 70 reads per base of the human transcriptome. A more detailed description can be found here. 19 The raw sequencing data are available at https://www.synapse.org/#!Synapse:syn4894912. For all our analysis (including differential expression, the construction of the SPIRON and the eQTL analysis), the human gene expression data were adjusted for covariates including sex, age, sample sources (the two places where the samples were collected) by regressing out their confounding effects.
In addition, 109 of these human brain samples were also analyzed with LC‐MS/MS, with a NanoAcquity UHPLC (Waters, Milford, FA) in combination with a Q‐Exactive Plus mass spectrometer (ThermoFisher Scientific, San Jose, CA). The mass spectrometer cycle was programmed to collect one full mass spectrum (MS) scan followed by 10 data dependent MS/MS scans. The MS scans in the range of 300‐1800 m/z were collected at a resolution of 70,000 at m/z 200 in profile mode and the MS/MS spectra were acquired at a resolution of 17,500 at m/z 200. Protein expression were quantified with the MaxQuant v1.5.2.8 with Thermo Foundation 2.0 with default parameters. A total of 3838 proteins were quantified and the data are available at https://www.synapse.org/#!Synapse:syn20801227. For differential expression, the protein expression data were adjusted for sex and age. As the samples for the proteomic data were all from the Mayo Clinic Brain Bank, no adjustment for sample source was necessary.
For mouse studies, two transgenic mouse models were used 19 : (1) CRND8 and APPPS1. CRND8 is a transgenic mouse strain that carries both the Swedish and Indiana mutations in the amyloid precursor protein App gene 25 ; APPPS1 mice have a human APP gene with the Swedish mutation (K670N and M671L) and a human PSEN1 gene with Delta exon 9 mutation. 26 Both strains over‐express APP. Across a time series of 3, 6, 9, 12, and 20 months, a total of 128 mouse forebrain samples were obtained. At each time point, the transgenic and non‐transgenic mice represent cases of amyloidosis and non‐transgenic wild type control samples, respectively. The covariates of each sample, such as sex, age, mouse models, being transgenic or not, are provided in Supplementary Table 1. Total RNA was extracted and a library was prepared with Illumina TrueSeq kits. Paired‐end sequencing with 101 base pair reads were performed using an Illumina HiSeq 2000 sequencer. Each sample was sequenced with approximately 100 million reads, translating to a sequencing depth of approximately 110 reads per base of the mouse transcriptome. The raw sequencing data are available at https://www.synapse.org/#!Synapse:syn3157182. For all the construction of SPIRON, the mouse gene expression data were adjusted for covariates, including sex, age, RIN, and the mouse models by regressing out their confounding effects.
3.1.2. IR detection and differential expression analysis
We developed a pipeline to identify IR from RNA‐seq data in four steps: (1) align reads to reference genome; (2) count reads that physically overlap with intronic regions; (3) calculate entropy for each intron; and (4) use a set of filters to identify IR events with high confidence. This pipeline takes aligned reads in BAM format and an intron coordination file in bed format as input. Short reads were aligned to reference genome and transcriptome using STAR 48 (version 2.4.2a). For the input of STAR, the versions of genome assemblies for humans and mice were GRCh38 and GRCm38, respectively, and the gene model versions were ENSEMBL 38.77 for humans and ENSEMBL 38.75 for mice. Other parameters were default. To avoid ambiguity, only introns that did not overlap with any exons of other splice isoforms/genes were considered in our study. Specifically, the human gene model contains 965,083 introns (including overlapping ones among splice isoforms of the same gene), from which 232,088 independent introns were identified; the mouse gene models contain 531,859 introns, from which 197,631 independent introns were identified. Because IR detection was performed for the independent introns and the independent introns are obtained based on annotated gene models, the number of detected IR events may change according to the annotation used. Taking the RefSeq and ENSEMBL gene models as an example, if a chromosome region is annotated as an intron in one model and as an exon in the other, it will affect the annotation of independent introns and the subsequent IR detection. If a chromosome region is annotated as an intron in both models, it will not affect IR detection. Reads that fell into introns were counted and converted to fragments per kilobase million (FPKM) values. Library size was calculated as the total number of exonic reads and was used for calculating FPKM of both introns and genes. Then, the number of raw counts, FPKM, the number of junction reads spanning exon‐intron boundary and the normalized entropy score (NE‐score) that measures the evenness of the distribution of reads across the intron region were used to filter for high confident IR events. Details of this pipeline were described in. 49 To reliably identify retention events, we applied strict thresholds to the above filters. An IR event was called if its number of reads ≥20, its FPKM ≥3, its NE‐score >0.9, and it has at least one junction read that spans the exon‐intron boundary. In this study, we added another filter: the ratio of its expression to its parental gene. This ratio was set to be higher than 0.2.
Based on the criteria above, we first identified IR events from RNA‐seq data. For each of the retained introns, we recorded the total number of reads mapped to it in each sample. Given there are n retained introns and m samples, we can obtain an n × m matrix containing the read counts for n introns in rows and m samples in columns. This matrix was then used as input for the edgeR method (version 3.28.1, with default parameters) to identify DEIs and differentially expressed genes (DEGs). We used an FDR‐corrected P‐value smaller than 0.05 and a fold change (FC) larger than 1.2 as the threshold parameters to call significant DEIs/DEGs.
3.1.3. Transcriptomic validation of human and mouse IR
For mouse and human samples, we custom‐designed Nanostring chips to validate retained introns. Using mouse samples, we first validated IR events using both RT‐PCR and Nanostring chips. We found Nanostring is more robust and therefore used it to validate IR in human samples. For Nanostring, ProbeSets were designed and assayed separately for each intron (Supplementary Table 2). The choice of introns to validate was again based on the overall expression of the sequence in the RNA‐seq data and found to be differentially expressed in AD samples. For each intron, in collaboration with NanoString, we designed Capture ProbeSets to target each exon‐intron boundary sequence. Reporter CodeSets were designed to hybridize within the adjacent intron. In mice, for each hybridization reaction, 100 ng of purified total RNA was used in each nCounter XT Gene Expression Assay and hybridized with the Reporter ProbeSet for 16 hours at 65°C. Hybridized reactions were processed on the Nanostring Prep Station with the high sensitivity setting and then imaged on the NanoString Digital Analyzer under a high resolution setting (280 FOVs). RNA samples from CRND8 transgenic and non‐transgenic mice at 12 and 20 months of age were used to validate retained introns events with Nanostring ProbeSets. In humans, the same protocol was used, except that 200 ng of total RNA was used.
3.1.4. Proteomic validation of human and mouse IR
We performed proteomic validation of IR according to Human Proteome Project Mass Spectrometry Data Interpretation Guidelines 2.1. 50 For human material, we performed quantitative proteomics on homogenate for a total of 266 samples from Brodmann area 10 (anterior prefrontal cortex) (see details at https://www.synapse.org/#!Synapse:syn5759470). Briefly, brain‐derived tryptic peptide mixtures were separated on a self‐packed C18 fused silica column (25 cm × 75 uM internal diameter; New Objective, Woburn, MA) by a NanoAcquity UHPLC (Waters, Milford, FA) and monitored on a Q‐Exactive Plus mass spectrometer (ThermoFisher Scientific, San Jose, CA). Elution was performed over a 120‐minute gradient. The MS scans (300‐1800 m/z range) were collected at a resolution of 70,000 at m/z 200 in profile mode and the MS/MS spectra were acquired at a resolution of 17,500 at m/z 200. Dynamic exclusion was set to exclude previous sequenced precursor ions for 30 seconds within a 10 ppm window. Precursor ions with +1 and +6 or higher charge states were excluded from sequencing. RAW data for the samples were analyzed using MaxQuant v1.5.2.8 with Thermo Foundation 2.0. The search engine Andromeda, a component of MaxQuant, was used to build and search a concatenated target‐decoy Uniprot human reference protein database (retrieved April 20, 2015; 90,411 target sequences), plus 245 contaminant proteins from the common repository of adventitious proteins and 11,006 intron‐retained proteins that were in silico 3‐frame‐translated from our generated intron‐retained transcripts by merging ENSEMBL transcripts with its retained introns. Only translated proteins with at least 30 amino acids were considered. Methionine oxidation, asparagine and glutamine deamidation, and protein N‐terminal acetylation were variable modifications; cysteine was assigned a fixed carbamidomethyl modification. Only fully tryptic peptides were considered with up to two missed cleavages in the database search. A precursor mass tolerance was set to ±20 ppm. Default values were used for other parameters. Following established guidelines, 50 only peptides with at least nine amino acids (AAs) were considered. The FDR for peptide spectral matches, protein identification based on peptides, and site decoy fraction were all set to 0.01. As peptide identification may vary with search engines, we also applied another method, namely, the JUMP search engine (v1.2.1, April 2016), 31 to test whether intronic peptides (if there were) could be replicated by a different method. For JUMP, carbamidomethyl was used as fixed modification for cysteine, and dynamic modification of methionine, asparagine, and glutamine was used. Precursor mass tolerance was set to 0.05 Da. Other parameters were default.
For mouse data, we searched the PRIDE data repository (http://www.ebi.ac.uk/pride/archive/) for brain‐specific datasets. 51 We identified a dataset (PXD001250), which is most suitable to our study because the proteome was resolved at the cell type‐ and brain‐region level, including cerebellum, prefrontal cortex, hippocampus, olfactory bulb, corpus callosum, striatum, thalamus, neurons, astrocytes, oligodendrocytes (for details see Sharma et al. 52 ). Briefly, mouse brain regions or cell types (acutely isolated or cultured) were lysed and proteins were digested using LysC and trypsin. LC‐MS/MS analysis was performed in a quadrupole Orbitrap mass spectrometer with a high field analyzer. A total of 270 RAW data files (including biological replicates) were generated. We downloaded all these RAW files and converted them to mzML files using the msconvert software in the Trans‐Proteomics Pipeline (TPP, version 4.8.0) and searched the raw mass spectral data using the Comet software (version: 2014.02 rev. 2) against the target‐decoy concatenated mouse UniProt database (55,276 proteins, retrieved on Jan. 2, 2016), appended by 5465 intron‐retained proteins that were in silico 3‐frame translated from custom‐created ENSEMBL transcripts generated by including retained intron sequences. Only translated proteins with at least 30 amino acids were considered. Following the mass spectra search pipeline used in the study where this dataset was originally generated and analyzed, 52 contaminant protein sequences were not considered. The contaminant signal is typically <1% of the total signal across all proteins quantified in a proteomic sample, so it is negligible. Cysteine carbamidomethylation was used as fixed modification, and N‐acetylation of proteins and oxidation of methionine were used as variable modifications. The precursor mass tolerance was set to ±20 ppm. Two missed enzymatic cleavages were allowed at most. Only peptides with at least nine AAs were considered. After Comet analysis, Peptide‐spectrum match (PSM) probability was calculated using the PeptideProphet software in the TPP software. As peptide identification results may vary between search engines, we further tested whether intron‐specific peptides (if there were any) identified by Comet could also be identified by a different method. We used another widely used tool, namely, X!Tandem (version 2017.2.1.4, with default parameters), to search the same mass spectral dataset against the protein sequence database.
3.1.5. Intron expression QTL analysis
The AMP‐AD consortium has performed genotype calling from whole genome sequencing data for the human samples. Quality controls (QCs) have been performed for the removal of SNPs with genotyping call rate <98%, minor allele frequency <0.02, Hardy‐Weinberg disequilibrium P <3.4 × 10−8 in controls, duplicate variants and multiallelic SNPs. The post‐QC SNP data were available at https://www.synapse.org/#!Synapse:syn10845773. We obtained this data for 80 AD and 76 elderly control samples from Mayo Clinic that have matched SNP and RNA‐seq data. Intron expression was log2‐transformed by following the conventional practice. We used the R package MatrixEQTL (version 2.3) for eQTL analysis. 53 The cisDist parameter that defines cis‐SNPs was set to 100,000 by default. For control samples, the intron expression was adjusted for sex, age, RIN, and sample source as control samples were collected from two different brain banks. Because AD samples were collected from only one brain bank, the intron expression was adjusted only for sex, age, and RIN. FDR = 0.05 was used as the threshold to identify significant eQTLs.
3.1.6. Constructing SPIRONs
Genes in the U1‐type major splicing pathway, the U12‐type minor or atypical splicing pathway and the spliceosome were first downloaded from the PathCards database (http://pathcards.genecards.org/). After removing redundant entries, a set of 192 RefSeq gene symbols was obtained. Of these, 165 and 173 genes were able to be one‐to‐one mapped to human and mouse ENSEMBL genes, respectively, and were used in our analysis. As a summary, the human intron expression data consist of 4535 IRs measured in 164 samples (84 Alzheimer's and 80 elderly controls without neurodegenerative diseases); the mouse data contain 4086 IRs in 128 samples. The expression of each intron and splicing gene was log2‐transformed and standardized to have zero mean and unit variance.
To build parsimonious models, the LASSO method 54 was used to select a subset of splicing genes that collectively explain the variance of expression values of retained introns. Due to the fact that LASSO is sensitive to variations in samples, we used a Monte Carlo approach to identify splicing factors that can robustly predict intron expression as follows. Let an n × k matrix X denote the expression matrix of k splicing factors in columns and n samples in rows. For an individual intron, let an n × 1 vector y record its expression values in n samples. This dataset was denoted by (X, y). We identified splicing factors for predicting intron expression in three steps. First, for each intron, we randomly selected 80% of the n samples without replacement as the training set, denoted by (Xtrain, ytrain). The remaining 20% of the samples were used as the test set, denoted by (Xtest, ytest). This 80% to 20% split was adopted by following the convention for sample partition as used in 5‐fold cross‐validation, where all samples are divided into 5 groups and 4 of the 5 groups (80%) are used to build a model with the remaining group (20%) used for testing. Using the training set, we built a linear model using LASSO with the penalty factor λ optimized by cross‐validation using the one standard error rule. The LASSO model was built using the R package glmnet (version 3.0‐2). The LASSO model was enabled by setting the parameter alpha to 1.0 in the glmnet function. Default parameters were used for the LASSO method. We then calculated the predictive performance in terms of R2 on the test set. R2 measures the fraction of the intron expression variance that can be explained by splicing factors. The regression coefficient associated with each splicing factor, denoted by β, was also recorded. In the second step, we repeated the above procedure 50 times, calculated the average R2, the average β, and the selection probability (the number of times selected by LASSO divided by 50) for each splicing factor. Third, we determined whether an intron will be included for building SPIRON: an intron will be chosen if its average R2 ≥ 0.50, namely, if the splicing pathway can explain 50% of the variance of intron expression considering the fact there are actually many other regulatory factors, such as DNA methylation, contributing to IR. 1 If an intron was selected, only those splicing factors with a selection probability ≥0.5 will be used. The averaged β between the splicing factor and the intron was used as the weight of edges in SPIRON. We applied the approach to both human and mouse data, and built SPIRONs on the AD and control samples separately.
To quantitatively compare the regulatory pattern of splicing pathways on introns between species, we calculated a conservation score for each splicing gene as follows. First, for each splicing gene in SPIRON network, we identified all its neighboring introns. The edge weights between the splicing gene and neighboring introns were extracted and collected into a vector, denoted as w h for human and w m for mouse. Second, we compared the distribution between w h and w m. Note that wh and wm denote the edge weight vectors between a splicing factor and its directly connected introns for the two species, human and mouse, respectively. For the same splicing factor, its directly connected introns in the two species are different. Therefore, we cannot directly compare the individual element values in wh and wm. However, we can compare the distribution of the values in wh and wm to investigate whether the correlation pattern between splicing factors and their connected introns are different or not for the two species. To do so, we calculated their distributions by discretizing w h and w m into 15‐dimensional vectors of density values, denoted as d h and d m, respectively. Third, we calculated the Pearson correlations between d h and d m as the conservation score of splicing factors between species. In doing so, we computed for each splicing factor a conservation score describing its similarity between its regulatory pattern for IR in humans and that in mice.
3.2. Results
3.2.1. Widespread IR in human and mouse AD brains
A total of 164 human temporal cortex poly‐A enriched RNA‐seq samples from 84 AD patients and 80 elderly controls without neurodegenerative diseases (described in 19 ) were used in this study. Our method for IR identification is described in Figure 1. At our chosen threshold (see Section 3.1, Methods), we identified a total of 4535 unique IR events (within a total of 2173 unique genes) across the human samples (Figure 2a). These retained introns and their expression level in terms of counts per million (CPM) are provided in Supplementary Table 3. A hotspot for IR appears on chr19, which is interesting because chr19 is small. A possible explanation is that the splicing of the genes on chr19 might be susceptible to repression if the splicing pathway does not function normally. Of the 2173 genes with IR, 2104 are protein‐coding in neXtProt, 55 representing 10.7% of the 19,587 predicted human protein‐coding genes recorded in the Human Proteome Project (Figure 3a). 56 This indicates that many proteins may be regulated by IR. 4
We then analyzed sequence features of the retained introns. Compared with non‐retained introns, retained introns were significantly shorter (P <1.0 × 10−6) (Figure 3b), and had significantly higher GC content (P <1.0 × 10−6) regardless of intron length (Figure 3c), which is consistent with a previous report. 8 Note that the number of non‐retained introns exceeds substantially that of retained introns. To control for the substantial difference in the numbers between the retained and non‐retained introns, we randomly sampled the same number of non‐retained introns as that of retained introns. We tested whether the GC content or length of the group of retained introns was significantly different from that of the group of non‐retained introns using the Mann‐Whitney U test. Of note, this test was conducted not for individual introns but for comparing the two groups of introns, that is, retained versus non‐retained. Consequently, this test was run only one time for GC content and once for intron length, therefore the resulting P‐values do not need to be adjusted for multiple testing. In addition, as the sampling of non‐retained introns is a random process, we repeated the sampling process 10 times and showed that the difference between retained and non‐retained introns is significant regardless of the random selection of non‐retained introns. Most of the human IR events were shared between the control and AD samples. Based on the IR threshold applied, we found five retained introns that were expressed only in AD brains but not in controls (Supplementary Table 4). Of the five, the most frequently retained intron is the one between exon 8 and 9 in the RTKN gene (chr2:74428737‐74428847), which is retained in 24% of AD samples (vs 0.0% of control samples).
IR is also common in mice; 4086 IR events were identified from 1736 unique genes (Supplementary Figure 1). These retained introns and their expression level in terms of CPM are provided in Supplementary Table 5. The number of IR events and the expression levels of retained introns vary across ages for both CRND8 and APPPS1 mice (Supplementary Figure 1a and 1b; Supplementary Table 6), suggesting that IR may be subject to development‐specific regulation. An intron (Chr17:34734207‐34734364) of C4b (a gene functioning in the complement system) shows the most prominent positive correlation with age (Supplementary Figure 1c). Similar to humans, IR in mice was identified in both AD and control brains, enriched in protein‐coding genes, and with higher GC content compared to non‐retained introns (Supplementary Figure 1c‐e).
Because brain cell types are heterogeneous, we tested whether intron‐retained genes show cell type specificity. We obtained brain cell type–specific genes for both humans and mice from McKenzie et al. 39 The genes that were specific to five brain cell types, namely, astrocyte, endothelial cell, microglia, neuron, and oligodendrocytes, were provided. The cell type specificity was calculated as the minimum FC in expression between the cell type of interest and each of the other cell types and the threshold of FC = 4 was used to identify cell type specific genes. 39 We found that 111 of the human IR genes and 70 of the mouse IR genes were cell type specific. Taking the IR gene SLCO1C1 as an example, its minimum FC of expression between astrocyte and the other cell types is 14, indicating that it was highly specifically expressed in astrocytes. The cell type–specific IR genes for humans and mice were provided (Supplementary Table 7).
We investigated the correlation between the expression of retained introns with that of their parental genes. For each intron, we fitted a linear regression model to correlate the expression with that of its parental gene. The statistical metric R2 (also called coefficient of determination) was used to quantify the correlation. The sign of the slope of the regression model indicates the direction of correlation (positive or negative). For both the human and mouse data, we found that the correlation between intron expression and the parental gene expression can be either positive or negative (Supplementary Figure 2), which is consistent with the observation that both positive and negative correlation may occur between introns and the parent genes in previous work. 40 Briefly, we found that most retained introns were positively correlated with the expression of the parental genes and the overall correlation was weak with the median of R2 being 0.36 and 0.015 for the positively and negatively correlated intron‐gene pairs, respectively, in humans, and 0.199 and 0.169 for the positively and negatively correlated intron‐gene pairs, respectively, in mice (Supplementary Figure 2). The factor underlying the direction of correlation between the expression of introns and that of the parental gene may be complex. It has been shown that the direction of correlation partially depends on gene functions. 40
We next asked whether IR was conserved between human and mice in the context of AD. We identified a set of 743 homologous genes whose introns were retained in both humans and mice (Supplementary Table 8). For each gene, we compared the DNA sequence of every retained intron in mice to that in humans using BLAST with default parameters (version 2.3.0+). We found that only 33 retained introns (from 31 unique genes) showed high sequence similarity (>80%) between humans and mice (Supplementary Table 9). One of the most conserved retained introns is from SRSF6 (Serine/Arginine‐Rich Splicing Factor 6), a gene whose transcripts are involved in mRNA splicing. The nucleotide sequences (350 bases) of its retained intron in human and mouse have a similarity of 93% (Supplementary Figure 3).
3.2.2. Experimental validation of IR by nanostring chip and RT‐PCR
We selected a subset of IR events identified from poly‐A rich RNA‐seq data for experimental validation. In human samples, we selected 30 introns from 26 genes which are both highly expressed and differentially expressed between elderly control and AD patients (FDR < 0.01). We custom‐designed a Nanostring chip (Section 3.1, Methods; Supplementary Table 2), and tested expression levels of these introns. We found 29 of 30 showed significantly higher expression (P < 0.01) compared to the negative control probes (Supplementary Figure 4). As an illustration, intronic counts for BAIAP2 (an innate immunity gene) and CELF1 (a splicing factor) are much higher than that of negative control probes (Figure 4a), suggesting our identified IRs are reliable. These Nanostring results support our identification of IR events from the human RNA‐seq data.
For mice, we selected 21 retained introns from eight genes using the same criteria as for humans. Due to probe design requirements, only 15 of them could be targeted by Nanostring probes. Of these 15 introns, 14 were validated based on Nanostring expression, with an IR from the Trem2 gene being the only one that could not be validated by this approach (Supplementary Figure 5). All of the 21 retained introns were also tested using RT‐PCR by designing primers to the exonic regions and visualizing the length of the PCR product. Except for two introns (one in Trem2 and one in Nr4a1), the other 19 introns were confirmed (Supplementary Figure 6). Figure 4b shows the retention of intron 20 in C4b and intron 15 in Per1 (component of the circadian clock). These Nanostring and RT‐PCR results validate our findings at a rate of >90%, supporting the reliability of our method.
3.2.3. Protein‐level expression of retained introns
Splice isoforms containing introns are rarely translated into proteins because they generally trigger the NMD pathway and are subsequently degraded. 4 To the best of our knowledge, there are only a few reported cases where introns have been observed to avoid the NMD pathway and be translated. 11 , 12 To test the hypothesis that a small percentage of our identified retained introns may escape NMD and be translated into proteins, we searched the tandem mass spectra data from mouse and human brain samples against the customized protein sequence databases that include both in silico translated intron‐specific proteins and all proteins from UniProt for humans and mice, respectively (Section 3.1, Methods).
For mice, we searched the mass spectra of 270 mouse brain region samples and identified 255 retained‐intron specific PSMs mapping to 14 unique non‐nested peptides from six proteins (FDR <0.01) (Supplementary Figure 7). The uniqueness of these detected peptides was validated using both the nextProt “peptide uniqueness checker” tool and PeptideAtlas. 57 The most confident identification was a novel protein isoform of the Farp1 gene (800 AAs), which was supported by nine non‐nested intron‐specific peptides (with 9‐32 AAs) resulting from a total of 244 PSMs (FDR <0.001) in 132 brain samples covering a wide range of regions and cell types, including cerebellum, prefrontal cortex, hippocampus, olfactory bulb, corpus callosum, striatum, thalamus, neurons, astrocytes, and oligodendrocytes. The significance of PSMs of these unique peptides and their detection in multiple samples provided evidence that the Farp1 intron was translated. The mass spectra of three peptides observed in an olfactory bulb sample are shown (Figure 4). As Farp1 is involved in synapse formation, 28 our finding implies that IR in this gene may be related to synapse function. Whether the IR of Farp1 is related to AD still needs to be investigated as synapse function can be unrelated to AD. For each of the other five proteins (Slc4a4, Rcbtb1, Rad23a, Plin4, and Dos), only one unique and intron‐specific peptide was detected (Supplementary Figure 7), not satisfying the PE1‐level evidence to claim expression of novel proteins based on the Human Proteome Project (HPP) protein discovery guidelines 50 (requiring two non‐nested proteotypic peptides of ε 9 AAs), but only making them potential proteins translated from intronic regions. Further, as peptide identification may vary between spectra search engines, we searched the same proteomic data using another commonly used approach, namely, X!Tandem, 58 and examined whether the peptides detected by Comet 59 could also be detected by X!Tandem in the same sample. The results are provided in Supplementary Table 10. We found that all the peptides of Dos, Farp1, Slc4a4 and Plin4 were also detected by X!Tandem, increasing the confidence of the translation of retained introns. For Rcbtb1 and Rad23a, their peptides were not detected by X!Tandem in the same samples as detected by Comet, suggesting that the translation of retained introns of the two genes is putative.
For humans, we searched an independent human brain proteomic dataset (266 samples, see details at https://www.synapse.org/#!Synapse:syn5759470) from the Mount Sinai Brain Bank project. We identified peptides unique to two intron‐retained protein isoforms: PLEC (involved in interlinking cytoskeleton molecules) and EIF2D (a translation initiation factor), using FDR < 0.01. For the PLEC isoform (174 AAs), two intronic peptides were uniquely detected (FDR < 0.01). One peptide of 19 AAs spans the exon‐intron boundary with two AAs in the intron; the other peptide arising from within the same intron is less stringent since it contains only eight AAs (Supplementary Figure 8). 50 For the EIF2D isoform (58 AAs), a peptide with nine AAs was detected fully inside the intron region, which does not meet the PE1‐level criterion of two unique non‐nested peptides. 50 The JUMP search engine was also used to attempt independent identification of the three IR peptides with the best MaxQuant match scores for those peptides. Unfortunately, the three peptide sequences were not detected. These results suggest that the evidence for translation of the introns in PLEC and EIF2D is weak.
3.2.4. Functional association of IR with AD
We investigated the biological functions of the 2173 human intron‐retained genes through Gene Ontology (GO) enrichment analysis (Supplementary Table 11). The most enriched GO biological processes include RNA splicing pathways (GO:0008380, P = 1.78 × 10−6), chromatin modification (GO:0016570, P = 5.24 × 10−5) and neurological functions such as Schwann cell differentiation (GO:0014037, P = 0.008), neurotrophin signaling pathway (GO:0038179, P = 0.004), and regulation of neuron projection development (GO:0010975, P = 0.01). The P values above were all Bonferroni‐corrected. RNA splicing and chromatin modification was reportedly associated with AD pathology. 22 , 60 Dysregulation of neurotrophin was suggested to be involved in memory loss, a main symptom of AD. 61
We tested for differential expression of retained introns between AD cases and controls (Section 3.1, Methods). We identified 2598 DEI retention events (FDR < 0.05) (Figure 2B, Supplementary Table 12) after adjusting for sex, age, RIN, and sample source, of which 2366 and 232 were up and down, respectively. The parental genes of the DEIs were enriched in functions associated with neurodevelopment or AD pathology, including Schwann cell differentiation (GO:0014037, FDR = 8.0 × 10−4), regulation of cell morphogenesis involved in differentiation (GO:0010769, FDR = 0.001), regulation of axonogenesis (GO:0050770, FDR = 0.001), and regulation of mRNA splicing (GO:0050684, FDR = 0.008) (Figure 5A).
We compared genes with DEIs to the differentially expressed genes (DEGs) identified using only exonic reads. This analysis showed that 63% of the parental genes of DEIs are not identified as DEGs based on exonic reads (Supplementary Figure 9). One possible reason for this could be that RNA‐seq data represent expression from a population of cells in which multiple isoforms of the same gene are expressed, with the exonic read counts from a lesser‐expressed isoform being obfuscated by the more highly expressed isoforms. When only the intronic reads are considered, differences in isoform expression are more apparent. TRAK1 (Trafficking Kinesin Protein 1) is an example, a gene involved in lysosome trafficking; one of its introns is highly differentially expressed even though the gene is not (Figure 5B).
To quantitatively assess how IR is related to AD severity, we correlated the expression level of retained introns with Braak scores, reflecting tau pathology severity. We used the 159 human samples from Mayo Clinic with Braak scores available for this analysis. The intron expression adjusted for sex, age, RIN, and sites was used. We found that 1907 of the 2598 DEIs were significantly correlated with Braak staging scores (FDR < 0.01) with absolute Pearson correlation values ranging from 0.30 to 0.72 (Supplementary Table 13). An example intron high correlating with the Braak score was in HMBOX1 (correlation = 0.72, P = 2.6 × 10−23 (Figure 5C), a transcription factor reported to regulate natural killer (NK) cell functions through suppressing the NKG2D/DAP10 signaling pathway. 62 Also, NK cells were shown to be associated with AD, 63 implying a relationship between HMBOX1 and AD. This finding suggests an association between IR and innate immunity, which in turn was reported to be associated with AD. 33
As IR may lead to NMD and thus reduce the protein expression, we analyzed whether the proteins encoded by the parental genes of the DEIs were also associated with AD by comparing the Mayo Clinic RNA‐seq data reported here and its matched proteomic data (Section 3.1, Methods). 41 We used the data from 109 samples with matched RNA‐seq and proteomic data. Protein expression for 366 parental genes of the DEIs was available in the proteomic data. The protein expression data were adjusted for sex and age by regressing our their effects. First, we examined whether the IR could lead to NMD in our data, because NMD is generally the major pathway that IR transcripts undergo. We calculated the log2 transformed FC of retained intron expression between AD and controls, denoted by log2(FCintron). The FC of protein expression between the same set of AD and controls was also calculated, denoted by log2(FCprotein). We found that most IR transcripts were likely degraded by NMD, as indicated by the finding that the protein expression for the gene with higher retained intron expression tends to decrease more than that for the same gene with lower retained intron expression (Figure 6A). For example, when intron expression is higher in AD samples (ie, log2[FCintron] >0), the value of log2(FCprotein) is mostly smaller than log2(FCintron) (ie, below the diagonal line). When intron expression is lower in AD (ie, log2[FCintron] <0), the value of log2(FCprotein) is mostly higher than log2(FCintron) (ie, above the diagonal line). This observation suggests that protein expression is reduced when the mRNA contains a higher level of intron expression, reflecting the activity of NMD. We illustrated this likely NMD mechanism with the TRIM9 gene as an example. As shown in Figure 6B, its mRNA expression level is similar between the AD and control samples. However, the intron expression level in the AD samples is significantly higher than that in the control samples, which results in reduced level of protein expression likely due to increased NMD. Second, we tested whether the proteins corresponding to the genes with IR were associated with AD. For the 366 proteins above, we performed differential expression between AD and control with the Mann Whitney U test. We found that 70 were significantly expressed (FDR <0.05), suggesting an association of the protein products of the IR genes with AD. The differential proteins were provided in Supplementary Table 14. Third, for these differentially expressed proteins, we examined how the protein expression level was affected by the expression of its parent gene as well as the retained intron. To this end, we built a linear regression model for protein expression where parent gene expression and intron expression were included as two variables. We found that protein expression was weakly correlated with the expression of parent genes and retained introns, with a Spearman correlation of 0.26 ± 0.12 across all these differentially expressed proteins.
3.2.5. Intron expression QTL in AD and control brains
To explore the genetic determinants of the expression of retained introns, we performed a genome‐wide retained intron expression QTL analysis (ieQTL) for AD and control samples separately. SNPs in cis (1000 kb upstream of the intron or within the intron) were analyzed (Figure 2C). The intron expression data for both AD and control samples was adjusted for sex, age, and RIN. As control samples were collected from two different brain banks, the intron expression was also adjusted for sample source (see details in Section 3.1, Methods). We ran eQTL analysis for AD and control samples separately. The Manhattan plot of all the significant associations is provided (Figure 7). In AD samples, we identified 2102 ieQTLs that were significantly associated with the expression of 277 introns (FDR < 0.05); in control samples, 1583 ieQTLs were identified with significant association with 199 introns. These numbers translate to approximately seven QTLs per intron for both AD and control data, suggesting that intron expression may be controlled by multiple factors. For intuitive understanding of how intron expression is correlated with genotypes, we plotted the expression of retained introns against the genotype using two examples of eQTLs (Figure 7B). Shown in the left panel is the association between intron chr3:150584242‐150585393 and its QTL rs2090916 in AD samples. Clearly, this intron showed the lowest, medium, and highest expression in the reference allele group (AA), heterozygous group (AG) and the alternative allele group (GG), respectively, suggesting a strong genetic control of the expression of retained introns. The second example of association between chr20:35545404‐35547261 and rs1010759 (Figure 7B, right) also suggests the possible genetic regulation of intron expression level.
We compared ieQTLs that were separately identified in the AD and control samples. We found that 932 ieQTLs were shared between the AD and control samples and that 1170 and 651 ieQTLs were specific to the AD and control samples, respectively (Figure 7C). This finding of condition‐specific ieQTLs suggests that ieQTLs provide a new window into the difference between the AD and control samples. Among the AD‐specific ieQTLs, the SNP rs2589949 (G >A mutation) was a significant one, strongly associated with the expression of the intron (chr15:90439399‐90440501) of the gene IQGAP1 (FDR = 1.94 × 10−9). Of interest, IQGAP1 has been shown to regulate spine density and cognitive processes. 64 Our analysis suggests that the IR and cis‐genetic variants might be associated with the function of IQGAP1. Another example is the intron of SH3TC2 (chr5:149004902‐149006880), a gene functioning in myelination. Its expression is significantly associated with rs11168078 (T >C mutation) (FDR = 5.60 × 10−10). For the above two examples, their associations in control samples are insignificant, suggesting that the regulatory network of intron expression in disease status may be disrupted and that ieQTL analysis may provide a new window into the molecular etiology of AD.
We further validated our identified ieQTLs. Because there is no available benchmark ieQTL data for validation, we chose to validate ieQTLs indirectly by comparing them with gene eQTLs. The reason is that intron expression reflects one aspect of gene expression and a proportion of ieQTLs should also be conventional eQTLs. We converted the SNP‐intron associations to SNP‐gene associations and tested whether SNP‐gene associations could be validated by benchmark eQTL data.
We performed the validation against two public databases. The first is for eQTL data, that is, SNP‐gene associations, from the ExSNP database (http://www.exsnp.org/Download). 42 We found that 26% and 31% (far above the baseline 0.0%) of the SNP‐gene associations of the corresponding ieQTLs identified in AD and control samples were able to be validated. The second eQTL dataset is from the GTEx database (version: v6p). We considered the significant SNP‐gene pairs identified in human brains (involving 10 brain regions, see details in Supplementary Table 15). We found that 35% and 43% (the baseline was near zero percentage) of the SNP‐gene associations in the AD and control data could be replicated. These results suggest that our ieQTLs are likely valid and many of them reflect traditional coding gene based eQTLs, though most seem to be specific to the retained introns.
3.2.6. SPIRONs and their association with AD‐related traits
Alternative splicing has been previously implicated in AD. 22 , 23 It has been reported that the expression of retained introns are partly regulated by a cooperative regulatory network consisting of the splicing factors in the splicing pathway. 40 , 44 , 45 , 46 Therefore, we hypothesized that the expression level of retained introns was correlated with that of splicing factors. The numbers of splicing genes annotated for mice and humans may vary depending on the database used. For example, 71 human splicing factors were annotated in the SpliceAid‐F database. 65 A total of 56 mouse splicing factors were annotated in the SFMetaDB database. 66 The PathCard database 67 has a more comprehensive annotation, containing a total of 192 splicing factors functioning in the major splicing pathway, in the minor splicing pathway or in the spliceosome. We used the splicing factors obtained from the PathCards database for both humans and mice (Section 3.1, Methods). The correlation between introns and splicing factors forms a network, which we call the SPIRON for brevity. This SPIRON was inferred by correlating the expression level of introns to that of splicing genes using a multivariate linear regression model with features selected by the LASSO method (Section 3.1, Methods). For both humans and mice, the gene and intron expression data were adjusted for sex, age, and RIN. Specifically for humans, the expression data were also adjusted for sample source for controls (Section 3.1, Methods). Specifically for mice, the expression data were also adjusted for mouse models. Regression coefficients of these models, denoted as β, between intron and splicing genes were used as edge weights in the network. To understanding the relevance of SPIRON to AD, we built SPIRON for the AD and control samples, respectively for both humans and mice, thus totaling four SPIRONs. The full network data of the four SPIRONs are provided in Supplementary Tables 16‐19. The distributions of edge weights of the networks are provided in Supplementary Figure 10. For visualization, we showed the four networks by keeping only the edge with weight >0.2 (Figure 8).
For humans, both the AD (Figure 8A, left) and control‐specific (Figure 8A, right) SPIRONs were highly structured with a number of co‐regulated introns forming modules that centered on a major splicing factor. From the SPIRONs, we found that most introns, although regulated by the orchestrated network of multiple splicing factors, appear to be strongly regulated by a major splicing factor. This finding was shown to be consistent when a different model was used to build SPIRON (Supplementary Figure 11 and Supplementary Figure 12), implying the robustness of the network. We observed several major splicing factors such as SNRNP70, PRPF40B, HSPA2, SRRM1, SRSF6, RBM5, THOC1, HNRNPH1, POLR2F, and ACIN1. SNRPNP70 is a component of the U1 snRNP complex, essential for recognizing 5′ splicing sites and recruiting proteins for assembling the spliceosome. It has been shown that SNRPNP70 knockdown or inhibition of U1 snRNP were associated with increased RNA splicing deficiency in AD pathogenesis. 23 PRPF40B is a splicing factor involved in pre‐mRNA splicing. HSPA2 is a heat shock protein that mediates folding of proteins, is associated with assembly of spliceosomes, and is involved in cellular stress response. 68 Notably, the PathCards database shows that HSPA2 has only low‐ranking evidence for being involved in the general splicing pathway, but it appears to be a major player in IR based on our network. Some of the major splicing factors such as SNRNP70, PRPF40B, SRRM1, and ACIN1 appear in both the AD‐ and control‐specific SPIRONs, suggesting the conservation of major splicing factors in different conditions. Regarding the regulation direction (the sign of weight), we found that most splicing factors were positively correlated with introns among the edges with weight >0.2 shown in Figure 8. Note that In the full human or mouse SPIRON, neither the positive correlation nor the negative correlation dominates in numbers. Likewise, we also found that neither upregulated nor downregulated splicing factors dominated in numbers in both humans and mice. Two examples of exceptions were PUF60 and BCAS2 that appeared to dominantly regulate IR in a negative manner. In contrast, SNRNP70 and RBM5 correlated mainly in a positive direction with their corresponding introns in both AD and control‐specific SPIRION. Other genes such as POLR2G (in AD SPIRON) and LSM4 (in control SPIRON) displayed both positive and negative correlations.
The mouse SPIRON showed similar patterns with the human SPIRON. For example, both the AD‐ and control‐specific SPIRONs were also highly structured with most introns correlated with a single splicing factor (Figure 8B). The major splicing factors such as Snrnp70, Prpf40b, Acin1, Srrm1, Thoc2, and Fus appeared in both networks. By comparison, we found that some of these major splicing factors such as Snrnp70, Prpf40b, Acin1, and Srrm1 occur in both the two human SPIRON and the two mouse SPIRON and the direction of regulation of them were also consistent between humans and mice, suggesting that the regulatory relationship between splicing factors and introns might be conserved between the two species. In addition to this similarity, there were differences between the human and mouse networks. For example, the HSPA2 was part of a robust module in the human but not the mouse network. In contrast, Rnps1 was correlated with a number of introns in mice but not in humans.
We tested whether the SPIRON between humans and mice were conserved. For each splicing factor, we computed the correlation of its regulatory patterns between humans and mice as its conservation score (Section 3.1, Methods). First, we compared the AD‐specific SPRION for the humans and mice. We found that, for 91 of the 152 homologous splicing factors in the SPIRON, their regulatory patterns for IR were likely conserved between humans and mice (FDR < 0.05; corresponding to correlation >0.55) (Figure 9A). One example is SRSF5, a component of the U1‐type spliceosome (Figure 9B). For splicing genes with uncorrelated patterns, it is interesting to find that their regulation directions were often opposite between species. For example, CSTF2 appears to correlate with retained introns in human in a predominantly negative manner but positively in mice (Figure 9C), while U2AF2 correlates positively with retained introns in humans but negatively in mice (Figure 9D). Second, we compared the control‐specific SPRION for the humans and mice. We found that the regulatory patterns of 81 splicing factors were likely conserved between humans and mice (Supplementary Figure 13).
Motivated by the previous report that RNA splicing is associated with AD, 22 we investigated the network alteration between the AD‐ and control‐specific SPIRONs. First, we tested whether the degree (ie, the number of correlated introns) of splicing factors was different between the two networks. We calculated the degree of each splicing factor based on the full AD‐ and control‐specific SPIRONs. We found that although the degree of splicing factors in the two networks was significantly correlated, the correlation coefficient (r = 0.58) was not high, with some splicing factors having a much higher degree in the AD‐specific SPIRON than in the control‐specific SPIRON and others having a much higher degree in the control‐specific SPIRON (Figure 9C, left). For example, PHF5A (PHD Finger Protein 5A) was connected with 112 introns in the control‐specific SPIRON while only 38 introns were connected to it in the AD‐specific SPIRON. This observation suggests that the degree of some splicing factors was differential. Second, we calculated the average β for each splicing factor over all its connected introns and compared it between the AD‐ and control‐specific SPIRONs (Figure 9C, right). We found that some of the splicing factors showed large differences in between the two networks. For example, the average β for SF3A1 is 0.028 in the control‐specific SPIRON while it was altered to –0.081 in the AD‐specific SPIRON. We also compared the degree and β for splicing factors between the two networks in mice and the results were similar (Supplementary Figure 14).
Next we analyzed whether the modules centered on the splicing factors were associated with human AD pathological traits. We used the Braak score as the trait. For each splicing factor, we identified the introns shared between the AD‐ and control‐specific networks. These shared introns together with the splicing factor were considered as a module. Following the established method to correlate modules with traits as described in the weighted gene co‐expression network analysis (WGCNA) method, 47 we considered only modules involving at least 20 genes and tested the correlation of the eigengene (ie, the first principal component [PC1] of the module) with the Braak score. We identified six modules with a significant Spearman correlation (denoted by r) (FDR < 0.01) and with an appreciable correlation (|r | > 0.5). For example, the PC1 for the SF3B4 module was significantly correlated with the Braak score (r = –0.55, FDR = 3.9 × 10−12) (Figure 8D, left). Another example was for the SNRPA1 module, which was significantly correlated with Braak score (Figure 8D, right). The correlations with Braak scores of the eigengenes for the remaining four modules (SNRPF, PCF11, HNRNPH1, PTBP1) are shown in Supplementary Figure 15. In summary, we showed that the network properties of degree or edge weight of a subset of splicing factors were altered between the two networks and identified modules associated with AD severity measured with Braak scores.
CONFLICT OF INTERESTS
The authors declare no competing financial interests.
Supporting information
Supplementary information
Supplementary information
Supplementary information
Supplementary information
Supplementary information
Supplementary information
Supplementary information
Supplementary information
Supplementary information
Supplementary information
Supplementary information
Supplementary information
Supplementary information
Supplementary information
Supplementary information
Supplementary information
Supplementary information
Supplementary information
Supplementary information
Supplementary information
ACKNOWLEDGMENTS
AMP‐AD University of Florida/Mayo Clinic/Institute for Systems Biology: For the human brain donations, we thank all patients and their families, without whom this work would not have been possible. We thank Thomas G. Beach (Banner Sun Health Institute, AZ) for sharing human tissue.
This work was supported by National Institute on Aging (AG046139‐01 to TEG, NET, NP, SGY, AG051504 to N.E.T); National Institute of Neurological Disorders and Stroke (R01 NS080820 to N.E.T.); the Accelerating Medicine Partnership for AD (U01AG046161); the Emory Alzheimer's Disease Research Center (P50 AG025688); National Institutes of Health grants (P30ES017885‐01A1 and U24CA210967 to GSO); and National Natural Science Foundation of China (61702556 to HDL).
Study data were provided by the following sources: The Mayo Clinic Alzheimer's Disease Genetic Studies, led by Dr. Nilufer Ertekin‐Taner and Dr. Steven G. Younkin, Mayo Clinic, Jacksonville, FL using samples from the Mayo Clinic Study of Aging, the Mayo Clinic Alzheimer's Disease Research Center, and the Mayo Clinic Brain Bank. Data collection was supported through funding by NIA grants P50 AG016574, R01 AG032990, U01 AG046139, R01 AG018023, U01 AG006576, U01 AG006786, R01 AG025711, R01 AG017216, R01 AG003949, NINDS grant R01 NS080820, CurePSP Foundation, and support from Mayo Foundation. Study data include samples collected through the Sun Health Research Institute Brain and Body Donation Program of Sun City, Arizona. The Brain and Body Donation Program is supported by the National Institute of Neurological Disorders and Stroke (U24 NS072026 National Brain and Tissue Resource for Parkinson's Disease and Related Disorders), the National Institute on Aging (P30 AG19610 Arizona Alzheimer's Disease Core Center), the Arizona Department of Health Services (contract 211002, Arizona Alzheimer's Research Center), the Arizona Biomedical Research Commission (contracts 4001, 0011, 05‐901 and 1001 to the Arizona Parkinson's Disease Consortium) and the Michael J. Fox Foundation for Parkinson's Research.
Li H‐D, Funk CC, McFarland K, et al. Integrative functional genomic analysis of intron retention in human and mouse brain with Alzheimer's disease. Alzheimer's Dement. 2021;17:984–1004. 10.1002/alz.12254
Hong‐Dong Li and Cory C. Funk contributed equally to this study.
REFERENCES
- 1. Wong JJL, Gao D, Nguyen TV, et al. Intron retention is regulated by altered MeCP2‐mediated splicing factor recruitment. Nat Commun. 2017;8:15134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wong JJ‐L, Ritchie W, Ebner OA, et al. Orchestrated intron retention regulates normal granulocyte differentiation. Cell. 2013;154:583‐595. [DOI] [PubMed] [Google Scholar]
- 3. Li H‐D, Menon R, Omenn G, Guan Y. The emerging era of genomic data integration for analyzing splice isoform function. Trends Genet. 2014;30:340‐347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ge Y, Porse BT. The functional consequences of intron retention: alternative splicing coupled to NMD as a regulator of gene expression. BioEssays. 2014;36:236‐243. [DOI] [PubMed] [Google Scholar]
- 5. Pimentel H, Parra M, Gee SL, Mohandas N, Pachter L, Conboy JG. A dynamic intron retention program enriched in RNA processing genes regulates gene expression during terminal erythropoiesis. Nucleic Acids Res. 2016;44:838‐851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jung H, Lee D, Lee J, et al. Intron retention is a widespread mechanism of tumor‐suppressor inactivation. Nat Genet. 2015;47:1242‐1248. [DOI] [PubMed] [Google Scholar]
- 7. Dvinge H, Bradley RK. Widespread intron retention diversifies most cancer transcriptomes. Genome Med. 2015;7:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Braunschweig U, Barbosa‐Morais NL, Pan Q, et al. Widespread intron retention in mammals functionally tunes transcriptomes. Genome Res. 2014;24:1774‐1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Burgess DJ. Alternative splicing: retaining introns to sculpt gene expression. Nat Rev Genet. 2014;15:707. [Google Scholar]
- 10. Rio DC. Regulation of Drosophila P element transposition. Trends Genet. 1991;7:282‐287. [DOI] [PubMed] [Google Scholar]
- 11. Forrest ST, Barringhaus KG, Perlegas D, Hammarskjold M‐L, McNamara CA. Intron retention generates a novel Id3 isoform that inhibits vascular lesion formation. J Biol Chem. 2004;279:32897‐32903. [DOI] [PubMed] [Google Scholar]
- 12. Kazen‐Gillespie KA, Ragsdale DS, D'Andrea MR, Mattei LN, Rogers KE, Isom LL. Cloning, localization, and functional expression of sodium channel beta1A subunits. J Biol Chem. 2000;275:1079‐1088. [DOI] [PubMed] [Google Scholar]
- 13. Dytrych L, Sherman DL, Gillespie CS, Brophy PJ. Two PDZ domain proteins encoded by the murine periaxin gene are the result of alternative intron retention and are differentially targeted in Schwann cells. J Biol Chem. 1998;273:5794‐5800. [DOI] [PubMed] [Google Scholar]
- 14. Lu F, Gladden AB, Diehl JA. An alternatively spliced cyclin D1 isoform, cyclin D1b, is a nuclear oncogene. Cancer Res. 2003;63:7056‐7061. [PubMed] [Google Scholar]
- 15. Green ID, Pinello N, Song R, et al. Macrophage development and activation involve coordinated intron retention in key inflammatory regulators. Nucleic Acids Res. 2020;48:6513‐6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Scotti MM, Swanson MS. RNA mis‐splicing in disease. Nat Rev Genet. 2016;17:19‐32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Querfurth HW, LaFerla FM. Alzheimer's disease. N Engl J Med. 2010;362:329‐344. [DOI] [PubMed] [Google Scholar]
- 18. Lambert J‐C, Ibrahim‐Verbaas CA, Harold D, et al. Meta‐analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet. 2013;45:1452‐1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Allen M, Carrasquillo MM, Funk C, et al. Human whole genome genotype and transcriptome data for Alzheimer's and other neurodegenerative diseases. Sci Data. 2016;3:160089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Allen M, Wang X, Burgess JD, et al. Conserved brain myelination networks are altered in Alzheimer's and other neurodegenerative diseases. Alzheimers Dement. 2018;14(3):352‐366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Allen M, Wang X, Serie DJ, et al. Divergent brain gene expression patterns associate with distinct cell‐specific tau neuropathology traits in progressive supranuclear palsy. Acta Neuropathol. 2018;136:709‐727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Love J, Hayden E, Rohn T. Alternative splicing in Alzheimer's disease. J Parkinsons Dis Alzheimer Dis. 2015;2:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bai B, Hales CM, Chen P‐C, et al. U1 small nuclear ribonucleoprotein complex and RNA splicing alterations in Alzheimer's disease. Proc Natl Acad Sci U S A. 2013;110:16562‐16567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Johnson ECB, Dammer EB, Duong DM, et al. Deep proteomic network analysis of Alzheimer's disease brain reveals alterations in RNA binding proteins and RNA splicing associated with disease. Mol Neurodegener. 2018;13:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chishti MA, Yang D‐S, Janus C, et al. Early‐onset amyloid deposition and cognitive deficits in transgenic mice expressing a double mutant form of amyloid precursor protein 695. J Biol Chem. 2001;276:21562‐21570. [DOI] [PubMed] [Google Scholar]
- 26. Xiong H, Callaghan D, Wodzinska J, et al. Biochemical and behavioral characterization of the double transgenic mouse model (APPswe/PS1dE9) of Alzheimer's disease. Neurosci Bull. 2011;27:221‐232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bjornson RD, Carriero NJ, Colangelo C, et al. X!!Tandem, an improved method for running X!Tandem in parallel on collections of commodity computers. J Proteome Res. 2008;7:293‐299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cheadle L, Biederer T. The novel synaptogenic protein Farp1 links postsynaptic cytoskeletal dynamics and transsynaptic organization. J Cell Biol. 2012;199:985‐1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhuang B, Su YS, Sockanathan S. FARP1 promotes the dendritic growth of spinal motor neuron subtypes through transmembrane Semaphorin6A and PlexinA4 signaling. Neuron. 2009;61:359‐372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li H‐D, Menon R, Govindarajoo B, et al. Functional networks of highest‐connected splice isoforms: from the Chromosome 17 Human Proteome Project. J Proteome Res. 2015;14:3484‐3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang X, Li Y, Wu Z, Wang H, Tan H, Peng J. JUMP: a tag‐based database search tool for peptide identification with high sensitivity and accuracy. Mol Cell Proteomics. 2014;13:3663‐3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mosher KI, Wyss‐Coray T. Microglial dysfunction in brain aging and Alzheimer's disease. Biochem Pharmacol. 2014;88:594‐604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Heneka MT, Golenbock DT, Latz E. Innate immunity in Alzheimer's disease. Nat Immunol. 2015;16:229‐236. [DOI] [PubMed] [Google Scholar]
- 34. Conway OJ, Carrasquillo MM, Wang X, et al. ABI3 and PLCG2 missense variants as risk factors for neurodegenerative diseases in Caucasians and African Americans. Mol Neurodegener. 2018;13:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang X, Allen M, Li S, et al. Deciphering cellular transcriptional alterations in Alzheimer's disease brains. Mol Neurodegener. 2020;15:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Johnson KA, Fox NC, Sperling RA, Klunk WE. Brain imaging in Alzheimer disease. Cold Spring Harb Perspect Med. 2012;2:a006213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Braak H, Braak E. Neuropathological stageing of Alzheimer‐related changes. Acta Neuropathol. 1991;82:239‐259. [DOI] [PubMed] [Google Scholar]
- 38. Zahn‐Zabal M, Michel PA, Gateau A, et al. The neXtProt knowledgebase in 2020: data, tools and usability improvements. Nucleic Acids Res. 2020;48:D328‐D334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. McKenzie AT, Wang M, Hauberg ME, et al. Brain cell type specific gene expression and co‐expression network architectures. Sci Rep. 2018;8:8868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ullrich S, Guigó R. Dynamic changes in intron retention are tightly associated with regulation of splicing factors and proliferative activity during B‐cell development. Nucleic Acids Res. 2020;48:1327‐1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Swarup V, Chang TS, Duong DM, et al. Identification of conserved proteomic networks in neurodegenerative dementia. Cell Rep. 2020;31:107807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yu C‐H, Pal LR, Moult J. Consensus genome‐wide expression quantitative trait loci and their relationship with human complex trait disease. OMICS. 2016;20:400‐414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. GTEx Consortium . The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318‐1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hsu TYT, Simon LM, Neill NJ, et al. The spliceosome is a therapeutic vulnerability in MYC‐driven cancer. Nature. 2015;525:384‐388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jacob AG, Smith CWJ. Intron retention as a component of regulated gene expression programs. Hum Genet. 2017;136:1043‐1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Saltzman AL, Kim YK, Pan Q, Fagnani MM, Maquat LE, Blencowe BJ. Regulation of multiple core spliceosomal proteins by alternative splicing‐coupled nonsense‐mediated mRNA decay. Mol Cell Biol. 2008;28:4320‐4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang B, Horvath S. A general framework for weighted gene co‐expression network analysis. Stat Appl Genet Mol Biol. 2005;4:17. [DOI] [PubMed] [Google Scholar]
- 48. Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA‐seq aligner. Bioinformatics. 2013;29:15‐21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li H‐D, Funk CC, Price ND. iREAD: a tool for intron retention detection from RNA‐seq data. BMC Genomics. 2020;21:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Deutsch EW, Overall CM, Van Eyk JE, et al. Human Proteome Project mass spectrometry data interpretation Guidelines 2.1. J Proteome Res. 2016;15:3961‐3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Perez‐Riverol Y, Csordas A, Bai J, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47:D442‐D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sharma K, Schmitt S, Bergner CG, et al. Cell type‐ and brain region‐resolved mouse brain proteome. Nat Neurosci. 2015;18:1819‐1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shabalin AA. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012;28:1353‐1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tibshirani R. Regression shrinkage and selection via the Lasso. J R Statist Soc B. 1996;58:267‐288. [Google Scholar]
- 55. Gaudet P, Michel P‐A, Zahn‐Zabal M, et al. The neXtProt knowledgebase on human proteins: current status. Nucleic Acids Res. 2015;43:D764‐D770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Omenn GS, Lane L, Lundberg EK, Overall CM, Deutsch EW. Progress on the HUPO draft Human Proteome: 2017 metrics of the human proteome Project. J Proteome Res. 2017;16:4281‐4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Desiere F, Deutsch EW, King NL, et al. The PeptideAtlas project. Nucleic Acids Res. 2006;34:D655‐D658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fenyö D, Beavis RC. A method for assessing the statistical significance of mass spectrometry‐based protein identifications using general scoring schemes. Anal Chem. 2003;75:768‐774. [DOI] [PubMed] [Google Scholar]
- 59. Eng JK, Jahan TA, Hoopmann MR. Comet: an open‐source MS/MS sequence database search tool. Proteomics. 2013;13:22‐24. [DOI] [PubMed] [Google Scholar]
- 60. Bahari‐Javan S, Sananbenesi F, Fischer A. Histone‐acetylation: a link between Alzheimer's disease and post‐traumatic stress disorder?. Front Neurosci. 2014;8:160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gold CA, Budson AE. Memory loss in Alzheimer's disease: implications for development of therapeutics. Expert Rev Neurother. 2008;8:1879‐1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wu L, Zhang C, Zhang J. HMBOX1 negatively regulates NK cell functions by suppressing the NKG2D/DAP10 signaling pathway. Cell Mol Immunol. 2011;8:433‐440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Jadidi‐Niaragh F, Shegarfi H, Naddafi F, Mirshafiey A. The role of natural killer cells in Alzheimer's disease. Scand J Immunol. 2012;76:451‐456. [DOI] [PubMed] [Google Scholar]
- 64. Gao C, Frausto SF, Guedea AL, et al. IQGAP1 regulates NR2A signaling, spine density, and cognitive processes. J Neurosci. 2011;31:8533‐8542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Giulietti M, Piva F, D'Antonio M, et al. SpliceAid‐F: a database of human splicing factors and their RNA‐binding sites. Nucleic Acids Res. 2013;41:D125‐D131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Li J, Tseng C‐S, Federico A, et al. SFMetaDB: a comprehensive annotation of mouse RNA splicing factor RNA‐Seq datasets. Database. 2017;2017:bax071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Frida B, Noam N, Gil S, et al. PathCards: multi‐source consolidation of human biological pathways. Database. 2015;2015:bav006. 10.1093/database/bav006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Padhi A, Ghaly MM, Ma L. Testis‐enriched heat shock protein A2 (HSPA2): adaptive advantages of the birds with internal testes over the mammals with testicular descent. Sci Rep. 2016;6:18770. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information
Supplementary information
Supplementary information
Supplementary information
Supplementary information
Supplementary information
Supplementary information
Supplementary information
Supplementary information
Supplementary information
Supplementary information
Supplementary information
Supplementary information
Supplementary information
Supplementary information
Supplementary information
Supplementary information
Supplementary information
Supplementary information
Supplementary information


