Abstract
Point‐of‐care (POC) diagnostic tests for the rapid detection of individuals infected with Mycobacterium leprae, the causative pathogen of leprosy, represent efficient tools to guide therapeutic and prophylactic treatment strategies in leprosy control programs, thus positively contributing to clinical outcome and reducing transmission of this infectious disease. Levels of antibodies directed against the M. leprae‐specific phenolic glycolipid I (PGL−I) closely correlate with an individual's bacterial load and a higher risk of developing leprosy. We describe herein the assembly of a set of PGL glycans carrying the characteristic phenol aglycon and featuring different methylation patterns. The PGL trisaccharides were applied to construct neoglycoproteins that were used to detect anti‐PGL IgM antibodies in leprosy patients. ELISAs and quantitative lateral‐flow assays based on up‐converting nanoparticles (UCP‐LFAs) showed that the generated PGL−I and PGL‐II trisaccharide neoglycoconjugates can be applied for the detection of anti M. leprae IgM antibodies in POC tests.
Keywords: carbohydrates, diagnosis, glycoconjugates, lateral flow assay, leprosy
Getting to the point of care: Trisaccharides of phenolic glycolipids (PGL) originating from M. leprae were evaluated for their diagnostic potential as neoglycoproteins in ELISA and an up‐converting particle lateral flow assay (UCP‐LFA). Two naturally occurring trisaccharides were found to be suitable for this purpose and can thus be used for future field studies in leprosy endemic areas.
Introduction
Leprosy is a chronic disease caused by Mycobacterium leprae, a bacillus with tropism for skin and peripheral nerves. [1] Although the disease can be cured with multidrug therapy (MDT), it still is a harsh reality in 145 countries mostly affecting individuals in poor socioeconomic circumstances. Its late‐ or misdiagnosis often leads to irreversible deformities and lifelong handicaps. [2] Leprosy's featuring aspect is the plateauing global annual number of new cases of roughly 200 000 including 10 % children, which indicates that transmission is ongoing. [3] Field‐friendly diagnostic tests to facilitate identification of infected individuals, will enable timely prophylactic‐ and therapeutic treatment, respectively. Overall, this will help prevent permanent leprosy‐associated disabilities and consequently support a higher quality of life, improved long‐term outcomes and reduced economic burden.
Phenolic glycolipid‐I (PGL−I, Figure 1) is a cell wall component of M. leprae which modulates the host immune response by limiting the production of nitric oxides synthase in a complement receptor 3 (CR3) dependent manner. [4] The binding to CR3 also enhances the invasion of dendritic cells and polymorphonuclear neutrophils, which further dampens the immune response. [5] Furthermore, macrophages infected with Mycobacterium marinum expressing PGL−I (as a proxy for live M. leprae) induce demyelination, a process leading to nerve damage in leprosy. [6] After infection a predominantly IgM antibody response can be generated against M. leprae PGL−I. [7] The level of anti‐PGL−I antibodies strongly correlates with bacterial load, [8] allowing identification of those who are most infectious. Moreover, antibody levels in young children can be used to indicate recent transmission rates in endemic areas. [9] Previously, we developed a low complexity, point‐of‐care (POC) lateral flow assay based on up‐converting particles (UCP‐LFA) that quantitatively measures anti‐PGL−I IgM levels in serum and fingerstick blood.[ 10 , 11 , 12 ] This assay is currently applied in field‐trials in Africa, to identify and treat M. leprae infected individuals, and evaluated for surveyance purposes of M. leprae‐infected red squirrels in the UK. [13]
Figure 1.
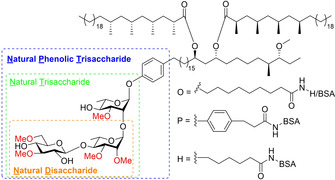
PGL−I and synthetic conjugates used for diagnostics.
For decades serological analyses for leprosy research have been performed using ND−O‐HSA (natural disaccharide linked via an octyl carboxylic acid linker to human serum albumin, Figure 1),[ 7 , 14 , 15 , 16 ] a conjugate bearing a synthetic disaccharide antigen. This glycoconjugate contains the terminal two sugars of the naturally occurring M. leprae PGL−I, which has been synthesized to serve as a convenient water‐soluble antigen for use in leprosy research. [17] The terminal 3,6‐di‐O‐methylglucose was found to be the most important structural determinant for antibody binding, [18] with disaccharides showing a higher sensitivity than monosaccharides. [19] Although in most sera of leprosy patients the disaccharide epitope was found to be effective for the detection of antibodies, it has been suggested that the structure of the native glycolipid needed to be more emulated to further improve the binding. [17] Therefore, the trisaccharide containing conjugate with the same octyl linker (natural trisaccharide linked via an octyl carboxylic acid linker to bovine serum albumin, NT−O‐BSA) was probed by Brennan and co‐workers. [20] While they found no discernable increase in sensitivity or specificity, [21] Izumi et al. found that a trisaccharide epitope with a phenol on the reducing end with a propyl linker (natural trisaccharide linked via a phenol linker to bovine serum albumin, NT−P‐BSA) did improve the sensitivity, as well as the antigenic specificity, compared to the disaccharide (ND−P‐BSA). [22] These findings were confirmed in a study, comparing ND−O‐BSA to NT−O‐BSA and NT−P‐BSA, demonstrating that the latter provided the best sensitivity and specificity. [23] Combined, these results indicate that the phenol on the reducing end of the saccharides can increase specific antibody binding. Two additional phenolic glycolipids, PGL‐II and PGL‐III, differing from PGL−I in the methylation pattern of the trisaccharide, are present in the cell wall of M. leprae as well, and can also modulate the host innate immune response against the mycobacterium.[ 24 , 25 , 26 ] Although M. leprae‐specific PGL−I has the strongest modulatory capacity, [4] Lowary and co‐workers have described that the methylation pattern in related PGLs originating from Mycobacterium tuberculosis and Mycobacterium kansasii play an important role in shaping the immune response against these PGLs.[ 27 , 28 , 29 , 30 ]
To investigate the binding of the M. leprae PGL‐trisaccharides with the natural phenol anomeric appendage to human antibodies, we here set out to synthesize a set of PGL trisaccharides (Figure 2). These PGLs, featuring different methylation patterns, are equipped with a C6‐linker attached to the phenol to increase accessibility for binding when conjugated to a carrier protein.[ 31 , 32 ] We here describe the assembly of the natural phenolic trisaccharides 1, 2 and 3 (NPT1, NPT2, NPT3, respectively), functionalized with a hexanoic acid linker for conjugation to BSA, to provide the NPT1‐H‐BSA, NPT2‐H‐BSA and NPT3‐H‐BSA conjugates. These conjugates were evaluated using ELISAs to detect IgM antibodies against M. leprae in a cohort of leprosy patient sera. Since leprosy frequently occurs in remote, low resource areas we additionally assessed the performance of these PGLs in UCP‐LFAs.
Figure 2.
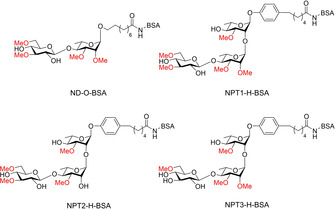
PGL‐conjugates assembled in this study.
Results and Discussion
Synthesis of glycoconjugates
Scheme 1 depicts the synthesis of the required NPT‐glycans. The strategy used for the synthesis of the trisaccharides is based on an approach we previously developed for the assembly of M. tuberculosis phenolic glycolipids in which iodophenol glycosides were generated and subsequently functionalized with a lipid tail through a Sonogashira coupling. [33] The synthetic approach applied here can thus also be applied for the total synthesis of the natural PGL−I, PGL‐II and PGL‐III without any modifications. We therefore used iodophenol rhamnose 1 as the first building block. It was found that a condensation of this acceptor with rhamnosyl donor 2 under the agency of N‐iodosuccinimide (NIS) and triflic acid (TfOH) led to partial iodination of the iodoaryl ring and we therefore switched to the use of the use the diphenylsulfoxide (Ph2SO)‐triflic anhydride (Tf2O) reagent combination to activate the thioglycoside. [34] Subsequent addition of acceptor 1 to the activated donor provided dirhamnoside 3 in 66 % yield. The C‐2’‐O‐benzoyl was then replaced for a methyl ether and subsequently the C‐4’‐O‐PMB was removed using a catalytic amount of HCl in HFIP, [35] resulting in disaccharide acceptor 6 in 86 % yield over 3 steps. This acceptor was coupled with donor 13 a using the Ph2SO/Tf2O activator delivering the fully protected trisaccharide 8 a in 93 % yield. Similarly, the condensation of 6 and donor 13 b delivered trisaccharide 8 c in quantitative yield.
Scheme 1.
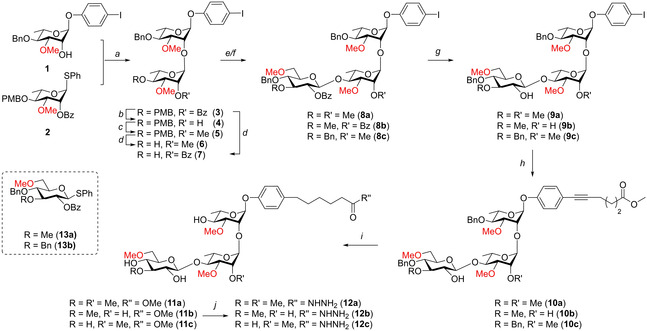
a) Ph2SO, Tf2O, TTBP, CH2Cl2, −60 °C, 66 %; b) Na, MeOH/THF, 97 %; c) NaH, MeI, DMF, 95 %; d) HCl/HFIP, HFIP/CH2Cl2, 93 % (6), 100 % (7); e) Donor 13 a, Ph2SO, Tf2O, TTBP, CH2Cl2 −60 °C, 93 % (8 a), 88 % (8 b); f) Donor 13 b, Ph2SO, Tf2O, TTBP, CH2Cl2 −60 °C, 100 % (8 c); g) Na, MeOH/THF, 99 % (9 a), 84 % (9 b), 97 % (9 c); h) Methyl hex‐5‐ynoate, Pd(PPh3)2Cl2, PPh3, CuI, Et3N, 99 % (10 a), 77 % (10 b), 82 % (10 c); i) Pd/C, H2, THF/MeOH, 100 % (11 a), 82 % (11 b), 90 % (11 c); j) N2H4 .H2O, EtOH, 89 % (12 a), 100 % (12 b). 84 % (12 c).
To synthesize trisaccharide 8 b, first disaccharide acceptor 7 was generated by removal of the PMB ether in 3 using a catalytic amount of HCl in HFIP to give the required alcohol in quantitative yield. Coupling of acceptor 3 to glucose donor 13 a then gave trisaccharide 8 b in 88 % yield. After the benzoyl protecting groups were removed from the trisaccharides, the iodoaryl glycosides were coupled to methyl hex‐5‐ynoate using a Sonogashira cross coupling. Global deprotection of the trisaccharides and reduction of the triple bond was accomplished by a single hydrogenation reaction to provide the target trisaccharide having a methyl ester spacer, in excellent yields. The methyl esters could then be transformed into hydrazides 12 a, 12 b and 12 c which could be used for conjugation to BSA.
Scheme 2A depicts the assembly of the ND−O‐disaccharide 21 which was required for comparison. The synthesis started with the coupling of rhamnose donor 2 with methyl 9‐hydroxynonanoate under the activation Ph2SO/Tf2O, to give spacer equipped rhamnose 14 in good yield. After the C‐2‐O‐benzoyl ester was removed and replaced for the required methyl ether, the C‐4‐PMB ether could be cleaved which furnished acceptor 17 in excellent yield. This acceptor could be coupled to glucose donor 13 a, using Ph2SO/Tf2O, after which the benzoate ester was cleaved by treatment with NaOMe and the benzyl ether removed using a hydrogenation step. Finally, the methyl ester of the linker was transformed into the corresponding hydrazide to provide the required ND−O‐disaccharide 21 in quantitative yield.
Scheme 2.
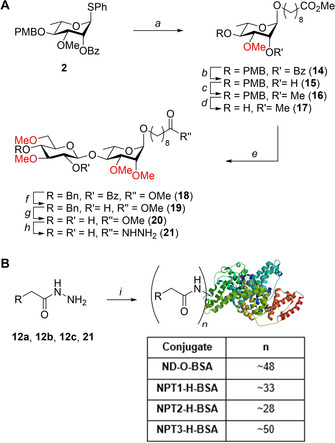
A: Disaccharide synthesis. a) Methyl 9‐hydroxynonanoate, Ph2SO, Tf2O, TTBP, CH2Cl2 −60 °C, 79 %; b), Na, MeOH/THF, 97 %; c), NaH, MeI, DMF, 91 %; d) HCl/HFIP, HFIP/CH2Cl2, 95 %, e) Donor 13 a, Ph2SO, Tf2O, TTBP, CH2Cl2 −60 °C, 84 %; f), Na, MeOH/THF, 82 %; g) Pd/C, H2, THF, 95 %; h) N2H4 .H2O, EtOH, 100 %. B) Conjugation of hydrazides to BSA. i) 1: HCl/dioxane, tBuONO, DMF, −30 °C, 2: BSA, Na2B4O7, NaHCO3, H2O, 0 °C.
The conjugation of the saccharides to BSA is depicted in Scheme 2B. First the hydrazides were transformed into the corresponding acyl azides using tert‐butyl nitrite and HCl in dioxane/DMF at −30 °C. After complete conversion the reaction was quenched and the resulting mixture was transferred to an ice‐cooled solution of BSA in aqueous Na2B4O7 and NaHCO3 (pH 9.2). After desalting and purification by means of gel filtration we obtained the conjugates NPT1‐H‐BSA, NPT2‐H BSA and NPT3‐H‐BSA having 33, 28 and 50 saccharides per BSA and the control conjugate ND−O‐BSA functionalized with 48 copies of the disaccharide on each protein, as revealed by SDS‐PAGE and MALDI‐TOF analyses.
Optimization of ELISA conditionsTo determine the optimal conditions of the synthesized PGL conjugates for application in ELISAs detecting human anti‐PGL−I IgM antibodies, both the coating concentration (PGL−I) and sample dilution (serum) were investigated. Three different coating concentrations (400, 200, and 100 ng/well) of ND−O‐BSA and NPT1‐H‐BSA were compared to the concentration used in the routine anti‐PGL−I IgM ELISAs based on ND−O‐HSA (200 ng/well). Coating concentration marginally affected the ELISA result (Figure S1 in the Supporting Information). Therefore, the concentration of the routine ELISA (200 ng/well) was used for all subsequent experiments in this study. To evaluate whether the same serum sample dilution as used for ND−O‐HSA in routine ELISAs (1 : 400) can be applied, serum dilutions ranging from a 1 : 10 to 1 : 16000 were analyzed in ELISAs using ND−O‐BSA and NPT1‐H‐BSA conjugates. Antibody titration curves were similar for all three synthetic PGL conjugates, therefore a sample dilution of 1 : 400 (previously applied for ND−O‐HSA) was used for all subsequent experiments (Figure S2).
Analysis of leprosy patients’ sera using NPT1‐H‐BSA in ELISA
Serum samples of leprosy patients with varying anti‐M. leprae PGL−I specific IgM levels (S1–S7), as pre‐determined using ND−O‐HSA, [8] as well as control samples from an leprosy endemic (S8) and non‐endemic region (S9) were assessed by ELISA using the newly generated NPT1‐H‐BSA and ND−O‐BSA conjugate as well as the previously used ND−O‐HSA conjugate (Figure 3). For both synthetic PGL conjugates, IgM levels highly correlated with those obtained using ND−O‐HSA as coating antigen (ND−O‐BSA: R 2=0.99; NPT1‐H‐BSA: R 2=0.98).
Figure 3.
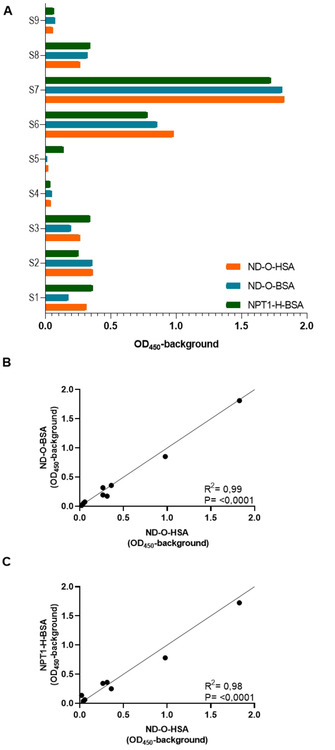
Correlation of two newly synthesized PGL conjugates (ND−O‐BSA and NPT1‐H‐BSA) with ND−O‐HSA in ELISA. Sera of leprosy patients (S1–S7) with various anti‐PGL−I IgM levels as pre‐determined from ND−O‐HSA and control samples from an endemic (S8) and non‐endemic region (S9) were tested in ELISAs using the three synthetic PGL conjugates. A) OD450 values with the background subtracted (OD450−background; y‐axis) as detected by using ND−O‐HSA (orange), ND−O‐BSA (blue) and NPT1‐H‐BSA (green) representing IgM antibody levels. B) Correlation of IgM binding (OD450−background) to ND−O‐HSA (x‐axis) and ND−O‐BSA (y‐axis). C) Correlation of IgM binding (OD450−background) to ND−O‐HSA (x‐axis) and NPT1‐H‐BSA (y‐axis).
Application of PGL conjugates in POC rapid test UCP‐LFA
The data obtained using ELISAs indicated that detection of IgM antibodies directed against both ND−O‐BSA and NPT1‐H‐BSA correlated highly with the ND−O‐HSA‐specific IgM levels. As leprosy diagnostics should allow POC testing in the field, these two synthetic conjugates were incorporated into the UCP‐LFA format.[ 8 , 12 , 13 ] Ten banked serum samples of leprosy patients were tested and similar results were obtained for the three different synthetic PGL conjugates (Figure S3). The correlation between the three synthetic PGL conjugates was further evaluated in the UCP‐LFA using a larger sample set of banked leprosy patients’ sera (n=92). Again the IgM levels directed against ND−O‐HSA significantly correlated with the IgM levels detected using the ND−O‐BSA (R 2=0.95) and NPT1‐H‐BSA UCP‐LFA (R 2=0.91; Figure 4). This indicates that the newly synthesized PGL−Is are well applicable to the UCP‐LFA format.
Figure 4.
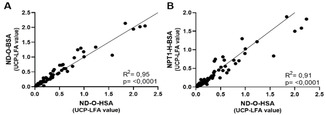
Correlation of ND−O‐HSA UCP‐LFA with ND−O‐BSA and NPT1‐H‐BSA UCP‐LFA. Correlation of the UCP‐LFA for the three different synthetic PGL conjugates was assessed using 92 samples. A) Correlation of the UCP‐LFA values representing the levels of IgM detected by using ND−O‐HSA UCP‐LFA (x‐axis) and ND−O‐BSA UCP‐LFA (y‐axis). B) Correlation of the UCP‐LFA values representing the levels of IgM detected by using the ND−O‐HSA UCP‐LFA (x‐axis) and NPT1‐H‐BSA UCP‐LFA (y‐axis).
Stability of ND‐O‐BSA and NPT1‐H‐BSA in UCP‐LFA
To assess the stability of the different PGL conjugates in the UCP‐LFA format, aliquots of the same 92 samples were tested one month after the first test time point. UCP‐LFA values of strips with NPT1‐H‐BSA measured one month after the first time point showed good correlation with the initial UCP‐LFA values (R 2=0.95), similar to ND−O‐HSA (R 2=0.96; Figure S4). For the ND−O‐BSA UCP‐LFA, more discrepancies were observed between the two timepoints, resulting in a reasonable correlation (R 2=0.64). This indicates that the NPT1‐H‐BSA conjugate was more stable in the UCP‐LFA format than ND−O‐BSA. Additionally, for a small set of five serum samples the stability was tested at seven different timepoints ranging from two months to thirteen months after production. Little variation was observed between the time points for test sera. Furthermore, analysis of the negative control serum sample at all time points indicated that no background signal developed in aging strips (Figure S5). These results thus show that UCP‐LFAs with the new conjugates, especially NPT1‐H‐BSA, have consistent results over time and remain stable for more than one year.
Assessment of NPT2‐H‐BSA and NPT3‐H‐BSA in ELISA and UCP‐LFA
Next, the two additional trisaccharide conjugates, NPT2‐H‐BSA and NPT3‐H‐BSA were also assessed in ELISA and UCP‐LFA alongside the earlier described PGL conjugates. Whereas NPT2‐H‐BSA showed a similar pattern as ND−O‐BSA and NPT1‐H‐BSA, considerably lower levels of IgM were binding to NPT3‐ H‐BSA both in ELISA and UCP‐LFA (Figure 5).
Figure 5.
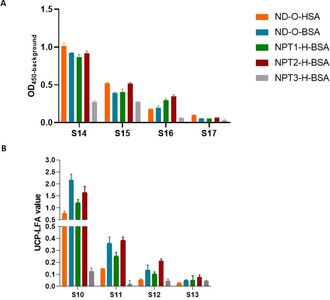
Comparison of IgM responses to five different synthetic PGLs in ELISA and UCP‐LFA. IgM responses to ND−O‐HSA (orange), ND−O‐BSA (blue), NPT1‐H‐BSA (green), NPT2‐H‐BSA (red), and NPT3‐H‐BSA were evaluated in A) ELISA and B) UCP‐LFA by using patient samples. A) OD450−background (y‐axis) represents the IgM antibody levels detected by ELISA against the five different synthetic PGLs. B) The UCP‐LFA values (y‐axis; arbitrary units) as determined by using the UCP‐LFA indicate the IgM antibody levels directed against the five different synthetic PGLs.
This result shows the importance of the methylation pattern of the trisaccharides and indicates that human anti‐PGL IgM binds to NPT3‐H‐BSA to a lesser extent. [36] In a larger sample set (n=210) NPT2‐H‐BSA UCP‐LFA values correlated very well with the UCP‐LFA values obtained using the ND−O‐BSA (R 2=0.87) and NPT1‐H‐BSA (R 2=0.92) UCP‐LFA (Figure S6). The correlation with NPT3‐H‐BSA was not assessed in this sample set, as only low IgM responses were observed for this UCP‐LFA (Figure 5).
Conclusion
In this study we have generated a set of BSA conjugates of M. leprae phenolic glycolipid trisaccharides connecting the glycans to the protein via a hexanoic acid linker attached to the anomeric phenol on the reducing end, and used these for detection of anti‐PGL IgM antibodies. The conjugates were evaluated as coating antigen both in ELISA and lateral flow format (UCP‐LFA) in order to investigate the influence of the phenol on the reducing end as well as the methylation pattern of the three glycoforms. The required glycans were successfully synthesized using a route based on thioglycosides and an iodophenol bearing rhamnoside, which allowed for the functionalization with a linker through a Sonogashira coupling, a strategy which will also be applied to the total synthesis of PGL−I, PGL‐II and PGL‐III. In ELISA the conjugates showed a high correlation with results obtained with ND−O‐HSA as coating antigen. Based on this data the conjugates were incorporated into the UCP‐LFA format suitable for POC testing in the field. A high correlation with the results of ND−O‐HSA was found here as well and therefore the conjugates are thought to be well applicable to the format. The stability of NPT1‐H‐BSA in the UCP‐LFA format was also assessed yielding consistent results over a period of 13 months, without development of any background signal within this time frame. This is advantageous for the application of this format in low resource areas where a cold chain is not always available. When the three trisaccharides NPT1, NPT2 and NPT3 were compared, significantly lower levels of IgM were found to bind to the latter trisaccharide. This is in line with previous assessments which determined the C‐3‐O‐methyl of the terminal glucose to be a highly important structural determinant for antibody binding. [18]
In summary, our data indicate that trisaccharide conjugates represent robust targets for the detection of anti‐PGL−I antibodies in point‐of‐care (POC) tests. The conjugates developed here can be used in future field studies in leprosy endemic areas.
Experimental Section
Study cohorts: HIV‐negative, treated and untreated leprosy patients and controls were recruited on a voluntary basis at the Dept. Dermatology, Erasmus MC, University Medical Center, Rotterdam, The Netherlands. This study was performed according to the Helsinki Declaration. All patients received treatment according to national guidelines. Ethical approval for the study was obtained from the ethical boards in The Netherlands (MEC‐2012‐589).
Synthesis: The experimental procedures and data of all newly synthesized compounds can be found in the supporting information.
Conjugation: [39] Hydrazide (100 equiv) was dissolved in DMF (0.05 M) and cooled to −30 °C (DCE bath, liquid N2). HCl in dioxane (400 equiv) was then added, followed by a solution of tBuONO in DMF (1 : 10, 800 equiv). The reaction was stirred until the starting material was converted to a higher running spot on TLC (MeOH/CH2Cl2 1 : 9) after which DIPEA (1000 equiv) was added. The cold solution was then transferred to a 0 °C solution of BSA (1.0 equiv) in borax buffer (0.1 mM, pH 9.2) and stirred overnight while slowly warming to RT. The buffered solution was diluted (1 : 14) with Milli‐Q and spun down using a 3 kDa MWCO filter. The retentate was diluted to 4 mL with Milli‐Q and transferred to a 10 kDa MWCO filter. After spinning down the new retentate was purified by means of gel filtration with sephadex G‐75 medium in Milli‐Q. The product in the void volume was lyophilized and used without further purification.
Quality control for synthesized conjugates: The conjugates were analyzed with SDS‐PAGE with BSA as a reference and stained with Coomassie Brilliant Blue to ensure no unconjugated protein was present. The sugar content was estimated using the phenol/sulfuric acid and protein concentrations were determined using a BCA kit (Pierce). Finally the amount of sugars per BSA was determined using MALDI‐TOF analysis (see the Supporting Information). The measurement procedure was as follows: 1 μL of sample solution (2 mg/mL in 7 : 3 MeCN:H2O+0.1 % TFA) was mixed together with 1 μL of 3,5‐dimethoxy‐4‐hydroxycinnamic acid and the dried‐droplet sample preparation method was applied. Spectra were assembled from 2000 shots in the linear mode with a 1 kHz laser.
PGL‐I ELISA: The PGL−I ELISA was performed as previously described. [8] Briefly, 200 ng synthetic PGL was coated per well in 50 μL in 0.1 M Na2CO3/NaHCO3 buffer (pH 9.6) at 4 °C overnight. After blocking with 200 μL PBS/1 %BSA/0.05 % Tween‐80 per well for 1 h, 50 μL of 1 : 400 diluted sample was added and incubated for 2 h at room temperature. Then, 50 μL per well of a 1 : 8000 dilution of anti‐human IgM‐HRP, (A6907, Sigma–Aldrich) in 0.05 %Tween 20/PBS was incubated for 2 h. In between each step the wells were washed 3 times with PBS/0.05 % Tween‐20. 50 μL of 3,3’,5,5’‐tetramethylbenzidine (TMB) was added and the color reaction was stopped using H2SO4 after 10–15 min. Absorbance was determined at a wavelength of 450 nm.
ND‐O‐HSA: The disaccharide epitope (3,6‐di‐O‐methyl‐β‐d‐glucopyranosyl(1→4)2,3‐di‐O‐methyl‐α‐l‐rhamnopyranoside) coupled to human serum albumin (designated ND−O‐HSA) was obtained through the Biodefense and Emerging Infections Research Resources Repository (https://www.beiresources.org/).
UCP‐LFAs: PGL−I lateral flow strips were produced as described earlier. [10] In short, the test line consists of 100 ng synthetic PGL−I and the flow control line of 100 ng Rabbit‐anti‐Goat (RαG; G4018, Sigma–Aldrich). Conjugates of UCP particles with goat anti‐human IgM (I0759, Sigma–Aldrich) at a concentration of 50 μg antibody per mg UCP were applied to the sample/conjugate pad at a density of 400 ng. Samples were diluted 50‐fold in high salt finger stick buffer supplemented with 1 % (v/v) Triton X‐100 (HSFS; 100 mM Tris pH 8, 270 mM NaCl, 1 % (w/v) BSA). 50 μL of diluted sample was added to microtiter plate wells before LF strips were placed in the corresponding wells. Immunochromatography was allowed to continue for at least 30 min until dry. LF strips were analyzed using a UCP dedicated benchtop reader (UPCON; Labrox, Finland). Test results are displayed as an arbitrary value with Test signal normalized to the flow‐control signal based on fluorescence units (RFUs) measured at the respective lines. Strips were stored in containers with silica dry packs at room temperature. Containers were sealed with parafilm.
Statistical analysis: Graphpad Prism version 8.00 for Windows was used to determine the correlation (R 2) between the different tests performed. The statistical significance level used was p≤0.05
Conflict of interest
The authors declare no conflict of interest.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary
Acknowledgements
We thank M. C. Araman for the MALDI‐TOF measurements and Dr. C. van Hees (ErasmusMC, Rotterdam) for providing the sera of leprosy patients. The Netherlands Organisation for Scientific Research (NWO) is acknowledged for financial support (TOP‐PUNT for G.A.M.).
J. H. M. van Dijk, A. van Hooij, L. M. Groot, J. Geboers, R. Moretti, E. Verhard-Seymonsbergen, D. de Jong, G. A. van der Marel, P. L. A. M. Corstjens, J. D. C. Codée, A. Geluk, ChemBioChem 2021, 22, 1487.
Contributor Information
Prof. Jeroen D. C. Codée, Email: jcodee@chem.leidenuniv.nl.
Annemieke Geluk, Email: a.geluk@lumc.nl.
References
- 1. Fischer M., JDDG J. Ger. Soc. Dermatol. 2017, 15, 801–827. [DOI] [PubMed] [Google Scholar]
- 2. Scollard D. M., Adams L. B., Gillis T. P., Krahenbuhl J. L., Truman R. W., Williams D. L., Clin. Microbiol. Rev. 2006, 19, 338–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barreto J. G., Frade M. A. C., Bernardes Filho F., da Silva M. B., Spencer J. S., Salgado C. G., Curr. Infect. Dis. Rep. 2017, 19, 23. [DOI] [PubMed] [Google Scholar]
- 4. Oldenburg R., Mayau V., Prandi J., Arbues A., Astarie-Dequeker C., Guilhot C., Werts C., Winter N., Demangel C., Front. Immunol. 2018, 9, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Doz-Deblauwe É., Carreras F., Arbues A., Remot A., Epardaud M., Malaga W., Mayau V., Prandi J., Astarie-Dequeker C., Guilhot C., Demangel C., Winter N., Front. Immunol. 2019, 10, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Madigan C. A., Cambier C. J., Kelly-Scumpia K. M., Scumpia P. O., Cheng T. Y., Zailaa J., Bloom B. R., Moody D. B., Smale S. T., Sagasti A., Modlin R. L., Ramakrishnan L., Cell 2017, 170, 973–985.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Spencer J. S., Kim H. J., Wheat W. H., Chatterjee D., Balagon M. V., Cellona R. V., Tan E. V., Gelber R., Saunderson P., Duthie M. S., Reece S. T., Burman W., Belknap R., Mac Kenzie W. R., Geluk A., Oskam L., Dockrell H. M., Brennan P. J., Clin. Vaccine Immunol. 2011, 18, 260–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Van Hooij A., Fat E. M. T. K., Van Den Eeden S. J. F., Wilson L., Da Silva M. B., Salgado C. G., Spencer J. S., Corstjens P. L. A. M., Geluk A., Sci. Rep. 2017, 7, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Van Beers S., Hatta M., Klatser P. R., Int. J. Lepr. Other Mycobact. Dis. 1999, 67, 243–249. [PubMed] [Google Scholar]
- 10. Corstjens P. L. A. M., van Hooij A., Tjon Kon Fat E. M., Alam K., Vrolijk L. B., Dlamini S., da Silva M. B., Spencer J. S., Salgado C. G., Richardus J. H., van Hees C. L. M., Geluk A., Clin. Biochem. 2019, 66, 76–82. [DOI] [PubMed] [Google Scholar]
- 11. Corstjens P. L. A. M., Van Hooij A., Tjon E. M., Fat K., Wilson L., Geluk A., Clin. Vaccine Immunol. 2016, 23, 515–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bobosha K., Tjon Kon Fat E. M., van den Eeden S. J. F., Bekele Y., van der Ploeg-van Schip J. J., de Dood C. J., Dijkman K., Franken K. L. M. C., Wilson L., Aseffa A., Spencer J. S., Ottenhoff T. H. M., Corstjens P. L. A. M., Geluk A., PLoS Neglected Trop. Dis. 2014, 8, 2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schilling A. K., van Hooij A., Corstjens P., Lurz P. W. W., DelPozo J., Stevenson K., Meredith A., Geluk A., Eur. J. Wildl. Res. 2019, 65, 1–5. [Google Scholar]
- 14. Bührer S. S., Smits H. L., Gussenhoven G. C., Van Ingen C. W., Klatser P. R., Am. J. Trop. Med. Hyg. 1998, 58, 133–136. [DOI] [PubMed] [Google Scholar]
- 15. Buhrer-Sékula S., Smits H. L., Gussenhoven G. C., Van Leeuwen J., Amador S., Fujiwara T., Klatser P. R., Oskam L., J. Clin. Microbiol. 2003, 41, 1991–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Spencer J. S., Brennan P. J., Lepr. Rev. 2011, 82, 344–357. [PubMed] [Google Scholar]
- 17. Chatterjee D., Cho S. N., Brennan P. J., Aspinall G. O., Carbohydr. Res. 1986, 156, 39–56. [DOI] [PubMed] [Google Scholar]
- 18. Fujiwara T., Hunter S. W., Cho S. N., Aspinall G. O., Brennan P. J., Infect. Immun. 1984, 43, 245–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chatterjee D., Douglas J. T., Cho S. N., Rea T. H., Gelber R. H., Aspinall G. O., Brennan P. J., Glycoconjugate J. 1985, 2, 187–208. [Google Scholar]
- 20. Fujiwara T., Aspinall G. O., Hunter S. W., Brennan P. J., Carbohydr. Res. 1987, 163, 41–52. [DOI] [PubMed] [Google Scholar]
- 21. Chatterjee D., Cho S. N., Stewart C., Douglas J. T., Fujiwara T., Brennan P. J., Carbohydr. Res. 1988, 183, 241–260. [DOI] [PubMed] [Google Scholar]
- 22. Fujiwara T., Izumi S., Agric. Biol. Chem. 1987, 51, 2539–2547. [Google Scholar]
- 23. Chanteau S., Cartel J. L., Roux J., Plichart R., Bach M. A., Chanteau S., J. Infect. Dis. 1988, 157, 770–776. [DOI] [PubMed] [Google Scholar]
- 24. Arbues A., Lugo-Villarino G., Neyrolles O., Guilhot C., Astarie-Dequeker C., Front. Cell. Infect. Microbiol. 2014, 4, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Arbués A., Malaga W., Constant P., Guilhot C., Prandi J., Astarie-Dequeker C., ACS Chem. Biol. 2016, 11, 2865–2875. [DOI] [PubMed] [Google Scholar]
- 26. Cambier C. J., O'Leary S. M., O'Sullivan M. P., Keane J., Ramakrishnan L., Immunity 2017, 47, 552–565.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Elsaidi H. R. H., Lowary T. L., Chem. Sci. 2015, 6, 3161–3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Elsaidi H. R. H., Lowary T. L., ChemBioChem 2014, 15, 1176–1182. [DOI] [PubMed] [Google Scholar]
- 29. Elsaidi H. R. H., Barreda D. R., Cairo C. W., Lowary T. L., ChemBioChem 2013, 14, 2153–2159. [DOI] [PubMed] [Google Scholar]
- 30. Zheng R. B., Jégouzo S. A. F., Joe M., Bai Y., Tran H. A., Shen K., Saupe J., Xia L., Ahmed M. F., Liu Y. H., Patil P. S., Tripathi A., Hung S. C., Taylor M. E., Lowary T. L., Drickamer K., ACS Chem. Biol. 2017, 12, 2990–3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lewallen D. M., Siler D., Iyer S. S., ChemBioChem 2009, 10, 1486–1489. [DOI] [PubMed] [Google Scholar]
- 32. Tessier M. B., Grant O. C., Heimburg-Molinaro J., Smith D., Jadey S., Gulick A. M., Glushka J., Deutscher S. L., Rittenhouse-Olson K., Woods R. J., PLoS One 2013, 8, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Barroso S., Castelli R., Baggelaar M. P., Geerdink D., Terhorst B., Casas-Arce E., Overkleeft H. S., Van Der Marel G. A., Codée J. D. C., Minnaard A. J., Angew. Chem. Int. Ed. 2012, 51, 11774–11777; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2012, 124, 11944–11947. [Google Scholar]
- 34. Codée J. D. C., Litjens R. E. J. N., Den Heeten R., Overkleeft H. S., Van Boom J. H., Van Der Marel G. A., Org. Lett. 2003, 5, 1519–1522. [DOI] [PubMed] [Google Scholar]
- 35. Volbeda A. G., Kistemaker H. A. V., Overkleeft H. S., Van Der Marel G. A., Filippov D. V., Codée J. D. C., J. Org. Chem. 2015, 80, 8796–8806. [DOI] [PubMed] [Google Scholar]
- 36.Although we cannot rule out the potential negative effect of the high glycan loading of the NPT3-H-BSA conjugate on antibody binding,[37] Fujiwara et al.[38] have shown that glycan loading had little effect on antibody binding to a NT−P-BSA conjugate with different degrees of loading (12–52 sugars/BSA).
- 37. Oyelaran O., Li Q., Farnsworth D., Gildersleeve J. C., J. Proteome Res. 2009, 8, 3529–3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fujiwara T., Izumi S., Wu Q., Japanese J. Lepr. 1991, 60, 132–138. [DOI] [PubMed] [Google Scholar]
- 39. Zhang J., Chatterjee D., Brennan P. J., Spencer J. S., Liav A., Bioorg. Med. Chem. Lett. 2010, 20, 3250–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary


