Abstract
Background: Osteomyelitis is an inflammatory process characterized by progressive bone destruction. Moreover, chronic bacterial osteomyelitis is regarded as a difficult-to-treat clinical entity due to its long-standing course and frequent infection recurrence. However, the role of genetic factors in the occurrence and development of bacterial osteomyelitis is poorly understood.
Methods: We performed a systematic review to assess the frequency of individual alleles and genotypes of single-nucleotide polymorphisms (SNPs) among patients with bacterial osteomyelitis and healthy people to identify whether the SNPs are associated with the risk of developing bacterial osteomyelitis. Then, gene ontology and Kyoto Encyclopedia of Gene and Genomes analyses were performed to identify the potential biological effects of these genes on the pathogenesis of bacterial osteomyelitis.
Result: Fourteen eligible studies containing 25 genes were analyzed. In this review, we discovered that the SNPs in IL1B, IL6, IL4, IL10, IL12B, IL1A, IFNG, TNF, PTGS2, CTSG, vitamin D receptor (VDR), MMP1, PLAT, and BAX increased the risk of bacterial osteomyelitis, whereas those in IL1RN and TLR2 could protect against osteomyelitis. The bioinformatic analysis indicated that these osteomyelitis-related genes were mainly enriched in inflammatory reaction pathways, suggesting that inflammation plays a vital role in the development of bacterial osteomyelitis. Furthermore, functional notation for 25 SNPs in 17 significant genes was performed using the RegulomeDB and NCBI databases. Four SNPs (rs1143627, rs16944, rs2430561, and rs2070874) had smaller scores from regulome analysis, implying significant biological function.
Conclusion: We systematically summarized several SNPs linked to bacterial osteomyelitis and discovered that these gene polymorphisms could be a genetic factor for bacterial osteomyelitis. Moreover, further large-scale cohort studies are needed to enhance our comprehensive understanding of the development of osteomyelitis to provide earlier individualized preventions and interventions for patients with osteomyelitis in clinical practice.
Keywords: osteomyelitis, genetic polymorphism, genotype, susceptibility, systematic review
Introduction
Osteomyelitis is an inflammatory process characterized by progressive bone destruction. It is a bone infection mainly caused by microorganism invasion, and Staphylococcus aureus is the bacterial pathogen frequently isolated from patients with posttraumatic and hematogenous osteomyelitis (Lew and Waldvogel, 2004; Olson and Horswill, 2013). Commonly, according to the etiology, osteomyelitis could be divided into three types: posttraumatic osteomyelitis, hematogenous osteomyelitis, and osteomyelitis caused by vascular insufficiency (Lew and Waldvogel, 2004). Posttraumatic osteomyelitis predominantly occurs following open traumatic fracture, skeleton surgery, or prosthetic joint replacement. Meanwhile, hematogenous osteomyelitis typically occurs in children, characterized by the spread of bacteria from a lesion to the bone through the bloodstream. Osteomyelitis secondary to vascular insufficiency particularly occurs in patients with diabetes or diabetic foot infection (Lew and Waldvogel, 2004). Chronic osteomyelitis is regarded as a difficult-to-treat clinical entity due to its long-standing course and frequent infection recurrence, with a high risk of morbidity and mortality (Valour et al., 2014). Patients with chronic osteomyelitis have a higher incidence of psychosocial impairment (Tseng et al., 2014) and have healthcare and economic burden (Kapadia et al., 2016). The pathogenesis of osteomyelitis is linked to both environmental and genetic factors. Currently, several pieces of evidence have suggested that genetic predisposition plays an essential role in the pathogenesis of osteomyelitis (Chen et al., 2017; Paludo et al., 2017). With rapid development and application of sequencing and genetic association analysis for complicated diseases, genetic variants that potentially contribute to the occurrence of osteomyelitis are widely investigated.
Single-nucleotide polymorphisms (SNPs) of DNA sequences are common in the population. Many SNPs in genes related to the occurrence of osteomyelitis have been widely reported. For example, TaqI (rs731236) and FokI (rs2228570) of the vitamin D receptor (VDR) gene polymorphism may contribute to the susceptibility of chronic osteomyelitis (Jiang et al., 2016). This review was conducted to examine the individual allele frequency or genotype of gene variants among patients with osteomyelitis to identify whether gene polymorphisms are associated with the probability of developing osteomyelitis.
Materials and Methods
This systematic review was performed based on the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines (Moher et al., 2009).
Literature Search Strategy
A systematic literature search was conducted using the PubMed, EMBASE, and Web of Science databases. The following terms— “genetic polymorphism,” “genetic variants,” “DNA polymorphism,” “single-nucleotide polymorphism,” “SNP,” “osteomyelitis,” and “bone infection” —were used to search all eligible studies on the relationship between SNPs and the risk of osteomyelitis and published until the end of December 2020. Additional studies were identified by screening the reference lists of the included studies. English articles were included in this review. Detailed search strategies are provided in Supplementary Table 1.
Inclusion and Exclusion Criteria
The selected studies fulfilled the following inclusion criteria:
Bacterial osteomyelitis was definitely diagnosed according to the standard criteria.
The study reported the association between SNPs and susceptibility to osteomyelitis.
Sufficient data could be extracted from the study.
The study was limited to case–control or cohort studies on humans.
The exclusion criteria were as follows:
The same study was duplicated or overlapped.
Case reports, letters, meta-analyses, reviews, or studies on animals were excluded.
The study reported the correlation between other types of genetic polymorphisms and osteomyelitis.
The study reported the association of gene polymorphism with non-bacterial osteomyelitis.
We screened the literature by title and abstract. For the abstracts that we could not fully access, we obtained the full text to complete the assessment before we decided to include or exclude them. Two reviewers independently screened the articles and discussed uncertain publications to resolve disagreements.
Quality Assessment
Two reviewers (XP.X. and JB.L.) independently conducted a quality methodological assessment of the studies included in the review based on the Newcastle–Ottawa Scale (NOS; Stang, 2010). The NOS scores ranged from zero stars to nine stars. Studies with NOS scores of below six stars were excluded in the review, and those with a score of at least six stars were considered good quality. We resolved disagreements by discussion or consultation with the third reviewer (F.G.), if necessary.
Biological Function Annotation of Targeted Genes and SNPs
The protein–protein interaction network of the genes included in the review was constructed using the Search Tool for the Retrieval of Interacting Genes (STRING, version 11.0) database (https://string-db.org/). Hub genes are discovered using Cytoscape (version 3.4.0). Gene ontology (GO) and Kyoto Encyclopedia of Gene and Genomes (KEGG) analyses were performed to analyze the cell components, biological processes, and pathway enrichment of these osteomyelitis-related genes based on the DAVID database (version 6.8, https://david.ncifcrf.gov/tools.jsp/). P < 0.05 were used to denote statistical significance, and ≥3 enriched genes were considered significant as well.
The potential biological functions of SNPs were evaluated using the RegulomeDB (version 2.0, https://www.regulomedb.org/regulome-search/) and NCBI (https://www.ncbi.nlm.nih.gov/) databases. The two databases could predict whether gene variants affect transcription factor binding and gene expression.
Data Extraction and Statistical Analysis
The data extracted from each included studies were as follows: first author, year of publication, nation, study design, number of cases and controls, name of gene SNP, distribution of genotype and allele frequency in cases and controls, genotype method, and Hardy–Weinberg equilibrium (HEW) in controls. Two reviewers independently extracted important data from each included study. The data from each study were as follows: odds ratios (ORs), 95% confidence interval (CI), p-value, and genetic model. P < 0.05 were used to denote statistical significance.
Results
Characteristics of the Included Studies
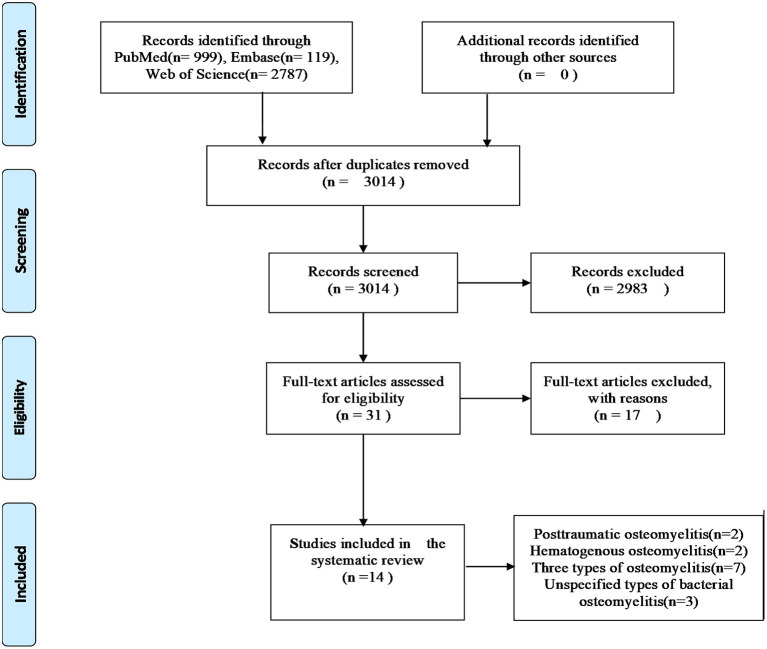
Based on our literature search strategies, 3,905 publications were obtained from three databases (Supplementary Table 1), of which 891 were removed because of duplication. Then, 2,983 of the remaining publications were excluded by browsing their titles and abstracts. The full texts of 31 publications were obtained, of which 17 were excluded because of the following reasons: nine studies focused on chronic non-bacterial osteomyelitis; in two studies, SNPs were not involved; two studies were not case–control studies; in three studies, controls were not healthy individuals; and one study had inconsistent data. Eventually, 14 studies fulfilled our inclusive criteria (including 1,248 cases and 1,712 controls). The document selection process is detailed in the flowchart (Moher et al., 2009), shown in Figure 1. Among the 14 included studies, two have reported the associations between SNPs and posttraumatic bacterial osteomyelitis (Wang et al., 2017; Jiang et al., 2020), and two studies have reported the relationship between SNPs and hematogenous bacterial osteomyelitis (Osman et al., 2015, 2016). Seven studies have reported the associations between SNPs and three types of osteomyelitis (posttraumatic, hematogenous, and vascular insufficiency-related bacterial osteomyelitis; Asensi et al., 2003; Montes et al., 2006; Valle-Garay et al., 2013; Jiang et al., 2016; Hou et al., 2018; Perez-Is et al., 2019; Zhao et al., 2020). The other three studies have reported the relationship of SNP with unspecified types of bacterial osteomyelitis (Ocana et al., 2007; Tsezou et al., 2008; Kong et al., 2017). This systematic review involved 1,248 patients with bacterial osteomyelitis, including 719 patients with posttraumatic bacterial osteomyelitis, 190 patients with hematogenous bacterial osteomyelitis, 98 patients with vascular insufficiency-related bacterial osteomyelitis, and 241 patients with unspecified types of bacterial osteomyelitis. The characteristics of selected studies are summarized in Table 1. The qualities of the included studies were evaluated using the NOS, and the scores are also presented in Table 1. It should be noted that among the five articles from the same authors, the cases and controls of each article shared their own data. The two articles in the team of Asensi et al. also shared their own data (Asensi et al., 2003; Perez-Is et al., 2019). All genotyped gene SNPs in 11 studies complied with the Hardy–Weinberg equilibrium (HWE) for healthy controls (p > 0.05), and three studies did not mention the HWE results.
Figure 1.
The detailed process of document selection.
Table 1.
Characteristics of included studies.
| References | Nation | SNPs | Case | Control | Types of case and control | Study design | Genotype method | Nos. |
|---|---|---|---|---|---|---|---|---|
| Zhao et al. (2020) | China | IFN-γ: rs2430561(874T/A) | 233 (185/48) | 200 (147/53) | Patients (P/H/V*:189/23/21) vs. healthy individuals | Case–control study | SNaPshot system | 7 |
| Perez-Is et al. (2019) | Spain | CTSG: rs45567233 | 329 (245/84) | 415 (295/120) | Patients (P/H/V:196/56/77) vs. healthy individuals | Case–control study | PCR-RFLP | 7 |
| Hou et al. (2018) | China | TNF-α: rs1799964, rs1800630, rs1799724, rs1800750,rs1800629, rs361525 | 233 (185/48) | 200 (147/53) | Patients (P/H/V:189/23/21) vs. healthy individuals | Case–control study | SNaPshot system | 7 |
| Jiang et al. (2016) | China | VDR: rs731236, rs1544410, rs2228570, rs7975232 | 233 (185/48) | 200(147/53) | Patients (P/H/V:189/23/21) vs. healthy individuals | Case–control study | SNaPshot system | 7 |
| Valle-Garay et al. (2013) | Spain | tPA: rs4646972, rs1799889 | 261 (186/75) | 299 (205/94) | Patients (P/H: 224 /37) vs. healthy individuals | Case–control study | PCR-RFLP | 8 |
| Montes et al. (2006) | Spain | TLR2: Arg753Gln TLR4: rs498670, rs498671 | 80 (54/26) | 155 | Patients (P/H:67/13) vs. healthy individuals | Case–control study | PCR-RFLP | 8 |
| Asensi et al. (2003) | Spain | IL-1α: rs1800587 IL-1β: rs1143634 TNF-α: rs 1800629 IL-6: rs1800795 | 52 | 109 | Patients (P/H:43/9) vs. healthy individuals | Case–control study | PCR-RFLP | 8 |
| Wang et al. (2017) | China | COX-2: rs20417, rs689466 | 189 (156/33) | 220 (167/53) | Patients (P:189) vs. healthy individuals | Case–control study | SNaPshot system | 7 |
| Jiang et al. (2020) | China | IL-1α: rs17561, rs1800587 IL-1β: rs16944, rs1143627, rs1143634, rs2853550 IL1RN: rs4251961, rs419598, rs315951 IL-4: rs2243248, rs2243250 IL-6: rs1800795, rs1800796, rs1800797 IL-8: rs4073, rs2227306, rs2227307 IL-10: rs3024491, rs3024496, rs1800871, rs1800872, rs1800896 IL17A: rs2275913 IF17F: rs763780 | 233 (185/48) | 200 (147/53) | Patients (P:189) vs. healthy individuals | Case–control study | SNaPshot system | 7 |
| Osman et al. (2016) | Saudi Arabia | IL-1β: rs16944, rs1143634 IL-1α: rs1800587 TLR2: rs3804099 TLR4: rs4986790, rs4986791 IL-1R: rs2234650 TNF-α: rs1800629, rs361525 IL1RN: rs315952 | 52 (22/30) | 103 (44/59) | Hematogenous osteomyelitis patients vs. healthy individuals | Case–control study | PCR | 6 |
| Osman et al. (2015) | Saudi Arabia | IL-4: rs2070874, rs2243248 IL-10: rs1800896, rs1800871, rs1800872 TGF-B1: rs1800470 IL-4R: rs1801275 IL-12B: rs3212227 IL-2: rs2069762 | 52 (22/30) | 103 (44/59) | Hematogenous osteomyelitis patients vs. healthy individuals | Case–control study | PCR | 6 |
| Kong et al. (2017) | China | MMP1: rs1799750, rs1144393 | 80 (41/40) | 81 (37/44) | Osteomyelitis patients vs. healthy individuals | Case–control study | PCR-RFLP | 6 |
| Ocana et al. (2007) | Spain | BAX: G(-248)A | 80 (53/27) | 220 | Osteomyelitis patients vs. healthy individuals | Case–control study | PCR-RFLP | 8 |
| Tsezou et al. (2008) | Greek | IL-1α: rs1800587 IL-4: rs2243248, rs2243250 IL-6: rs1800795 | 81 (42/39) | 110 (62/48) | Osteomyelitis patients vs. healthy individuals | Case–control study | PCR-SSP assay | 6 |
P/H/V represents the number of patients with posttraumatic osteomyelitis, or hematogenous osteomyelitis, or vascular insufficiency-related osteomyelitis.
Summary of the Outcomes
According to the included studies, we collected 25 SNPs in 17 significant genes (Table 2), and we classified the protein products encoded by the 17 significant genes into the following two categories.
Table 2.
Summary of significant genes and polymorphisms of included studies.
| References | Gene | SNPs | Genotype | Unadjusted p-value* | OR (95%) | Risk or protective factor |
|---|---|---|---|---|---|---|
| Jiang et al. (2020) | IL-1β | rs16944 | GG+AG vs. AA | 0.017 | 1.854 (0.988–1.741) | Risk factor |
| GG vs. AA | 0.041 | 1.831 (1.026–3.267) | Risk factor | |||
| AG vs. AA | 0.022 | 1.869 (1.093–3.194) | Risk factor | |||
| rs1143627 | TT+CT vs. CC | 0.032 | 1.735 (1.050–2.868) | Risk factor | ||
| TT vs. CC | 0.040 | 1.839 (1.029–3.285) | Risk factor | |||
| Osman et al. (2016) | IL-1β | rs16944 | GG vs. AG+AA | 0.005 | 0.34 (0.16–0.73) | Protective factor |
| AA vs. GG+GA | 0.005 | 4.11 (1.51–11.21) | Risk factor | |||
| Asensi et al. (2003) | IL-1β | rs1143634 | TT vs. CC+CT | 0.014 | 6.98 (1.21–52.14) | Risk factor |
| Jiang et al. (2020) | IL1RN | rs4251961 | C vs. T | 0.01 | 0.519 (0.313–0.861) | Protective factor |
| CC+CT vs. TT | 0.005 | 0.446 (0.254–0.781) | Protective factor | |||
| CT vs. TT | 0.003 | 0.409 (0.227–0.737) | Protective factor | |||
| Jiang et al. (2020) | IL-6 | rs1800796 | CC+CG vs. GG | 0.029 | 4.184 (1.154–15.165) | Risk factor |
| CC vs. GG | 0.026 | 4.378 (1.197–16.007) | Risk factor | |||
| CG vs. GG | 0.046 | 3.834 (1.024–14.347) | Risk factor | |||
| Tsezou et al. (2008) | IL-6: | rs1800795 | CC+CG vs. GG | <0.01 | 2.05 (1.14–3.67) | Risk factor |
| Osman et al. (2015) | IL-4: | rs2070874 | C vs. T | 0.01 | 2.1 (1.92–3.71) | Risk factor |
| CC vs. TT+TC | 0.009 | 2.53 (1.25–5.13) | Risk factor | |||
| CT vs. CC+TT | 0.041 | 0.45 (0.21–0.97) | Protective factor | |||
| rs2243248 | GT vs. GG+TT | 0.006 | 9.18 (1.87–45.0) | Risk factor | ||
| Tsezou et al. (2008) | IL-4: | rs2243248 | TT+TG vs. GG | <0.01 | 21.33 (9.49–47.96) | Risk factor |
| rs2243250 | TT+TC vs. CC | <0.01 | 10.09 (5.15–19.75) | Risk factor | ||
| Osman et al. (2015) | IL-10 | rs1800871 | A vs. G | 0.044 | 1.7 (1.01–2.98) | Risk factor |
| Osman et al. (2015) | IL-12B | rs3212227 | GG vs. TT+TG | 0.049 | 2.04 (1.0–4.16) | Risk factor |
| Tsezou et al. (2008) | IL-1α: | rs1800587 | TT+TC vs. CC | <0.01 | 3.33 (1.78–6.24) | Risk factor |
| Asensi et al. (2003) | IL-1α | rs1800587 | TT vs. CC+CT | 0.008 | 3.7 (1.35–10.34) | Risk factor |
| Zhao et al. (2020) | IFN-γ | rs2430561 | A vs. T | 0.013 | 1.742 (1.122–2.705) | Risk factor |
| AA + AT vs. TT | 0.017 | 1.820 (1.111–2.983) | Risk factor | |||
| AT vs. TT | 0.029 | 1.781 (1.057–3.002) | Risk factor | |||
| Hou et al. (2018) | TNF-α | rs1799964 | T vs. A | 0.035 | 1.466 (1.025–2.095) | Risk factor |
| TT vs. AA+AT | 0.048 | 1.516 (1.004–2.290) | Risk factor | |||
| Osman et al. (2016) | TLR2 | rs3804099 | T vs. C | 0.004 | 0.56 (0.34–0.94) | Protective factor |
| TT vs. CC+CT | 0.011 | 0.23 (0.08–0.71) | Protective factor | |||
| Montes et al. (2006) | TLR4 | rs4986790 | GG vs. AA+AG | 0.038 | NA | |
| rs4986791 | TT vs. CC+CT | 0.038 | NA | |||
| Wang et al. (2017) | COX-2 | rs689466 | GG vs. AA+AG | 0.018 | 1.74 (1.098–2.755) | Risk factor |
| Perez-Is et al. (2019) | CTSG | rs45567233 | G vs. A | 0.01 | 1.78 (1.14–2.78) | Risk factor |
| AG vsGG+AA | 0.015 | 1.78 (1.11–2.84) | Risk factor | |||
| Jiang et al. (2016) | VDR | rs731236 | C vs. T | 0.044 | 1.830 (1.009–3.319) | Risk factor |
| CC+CT vs. TT | 0.05 | 1.887 (1.001–3.319) | Risk factor | |||
| rs2228570 | C vs. T | 0.029 | 1.347 (1.031–1.761) | Risk factor | ||
| CC+CT vs. TT | 0.042 | 1.594 (1.016–2.500) | Risk factor | |||
| CC vs. TT | 0.034 | 1.803 (1.046–3.106) | Risk factor | |||
| Kong et al. (2017) | MMP1 | rs1144393 | G vs. A | 0.033 | 1.622 (1.038–2.536) | Risk factor |
| GG vs. AA | 0.024 | 2.792 (1.127–6.917) | Risk factor | |||
| rs1799750 | 2G vs. 1G | 0.014 | 1.735 (1.115–2.701) | Risk factor | ||
| 2G/2G vs. 1G/2G +1G/1G | 0.025 | 2.605 (1.116–6.082) | Risk factor | |||
| Valle-Garay et al. (2013) | tPA | rs4646972 | I vs. D | 0.001 | 1.49 (1.17–1.91) | Risk factor |
| I/I vs. I/D+D/D | <0.0001 | 2.59 (1.74–3.87) | Risk factor | |||
| Ocana et al. (2007) | BAX | G(-248)A | A vs. G | 0.028 | 1.81 (1.06–3.07) | Risk factor |
No special significance can be ascribed to associations with p < 0.05.
Cytokine-Related Proteins
In seven case–control studies, 14 SNPs in nine genes encoding cytokine-related proteins (IL-1α, IL-1β, IL-1RN, IL-4, IL-6, IL-10, IL-12β, IFN-γ, and TNF-α) were investigated. Recently, Jiang et al. (2020) have reported a significant statistical difference in the genotype distribution of IL1B genes rs16944 and rs1143627 between patients with osteomyelitis and controls and revealed that the GG and AG genotypes of rs16944 and the TT and CT genotypes of rs1143627 were linked to the risk of posttraumatic osteomyelitis. Additionally, the CC and CG genotypes of rs1800796 located on the IL6 gene were associated with an increased risk of posttraumatic osteomyelitis. However, the mutant C allele and CT genotype of rs4251961 within IL1RN were considered a protective factor against posttraumatic osteomyelitis. Additionally, Asensi et al. (2003) have revealed that the TT genotype within IL1A rs1800587 and IL1B rs1143634 might be associated with the occurrence of osteomyelitis. Among patients with osteomyelitis, the TT genotype in rs1800587 was significantly associated with a decreased age at which the diagnosis of osteomyelitis is made. Tsezou et al. (2008) have reported that the results of dominant genetic models used for rs1800587, rs2243248, rs2243250, and rs1800795 demonstrated that IL1A, IL4, and IL6 SNPs could contribute to the genetic pathologies of osteomyelitis. The result of IL1A rs1800587 regarded as a risk factor for osteomyelitis was consistent with the outcome reported by Asensi et al. (2003). However, no significant difference was observed between patients with osteomyelitis and healthy controls regarding the individual alleles and genotypes of IL-1α rs1800587 in the studies of Jiang et al. (2020) and Osman et al. (2016). Osman et al. (2015) have reported that the population with the C allele or CC genotype might increase the susceptibility to hematogenous osteomyelitis, whereas the T allele or CT genotype could act as a protector factor. A heterozygous genetic model of rs2243248 has demonstrated that the mutant G allele contributed to hematogenous osteomyelitis, although the distribution difference of allele G and T frequencies among cases and controls was not statistically significant (p > 0.05). In addition, the allele A of rs1800871 within the IL10 gene and the GG genotype of IL12B gene rs3212227 were identified to contribute to hematogenous osteomyelitis. Osman et al. (2016) have also revealed that the AA genotype of rs16944 in IL1β could be a risk factor, whereas the allele G and GG genotype of this SNP were considered a protector against hematogenous osteomyelitis among Saudis. However, this result was contrary to the conclusion of the study by Jiang et al. (2020). Zhao et al. (2020) have recently reported that the mutant allele A in rs2430561 might be a risk factor of posttraumatic osteomyelitis, and individuals with the AT genotype in this gene might have a higher risk of developing posttraumatic osteomyelitis. Hou et al. (2018) have first reported that individuals with the TT genotype of the rs1799964 SNP in TNF might have a higher risk of developing extremity chronical osteomyelitis in China. However, the results of a study (Asensi et al., 2003) indicated that rs1799964 in TNF is not associated with the susceptibility to the development of chronic bacterial osteomyelitis.
Protein, Receptor, and Enzyme
In another seven case–control studies, 11 SNPs in eight genes encoding a protein (BAX), receptors (VDR, TLR2, and TLR4), and enzymes (CTSG, COX-2, MMP-1, and t-PA) were investigated. Ocana et al. (2007) have found that the frequency of the mutant A allele at position 248 within the BAX gene was higher in patients with osteomyelitis than that in healthy controls, which was linked to the lower expression of BAX and prolonged survival of peripheral blood neutrophils. Jiang et al. (2016) have identified that the frequencies of the mutant C allele of rs731236 (TaqI) and rs2228570 (FokI) were higher in patients with chronic osteomyelitis than those in healthy individuals. The result of the dominant genetic model of rs731236 and the dominant and homozygous genetic models of rs2228570 has suggested that VDR SNPs are significantly linked to the susceptibility of developing chronic osteomyelitis. Different genotypes in rs731236 (TaqI) and rs2228570 (FokI) polymorphism were significantly associated with serum TNF-α levels in patients with osteomyelitis. Osman et al. (2016) have found that the mutant T allele and TT genotype of rs3804099 in the TLR2 gene might protect against hematogenous osteomyelitis in the Saudi population. Montes et al. (2006) have demonstrated that although the contribution of the A allele of rs498670 and T allele of rs498671 in TLR4 did not differ between patients and controls, the outcomes of the recessive genetic model of rs498670 (GG genotype) and rs498671 (TT genotype) revealed that individuals with GG or TT would have an increased susceptibility to osteomyelitis. Perez-Is et al. (2019) have reported that the G allele of rs45567233 situated in CTSG was more frequent in patients with osteomyelitis than that in controls. The result has suggested that the G allele could be a risk factor, and people with the AG genotype would elevate the risk of osteomyelitis in a Spanish population. The association of the rs45567233 polymorphism with the susceptibility of osteomyelitis might be achieved by elevating serum CTSG activity and lactoferrin levels. Wang et al. (2017) have concluded that the G allele and GG genotype of rs689466 located in the PTGS2 gene (COX-2) could be considered a risk factor contributing to the onset of posttraumatic osteomyelitis. Serum C-reactive protein (p = 0.017) and IL-6 (p = 0.006) levels were significantly higher in patients with posttraumatic osteomyelitis accompanied with the GG genotype, instead of the CG genotype. Kong et al. (2017) have concluded that the G allele of rs1144393 in the MMP1 gene was regarded as a genetic risk factor and carriers of the GG genotype have an increased risk of osteomyelitis.
Bioinformatics Analysis
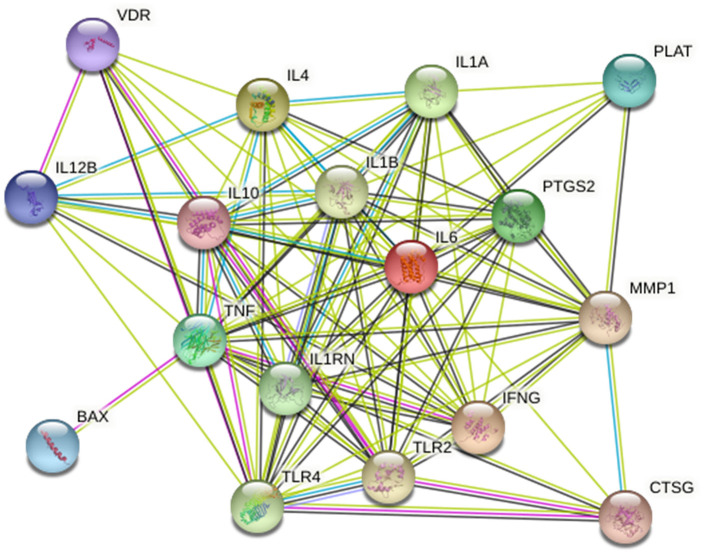
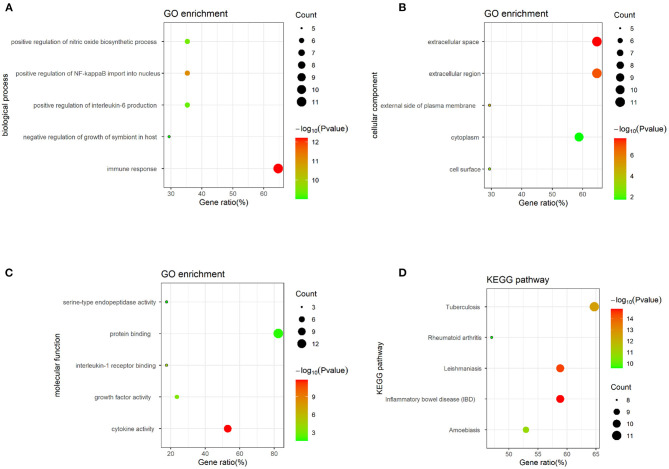
Twenty-five osteomyelitis-related genes were presented in the publications included in this review. The proteins encoded by osteomyelitis-related genes that were extracted from the included publications exhibited significant correlations (Figure 2). Among these genes, the node degree of TNF was the highest (Table 3). Additionally, the results of the GO and KEEG analysis of these osteomyelitis-related genes are shown in Figure 3. For biological processes, these genes were significantly enriched in the following terms: “immune response,” “positive regulation of NF-kappa B import into nucleus,” “positive regulation of nitric oxide biosynthetic process,” “positive regulation of interleukin-6 production,” and “negative regulation of growth of symbiont in host.” For cell components, these genes were enriched in the following terms: “extracellular space,” “extracellular region,” “external side of plasma membrane,” “cell surface,” and “cytoplasm.” For molecular functions, these genes were mainly enriched in the following terms: “cytokine activity,” “interleukin-1 receptor binding,” “growth factor activity,” “protein binding,” and “serine-type endopeptidase activity.” According to the result of the KEEG analysis, these genes mainly participated in the pathways including inflammatory bowel disease (IBD), leishmaniasis, tuberculosis, amoebiasis, and rheumatoid arthritis. The results indicated that inflammation is involved in the pathogenesis of osteomyelitis. In addition, 25 SNPs of 17 genes were reported to be significantly associated with the risk of osteomyelitis (Table 2). Many SNPs were mainly located in the intron or promotor region. According to regulome analysis, IL1B rs1143627, IL1B rs16944, IFNG rs2430561, and IL4 rs2070874 had scores of 1b, 1f, 2b, and 2b, respectively (Table 4). Smaller scores imply that SNPs have a greater biological functional significance.
Figure 2.
Protein–protein interaction network of osteomyelitis-related genes.
Table 3.
Hub genes among the PPI network.
| Gene symbol | Degree | Betweenness |
|---|---|---|
| TNF | 22 | 52.46152 |
| TLR4 | 21 | 10.46152 |
| CXCL8 | 21 | 10.46152 |
| IL6 | 21 | 10.46152 |
| TLR2 | 20 | 8.44535 |
| IL17A | 20 | 4.96152 |
| IFNG | 20 | 4.96152 |
| IL4 | 20 | 4.96152 |
| IL10 | 20 | 4.96152 |
Figure 3.
GO and KEGG analysis of osteomyelitis-related genes, containing (A) biological process, (B) cell component, (C) molecular function, and (D) biological pathway.
Table 4.
RegulomeDB score for all of these 23 selected SNPs.
| Chrom | Gene | SNP_ID | Functional consequence | Score ranking | Supporting data |
|---|---|---|---|---|---|
| chr2 | IL-1β | rs1143627 | 5′-UTR variant | 1b | eQTL+ TF binding + any motif + DNase Footprint + DNase peak |
| chr2 | IL-1β | rs16944 | Upstream transcript variant | 1f | eQTL+TF binding/DNase peak |
| chr12 | IFN-γ | rs2430561 | Intron variant | 2b | TF binding + any motif + DNase Footprint + DNase peak |
| chr5 | IL-4 | rs2070874 | 5′-UTR variant | 2b | TF binding + any motif + DNase Footprint + DNase peak |
| chr1 | COX2 | rs689466 | Upstream variant | 3a | TF binding + any motif + DNase peak |
| chr1 | IL-10 | rs1800871 | Upstream transcript variant | 3a | TF binding + any motif + DNase peak |
| chr6 | TNF-α | rs1799964 | Upstream transcript variant | 3a | TF binding + any motif + DNase peak |
| chr11 | MMP1 | rs1144393 | Upstream variant | 4 | TF binding + DNase peak |
| chr11 | MMP1 | rs1799750 | Upstream variant | 4 | TF binding + DNase peak |
| chr12 | VDR | rs731236 | Synonymous variant | 4 | TF binding + DNase peak |
| chr14 | CTSG | rs45567233 | Missense variant | 4 | TF binding + DNase peak |
| chr2 | IL1RN | rs4251961 | Intron variant | 4 | TF binding + DNase peak |
| chr5 | IL-4 | rs2243248 | Upstream transcript variant | 4 | TF binding + DNase peak |
| chr5 | IL-4 | rs2243250 | Upstream transcript variant | 4 | TF binding + DNase peak |
| chr7 | IL-6 | rs1800796 | 5′-UTR variant | 4 | TF binding + DNase peak |
| chr7 | IL-6 | rs1800795 | Intron variant | 4 | TF binding + DNase peak |
| chr12 | VDR | rs2228570 | Missense variant | 5 | TF binding or DNase peak |
| chr2 | IL-1α | rs1800587 | 5′-UTR variant | 5 | TF binding or DNase peak |
| chr2 | IL-1β | rs1143634 | Synonymous variant | 5 | TF binding or DNase peak |
| chr9 | TLR4 | rs4986790 | Missense variant | 6 | Other |
| chr4 | TLR2 | rs3804099 | Synonymous variant | 7 | Other |
| chr5 | IL12B | rs3212227 | 3′-UTR variant | 7 | Other |
| chr9 | TLR4 | rs4986791 | Missense variant | 7 | Other |
Discussion
Bacterial osteomyelitis contains a complex inflammatory reaction caused by invading microorganisms. S. aureus is the bacterial pathogen frequently associated with posttraumatic and hematogenous osteomyelitis (Lew and Waldvogel, 2004). Despite appropriate treatments through medications and surgery, up to 30% of osteomyelitis cases become chronic, resulting in serious disability and economic burden (Lew and Waldvogel, 2004). To solve the problem of intractable chronic osteomyelitis disease, exploring the etiology and pathology of osteomyelitis is necessary.
The occurrence of bacterial osteomyelitis is a complicated process caused by both genetic and environmental factors. Much attention has been paid to exploring the close association between host factors in terms of gene polymorphisms and the risk of osteomyelitis. This review was conducted to summarize the SNPs of 25 genes linked to the susceptibility of osteomyelitis based on published literature. According to our review, rs1143627 and rs1143634 in IL1B, rs1800796 and rs1800795 in IL6, rs2243248 and rs2243250 in IL4, rs1800871 in IL10, rs3212227 in IL12B, rs1800587 in IL1A, rs2430561 in IFNG, rs1799964 in TNF, rs689466 in PTGS2, rs45567233 in CTSG, rs731236 and rs2228570 in VDR, rs1144393 and rs1799750 in MMP1, and rs4646972 in PLAT(t-PA) and BAX-248G/A increased the risk of osteomyelitis, whereas rs4251961 in IL1RN, rs380099 in TLR2, and the CT genotype of rs2070874 in IL4 could protect against osteomyelitis. However, the results of Jiang et al. (2020) have suggested that the GG or GA genotype of IL1B rs16944 was a risk factor of osteomyelitis, which was inconsistent with the results of the study by Osman et al. (2016). The inconsistencies could be attributed to the insufficient sample size and population from different countries. Therefore, further research should be conducted to identify the link between IL1B gene polymorphism and osteomyelitis to clarify the etiology and pathogenesis of osteomyelitis.
Many genes collected from the publications included in this review can regulate the immune system and, therefore, contribute to host defense against pathogenic microorganisms in the bodily tissues and blood (Hill, 2012). Pathogen molecular patterns binding to pattern recognition receptors, such as Toll-like receptors (i.e., TLR2 and TLR4), can initiate inflammatory reactions of innate immune cells and induce the expression of pro-inflammatory cytokines (such as IL-1β and TNF-α). The activation of TLRs expressed in bone cells could influence osteoclast differentiation and activities in a complicated manner. TLRs expressed in early osteoclast precursors inhibit the differentiation of these cells, whereas the activation of TLR expressed in osteoblasts triggers the secretion of osteoclastogenic cytokines, including RANKL and TNF-α, which contribute to osteoclast differentiation and activation (Bar-Shavit, 2008). Yoshii et al. (2002) reported that pro-inflammatory IL-1β, IL-6, IL-4, and TNF-α levels in a locally infected bone increased during the infection period in a murine model of osteomyelitis, and IL-1β and IL-6 might contribute to bone damage during the earlier period of infection. IL-1R signaling contributes to bone destruction during osteomyelitis, whereas it also plays an important role in repressing local bacterial replication during bone infection (Putnam et al., 2019). IL-1β-activated osteoclasts exhibit strong absorbing ability and high H+ release (Shiratori et al., 2018). IL-10 is regarded as an immune modulatory cytokine that mitigates damage by decreasing the expression of inflammatory cytokines. IL-10 promoter polymorphisms seemed to be associated with the pathogenesis of chronic non-bacterial osteomyelitis (auto-inflammatory disorder) through the involvement of IL-10 dysfunction (Hofmann et al., 2011). Neutrophils are the first-line innate immune defense against many microbial infections. Meanwhile, the elimination of neutrophils through apoptosis or taken in by macrophages could alleviate the destructive nature of inflammation and promote resolution of the inflammation (Savill et al., 2002). BAX-α is a proapoptotic protein, and bcl-2 is an antiapoptotic protein (Oltvai et al., 1993). A high BAX-α/bcl-2 ratio would lead to apoptosis of leukemic cells (Pepper et al., 1998). BAX gene mutation could influence protein expression and biological function (Addeo et al., 2007). IFN-γ secreted by immunocytes in response to bacterial invasion strengthens antigen presentation and the phagocytic abilities of macrophages (Gomez et al., 2015). Meta-analyses of the association between IFN-γ+874T/A and susceptibility to leukemia (Wu et al., 2016), hepatocellular carcinoma (Zhou et al., 2015), and asthma (Nie et al., 2014) have been conducted. Matrix metalloproteinases (MMPs), a family of enzymes, play an important role in the degradation and rebuild of extracellular matrix under both normal physiological and pathological conditions. MMPs are involved in matrix degradation and joint destruction in arthritis diseases (Pap et al., 2000; Tetlow et al., 2001). The expression of inducible MMPs is increased by stimulating inflammatory mediators, such as TNF-α and IL-1 (Nagase and Woessner, 1999). A population-based study has discovered that the MMP1–1607(1G/2G) polymorphism might be associated with reduced bone mineral density at the distal radius in postmenopausal women (Yamada et al., 2002). Cathepsin G (CTSG) is a serine protease stored in the neutrophil azurophilic granules and has antimicrobial properties (Miyasaki et al., 1995). In addition, CTSG could activate extracellular MMPs at the site of inflammation, causing the degradation of extracellular matrix components (Baggiolini et al., 1978; Korkmaz et al., 2008). CTSG also activates osteoclast precursors by stimulating the expression of RANKL, enhancing mammary tumor-induced osteolysis (Beaujouin and Liaudet-Coopman, 2008). Vitamin D is essential in calcium homeostasis and bone metabolism. In addition, it participates in the regulation of inflammatory reactions (Yin and Agrawal, 2014). VDR is coded by the VDR gene located on chromosome 12. In addition, VDR gene polymorphisms affect the biological function of VDR. The gene polymorphisms TaqI, BsmI, FokI, and ApaI were most frequently investigated in many skeletal diseases. A case–control study has reported that the VDR FokI polymorphism was linked to the risk of osteoporosis in postmenopausal women (Wu et al., 2019). Recently, the results of a meta-analysis have suggested that VDR BsmI and TaqI polymorphisms were associated with the susceptibility to osteoarthritis in the spine (Cezar-Dos-Santos et al., 2020). Simultaneously, our bioinformatics analysis discovered that these 25 genes were associated with immune responses (biological process), extracellular space (cellular component), and cytokine activity (molecular function). Interestedly, these osteomyelitis-related genes were enriched in the disease pathways, including IBD. Inflammation participates in the pathogenesis of bacterial osteomyelitis from the aspect of bioinformatics analysis.
Four SNPs (IL1B rs1143627, IL1B rs16944, IFNG rs2430561, and IL4 rs2070874) had smaller scores from the regulome analysis, implying that these SNPs had significant biological functions. rs1143627 and rs2070874 were located in the 5′-UTR region of IL1B and IL4 gene, respectively. rs16944 is located in an intron of the IL1B gene, and rs2430561 is located in the IFNG promoter region. Thus, we can hypothesize that these SNPs contribute to the risk of osteomyelitis, possibly by affecting transcription factors or other molecules binding to the motif.
This review summarized a series of genes and SNPs associated with osteomyelitis. Besides, the emergence of genome-wide association studies (GWAS) provides many opportunities to identify alleles associated with complex diseases (Altshuler et al., 2008). A GWAS from the UK Biobank (http://geneatlas.roslin.ed.ac.uk/; Bycroft et al., 2018) involved 452,264 individuals, including 698 patients with osteomyelitis. The subjects were from the UK and were between 40 and 69 years old. SNP genotyping was identified using the UK Biobank AXIOM array. However, unfortunately, we have not found any published data about the relationship between several SNPs and the risk of osteomyelitis using GWAS.
Genetic variants exist among individuals, but the influence of these genetic polymorphisms on clinical significance or phenotypic diversity has not been known yet. Lappalainen et al. (2013) have demonstrated that genetic variation could affect the occurrence and development of diseases by regulating gene expression. Thus, elucidating how genotype varieties clinically affect phenotypes in complicated diseases remains challenging. Exploring the role of genetics in phenotypes and diseases and its potential interactions with other factors is of great significance for understanding pathogenesis of diseases. It is hoped that this will provide more opportunities for drug development and personalized treatments. Note that human genetics is a valuable tool for the therapeutic hypothesis in drug development. Plenge et al. (2013) have provided empirical examples of drug–gene pairs and objective criteria to highlight the role of genetic findings for drug discovery in the future. For example, anakinra (interleukin-1 receptor antagonist) treatment is effective in patients with adult-onset Still disease in the preliminary experience (Lequerre et al., 2008). Similarly, according to our summary of genetic SNPs associated with bacterial osteomyelitis, some inflammatory cytokine-related inhibitors can potentially be used to test therapeutic hypotheses for the development of drugs and treatment of bacterial osteomyelitis, such as ustekinumab (anti-IL12 monoclonal antibody), tocilizumab (anti-IL6R monoclonal antibody), and anakinra (interleukin-1 receptor antagonist).
In this review, we summarized the association of gene polymorphisms with the susceptibility to osteomyelitis. However, this review has many limitations as well. The primary limitations of this review include the following: (1) the limited number of available studies and individuals selected for this review; (2) the low statistical efficacy caused by the limited sample size; (3) the limitation of ethnic diversity; (4) three different types of osteomyelitis that were not analyzed separately; and (5) the limited amount of data extracted from few studies, preventing us from conducting more quantitative analysis (meta-analysis).
Conclusion
This review summarized the association between gene polymorphisms and the increasing risk of osteomyelitis. According to this review, rs1143627 and rs1143634 in IL-1B, rs1800796 and rs1800795 in IL6, rs2243248 and rs2243250 in IL4, rs1800871 in IL10, rs3212227 in IL12B, rs1800587 in IL1A, rs2430561 in IFNG, rs1799964 in TNF, rs689466 in PTGS2, rs45567233 in CTSG, rs731236 and rs2228570 in VDR, rs1144393 and rs1799750 in MMP1, and rs4646972 in PLAT(t-PA) and BAX-248G/A increased the risk of osteomyelitis, whereas rs4251961 in IL1RN, rs380099 in TLR2, and the CT genotype, not the CC genotype, of rs2070874 in IL4 could protect against osteomyelitis. However, due to the small sample size included in this review, we cannot draw a definitive conclusion on the correlation between genetic polymorphisms and susceptibility to osteomyelitis. Therefore, large-scale prospective studies should be conducted to further illustrate the relationship between SNPs and the risk of developing osteomyelitis. Meanwhile, investigating how genetic diversity influences clinical phenotypes is also necessary to better understand the role of genetic factors in the pathogenesis of osteomyelitis and to provide more personalized preventions and interventions for osteomyelitis in clinical practice.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
XX and JL designed the study and collected the data. XX drafted the manuscript. FG, KZ, ZSu, QW, ZSui, and PZ contributed to the writing. TY provided critical feedback and contributed to the review of the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This study was supported by the National Natural Science Foundation of China (Grant No. 31970090).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.654792/full#supplementary-material
References
- Addeo R., Crisci S., D'Angelo V., Vincenzi B., Casale F., Pettinato G., et al. (2007). Bax mutation and overexpression inversely correlate with immature phenotype and prognosis of childhood germ cell tumors. Oncol. Rep. 17, 1155–1161. 10.3892/or.17.5.1155 [DOI] [PubMed] [Google Scholar]
- Altshuler D., Daly M. J., Lander E. S. (2008). Genetic mapping in human disease. Science 322, 881–888. 10.1126/science.1156409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asensi V., Alvarez V., Valle E., Meana A., Fierer J., Coto E., et al. (2003). IL-1 alpha (-889) promoter polymorphism is a risk factor for osteomyelitis. Am. J. Med. Genet. A 119A, 132–136. 10.1002/ajmg.a.20137 [DOI] [PubMed] [Google Scholar]
- Baggiolini M., Bretz U., Dewald B., Feigenson M. E. (1978). The polymorphonuclear leukocyte. Agents Actions 8, 3–10. 10.1007/BF01972395 [DOI] [PubMed] [Google Scholar]
- Bar-Shavit Z. (2008). Taking a toll on the bones: regulation of bone metabolism by innate immune regulators. Autoimmunity 41, 195–203. 10.1080/08916930701694469 [DOI] [PubMed] [Google Scholar]
- Beaujouin M., Liaudet-Coopman E. (2008). Cathepsin D overexpressed by cancer cells can enhance apoptosis-dependent chemo-sensitivity independently of its catalytic activity. Adv. Exp. Med. Biol. 617, 453–461. 10.1007/978-0-387-69080-3_44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bycroft C., Freeman C., Petkova D., Band G., Elliott L. T., Sharp K., et al. (2018). The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209. 10.1038/s41586-018-0579-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cezar-Dos-Santos F., Brandao Berti F. C., Martins Okuyama N. C., Trugilo K. P., Colado Simao A. N., Ehara Watanabe M. A., et al. (2020). FOXP3 genetic variants do not impact circulating and cervical interleukin-10 levels in human papillomavirus infection in women. Viral Immunol. 33, 652–655. 10.1089/vim.2020.0040 [DOI] [PubMed] [Google Scholar]
- Chen X., Jiao J., He X., Zhang J., Wang H., Xu Y., et al. (2017). CHI3L1 regulation of inflammation and the effects on osteogenesis in a Staphylococcus aureus-induced murine model of osteomyelitis. FEBS J. 284, 1738–1747. 10.1111/febs.14082 [DOI] [PubMed] [Google Scholar]
- Gomez J. C., Yamada M., Martin J. R., Dang H., Brickey W. J., Bergmeier W., et al. (2015). Mechanisms of interferon-gamma production by neutrophils and its function during Streptococcus pneumoniae pneumonia. Am. J. Respir. Cell Mol. Biol. 52, 349–364. 10.1165/rcmb.2013-0316OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill A. V. (2012). Evolution, revolution and heresy in the genetics of infectious disease susceptibility. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 840–849. 10.1098/rstb.2011.0275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann S. R., Schwarz T., Moller J. C., Morbach H., Schnabel A., Rosen-Wolff A., et al. (2011). Chronic non-bacterial osteomyelitis is associated with impaired Sp1 signaling, reduced IL10 promoter phosphorylation, and reduced myeloid IL-10 expression. Clin. Immunol. 141, 317–327. 10.1016/j.clim.2011.08.012 [DOI] [PubMed] [Google Scholar]
- Hou Y., Bai L., Jiang N., Yao Z., Xue L., Yu B. (2018). Screening of TNF-alpha gene polymorphisms in patients with extremity chronic osteomyelitis in China. Per. Med. 15, 395–401. 10.2217/pme-2018-0017 [DOI] [PubMed] [Google Scholar]
- Jiang N., Li S. Y., Ma Y. F., Hu Y. J., Lin Q. R., Yu B. (2020). Associations between interleukin gene polymorphisms and risks of developing extremity posttraumatic osteomyelitis in chinese han population. Mediators Inflamm. 2020:3278081. 10.1155/2020/3278081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N., Zhao X. Q., Qin C. H., Hu Y. J., Wang L., Xie G. P., et al. (2016). Association of vitamin D receptor gene TaqI, BsmI, FokI and ApaI polymorphisms and susceptibility to extremity chronic osteomyelitis in Chinese population. Injury 47, 1655–1660. 10.1016/j.injury.2016.06.005 [DOI] [PubMed] [Google Scholar]
- Kapadia B. H., Berg R. A., Daley J. A., Fritz J., Bhave A., Mont M. A. (2016). Periprosthetic joint infection. Lancet 387, 386–394. 10.1016/S0140-6736(14)61798-0 [DOI] [PubMed] [Google Scholar]
- Kong Q., Jin Y., Yan S., Wang Y., Zhao J., Feng Z., et al. (2017). Examining the association of MMP-1 gene−1607 (2G/1G) and−519 (A/G) polymorphisms with the risk of osteomyelitis: a case-control study. Medicine 96:e4969. 10.1097/MD.0000000000004969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkmaz B., Moreau T., Gauthier F. (2008). Neutrophil elastase, proteinase 3 and cathepsin G: physicochemical properties, activity and physiopathological functions. Biochimie 90, 227–242. 10.1016/j.biochi.2007.10.009 [DOI] [PubMed] [Google Scholar]
- Lappalainen T., Sammeth M., Friedlander M. R., t Hoen P. A., Monlong J., Rivas M. A., et al. (2013). Transcriptome and genome sequencing uncovers functional variation in humans. Nature 501, 506–511. 10.1038/nature12531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lequerre T., Quartier P., Rosellini D., Alaoui F., De Bandt M., Mejjad O., et al. (2008). Interleukin-1 receptor antagonist (anakinra) treatment in patients with systemic-onset juvenile idiopathic arthritis or adult onset Still disease: preliminary experience in France. Ann. Rheum. Dis. 67, 302–308. 10.1136/ard.2007.076034 [DOI] [PubMed] [Google Scholar]
- Lew D. P., Waldvogel F. A. (2004). Osteomyelitis. Lancet 364, 369–379. 10.1016/S0140-6736(04)16727-5 [DOI] [PubMed] [Google Scholar]
- Miyasaki K. T., Qu X. D., Harwig S. S., Cho Y., Lehrer R. I. (1995). Identification of CG-1, a natural peptide antibiotic derived from human neutrophil cathepsin G. Adv. Dent. Res. 9, 63–66. 10.1177/08959374950090011201 [DOI] [PubMed] [Google Scholar]
- Moher D., Liberati A., Tetzlaff J., Altman D. G., Group P. (2009). Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ 339:b2535. 10.1136/bmj.b2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montes A. H., Asensi V., Alvarez V., Valle E., Ocana M. G., Meana A., et al. (2006). The Toll-like receptor 4 (Asp299Gly) polymorphism is a risk factor for Gram-negative and haematogenous osteomyelitis. Clin. Exp. Immunol. 143, 404–413. 10.1111/j.1365-2249.2005.03002.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagase H., Woessner J. F., Jr. (1999). Matrix metalloproteinases. J. Biol. Chem. 274, 21491–21494. 10.1074/jbc.274.31.21491 [DOI] [PubMed] [Google Scholar]
- Nie W., Meng L., Wang X., Xiu Q. (2014). Interferon-gamma +874A/T polymorphism is associated with asthma risk: a meta-analysis. J. Invest. Allergol. Clin. Immunol. 24, 324–330. [PubMed] [Google Scholar]
- Ocana M. G., Valle-Garay E., Montes A. H., Meana A., Carton J. A., Fierer J., et al. (2007). Bax gene G(-248)A promoter polymorphism is associated with increased lifespan of the neutrophils of patients with osteomyelitis. Genet. Med. 9, 249–255. 10.1097/GIM.0b013e318039b23d [DOI] [PubMed] [Google Scholar]
- Olson M. E., Horswill A. R. (2013). Staphylococcus aureus osteomyelitis: bad to the bone. Cell Host Microbe 13, 629–631. 10.1016/j.chom.2013.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oltvai Z. N., Milliman C. L., Korsmeyer S. J. (1993). BCL-2 heterodimerizes in vivo with a conserved homolog, Bax, that accelerates programmed cell death. Cell 74, 609–619. 10.1016/0092-8674(93)90509-O [DOI] [PubMed] [Google Scholar]
- Osman A. E., Mubasher M., ElSheikh N. E., AlHarthi H., AlAlallah I. A., Elbeshir A. A., et al. (2015). Investigation of polymorphisms in anti-inflammatory cytokine genes in hematogenous osteomyelitis. Genet. Mol. Res. 14, 16981–16986. 10.4238/2015.December.15.4 [DOI] [PubMed] [Google Scholar]
- Osman A. E., Mubasher M., ElSheikh N. E., AlHarthi H., AlZahrani M. S., Ahmed N., et al. (2016). Association of single nucleotide polymorphisms in pro-inflammatory cytokine and toll-like receptor genes with pediatric hematogenous osteomyelitis. Genet. Mol. Res. 15:gmr.15027718. 10.4238/gmr.15027718 [DOI] [PubMed] [Google Scholar]
- Paludo E., Ibelli A. M. G., Peixoto J. O., Tavernari F. C., Lima-Rosa C. A. V., Pandolfi J. R. C., et al. (2017). The involvement of RUNX2 and SPARC genes in the bacterial chondronecrosis with osteomyelitis in broilers. Animal 11, 1063–1070. 10.1017/S1751731116002433 [DOI] [PubMed] [Google Scholar]
- Pap T., Shigeyama Y., Kuchen S., Fernihough J. K., Simmen B., Gay R. E., et al. (2000). Differential expression pattern of membrane-type matrix metalloproteinases in rheumatoid arthritis. Arthritis Rheum. 43, 1226–1232. [DOI] [PubMed] [Google Scholar]
- Pepper C., Hoy T., Bentley P. (1998). Elevated Bcl-2/Bax are a consistent feature of apoptosis resistance in B-cell chronic lymphocytic leukaemia and are correlated with in vivo chemoresistance. Leuk. Lymphoma 28, 355–361. 10.3109/10428199809092690 [DOI] [PubMed] [Google Scholar]
- Perez-Is L., Ocana M. G., Montes A. H., Carton J. A., Alvarez V., Meana A., et al. (2019). The N125S polymorphism in the cathepsin G gene (rs45567233) is associated with susceptibility to osteomyelitis in a Spanish population. PLoS ONE 14:e0220022. 10.1371/journal.pone.0220022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plenge R. M., Scolnick E. M., Altshuler D. (2013). Validating therapeutic targets through human genetics. Nat. Rev. Drug Discov. 12, 581–594. 10.1038/nrd4051 [DOI] [PubMed] [Google Scholar]
- Putnam N. E., Fulbright L. E., Curry J. M., Ford C. A., Petronglo J. R., Hendrix A. S., et al. (2019). MyD88 and IL-1R signaling drive antibacterial immunity and osteoclast-driven bone loss during Staphylococcus aureus osteomyelitis. PLoS Pathog. 15:e1007744. 10.1371/journal.ppat.1007744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savill J., Dransfield I., Gregory C., Haslett C. (2002). A blast from the past: clearance of apoptotic cells regulates immune responses. Nat. Rev. Immunol. 2, 965–975. 10.1038/nri957 [DOI] [PubMed] [Google Scholar]
- Shiratori T., Kyumoto-Nakamura Y., Kukita A., Uehara N., Zhang J., Koda K., et al. (2018). IL-1β induces pathologically activated osteoclasts bearing extremely high levels of resorbing activity: a possible pathological subpopulation of osteoclasts, accompanied by suppressed expression of Kindlin-3 and Talin-1. J. Immunol. 200, 218–228. 10.4049/jimmunol.1602035 [DOI] [PubMed] [Google Scholar]
- Stang A. (2010). Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur. J. Epidemiol. 25, 603–605. 10.1007/s10654-010-9491-z [DOI] [PubMed] [Google Scholar]
- Tetlow L. C., Adlam D. J., Woolley D. E. (2001). Matrix metalloproteinase and proinflammatory cytokine production by chondrocytes of human osteoarthritic cartilage: associations with degenerative changes. Arthritis Rheum. 44, 585–594. [DOI] [PubMed] [Google Scholar]
- Tseng C. H., Huang W. S., Muo C. H., Chang Y. J., Kao C. H. (2014). Increased depression risk among patients with chronic osteomyelitis. J. Psychosom. Res. 77, 535–540. 10.1016/j.jpsychores.2014.09.008 [DOI] [PubMed] [Google Scholar]
- Tsezou A., Poultsides L., Kostopoulou F., Zintzaras E., Satra M., Kitsiou-Tzeli S., et al. (2008). Influence of interleukin 1α (IL-1α), IL-4, and IL-6 polymorphisms on genetic susceptibility to chronic osteomyelitis. Clin. Vaccine Immunol. 15, 1888–1890. 10.1128/CVI.00209-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle-Garay E., Montes A. H., Corte J. R., Meana A., Fierer J., Asensi V. (2013). tPA Alu (I/D) polymorphism associates with bacterial osteomyelitis. J. Infect. Dis. 208, 218–223. 10.1093/infdis/jit158 [DOI] [PubMed] [Google Scholar]
- Valour F., Bouaziz A., Karsenty J., Ader F., Lustig S., Laurent F., et al. (2014). Determinants of methicillin-susceptible Staphylococcus aureus native bone and joint infection treatment failure: a retrospective cohort study. BMC Infect. Dis. 14:443. 10.1186/1471-2334-14-443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Jiang N., Lin Q. R., Qin C. H., Hu Y. J., Yu B. (2017). Cyclooxygenase-2 (COX-2) polymorphism rs689466 may contribute to the increased susceptibility to post-traumatic osteomyelitis in Chinese population. Infect. Dis. 49, 817–823. 10.1080/23744235.2017.1347816 [DOI] [PubMed] [Google Scholar]
- Wu F., Zhou D., Shen G., Cui Y., Lv Q., Wei F. (2019). Association of VDR and OPG gene polymorphism with osteoporosis risk in Chinese postmenopausal women. Climacteric 22, 208–212. 10.1080/13697137.2018.1554643 [DOI] [PubMed] [Google Scholar]
- Wu Z., Sun Y., Zhu S., Tang S., Liu C., Qin W. (2016). Association of interferon gamma +874T/A polymorphism and leukemia risk: a meta-analysis. Medicine 95:e3129. 10.1097/MD.0000000000003129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y., Ando F., Niino N., Shimokata H. (2002). Association of a polymorphism of the matrix metalloproteinase-1 gene with bone mineral density. Matrix Biol. 21, 389–392. 10.1016/S0945-053X(02)00030-6 [DOI] [PubMed] [Google Scholar]
- Yin K., Agrawal D. K. (2014). Vitamin D and inflammatory diseases. J. Inflamm. Res. 7, 69–87. 10.2147/JIR.S63898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii T., Magara S., Miyai D., Nishimura H., Kuroki E., Furudoi S., et al. (2002). Local levels of interleukin-1beta,−4,−6 and tumor necrosis factor alpha in an experimental model of murine osteomyelitis due to staphylococcus aureus. Cytokine 19, 59–65. 10.1006/cyto.2002.1039 [DOI] [PubMed] [Google Scholar]
- Zhao X. Q., Jiang N., Hu Y. J., Yu B. (2020). IFN-gamma +874T/A polymorphism increases susceptibility to post-traumatic osteomyelitis. Int. J. Immunogenet. 47, 163–168. 10.1111/iji.12462 [DOI] [PubMed] [Google Scholar]
- Zhou H., Wang L., Li X., Song J., Jiang T., Wu X., et al. (2015). Interferon-gamma +874A/T polymorphism and hepatocellular carcinoma risk: a meta-analysis. Med. Sci. Monit. 21, 689–693. 10.12659/MSM.892885 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.