Abstract
Low concentrations of type‐I interferon (IFN) in blood seem to be associated with more severe forms of Coronavirus disease 2019 (COVID‐19). However, following the type‐I interferon response (IR) in early stage disease is a major challenge. We evaluated detection of a molecular interferon signature on a FilmArray® system, which includes PCR assays for four interferon stimulated genes. We analyzed three types of patient populations: (i) children admitted to a pediatric emergency unit for fever and suspected infection, (ii) ICU‐admitted patients with severe COVID‐19, and (iii) healthcare workers with mild COVID‐19. The results were compared to the reference tools, that is, molecular signature assessed with Nanostring® and IFN‐α2 quantification by SIMOA® (Single MOlecule Array). A strong correlation was observed between the IR measured by the FilmArray®, Nanostring®, and SIMOA® platforms (r‐Spearman 0.996 and 0.838, respectively). The FilmArray® panel could be used in the COVID‐19 pandemic to evaluate the IR in 45‐min with 2 min hand‐on‐time at hospitalization and to monitor the IR in future clinical trials.
Keywords: COVID‐19, Clinical application, FilmArray®, Interferon, In vitro diagnostic
The type‐I Interferon response impairment increases the risk of severe forms of viral infections (like in COVID‐19 disease). We proposed a new integrated tool allowing the fast assessment of type‐I Interferon response (based on interferon stimulated gene expression measurement) to improve patient management.
Introduction
Type‐I interferons (IFN‐I) are a group of cytokines with broad‐spectrum antiviral activities, controlling immune cell proliferation and modulating the overall immune response [1]. IFN‐Is are key cytokines involved in the response to viral infection and recombinant interferons are used therapeutically during treatment of several immune pathological conditions [2, 3]. An antiviral response is induced upon viral infections and across many cell types which express protein and nucleic acid sensors. Signal transduction from these sensors activates the transcription of IFN genes. IFNs are secreted and bind to specific cell surface receptors. Upon receptor binding and activation, IFNs induce the transcription of hundreds of interferon‐stimulated genes (ISGs). Most ISGs are involved in inflammation, cell‐to‐cell signaling, and immunomodulation.
Coronavirus disease 2019 (COVID‐19) is currently pandemic and characterized, in severe cases, by a cytokine storm associated with defective IFN production [4, 5, 6]. There are several causes to this impairment, some of which are known as such and include autoantibody production [7], or specific genetic mutations [8]. Whatever the origin of the type I IFN deficit, it is associated with more severe forms of COVID‐19 disease [4, 5, 9, 10]. Consequently, there is an urgent need to identify patients with an impaired IFN‐I response to propose alternative therapeutics such as recombinant IFN‐type I administration [11].
IFNs are secreted at very low concentrations during the course of infection. Most of the ELISA platforms used by the routine diagnostic laboratory are not able to precisely quantify these proteins. Only an ultrasensitive digital ELISA (SIMOA®) makes it possible to detect and enumerate the IFN‐α2 protein. Consequently, IFN‐α2 detection in clinical specimens remains a challenging task. Alternative solutions have therefore been proposed to monitor the production of these cytokines. The quantification of ISGs transcription (ISG score) in whole blood provides a surrogate estimate of the cellular response to the IFN. We recently reported that IFN‐α2 protein dosage and ISG score are correlated and allow the discrimination between viral and bacterial infection in samples of pediatric origin [12]. The application of this score, measured with a Nanostring® platform, is already used to explore type‐I interferonopathies in the context of autoimmunity [13, 14, 15]. Nanostring® and SIMOA® platforms require complex hand‐on activities and have an extended time‐to‐results. Rapid and sensitive monitoring of an IFN response requires a simpler and faster tool, more adapted to routine clinical practice. The aim of this proof‐of‐concept study is to evaluate a prototype multiplex interferon transcription profiling panel for its ability to semiquantify four interferon‐related mRNA markers directly from blood, using the BioFire FilmArray® PCR System. The FilmArray® System is a sample‐to‐answer, semiquantitative, multiplex PCR test that requires approximately 2 min hands‐on‐time for whole blood samples and results are available in less than 1 h. Several FDA‐cleared and CE‐marked diagnostic panels are in use on the FilmArray® System [16]. Beyond its capacity to detect various pathogens simultaneously, this system has already demonstrated its ability to monitor host response during sepsis [17]. In the present study, using samples from a pediatric cohort (exploratory cohort) and a subset‐cohort of COVID‐19 patients (validation cohort), we compared the performance of three platforms (molecular signature assessed with Nanostring® [18], IFN‐α2 quantification by SIMOA® [19], and FilmArray®) in order to assess the relevance of either of these tests to help stratify patients for whom an IFN support treatment would be considered.
Results and discussion
Correlation of FilmArray® interferon response panel with reference tools
The quantification of ISGs transcription (IFN signatures) from whole blood provides a surrogate estimate of the exposure of cells to type‐I IFN. An IFN‐based signature of four genes (IFI27, IFIT1, RSAD2, and IFI44L) was evaluated. We used this FilmArray® prototype to measure ISG score (i) on samples selected from the ANTOINE pediatric patients cohort with proven bacterial or viral infections (n = 86) previously tested with Nanostring® and SIMOA® platforms [12], (ii) on samples collected from severe SARS‐CoV‐2 infected patients hospitalized in Intensive Care Unit (ICU) (n = 25), (iii) a total of 21 SARS‐CoV‐2 positive samples collected from healthcare workers with mild symptoms (n = 7). The FilmArray® prototype uses four ISGs from the six (IFI27, IFI44L, RSAD2, IFIT1, SIGLEC1, and ISG15) included in the Nanostring® IFN‐based signature [12]. We assessed the correlation of the FilmArray® pouch prototype with the solutions currently published and available to monitor the IFN response. The four ISG‐based IFN score using the FilmArray® pouch prototype with the six ISG‐based Nanostring® signature were strongly correlated whatever the infectious context or the severity of the disease (n = 118, ρ‐spearman [IC95%]: 0.991 [0.987; 0.994], Fig. 1A). In the same way, the four ISG‐based IFN score using the FilmArray® pouch prototype were also found to be well correlated with the IFN‐α2 protein quantification (n = 78, 0.832 [0.746; 0.891] respectively, Fig. 1B). We therefore suggest that the FilmArray® test could be a suitable tool for monitoring the type‐I interferon response.
Figure 1.
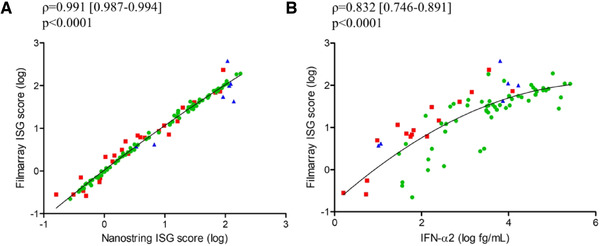
Type‐I Interferon quantification using FilmArray® platform. (A) Spearman correlation [IC95%] between ISG score as defined by a 6‐interferon stimulated gene transcriptional signature using Nanostring® platform and ISG score as defined by a 4‐interferon stimulated gene transcriptional signature using FilmArray® technology, obtained from whole blood from (i) febrile infants (n = 86 patients from one experiment, green circle) with proven infections attending pediatric emergency departments, (ii) patients with severe infection caused by SARS‐CoV2 hospitalized in the ICU (n = 26 patients from one experiment, red square), and (iii) healthcare workers with COVID‐19 with mild symptoms (n = 7 patients from one experiment, blue triangle) (ρ = 0.991). (B) Spearman correlation between ISG score as defined by a 4‐interferon stimulated gene transcriptional signature using FilmArray® technology and IFN‐α2 concentrations measured by SIMOA® platform, obtained from whole blood from (i) febrile infants (n = 56 patients from one experiment, green circle) with proven infections attending pediatric emergency departments, (ii) patients with severe infection caused by SARS‐CoV2 hospitalized in the ICU (n = 16 patients from one experiment, red square), and (iii) healthcare workers with COVID‐19 with mild symptoms (n = 7 patients from one experiment, blue triangle) (ρ = 0.832). Patients are represented by different forms and nonlinear regression (second order polynomial, quadratic) is represented by a black line. IFN (Interferon), ISG (Interferon Stimulated Genes).
New faster tool to monitor the IFN‐I response in COVID patients at ICU admission
We assessed the ISG score using the FilmArray® tool in severe SARS‐CoV‐2 infected patients at ±5 days after ICU admission (n = 16). Setting a cut‐off of 3.2 arbitrary units with the FilmArray® prototype, we were able to segregate the COVID‐19 patients with an impairment of their type‐I IFN response from IFN‐positive patients, as previously observed using IFN‐α2 quantification with SIMOA® platform (Fig. 2) [4]. In addition, ISG scores for healthy volunteers (n = 10) were not statistically different compared to COVID‐19 patients with an altered type I IFN response at the moment of their ICU admission (Fig. 2). Consequently, the FilmArray® prototype is able to detect an IFN‐I response in COVID‐19 patients and to reveal the impairment of type‐I IFN response for some patients.
Figure 2.
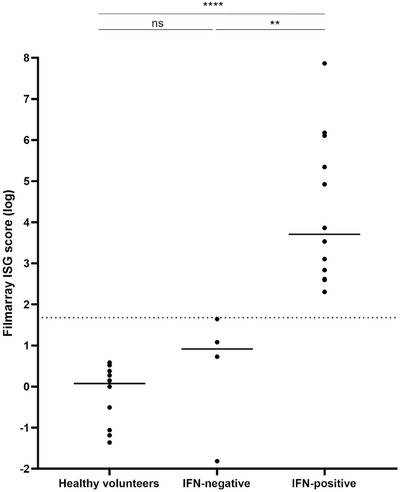
Type‐I IFN assessment in COVID‐19 critically ill patients using FilmArray® platform. ISG score as defined by a 4 interferon stimulated gene transcriptional signature was determined by IFN pouch FilmArray® panel, obtained from whole blood from selected patients severely infected by SARS‐CoV‐2 at ±5 days after admission in the ICU (n = 16 patients from one experiment) and healthy volunteers (n = 10 donors from one experiment). COVID‐19 critically‐ill patients are represented by a black dot, and these samples are stratified according to their impairment of IFN response. The healthy volunteers (n = 10 donors from one experiment) are represented by a black dot also. The dashed line represents the arbitrary threshold of the ISG score set to separate patients with a positive (n = 4 patients from one experiment) or negative (n = 12 patients from one experiment) IFN response. The box‐and‐whisker plots represent the median and min‐max; Mann–Whitney U test **p < 0.01; ****p < 0.0001. IFN (Interferon), ISG (Interferon Stimulated Genes).
Prospective studies are needed to determine if patients management could be optimized by monitoring of the IFN response during ICU admission. Finally, it would be interesting to perform a larger prospective study to investigate whether this score can predict severity of the COVID‐19 disease as previously suggested [5, 10, 20] or if its correlated to the viral load.
Concluding remarks
We have shown that the FilmArray® tool can be used to adequately track, the type‐I interferon response. Preliminary data show that this tool can rapidly identify COVID‐19 patients as soon as ICU admission with an impairment of type‐I interferon. The assay works directly from whole blood, and seems to do as well as the more complex SIMOA® and Nanostring® tests. We must keep in mind that ISGs are an indirect marker of IFN‐α secretion, mainly produced in viral context but can be also induced by other types of IFN.
Furthermore, the IFN‐I response is also dysregulated in some autoimmune diseases and the monitoring of type‐I IFN is needed for diagnosis and treatment follow‐up. FilmArray® could be a suitable option and needs to be evaluated in further studies.
Materials and methods
Participants
Serum or plasma samples and PAXgene™ tubes were collected from (i) children admitted to a pediatric emergency unit for reason of fever with proven viral of bacterial infection (median age, 21 months; 47% male; 51% bacterial infection; 49% viral infection); (ii) patients with severe infection caused by SARS‐CoV‐2 admitted in the ICU of the University Hospital of Lyon (Hôpital de la Croix Rousse, Hospices Civils de Lyon), France (median age, 70 years; 88% male; median delay between symptom onset and ICU admission, 13 days); (iii) healthcare workers with mild COVID‐19 symptoms (median age, 43 years; 0% male; median delay between symptom onset and protocol inclusion, 2 days); and (iv) healthy volunteers. These studies were approved by ethical committees for biomedical research (see details in Supplementary methods). Diagnosis of COVID‐19 was confirmed in all patients by qRT‐PCR (cobas® SARS‐CoV‐2 Test, Roche Diagnostics). Concomitantly, blood samples from 25 healthy volunteers were independently obtained from EFS (Etablissement Français du Sang, Lyon, France, see details in Supplementary methods concerning the legislation).
IFN‐α protein measurement
Serum IFN‐α2 concentrations (fg/mL) were determined by single molecule array (SIMOA®) using a commercial kit for IFN‐α2 quantification (Quanterix™, Lexington, MA). The assay was based on a three‐step protocol using an HD‐1 Analyzer (Quanterix) and required 3 h of processing time from sample to results [12, 19].
IFN score assessment
PAXgeneTM blood samples were stabilized for at least 2 h at room temperature after collection, and frozen at −80°C. Total RNA was extracted using the PAXgene™ Blood RNA kit (PreAnalytix), following the manufacturer's guidelines. The RNA quantity and quality were determined using a Nanodrop (Thermo Scientific) and Bioanalyser 2100 (Agilent) respectively, in accordance with the manufacturer's instructions. mRNA quantification of 6 ISGs (interferon alpha inducible protein 27 (IFI27), interferon induced protein 44 like (IFI44L), Interferon Induced Protein With Tetratricopeptide Repeats 1 (IFIT1), ISG15 Ubiquitin Like Modifier (ISG15), Radical S‐Adenosyl Methionine Domain Containing 2 (RSAD2), Sialic Acid Binding Ig Like Lectin 1 (SIGLEC1), and 3 housekeeping genes (Actin Beta (ACTB), Hypoxanthine Phosphoribosyltransferase 1 (HPRT1), RNA Polymerase II Subunit A (POLR2A)), was performed using Nanostring® technology (Nanostring Technologies©, WA), requiring around 24 h from sample to results [18]. Data standardization was obtained using the geometric mean of internal control and housekeeping genes count number. The ISG score was calculated as previously described [14].
FilmArray® IFN panel
The first prototype of the IFN pouch encompasses four ISGs previously measured with Nanostring© platform (IFI27, IFI44L, IFIT1, RSAD2) and three housekeeping genes (hypoxanthine phosphoribosyltransferase 1 (HPRT1), peptidylprolyl isomerase B (PPIB), 2,4‐dienoyl‐CoA reductase 1 (DECR1)), for signal normalization. One hundred microliters of PAXgeneTM blood samples were tested with the IFN prototype [17] according to manufacturer's instructions. In brief, the pouches were hydrated with the hydration solution supplied with the kit. The PAXgeneTM blood samples were mixed with 800 μL of the Sample Buffer provided with the kit and directly injected into the pouch and ran on FilmArray® 2.0 and FilmArray® Torch instruments (BioFire Diagnostics, LLC., Salt Lake City, UT). Results were delivered in less than 1 h. Using a research version of the instrument, real‐time quantification cycle (Cq) values and postamplification melt peaks were measured for each assay. Normalized expression values of each assay were then computed using the internal reference genes. ISG score was calculated in the same way as for the Nanostring® platforms.
Statistical analysis
Nonparametric Mann–Whitney tests and Spearman's correlation were performed for all parameters using the GraphPad Prism 5.02 software. A p‐value of <0.05 was considered statistically significant. Correlations are only performed on dually detected samples.
Peer review
The peer review history for this article is available at https://publons.com/publon/10.1002/eji.202048978.
Conflict of Interest
MM, FM, and KBP are employees of bioMérieux SA, an in vitro diagnostic company. MH and AH are employees of BioFire Diagnostics, a bioMérieux SA company. All other authors declare that they have no commercial or financial conflict of interest.
Abbreviations
- COVID‐19
Coronavirus disease 2019
- ICU
Intensive Care Unit
- IFN‐I
type‐I Interferon
- IR
interferon response
- ISGs
interferon‐stimulated genes
- SIMOA®
Single Molecule Array
Supporting information
Supporting Information
Acknowledgments
This project has received funding from the European Union's Horizon 2020 research and innovation program under Grant agreement No. 668303. The authors would like to thank the HCL Covid Task Force for their helpful advice as well as the Hospices Civils de Lyon and the Fondation des Hospices Civils de Lyon for their support, as well as all collaborators included in the three following study groups. We thank Alex Van Belkum for language editing and critical reading of the manuscript.
ANTOINE Study group
François Bartolo, Romain Basmaci, Lucille Boisselier, Coralie Bouchiat‐Sarabi, Claire Capella, Jean‐sebastien Casalegno, Valérie Cheynet, Christelle Compagnon, Romain Deshayes de Cambronne, Nina Droz, Laurence Generenaz, Yves Gillet, Audrey Guichard, Laure Hees, Ellia Mezgueldi, William Mouton, Guy Oriol, Antoine Ouziel, Alexandre Pachot, Sylvie Pons, Aurelie Portefaix, Philippe Rebaud, Chadia Toumi.
COVID HCL Study group
Florence Ader, Alexandre Gaymard, Marine Villard, Bruno Lina, Laurent Bitker, Agathe Becker, Nicholas Benech, Pierre Chauvelot, Christian Chidiac, Anne Conrad, Tristan Ferry, Patrick Miailhes, Thomas Perpoint, Marielle Perry, Cécile Pouderoux, Sandrine Roux, Claire Triffault‐Fillit, Florent Valour, Yonis Hodane, Louis Chauvelot, Paul Chabert, Judith Provoost, Guillaume David, Laure Folliet, Pierre Lecam, Geneviève Billaud, Maude Bouscambert, Vanessa Escuret, Emilie Frobert, Antonin Bal, Grégory Destras, Laurence Josset, Florence Morfin, Clément Munier, Martine Valette, Fabienne Venet, Lorna Garnier, Rémi Pescarmona, Christine Lombard, Thierry Walzer
COVID‐SER Study group
Jérôme Adnot, Dulce Alfaiate, Antonin Bal, Alain Bergeret, André Boibieux, Florent Bonnet, Florence Brunel‐Dalmas, Eurydice Caire, Pierre Chiarello, Laurent Cotte, Constance d'Aubarede, François Durupt, Vanessa Escuret, Pascal Fascia, Juliette Fontaine, Lucie Gaillot‐Durand, Myriam Gillet, Matthieu Godinot, François Gueyffier, Nicolas Guibert, Matthieu Lahousse, Bruno Lina, Hélène Lozano, Djamila Makhloufi, Amélie Massardier‐Pilonchéry, Marie‐Paule Milon, Frédéric Moll, David Narbey, Julie‐Anne Nazare, Fatima Oria, Adèle Paul, Marielle Perry, Gaëlle Bourgeois, Barbara Charbotel, Virginie Pitiot, Mélanie Prudent, Muriel Rabilloud, Audrey Samperiz, Isabelle Schlienger, Chantal Simon, Mary‐Anne Trabaud.
All authors were involved in the analysis and interpretation of data as well as in drafting the manuscript or revising it critically for important intellectual content. MM, SV, AB, FM, STA, AH, MH, and KBP made substantial contributions to the conception and design of the study. JCR, JBF, and EJ carried out patient recruitment. AH, MH, MM, FM, and KBP performed FilmArray® panel design. MM and MP designed and performed the experiments. MM performed the data analyses. MM, STA, KBP, and FM performed data interpretations. MM wrote the paper. All authors read and approved the final manuscript.
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1. Theofilopoulos, A. N. , Baccala, R. , Beutler, B. and Kono, D. H. Type I interferons (alpha/beta) in immunity and autoimmunity. Annu. Rev. Immunol. 2005. 23: 307–336. [DOI] [PubMed] [Google Scholar]
- 2. McNab, F. , Mayer‐Barber, K. , Sher, A. , Wack, A. and O'Garra, A. Type I interferons in infectious disease. Nat. Rev. Immunol. 2015. 15: 87–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Psarras, A. , Emery, P. and Vital, E. M. Type I interferon‐mediated autoimmune diseases: pathogenesis, diagnosis and targeted therapy. Rheumatology 2017. 56: 1662–1675. [DOI] [PubMed] [Google Scholar]
- 4. Trouillet‐Assant, S. , Viel, S. , Gaymard, A. , Pons, S. , Richard, J. C. , Perret, M. , Villard, M. et al., Type I IFN immunoprofiling in COVID‐19 patients. J. Allergy Clin. Immunol. 2020. 146: 206–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hadjadj, J. , Yatim, N. , Barnabei, L. , Corneau, A. , Boussier, J. , Smith, N. , Péré, H. et al., Impaired type I interferon activity and inflammatory responses in severe COVID‐19 patients. Science 2020. 369: 718–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Channappanavar, R. , Fehr, A. R. , Vijay, R. , Mack, M. , Zhao, J. , Meyerholz, D. K. and Perlman, S. Dysregulated Type I interferon and inflammatory monocyte‐macrophage responses cause lethal pneumonia in SARS‐CoV‐infected mice. Cell Host Microbe 2016. 19: 181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bastard, P. , Rosen, L. B. , Zhang, Q. , Michailidis, E. , Hoffmann, H. H. , Zhang, Y. , Dorgham, K. et al., Auto‐antibodies against type I IFNs in patients with life‐threatening COVID‐19. Science 2020. 370: eabd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang, Q. , Bastard, P. , Liu, Z. , Le Pen, J. , Moncada‐Velez, M. , Chen, J. , Ogishi, M. et al., Inborn errors of type I IFN immunity in patients with life‐threatening COVID‐19. Science 2020. 370: eabd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang, N. , Zhan, Y. , Zhu, L. , Hou, Z. , Liu, F. , Song, P. , Qiu, F. et al., Retrospective multicenter cohort study shows early interferon therapy is associated with favorable clinical responses in COVID‐19 patients. Cell Host Microbe 2020. 28: 454–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lee, J. S. , Park, S. , Jeong, H. W. , Ahn, J. Y. , Choi, S. J. , Lee, H. , Choi, B. et al., Immunophenotyping of COVID‐19 and influenza highlights the role of type I interferons in development of severe COVID‐19. Sci. Immunol. 2020, 5:eabd1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhou, Q. , Chen, V. , Shannon, C. P. , Wei, X. S. , Xiang, X. , Wang, X. , Wang, Z. H. et al., Interferon‐α2b Treatment for COVID‐19. Front. Immunol. 2020. 11: 1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Trouillet‐Assant, S. , Viel, S. , Ouziel, A. , Boisselier, L. , Rebaud, P. , Basmaci, R. , Droz, N. et al., Type I interferon in children with viral or bacterial infections. Clin. Chem. 2020. 66: 802–808. [DOI] [PubMed] [Google Scholar]
- 13. Rice, G. I. , Melki, I. , Frémond, M. L. , Briggs, T. A. , Rodero, M. P. , Kitabayashi, N. , Oojageer, A. et al., Assessment of type I interferon signaling in pediatric inflammatory disease. J. Clin. Immunol. 2017. 37: 123–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pescarmona, R. , Belot, A. , Villard, M. , Besson, L. , Lopez, J. , Mosnier, I. , Mathieu, A. L. et al., Comparison of RT‐qPCR and Nanostring in the measurement of blood interferon response for the diagnosis of type I interferonopathies. Cytokine 2019. 113: 446–452. [DOI] [PubMed] [Google Scholar]
- 15. Crow, Y. J. and Manel, N. Aicardi‐Goutières syndrome and the type I interferonopathies. Nat. Rev. Immunol. 2015. 15: 429–440. [DOI] [PubMed] [Google Scholar]
- 16. Poritz, M. A. , Blaschke, A. J. , Byington, C. L. , Meyers, L. , Nilsson, K. , Jones, D. E. , Thatcher, S. A. et al., FilmArray, an automated nested multiplex PCR system for multi‐pathogen detection: development and application to respiratory tract infection. PLoS One 2011. 6: e26047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tawfik, D. M. , Vachot, L. , Bocquet, A. , Venet, F. , Rimmelé, T. , Monneret, G. , Blein, S. et al., Immune profiling panel: a proof‐of‐concept study of a new multiplex molecular tool to assess the immune status of critically ill patients. J. Infect. Dis. 2020. 222: S84–S95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tsang, H. F. , Xue, V. W. , Koh, S. P. , Chiu, Y. M. , Ng, L. P. and Wong, S. C. NanoString, a novel digital color‐coded barcode technology: current and future applications in molecular diagnostics. Expert Rev. Mol. Diagn. 2017, 17: 95–103. [DOI] [PubMed] [Google Scholar]
- 19. Wilson, D. H. , Rissin, D. M. , Kan, C. W. , Fournier, D. R. , Piech, T. , Campbell, T. G. , Meyer, R. E. et al., The Simoa HD‐1 analyzer: a novel fully automated digital immunoassay analyzer with single‐molecule sensitivity and multiplexing. J. Lab. Automat. 2016. 21: 533–547. [DOI] [PubMed] [Google Scholar]
- 20. Wang, Z. , Pan, H. and Jiang, B. Type I IFN deficiency: an immunological characteristic of severe COVID‐19 patients. Signal Transduct. Target. Ther. 2020. 5: 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


