Abstract
Nonalcoholic fatty liver disease (NAFLD) is the most common form of pediatric liver disease in the United States, and often associated with obesity and metabolic syndrome. NAFLD comprises a broad spectrum of liver diseases, from hepatic steatosis to steatohepatitis, fibrosis and cirrhosis. Disease progression is considered a multi-modal process of liver injury. The intestinal microbiome has been implicated in several aspects of NAFLD pathophysiology. Pediatric studies associating the intestinal microbiome with NAFLD have been limited in number and complicated by inconsistencies in study design and approach. Nevertheless, they provide support for involvement of the intestinal microbiome in NAFLD development and progression and point to common mechanisms shared by microbiome-associated inflammatory diseases with potential to inform future therapeutic intervention.
I. NAFLD
Nonalcoholic fatty liver disease (NAFLD) is the most common cause of pediatric liver disease in the US (1) and the incidence is increasing rapidly (2). Although the specific histologic criteria may differ between adults and children, diagnosis requires that minimally 5% of hepatocytes exhibit steatosis, with the exclusion of other identifiable liver disease that may cause steatosis (3). While NAFLD is often associated with obesity, not every patient with NAFLD is obese, and most individuals with obesity do not develop NAFLD (4). A subset of patients with hepatic steatosis will progress and develop nonalcoholic steatohepatitis (NASH), with some progressing further to develop fibrosis, cirrhosis, and liver failure. The term NAFLD is used to describe the entire broad spectrum of liver diseases that range from hepatic steatosis to steatohepatitis, fibrosis, and cirrhosis. Disease progression is considered a multi-hit process (Figure 1). The “two hit NAFLD” hypothesis was first suggested by Day and James (5) with the initial “hit” resulting in the development of hepatic steatosis, while the second “hit” involves oxidative stress and lipid peroxidation, ultimately causing steatohepatitis. Multiple mechanisms have been proposed for each of these “hits”, suggesting that NAFLD may be a “final common outcome” from an accumulated series of diverse insults to the liver. Recently, the intestinal microbiome has been implicated in contributing to each “hit”, capable of both triggering and exacerbating NAFLD pathophysiology. In this review, we will examine connections between the intestinal microbiome and the liver, discuss recent pediatric studies that reveal potential mechanisms for how the microbiome influences NAFLD, and then describe evidence to support these mechanisms as explored through animal models of NAFLD.
Figure 1. Development and progression of NAFLD: A multi-hit process.
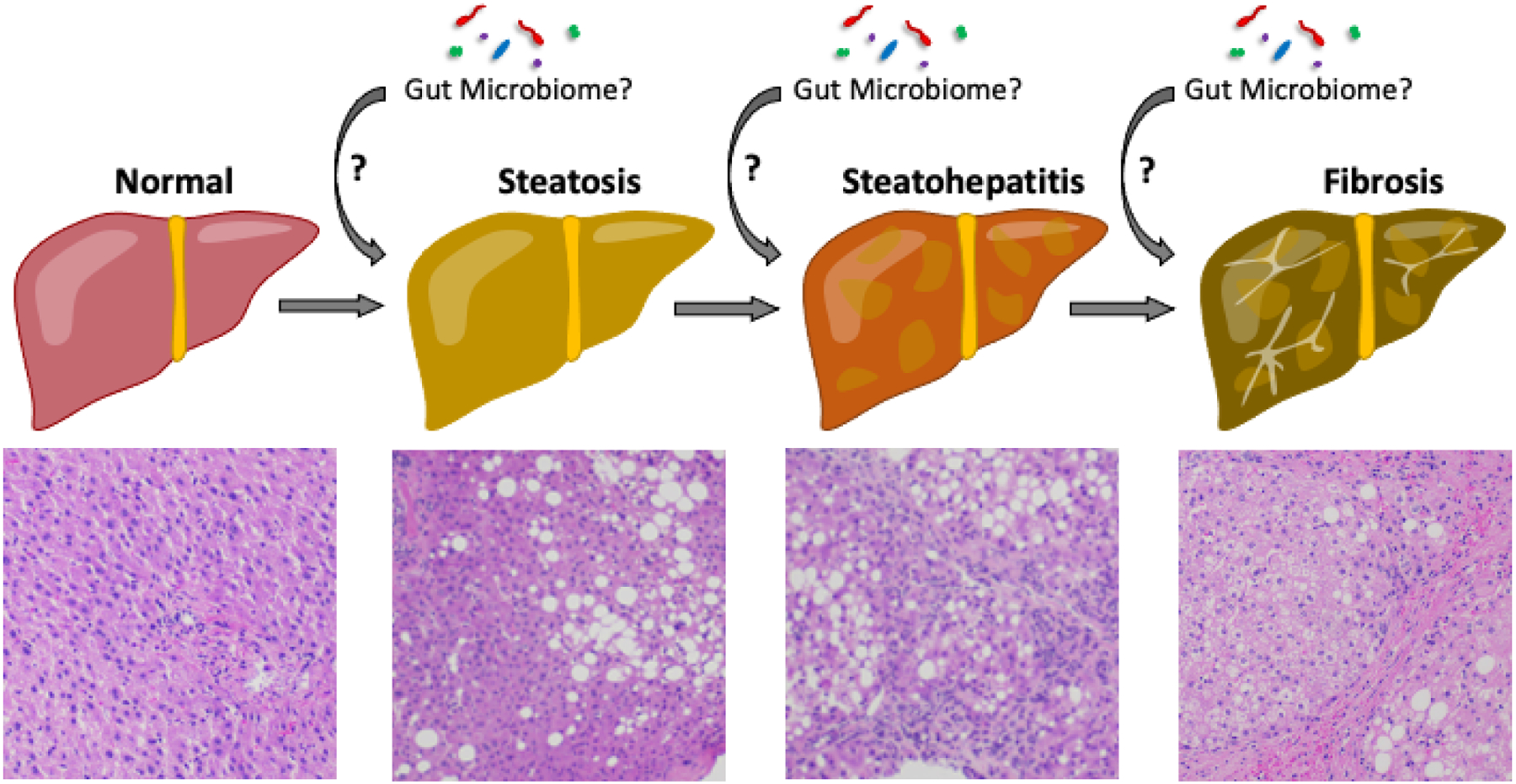
The development of NAFLD is considered to be the result of multiple injuries to the liver. The first “hit” triggers the development of steatosis. A subset of patients will experience additional “hits”, resulting in steatohepatitis, then fibrosis, and ultimately even cirrhosis. Many different mechanisms have been proposed for each step in NAFLD progression. Evidence from human and animal studies suggest that the microbiome could be instrumental in each step of NAFLD development and progression.
II. Intestinal microbiome-background
The intestinal tract is home to a vast microbial ecosystem that lives in a symbiotic relationship with its host (6–8). It is clear that the intestinal microbiota has a profound impact on human health and disease, but the mechanisms by which this occurs are not completely understood (7, 9). While initial studies focused on interactions that specifically influenced the intestinal environment, it is clear that these interactions can also have broad systemic effects, including impact on the liver and liver function.
The intestinal microbial ecosystem begins to establish itself at birth; evolution to an adult composition is shaped by the host environment (e.g. genetics, pH, oxygen, immune effectors, host-derived nutrition) and external influences, including diet and medications (antibiotics and xenobiotics) (reviewed in (10–13). In addition, bacteria engage in competition within the intestinal ecosystem for limited physical and metabolic/nutritional niches through a variety of fitness attributes and production of bacteriocins and other antibiotics (14, 15). Direct interactions with the host include sampling of the gut environment by dendritic cells and macrophages, immune antigen presenting cells (APCs) which traffic to local immune sites and present bacterial antigens to lymphocytes, generating local immune responses that, in the healthy host, are predominantly regulatory. Immune responses generated at the intestinal mucosal surface exert systemic effects as immune cells can traffic to other mucosal surfaces throughout the body. Indirect interactions with the host are more extensive, with the shedding of bacterial byproducts (e.g. lipopolysaccharide (LPS); flagellin; lipoteichoic acid (LTA)) and the production of bacterial metabolites, including dietary fermentation byproducts. These byproducts impact the luminal microenvironment by providing energy to the mucosal epithelium, maintaining mucosal epithelial integrity, stimulating immune responses, inducing antimicrobial peptide expression, and regulating microbial competition and expression of enteric pathogen virulence factors (reviewed in (7). Microbial byproducts and metabolites that reach the circulation can also have profound systemic effects, broadly influencing host physiology, immunology, and metabolism (7, 9). Therefore, disruption of the microbiome, described as dysbiosis, can have wide ranging impact on host physiology and immunology, resulting in the onset and exacerbation of diverse disease processes (reviewed in (16).
III. Gut-liver connection
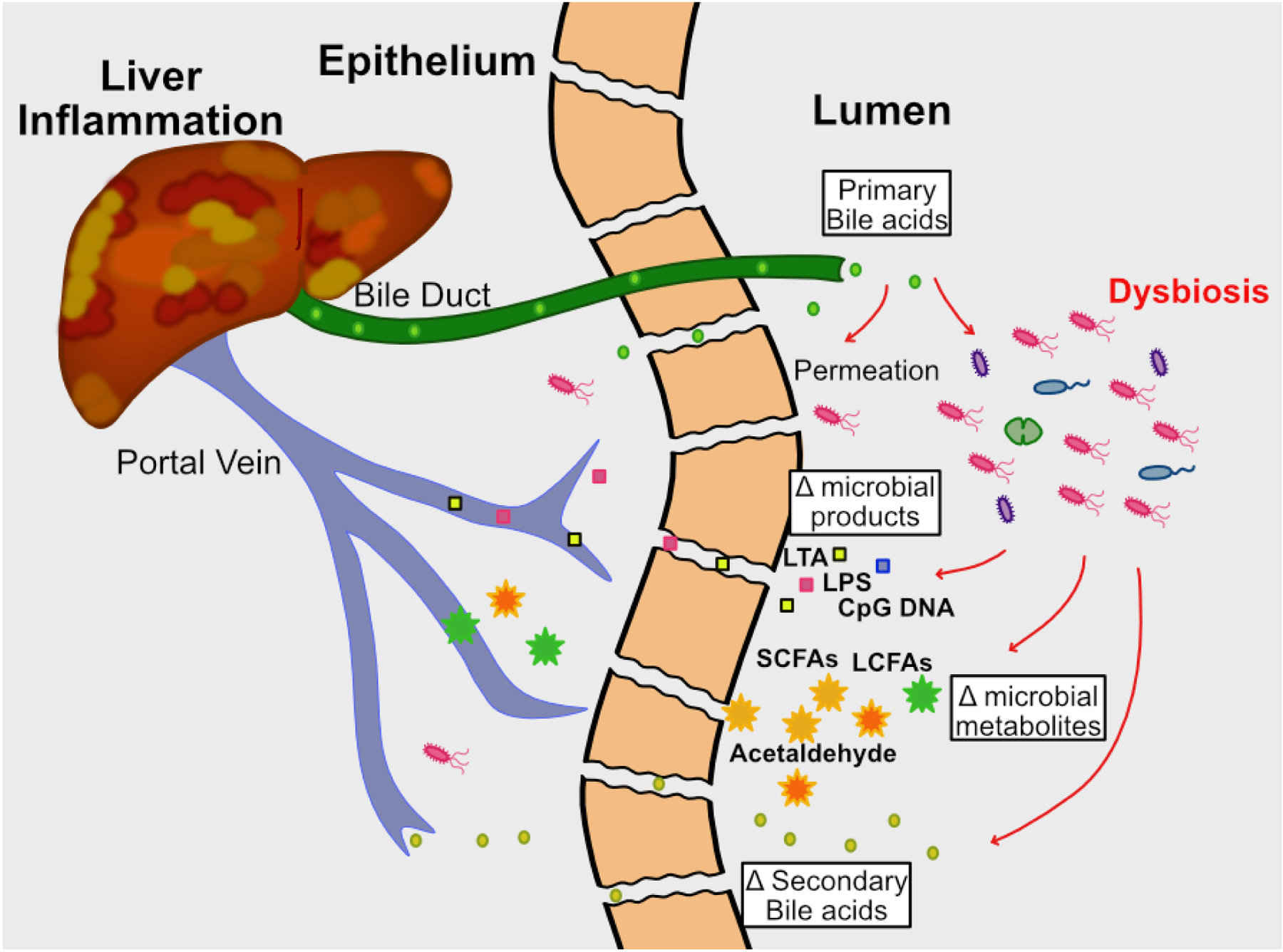
There are multiple interdependent interactions between the intestinal microbiome and the liver. Enterohepatic circulation entails communication between the liver and gut, whereby the liver secretes bile/bile acids and other metabolites through the common bile duct into the small intestine. The majority of primary bile acids are reabsorbed in the small intestine and returned to the liver through the portal vein. Commensal bacteria further metabolize the remaining primary bile acids into secondary bile acids, which, along with other microbial products (e.g. microbe-associated molecular patterns (MAMPs) such as flagellin and LPS) and dietary metabolites, can be reabsorbed by the epithelium and returned to the liver by the hepatic portal system, provided they are not excreted (Figure 2a). Intact bacteria also gain access to the liver through the portal vein. The liver is considered a microbial “firewall” for both pathogens and commensals, clearing bacteria from circulation and preventing systemic spread and infection (17, 18). The majority of bacteria are cleared from the liver after being engulfed by Kupffer cells, resident liver macrophages. Disruption of this bidirectional communication, through either damage or disease of the intestinal tract or liver, will likely result in reciprocal organ dysfunction (Figure 2b). Liver dysfunction will alter the composition of bile and metabolic components delivered to the small intestine, thus changing the intestinal microenvironment and resulting in perturbed microbial composition ((19). Reciprocally, intestinal dysbiosis will also generate altered microbial metabolites (SCFA’s, acetaldehyde, secondary bile acids) and bacterial products (LPS, LTA, CpG DNA and other MAMPs) that are delivered to the liver via the portal vein, resulting in altered hepatic function (19). Furthermore, intestinal dysbiosis driven by diet, genetics, or xenobiotics can loosen tight junctions, allowing increased transport of bacteria and altered bacterial byproducts to the portal circulation. Thus, dysbiosis associated with liver disease may be both a cause and an effect of liver damage (Figure 2b).
Figure 2. The gut-liver axis in health and disease.
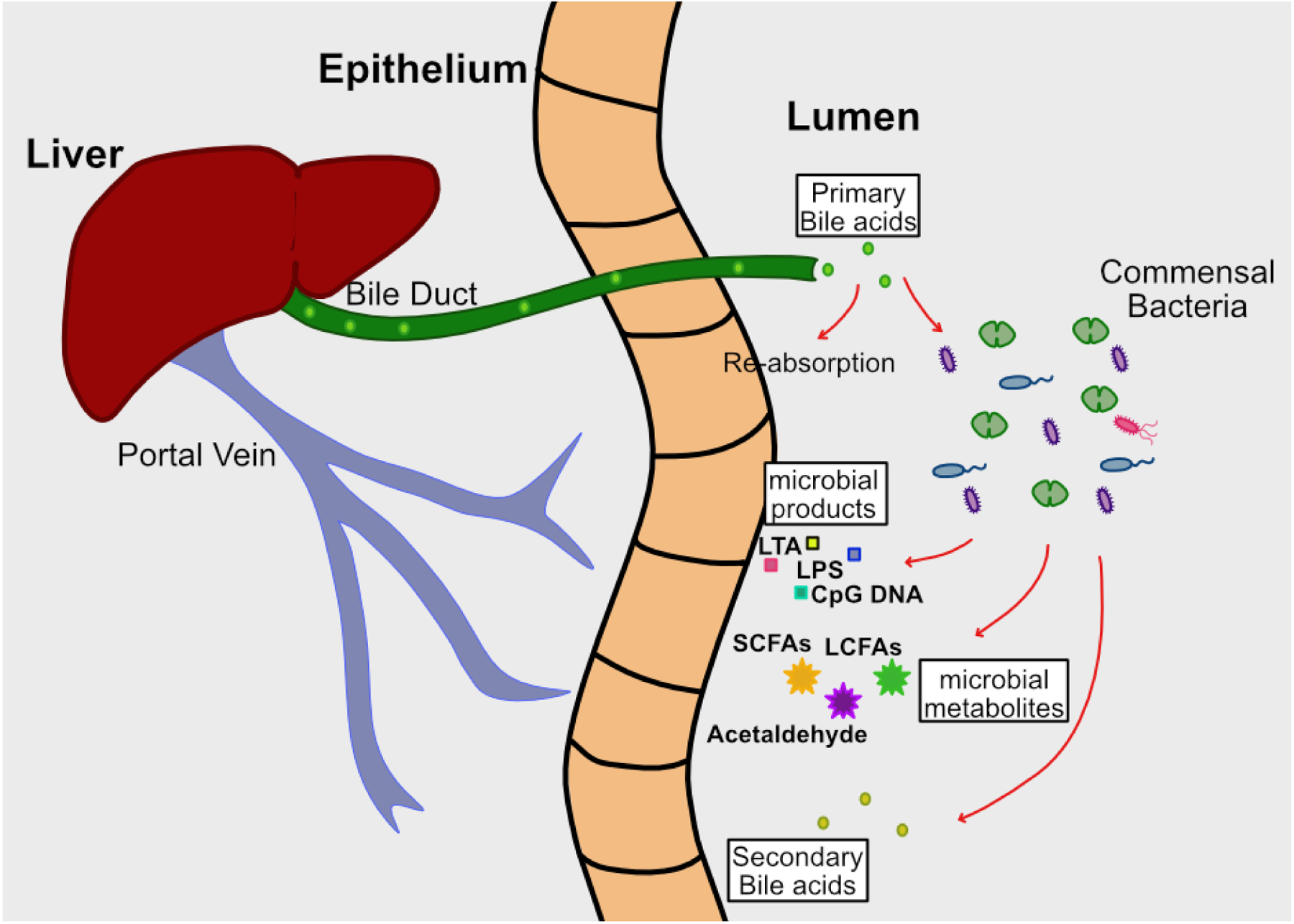
2a. During homeostasis, the liver secretes bile acids into the small intestine. The majority are reabsorbed in the proximal small intestine and returned to the liver via the hepatic-portal system. The remainder are metabolized by the microbiota into secondary bile acids, and along with other bacterial products and dietary metabolites, are absorbed by the intestinal epithelium and returned to the liver via the portal vein. Commensal bacteria that enter the liver via the portal circulation are engulfed and eliminated by Kupffer cells.
2b. When microbial homeostasis is disrupted, the composition of the microbiota changes (dysbiosis), resulting in altered metabolite production. This leads to loosening of tight junctions between epithelial cells, which results in increased intestinal permeability. There is greater movement of bacterial products, metabolites, and live bacteria into the portal circulation and ultimately the liver, leading to inflammation and hepatocyte damage.
IV. Mechanisms by which microbiota may influence NAFLD pathogenesis
Both human and animal studies have generated several hypotheses related to mechanisms that involve the gut microbiota in each step of NAFLD pathogenesis.
Microbiome and steatosis
The initial step in the development of NAFLD is hepatic steatosis. There are several ways the intestinal microbiome has been implicated in the development of steatosis. A number of mouse models have used choline deficiency to cause NAFLD (20). Choline is necessary for hepatic lipid mobilization, and loss of choline through dietary limitation or metabolism by the microbiota can lead to hepatic steatosis (20, 21). While limited evidence for similar mechanisms has been shown in human studies in adults (21), pediatric studies do not provide evidence that support choline deficiency as a primary NAFLD mechanism. Similarly, alterations in bile acids have been implicated in both animal models of NAFLD and in adult human studies but have not yet been implicated by pediatric studies (22, 23). Clearly, bile acid signaling through the farnesoid X receptor (FXR) has been shown to be an integral signaling mechanism in metabolism and has been implicated broadly in metabolic disease (reviewed in (24).
As previously mentioned, the microbiome is involved in energy harvest and fat storage. Studies in mice showed that the microbiota promotes absorption of dietary monosaccharides coupled with increased activation of lipogenic enzymes, thus inducing de novo lipogenesis (25). More recent work investigating the hepatic consequences of fructose consumption has shown that gut microbiome fructose fermentation also results in acetate production, which, after conversion to acetyl-CoA, stimulates de novo hepatic lipogenesis (26). Interestingly, acetate is also a byproduct of bacterial metabolism of ethanol (reviewed in (27)), demonstrating overlapping mechanisms between alcoholic liver disease and NAFLD in the development of hepatic steatosis. Early work in mice suggested that endogenous alcohol production by intestinal bacteria may be a contributor to the pathogenesis of NAFLD in the context of obesity. Cope et al. showed that ob/ob mice had an age-related increase in breath ethanol content that was not present in lean mice and that was blunted by the addition of neomycin (28). Later, Nair et al. (29) measured breath ethanol concentration in 21 patients with biopsy-proven NASH and in 10 healthy controls. Higher breath ethanol was observed in women vs men and with more severe obesity. However, there was not a between group difference for patients with NASH vs controls. A limited number of studies have shown increased circulating ethanol and ethanol metabolites in NAFLD, which could arise from microbial fermentation in the GI tract (30, 31); reviewed in (32). A high alcohol producing Klebsiella pneumoniae strain was isolated from a patient with severe NASH secondary to auto-brewery syndrome and was also found associated with NAFLD in a separate cohort of adult patients. Association of mice with this bacterial strain was found to accelerate the development of NAFLD in mice when fed a high fat diet (31), suggesting that endogenous microbial alcohol production is one potential mechanism that may contribute to hepatic steatosis in some non-alcoholics. In addition to the role of increased acetate noted above, the microbiota and mucosal epithelium have the capability to metabolize microbial-produced alcohol into acetaldehyde, which may increase gut permeability through loosening epithelial tight junctions (33).
Microbiome and steatohepatitis/fibrosis
Dysbiosis, leading to increased intestinal permeability or “leaky gut”, and systemic inflammation have long been implicated as contributing to several diseases, including obesity and NAFLD (34). Loss of homeostasis at the intestinal mucosal surface, whether through genetics, diet, or disease, is characterized by dysbiosis (reviewed in (10)). Alterations in bacterial colonization will result in changes of the dominant bacterial products, dietary metabolites, and secondary bile acids generated in the gut. These changes, as noted above, can lead to the loosening of epithelial tight junctions, breakdown of mucosal integrity, and result in increased intestinal permeability, with increased transport of bacteria and bacterial products to the liver via the portal circulation, ultimately affecting liver function (Figure 2b). MAMPs are immune active bacterial products and include LPS, flagellin, and CpG DNA, which interact with toll-like receptors (TLRs) on immune cells. In the liver, they interact with Kupffer cells, driving inflammatory responses that result in hepatocyte damage and steatohepatitis (reviewed in (35). Disrupting MAMP-TLR interaction by deleting MyD88, an essential TLR signaling pathway component, prevents the development of liver disease in a high fat diet (HFD) mouse model of NAFLD (36). More specifically, interaction between endotoxin (LPS, from Gram-negative bacteria) and TLR4 has been implicated in the development of NAFLD in a distinct HFD mouse model. In this study, germ-free mice were mono-associated with either one of 3 non-virulent LPS-producing bacterial strains isolated from human subjects with obesity and NAFLD, or a low endotoxin bacterial strain, and then fed a HFD. Mice colonized with high endotoxin-producing strains developed NAFLD, while those colonized with a low endotoxin-producer did not. When TLR4 deficient mice were used, the development of NAFLD was prevented (37). Similarly, MAMP-TLR interactions have been implicated in promoting liver fibrosis. Work by Seki et al (38), showed that LPS-TLR4 interaction on hepatic stellate cells (HSCs) stimulated chemokine secretion, attracting and activating Kupffer cells and modifying TGF-beta signaling in HSCs, inducing liver fibrosis. Additional studies showed that disruption of the TLR pathway prevented the development of fibrosis in mouse models of NAFLD (39, 40). The progression of dysbiosis, “leaky gut”, and increased liver exposure and response to bacteria and bacterial products has been invoked in several chronic liver diseases and could be considered part of a “final common pathway” of microbiome-associated liver injury, that is not specific to NAFLD.
V. Evidence for microbiome involvement in NAFLD
Mouse models have provided significant evidence for gut microbiome involvement in obesity. Evidence from germ-free (GF) mice suggested resistance to developing obesity in the absence of intestinal bacteria (41). Other studies showed a role for the microbiota in energy harvest and fat storage (41, 42). Fecal microbiota transplant experiments showed that the transfer of fecal samples from individuals with obesity resulted in obesity in recipient GF mice, while transfer of samples from lean individuals did not (42). The development of obesity in a number of mouse models has been associated with increased systemic inflammation and metabolic syndrome (43). These results provoked questions of whether microbiota-driven inflammation could further promote liver damage, resulting in NAFLD. Mouse models of steatosis and NAFLD have been used to provide compelling evidence that the microbiome is an integral contributor to the development of NAFLD. Work by Le Roy et al. demonstrated that hepatic steatosis could be induced in germ-free mice via fecal transplant from hyperglycemic mice (44). More recent work demonstrated that microbiota ablation prevented the development of NAFLD in diet-induced obesity mouse models (36). Concurrently, studies in adult and pediatric human subjects investigated associations between the gut microbiome and NAFLD. In this review, we focus on pediatric studies.
VI. Microbiome in pediatric NAFLD
To date there have been five studies of the relationship between the intestinal microbiome in children with NAFLD. The demographics for these studies are shown in Table 1. Many of these studies explored the previously outlined putative mechanisms for microbiome involvement in NAFLD, including the role of proinflammatory bacterial species, alcohol production, and energy harvest.
Table 1.
Characteristics of Pediatric Studies of Microbiome and NAFLD
| Study | Ages | Cases | Controls | Findings | ||
|---|---|---|---|---|---|---|
| Diagnosis | N | Definition | N | |||
| Zhu (25) | Liver Histology | 22 | Normal weight | 16 | NASH: Prevotella enterotype, increased serum ethanol | |
| Obesity with normal ALT | 25 | |||||
| Michail (26) | Mean 13 | Ultrasound and ALT | 13 | Normal weight | 26 | NAFLD: increased Prevotella, increased fecal ethanol |
| Obesity with normal ALT | 11 | |||||
| Del Chierico (27) | 7–16 | Liver Histology | 53 | Healthy | 54 |
NASH: lower α-diversitv NAFLD: lower Oscillospira, no difference in ethanol |
| Obesity | 8 | |||||
| Stanislawski (28) | 12–19 | MRI Hepatic Fat Fraction | 8 | MRI Hepatic Fat Fraction | 99 | Hepatic Steatosis: higher with greater Paraprevotella and Sutterella; lower with greater Oscillospira and Varibaculum |
| Schwimmer (29) | 8–17 | Liver Histology | 87 | Obesity with normal liver MRI Proton Density Fat Fraction and ALT | 37 |
NAFLD: lower α-diversitv, greater LPS biosynthesis NASH: higher Lactobacillus and Oribacterium; greater LPS biosynthesis Fibrosis: higher stage with more Lactobacillus; lower stage with more Akkermansia; greater flagellar assembly enrichment with higher stage |
In 2013, Zhu et al compared 22 children with NASH to 25 children with obesity but normal ALT (30). Some of the controls with obesity had abnormal liver ultrasound studies suggestive of hepatic steatosis. They also had a control group of 16 children with normal weight. They found that an enterotype enriched in Prevotella was associated with NASH. They measured serum ethanol levels in a subset of the participants (cases with NASH (n=13), controls with obesity (n=7), controls with normal weight (n=10)) and found that serum ethanol levels were the same in children with normal weight and children with obesity but approximately 30% higher in children with NASH. This was attributed to higher abundance of Escherichia species in children with NASH relative to either control group. The role of Escherichia in elevated alcohol levels was ascribed to the ability of Escherichia and other Enterobacteriaceae in their ability to produce alcohol during anaerobic fermentation.
In 2015, Michail also used a 3-group study design (45). They evaluated 13 children with NAFLD assigned by a combination of obesity with elevated ALT and abnormal liver ultrasound. The control groups were 11 children with obesity but normal ALT and liver ultrasound, and 26 “healthy” children as determined by having a normal BMI. Notably the BMI in children with NAFLD was much higher than in the obesity control group (40.8 vs 31.2 Kg/m2). At the genus level, Prevotella was more abundant in the children with NAFLD than in either control group. In addition, fecal ethanol concentration was greater in children with NAFLD than in either control group.
In 2017, Del Chierico et al studied 53 children with NAFLD, 8 children with obesity without NAFLD, and 54 controls with normal weight (46). They further divided the NAFLD group into approximately half with and half without NASH. In children with NASH, alpha-diversity was lower than in controls but was greater than children with NAFLD but not NASH. Alpha-diversity, as applied to studies of the microbiome and intestinal microbial ecology, is the bacterial species diversity within an individual sample. Measurements of alpha-diversity may be influenced by both bacterial richness (number of different species) and evenness (species relative abundance). Prevotella abundance was not significantly different between the 4 groups studied. At the genus level, only Oscillospira differed by groups, with greatest abundance in healthy controls and lowest in patients with NAFLD without a difference by NASH status. Ethanol was assessed in stool; mean values were not significantly different between patients with NASH versus controls. However, ethanol was elevated in 17% of patients with NAFLD and 25% of controls.
In 2018, Stanislawski et al studied 107 adolescents from an original birth cohort of 604 infants using MRI measured hepatic fat fraction to classify the participants as having (n=8) or not having NAFLD (n=99) (47). The small number of participants with NAFLD limited the ability to assess microbiome differences by NAFLD status. The investigators did find relationships with hepatic fat fraction as a continuous measure. They found that alpha-diversity was significantly inversely correlated with hepatic fat fraction in multivariate analysis. Using random forest analysis they reported that the taxa Paraprevotella and Sutterella were positively associated with hepatic fat fraction, and that Oscillospira and Varibaculum were inversely associated with hepatic fat fraction.
In 2019, Schwimmer et al published a case-control study of children with NAFLD versus children with obesity who did not have NAFLD (48). NAFLD was based upon clinical evaluation and liver histology. The absence of NAFLD in children with obesity was demonstrated through clinical history, laboratory studies, and liver MRI proton density fat fraction. This yielded groups that were well-matched for BMI and percent body fat. At the community level, alpha-diversity was significantly lower in children with NAFLD. When evaluating the relationship with disease severity, among children with NAFLD at the genus level, Lactobacillus and Oribacterium were higher in patients with NASH, while Oscillibacter, Lactonifactor, Akkermansia, and Enterococcus were higher in patients with NAFLD but not NASH. With respect to the severity of fibrosis, Lactobacillus was more prevalent in children with moderate-to-severe fibrosis, and Akkermansia was more prevalent in children with absent-to-mild fibrosis. In addition, children with severe fibrosis were more likely to have high Prevotella abundance. LPS biosynthesis was significantly enriched in children with NAFLD compared to controls and was further enriched in those children with NASH. Flagellar assembly was also enriched in children with NAFLD compared to controls, and further enriched in those children with moderate-to-severe fibrosis versus absent-to-mild fibrosis. Unlike the findings of Zhu et al, however, this study did not observe a difference in serum alcohol levels between cases and controls or associated with the severity of NAFLD. Further, there were no evident differences in the abundance of gene pathways associated with alcohol metabolism.
The pediatric studies to date consistently show that NAFLD is associated with dysbiosis and loss of alpha-diversity of the intestinal microbiome. However, the differences in study design make it difficult to compare results and draw firm conclusions related to bacterial composition. In addition, geographic, race/ethnicity, genetic and dietary differences may be contributing to the microbiome variations between study populations, further complicating study interpretation. Thus, we are left with several controversies, including the involvement of microbial alcohol production, and the importance of abundance of specific bacterial taxa, such as Prevotella and Escherichia. Prevotella abundance has been commonly associated with disease severity in both pediatric and adult NAFLD (49), but also identified as an important contributor to metabolic health (50). Recent work has shown that Prevotella are a highly genomically diverse species, whose abundance in the gut are driven by their ability to utilize distinct polysaccharides and are overall underrepresented in the microbiomes of Westernized populations. Functional distinctions between members of a single species are lost when analysis is limited to 16S rDNA sequencing (51–53) (54). It is likely that greater focus on the microbial genetic and metabolic potential, through examining the microbial metagenome and metatranscriptome, could yield more consistent results with greater relevance to disease pathophysiology and better direct investigation of disease mechanism.
VII. Commonality among diseases with increased intestinal permeability
A diverse set of mechanisms drives dysbiosis, leaky gut and the common pathway of liver injury. Interestingly, other pediatric diseases are also associated with dysbiosis and increased gut permeability, including inflammatory bowel diseases (IBD) such as Crohn’s disease, and type 1 diabetes mellitus (T1DM). Patients with these diseases tend not to have obesity, however there is evidence from recent studies that there is a higher incidence of NAFLD with both IBD (55) and T1DM (56). Similar to NAFLD, human and animal studies have demonstrated a significant role for the intestinal microbiome in disease pathophysiology of chronic inflammatory diseases (16, 34) including IBD and T1DM (reviewed in (57, 58). This suggests the importance of both the microbiome and that of additional “hits”, whether genetic or environmental, that drive the pathophysiology of these diverse diseases. It also supports the investigation of microbiome-targeted interventions that could modify or ameliorate disease severity in several dysbiosis-associated syndromes (reviewed in (59, 60).
VIII. Conclusions
Reciprocal interactions between the intestinal tract and the liver are critical to gut health and metabolic homeostasis. Disruption of the gut-liver interface can lead to metabolic derangement and liver damage. Studies in mice have clearly shown that the gut microbiome has an integral role in causing and exacerbating hepatic steatosis, steatohepatitis, and hepatic fibrosis in NAFLD models, through diverse mechanisms and often in the context of a diet-induced disease model. Dysbiosis is a consistent finding in pediatric and adult patients with NAFLD. However, the specific findings with regard to microbial composition have varied widely between studies and cause-effect relationships are, as yet, undetermined. Further studies using more consistent experimental design and focus on functional implications of microbial dysbiosis could better refine our understanding of the role of the microbiome in NAFLD pathogenesis. Nevertheless, the results of current studies may give rise to new testable mechanisms that may inform our understanding of human disease and lead to novel microbiome-based interventions. Overlap between diverse diseases associated with dysbiosis ultimately may suggest common microbiome-based mechanisms and possible common approaches for intervention.
Acknowledgements
We thank Kevin Jennings for providing the illustration for Figure 1; Dr. Rachel Brody for providing the histology images for Figure 1; and Dr. Kaitlin Johnson for providing the illustrations for Figure 2. This work was supported by R01DK088831 (N.H.S), UL1TR000100 of Clinical and Translation Service Award funding and grant UL1TR001442 of Clinical and Translational Service Award funding (J.B.S).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
REFERENCES
- 1.Schwimmer JB et al. , Prevalence of fatty liver in children and adolescents. Pediatrics 118, 1388–1393 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Sahota AK et al. , Incidence of Nonalcoholic Fatty Liver Disease in Children: 2009–2018. Pediatrics 146, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwimmer JB et al. , Histopathology of pediatric nonalcoholic fatty liver disease. Hepatology 42, 641–649 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Yu EL et al. , Prevalence of Nonalcoholic Fatty Liver Disease in Children with Obesity. J Pediatr 207, 64–70 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Day CP, James OF, Steatohepatitis: a tale of two “hits”? Gastroenterology 114, 842–845 (1998). [DOI] [PubMed] [Google Scholar]
- 6.Turnbaugh PJ et al. , The human microbiome project. Nature 449, 804–810 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Round JL, Mazmanian SK, The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol 9, 313–323 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Human Microbiome Project C, Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan Y, Pedersen O, Gut microbiota in human metabolic health and disease. Nat Rev Microbiol 19, 55–71 (2021). [DOI] [PubMed] [Google Scholar]
- 10.Bevins CL, Salzman NH, The potter’s wheel: the host’s role in sculpting its microbiota. Cell Mol Life Sci 68, 3675–3685 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu GD et al. , Linking long-term dietary patterns with gut microbial enterotypes. Science 334, 105–108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dominguez-Bello MG, Godoy-Vitorino F, Knight R, Blaser MJ, Role of the microbiome in human development. Gut 68, 1108–1114 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanes C et al. , Role of dietary fiber in the recovery of the human gut microbiome and its metabolome. Cell Host Microbe, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dobson A, Cotter PD, Ross RP, Hill C, Bacteriocin production: a probiotic trait? Appl Environ Microbiol 78, 1–6 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chung LK, Raffatellu M, pros GI: Antimicrobial defense in the gastrointestinal tract. Semin Cell Dev Biol 88, 129–137 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu WH, Zegarra-Ruiz DF, Diehl GE, Intestinal Microbes in Autoimmune and Inflammatory Disease. Front Immunol 11, 597966 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McDonald B et al. , Programing of an Intravascular Immune Firewall by the Gut Microbiota Protects against Pathogen Dissemination during Infection. Cell Host Microbe 28, 660–668 e664 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Balmer ML et al. , The liver may act as a firewall mediating mutualism between the host and its gut commensal microbiota. Sci Transl Med 6, 237ra266 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Jones RM, Neish AS, Gut Microbiota in Intestinal and Liver Disease. Annu Rev Pathol 16, 251–275 (2021). [DOI] [PubMed] [Google Scholar]
- 20.Koteish A, Mae Diehl A, Animal models of steatohepatitis. Best Pract Res Clin Gastroenterol 16, 679–690 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Spencer MD et al. , Association between composition of the human gastrointestinal microbiome and development of fatty liver with choline deficiency. Gastroenterology 140, 976–986 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nimer N et al. , Bile acids profile, histopathological indices and genetic variants for non-alcoholic fatty liver disease progression. Metabolism 116, 154457 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee G et al. , Distinct signatures of gut microbiome and metabolites associated with significant fibrosis in non-obese NAFLD. Nat Commun 11, 4982 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chavez-Talavera O, Tailleux A, Lefebvre P, Staels B, Bile Acid Control of Metabolism and Inflammation in Obesity, Type 2 Diabetes, Dyslipidemia, and Nonalcoholic Fatty Liver Disease. Gastroenterology 152, 1679–1694 e1673 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Backhed F et al. , The gut microbiota as an environmental factor that regulates fat storage. Proceedings of the National Academy of Sciences of the United States of America 101, 15718–15723 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao S et al. , Dietary fructose feeds hepatic lipogenesis via microbiota-derived acetate. Nature 579, 586–591 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; ** Identifies mechanism of hepatic de novo lipogenesis through interaction of dietary fructose and the intestinal microbiota
- 27.Zakhari S, Overview: how is alcohol metabolized by the body? Alcohol Res Health 29, 245–254 (2006). [PMC free article] [PubMed] [Google Scholar]
- 28.Cope K, Risby T, Diehl AM, Increased gastrointestinal ethanol production in obese mice: implications for fatty liver disease pathogenesis. Gastroenterology 119, 1340–1347 (2000). [DOI] [PubMed] [Google Scholar]
- 29.Nair S, Cope K, Risby TH, Diehl AM, Obesity and female gender increase breath ethanol concentration: potential implications for the pathogenesis of nonalcoholic steatohepatitis. Am J Gastroenterol 96, 1200–1204 (2001). [DOI] [PubMed] [Google Scholar]
- 30.Zhu L et al. , Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: a connection between endogenous alcohol and NASH. Hepatology 57, 601–609 (2013). [DOI] [PubMed] [Google Scholar]; * First study demonstrating dysbiosis in children with NAFLD/NASH
- 31.Yuan J et al. , Fatty Liver Disease Caused by High-Alcohol-Producing Klebsiella pneumoniae. Cell Metab 30, 675–688 e677 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Boursier J, Diehl AM, Nonalcoholic Fatty Liver Disease and the Gut Microbiome. Clin Liver Dis 20, 263–275 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Basuroy S, Sheth P, Mansbach CM, Rao RK, Acetaldehyde disrupts tight junctions and adherens junctions in human colonic mucosa: protection by EGF and L-glutamine. Am J Physiol Gastrointest Liver Physiol 289, G367–375 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Chassaing B, Aitken JD, Gewirtz AT, Vijay-Kumar M, Gut microbiota drives metabolic disease in immunologically altered mice. Adv Immunol 116, 93–112 (2012). [DOI] [PubMed] [Google Scholar]
- 35.Roh YS, Seki E, Toll-like receptors in alcoholic liver disease, non-alcoholic steatohepatitis and carcinogenesis. J Gastroenterol Hepatol 28 Suppl 1, 38–42 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tran HQ et al. , “Western Diet”-Induced Adipose Inflammation Requires a Complex Gut Microbiota. Cell Mol Gastroenterol Hepatol 9, 313–333 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; * Demonstrated the role of pathobionts and microbiota-innate immune signaling in metabolic syndrome and hepatic steatosis.
- 37.Fei N et al. , Endotoxin Producers Overgrowing in Human Gut Microbiota as the Causative Agents for Nonalcoholic Fatty Liver Disease. mBio 11, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; * Demonstrates mechanism by which LPS production by members of the microbiome causes NAFLD in a mouse model
- 38.Seki E et al. , TLR4 enhances TGF-beta signaling and hepatic fibrosis. Nature Medicine 13, 1324–1332 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Csak T et al. , Deficiency in myeloid differentiation factor-2 and toll-like receptor 4 expression attenuates nonalcoholic steatohepatitis and fibrosis in mice. Am J Physiol Gastrointest Liver Physiol 300, G433–441 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miura K et al. , Toll-like receptor 9 promotes steatohepatitis by induction of interleukin-1beta in mice. Gastroenterology 139, 323–334 e327 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Backhed F, Manchester JK, Semenkovich CF, Gordon JI, Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci U S A 104, 979–984 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Turnbaugh PJ et al. , An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Chassaing B, Etienne-Mesmin L, Gewirtz AT, Microbiota-liver axis in hepatic disease. Hepatology 59, 328–339 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Le Roy T et al. , Intestinal microbiota determines development of non-alcoholic fatty liver disease in mice. Gut 62, 1787–1794 (2013). [DOI] [PubMed] [Google Scholar]
- 45.Michail S et al. , Altered gut microbial energy and metabolism in children with non-alcoholic fatty liver disease. FEMS Microbiol Ecol 91, 1–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Del Chierico F et al. , Gut microbiota profiling of pediatric nonalcoholic fatty liver disease and obese patients unveiled by an integrated meta-omics-based approach. Hepatology 65, 451–464 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Stanislawski MA et al. , Gut microbiota in adolescents and the association with fatty liver: the EPOCH study. Pediatr Res 84, 219–227 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schwimmer JB et al. , Microbiome Signatures Associated With Steatohepatitis and Moderate to Severe Fibrosis in Children With Nonalcoholic Fatty Liver Disease. Gastroenterology 157, 1109–1122 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; ** Largest study of the microbiome in pediatric NAFLD with matched obese control group, showing dysbiosis and associating abundance of genes encoding bacterial inflammatory products with disease presence and severity.
- 49.Dong TS et al. , A Microbial Signature Identifies Advanced Fibrosis in Patients with Chronic Liver Disease Mainly Due to NAFLD. Sci Rep 10, 2771 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Asnicar F et al. , Microbiome connections with host metabolism and habitual diet from 1,098 deeply phenotyped individuals. Nat Med, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]; * Large scale cohort study of diet, phenotype and microbiome, showing associations between specific microbes, nutrients, and metabolic health and disease.
- 51.De Filippis F et al. , Distinct Genetic and Functional Traits of Human Intestinal Prevotella copri Strains Are Associated with Different Habitual Diets. Cell Host Microbe 25, 444–453 e443 (2019). [DOI] [PubMed] [Google Scholar]
- 52.Galvez EJC et al. , Distinct Polysaccharide Utilization Determines Interspecies Competition between Intestinal Prevotella spp. Cell Host Microbe 28, 838–852 e836 (2020). [DOI] [PubMed] [Google Scholar]
- 53.Fehlner-Peach H et al. , Distinct Polysaccharide Utilization Profiles of Human Intestinal Prevotella copri Isolates. Cell Host Microbe 26, 680–690 e685 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tett A et al. , The Prevotella copri Complex Comprises Four Distinct Clades Underrepresented in Westernized Populations. Cell Host Microbe, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin A, Roth H, Anyane-Yeboa A, Rubin DT, Paul S, Prevalence of Nonalcoholic Fatty Liver Disease in Patients With Inflammatory Bowel Disease: A Systematic Review and Meta-analysis. Inflamm Bowel Dis, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.de Vries M, Westerink J, Kaasjager K, de Valk HW, Prevalence of Nonalcoholic Fatty Liver Disease (NAFLD) in Patients With Type 1 Diabetes Mellitus: A Systematic Review and Meta-Analysis. J Clin Endocrinol Metab 105, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chang JT, Pathophysiology of Inflammatory Bowel Diseases. N Engl J Med 383, 2652–2664 (2020). [DOI] [PubMed] [Google Scholar]
- 58.Zheng P, Li Z, Zhou Z, Gut microbiome in type 1 diabetes: A comprehensive review. Diabetes Metab Res Rev 34, e3043 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marietta E, Mangalam AK, Taneja V, Murray JA, Intestinal Dysbiosis in, and Enteral Bacterial Therapies for, Systemic Autoimmune Diseases. Front Immunol 11, 573079 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu W et al. , A bridge for short-chain fatty acids to affect inflammatory bowel disease, type 1 diabetes, and non-alcoholic fatty liver disease positively: by changing gut barrier. Eur J Nutr, (2020). [DOI] [PubMed] [Google Scholar]


