Abstract
Vibrio cholerae is the causative agent of cholera, a notorious diarrheal disease that is typically transmitted via contaminated drinking water. The current pandemic agent, the El Tor biotype, has undergone several genetic changes that include horizontal acquisition of two genomic islands (VSP-I and VSP-II). VSP presence strongly correlates with pandemicity; however, the contribution of these islands to V. cholerae’s life cycle, particularly the 26-kb VSP-II, remains poorly understood. VSP-II-encoded genes are not expressed under standard laboratory conditions, suggesting that their induction requires an unknown signal from the host or environment. One signal that bacteria encounter under both host and environmental conditions is metal limitation. While studying V. cholerae’s zinc-starvation response in vitro, we noticed that a mutant constitutively expressing zinc starvation genes (Δzur) congregates at the bottom of a culture tube when grown in a nutrient-poor medium. Using transposon mutagenesis, we found that flagellar motility, chemotaxis, and VSP-II encoded genes were required for congregation. The VSP-II genes encode an AraC-like transcriptional activator (VerA) and a methyl-accepting chemotaxis protein (AerB). Using RNA-seq and lacZ transcriptional reporters, we show that VerA is a novel Zur target and an activator of the nearby AerB chemoreceptor. AerB interfaces with the chemotaxis system to drive oxygen-dependent congregation and energy taxis. Importantly, this work suggests a functional link between VSP-II, zinc-starved environments, and energy taxis, yielding insights into the role of VSP-II in a metal-limited host or aquatic reservoir.
Author summary
The Vibrio Seventh Pandemic island was horizontally acquired by the El Tor pandemic strain, but its role in pathogenicity or environmental persistence is unknown. A major barrier to VSP-II study was the lack of stimuli favoring its expression. We show that zinc starvation induces expression of island components and describe a transcriptional network that activates a VSP-II encoded energy taxis receptor. Importantly, energy taxis may enable V. cholerae to locate more favorable microenvironments, possibly to colonize anoxic portions of the gut or environmental sediments.
Introduction
The Gram-negative bacterium Vibrio cholerae, the causative agent of cholera [1], is well-adapted to two distinct lifestyles: as a colonizer of macroinvertebrates in the aquatic environment and as a potentially lethal pathogen inside the human intestine [2]. V. cholerae persists in aquatic reservoirs by colonizing a variety of (mostly chitinous) biotic surfaces, such as copepods [3–6], shellfish [7,8], and arthropods [9,10]. V. cholerae is ingested via contaminated drinking water or, less commonly, via undercooked seafood [11,12]. Once inside the human host, pathogenic varieties of V. cholerae (typically O1 and O139 serovars [13]) rely on virulence factors to establish infection; the toxin co-regulated pilus (TCP) facilitates attachment to the intestinal wall [14,15] and cholera toxin (CTX) secretion ultimately drives efflux of water and salts from the intestinal epithelium [16]. CTX additionally promotes nutrient competition via depletion of free (i.e., not heme-bound) iron in the intestine [17].
The current (seventh) cholera pandemic agent, the O1 serovar El Tor biotype, arose from a non-pathogenic precursor via acquisition of TCP and CTX virulence factors [18]. Unlike its pandemic predecessor (the classical biotype), El Tor underwent several changes that include, among others [19], the development of resistance against the antimicrobial peptide polymyxin B [20,21] and horizontal acquisition of two genomic islands (VSP-I and VSP-II) [22,23]. Presence of VSP-I and -II strongly correlate with pandemicity; however, only genes encoded on VSP-I have been directly linked to increased fitness in a host [24]. VSP-II is a poorly understood 26-kb island that contains 30 annotated ORFs spanning vc0490-vc0516 [25], only two of which have validated functions: an integrase (vc0516, [26]) and a peptidoglycan endopeptidase (vc0503, [27]). The remaining uncharacterized genes are predicted to encode transcriptional regulators (VC0497, VC0513), ribonuclease H (VC0498), a type IV pilin (VC0502), a DNA repair protein (VC0510), methyl-accepting chemotaxis proteins (VC0512, VC0514), a cyclic di-GMP phosphodiesterase (VC0515), and hypothetical proteins [23]. It is unclear if or how VSP-II enhances the pathogenicity or environmental fitness of El Tor. Intriguingly, VSP-II genes are not expressed under standard laboratory conditions [28], suggesting that their induction requires an unknown signal from the host or environment.
One signal that bacteria encounter under both host and environmental conditions is metal limitation. Bacteria must acquire divalent zinc cofactors from their surroundings to perform essential cellular processes; however, vertebrate hosts actively sequester zinc and other essential transition metals to limit bacterial growth (i.e. nutritional immunity) [29–32]. In the environment, V. cholerae frequently colonize the chitinous exoskeletons of aquatic and marine invertebrates and exposure to chitin oligomers has been suggested to induce zinc and iron starvation in V. cholerae [33]. In order to cope with zinc starvation stress, V. cholerae encodes a set of genes under the control of the well-conserved Zur repressor. When zinc availability is low, Zur dissociates from a conserved promoter sequence, allowing for transcription of downstream genes. V. cholerae genes containing a Zur binding region include those encoding zinc import systems (ZnuABC and ZrgABCDE) [34], ribosomal proteins (RpmE2, RpmJ) [35,36], a GTP cyclohydrolase (RibA) [35], and the VSP-II-encoded peptidoglycan endopeptidase (ShyB) [27].
Here, we show that many genes of the VSP-II island are expressed during zinc starvation. These findings stemmed from an initial observation that a V. cholerae Δzur mutant accumulated at the bottom of nutrient-poor liquid cultures. We hypothesized that this behavior was mediated by unidentified members of the Zur regulon. Using a transposon mutagenesis screen and RNA-seq, we identified Zur-regulated congregation factors encoded on VSP-II. These included the transcriptional activator VerA (Vibrio energy taxis regulator A) encoded within the vc0513-vc0515 operon. VerA activates expression of the AerB (aerotaxis B) chemotaxis receptor encoded by vc0512. We show that AerB mediates oxygen-dependent congregation and energy taxis. Importantly, these results implicate a role for VSP-II genes in chemotactic movement during zinc starvation.
Results
The V. cholerae Δzur mutant congregates in minimal medium
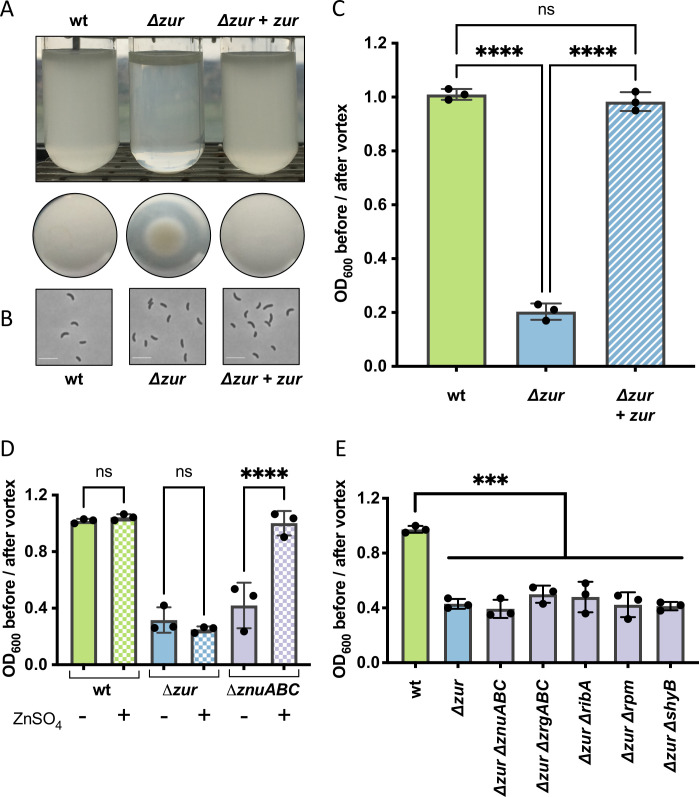
We noticed serendipitously that a V. cholerae N16961 Δzur mutant (but not the wild type) accumulated at the bottom of a culture tube when grown shaking overnight in M9 minimal medium (Fig 1A). A similar result was observed in static overnight cultures (S1A Fig). Microscopic inspection of Δzur cells transferred to an agar pad revealed mostly individual cells with no obvious changes in morphology (Fig 1B); the lack of strong cell-to-cell interactions holding these “congregates” together is consistent with the ease at which the pellet was dispersed by agitation (S1 Movie). V. cholerae aggregations in liquid culture are reportedly mediated by numerous mechanisms (e.g. quorum sensing, attachment pili, and O-antigen synthesis [37–44]) and stimuli (e.g. autoinducers, calcium ions [37], cationic polymers [43]), but none thus far have been tied to zinc homeostasis. We therefore sought to identify factors that were required for Δzur to congregate. Congregation (quantified as the ratio of optical densities (OD600nm) in the supernatant before and after vortexing) was alleviated by complementing zur in trans, excluding polar effects resulting from zur deletion (Fig 1C). We next examined the role of zinc availability on congregation. Since metals can absorb to the surface of borosilicate glass culture tubes [45], we instead grew V. cholerae in plastic tubes and noted that Δzur still congregated at the bottom (S1B Fig), indicating that this phenotype is not linked to the properties of the culture vessel. Imposing zinc starvation via deletion of genes encoding V. cholerae’s primary zinc importer ZnuABC caused cells to congregate similarly to the Δzur mutant. Congregation of ΔznuABC (which still elaborates the low-affinity zinc transporter ZrgABC [34]) was reversed by zinc supplementation (Fig 1D). In contrast, the Δzur mutant, which constitutively expresses zinc starvation genes, congregated in both the presence and absence of exogenous zinc. These data indicate that congregation occurs in minimal medium when the Zur regulon is induced (i.e., during zinc deficiency or in a zur deletion strain) and is not a direct consequence of zinc availability per se. Surprisingly, none of the annotated members of the Zur regulon were required for congregation (Figs 1E and S1C), suggesting that there may be other Zur-regulated congregation genes yet to be identified.
Fig 1. A V. cholerae Δzur mutant congregates in M9 minimal medium.
(A-C) Wild-type, Δzur, and Δzur carrying an integrated, IPTG-inducible copy of zur (denoted + zur) were grown overnight at 30°C in M9 minimal medium supplemented with glucose (0.2%) and inducer (IPTG, 500 μM). (A) Representative side and bottom-view photos of overnight cultures are shown. (B) Cells collected from the bottom of the tube were imaged on an 0.8% agarose pad. Scale bar, 5 μM. (C) Congregation was quantified by measuring the optical density (at 600 nm) of the culture supernatant before and after a brief vortex. A ratio close to 1 represents a homogenous culture, a ratio closer to 0 indicates congregation. (D) Congregation was measured in wild type, Δzur, and a zinc importer mutant (ΔznuABC) grown in M9 glucose (0.2%) in the absence (solid bars) or presence (checkered bars) of exogenous zinc (ZnSO4, 1 μM). (E) Congregation in M9 glucose (0.2%) was measured in wild type, Δzur, and Δzur lacking components of the zinc starvation response (znuABC, zrgABC, ribA, rpmE2/rpmJ2, or shyB). For all plots, the shown raw data points are biological replicates, error bars represent standard deviation, and asterisks denote statistical difference via Ordinary one-way ANOVA test (****, p < 0.0001; ***, p < 0.001; n.s., not significant).
Δzur congregation requires motility, chemotaxis, and VSP-II encoded proteins
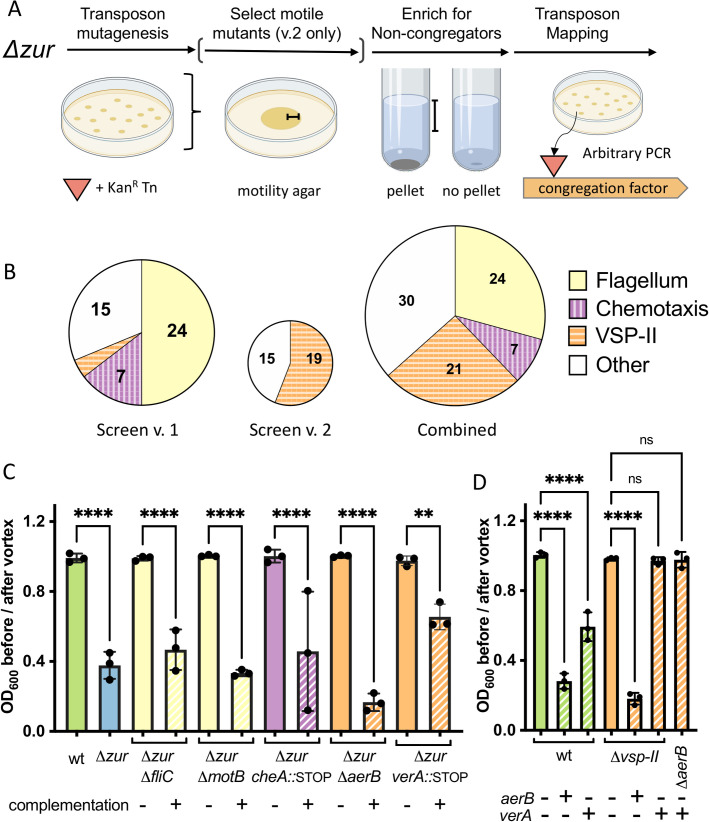
We reasoned that we could leverage the Δzur congregation phenotype to identify novel components of V. cholerae’s zinc starvation response. To find such Zur-regulated “congregation factors”, we subjected the Δzur mutant to transposon mutagenesis and screened for insertions that prevent congregation (see Methods for details) (Fig 2A). Δzur transposon libraries were inoculated into M9 minimal medium and repeatedly sub-cultured until no pellet formed. Transposon insertions sites were identified using arbitrary PCR on isolated colonies [46]. The insertions that prevented congregation overwhelmingly mapped to loci encoding motility and chemotaxis genes (Fig 2B). Twenty-four out of 48 recovered transposon mutants were disrupted in flagellar components or motility regulators. We reconstituted these types of mutations in Δzur by inactivating flagellum assembly (major flagellin subunit, fliC) or rotation (motor protein, motB). Both Δzur ΔfliC and Δzur ΔmotB failed to form a pellet and congregation could be restored by complementing each of these genes in trans (Fig 2C). These data suggest that Δzur congregation is a motility-dependent process. Additionally, seven transposons inserted within genes encoding parts of V. cholerae’s chemotaxis machinery (che-2) (Fig 2B); this system modulates bacterial movement in response to a chemical gradient. Mutating a component of this chemotactic phosphorelay (cheA::STOP) was sufficient to prevent congregation in Δzur, while trans expression of cheA restored pellet formation to the Δzur cheA::STOP mutant (Fig 2C). Deletion of other che-2 open reading frames also prevented Δzur from congregating (S1D Fig). Collectively, these data suggest that motility and chemotaxis are required for Δzur congregation in minimal medium.
Fig 2. Δzur congregation requires motility and components of VSP-II.
(A) Δzur was mutagenized with mariner transposons to generate a library of insertion mutants (see Methods for details). Non-congregating mutants within the library were enriched via repeated subculturing of the supernatant until no pellet formed. Brackets indicate harvested zones. Transposon insertions were mapped using arbitrary PCR. In a modified version of this screen (v.2), the transposon library was pre-filtered to select for motile mutants on soft agar (0.3%). Schema created with BioRender.com. (B) Transposon insertions mapped to genes encoding flagellar components and regulators (24 hits, yellow), chemotaxis proteins (7 hits, purple/vertical lines), and VSP-II genes (21 hits, orange/horizontal lines). (C) Select motility (fliC, motB), chemotaxis (cheA), and VSP-II genes (vc0512/aerB, vc0513/verA) were mutated in a Δzur background (solid bars) and complemented back in trans (+) under an IPTG inducible promoter integrated within chromosomal lacZ. (Note: Piptg-verA on a multicopy plasmid was used for complementing the Δzur verA::STOP mutant. These cultures were grown with kanamycin to ensure retention of either the empty or verA-expressing plasmid). Congregation in M9 glucose (0.2%) supplemented with inducer (IPTG, 100 μM) was quantified by measuring the optical density (at 600 nm) of the culture supernatant before and after a brief vortex. (D) Integrated, chromosomal copies of aerB or verA were overexpressed in wild-type and Δvsp-II (or a ΔaerB) backgrounds in M9. Ten and 200 μM of IPTG were used for aerB and verA induction, respectively. For all plots, the shown raw data points are biological replicates, error bars represent standard deviation, and asterisks denote statistical difference via Ordinary one-way ANOVA test (****, p < 0.0001; **, p < 0.01; n.s., not significant).
We noted that the Δzur phenotype resembles aggregation in E. coli rough mutants, which have reduced expression of lipopolysaccharides [47]. We observed similar aggregation in V. cholerae rough mutants (vc2205::kan), but this aggregation did not require motility to form and is therefore mediated by a distinct mechanism (S1E Fig). We anticipated initially that Δzur pellet formation was a group behavior that may require processes associated with surface attachment (e.g., biofilm formation, attachment pili) or cellular communication (e.g., quorum sensing); however, such mutants were not identified by the transposon screen. We thus separately assessed this in a targeted fashion by testing whether Δzur still congregates when deficient in biofilm formation (ΔvspL) or type IV pili attachment (Δ4: ΔtcpA ΔmshA ΔpilA, and orphan pilin Δvc0502). Consistent with these processes not answering our screen, biofilm and type IV pili encoding genes were not required for Δzur to congregate (S1F Fig). Notably, N16961 contains an authentic frameshift mutation in the quorum sensing gene hapR [48,49]; however, a repair to hapR [50] did not alter congregation dynamics in Δzur (S1G Fig). We additionally demonstrated that other quorum sensing genes (ΔcsqA, ΔcsqS, Δtdh, ΔluxQ, or ΔluxS) were dispensable for this phenotype (S1F Fig). Taken together, these data indicate that Δzur pellet formation is not a clumping phenomenon driven by typical colonization and congregation factors, but rather a chemotaxis/motility-mediated assembly in the lower strata of a growth medium column.
Since congregation appeared to require induction of the Zur regulon, we were surprised that the transposon screen was not strongly answered by genes with an obvious Zur binding site in their promoters. We reasoned, however, that our screen did not reach saturation due to the large number of motility genes encoded in the V. cholerae genome. We therefore refined the screen by pre-selecting for mutants that retained motility on soft agar, followed by a subsequent screen for loss of pellet formation in the motile subset of the mutant pool, as described above (Fig 2A). Interestingly, 19 of the 34 transposon insertions answering this screen mapped to the Vibrio Seventh Pandemic island (VSP-II) (Figs 2B and S2A and S2B), a horizontally acquired genomic region that is strongly associated with the El Tor biotype and the current (seventh) cholera pandemic. Transposons concentrated in a section of VSP-II that encodes a putative AraC-like transcriptional activator (VC0513, henceforth “VerA”), two ligand-sensing chemotaxis proteins (VC0512, formerly Aer-1 is henceforth referred to as “AerB”, and VC0514), and a cyclic di-GMP phosphodiesterase (VC0515). Notably, the verA/vc0513-vc0515 operon is preceded by a canonical Zur binding site and is thus a novel candidate for Zur-dependent regulation.
To validate the VSP-II genes’ involvement in Δzur congregation, we inactivated each gene in a Δzur background through either clean deletion or through insertion of a premature stop codon. ΔaerB and verA::STOP mutations prevented Δzur from congregating, whereas respective complementation with aerB and verA restored the pellet (Fig 2C). We noted that although verA and aerB were both required for congregation, transposon hits were concentrated in verA. Since a Δzur verA::STOP mutant yields a significantly increased swarm diameter on soft agar (S2C Fig), we speculate that verA insertions were overrepresented in the motile subset of our transposon library. We additionally tested a deletion of the entire VSP-I island and mutations in all other open-reading frames on VSP-II (including vc0514 and vc0515), none of which were required for Δzur congregation under the conditions tested (S1H Fig).
To determine if either verA or aerB are sufficient to generate congregates, we overexpressed each gene in a wild-type and a Δvsp-II background. Both aerB and verA overexpression caused the wild-type to congregate, but only the aerB chemoreceptor triggered congregation in a strain lacking other VSP-II genes (Fig 2D). These data suggest that AerB drives the observed pellet formation and raise the possibility that VerA functions as a transcriptional activator of aerB. Altogether, these two screens indicate that pellet formation in Δzur is driven by chemotactic flagellar movement, with assistance from a VSP-II encoded transcriptional activator (VerA) and chemoreceptor (AerB). These results were intriguing given that very little is known about the regulation or function of VSP-II encoded genes.
Several VSP-II genes are significantly upregulated in a Δzur mutant
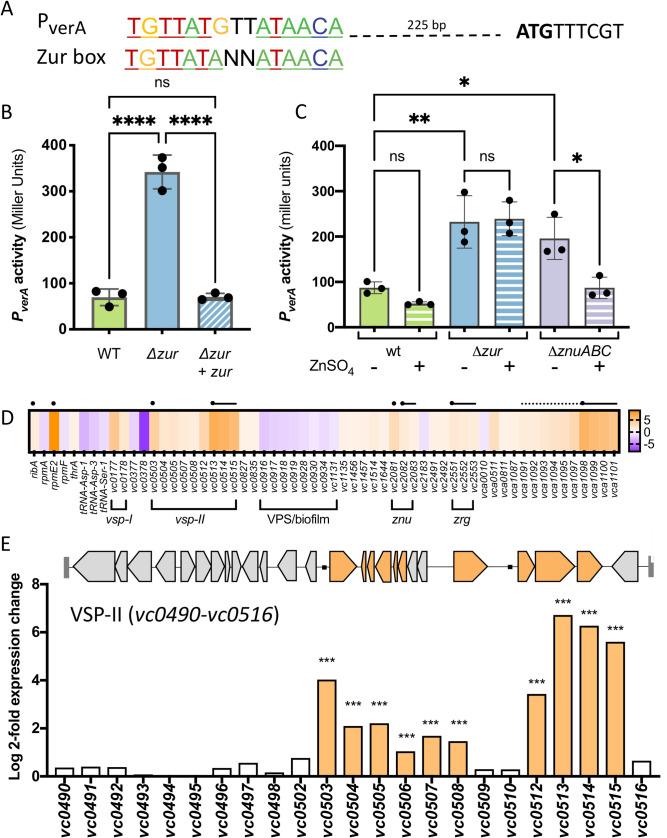
Prior inquiry into VSP-II function was made difficult by a lack of native gene expression under laboratory conditions; thus, we prioritized mapping the transcriptional networks embedded in this island. We predicted that the VSP-II genes of interest are expressed in Δzur. Consistent with this idea, the verA promoter region contains a highly conserved Zur-binding sequence approximately 200 bp upstream of the mapped transcription start site (determined by 5’-RACE in a Δzur mutant, Figs 3A and S3). Although the distance between the Zur box and transcriptional start site is greater than that observed for most V. cholerae Zur targets, equivalent or greater distances are noted for the Zur-regulated ribA (140–210 bp upstream of the ORF) and zbp (~380 bp upstream of ORF) in closely related Vibrio spp., respectively [35]. To verify regulation by Zur, we measured verA promoter activity via a lacZ transcriptional fusion (PverA-lacZ). LacZ encodes β-galactosidase (LacZ) and, when expressed, produces a colorimetric readout in the presence of a cleavable substrate (e.g., ONPG). As expected, transcription from the verA promoter in zinc-rich LB medium was robust in Δzur relative to wild-type or a zur complemented strain (Fig 3B). This data indicates that Zur negatively regulates verA transcription in a rich medium. We also tested PverA-lacZ expression in M9 minimal medium in a wild-type, Δzur, and ΔznuABC background and noted that promoter activity corresponded to the conditions in Fig 1D that triggered congregation (Fig 3C). PverA activity was low in the wild-type background, suggesting that V. cholerae is not zinc-deficient in our M9 liquid culture. In contrast, PverA activity was robust in both Δzur and a mutant deficient in zinc uptake (ΔznuABC). Consistent with Zur’s zinc sensing function, Δzur PverA-lacZ strain retained high levels of β-galactosidase activity regardless of zinc availability, whereas PverA-lacZ in ΔznuABC was repressible with exogenous zinc. These data, in conjunction with the highly conserved Zur binding site, suggest that the VerA-encoding vc0513-vc0515 operon is a novel component of the Zur-regulated zinc starvation response in N16961.
Fig 3. Several VPS-II genes are upregulated in a Δzur mutant.
(A) A predicted Zur-binding site (TGTTATGTTATAACA) located approximately 200 bp upstream of the verA open reading frame was aligned with the Vibrionacae Zur binding consensus sequence [35]. The predicted start codon (ATG) is indicated in bold. (B) PverA-lacZ transcriptional reporters were introduced into a wild-type and Δzur background, paired with either an empty vector or IPTG-inducible copy of zur (+ zur). Strains were grown overnight and diluted 1:100 into LB with kanamycin and inducer (IPTG, 200 μM). After 3 hours of growth at 37°C (to mid/late exponential phase), promoter activity was quantified in Miller Units by measuring β-galactosidase (LacZ) activity against an ONPG chromogenic substrate (see Methods for more details). (C) Wild-type, Δzur, and ΔznuABC mutants carrying the PverA-lacZ reporter were grown in M9 minimal medium in the presence (+) and absence (-) of exogenous zinc (ZnSO4, 1 μM). After overnight growth (~16 h), promoter activity was measured in Miller units. For bar graphs, raw data points represent biological replicates, error bars represent standard deviation, and asterisks denote statistical difference via Ordinary one-way ANOVA (****, p < 0.0001; **, p < 0.01; *, p < 0.05; and n.s., not significant). (D-E) RNA was isolated from wild-type and Δzur cells at mid-log phase and prepared for RNA-seq (see Methods and Materials). Genes with significant differential expression in Δzur (log 2-fold change > 1, adjusted p-value <0.05) relative to wild-type N16961 are shown. (D) The heat map indicates increased (orange) or decreased (purple) expression relative to the wild-type strain. Black circles represent putative Zur binding sites and lines correspond to likely operons. (E) Log 2-fold expression changes for all VSP-II genes (vc0490-vc0516) are shown alongside a schematic of VSP-II open reading frames. Significantly upregulated genes are shown in orange.
Global transcriptomic studies of the Zur regulon have been conducted in a number of bacteria, but none thus far have been reported in the Vibrio genera [51–68]. We thus performed an RNA-seq experiment comparing transcript abundance in wild-type N16961 and Δzur to assess V. cholerae’s Zur regulon more comprehensively (including indirect effects). To ensure sufficient repression of Zur targets in the wild-type, cells were grown to mid-log phase in LB medium. Analyses identified 58 differentially expressed genes in Δzur (log 2-fold change >1, adjusted p-value <0.05) (Figs 3D and 3E and S4 and S1 Table). Seven promoters (situated in proximity to 23 of the 42 upregulated genes) contained an upstream canonical Zur-binding site. Among them were known or inferred (based on E. coli) Zur regulon components, including genes that encode zinc uptake systems (ZnuABC, ZrgACD), an alternative ribosomal protein (RpmE2), and a GTP cyclohydrolase (RibA). This transcriptomic analysis also uncovered what appears to be a bidirectional promoter with a Zur box: this locus encodes a strongly upregulated ABC-type transporter (vca1098-vca1101) in one direction and upregulated portions of the chemotaxis-3 (che-3) cluster (vca1091-vca1095, vca1097) in the other. Using a lacZ transcriptional reporter, we verified that the ABC-type transporter is indeed Zur-regulated (S5 Fig). Neither the transporter nor the che-3 cluster (vca1090-vca1097), however, were required for Δzur congregation (S1C Fig). We observed a striking cluster of ten up-regulated genes on VSP-II (comprising 33% of the annotated open reading frames on VSP-II), including the previously characterized peptidoglycan hydrolase ShyB (encoded by vc0503) and the verA/vc0513-vc0515 operon (Fig 3D and 3E). These results are consistent with both our transcriptional fusion data and the transposon screen.
The RNA-seq analysis also identified 36 differentially expressed genes that lacked canonical Zur binding sites (Figs 3D and 3E and S4 and S1 Table). Nineteen of these genes were significantly up-regulated in Δzur, including several genes on VSP-II (vc0504-vc0508 and vc0512) and VSP-I (vspR/vc0177, capV/vc0178). Other upregulated transcripts in Δzur encode for cholera toxin (ctxA/B), the toxin co-regulated pilus biosynthesis proteins (tcpT/H), and a chitin binding protein (gbpA). Seventeen genes were significantly down-regulated in Δzur, many of which were related to vibrio polysaccharide (VPS) synthesis and biofilm formation [69]. Thus, a zur deletion affects numerous genes indirectly, possibly through Zur-dependent secondary regulators (e.g. VC0515, cyclic di-GMP phosphodiesterase; VerA, AraC-like transcriptional regulator), via secondary responses to the influx of zinc that the Δzur mutant is expected to experience, or via Zur-dependent small RNA interference. We did conduct a perfunctory analysis of small RNAs and encourage interested research communities to utilize our data deposited in NCBI GEO (GSE173966) to pursue additional lines of inquiry.
VerA is a Zur-regulated transcriptional activator of aerB
Our mutational analyses above raised the possibility that the putative methyl accepting chemotaxis protein (MCP) AerB is controlled by the transcriptional activator VerA. Given the importance of AraC-family regulators in governing V. cholerae’s host-associated behaviors (e.g., ToxT, intestinal colonization and virulence [70–73]; Tfos, chitin-induced natural competence [74–76]) we sought to characterize the full VerA regulon. We performed an RNA-seq experiment comparing transcript abundance in N16961 overexpressing verA, relative to an empty vector control. Surprisingly, only three other genes were significantly upregulated (log 2-fold change >1, adjusted p-value <0.05): vc0512/aerB, vc0514, and vc0515 (Fig 4A and S2 Table). We validated these findings using lacZ transcriptional reporters. Plasmid-mediated verA overexpression was sufficient to induce PverA-lacZ in rich LB medium, suggesting that this operon is autoregulated by VerA (Fig 4B). To remove the autoregulatory effect of native VerA from our analysis, we performed additional measurements in a parent strain lacking VSP-II (and thus native verA). These data suggest that loss of Zur binding may lead to only a small increase in verA transcription, which is further amplified by a VerA-dependent positive feedback loop.
Fig 4. VerA is an AraC-like transcriptional activator that positively regulates aerB and the vc0513-vc0515 operon.
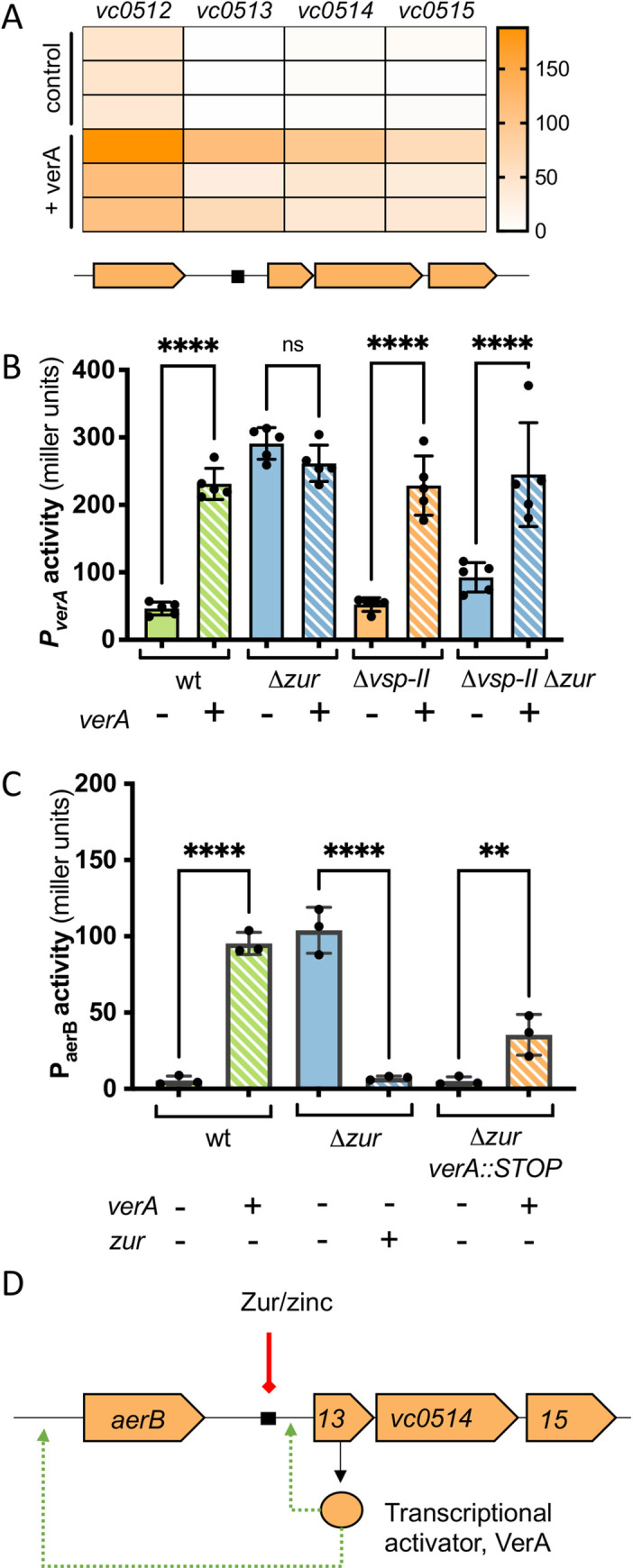
(A) Overnight cultures of wild-type N16961 carrying either an IPTG-inducible copy of verA (+ verA) or empty vector (control) were diluted 1:100 in fresh LB containing kanamycin and IPTG (1 mM). RNA was isolated from cells at mid-log phase and prepared for RNA-seq (see Methods and Materials). Heat map shows normalized expression values for differentially expressed genes across three biological replicates. (B-C) Overnight cultures of strains carrying lacZ transcriptional reporters were diluted 1:100 in LB and grown for 3 h at 37°C. Kanamycin and inducer IPTG (500 μM) were included in the growth medium for retention and trans expression (+) of verA or zur from an IPTG-inducible promoter. Promoter activity (in Miller Units) was measured via β-galactosidase assays (See Methods and Materials). (B) PverA-lacZ activity was measured in wild-type, Δzur, Δvsp-II, and Δzur Δvsp-II strains carrying a plasmid-borne, IPTG-inducible copy of verA (+, striped bars) or an empty vector control (-, solid bars). (C) Activity from a PaerB-lacZ reporter was measured in wild-type, Δzur, and Δzur verA::STOP backgrounds harboring a plasmid-borne, IPTG-inducible copy of verA or zur (+, striped bars) or an empty vector control (-, solid bars). (D) Proposed model for Zur repression of the verA promoter (solid line, red) via a conserved Zur binding site (black box) and subsequent VerA-dependent activation (dashed arrow, green) of the aerB and verA promoters.
Although the aerB promoter lacks a conserved Zur binding site, our transcriptomic data suggests that Zur-regulated VerA promotes aerB transcription. To verify this, we constructed a PaerB-lacZ transcriptional reporter. Our initial attempt using a small (400 bp) promoter fragment did not yield detectable signal under inducing conditions (S6 Fig). 5’-RACE mapping of the transcription start indicated that aerB is part of a much longer transcript (extending >1 kb upstream of the start codon). Thus, we designed a new reporter construct to include this entire region. PaerB activity in standard LB medium fell below our threshold for detection, but we found that VerA overexpression was sufficient to activate the aerB promoter (Fig 4C). PaerB was also strongly induced in a Δzur strain background–consistent with our initial RNA-seq–but only if the strain also carried a native or trans copy of verA. These data indicate that aerB expression is dependent upon VerA-mediated activation. In summary, we found that VerA is a Zur-regulated, transcriptional activator that upregulates four genes (aerB, verA/vc0513-vc0515) (Fig 4D).
AerB mediates energy taxis and pellet formation
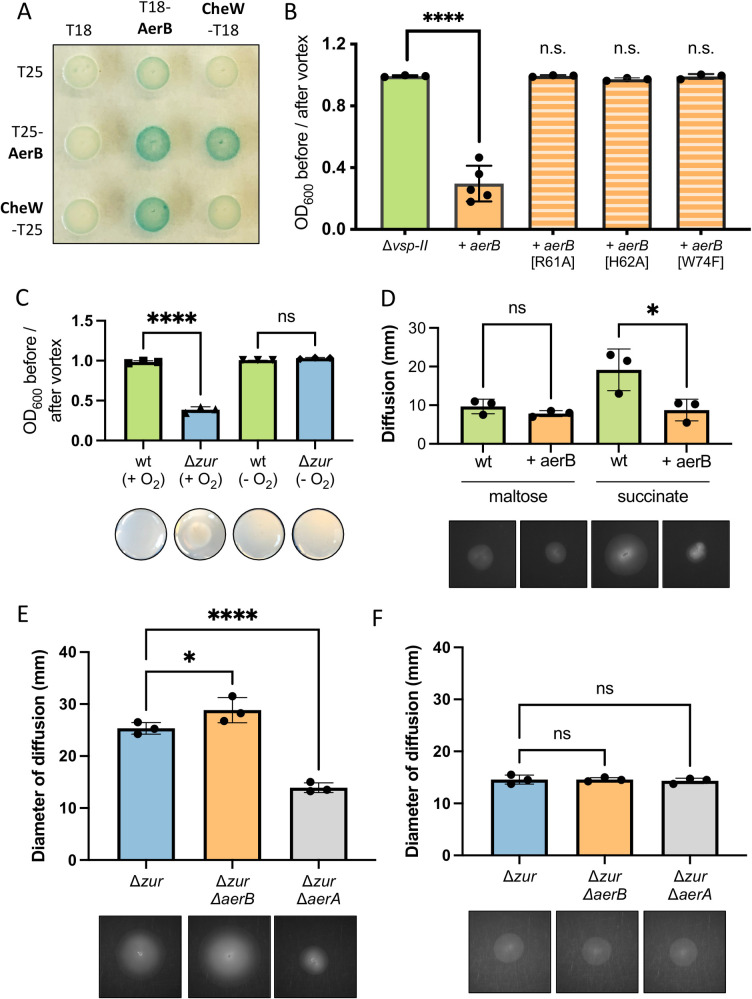
VerA-mediated induction of AerB drives V. cholerae to congregate in minimal medium. AerB is predicted to encode an MCP that senses concentration gradients of a particular ligand (either an attractant or repellent) and relays that signal via Che proteins that alter flagellar rotation [77]. To determine if AerB indeed functions as a chemotaxis receptor, we first tested whether AerB interacts with the chemotaxis coupling protein, CheW. In a bacterial two hybrid assay, AerB and CheW were each fused with one domain of the adenylate cyclase (AC) protein (T18 or T25) and co-transformed into an E. coli strain. If the proteins of interest interact, the proximal AC domains will synthesize cAMP and induce lacZ expression via a cAMP-CAP promoter; thus, a positive protein interaction will yield blue colonies in the presence of X-gal. E. coli co-transformed with T18-AerB and CheW-T25 (or the reciprocal tags) yielded bright blue spots (Fig 5A). We additionally detected strong protein-protein interaction between T18-AerB and T25-AerB, indicating that our chemoreceptor can dimerize (or oligomerize) like other MCPs [78]. To confirm that AerB’s MCP signaling domain is required for congregation, we next mutated a glycine residue within the highly conserved C-terminal hairpin loop (R-A-G-E/D-X-G) [79] of AerB (S7 Fig), which is required for in vitro signal generation in other MCPs [80]. A Δvsp-II strain expressing this AerB[G385C] mutant was unable to congregate (Fig 5B), consistent with MCP function. Together, these data suggest that AerB indeed functions as a chemotaxis receptor.
Fig 5. AerB encodes a methyl-accepting chemotaxis protein involved in energy taxis.
(A) In a bacterial two-hybrid assay, E. coli BTH101 was co-transformed with vectors carrying one domain of adenylate cyclase (T18 or T25) or an adenylate cyclase fusion with a protein of interest: CheW-T(18/25) or T(18/25)-AerB. Co-transformants were spotted onto an LB agar containing kanamycin and ampicillin (for selection), X-gal (for blue-white detection), and inducer (IPTG, 500 μM). Plates were incubated overnight at 30°C and for an additional day at room temperature. Blue color signifies positive protein-protein interactions. (B) Δvsp-II strains carrying an integrated, IPTG-inducible copy of either aerB or aerB point mutants (G385C, R61A, H62A, or W74F) were grown shaking overnight in M9 minimal medium supplemented with glucose (0.2%) and inducer (IPTG, 10 μM) at 30°C. Congregation was quantified by measuring the optical density (at 600 nm) of the culture supernatant before and after a brief vortex. (C) Wild-type and Δzur were grown overnight in 5 mL M9 minimal medium plus glucose (0.5%) and a terminal electron acceptor (fumarate, 50 mM). Cultures were grown aerobically (+ O2) or anoxically (- O2) shaking overnight at 30°C. Congregation was quantified as described above and representative images of the bottom of each culture tube are shown. (D) Wild-type and a strain carrying an inducible chromosomal copy of aerA (+ aerA) were grown overnight in LB medium and washed thrice in M9 minimal medium lacking a carbon source. A sterile toothpick was used to inoculate cells into M9 soft agar (0.3%) containing inducer (IPTG, 10 μM) and either succinate (30 mM) or maltose (0.1 mM) as a carbon source. The diameter of diffusion (mm) was measured following a 48-hr incubation at 30°C and representative diffusion patterns are shown for each strain. (E-F) Motility of Δzur, Δzur ΔaerB, and Δzur ΔaerA were measured as described above in (E) succinate and (F) maltose plates. Representative swarm diameters are shown. For all bar graphs, raw data points represent biological replicates, error bars represent standard deviation, and asterisks denote statistical difference via Ordinary one-way ANOVA test (****, p < 0.0001; *, p < 0.05; n.s., not significant).
Intriguingly, the chemical ligands for AerB and the vast majority of V. cholerae’s 46 encoded MCPs are yet to be determined [81]. The AerB N-terminus harbors a PAS domain [82], a protein family that typically senses light, oxygen, redox stress, or electron acceptors [83]. We hypothesized that this PAS-containing chemoreceptor mediates energy taxis, perhaps along the oxygen gradient in our vertical culture tubes. We first tested whether oxygen was required for Δzur to congregate. Wild-type and Δzur were cultured in both aerobic and anoxic tubes in M9 minimal medium with the terminal electron acceptor fumarate to enable anaerobic glucose respiration. In contrast with the aerobic cultures, Δzur did not congregate under anoxic conditions (Fig 5C). A similar result was observed under glucose-fermenting conditions (i.e., when fumarate was omitted from the medium) (S8 Fig). These data indicate that the chemotaxis-driven congregation in Δzur is oxygen-dependent.
AerB shares 31% amino acid identity with V. cholerae’s primary aerotaxis receptor Aer-2 (renamed here to AerA, as numbers in bacterial gene names can be confused with mutant alleles) (S7A Fig), which exhibits a positive response to oxygen [84]. Other homologs include E. coli’s AerEC (B0372, 31% identity), which positively responds to oxygen via sensing the electron acceptor FAD [83,85,86]. Alignment of AerB with additional orthologs from Azospirillum brasilense [87] and Shewanella oneidensis [88] revealed conservation of a critical FAD-binding tryptophan residue, among others (S7B Fig). The corresponding amino acids were mutated in aerB and each mutant was expressed in a Δvsp-II background to determine whether they still promoted congregation. Strains expressing the W74F mutant failed to congregate, suggesting that this highly conserved FAD-binding residue is essential for function (Fig 5B). Two additional AerB mutations (R61A and H62A), corresponding to E. coli FAD-binding residues, were also unable to congregate. The requirement for oxygen and these putative FAD-binding residues suggests that AerB may bind FAD or a similar ligand to facilitate energy taxis.
Since aerB-expressing cells congregated at the bottom of the culture tube, we hypothesized that AerB mediates a negative response to oxygen, in contrast to the positive response mediated by AerA and AerEC. To further interrogate energy taxis, we examined the swarming dynamics of V. cholerae in an established aerotaxis assay, which uses soft agar with carbon sources that vary in their ability to accentuate aerotaxis behavior [84,85]. Succinate, for example, can only be catabolized via respiration, which consumes oxygen, thereby generating an O2 gradient that increases with distance from the inoculation site. Since no other classical attractants/repellents are present, motility on succinate plates reveals aerotaxis as the primary taxis behavior [85]. In contrast, maltose agar provides other cues for chemotaxis (including chemotaxis towards maltose itself), obscuring an aerotactic response. The diameter of diffusion in succinate and maltose (at 30°C) was measured two days post-inoculation. Induction of aerB in a wild-type background significantly reduced the swarming diameter in succinate (Fig 5D). In contrast, aerB induction did not affect the swarming diameter in maltose plates. These data suggest that AerB promotes a negative chemotactic response to (or in the presence of) oxygen gradients.
Additional assays were performed in a Δzur background to ensure robust expression of aerB from the native promoter. aerA mutants were included as a control. Compared to Δzur, the diameter of Δzur ΔaerB migration on succinate was subtly, but significantly, increased (Figs 5E and S9A and S9B). This is consistent with AerB promoting a negative response to oxygen. Conversely, Δzur ΔaerA showed a significant decrease in swarming ability, consistent with previous reports that showed AerA promoting a positive response to oxygen [84]. There were no significant differences between the swarm diameter of Δzur and the aer mutants on maltose plates (Figs 5F and S9C and S9D). These assays were additionally performed in a wild-type background and aerB showed no effect on swarming behavior (S9 Fig), consistent with lack of aerB transcriptional expression in wild-type background. These results are consistent with AerB functioning as an energy taxis receptor that is either dependent on, or responsive, to oxygen.
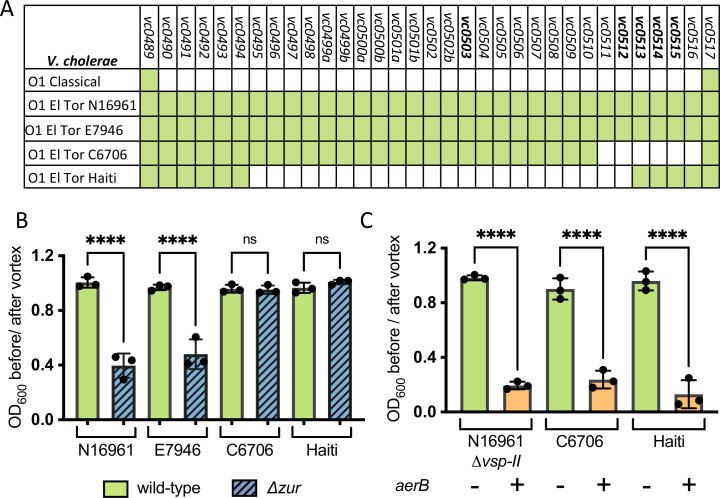
Ectopic AerB induces congregation in El Tor strains with atypical VSP-II islands
Although the VSP-II island is strongly correlated with the 7th pandemic strain, variants of this island have been detected in other El Tor isolates (Fig 6A). The Zur-regulated VSP-II genes characterized in this study appear to be in a hotspot for island variation: C6706 (Peru, 1991) lacks vc0511-vc0515 while the Haiti strain (2010) lacks vc0495-vc0512. We predicted that only El Tor Δzur strains with prototypical islands (harboring aerB/vc0512 and verA/vc0513) will congregate in minimal medium. As expected, the prototypical VSP-II strains (N16961 and E7946) lacking zur congregated in overnight culture (Fig 6B). This suggests that Δzur congregation is likely not due to strain-specific variations outside of VSP-II. In contrast, neither the C6706 nor the Haiti mutant congregated in minimal medium, presumably due to the absence of aerB. To test whether we could promote congregation in these VSP-II variants, we expressed a chromosomally integrated, inducible copy of aerB in C6706 and Haiti. Similar to the N16961 Δvsp-II control, C6706 and Haiti expressing aerB congregated in minimal medium (Fig 6C). These data suggest that AerB’s interaction partners are conserved in other V. cholerae El Tor isolates.
Fig 6. Ectopic AerB expression causes atypical El Tor strains to congregate.
(A) Presence (green) or absence (white) of VSP-II open reading frames in V. cholerae strains with prototypical (N16961, E7946) and variant (Haiti, C6706) VSP-II islands are shown. (B) Wild-type or Δzur varieties of V. cholerae (N16961, E7946, C6706, Haiti) were grown overnight (~15 hr) shaking at 30°C in M9 minimal medium with glucose (0.2%). Congregation was quantified by measuring the optical density (at 600 nm) of the culture supernatant before and after a brief vortex. (C) V. cholerae strains with (+) or without (-) an IPTG-inducible, chromosomal copy of aerB were grown as described in B but with the addition of IPTG (10 uM). For all bar graphs, raw data points represent biological replicates, error bars represent standard deviation, and asterisks denote statistical difference via Ordinary one-way ANOVA (****, p < 0.0001; n.s., not significant).
A model for oxygen-dependent V. cholerae congregation in zinc starved environments
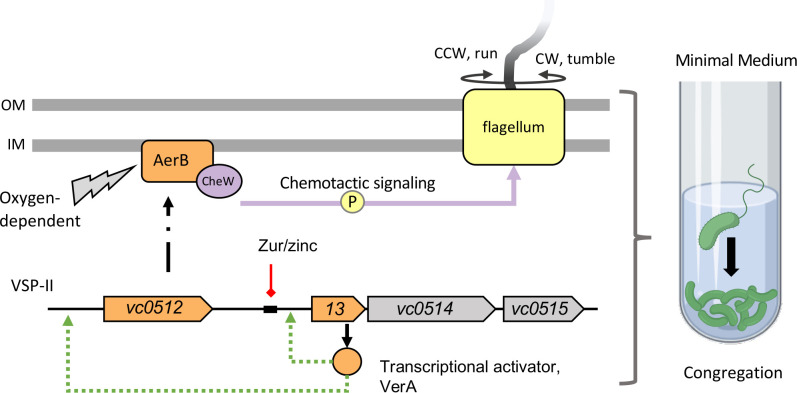
In summary, we propose the following model for Δzur congregation in M9 minimal medium (Fig 7). In zinc rich conditions, Zur acts as a repressor of the VerA-encoding vc0513-vc0515 operon on VSP-II. In the absence of Zur or in zinc starvation, the VerA transcriptional activator induces expression of its own operon and the nearby aerB. AerB serves as a receptor for oxygen-dependent energy taxis and relays changes in signal concentration to the core chemotaxis machinery and the flagellum. This results in cells congregating at the bottom of the culture tube in an oxygen-dependent manner.
Fig 7. Proposed model of Zur-regulation of VSP-II encoded genes and its effect on chemotaxis.
Zur forms a complex with divalent zinc ions and binds with high affinity to specific DNA sequences (black rectangle), repressing transcription of downstream genes (red line). In the absence of zur or during zinc starvation, VSP-II genes, including the vc0513-vc0515 operon are derepressed. The vc0513-encoded transcriptional activator VerA induces transcription (green arrow) of aerB, which encodes a chemotaxis receptor. AerB interacts with the chemotaxis coupling protein CheW (purple) and mediates a signal relay that alters flagellar (yellow) rotation and cell reorientation. AerB induction in minimal medium causes V. cholerae to congregate in an oxygen-dependent manner away from the air-liquid interface. Model created with BioRender.com.
Discussion
The mysterious Vibrio Seventh Pandemic Island (VSP-II) present in the El Tor biotype has largely evaded characterization due to lack of knowledge of stimuli that favor its induction. We report that Zur, the transcriptional repressor of the zinc starvation response, is a direct and indirect regulator of other VSP-II genes. Novel Zur targets reported here include the vc0513-vc0515 operon, which encodes the VerA transcriptional activator that increases expression of VSP-II chemotaxis and motility-related genes. One of these secondary targets, AerB, encodes a chemoreceptor involved in energy taxis.
The role of zinc availability in VSP-II induction
It has long been suspected that the VSP islands function as either pathogenicity or environmental persistence islands. Recent work interestingly suggests that VSP-I may function as a phage defense system [89,90]. In contrast, we and others have not yet identified a set of growth conditions under which VSP-II confers a fitness benefit [23,25,91,92] (S10 Fig). Given the robust expression of VSP-II loci in the absence of Zur, we propose two contexts where V. cholerae may encounter zinc starvation and express these island-encoded genes: within the human host, and/or on (chitinous) biotic surfaces in aquatic reservoirs.
The human host is a well-studied example of a metal-limited environment. Vertebrate hosts sequester desirable metal cofactors (e.g. zinc) in order to restrict the growth of potentially harmful bacteria (i.e. nutritional immunity, [29–32]). Pathogens lacking zinc acquisition systems often exhibit colonization defects in vivo [34,93–100], potentially because they are unable to compete against the microbiota for precious metal cofactors [101]. Induction of zinc starvation genes in pathogenic V. cholerae appears to be dependent upon the in vivo model used; for example, the primary zinc importer and the vc0513-vc0515 operon are upregulated in a mouse but not in a rabbit model (relative to LB) [28]. In terms of fitness, in vivo infection assays showed that loss of V. cholerae’s zinc importers led to modest colonization defects in both mouse [34] and rabbit infection models [102]; however, the latter Tn-Seq analysis performed in a rabbit model did not detect any significant fitness defects in VSP-II mutants [102]. In summary, although VSP-II genes may be expressed in certain animal models, it remains unclear if the island confers any advantage during host infection.
More generally, V. cholerae experiences metal starvation within thick bacterial communities and thus metal transporters and regulators contribute optimal V. cholerae biofilm formation [103]. Zur-regulated genes (including vc0503 and vc0513-vc0515) are reportedly induced by exposure to chitin oligomers [33], raising the possibility that V. cholerae is zinc-limited while colonizing copepods or crustaceans in the environment. It is thus plausible that the Zur-regulated VSP-II genes may be involved in V. cholerae’s chemotactic movements within the aquatic reservoir.
VSP-II-encoded genes facilitate chemotactic responses
Connections between zinc homeostasis and altered motility patterns have been reported in other bacteria, but these phenotypes appear to be indirect consequences of zinc availability rather than Zur repression of secondary transcriptional regulators [97,104–106]. The VerA-regulated chemoreceptor, AerB, generates congregation in liquid culture and appears to mediate energy taxis. This is in apparent contradiction with a report that did not find a role for AerB (referred to as Aer-1) in aerotaxis [84]; however, this may be explained by a lack of native aerB expression under their experimental conditions. aerA expression, on the other hand, does not appear to be regulated by Zur and native levels are sufficient to alter motility of the wild-type on soft agar. The balanced action of aerotactic responses conferred by AerA and AerB may function analogously to the Aer and Tsr receptors [107], which enable E. coli to navigate to an optimum oxygen concentration. However, we do not exclude the possibility that the AerB chemotactic response is more complex than the model proposed here.
Although the role of chemotaxis in autoaggregation has not been previously reported in V. cholerae, this phenomenon has been characterized in several distantly related bacteria [108]. A. brasilense, for example, aggregates in response to oxygen/redox stress via a PAS-containing chemoreceptor homologous to AerB (33% amino acid identity, S7A Fig) [87,109,110]. As a second example, Shewanella oneidensis “congregates” around insoluble electron acceptors [111] via an MCP with a PAS domain (SO_1385, 39% amino acid identity to AerB), an MCP with a Ca2+-sensing Cache domain (SO_2240, 39% amino acid identity to VC0514), and a protein involved in extracellular electron transport (CymA, SO_4591) [88]. This study, along with a correlogy analysis of VSP genes [90], suggests that the AerB/VC0512 and VC0514 MCPs may be functionally linked. Future work will investigate the relationship between these VerA-regulated chemotaxis receptors.
The energy taxis system described here may enable V. cholerae to avoid redox stress in low-zinc environments, since deletion of zinc importer systems is associated with heightened redox susceptibility in E. coli [98]. We speculate that AerB may allow V. cholerae to colonize other niches within the host (e.g., anaerobic parts of the gut), similar to a redox-repellent chemotaxis system in Helicobacter pylori that enables gland colonization in vivo [112,113]. Alternatively, this chemotaxis system may allow V. cholerae to exploit different niches within the aquatic reservoir (e.g., anoxic sediments with chitin detritus); however, each of these biologically relevant conditions are difficult to recapitulate in vitro.
Chemotaxis enhances virulence in a number of enteric pathogens, but this does not seem to generally hold true for V. cholerae [114]. In an infection model, non-chemotactic (counter-clockwise biased) mutants outcompeted wild-type V. cholerae and aberrantly colonized parts of the upper small intestine [115], suggesting that chemotaxis is dispensable and possibly deleterious for host pathogenesis. V. cholerae appears to broadly downregulate chemotaxis genes in a mouse infection model [28] and in stool shed from human patients [116]. Intriguingly, this decrease may be mediated in part by VSP-I; the island-encoded DncV synthesizes a cyclic AMP-GMP signaling molecule that decreases expression of chemotaxis genes and enhances virulence [24]. Specific chemoreceptors, however, are upregulated within a host and/or enhance virulence (see [117] for a review). Given the conflicting roles for chemotaxis within a host and the lack of evidence for VSP-II’s role during infection, we alternatively suggest that VSP-II encoded chemotaxis genes may serve a purpose in an aquatic environment with oxygen and nutrient gradients.
Stress and starvation responses can be co-opted by acquired genetic elements
We report that targets of the Zur-regulated VerA appear to be restricted to VSP-II, at least under the conditions tested. This restriction is logical given that transcriptional activators require specific DNA-binding sequences, and these may not be present in the native chromosome of a horizontal transfer recipient. Zur control of secondary regulators, including those that impact gene expression more broadly via signaling molecules (i.e., cyclic di-GMP phosphodiesterases like VC0515) may function to expand the complexity and tunability of the Zur-regulon in response to zinc availability and compounding environmental signals.
Horizontal acquisition of genomic islands can help bacteria (and pathogens) evolve in specific niches. VSP-II retains the ability to excise to a circular intermediate in N16961, indicating the potential for future horizontal transfer events [26]. We observed that 33% of the ORFs on the prototypical VSP-II island are expressed in the absence of Zur. Intriguingly, genomic island “desilencing” in response to zinc starvation has been reported in diverse bacterium, including Mycobacterium avium ssp. Paratuberculosis [67] and Cupriavidus metallidurans [55]. We note in this study that other El Tor isolates lack some Zur-regulated components of VSP-II. Given that the emergence of VSP-II containing 7th pandemic strains is recent on an evolutionary timescale, the contents of these islands may still be undergoing selection.
In summary, investigation of our Zur-associated congregation phenotype enabled identification of novel components of the zinc starvation response present on the El Tor Vibrio Seventh Pandemic Island -II (VSP-II). Further characterization of these island-encoded genes may aid in establishing VSP-II’s role as either a pathogenicity or environmental persistence island.
Methods
Bacterial growth conditions
Bacterial strains were grown by shaking (200 rpm) in 5 mL of LB medium at 30°C (for V. cholerae and E. coli BTH101) or 37°C (for other E. coli) in borosilicate glass tubes, unless otherwise specified. M9 minimal medium with glucose (0.2%) was prepared with ultrapure Mili-Q water to minimize metal contamination. Antibiotics, where appropriate, were used at the following concentrations: streptomycin, 200 μg ml−1; ampicillin, 100 μg ml−1, and kanamycin, 50 μg ml−1. IPTG was added to induce Piptg promoters at indicated concentrations.
Plasmid and strain construction
For all cloning procedures, N16961 gDNA was amplified via Q5 DNA polymerase (NEB) with the oligos summarized in S3 Table. Fragments were Gibson assembled [118] into restriction-digested plasmids. For gene deletions, 700 bp flanking regions were assembled into XbaI-digested pCVD442 (AmpR). For complementation experiments, genes of interest were amplified with a strong ribosome binding site and assembled into SmaI-digested pHLmob (kanR) or pTD101 (AmpR) downstream of an IPTG-inducible promoter. lacZ transcriptional reporters were built by amplifying the desired promoter region and assembling into NheI-digested pAM325 (KanR). The resulting promoter-lacZ fusions were amplified for assembly into StuI-digested pJL1 (AmpR). Cloning for bacterial two-hybrid assays are described in a separate section below. All assemblies were initially transformed into DH5alpha λpir and subsequently into an E. coli donor strain (MFD λpir or SM10 λpir).
For conjugations into V. cholerae, stationary phase recipients and MFD λpir donor strains were washed of antibiotics, mixed in equal ratios, and spotted onto an LB DAP plate. After a 4 hr incubation at 37°C, cells were streaked onto an LB plate with ampicillin or kanamycin to select for transconjugants. Conjugations using SM10 λpir donors were performed in the absence of DAP and with the addition of streptomycin to selective plates. pHLmob transconjugants were purified on an additional kanamycin LB agar plate. Integration vectors (pCVD442, pTD101, and pJL1) were cured through two rounds of purification on salt-free sucrose (10%) agar. Gene deletions or STOP codon replacements introduced by pCVD442 were verified by PCR using the oligos indicated in S3 Table. Successful integration of lacZ targeting vectors (pTD101, pJL1) were identified by blue-white screening on plates containing 5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal, 40 μg ml−1). pJL-1 vectors were additionally checked via PCR. All strains used in this study are summarized in S3 Table. V. cholerae strains were derived from N16961, unless otherwise indicated as E7946 [119], C6706 (not strepR) [14], or Haiti [19]. The N16961 accession numbers for genes referenced in this study are as follows: zur/vc0378, znuABC/vc2081-vc2083, zrgABC/vc2551-2553, ribA/vc1263, rpmE2/vc0878, rpmJ2/vc0879, shyB/vc0503, aerB/vc0512, verA/vc0513, fliC/vc2199, motB/vc0893, cheA-2/vc2063, cheW-1/vc2059, cheZ/vc2064, cheY/vc2065, vpsL/vc0934, csqS/vca0522, csqA/vc0523, tdh/vca0885, luxS/vc0557, luxQ/vca0736, and aerA/vca0658.
Site-directed mutagenesis
Site-directed mutagenesis was performed using NEB kit #E0554S according to manufacturer instructions. A pTD101 plasmid carrying aerB was used as the template for Q5 amplification with the following mutagenic primer pairs: R61A, SM-1294/1295; and H62A, SM-1296/1297 (S3 Table). Products were purified and treated with kinase, ligase, and Dpn1 at 37°C for 30 minutes. This reaction mixture was transformed into DH5α ƛpir. Mutations were confirmed via Sanger Sequencing. aerB fragments containing W74F (SM-1306) and G395C (SM-1307) were chemically synthesized by Integrated DNA Technologies (IDT) and assembled into pTD101; sequences are listed in S3 Table.
Congregation assays
Bacterial congregation was quantified by measuring absorbance (OD600) in a spectrophotometer of the culture supernatant before and after a brief vortex (5 seconds). Congregation score represents the ratio of before and after pellet disruption; a ratio closer to one indicates that the culture is homogenous, a ratio closer to zero indicates that the cells are concentrated at the bottom of the culture tube.
Transposon mutagenesis screen & Arbitrary PCR
V. cholerae N16961 Δzur was mutagenized with Himar1 mariner transposons via an SM10 λpir donor strain carrying pSC189 [120]. Four independent Δzur transposon libraries were generated, as previously described [27]. Each library was separately harvested from the plate using sterile rubber scrapers, vortexed into LB, and preserved in glycerol at -80°C. Individual culture tubes containing 5 mL of M9 minimal medium with glucose (0.2%) and kanamycin were inoculated with transposon libraries. Overnight cultures were back-diluted 1000-fold into fresh medium and incubated overnight; this process was repeated until no visible pellet had formed. Isolated colonies were tested to verify that they did not generate a pellet. The second screen was performed identically to the first, except that cultures were first inoculated into a M9 motility plate (0.3% agar) and allowed to migrate for 48 hours at 30°C degrees. Scrapings from the outer zone (collected with a 1 mL pipette tip) were inoculated a culture tube containing M9 minimal medium. For both screens, the transposon insertion site for each isogenic colony was identified by arbitrary PCR [46]. As described elsewhere, this technique amplifies the chromosomal DNA adjacent to the mariner transposon. Amplicons were Sanger sequenced at the Cornell Institute of Biotechnology, and regions of high-quality were aligned to the N16961 reference genome using BLAST [121].
RNA-seq and analysis
Overnight cultures of wild-type N16961 and the Δzur mutant were diluted 1:100 into LB and grown shaking at 37°C until cells reached mid-log phase (optical density at 600 nm [OD600], 0.5). RNA was extracted using mirVana miRNA Isolation Kit (Invitrogen, AM1560). Genomic DNA contamination was removed through two DNAfree (Ambion) treatments each followed by glass fiber column purification. Library preparations, Illumina sequencing, and data analysis were performed by GENEWIZ (South Plainfield, NJ). Differentially expressed genes were those with log 2-fold change >1 and an adjusted p-value <0.05. Raw and analyzed data files were deposited in the NCBI GEO database (GSE173966).
For VC0513 overexpression, wild-type N16961 carrying either pHLmob or pHLmob(Piptg-vc0513) was sub-cultured into LB kanamycin IPTG (500 μM) and grown at 37°C for 3 hour (~ mid-log phase). Total RNA isolations and DNase treatments were performed as described above. Library preparations, Illumina sequencing, and data analysis (using DESeq2 [122]) were performed by the Cornell Transcriptional Expression Facility. Differentially expressed genes were those with log 2-fold change >1 and an adjusted p-value <0.05. Raw and analyzed data files were deposited in the NCBI GEO database (GSE174028).
5’-Rapid Amplification of cDNA Ends (5’-RACE)
Transcription start sites were identified with 5′-RACE. To obtain vc0512, vc0513, and vca1098 transcripts, the Δzur mutant was grown in LB at 37°C until cells reached mid-log phase (optical density at 600 nm [OD600], 0.5). RNA extractions and DNAse treatments were performed as described for RNAseq. PCR was performed to check for genomic DNA contamination; no amplicons were detected within 34 cycles. Reverse transcription was performed with the Template Switching Reverse Transcriptase enzyme mix (NEB #M0466) according to manufacturer protocols using gene specific primers (vc0512, SM-1133; vc0513, SM-1131; vca1098, SM-1129) and the Template Switching Oligo (TSO). PCR Amplification of 5’-transcripts was performed with diluted cDNA, Q5 Hot Start High-Fidelity Master Mix (NEB #M0494), TSO-specific primer, and gene-specific primers (vc0512, SM-1134; vc0513, SM-1132, vca1098, SM-1130). Products were sanger sequenced using the following primers: SM-1134, SM-1156, and SM1157 for vc0512, SM-1132 for vc0513, and SM-1130 for vca1098. Primer sequences are listed in S3 Table.
β-galactosidase activity measurements
V. cholerae strains carrying promoter-lacZ fusions were grown overnight in LB at 30°C, with kanamycin for plasmid (pHLmob) maintenance. Strains were diluted 1:100 into LB containing kanamycin and IPTG (1 mM) and grown shaking at 37°C. Exponential phase cells were harvested (~ 3hr) and β-galactosidase activity against an ortho-Nitrophenyl-β-galactoside substrate (ONPG) substrate was quantified as described elsewhere [123,124].
Motility assays
Motility plates (0.3% agar) were prepared with M9 minimal medium with variable carbon sources (succinate, 30 mM; and maltose, 0.1 mM; glucose, 0.2%). Strains were grown overnight in LB medium and washed three times in M9 without a carbon source. Plates were inoculated via toothpick stabs and incubated 30°C for 48-hr. The migration diameter (mm) was recorded.
Bacterial two hybrid assays
Protein-protein interactions were detected using the BACTH bacterial two hybrid system [125]. cheW and aerB (excluding transmembrane domains and native start/stop codons) were cloned into SmaI-digested pUT18(C) (KanR) or pK(N)T25 (AmpR) expression vectors to yield N-terminal T(18/25)-Aer or C-terminal CheW-T(18/25) fusions. Electrocompetent E. coli BTH101 were co-transformed with a pUT18 and pKT25 vector that carried either: an unfused adenylate cyclase domain (T18 or T25), the CheW-T(18/25) fusion or the T(18/25)-AerB fusion. Following 1 hour of outgrowth in SOC at 30°C, 10 μL of concentrated outgrowth was spotted onto LB agar containing kanamycin and ampicillin (for selection), X-gal (for blue-white detection), and inducer (IPTG, 500 μM). Plates were incubated overnight at 30°C and for an additional day at room temperature before being imaged.
Anaerobic cultures
5 mL of M9 minimal medium without MgSO4, CaCl2, or carbon source were added to glass culture tubes. Tubes were sealed with rubber stoppers, crimped, purged for 10 cycles (20 sec vacuum, 20 sec N2 purge), and autoclaved (gravity, 20 min). Post-autoclaving, the medium was amended with sterile solutions of MgSO4 (to 2 mM), CaCl2 (to 0.1 mM), glucose (to 0.5%) and with or without fumarate (to 50mM) using sterile syringes and needles. Tubes were injected with a V. cholerae cell suspension (1:100) and grown overnight shaking at 30°C. Aerobic tubes containing M9 glucose (0.5%) with or without fumarate were included as a control. Congregation was measured via spectrophotometry, as described above.
Supporting information
(A-H) All strains were grown overnight in M9 minimal medium plus glucose (0.2%). All cultures were grown shaking (200 rpm), with the exception of static growth tested in panel (A). All cultures were grown in borosilicate glass tubes, with the exception of plastic tubes used in panel (B). Congregation was quantified by measuring the optical density (at 600 nm) of the culture supernatant before and after a brief vortex. The following mutants were tested in a Δzur background: (C) other putative Zur-regulatory targets (ABC-type transporter, Δvca1098-vca1101; che-III cluster, Δvca1090-vca1097), (D) chemotaxis genes (cheA::STOP, ΔcheY, ΔcheZ), (F) biofilm formation genes (ΔvspL), type IV pili (ΔtcpA, ΔmshA, ΔpilA, and Δvc0502), quorum sensing genes (ΔcsqA, ΔcsqS, Δtdh, ΔluxS, or ΔluxQ), (G) N16961 hapRrepaired, (H) the Vibrio Seventh Pandemic (VSP) island -I (Δvc0175-vc0185), and regions of VSP-II (“ΔVSP-II”, Δvc0491-vc0515; Δvc0490-vc0510, Δvc0511, Δvc0512, Δvc0513 or vc0513::STOP, Δvc0514 or vc0514::STOP, Δvc0515 or vc0515::STOP, or Δvc0516). (E) Congregation was also measured in a rough mutant (vc0225::STOP) and a rough mutant harboring deletions for ΔfliC, ΔmotB, or Δvsp-II. Data points represent biological replicates, error bars represent standard deviation, and asterisks denote statistical difference relative to the wild-type strain via (A,B,G) unpaired t-test or (C-F, H) Ordinary one-way ANOVA (****, p < 0.0001; ***, p < 0.001; *, p < 0.05).
(TIFF)
(A) Table indicating the number of transposon insertions within motility, chemotaxis, and VSP-II genes for each of the screens (without pre-selection, v.1; with pre-selection of motile mutants, v.2) described in Fig 2. (B) Approximate location of transposon insertions (triangles) determined by arbitrary PCR [46] and Sanger sequencing are shown. (C) Strains carrying either an empty vector (-) or complementation vector (+) were grown overnight in LB medium with kanamycin. Strains were washed thrice with M9 minimal medium. A sterile toothpick was used to inoculate cells into M9 soft agar (0.3%) containing glucose (0.2%), kanamycin, and inducer (IPTG, 500 μM). The diameter of diffusion (mm) was measured following a 48-hr incubation at 30°C. Raw data points represent biological replicates, error bars represent standard deviation, and asterisks denote statistical difference via Ordinary one-way ANOVA test (****, p < 0.0001; **, p < 0.01).
(TIFF)
Diagram of the verA promoter region annotated with theh following features: predicted Zur box (red), predicted -10 and -35 regions (purple) [126], suggested start codon (ATG, green).
(TIFF)
(A-B) Volcano plots showing log 2-fold changes in gene expression in Δzur relative to wild-type; positive values represent up-regulation in Δzur and negative values represent down-regulation in Δzur. The y-axis denotes the negative log inverse of the p-value. Differentially expressed genes (log 2-fold change >1, adjusted p-value < 0.05) are denoted in red and are labeled with gene identifiers. Panel (B) shows the subset of genes within the blue box in Panel (A).
(TIFF)
(A) Diagram of the vca1098 promoter region annotated with the following features: predicted Zur box, red; predicted -10 region, purple [126]; transcription start site, +1 (5’-RACE); predicted ribosome binding site (RBS), yellow; proposed start codon (ATG), green. Asterisks indicate Zur box nucleotides (region “a” or “b”) that were altered in the mutant reporters described below. (B) vca1098 promoter lacZ transcriptional reporters (Pvca1098-lacZ, solid bars) or mutated versions (Pvca1098Zur box* a or b-lacZ, striped bars) were inserted into a wild-type or Δzur background harboring a plasmid-borne, IPTG-inducible copy of zur (+) or empty vector control (-). Strains were grown overnight in LB and kanamycin, diluted 1:100 in fresh media containing inducer (IPTG, 400 μM), and grown for 3 hours at 37°C. Promoter activity (in Miller Units) was measured via β-galactosidase assays (See Methods and Materials). (C) Wild-type and Δzur strains carrying Pvca1098-lacZ or mutant derivatives were streaked onto M9 minimal medium agar with glucose (0.2%), X-gal, and with or without added zinc (ZnSO4, 10 μM). Plates were incubated overnight at 30°C and then for an additional day at room temperature. vca1098 promoter activity is signified by a blue colony color.
(TIFF)
(A) Schematics for two attempted PaerB-lacZ reporters containing either 400 bp or 1,314 bp of the promoter region are shown. (B) The PaerB400 bp-lacZ and PaerB 1,314 bp-lacZ reporters were integrated into a wild-type or Δzur background and were struck onto LB X-gal plates. Plates were incubated overnight at 30°C and then for an additional day at room temperature. PaerB expression is indicated by a blue colony color.
(TIFF)
(A) Results of protein BLAST [121] and (B) Clustal Omega alignment [127] of AerB (VC0512) with homologs from V. cholerae (AerA/VCA0658), E. coli (Aer/B3072), A. brasiliensis (AerC/AKM58_23950), and S. oneidensis (SO_1385). Conserved ligand binding and MCP residues targeted for mutation are indicated by orange and pink arrows, respectively.
(TIFF)
Wild-type and Δzur were grown overnight in 5 mL M9 minimal medium plus glucose (0.5%) fermentatively (without a terminal electron acceptor) and cultured under aerobic (+ O2) or anoxic (- O2) conditions (see Methods for details). Tubes were grown shaking overnight at 30°C and congregation was quantified via spectrophotometry as described previously. All data points represent biological replicates, error bars represent standard deviation, and asterisks denote statistical difference via Ordinary one-way ANOVA test (****, p < 0.0001; n.s., not significant).
(TIFF)
(A-D) The indicated strains were grown overnight in LB medium and washed thrice in M9 minimal medium lacking a carbon source. A sterile toothpick was used to inoculate cells into M9 soft agar (0.3%) with either (A-B) succinate (30 mM) or (C-D) maltose (0.1 mM) as a carbon source. The diameter of diffusion (mm) was measured following a 48-h incubation at 30°C and representative swarms are shown (A,C). Note: Data for Δzur, Δzur ΔaerB, and Δzur ΔaerA are the same as shown in Fig 5 and are shown here for comparison with a wild-type background. All data points represent biological replicates, error bars represent standard deviation, and asterisks denote statistical difference via Ordinary one-way ANOVA test (****, p < 0.0001; ***, p < 0.001, and n.s., not significant).
(TIFF)
Wild-type, Δvsp-I, and Δvsp-II were grown overnight in M9 minimal medium with glucose (0.2%) at 30°C. Cultures were washed twice and diluted 1:100 into (A) fresh M9 minimal medium glucose (0.2%), (B) plus the zinc-specific chelator TPEN (250 nM), or (C) plus TPEN and exogenous zinc (ZnSO4, 1 μM). Growth at 30°C of each 200-μl culture in a 100-well plate was monitored by optical density at 600 nm (OD600) on a Bioscreen C plate reader (Growth Curves America).
(TIFF)
Δzur was grown overnight shaking (200 rpm) in M9 minimal medium with glucose (0.2%) at 30°C. The culture was manually agitated to disturb the pellet.
(MOV)
Transcript abundances in Δzur relative to wild-type was measured using RNA-seq (see Methods for details). Gene ID’s and putative ontology [82] are shown for all significant (adjusted p-value < 0.05) differentially expressed (log 2-fold change > 1) genes. Positive values represent up-regulation and negative values represent down-regulation in Δzur relative to the wild-type. Superscripts denote (a) a nearby canonical Zur box, (b) location on VSP-I or (c) location on VSP-II.
(XLSX)
Transcript abundances in a strain overexpressing VerA (VC0513) relative to an empty-vector control were measured using RNA-seq (see Methods for details). Gene ID’s and descriptions [82] are shown for all significant (adjusted p-value < 0.05) differentially expressed (log 2-fold change > 1) genes.
(XLSX)
Strains used in this study are listed with unique identifiers (SGM-#). For E. coli donor strains, the primers or gene block used to construct each plasmid are listed in the “Oligos” column. Genetic changes introduced into V. cholerae were screened using the method indicated in the “confirmation” column: either via PCR using the indicated primers (SM-#), via purification on kanamycin plates, or via blue-white screening on X-gal plates.
(XLSX)
Acknowledgments
We thank Sean Murphy and Dr. Daniel Buckley for providing assistance with anoxic culturing, and Dr. John Mekalanos and Dr. Melanie Blokesch for generously sharing V. cholerae strains. We thank members of the Dörr lab for helpful discussions.
Data Availability
Raw RNA-seq files are available from the GEO database (GSE173966, GSE174028). All other relevant data are shown as raw data points within the manuscript and its Supporting Information files.
Funding Statement
National Institute of Health grant R01GM130971 to TD. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Clemens JD, Nair GB, Ahmed T, Qadri F, Holmgren J. Cholera. The Lancet. Elsevier Ltd; 2017;390: 1539–1549. doi: 10.1016/S0140-6736(17)30559-7 [DOI] [PubMed] [Google Scholar]
- 2.Reidl J, Klose KE. Vibrio cholerae and cholera: out of the water and into the host. FEMS Microbiol Rev. 2002;26: 125–139. doi: 10.1111/j.1574-6976.2002.tb00605.x [DOI] [PubMed] [Google Scholar]
- 3.Huq A, Small EB, West PA, Huq MI, Rahman R, Colwell RR. Ecological relationships between Vibrio cholerae and planktonic crustacean copepods. Appl Environ Microbiol. American Society for Microbiology; 1983;45: 275–283. doi: 10.1128/aem.45.1.275-283.1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Magny GC, Mozumder PK, Grim CJ, Hasan NA, Naser MN, Alam M, et al. Role of Zooplankton Diversity in Vibrio cholerae Population Dynamics and in the Incidence of Cholera in the Bangladesh Sundarbans. Appl Environ Microbiol. American Society for Microbiology; 2011;77: 6125–6132. doi: 10.1128/AEM.01472-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tamplin ML, Gauzens AL, Huq A, Sack DA, Colwell RR. Attachment of Vibrio cholerae serogroup O1 to zooplankton and phytoplankton of Bangladesh waters. Appl Environ Microbiol. American Society for Microbiology; 1990;56: 1977–1980. doi: 10.1128/aem.56.6.1977-1980.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sochard MR, Wilson DF, Austin B, Colwell RR. Bacteria associated with the surface and gut of marine copepods. Appl Environ Microbiol. American Society for Microbiology; 1979;37: 750–759. doi: 10.1128/aem.37.4.750-759.1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Twedt RM, Madden JM, Hunt JM, Francis DW, Peeler JT, Duran AP, et al. Characterization of Vibrio cholerae isolated from oysters. Appl Environ Microbiol. American Society for Microbiology; 1981;41: 1475–1478. doi: 10.1128/aem.41.6.1475-1478.1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hood MA, Ness GE, Rodrick GE. Isolation of Vibrio cholerae serotype O1 from the eastern oyster, Crassostrea virginica. Appl Environ Microbiol. American Society for Microbiology; 1981;41: 559–560. doi: 10.1128/aem.41.2.559-560.1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Purdy AE, Watnick PI. Spatially selective colonization of the arthropod intestine through activation of Vibrio cholerae biofilm formation. Proc Natl Acad Sci USA. National Academy of Sciences; 2011;108: 19737–19742. doi: 10.1073/pnas.1111530108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Broza M, Halpern M. Chironomid egg masses and Vibrio cholerae. Nature. Nature Publishing Group; 2001;412: 40–40. doi: 10.1038/35083691 [DOI] [PubMed] [Google Scholar]
- 11.DePAOLA A. Vibrio cholerae in Marine Foods and Environmental Waters: A Literature Review. Journal of Food Science. John Wiley & Sons, Ltd; 1981;46: 66–70. doi: 10.1111/j.1365-2621.1981.tb14532.x [DOI] [Google Scholar]
- 12.Kaysner CA, Hill WE. Vibrio cholerae and Cholera: Molecular to Global Perspectives [Internet]. Wachsmuth IK, Blake PA, Olsvik Ø, editors. Washington, DC, USA: John Wiley & Sons, Ltd; 1994. pp. 27–39. doi: 10.1128/9781555818364.ch2 [DOI] [Google Scholar]
- 13.Chatterjee SN, Chaudhuri K. Lipopolysaccharides of Vibrio cholerae: I. Physical and chemical characterization. Biochimica et Biophysica Acta (BBA)—Molecular Basis of Disease. Elsevier; 2003;1639: 65–79. doi: 10.1016/j.bbadis.2003.08.004 [DOI] [PubMed] [Google Scholar]
- 14.Thelin KH, Taylor RK. Toxin-coregulated pilus, but not mannose-sensitive hemagglutinin, is required for colonization by Vibrio cholerae O1 El Tor biotype and O139 strains. Infection and Immunity. American Society for Microbiology Journals; 1996;64: 2853–2856. doi: 10.1128/iai.64.7.2853-2856.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krebs SJ, Taylor RK. Protection and Attachment of Vibrio cholerae Mediated by the Toxin-Coregulated Pilus in the Infant Mouse Model. Journal of Bacteriology. American Society for Microbiology Journals; 2011;193: 5260–5270. doi: 10.1128/JB.00378-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanchez J, Holmgren J. Cholera toxin structure, gene regulation and pathophysiological and immunological aspects. Cell Mol Life Sci. SP Birkhäuser Verlag Basel; 2008;65: 1347–1360. doi: 10.1007/s00018-008-7496-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rivera-Chávez F, Mekalanos JJ. Cholera toxin promotes pathogen acquisition of host-derived nutrients. Nature. Nature Publishing Group; 2019;572: 244–248. doi: 10.1038/s41586-019-1453-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu D, Liu B, Feng L, Ding P, Guo X, Wang M, et al. Origins of the current seventh cholera pandemic. Proc Natl Acad Sci USA. 2016;113: E7730–E7739. doi: 10.1073/pnas.1608732113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Son MS, Megli CJ, Kovacikova G, Qadri F, Taylor RK. Characterization of Vibrio cholerae O1 El Tor biotype variant clinical isolates from Bangladesh and Haiti, including a molecular genetic analysis of virulence genes. J Clin Microbiol. American Society for Microbiology Journals; 2011;49: 3739–3749. doi: 10.1128/JCM.01286-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henderson JC, Herrera CM, Trent MS. AlmG, responsible for polymyxin resistance in pandemic Vibrio cholerae, is a glycyltransferase distantly related to lipid A late acyltransferases. J Biol Chem. American Society for Biochemistry and Molecular Biology; 2017;292: 21205–21215. doi: 10.1074/jbc.RA117.000131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hankins JV, Madsen JA, Giles DK, Brodbelt JS, Trent MS. Amino acid addition to Vibrio cholerae LPS establishes a link between surface remodeling in Gram-positive and Gram-negative bacteria. Proc Natl Acad Sci USA. National Academy of Sciences; 2012;109: 8722–8727. doi: 10.1073/pnas.1201313109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dziejman M, Balon E, Boyd D, Fraser CM, Heidelberg JF, Mekalanos JJ. Comparative genomic analysis of Vibrio cholerae: Genes that correlate with cholera endemic and pandemic disease. Proc Natl Acad Sci USA. 2002;99: 1556–1561. doi: 10.1073/pnas.042667999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Shea YA, Finnan S, Reen JF, Morrissey JP, OGara F, Boyd EF. The Vibrio seventh pandemic island-II is a 26.9 kb genomic island present in Vibrio cholerae El Tor and O139 serogroup isolates that shows homology to a 43.4 kb genomic island in V. vulnificus. Microbiology. 2004;150: 4053–4063. doi: 10.1099/mic.0.27172-0 [DOI] [PubMed] [Google Scholar]
- 24.Davies BW, Bogard RW, Young TS, Mekalanos JJ. Coordinated Regulation of Accessory Genetic Elements Produces Cyclic Di-Nucleotides for V. cholerae Virulence. Cell. Elsevier; 2012;149: 358–370. doi: 10.1016/j.cell.2012.01.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taviani E, Grim CJ, Choi J, Chun J, Haley BJ, Hasan NA, et al. Discovery of novel Vibrio cholerae VSP-II genomic islands using comparative genomic analysis. FEMS Microbiology Letters. 2010;308: 130–137. doi: 10.1111/j.1574-6968.2010.02008.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murphy RA, Boyd EF. Three Pathogenicity Islands of Vibrio cholerae Can Excise from the Chromosome and Form Circular Intermediates. Journal of Bacteriology. 2008;190: 636–647. doi: 10.1128/JB.00562-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murphy SG, Alvarez L, Adams MC, Liu S, Chappie JS, Cava F, et al. Endopeptidase Regulation as a Novel Function of the Zur-Dependent Zinc Starvation Response. Salama NR, editor. mBio. 2019;10: e02620–18. doi: 10.1128/mBio.02620-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mandlik A, Livny J, Robins WP, Ritchie JM, Mekalanos JJ, Waldor MK. RNA-Seq-Based Monitoring of Infection-Linked Changes in Vibrio cholerae Gene Expression. Cell Host & Microbe. Cell Press; 2011;10: 165–174. doi: 10.1016/j.chom.2011.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palmer LD, Skaar EP. Transition Metals and Virulence in Bacteria. Annu Rev Genet. Annual Reviews; 2016;50: 67–91. doi: 10.1146/annurev-genet-120215-035146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kehl-Fie TE, Skaar EP. Nutritional immunity beyond iron: a role for manganese and zinc. Current Opinion in Chemical Biology. Elsevier Ltd; 2010;14: 218–224. doi: 10.1016/j.cbpa.2009.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hood MI, Skaar EP. Nutritional immunity: transition metals at the pathogen–host interface. Nat Rev Microbiol. Nature Publishing Group; 2012;10: 525–537. doi: 10.1038/nrmicro2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hennigar SR, McClung JP. Nutritional Immunity: Starving Pathogens of Trace Minerals. American Journal of Lifestyle Medicine. SAGE PublicationsSage CA: Los Angeles, CA; 2016;10: 170–173. doi: 10.1177/1559827616629117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meibom KL, Li XB, Nielsen AT, Wu C-Y, Roseman S, Schoolnik GK. The Vibrio cholerae chitin utilization program. Proc Natl Acad Sci USA. National Academy of Sciences; 2004;101: 2524–2529. doi: 10.1073/pnas.0308707101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheng Y, Fan F, Jensen O, Zhong Z, Kan B, Wang H, et al. Dual Zinc Transporter Systems in Vibrio cholerae Promote Competitive Advantages over Gut Microbiome. Infection and Immunity. 2015;83: 3902–3908. doi: 10.1128/IAI.00447-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Novichkov PS, Kazakov AE, Ravcheev DA, Leyn SA, Kovaleva GY, Sutormin RA, et al. RegPrecise 3.0 –A resource for genome-scale exploration of transcriptional regulation in bacteria. BMC Genomics. BioMed Central; 2013;14: 745. doi: 10.1186/1471-2164-14-745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Panina EM, Mironov AA, Gelfand MS. Comparative genomics of bacterial zinc regulons: Enhanced ion transport, pathogenesis, and rearrangement of ribosomal proteins. Proc Natl Acad Sci USA. National Academy of Sciences; 2011;100: 9912–9917. doi: 10.1073/pnas.1733691100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jemielita M, Wingreen NS, Bassler BL. Quorum sensing controls Vibrio cholerae multicellular aggregate formation. eLife. eLife Sciences Publications Limited; 2018;7: e1002210. doi: 10.7554/eLife.42057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taylor RK, Miller VL, Furlong DB, Mekalanos JJ. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc Natl Acad Sci USA. National Academy of Sciences; 1987;84: 2833–2837. doi: 10.1073/pnas.84.9.2833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun D, Lafferty MJ, Peek JA, Taylor RK. Domains within the Vibrio cholerae toxin coregulated pilin subunit that mediate bacterial colonization. Gene. Elsevier; 1997;192: 79–85. doi: 10.1016/s0378-1119(97)00007-3 [DOI] [PubMed] [Google Scholar]
- 40.Kirn TJ, Lafferty MJ, Sandoe CMP, Taylor RK. Delineation of pilin domains required for bacterial association into microcolonies and intestinal colonization by Vibrio cholerae. Molecular Microbiology. John Wiley & Sons, Ltd; 2000;35: 896–910. doi: 10.1046/j.1365-2958.2000.01764.x [DOI] [PubMed] [Google Scholar]
- 41.Adams DW, Stutzmann S, Stoudmann C, Blokesch M. DNA-uptake pili of Vibrio cholerae are required for chitin colonization and capable of kin recognition via sequence-specific self-interaction. 2019;4: 1545–1557. doi: 10.1038/s41564-019-0479-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trunk T, Khalil HS, Leo JC. Bacterial autoaggregation. AIMS Microbiology. AIMS Press; 2018;4: 140–164. doi: 10.3934/microbiol.2018.1.140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perez-Soto N, Creese O, Fernandez-Trillo F, Krachler A-M. Aggregation of Vibrio cholerae by Cationic Polymers Enhances Quorum Sensing but Overrides Biofilm Dissipation in Response to Autoinduction. ACS Chemical Biology. American Chemical Society; 2018;13: 3021–3029. doi: 10.1021/acschembio.8b00815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zamorano-Sánchez D, Xian W, Lee CK, Salinas M, Thongsomboon W, Cegelski L, et al. Functional Specialization in Vibrio cholerae Diguanylate Cyclases: Distinct Modes of Motility Suppression and c-di-GMP Production. mBio. American Society for Microbiology; 2019;10. doi: 10.1128/mBio.00670-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Struempler AW. Adsorption characteristics of silver, lead, cadmium, zinc, and nickel on borosilicate glass, polyethylene, and polypropylene container surfaces. Anal Chem. 2002;45: 2251–2254. doi: 10.1021/ac60335a014 [DOI] [Google Scholar]
- 46.O’Toole GA, Pratt LA, Watnick PI, Newman DK, Weaver VB, Kolter R. Genetic approaches to study of biofilms. Methods Enzymol. Academic Press; 1999;310: 91–109. doi: 10.1016/s0076-6879(99)10008-9 [DOI] [PubMed] [Google Scholar]
- 47.Nakao R, Ramstedt M, Wai SN, Uhlin BE. Enhanced Biofilm Formation by Escherichia coli LPS Mutants Defective in Hep Biosynthesis. Dobrindt U, editor. PLOS ONE. Public Library of Science; 2012;7: e51241. doi: 10.1371/journal.pone.0051241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heidelberg JF, Eisen JA, Nelson WC, Clayton RA, Gwinn ML, Dodson RJ, et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. Nature Publishing Group; 2000;406: 477–483. doi: 10.1038/35020000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Joelsson A, Liu Z, Zhu J. Genetic and Phenotypic Diversity of Quorum-Sensing Systems in Clinical and Environmental Isolates of Vibrio cholerae. Infection and Immunity. American Society for Microbiology Journals; 2006;74: 1141–1147. doi: 10.1128/IAI.74.2.1141-1147.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stutzmann S, Blokesch M, D’Orazio SEF. Circulation of a Quorum-Sensing-Impaired Variant of Vibrio cholerae Strain C6706 Masks Important Phenotypes. D’Orazio SEF, editor. mSphere. American Society for Microbiology Journals; 2016;1: e00098–16. doi: 10.1128/mSphere.00098-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hensley MP, Gunasekera TS, Easton JA, Sigdel TK, Sugarbaker SA, Klingbeil L, et al. Characterization of Zn(II)-responsive ribosomal proteins YkgM and L31 in E. coli. Journal of Inorganic Biochemistry. Elsevier; 2012;111: 164–172. doi: 10.1016/j.jinorgbio.2011.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sigdel TK, Easton JA, Crowder MW. Transcriptional response of Escherichia coli to TPEN. Journal of Bacteriology. 2006;188: 6709–6713. doi: 10.1128/JB.00680-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kallifidas D, Ben Pascoe, Owen GA, Strain-Damerell CM, Hong H-J, Paget MSB. The Zinc-Responsive Regulator Zur Controls Expression of the Coelibactin Gene Cluster in Streptomyces coelicolor. Journal of Bacteriology. American Society for Microbiology Journals; 2010;192: 608–611. doi: 10.1128/JB.01022-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gaballa A, Wang T, Ye RW, Helmann JD. Functional analysis of the Bacillus subtilis Zur regulon. Journal of Bacteriology. 2002;184: 6508–6514. doi: 10.1128/JB.184.23.6508-6514.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bütof L, Schmidt-Vogler C, Herzberg M, Große C, Nies DH, DiRita VJ. The Components of the Unique Zur Regulon of Cupriavidus metallidurans Mediate Cytoplasmic Zinc Handling. DiRita VJ, editor. Journal of Bacteriology. American Society for Microbiology Journals; 2017;199: e00372–17. doi: 10.1128/JB.00372-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pawlik M-C, Hubert K, Joseph B, Claus H, Schoen C, Vogel U. The zinc-responsive regulon of Neisseria meningitidis comprises seventeen genes under control of a Zur element. Journal of Bacteriology. American Society for Microbiology; 2012;194: 6594–6603. doi: 10.1128/JB.01091-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Neupane DP, Jacquez B, Sundararajan A, Ramaraj T, Schilkey FD, Yukl ET. Zinc-Dependent Transcriptional Regulation in Paracoccus denitrificans. Front Microbiol. Frontiers; 2017;8: 123. doi: 10.3389/fmicb.2017.00569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mazzon RR, Braz VS, da Silva Neto JF, do Valle Marques M. Analysis of the Caulobacter crescentus Zur regulon reveals novel insights in zinc acquisition by TonB-dependent outer membrane proteins. BMC Genomics. BioMed Central; 2014;15: 1–14. doi: 10.1186/1471-2164-15-734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moreau GB, Qin A, Mann BJ, Stock AM. Zinc Acquisition Mechanisms Differ between Environmental and Virulent Francisella Species. Stock AM, editor. Journal of Bacteriology. American Society for Microbiology Journals; 2018;200: e00587–17. doi: 10.1128/JB.00587-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mortensen BL, Rathi S, Chazin WJ, Skaar EP. Acinetobacter baumannii Response to Host-Mediated Zinc Limitation Requires the Transcriptional Regulator Zur. Journal of Bacteriology. American Society for Microbiology Journals; 2014;196: 2616–2626. doi: 10.1128/JB.01650-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li Y, Qiu Y, Gao H, Guo Z, Han Y, Song Y, et al. Characterization of Zur-dependent genes and direct Zur targets in Yersinia pestis. BMC Microbiol. BioMed Central; 2009;9: 1–13. doi: 10.1186/1471-2180-9-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lim CK, Hassan KA, Penesyan A, Loper JE, Paulsen IT. The effect of zinc limitation on the transcriptome of Pseudomonas protegens Pf-5. Environmental Microbiology. John Wiley & Sons, Ltd; 2013;15: 702–715. doi: 10.1111/j.1462-2920.2012.02849.x [DOI] [PubMed] [Google Scholar]
- 63.Pederick VG, Eijkelkamp BA, Begg SL, Ween MP, McAllister LJ, Paton JC, et al. ZnuA and zinc homeostasis in Pseudomonas aeruginosa. Scientific Reports 2015 5. 2015;5: 1–14. doi: 10.1038/srep13139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mastropasqua MC, D’Orazio M, Cerasi M, Pacello F, Gismondi A, Canini A, et al. Growth of Pseudomonas aeruginosa in zinc poor environments is promoted by a nicotianamine-related metallophore. Molecular Microbiology. John Wiley & Sons, Ltd; 2017;106: 543–561. doi: 10.1111/mmi.13834 [DOI] [PubMed] [Google Scholar]
- 65.Latorre M, Low M, Gárate E, Reyes-Jara A, Murray BE, Cambiazo V, et al. Interplay between copper and zinc homeostasis through the transcriptional regulator Zur in Enterococcus faecalis. Metallomics. The Royal Society of Chemistry; 2015;7: 1137–1145. doi: 10.1039/C5MT00043B [DOI] [PubMed] [Google Scholar]
- 66.Schröder J, Jochmann N, Rodionov DA, Tauch A. The Zur regulon of Corynebacterium glutamicum ATCC 13032. BMC Genomics. BioMed Central; 2010;11: 1–18. doi: 10.1186/1471-2164-11-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Eckelt E, Jarek M, Frömke C, Meens J, Goethe R. Identification of a lineage specific zinc responsive genomic island in Mycobacterium avium ssp. paratuberculosis. BMC Genomics. BioMed Central; 2014;15: 1–15. doi: 10.1186/1471-2164-15-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Owen GA, Pascoe B, Kallifidas D, Paget MSB. Zinc-responsive regulation of alternative ribosomal protein genes in Streptomyces coelicolor involves zur and sigmaR. Journal of Bacteriology. 2007;189: 4078–4086. doi: 10.1128/JB.01901-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fong JCN, Syed KA, Klose KE, Yildiz FH. Role of Vibrio polysaccharide (vps) genes in VPS production, biofilm formation and Vibrio cholerae pathogenesis. Microbiology. 2010;156: 2757–2769. doi: 10.1099/mic.0.040196-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.DiRita VJ. Co-ordinate expression of virulence genes by ToxR in Vibrio cholerae. Molecular Microbiology. John Wiley & Sons, Ltd; 1992;6: 451–458. doi: 10.1111/j.1365-2958.1992.tb01489.x [DOI] [PubMed] [Google Scholar]
- 71.DiRita VJ, Parsot C, Jander G, Mekalanos JJ. Regulatory cascade controls virulence in Vibrio cholerae. Proc Natl Acad Sci USA. National Academy of Sciences; 1991;88: 5403–5407. doi: 10.1073/pnas.88.12.5403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Higgins DE, Nazareno E, DiRita VJ. The virulence gene activator ToxT from Vibrio cholerae is a member of the AraC family of transcriptional activators. Journal of Bacteriology. American Society for Microbiology Journals; 1992;174: 6974–6980. doi: 10.1128/jb.174.21.6974-6980.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weber GG, Klose KE. The complexity of ToxT-dependent transcription in Vibrio cholerae. The Indian Journal of Medical Research. Wolters Kluwer—Medknow Publications; 2011;133: 201–206. [PMC free article] [PubMed] [Google Scholar]
- 74.Metzger LC, Blokesch M. Regulation of competence-mediated horizontal gene transfer in the natural habitat of Vibrio cholerae. Current Opinion in Microbiology. Elsevier Current Trends; 2016;30: 1–7. doi: 10.1016/j.mib.2015.10.007 [DOI] [PubMed] [Google Scholar]
- 75.Yamamoto S, Mitobe J, Ishikawa T, Wai SN, Ohnishi M, Watanabe H, et al. Regulation of natural competence by the orphan two-component system sensor kinase ChiS involves a non-canonical transmembrane regulator in Vibrio cholerae. Molecular Microbiology. John Wiley & Sons, Ltd; 2014;91: 326–347. doi: 10.1111/mmi.12462 [DOI] [PubMed] [Google Scholar]
- 76.Dalia AB, Lazinski DW, Camilli A. Identification of a membrane-bound transcriptional regulator that links chitin and natural competence in Vibrio cholerae. mBio. American Society for Microbiology; 2014;5: e01028–13. doi: 10.1128/mBio.01028-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ud-Din AIMS, Roujeinikova A. Methyl-accepting chemotaxis proteins: a core sensing element in prokaryotes and archaea. Cell Mol Life Sci. Springer International Publishing; 2017;74: 3293–3303. doi: 10.1007/s00018-017-2514-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ringgaard S, Yang W, Alvarado A, Schirner K, Briegel A, DiRita VJ. Chemotaxis Arrays in Vibrio Species and Their Intracellular Positioning by the ParC/ParP System. DiRita VJ, editor. Journal of Bacteriology. American Society for Microbiology Journals; 2018;200: 267–17. doi: 10.1128/JB.00793-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Alexander RP, Zhulin IB. Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors. Proc Natl Acad Sci USA. National Academy of Sciences; 2007;104: 2885–2890. doi: 10.1073/pnas.0609359104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Matthew D Coleman, Randal B Bass, Ryan S Mehan A, Falke JJ. Conserved Glycine Residues in the Cytoplasmic Domain of the Aspartate Receptor Play Essential Roles in Kinase Coupling and On−Off Switching. Biochemistry. American Chemical Society; 2005;44: 7687–7695. doi: 10.1021/bi0501479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Boin MA, Austin MJ, Häse CC. Chemotaxis in Vibrio cholerae. FEMS Microbiology Letters. 2004;239: 1–8. doi: 10.1016/j.femsle.2004.08.039 [DOI] [PubMed] [Google Scholar]
- 82.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. Oxford University Press; 2004;32: D277–D280. doi: 10.1093/nar/gkh063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Taylor BL. Aer on the inside looking out: paradigm for a PAS–HAMP role in sensing oxygen, redox and energy. Molecular Microbiology. John Wiley & Sons, Ltd; 2007;65: 1415–1424. doi: 10.1111/j.1365-2958.2007.05889.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Boin MA, Häse CC. Characterization of Vibrio cholerae aerotaxis. FEMS Microbiology Letters. 2007;276: 193–201. doi: 10.1111/j.1574-6968.2007.00931.x [DOI] [PubMed] [Google Scholar]
- 85.Bibikov SI, Biran R, Rudd KE, Parkinson JS. A signal transducer for aerotaxis in Escherichia coli. Journal of Bacteriology. American Society for Microbiology Journals; 1997;179: 4075–4079. doi: 10.1128/jb.179.12.4075-4079.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bibikov SI, Barnes LA, Gitin Y, Parkinson JS. Domain organization and flavin adenine dinucleotide-binding determinants in the aerotaxis signal transducer Aer of Escherichia coli. Proc Natl Acad Sci USA. National Academy of Sciences; 2000;97: 5830–5835. doi: 10.1073/pnas.100118697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xie Z, Ulrich LE, Zhulin IB, Alexandre G. PAS domain containing chemoreceptor couples dynamic changes in metabolism with chemotaxis. Proc Natl Acad Sci USA. National Academy of Sciences; 2010;107: 2235–2240. doi: 10.1073/pnas.0910055107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Harris HW, El-Naggar MY, Nealson KH. Shewanella oneidensis MR-1 chemotaxis proteins and electron-transport chain components essential for congregation near insoluble electron acceptors. Biochemical Society Transactions. 2012;40: 1167–1177. doi: 10.1042/BST20120232 [DOI] [PubMed] [Google Scholar]
- 89.Cohen D, Melamed S, Millman A, Shulman G, Oppenheimer-Shaanan Y, Kacen A, et al. Cyclic GMP–AMP signalling protects bacteria against viral infection. Nature. Nature Publishing Group; 2019;574: 691–695. doi: 10.1038/s41586-019-1605-5 [DOI] [PubMed] [Google Scholar]
- 90.Severin GB, Hsueh BY, Elg CA, Dover JA, Rhoades CR, Wessel AJ, et al. A Broadly Conserved Deoxycytidine Deaminase Protects Bacteria from Phage Infection. bioRxiv. Cold Spring Harbor Laboratory; 2021;: 2021.03.31.437871. doi: 10.1101/2021.03.31.437871 [DOI] [Google Scholar]
- 91.Sakib SN, Reddi G, Almagro-Moreno S, DiRita VJ. Environmental Role of Pathogenic Traits in Vibrio cholerae. DiRita VJ, editor. Journal of Bacteriology. American Society for Microbiology Journals; 2018;200: 2123–17. doi: 10.1128/JB.00795-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pant A, Bag S, Saha B, Verma J, Kumar P, Banerjee S, et al. Molecular insights into the genome dynamics and interactions between core and acquired genomes of Vibrio cholerae. Proc Natl Acad Sci USA. 2020;117: 23762–23773. doi: 10.1073/pnas.2006283117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Davis LM, Kakuda T, DiRita VJ. A Campylobacter jejuni znuA Orthologue Is Essential for Growth in Low-Zinc Environments and Chick Colonization. Journal of Bacteriology. American Society for Microbiology Journals; 2009;191: 1631–1640. doi: 10.1128/JB.01394-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ammendola S, Pasquali P, Pistoia C, Petrucci P, Petrarca P, Rotilio G, et al. High-Affinity Zn2+ Uptake System ZnuABC Is Required for Bacterial Zinc Homeostasis in Intracellular Environments and Contributes to the Virulence of Salmonella enterica. Infection and Immunity. American Society for Microbiology Journals; 2007;75: 5867–5876. doi: 10.1128/IAI.00559-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Campoy S, Jara M, Busquets N, de Rozas AMP, Badiola I, Barbé J. Role of the High-Affinity Zinc Uptake znuABC System in Salmonella enterica Serovar Typhimurium Virulence. Infection and Immunity. American Society for Microbiology Journals; 2002;70: 4721–4725. doi: 10.1128/IAI.70.8.4721-4725.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bobrov AG, Kirillina O, Fosso MY, Fetherston JD, Miller MC, VanCleave TT, et al. Zinc transporters YbtX and ZnuABC are required for the virulence of Yersinia pestis in bubonic and pneumonic plague in mice. Metallomics. The Royal Society of Chemistry; 2017;9: 757–772. doi: 10.1039/c7mt00126f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nielubowicz GR, Smith SN, Mobley HLT. Zinc uptake contributes to motility and provides a competitive advantage to Proteus mirabilis during experimental urinary tract infection. Infection and Immunity. 2010;78: 2823–2833. doi: 10.1128/IAI.01220-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sabri M, Houle S, Dozois CM. Roles of the extraintestinal pathogenic Escherichia coli ZnuACB and ZupT zinc transporters during urinary tract infection. Infection and Immunity. 2009;77: 1155–1164. doi: 10.1128/IAI.01082-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Corbett D, Wang J, Schuler S, Lopez-Castejon G, Glenn S, Brough D, et al. Two Zinc Uptake Systems Contribute to the Full Virulence of Listeria monocytogenes during Growth In Vitro and In Vivo. Bäumler AJ, editor. Infection and Immunity. American Society for Microbiology Journals; 2012;80: 14–21. doi: 10.1128/IAI.05904-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hood MI, Mortensen BL, Moore JL, Zhang Y, Kehl-Fie TE, Sugitani N, et al. Identification of an Acinetobacter baumannii Zinc Acquisition System that Facilitates Resistance to Calprotectin-mediated Zinc Sequestration. PLoS Pathog. Public Library of Science; 2012;8: e1003068. doi: 10.1371/journal.ppat.1003068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gielda LM, DiRita VJ. Zinc competition among the intestinal microbiota. mBio. 2012;3: e00171–12. doi: 10.1128/mBio.00171-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kamp HD, Patimalla-Dipali B, Lazinski DW, Wallace-Gadsden F, Camilli A. Gene Fitness Landscapes of Vibrio cholerae at Important Stages of Its Life Cycle. PLoS Pathog. Public Library of Science; 2013;9: e1003800. doi: 10.1371/journal.ppat.1003800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mueller RS, McDougald D, Cusumano D, Sodhi N, Kjelleberg S, Azam F, et al. Vibrio cholerae strains possess multiple strategies for abiotic and biotic surface colonization. Journal of Bacteriology. American Society for Microbiology Journals; 2007;189: 5348–5360. doi: 10.1128/JB.01867-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lai HC, Gygi D, Fraser GM, Hughes C. A swarming-defective mutant of Proteus mirabilis lacking a putative cation-transporting membrane P-type ATPase. Microbiology. 1998;144: 1957–1961. doi: 10.1099/00221287-144-7-1957 [DOI] [PubMed] [Google Scholar]
- 105.Ammendola S, D’Amico Y, Chirullo B, Drumo R, Civardelli D, Pasquali P, et al. Zinc is required to ensure the expression of flagella and the ability to form biofilms in Salmonella enterica sv Typhimurium. Metallomics. Royal Society of Chemistry; 2016;8: 1131–1140. doi: 10.1039/c6mt00108d [DOI] [PubMed] [Google Scholar]
- 106.Yeo J, Dippel AB, Wang XC, Hammond MC. In Vivo Biochemistry: Single-Cell Dynamics of Cyclic Di-GMP in Escherichia coli in Response to Zinc Overload. Biochemistry. 2017;57: 108–116. doi: 10.1021/acs.biochem.7b00696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rebbapragada A, Johnson MS, Harding GP, Zuccarelli AJ, Fletcher HM, Zhulin IB, et al. The Aer protein and the serine chemoreceptor Tsr independently sense intracellular energy levels and transduce oxygen, redox, and energy signals for Escherichia coli behavior. Proc Natl Acad Sci USA. National Academy of Sciences; 1997;94: 10541–10546. doi: 10.1073/pnas.94.20.10541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Alexandre G. Chemotaxis Control of Transient Cell Aggregation. Margolin W, editor. Journal of Bacteriology. American Society for Microbiology Journals; 2015;197: 3230–3237. doi: 10.1128/JB.00121-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bible AN, Stephens BB, Ortega DR, Xie Z, Alexandre G. Function of a Chemotaxis-Like Signal Transduction Pathway in Modulating Motility, Cell Clumping, and Cell Length in the Alphaproteobacterium Azospirillum brasilense. Journal of Bacteriology. American Society for Microbiology Journals; 2008;190: 6365–6375. doi: 10.1128/JB.00734-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Russell MH, Bible AN, Fang X, Gooding JR, Campagna SR, Gomelsky M, et al. Integration of the Second Messenger c-di-GMP into the Chemotactic Signaling Pathway. Greenberg EP, Harwood CS, editors. mBio. American Society for Microbiology; 2013;4: 1024–13. doi: 10.1128/mBio.00001-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Harris HW, Sánchez-Andrea I, McLean JS, Salas EC, Tran W, El-Naggar MY, et al. Redox Sensing within the Genus Shewanella. Front Microbiol. Frontiers; 2018;8. doi: 10.3389/fmicb.2017.02568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Collins KD, Andermann TM, Draper J, Sanders L, Williams SM, Araghi C, et al. The Helicobacter pylori CZB Cytoplasmic Chemoreceptor TlpD Forms an Autonomous Polar Chemotaxis Signaling Complex That Mediates a Tactic Response to Oxidative Stress. Journal of Bacteriology. American Society for Microbiology Journals; 2016;198: 1563–1575. doi: 10.1128/JB.00071-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Collins KD, Hu S, Grasberger H, Kao JY, Ottemann KM. Chemotaxis Allows Bacteria To Overcome Host-Generated Reactive Oxygen Species That Constrain Gland Colonization. Infection and Immunity. American Society for Microbiology Journals; 2018;86. doi: 10.1128/IAI.00878-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Butler SM, Camilli A. Going against the grain: chemotaxis and infection in Vibrio cholerae. Nat Rev Microbiol. Nature Publishing Group; 2005;3: 611–620. doi: 10.1038/nrmicro1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Butler SM, Camilli A. Both chemotaxis and net motility greatly influence the infectivity of Vibrio cholerae. Proc Natl Acad Sci USA. 2004;101: 5018–5023. doi: 10.1073/pnas.0308052101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Merrell DS, Butler SM, Qadri F, Dolganov NA, Alam A, Cohen MB, et al. Host-induced epidemic spread of the cholera bacterium. Nature. Nature Publishing Group; 2002;417: 642–645. doi: 10.1038/nature00778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Matilla MA, Krell T. The effect of bacterial chemotaxis on host infection and pathogenicity. FEMS Microbiol Rev. 2017;42. doi: 10.1093/femsre/fux052 [DOI] [PubMed] [Google Scholar]
- 118.Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA III, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. Nature Publishing Group; 2009;6: 343–345. doi: 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- 119.Miller VL, DiRita VJ, Mekalanos JJ. Identification of toxS, a regulatory gene whose product enhances toxR-mediated activation of the cholera toxin promoter. Journal of Bacteriology. American Society for Microbiology Journals; 1989;171: 1288–1293. doi: 10.1128/jb.171.3.1288-1293.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chiang SL, Rubin EJ. Construction of a mariner-based transposon for epitope-tagging and genomic targeting. Gene. Elsevier; 2002;296: 179–185. doi: 10.1016/s0378-1119(02)00856-9 [DOI] [PubMed] [Google Scholar]
- 121.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. Academic Press; 1990;215: 403–410. doi: 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 122.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. BioMed Central; 2014;15: 1–21. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Miller J. Experiments in molecular genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 124.Zhang X, Bremer H. Control of the Escherichia coli rrnB P1 promoter strength by ppGpp. J Biol Chem. American Society for Biochemistry and Molecular Biology; 1995;270: 11181–11189. doi: 10.1074/jbc.270.19.11181 [DOI] [PubMed] [Google Scholar]
- 125.Karimova G, Pidoux J, Ullmann A, Ladant D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci USA. National Academy of Sciences; 1998;95: 5752–5756. doi: 10.1073/pnas.95.10.5752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Solovyev V, Salamov A. Automatic annotation of microbial genomes and metagenomic sequences. Metagenomics and Its Application in Agriculture. 2011. pp. 61–78. [Google Scholar]
- 127.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. Oxford University Press; 2007;23: 2947–2948. doi: 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]