Abstract
Seed shattering refers to the natural shedding of seeds when they ripe, a phenomenon typically observed in wild and weedy plant species. The timing and extent of this phenomenon varies considerably among plant species. Seed shattering is primarily a genetically controlled trait; however, it is significantly influenced by environmental conditions, management practices and their interactions, especially in agro-ecosystems. This trait is undesirable in domesticated crops where consistent efforts have been made to minimize it through conventional and molecular breeding approaches. However, this evolutionary trait serves as an important fitness and survival mechanism for most weeds that utilize it to ensure efficient dispersal of their seeds, paving the way for persistent soil seedbank development and sustained future populations. Weeds have continuously evolved variations in seed shattering as an adaptation under changing management regimes. High seed retention is common in many cropping weeds where weed maturity coincides with crop harvest, facilitating seed dispersal through harvesting operations, though some weeds have notoriously high seed shattering before crop harvest. However, high seed retention in some of the most problematic agricultural weed species such as annual ryegrass (Lolium rigidum), wild radish (Raphanus raphanistrum), and weedy amaranths (Amaranthus spp.) provides an opportunity to implement innovative weed management approaches such as harvest weed seed control, which aims at capturing and destroying weed seeds retained at crop harvest. The integration of such management options with other practices is important to avoid the rapid evolution of high seed shattering in target weed species. Advances in genetics and molecular biology have shown promise for reducing seed shattering in important crops, which could be exploited for manipulating seed shattering in weed species. Future research should focus on developing a better understanding of various seed shattering mechanisms in plants in relation to changing climatic and management regimes.
Keywords: weed seed dispersal, seedbank, harvest weed seed control, weed evolutionary adaptation, crop improvement
Introduction
Plants constantly evolve and adapt in the wild, shaped by natural selection (Darwin, 1859; Lenski, 2017). During the domestication of wild species, humans have intervened and accelerated the selection process for desired plant traits through artificial selection (Gregory, 2009). This has led to the loss of several adaptive traits in plants that are vital for persistence under natural conditions (Pickersgill, 2007; Flint-Garcia, 2013). For example, traits such as non-synchronous flowering, non-uniform seed maturity, seed shattering and seed dormancy are all important traits for wild plant populations in natural environments (Kantar et al., 2017). These traits allow wild plants to germinate, grow and reproduce under conditions that are conducive to their growth and development (Pickersgill, 2007). Among these traits, seed shattering, i.e., the capacity of a plant to shed its seeds, is essential for the dispersal and persistence of the offspring in many wild species (Dong and Wang, 2015). Shattering can occur over a period of a few to several days, increasing the chances that a significant proportion of the produced seeds are dispersed away from the mother plants and new niches are occupied (Delouche et al., 2007). Thus, seed shattering minimizes intra-population competition and increases species fitness (Thurber, 2012; Di Vittori et al., 2019).
In domesticated crops, seed shattering is an unfavorable trait due to its detrimental impact on harvestable grain yield (Serebrenik, 2013; Table 1). Domestication has selected for crops with almost no seed shattering ability, especially in those crops grown for grain production (Harlan et al., 1973). Some level of seed shattering is present and even preferred in pasture grasses and legumes as a specialized adaptation that ensures self-seeding and pasture regeneration (Dong and Wang, 2015). Many cultivated crops, if left as “wild populations,” revert to shattering phenotypes through back mutation (endoferality) as evident in wild rice (Oryza sativa) (Vigueira et al., 2013, 2019), or through continued introgression (exoferality) as in shattercane (Sorghum bicolor ssp. drummondii) (Ejeta and Grenier, 2005). This indicates that the shattering habit might be complementary for the persistence of previously domesticated crop species in undisturbed natural ecosystems (Di Vittori et al., 2019).
TABLE 1.
Seed loss due to shattering documented in cultivated crops.
| Family | Crop | Scientific name | Loss due to shattering (%, unless mentioned otherwise) | References |
| Poaceae | Oat | Avena sativa | 12–50 | Clarke, 1981 |
| Barley (rainfed) | Hordeum vulgare | 0–34 | Platt and Wells, 1949 | |
| Rice | Oryza sativa | 1–5 | Niruntrayakul et al., 2009 | |
| 28 g/plant (greenhouse); 61 g/plant (growth chamber) | Thurber et al., 2013 | |||
| Dallisgrass | Paspalum dilatatum | 30 | Bennett and Marchbanks, 1969 | |
| Bahiagrass | Paspalum notatum | 36–50 | Correa, 1974 | |
| Brassicaceae | Indian mustard | Brassica juncea | 4–7 | Gan et al., 2008 |
| Canola | Brassica napus | 6 | Gulden et al., 2003 | |
| 8 | Gan et al., 2008 | |||
| 50 | Price et al., 1996 | |||
| Yellow mustard | Sinapis alba | 5 | Gan et al., 2008 | |
| Rape mustard | Brassica rapa | 2 | Gan et al., 2008 | |
| Fabaceae | Chickpea | Cicer arietinum | 65 | Murgia et al., 2017 |
| Soybean | Glycine max | 5–10 | Davidson, 2014 | |
| 21 | Tukamuhabwa et al., 2002 | |||
| 37 | Philbrook and Oplinger, 1989 | |||
| 34–99 | Tiwari and Bhatnagar, 1991 |
Seed shattering is a highly diverse trait in weedy and wild species, e.g., in Italian ryegrass (Maity et al., 2021), influenced by years of selection (Vigueira et al., 2013; Table 2). Shattering of seed and its effective dispersal enable the weeds to survive and persist in natural as well as agricultural landscapes (Thurber, 2012). However, shattering can lead to substantial crop yield loss in commercial agriculture. In this review, the significance of seed shattering in crops and weeds, mechanisms of seed shattering and how different factors influence this important trait are discussed. A snapshot of how the recent developments in plant physiology, genetics and genomics have contributed to our understanding of this complex trait is also presented. The synthesis of knowledge on this important aspect of plant evolutionary biology is beneficial for crop improvement as well as weed management in modern agriculture.
TABLE 2.
Seed shattering values at crop harvest for major weeds in global cropping systems.
| Family | Scientific name | Common name* | Seed shattering (%) prior to main crop harvest** | Country/state or province | References |
| Amaranthaceae | Amaranthus tuberculatus | Tall waterhemp | 1–5 | United States/Nebraska, Missouri, Wisconsin, and Illinois | Schwartz et al., 2016 |
| Amaranthus palmeri | Palmer amaranth | 1–5 | United States/Arkansas, Tennessee, Illinois, Missouri, and Nebraska | Schwartz et al., 2016 | |
| 10 | United States/Puerto Rico | Green et al., 2016 | |||
| Amaranthus retroflexus | Redroot pigweed | 48 | United States/Virginia | Haring et al., 2017 | |
| 44 | Canada/Alberta | Beckie et al., 2017 | |||
| Amaranthaceae | Chenopodium album | Common lambsquarters | 10 | Alberta, Canada | Beckie et al., 2017 |
| 9 | United States/Virginia | Haring et al., 2017 | |||
| 50 | United States/Minnesota | Forcella et al., 1996 | |||
| Bassia scoparia | Kochia | 0 | Canada/Saskatchewan | Burton et al., 2017 | |
| 0 | Canada/Alberta and Saskatchewan | Tidemann et al., 2017 | |||
| 0 | Canada/Saskatchewan | Burton et al., 2017 | |||
| 0 | Canada/Alberta | Beckie et al., 2017 | |||
| Asteraceae | Ambrosia artemisiifolia | Common ragweed | 38 | United States/Virginia | Haring et al., 2017 |
| Ambrosia trifida | Giant ragweed | 20 | United States/Minnesota | Goplen et al., 2016 | |
| 40 | United States/Virginia | Haring et al., 2017 | |||
| Conyza bonariensis | Horseweed/flaxleaf fleabane | 7–81 | Australia/Queensland and New South Wales | Widderick et al., 2014 | |
| Sonchus asper | Prickly sowthistle | 92 | Canada/Alberta | Beckie et al., 2017 | |
| Sonchus oleraceus | Common sowthistle | 46–62 | Australia/Queensland and New South Wales | Widderick et al., 2014 | |
| Brassicaceae | Brassica napus | Canola | 2 | Canada/Alberta and Saskatchewan | Tidemann et al., 2017 |
| Raphanus raphanistrum | Wild radish | 1 | Australia/Western Australia Canada/Alberta | Burton et al., 2017; Walsh and Powles, 2014 | |
| 10 | Australia/Western Australia | Walsh and Powles, 2014 | |||
| Rapistrun rugosum | Turnip weed | 0–81 | Australia/Queensland and New South Wales | Widderick et al., 2014 | |
| Sinapis arvensis | Wild mustard | 30 | Canada/Saskatchewan Canada/Alberta | Beckie et al., 2017; Burton et al., 2017 | |
| 100 | United States/Minnesota | Forcella et al., 1996 | |||
| Sisymbrium thellungii | African turnip weed | 0 | Australia/Queensland and New South Wales | Widderick et al., 2014 | |
| Malvaceae | Hibiscus trionum | Flower of an hour/Bladder ketmia | 45–79 | Australia/Queensland and New South Wales | Widderick et al., 2014 |
| Malva neglecta | Buttonweed | 0 | Canada/Alberta | Beckie et al., 2017 | |
| Poaceae | Aegilops cylindrica | Jointed goatgrass | 30 | Australia/Western Australia | Walsh and Powles, 2014 |
| 25 | United States/Colorado | Soni et al., 2020 | |||
| Alopecurus myosuroides | Slender meadow foxtail | 40–90 | United Kingdom | Walsh et al., 2018 | |
| Avena fatua | Wild oat | 61 | Canada/Alberta | Beckie et al., 2017 | |
| 22–30 | Canada/Saskatchewan | Burton et al., 2017 | |||
| 16–31 | Western Australia, Australia | Walsh and Powles, 2014; Widderick et al., 2014 | |||
| Avena fatua; Avena sterilis | Wild oat | 80–96 | Spain and United Kingdom | Barroso et al., 2006 | |
| Bromus tectorum | Cheatgrass/downy brome | 33 | Australia/Western Australia | Walsh and Powles, 2014 | |
| 25 | United States/Colorado | Soni et al., 2020 | |||
| Chloris virgata | Rhodesgrass | 29–53 | Australia/Queensland and New South Wales | Widderick et al., 2014 | |
| Digitaria sanguinalis | Large crabgrass | 77 | United States/Virginia | Haring et al., 2017 | |
| Echinochloa colona | Jungle rice | 5–91 | Australia/Queensland and New South Wales | Widderick et al., 2014 | |
| 59–68 | United States/Arkansas | Schwartz-Lazaro et al., 2017 | |||
| 67 | United States/Puerto Rico | Green et al., 2016 | |||
| Lolium rigidum | Rigid ryegrass | 15 | Australia/Western Australia | Walsh and Powles, 2014 | |
| 15 | Spain/Catalonia | Blanco-Moreno et al., 2004 | |||
| Lolium perenne ssp. multiflorum | Italian ryegrass | 4.8–54 | United States/Texas | Maity et al., 2021 | |
| Oryza sativa | Red rice | 15–87 | United States/Arkansas | Burgos et al., 2014 | |
| Secale cereale | Feral rye | 25 | United States/Colorado | Soni et al., 2020 | |
| 10 | Spain/Catalonia | Blanco-Moreno et al., 2004 | |||
| Setaria faberi | Giant foxtail | 68 | United States/Virginia | Haring et al., 2017 | |
| 60 | United States/Minnesota | Forcella et al., 1996 | |||
| Setaria viridis | Green foxtail | 6 | Canada/Saskatchewan; Canada/Alberta | Beckie et al., 2017; Burton et al., 2016, 2017 | |
| 80 | United States/Virginia | Haring et al., 2017 | |||
| Sorghum halepense | Johnsongrass | 0–50 | United States/Texas | Young, 2021 | |
| 0–30 | United States/Arkansas | Young, 2021 | |||
| Polygonaceae | Polygonum convolvulus | Black bindweed | 18 | Canada/Saskatchewan | Burton et al., 2017 |
| Rubiaceae | Galium spp. | Cleavers | 26 | Canada/Alberta | Beckie et al., 2017 |
| Galium spurium + aparine | Cleavers | 2–4 | Canada/Saskatchewan | Burton et al., 2017 |
*Common names are according to the list approved by Weed Science Society of America (WSSA). **A substantial portion of this data was obtained from Walsh et al. (2018).
Factors Controlling Seed Shattering in Plants
Seed shattering in plants is regulated by complex physiological and genetic mechanisms (Zhao et al., 2019), in conjunction with environmental factors. Some of these mechanisms are fairly well understood in domesticated crops, whereas little is known for most wild and weedy species.
Physiological Control
The first step in seed or pod shattering is the formation of an abscission layer at the point where the seeds or pods are attached to the plants. Though the fundamental mechanism of abscission is the same for many crops, it varies with the type of tissue, as it may be the spikelet in cereals or a pod in legumes (Dong and Wang, 2015). Two main series of events occur during the process of abscission: the first is the disintegration of the entire or a portion of the cell wall as a result of biochemical changes, which is then followed by the mechanical tearing of the abscission layer (Pfeiffer, 1928). In the first event, the cells in abscission layers become elongated and eventually collapse after plasmolysis. In the second event, a sudden disruption of the abscission cells occurs due to enzymatic deterioration, resulting in the tearing of the abscission layer (Pfeiffer, 1928).
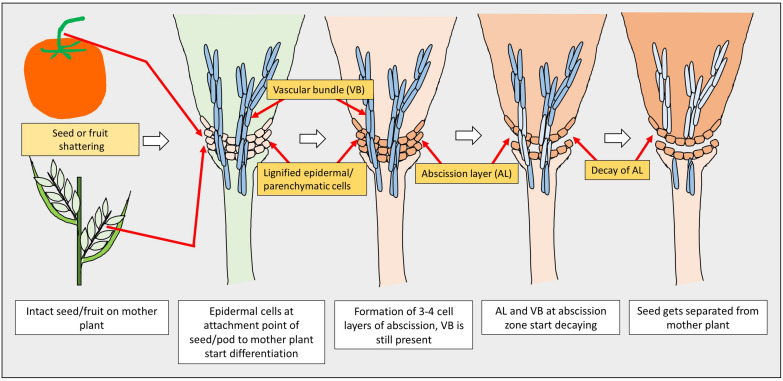
A model of seed shattering in monocot or fruit dehiscence in dicot is presented in Figure 1. In monocots, seed shattering is triggered by the formation of an abscission layer at the attachment point between the lemma and pedicel by cell wall thickening and lignification (Harlan and DeWet, 1965; Elgersma et al., 1988; Fuller and Allaby, 2009). Swelling and dissolving of the middle lamella between adjacent cell walls in the abscission layer allows for grain release (Htun et al., 2014). The structure and stage of formation and the anatomical location of the abscission layer may vary among plant species. In rice, development of an abscission layer between the spikelet and rachilla, followed by its degradation leads to seed shattering (Zheng et al., 2007; Fuller and Qin, 2008). Examination of the spikelet bases between domesticated rice and the wild shattering types revealed that domesticated spikelet bases are characterized by a dimpled appearance and possess less symmetrical scars, whereas the wild types had a smooth scar with a straight profile at the spikelet bases (Li et al., 2006; Fuller et al., 2009). In Lolium spp., the abscission layer is present at the attachment point of lemma and palea to the rachilla (Elgersma et al., 1988). The abscission layer is easily identifiable as the cells present in it are smaller than the parenchymatous cells in the rachilla. In perennial ryegrass (Lolium perenne L.), this layer usually consists of 4–8 cell layers (Elgersma et al., 1988). In bahiagrass (Paspalum notatum Fluegge), cells in the abscission layer were larger, more prominent and present in five to seven layers. The dimension of the dehiscence zone or abscission layer shows positive correlation with shattering resistance, as reported by Child et al. (2003) in Brassica napus. These cells eventually lost their wall, leading to shattering. In wild and weedy species, development of the abscission layer has been shown to occur at a much faster rate compared to their cultivated counterparts (Li et al., 2006). In wild rice, the abscission layer forms before flowering and begins degradation during the course of flowering, whereas in cultivated rice the abscission layers remain intact and show no sign of degradation even after flowering (Carrie et al., 2011). In dalliagrass (Paspalum dilatatum Poir), the abscission layer was identifiable between early booting and booting stages (Burson et al., 1978).
FIGURE 1.
Diagram showing stages of seed shattering in monocot and dicot plants.
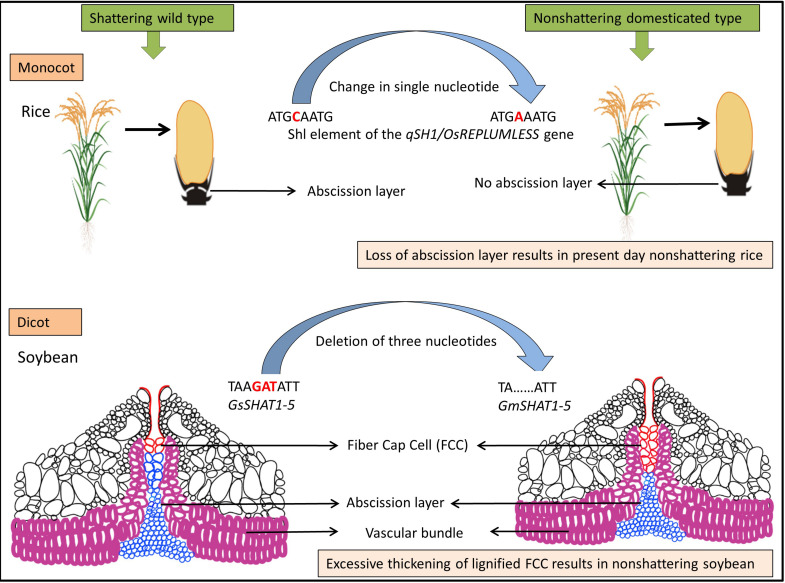
In dicots, studies on the mechanisms responsible for seed shattering (more appropriately, dehiscence of pod in legumes and siliqua in crucifers) are meager compared to that of monocots (Lin et al., 2012; Dong and Wang, 2015). Most relevant studies on pod dehiscence (development of abscission zones along the pod valve margin) have been conducted in soybean and French bean (Phaseolus vulgaris L.) (Romkaew et al., 2008; Dong et al., 2014; Murgia et al., 2017). Dehiscence in less domesticated crops begins long before the actual dehiscence, sometimes as early as the fertilization of the ovule (Ferrándiz et al., 1999). Pod dehiscence in dicots is induced by the formation of a specific dehiscence (or abscission) zone along the pod (Dong and Wang, 2015; Figure 2). The cells at the abscission zone start differentiating into lignified and separation layers during pod development, which then auto-degrade before pod dehiscence (Seymour et al., 2013). Lignification is a complex process involving the deposition of lignins on the extracellular polysaccharidic matrix (Ros, 1997), and a higher degree of lignification in the abscission layer cells indicates more shattering (Lee et al., 2018). The degree of lignification of the inner layer of the pod wall determined the extent of pod dehiscence in common bean (Murgia et al., 2017) and soybean (Funatsuki et al., 2014). In addition to lignin, other main fibers of the plant secondary cell wall such as cellulose and hemi-cellulose, alone or in combination provide strength and structural integrity to cell walls, which directly affect shattering (Baucher et al., 1998). Suanum et al. (2016) observed in yardlong bean (Vigna unguiculata ssp. sesquipedalis) and wild cowpea [Vigna unguiculata (L.) Walp.] that cellulose, hemi-cellulose and lignin contents in pods are highly correlated with pod dehiscence. The non-shattering genotypes have several layers of thickened fiber cap cells compared to the shattering types (Figure 2).
FIGURE 2.
Model for evolution of shattering resistance in cultivated rice [modified from Gasser and Simon (2011)] and dehiscence resistance in cultivated soybean [modified from Dong et al. (2014)].
Seed shattering in monocots and dicots is determined by a complex plant signaling network involving hormones (Vivian-Smith and Koltunow, 1999). Thickening, swelling, and dissolving of the cell layers in the abscission zones across plant parts are accomplished by up- and down-regulations of certain gene(s) triggering the production and activity of specific enzyme(s) and plant hormone(s). An increase in β-1,4-glucanase or cellulase activity has been observed during pod dehiscence in canola (B. napus) (Roberts et al., 2002), whereas increasing polygalacturonase activity is correlated with shedding of fruits in oil palm (Elaeis guineensis) (Henderson et al., 2001). A number of proteins such as expansin and chitinase (a Pathogenesis-Related Protein) are reported to directly influence the abscission process in various plant parts across multiple plant species (reviewed in Roberts et al., 2002). In addition to gibberellins (GA), abscisic acid (ABA) and cytokinin (CYT), ethylene (ETH) and auxin (IAA) concentrations in the abscission or dehiscence zones are also known to play a major role in determining seed shattering or pod dehiscence (Addicott, 1970; González-Carranza et al., 1998). RNA-sequencing and expression analysis show that the specific ABA-responsive 9-cis-epoxycarotenoid dioxygenase (NCED) gene, a key gene for ABA biosynthesis, and ABA concentration increase prior to and during abscission process and show a potential signal transduction network among the plant hormones involved in seed shattering (Lang et al., 2021). However, several studies suggest ETH as the primary regulator of seed shattering and ABA’s critical role depends on its interaction with IAA and ETH, suggesting an intermediary role of ABA in organ abscission (Marciniak et al., 2018). Cellulase activity shows a high positive correlation with the level of IAA, leading to rapid abscission (Chauvaux et al., 1997). Specifically, high concentration of auxins negatively influences seed shattering. Application of IAA on mature silique retarded the cellulase activity and pod dehiscence (Chauvaux et al., 1997), whereas ETH promoted the formation of the dehiscence zone (Ferrándiz, 2002). However, depending on the species, stage of application, and biochemical form, IAA can accelerate the abscission process (Addicott and Lynch, 1951). In Arabidopsis, studies have established correlation between dehiscence zone development and low levels of IAA (Heisler et al., 2001; Martinez and Vera, 2009). The commonly accepted model of abscission induction in plant organs involves the decrease of IAA levels and increase of ABA, GA, and ETH levels (Meir et al., 2010; Nakano and Ito, 2013; Marciniak et al., 2018).
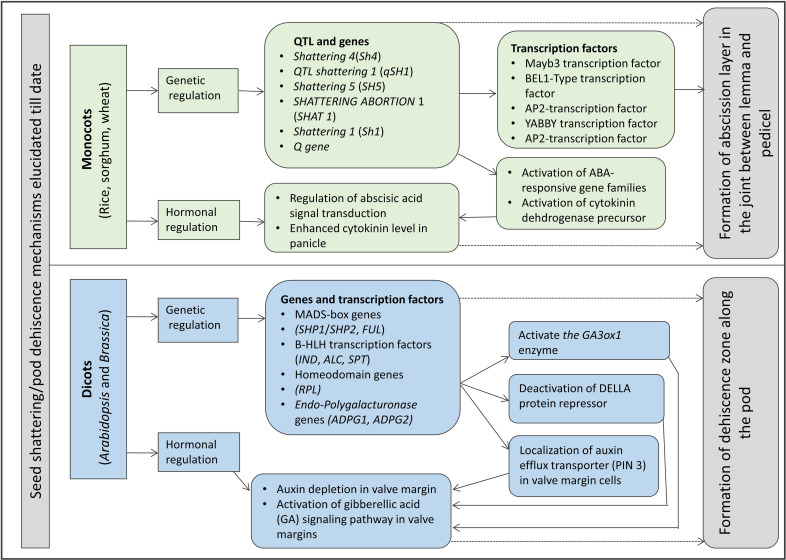
Genetic Control
The genetic mechanisms underlying seed shattering are regulated by a complex network of genes and their interactions (Dong and Wang, 2015; Figure 3). Various investigations aiming at deciphering the genetic mechanisms of seed shattering have indicated the parallel evolution of the non-shattering trait in cereals (Paterson et al., 1995; Konishi et al., 2006; Li et al., 2006; Lin et al., 2012; Tang et al., 2013; Fu et al., 2018). Reports suggest that seed shattering is usually a dominant trait, governed by few, recessive genes across species (Ladizinsky, 1985); e.g., four in rice (Tang and Morishima, 1989), two in common and durum wheat (Love and Craig, 1919), one is cowpea (Aliboh et al., 1997), and two in turnip rape (Mongkolporn et al., 2003; Table 3). The major seed shattering gene in sorghum (Sh1; that encodes a YABBY transcription factor) and its orthologs in rice induce seed shattering through one common mechanism, i.e., formation of an abscission layer between the pedicel and spikelet (Lin et al., 2012; Li et al., 2019). The loss-of-function mutation in these genes is independently selected for non-shattering in domesticated sorghum, rice (Lv et al., 2018) and corn (Zea mays L.) (Paterson et al., 1995; Lin et al., 2012). Konishi et al. (2006) reported that a single nucleotide change resulted in a non-shattering trait in domestic rice. In soybean, three nucleotides in the GsSHAT1-5 gene lead to a non-shattering type (Dong et al., 2014; Figure 3).
FIGURE 3.
A schematic of genetic mechanisms underlying pod and seed shattering in crop plants.
TABLE 3.
Inheritance pattern of seed shattering trait in selected plant species.
| Plant species | Scenarios | Genetic control/Inheritance pattern | References |
| Rice | Oryza sativa, weedy strains X cultivated strains | Four genes with segregation patterns ranging from monogenic to continuous, depending on the crosses | Tang and Morishima, 1989 |
| Common and durum wheat | Triticum vulgare X T. durum | One gene with dominant gene action, shattering is dominant to non-shattering | Love and Craig, 1919 |
| Einkorn wheat | Triticum monococcum X T. boeoticum | Two recessive genes with additive action, shattering is dominant to non-shattering | Sharma and Waines, 1980 |
| Ryegrass | Lolium temulentum X L. persicum | Two recessive genes with additive action, shattering is dominant to non-shattering | Senda et al., 2006 |
| Foxtail millet | Setaria viridis X S. italica | Two genes with additive action, hybrids with 0 or 1 allele from the shedding parent show no shedding, but with 2 or more alleles show shedding | Darmency and Pernes, 1987 |
| Pearl millet | Pennisetum mollissimum X P. glaucum | One gene with dominant gene action, shattering is dominant to non-shattering | Poncet et al., 1998 |
| Buckwheat | Fagopyrum homotropicum X F. esculentum | Three complementary dominant genes | Wang et al., 2005 |
| Cowpea | Vigna unguiculata, wild X cultivated | Monogenic dominance of pod shattering over non-shattering | Aliboh et al., 1997 |
| Turnip rape | Brassica rapa, shatter-susceptible X shatter-resistant | Two recessive major genes with a dominant epistasis effect | Mongkolporn et al., 2003 |
The genus Medicago is known for its pod coiling mechanism of seed dispersal, which is highly correlated with the increased valve margin lignification mechanism of the members of Brassicaceae (Fourquin et al., 2013). Genetic analysis elucidated that the pod coiling mechanism is governed by a minor protein sequence of SHATTERPROOF (SHP) orthologs (Ferrándiz and Fourquin, 2014). In rice, qSH1 [the major Quantitative Trait Loci (QTL) on chromosome 1 that controls seed shattering] is required for the formation of the abscission layer in the pedicel. It encodes a BEL-1 type homeobox transcription factor that is homologous to the RPL transcription factor of Arabidopsis, which is required for the development of replum cells in silique wall margin (Konishi et al., 2006). Suanum et al. (2016) indicated that the major QTL for the fibers such as cellulose, hemi-cellulose and lignin in pods of yardlong bean and wild cowpea are co-located with the major pod shattering QTL. Furthermore, over-expression analysis of the NAC and SHAT-5 genes in soybean revealed that shattering-resistant lines had increased biosynthesis of a secondary wall that resulted in thickening of lignified fiber cap cells (Dong et al., 2014). These findings suggest that a unique convergent mechanism is involved in seed shattering across distantly related genera.
In recent years, bi-parental mapping and genome-wide approaches have enabled dissection of the complex genetic control of seed shattering (Table 4). Specific genes and transcription factors regulating morphological and anatomical mechanisms governing seed shattering have been identified in both monocot and dicot species (Table 5). Various studies have found QTL explaining up to 50% of phenotypic variance for seed shattering (Funatsuki et al., 2006; Subudhi et al., 2014; Table 4). Recently, fourteen candidate gene derived polymorphic EST-SSR markers specific for abscission zone development and seed shattering were developed in Elymus nutans (Zhao et al., 2019). Though the abscission layer formation in plants appears to be controlled by a few major genes (McWilliam, 1980), the final quantum of seed shattering is a highly environment-dependent event, which varies significantly among cultivars, geography and seasons (Konishi et al., 2006), suggesting that seed shattering is a complex, polygenic trait (Zhou et al., 2012).
TABLE 4.
The specific Quantitative Trait Locus/Loci (QTL) reported to influence seed/pod shattering in food crops.
| Crop | Mapping Population | Markers | Identified QTL | Linkage group | Phenotypic variation | References |
| Monocot | ||||||
| Hybrid Elymus | 164 backcross progenies derived from creeping × basin wildrye hybrid and a true creeping wildrye tester | - | One pleiotropic QTL | 6 | 43.1% | Larson and Kellogg, 2009 |
| Rice | 120 DH lines derived from a cross between Cheongchenong × Nagdong | 217 SSR markers | 3 QTL for pulling strength and 4 QTL for bending strength | 1,2,4,6,9,10 | 5–14% | Lee et al., 2016 |
| 198 F7:8 RILs derived from the cross Bengal × PSRR-1 and 174 F8:9 RILs derived from the cross Cypress × PSRR-1 | SSR markers | Two QTL were consistent across the populations | 4 and 10 | 61.9% | Subudhi et al., 2014 | |
| CSSLs and NILs of Japonica rice landrace Jiucaiquing in IR-26 background | 192 SSR markers | Four QTL | 1, 3, 6 and 11 | - | Cheng et al., 2016 | |
| Dicot | ||||||
| Abyssinian Cabbage | 229 F2 lines derived from BC 73526 × BC 73524 | 6,464 DArT-Seq Markers | Five QTL | B1, B3, B8 and C5 | 3.75–5.27% | Raman et al., 2017 |
| Azuki bean | 188 F2 lines derived from a cross between JP 110658 × JP 109685 | 316 SSR markers | Two QTL | 4 and 9 | 6.4–18.2% | Kaga et al., 2008 |
| Canola | 126 DH lines derived from BLN2762 × Surpass 400 | DArT-Seq markers | Twelve QTL | 3, 4, 6, 7,8,9 | 57% | Raman et al., 2014 |
| Unstructured diversity panel of 143 accessions and two structured populations (96 DH lines and 124 F2 progenies) | - | Two QTL consistently detected across the populations and environments | A06 and A09 | - | Liu et al., 2016 | |
| Cowpea | 159 RILs derived from a cross between 524B × 219-01 | 202 SSR markers | Four QTL | 1 to 10 | 6.4 to 17.2% | Andargie et al., 2011 |
| 215 RILs derived from a cross between IT99K-573-1-1 × TVNu-1158 | 51,128 SNPs | Two QTL | 3 and 5 | 68% | Lo et al., 2018 | |
| Soybean | 104 RILs derived from the cross Toyomusume × Hayahikari and 96 F2 lines derived from the cross Toyomusume × HC1-F7-57 | 178 SSR markers | One major QTL detected across the populations | 10 | 50% | Funatsuki et al., 2006 |
TABLE 5.
Genes and transcription factors reported to control seed shattering in a number of plant species.
| Plant species | Gene/Transcription factor | Mechanism | Nature of allele for breeding | References |
| Arabidopsis | SHP1 and SHP 2 | Lignification of silique valve margin and the adjacent cells | Loss of function | Liljegren et al., 2000 |
| FUL | Lignification of silique valve margin and the adjacent cells | Ectopic expression | Liljegren et al., 2000 | |
| ALC | Encode a protein related to the myc/bHLH family of transcription factors which promotes separation of the valve cells from the replum | Loss of function | Rajani and Sundaresan, 2001 | |
| RPL | Encodes a homeodomain protein that prevents development of replum cells into silique valve margin | Loss of function | Roeder et al., 2003 | |
| STK or AGL 11 | Promotes the proper development of funicules by forming a clear abscission zone | Loss of function | Pinyopich et al., 2003 | |
| IND | Encodes a basic helix-loop-helix protein involved in patterning of the fruit cell types required for seed dispersal | Loss of function | Liljegren et al., 2004 | |
| NST 1 and NST 3 | Encodes protein which promote secondary walls synthesis in valve margins are required for dehiscence | Loss of function | Mitsuda and Ohme-Takagi, 2008 | |
| Rice | SHAT 1 | Encodes a transcription factor (APETALA2) required for the formation of abscission zone pedicel and spikelet | Loss of function | Zhou et al., 2012 |
| SSH1 | Encodes an APETALA2-like transcription factor SUPERNUMERARY BRACT required for the formation of abscission zone pedicel and spikelet | Loss of function | Jiang et al., 2019 | |
| qCSS3 | Improves seed shattering resistance | Loss of function | Tsujimura et al., 2019 | |
| OsGRF4 | Improves seed shattering resistance by differential abscission zone formation | High expression | Sun et al., 2016 | |
| Sorghum | Sh 1 | Encodes a transcription factor YAABY required for the formation of abscission zone | Loss of function | Olsen, 2012 |
| SpWRKY | Suppress the downstream cell wall biosynthesis genes to allow deposition of lignin that initiates abscission zone formation in the seed pedicel junction | Loss of function | Tang et al., 2013 | |
| Soybean | Pdh1 | Encodes a dirigent-like protein which promotes pod dehiscence by increasing the torsion of dried pod walls under low humidity | Loss of function | Funatsuki et al., 2014 |
| SHAT1-5 | promotes the significant thickening of fiber cap cells | Over expression | Dong et al., 2014 | |
| Wheat | Q | encodes a member of AP2-family transcription factor which confers the free threshing character | Ectopic expression | Simons et al., 2006 |
Environmental Control
Seed shattering in plants is strongly influenced by genotype × environmental (G × E) interactions (Liu et al., 2016). Though seed shattering is genetically controlled, the degree of shattering is influenced by the environmental conditions that plants experience during their growth (Tiwari and Bhatnagar, 1989; Thurber, 2012). Specifically, temperature, humidity and moisture appear to influence seed shattering. High temperature conditions are shown to increase seed shattering in rice (Ji et al., 2006; Thurber et al., 2010), chickpea (Cicer arietinum L.) (Van Gastel et al., 2007), birdsfoot trefoil (Lotus corniculatus L.) (Garcia-Diaz and Steiner, 2000), and soybean (Tsuchiya, 1987). Low humidity in canola (Brassica napus L.) (Tsuchiya, 1987; Gan et al., 2008) and soybean (Tiwari and Bhatnagar, 1989), high precipitation in canola (Vera et al., 2007) and soybean (Tiwari and Bhatnagar, 1989), and high wind in oilseed crops (Vera et al., 2007; Gan et al., 2008) have been shown to increase seed shattering. Conditions such as low humidity, high temperature, rapid temperature changes, wetting and drying, etc., which reduce the level of seed/pod moisture content, may ultimately induce pod shattering in soybean (Buckovic, 1952; Tsuchiya, 1987). The rate of moisture loss differs between two adjacent tissue layers of the abscission zone at the sutures, increasing the tension between the individual layers, eventually resulting in separation of the two valves of the pod leading to seed shattering (Buckovic, 1952). Some environmental conditions indirectly alter seed shattering window, by influencing physiological maturity. For instance, high temperature conditions during reproductive transition can induce early flowering (Maity et al., 2012; Pope et al., 2013), which can in turn result in early seed or fruit shedding. The enzymatic and biochemical mechanisms (discussed in section “Genetic Control”) that govern seed development and shattering are reported to be highly sensitive to environmental stresses (reviewed in Maity et al., 2016). For example, cellulase (Wang et al., 2011) and polygalacturonase (Yoshida et al., 1984), two important enzymes associated with seed shattering, are highly responsive to temperature stress. Consequently, seed shattering is influenced by environmental factors influencing at cellular levels, leading to visible phenotypic changes.
Plant acclimatization to different environments can also play a significant role on the extent of seed shattering. For example, Burton et al. (2017) indicated that seed shattering is less in early maturing crops. Elgersma et al. (1988) reported that crops with erect growth habit are prone to shedding seeds prior to harvest, because in a crop with prostrate growth habit, the seed heads are somewhat protected against wind. Plant morphological characteristics such as vascular structure, pod structure or vascular bundle size can influence seed shattering (Summers et al., 2003). Further, seed moisture content can also affect pod shattering, as shown in chickpea by Margheim et al. (2004).
Seed Shattering in Domesticated Crops
Seed shattering (or pod dehiscence in legumes, fruit shedding or spikelet shedding of grass spikes/panicles) is the first step in the process of seed dispersal (Harlan et al., 1973; Zhou et al., 2012). Seed shattering is an important weedy trait, and is a key trait that differentiates cultivated and wild plants (Onishim et al., 2007). In addition to causing grain yield loss, presence of substantial seed shattering in feral and de-domesticated populations of cultivated types can be a concern as they favor dispersal. Repeated use of weedy and wild relatives of crop cultivars as genetic resources for improving various traits in crop breeding program might have led to introgression of seed shattering gene(s) or QTL(s) in cultivated types, leading to rapid appearance of ferality and/or de-domestication (Vigueira et al., 2013). In crops, high seed retention or reduced seed shattering has always been a high priority (Hillman and Davies, 1990, 1999). This trait has been selected independently in several species across diverse geographical regions (Di Vittori et al., 2019), and is highly influenced by environmental conditions (Ji et al., 2006; Thurber, 2012).
During domestication, some plant traits have been lost, altered or accumulated over many generations such that cultivated types are genetically distinguishable from their wild ancestors. These collective changes are known as domestication syndrome (Hawkes, 1983; Hammer, 1984; Harlan, 1992). Reduced seed shattering, altered seed dispersal mechanisms, low dormancy, early maturity, decrease in seed phenol or tannin content, thick seed coat, alteration in seed size, seed color, etc. are some notable traits associated with domestication syndrome (Doebley et al., 2006). An analysis by Meyer et al. (2012) on the occurrence of important domestication syndrome traits in 203 crops found that selection for seed retention or reduced seed shattering started about 10,000 years ago. Since the beginning of domestication, seed retention has been considered a valuable trait, and consequently selection has been made against shattering over the years by farmers and plant breeders. However, seed shattering still exists in cultivated crops, contributing significantly to yield losses (Serebrenik, 2013; Table 1). Though modern crop cultivars have substantially low inherent and environment-induced (wind, rain, etc.) seed shattering, this trait could not be completely eliminated in several crops (Gepts and Debouck, 1991; Li et al., 2006; Di Vittori et al., 2019). For instance, weedy amaranths (e.g., Palmer amaranth) exhibit seed shattering (e.g., Schwartz-Lazaro et al., 2017), whereas the grain amaranths are bred as non-shattering types (Brenner, 2002). This is true for many other genera such as Helianthus (Burke et al., 2002) and Linum (Fu, 2011).
The extent of seed shattering highly varies across domesticated crop species, as influenced by the selection intensity imposed during domestication (Dong and Wang, 2015). Seed shattering has been widely studied in some plant families such as Brassicaceae [e.g., Brassica napus (Gulden et al., 2003); Arabidopsis thaliana (L.) Heynh. (Di Vittori et al., 2019)], Poaceae [e.g., rice; Oryza sativa (L.) (Vigueira et al., 2013)], and Fabaceae [e.g., French bean (Dong et al., 2014)]. Species with high fecundity levels tend to shatter a higher number of seeds (Boelt and Studer, 2010). Moreover, small-seeded biotypes are known to shatter more seed compared to large-seeded types (Sun et al., 2016). Some crop species such as range/pasture species are bred to retain some level of seed shattering to maintain a persistent seedbank for natural regeneration in long-term pastures. Moreover, seed retention is not considered a primary breeding target for forage species because it is suggested that the photosynthates required for high seed retention would reduce the volume of biomass production (Boelt and Studer, 2010; Huff, 2010; Humphreys et al., 2010), though there are exceptions (Griffiths, 1965; Saeidnia et al., 2017). However, when forage grass species are grown as annual pastures, seed shattering can be problematic since only a short pasture phase (1 to 2 years) is required or seed is to be harvested (Lemke et al., 2003). Meyer et al. (2012) estimated that seed shattering occurs at an average of 16% across different crops.
Seed Shattering in Wild and Weedy Species
Weeds have a tremendous ability to adapt to various selection pressures in agroecosystems (Charbonneau et al., 2018; Huang et al., 2018). Some of the notable adaptive traits in weedy plants include rapid growth habit, short life cycle, efficient seed dispersal and seed dormancy (Baker, 1965). Seed shattering has also been recognized as an essential adaptive trait that favors seed dispersal, seedbank establishment and weediness in many species (Constantin, 1960; Delouche et al., 2007; Burton et al., 2017). Most weeds are prolific seed producers and have the ability to distribute seed shattering over a long duration following physiological maturity (Burton et al., 2017). Seed shattering, however, greatly varies among different weed species, their biotypes and environmental conditions (Table 2). Seed shattering is genetically controlled, but is largely regulated by environmental conditions and agronomic practices (Shirtliffe et al., 2000; Walsh and Powles, 2014).
In arable weeds, seed shattering phenology can be highly variable, and is largely shaped by production practices. In mechanically harvested systems, for example, many annual weed species retain majority of their seeds till crop harvest so that the seed can be harvested and spread across the field by the harvest machinery (Walsh and Powles, 2014), which allows for the persistence of the species in crop fields for years (Shivrain et al., 2010). In many weeds, some level of seed retention at harvest may contaminate harvested crop seed, allowing for long-distance dispersal (Wilson et al., 2016). For example, Chinese sprangletop (Leptochloa chinensis L.) in northern Italy was presumed to have been introduced via contaminated rice seed from non-European countries (Benvenuti, 2004). Conversely, weeds may shatter the majority of their seed before crop harvest as an evolutionary adaptation. This adaptation can also be a direct response to harvest weed seed control (HWSC) technology in which all the seeds retained by weeds are captured at crop harvest and destroyed (Walsh et al., 2013; Walsh and Powles, 2014). It is important to note that HWSC is widely adopted only in Australia and the evidence of enhanced seed shattering as an adaptive mechanism against HWSC is still limited (Walsh et al., 2018). In this section, we specifically highlight four arable weed species that are known to exhibit high levels of shattering, to offer valuable insights on the field implications of this trait.
Shattercane
Shattercane (Sorghum bicolor) is a troublesome weed in summer row crops and is a weedy relative of cultivated sorghum (Defelice, 2006; Ohadi et al., 2018). The name shattercane derives from the habit that this race shows profuse seed shattering at physiological seed maturity stage (Defelice, 2006), even with only a slight breeze (Clark and Rosenow, 1992). Individual panicles of shattercane produce about 1,500-2,000 seeds (Roeth et al., 1994; Kegode, 1995), which typically shatter before crop harvest, ensuring seedbank replenishment before they are harvested with the main crop and removed (Dong and Wang, 2015). Kegode (1995) noted that about one-third of all biotypes of shattercane (especially the open-panicle types) drop seed when panicles mature (Kegode, 1995). The shattered seeds can remain viable in the soil seedbank for up to 13 years (Burnside et al., 1997). According to a survey conducted by Roeth et al. (1994) in Nebraska, the top four inches of soil in fields infested with shattercane contained up to 57 million viable seeds per hectare.
Weedy Rice
Weedy rice (Oryza sativa f. spontanea) is a common and troublesome weed of cultivated rice (Burgos et al., 2008). Weedy rice is morphologically very diverse and tends to shed seeds from the panicle before the harvest of cultivated rice (Gross et al., 2010; Nadir et al., 2017). Chin et al. (1999) reported a 19–56% seed shattering in weedy rice at 8–15 days after rice flowering in Vietnam. In Italy, Ferrero and Vidotto (1999) documented 65% weedy rice shattering at 30 days after rice flowering. Apart from the common weedy rice, Oryza rufipogon, a wild ancestor of cultivated rice which is native to the tropical wetlands of South Asia also shows a high degree of seed shattering (Vigueira et al., 2019). The selection pressure during the course of evolution across the world has resulted in co-evolution of modern non-shattering rice (Li et al., 2006; Di Vittori et al., 2019). However, limited efforts in maintaining the domesticated traits have sometimes culminated in the reversion of domesticated type to wild forms through de-domestication (Vigueira et al., 2013; Kanapeckas et al., 2016). For example, seed shattering in feral weedy rice was acquired during de-domestication (Kanapeckas et al., 2016). Studies on the molecular dissection of seed shattering in domesticated rice have identified different QTL such as sh3, sh4, and sh8 (Li et al., 2006; Vigueira et al., 2013).
Wild Oat
Wild oat (especially Avena fatua) is a widespread and competitive weed with a staggered germination pattern, making it a troublesome weed in major winter cereals in many parts of the world (Bullied et al., 2003; Beckie et al., 2012). Wild oats show high levels of seed shattering (Barroso et al., 2006), and seed can remain viable in the soil for up to 18 years (Gonzalez-Andujar and Perry, 1995). The extent of shattering could differ among different Avena spp. Bervillé et al. (2005) found that in A. fatua the abscission layer forms at the base of individual florets whereas in A. sterilis, the layer is developed at the base of an entire spikelet, leading to differences in shattering levels. Mahajan and Chauhan (2021) reported shattering differences between the two species in Queensland, Australia. When localized accessions of wild and cultivated Avena spp. are grown together, wild oat seeds matured faster than the cultivated crops and shattered before crop harvest (Maxwell et al., 2007). Seed shattering in wild oat appears to widely vary across geographical locations (Metz, 1969; Wilson, 1970; Feldman and Reed, 1974; Wilson and Cussans, 1975). For example, wild oat seed shattering prior to wheat harvest was reported to be at 22–20% in Saskatchewan, Canada (Burton et al., 2017), and even at 99% in the United Kingdom (Wilson, 1970). Shirtliffe et al. (2000, 2002) indicated that seed shattering pattern in wild oat can be predicted using thermal time, which can inform suitable harvest timing to maximize wild oat seed capture at harvest for subsequent destruction.
Wild Sunflower
Wild sunflower (Helianthus annuus) phenotypically resembles cultivated sunflower, but with a high potential for seed shattering and dispersal (Burke et al., 2002). Shattering in wild sunflower is augmented by the convex floral disc (i.e., high depth:width ratio) due to continued growth of the capitulum. The non-shattering crop types, in contrast, have a relatively flat head (i.e., low depth:width ratio) at maturity (Burke et al., 2002). A considerable density of volunteer sunflower plants can be found in sunflower production fields due to the presence of shattering in cultivated types, leading to yield loss (Reagon and Snow, 2006). The volunteers may arise from the shattered seeds from the same field or nearby fields, leading to competition with the cash crop and significant crop yield loss. Crop volunteers that are commonly found at field edges, alleys, etc. due to unaccounted seed shattering represent a possible channel for gene flow between the cultivated and the common wild sunflower (Massinga et al., 2003; Reagon and Snow, 2006).
Implications of Seed Shattering
Crop Improvement
Seed shattering is a detrimental trait in domesticated crops and consistent efforts have been made to eliminate this trait in breeding lines. Advances in molecular technologies have allowed us to develop an improved understanding of the genetic control of this trait in different crop species (Tables 4, 5). The identification of major QTL controlling seed shattering facilitates marker assisted selection (MAS) for improved crop lines with less shattering potential. For instance, EST-SSRs (expressed sequence tag-derived simple sequence repeats) were utilized in breeding for shattering tolerance in wild rye (Elymus nutans) (Zhao et al., 2019). The seed shattering-related genes identified in Arabidopsis and their orthologs in monocot species could be harnessed for reducing shattering potential (Dong and Wang, 2015). This approach has been utilized in a number of Brassica crops (Chandler et al., 2005; Østergaard et al., 2006; Kord et al., 2015). The successful expression of Arabidopsis genes in oilseed rape (Brassica juncea.) could be attributed to the similar genetic network governing the development of silique valve margin in both species (Østergaard et al., 2006), which remain highly conserved during evolution (Martinez-Anduijar et al., 2012).
Apart from the MAS-based approach, opportunities also exist for introducing shattering tolerance through gene editing/transgenic means. In this context, targeted gene editing technologies, particularly type II Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR)/CRISPR-associated protein 9 (Cas9) could be a potential functional genomics approach for knockdown of gene(s) governing seed shattering in crop plants (Bortesi and Fischer, 2015). A proof of concept for CRISPR-based gene editing for knocking down the ALCATRAZ (ALC) gene involved in valve margin development has been demonstrated in canola (Brassica napus) (Braatz et al., 2017). They transformed the tetraploid oilseed rape (Brassica napus) with a CRISPR-Cas9 construct targeting two ALC homoeologs and created a transgenic T1 plant with four alc mutant alleles. They did not find any wild-type alleles in the T2 generation and all the mutations were stably inherited from T1 to the T2 progeny, which proved that the T1 was a non-chimeric double heterozygote. These promising results indicate that precise nucleotide changes in genes encoding for abscission zone development and valve margin lignification could improve seed and pod shattering resistance in crop plants.
Crop Management
In crop species that lack a distinct non-shattering system, a number of agronomic and physiological interventions were tested and practiced for reducing seed shattering. In cultivated crops, the adjustment of harvest time based on seed moisture content and the development of abscission zone in reproductive parts is a primary approach to reduce grain yield loss (Silberstein et al., 2010; Shaheb et al., 2015; Xangsayasane et al., 2019). In several species such as Festulolium, adhesive preparations or film forming agents applied at the milk-ripe stage when seed moisture content is not less than 60–65% significantly reduced seed shattering (Obraztsov et al., 2018). Cutting seedheads before harvest maturity and allowing them to dry before threshing is another tactic to reduce seed shattering in a number of species such as oilseed Camelina (Sintim et al., 2016). Sweating, a variant of swathing, is the practice of placing freshly cut seedheads of grasses in heaps or under a cover for about 3 days before threshing the seeds in order to reduce seed shattering in the field (Hopkinson et al., 2003).
Various chemicals or hormones have been used in several species to reduce seed shattering. For example, Ascophyllum nodosum based biostimulant (Sealicit) has been shown to reduce pod shattering and yield loss in oilseed rape (Łangowski et al., 2019). In soybean, plant hormones such as gibberellic acid and nutrient complexes are reported to lower seed shattering (Gulluoglu et al., 2006). However, paclobutrazol, a known antagonist of the plant hormone gibberellin, is reported to improve seed yield in sesame, in part by reducing seed shattering (Mehmood et al., 2021). In birdsfoot trefoil, the use of desiccant sprays (di-n-butyl phthalate, pentachlorophenol, and endothal) were shown to reduce seed shattering (Wiggans et al., 1956).
Weed Management
The tendency of weed species to either shatter or retain their seeds until the harvest of crops that they infest has great implications for weed population dynamics and management. It is speculated that many weed species, especially in grain crops, have evolved high seed retention potential at crop harvest, which facilitates seed dispersal by harvest machinery and contamination with grain. However, a suite of technologies, collectively known as harvest weed seed control (HWSC) were developed in Australia to capture weed seed at harvest and destroy them, minimizing their dispersal into the field (Walsh et al., 2018). This way, a weed’s ability to retain a high amount of seeds at crop harvest for facilitated dispersal is utilized against them by preventing the seeds from entering the soil seedbank. The efficacy of this system is directly related to the proportion of seeds retained at crop harvest. Significant variations are observed across weed species, cropping systems and climates regarding weed seed retention levels; sowing time adjustment and early-maturing cultivars may facilitate more success with HWSC (Walsh et al., 2018). The agronomic, physiological, hormonal and chemical interventions described above (section “Crop Management”) can be utilized for manipulating seed shattering phenology in weeds and improving seed retention at harvest. However, weeds can evolve to escape HWSC tactics. Ashworth et al. (2016) showed, via recurrent selection, that Raphanus raphanistrum (wild radish) has the potential to exhibit early maturity in order to avoid harvest time weed management operations. Sun et al. (2021) further evaluated the early-flowering biotype selected by Ashworth et al. (2016) and confirmed that plants with the early-flowering phenotype retain more pods below the typical wheat harvest height.
Additionally, there are opportunities to employ genetic tools to reduce seed shattering in some of the most problematic weeds with higher seed shattering rates. For example, Yan et al. (2017) proposed a novel approach to partially silence the expression of the seed-shattering gene SH4 using artificial micro RNA and antisense RNA techniques in weedy rice. However, research efforts in this regard are very limited.
Conclusion and Future Research Needs
The productivity and economic gains in most food crops are assessed by their seed/grain yield. Besides the genetic potential of a crop to produce a high number of seeds, retention of the seed after physiological maturity till harvest is extremely important. Therefore, consistent breeding efforts have been made to minimize seed shattering in cultivated crops. However, this unique biological trait is highly prevalent in most weed species. Human-driven manipulations have minimized seed shattering in food crops, but is still present at a significant level in many crop species. Knowledge on the physiological and genetic control of seed shattering in plants is useful not only for successful weed management, but also for crop improvement. Yet, there are several unexplored aspects of this important plant trait, especially in an agricultural context. Future research should endeavor to better understand the ecology, physiology and genetics of seed shattering. In particular, seed shattering potential of various agronomically important weed species and the influence of different environmental factors need more research attention. This knowledge will help design and sustain innovative HWSC strategies. Further, potential changes to seed shattering patterns as influenced by adaptive evolution under various management and climate change scenarios warrant adequate investigation.
Author Contributions
MB conceived the manuscript. AM, AL, and DJ wrote the first draft of the manuscript. All authors edited and revised the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. Funding from the USDA-NIFA Foundational Program (Grant #2016-67013-24810) awarded to MB is gratefully acknowledged.
References
- Addicott F. T. (1970). Plant hormones in the control of abscission. Biol. Rev. 45 485–524. 10.1111/j.1469-185x.1970.tb01175.x [DOI] [Google Scholar]
- Addicott F. T., Lynch R. S. (1951). Acceleration and retardation of abscission by indoleacetic acid. Science 114 688–689. 10.1126/science.114.2974.688 [DOI] [PubMed] [Google Scholar]
- Aliboh V. O., Kehinde O. B., Fowale I. (1997). Inheritance of leaf mark, pod dehiscence and dry pod colour in crosses between wild and cultivated cowpeas. Afr. Crop Sci. J. 5 283–288. [Google Scholar]
- Andargie M., Pasquet R. S., Gowda B. S., Muluvi G. M., Timko M. P. (2011). Construction of a SSR-based genetic map and identification of QTL for domestication traits using recombinant inbred lines from a cross between wild and cultivated cowpea (V. unguiculata L.) Walp. Mol. Breed. 28 413–420. 10.1007/s11032-011-9598-2 [DOI] [Google Scholar]
- Ashworth M. B., Walsh M. J., Flower K. C., Vila-Aiub M. M., Powles S. B. (2016). Directional selection for flowering time leads to adaptive evolution in Raphanus raphanistrum (Wild radish). Evol. Appl. 9 619–629. 10.1111/eva.12350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker H. G. (1965). “Characteristics and mode of origin of weeds,” in The genetics of colonizing species, eds Baker H. G., Stebbins G. L. (New York, USA: Academic Press; ), 147–172. 10.2307/4041186 [DOI] [Google Scholar]
- Barroso J., Navarrete L., Sánchez Del Arco M. J., Fernandez-Quintanilla C., Lutman P. J. W., Perry N. H., et al. (2006). Dispersal of avena fatua and avena sterilis patches by natural dissemination, soil tillage and combine harvesters. Weed Res. 46 118–128. 10.1111/j.1365-3180.2006.00500.x [DOI] [Google Scholar]
- Baucher M., Monties B., Montagu M. V., Boerjan W. (1998). Biosynthesis and genetic engineering of lignin. Crit. Rev. Plant Sci. 1 125–197. 10.1080/07352689891304203 [DOI] [Google Scholar]
- Beckie H., Blackshaw R., Harker K. N., Tidemann B. (2017). Weed seed shatter in spring wheat in Alberta. Can. J. Plant Sci. 98 107–114. [Google Scholar]
- Beckie H. J., Francis A., Hall L. M. (2012). The biology of canadian weeds. 27. Avena fatua L. (updated). Can. J. Plant Sci. 92 1329–1357. 10.4141/cjps2012-005 [DOI] [Google Scholar]
- Bennett H. W., Marchbanks W. W. (1969). Seed drying and viability in dallisgrass. Agron. J. 61 175–177. 10.2134/agronj1969.00021962006100020001x [DOI] [Google Scholar]
- Benvenuti S. (2004). Weed dynamics in the mediterranean urban ecosystem: ecology, biodiversity and management. Weed Res. 44 341–354. 10.1111/j.1365-3180.2004.00410.x [DOI] [Google Scholar]
- Bervillé A., Breton C., Cunliffe K., Darmency H., Good G. A., Gressel J., et al. (2005). “Issues of ferality or potential for ferality in oats, olives, the vigna group, ryegrass species, safflower, and sugarcane,” in Crop ferality and volunteerism (Boca Raton: CRC Press; ), 231–255. 10.1201/9781420037999.ch15 [DOI] [Google Scholar]
- Blanco-Moreno J. M., Chamorro L., Masalles R. M., Recasens J., Sans F. X. (2004). Spatial distribution of Lolium rigidum seedlings following seed dispersal by combine harvesters. Weed Res. 44 375–387. 10.1111/j.1365-3180.2004.00412.x [DOI] [Google Scholar]
- Boelt B., Studer B. (2010). “Breeding for Grass Seed Yield in Fodder Crops and Amenity Grasses,” in Handbook of Plant Breeding, eds Boller B., Posselt U. K., Veronesi F. (New York: Springer; ), 161–174. 10.1007/978-1-4419-0760-8_7 [DOI] [Google Scholar]
- Bortesi L., Fischer R. (2015). The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 33 41–52. 10.1016/j.biotechadv.2014.12.006 [DOI] [PubMed] [Google Scholar]
- Braatz J., Harloff H. J., Mascher M., Stein N., Himmelbach A., Jung C. (2017). CRISPR-Cas9 targeted mutagenesis leads to simultaneous modification of different homoeologous gene copies in polyploid oilseed rape (Brassica napus). Plant Physiol. 174 935–942. 10.1104/pp.17.00426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner D. M. (2002). “Non-shattering grain amaranth populations,” in Trends in new crops and new uses, eds Janick J., Whipkey A. (Alexandria: ASHS Press; ), 104–106. [Google Scholar]
- Buckovic R. G. (1952). Some of the morphologic and agro-nomic factors associated with pod dehiscence in Lotus corniculatus. M.S. Thesis. Corvallis: Oregon State College Library. [Google Scholar]
- Bullied W., Marginet A., Van Acker R. C. (2003). Conventional and conservation-tillage systems influence emergence periodicity of annual weed species in canola. Weed Sci. 51 886–897. 10.1614/p2002-117 [DOI] [Google Scholar]
- Burgos N. R., Norsworthy J. K., Scott R. C., Smith K. L. (2008). Red rice (Oryza sativa) status after 5 years of imidazolinone-resistant rice technology in Arkansas. Weed Technol. 22 200–208. 10.1614/wt-07-075.1 [DOI] [Google Scholar]
- Burgos N. R., Singh V., Tseng T. M., Black H., Young N. D., Huang Z., et al. (2014). The impact of herbicide-resistant rice technology on phenotypic diversity and population structure of United States weedy rice. Plant Physiol. 66 1208–1220. 10.1104/pp.114.242719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke J. M., Tang S., Knapp S. J., Rieseberg L. H. (2002). Genetic analysis of sunflower domestication. Genetics 161 1257–1267. 10.1093/genetics/161.3.1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnside O. C., Wicks G. A., Fenster C. R. (1997). Longevity of shatter cane seed in soil across Nebraska. Weed Sci. 17 139–143. 10.1111/j.1365-3180.1977.tb00457.x [DOI] [Google Scholar]
- Burson B. L., Correa J., Potts H. C. (1978). Anatomical study of seed shattering in bahiagrass and dallisgrass. Crop Sci. 18 122–125. 10.2135/cropsci1978.0011183x001800010032x [DOI] [Google Scholar]
- Burton N. R., Beckie H. J., Willenborg C. J., Shirtliffe S. J., Schoenau J. J., Johnson E. N. (2016). Evaluating seed shatter of economically important weed species. Weed Sci. 64 673–682. 10.1614/WS-D-16-00081.1 [DOI] [Google Scholar]
- Burton N. R., Beckie H. J., Willenborg C. J., Shirtliffe S. J., Schoenau J. J., Johnson E. N., et al. (2017). Seed shatter of six economically important weed species in producer fields in Saskatchewan. Can. J. Plant Sci. 9 266–276. [Google Scholar]
- Carrie T. S., Hepler P. K., Caicedo Ana L. (2011). Timing is everything: early degradation of abscission layer is associated with increased seed shattering in US weedy rice. BMC Plant Biol. 11:14. 10.1186/1471-2229-11-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler J., Corbesier L., Spielmann P., Dettendorfer J., Stahl D., Apel K., et al. (2005). Modulating flowering time and prevention of pod shatter in oilseed rape. Mol. Breed. 15 87–94. 10.1007/s11032-004-2735-4 [DOI] [Google Scholar]
- Charbonneau A., Tack D., Lale A., Goldston J., Caple M., Conner E., et al. (2018). Weed evolution: Genetic differentiation among wild, weedy, and crop radish. Evol. Appl. 10 1964–1974. 10.1111/eva.12699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauvaux N., Child R., John K., Ulvskov P., Borkhardt B., Prinsen E., et al. (1997). The role of auxin in cell separation in the dehiscence zone of oilseed rape pods. J. Exp. Bot. 48 1423–1429. 10.1093/jxb/48.7.1423 12432039 [DOI] [Google Scholar]
- Cheng J., He Y., Zhan C., Yang B., Xu E., Zhang H., et al. (2016). Identification and characterization of quantitative trait loci for shattering in Japonica rice landrace Jiucaiqing from Taihu Lake valley, China. Plant Genom. 9 1–9. [DOI] [PubMed] [Google Scholar]
- Child R. D., Summers J. E., Babij J., Farrent J. W., Bruce D. M. (2003). Increased resistance to pod shatter is associated with changes in the vascular structure in pods of a resynthesized Brassica napus line. J. Exp. Bot. 54 1919–1930. 10.1093/jxb/erg209 [DOI] [PubMed] [Google Scholar]
- Chin V. C., Hach C. V., Thanh N. C., Tai N. T. (1999). “Weedy rice situation in Vietnam,” in Report of Global workshop on red rice control (Varadero, Cuba: Plant Production and Protection Division, FOA; ), 67–74. [Google Scholar]
- Clark L. E., Rosenow D. T. (1992). Off-type sorghum plants. College Station, TX: Texas Agricultural Experiment Station Pub. 885. [Google Scholar]
- Clarke J. M. (1981). Effect of delayed harvest on shattering losses in oats, barley and wheat. Can. J. Plant Sci. 61 25–28. 10.4141/cjps81-004 [DOI] [Google Scholar]
- Constantin M. J. (1960). Characteristics of red rice in Louisiana. [Ph.D Desertation]. Lousiana: Louisiana State University. [Google Scholar]
- Correa M. A. S. (1974). Some aspects of seed maturation in bahiagrass (Paspalum notatum Fluegge). [M.S. Thesis]. Mississippi: Mississippi State University. [Google Scholar]
- Darmency H., Pernes J. (1987). An inheritance study of domestication in foxtail millet using an interspecific cross. Plant Breed. 99 30–33. 10.1111/j.1439-0523.1987.tb01146.x [DOI] [Google Scholar]
- Darwin C. (1859). On the origin of species by means of natural selection, or the preservation of favoured races in the struggle for life. London: John Murray. [PMC free article] [PubMed] [Google Scholar]
- Davidson D. (2014). How to prevent soybeans from shattering during harvest. Available online at: http://www.ilsoyadvisor.com (accessed December 15, 2020) [Google Scholar]
- Defelice M. S. (2006). Shattercane, Sorghum Bicolor (L.) Moench Ssp. Drummondii (Nees ex Steud.) De Wet ex Davidse— black sheep of the family. Weed Tech. 20 1076–1083. 10.1614/wt-06-051.1 [DOI] [Google Scholar]
- Delouche J. C., Burgos N. R., Gealy D. R., Zorrilla de San M. G., Labrada R., Larinde M., et al. (2007). Weedy rices- origin, biology, ecology and control. FAO Plant production and protection paper-188. Rome, Italy: Food and Agriculture Organization. [Google Scholar]
- Di Vittori V., Gioia T., Rodriguez M., Bellucci E., Bitocchi E., Nanni L., et al. (2019). Convergent evolution of the seed shattering trait. Genes 10:68. 10.3390/genes10010068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J. F., Gaut B. S., Smith B. D. (2006). The molecular genetics of crop domestication. Cell 12 1309–1321. 10.1016/j.cell.2006.12.006 [DOI] [PubMed] [Google Scholar]
- Dong Y., Wang Y. Z. (2015). Seed shattering: from models to crops. Front. Plant Sci. 6:476. 10.3389/fpls.2015.0047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y., Yang X., Liu J., Wang B. H., Liu B. L., Wang Y. Z. (2014). Pod shattering resistance associated with domestication is mediated by a NAC gene in soybean. Nat. Commun. 5:3352. [DOI] [PubMed] [Google Scholar]
- Ejeta G., Grenier C. (2005). “Sorghum and its weedy hybrids,” in Crop Ferality and Volunteerism, ed. Gressel J. (Boca Raton, FL: CRC Press; ), 123–135. 10.1201/9781420037999.ch8 [DOI] [Google Scholar]
- Elgersma A., Leeuwangh J. E., Wilms H. J. (1988). Abscission and seed shattering in perennial ryegrass (Lolium perenne L.). Euphytica 39 51–57. 10.2478/v10129-011-0046-6 [DOI] [Google Scholar]
- Feldman M., Reed W. B. (1974). “Distribution of wild oat seeds during cereal crop swathing and combining,” in Proceedings of 1974 Annual Meeting of Canadian Society of Agricultural Engineering (Sainte Foy, Canada: Laval University; ), 303. [Google Scholar]
- Ferrándiz C. (2002). Regulation of fruit dehiscence in Arabidopsis. J. Expt. Bot. 53 2031–2038. 10.1093/jxb/erf082 [DOI] [PubMed] [Google Scholar]
- Ferrándiz C., Fourquin C. (2014). Role of the FUL-SHP network in the evolution of fruit morphology and function. J. Expt. Bot. 65 4505–4513. 10.1093/jxb/ert479 [DOI] [PubMed] [Google Scholar]
- Ferrándiz C., Pelaz S., Yanofsky M. F. (1999). Control of carpel and fruit development in Arabidopsis. Annu. Rev. Biochem. 68 321–354. 10.1146/annurev.biochem.68.1.321 [DOI] [PubMed] [Google Scholar]
- Ferrero A., Vidotto F. (1999). “Red rice control in rice pre-and post-planting,” in Report of Global workshop on red rice control (Varadero, Cuba: Plant Production and Protection Division, FAO; ), 155. [Google Scholar]
- Flint-Garcia S. A. (2013). Genetics and consequences of crop domestication. J. Agric. Food Chem. 61 8267–8276. 10.1021/jf305511d [DOI] [PubMed] [Google Scholar]
- Forcella F., Peterson D. H., Barbour J. C. (1996). Timing and measurement of weed seed shed in corn (Zea mays). Weed Technol. 10 535–543. 10.1017/s0890037x00040409 [DOI] [Google Scholar]
- Fourquin C., Cerro C., Victoria F. C., Vialette-Guiraud A., Oliveira A. C., Ferrandiz C. (2013). A change in SHATTERPROOF protein lies at the origin of a fruit morphological novelty and a new strategy for seed dispersal in Medicago genus. Plant Physiol. 162 907–917. 10.1104/pp.113.217570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y. B. (2011). Genetic evidence for early flax domestication with capsular dehiscence. Genet. Resour. Crop Evol. 58 1119–1128. 10.1007/s10722-010-9650-9 [DOI] [Google Scholar]
- Fu Z., Song J., Zhao J., Jameson P. E. (2018). Identification and expression of genes associated with the abscission layer controlling seed shattering in Lolium perenne. AoB Plants 11:ly076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller D., Qin L. (2008). Immature rice and its archaeobotanical recognition: a reply to Pan. Antiquity 82:316. [Google Scholar]
- Fuller D. Q., Allaby R. (2009). “Seed dispersal and crop domestication: shattering, germination and seasonality in evolution under cultivation,” in Annual Plant Reviews, Fruit development and seed dispersal, ed. Ostergaard L. (Hoboken, NJ: John Wiley & Sons, Inc; ), 354. [Google Scholar]
- Fuller D. Q., Qin L., Zheng Y., Zhao Z., Chen X., Hosoya L. A., et al. (2009). The domestication process and domestication rate in rice: spikelet bases from the Lower Yangtze. Science 323 1607–1610. 10.1126/science.1166605 [DOI] [PubMed] [Google Scholar]
- Funatsuki H., Ishimoto M., Tsuji H., Kawaguchi K., Hajika M., Fujino K. (2006). Simple sequence repeat markers linked to a major QTL controlling pod shattering in soybean. Plant. Breed. 125 195–197. 10.1111/j.1439-0523.2006.01199.x [DOI] [Google Scholar]
- Funatsuki H., Suzuki M., Hirose A., Inaba H., Yamada T., Hajika M., et al. (2014). Molecular basis of a shattering resistance boosting global dissemination of soybean. Proc. Nat. Acad. Sci. USA 111 17797–17802. 10.1073/pnas.1417282111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan Y., Malhi S. S., Brandt S. A., McDonald C. L. (2008). Assessment of seed shattering resistance and yield loss in five oilseed crops. Can. J. Plant Sci. 88 267–270. 10.4141/cjps07028 [DOI] [Google Scholar]
- Garcia-Diaz C. A., Steiner J. J. (2000). Birdsfoot trefoil seed production: III. Seed shatter and optimal harvest time. Crop Sci. 40 457–462. 10.2135/cropsci2000.402457x [DOI] [Google Scholar]
- Gasser C. S., Simon M. K. (2011). Seed dispersal: same gene, different organs. Curr. Bio. 21 R546–R548. [DOI] [PubMed] [Google Scholar]
- Gepts P., Debouck D. G. (1991). “Origin, domestication, and evolution of the common bean (Phaseolus vulgaris L.),” in Common Beans: Research for Crop Improvement, eds Voysest O., Van Schoonhoven A. (Oxon, UK: CABI; ), 7–53. [Google Scholar]
- Gonzalez-Andujar J. L., Perry J. N. (1995). Models for the herbicidal control of the seed bank of Avena sterilis: the effects of spatial and temporal heterogeneity and of dispersal. J. Appl. Ecol. 32 578–587. 10.2307/2404654 [DOI] [Google Scholar]
- González-Carranza Z. H., Lozoya-Gloria E., Roberts J. A. (1998). Recent developments in abscission: shedding light on the shedding process. Trends. Plant. Sci. 3 10–14. 10.1016/s1360-1385(97)01132-1 [DOI] [Google Scholar]
- Goplen J. J., Sheafier C. C., Becker R. L., Coulter J. A., Breitenbach F. R., Behnken L. M., et al. (2016). Giant ragweed (Ambrosia trifida) seed production and retention in soybean and field Margins. Weed Technol. 30 246–253. 10.1614/wt-d-15-00116.1 [DOI] [Google Scholar]
- Green J. K., Norsworthy J. K., Palhano M. G., Meyer C. J., Martin S. M., Schwartz L. M. (2016). Seed retention of Palmer amaranth and barnyard grass in soybean. San Juan, PR. Available online at: http://wssaabstracts.com/public/38/proceedings.html. (accessed December 19, 2020) [Google Scholar]
- Gregory T. R. (2009). Artificial selection and domestication: modern lessons from Darwin’s enduring analogy. Evolution (N. Y.) 2:27. [Google Scholar]
- Griffiths D. J. (1965). Breeding for higher seed yields from herbage varieties. J. Nat. Inst. Agric. Bot. 10 320–331. [Google Scholar]
- Gross B. L., Reagon M., Hsu S. C., Caicedo A. L., Jia Y., Olsen K. M., et al. (2010). Seeing red: the origin of grain pigmentation in US weedy rice. Mol. Ecol. 19 3380–3393. 10.1111/j.1365-294x.2010.04707.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulden R. H., Shirtliffe S. J., Thomas A. J. (2003). Harvest losses of canola (Brassica napus) cause large seed bank inputs. Weed Sci. 51 83–86. 10.1614/0043-1745(2003)051[0083:hlocbn]2.0.co;2 [DOI] [Google Scholar]
- Gulluoglu L., Arioglu H., Arslan M. (2006). Effects of some plant growth regulators and nutrient complexes on pod shattering and yield losses of soybean under hot and dry conditions. Asian J. Plant Sci. 5 368–372. 10.3923/ajps.2006.368.372 [DOI] [Google Scholar]
- Hammer K. (1984). Das Domestikation ssyndrom. Kulturpflanze. 32 11–34. [Google Scholar]
- Haring S. C., Flessner M. L., Everman W. J., Mirsky S. B. (2017). The role of late-season weather events on seed shattering of redroot pigweed (Amaranthus retroflexus) and common ragweed (Ambrosia artemisiifolia). Weed Sci. Soc. 5:175. [Google Scholar]
- Harlan J. R. (1992). Crops and ancient man. Madison: American Society for Agronomy. [Google Scholar]
- Harlan J. R., De Wet J. M. J., Price E. G. (1973). Comparative evolution of cereals. Evolution 27 311–325. 10.2307/2406971 [DOI] [PubMed] [Google Scholar]
- Harlan J. R., DeWet J. M. (1965). Some thoughts about weeds. Eco. Bot. 19 16–24. 10.1007/bf02971181 [DOI] [Google Scholar]
- Hawkes J. G. (1983). The diversity of crop plants. Cambridge, MA: Harvard University Press. [Google Scholar]
- Heisler M., Atkinson A., Bylstra Y., Walsh R., Smyth D. (2001). SPATULA, a gene that controls development of carpel margin tissues in Arabidopsis, encodes a bHLH protein. Development 128 1089–1098. 10.1242/dev.128.7.1089 [DOI] [PubMed] [Google Scholar]
- Henderson J., Davies H. A., Heyes S. J., Osborne D. J. (2001). The study of a monocotyledon abscission zone using microscopic, chemical, enzymatic and solid state 13C CP/MAS NMR analyses. Phytochemistry 56 131–139. 10.1016/s0031-9422(00)00447-7 [DOI] [PubMed] [Google Scholar]
- Hillman G. C., Davies M. S. (1990). Domestication rates in wild wheats and barley under primitive cultivation. Biol. J. Linn. Soc. 39 39–78. 10.1111/j.1095-8312.1990.tb01611.x [DOI] [Google Scholar]
- Hillman G. C., Davies M. S. (1999). “Domestication rate in wild wheats and barley under primitive cultivation: preliminary results and archaeological implications of field measurements of selection coefficient,” in Prehistory of Agriculture. New Experimental and Ethnographic Approaches, ed. Anderson P. C. (Los Angeles: University of California; ), 70–102. 10.2307/j.ctvhhhg2j.14 [DOI] [Google Scholar]
- Hopkinson J. M., English B. H., Harty R. L. (2003). Sweating of panicoid tropical pasture grass seeds. Seed Sci. Tech. 31 367–377. 10.15258/sst.2003.31.2.13 [DOI] [Google Scholar]
- Htun T. M., Inoue C., Chhourn O., Ishii T., Ishikawa R. (2014). Effect of quantitative trait loci for seed shattering on abscission layer formation in Asian wild rice Oryza rufipogon. Breed Sci. 64 199–205. 10.1270/jsbbs.64.199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z., Kelly S., Matsuo R., Li L.-F., Olsen K. M., Jia Y., et al. (2018). The role of standing variation in the evolution of weediness traits in South Asian weedy rice (Oryza spp.). G3 (Bethesda) 8 3679–3690. 10.1534/g3.118.200605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huff D. R. (2010). “Bluegrasses,” in Fodder crops and amenity grasses. Handbook of plant breeding, eds Boiler B., Veronesi F., Posselt U. (New York: Springer; ), 345–379. 10.1007/978-1-4419-0760-8_15 [DOI] [Google Scholar]
- Humphreys M., Feuerstein U., Vandewalle M., Baert J. (2010). “Ryegrasses,” in Fodder crops and amenity grasses. Handbook of plant breeding, eds Boiler B., Veronesi F., Posselt U. (New York: Springer; ), 211–260. 10.1007/978-1-4419-0760-8_10 [DOI] [Google Scholar]
- Ji H. S., Chu S. H., Jiang W., Cho Y. I., Hahn J. H., Eun M. Y., et al. (2006). Characterization and mapping of a shattering mutant in rice that corresponds to a block of domestication genes. Genetics 173 995–1005. 10.1534/genetics.105.054031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Ma X., Zhao S., Tang Y., Liu F., Gu P., et al. (2019). The APETALA2-like transcription factor supernumerary bract controls rice seed shattering and seed size. Plant cell 31 17–36. 10.1105/tpc.18.00304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaga A., Isemura T., Tomooka N., Vaughan D. A. (2008). The genetics of domestication of the azuki bean (Vigna angularis). Genetics 178 1013–1036. 10.1534/genetics.107.078451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanapeckas K. L., Vigueira C. C., Ortiz A., Gettler K. A., Burgos N. R., Fischer A. J., et al. (2016). Escape to ferality: The endoferal origin of weedy rice from crop rice through de-domestication. PLoS One 11:e0162676. 10.1371/journal.pone.0162676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantar M. B., Nashoba A. R., Anderson J. E., Blackman B. K., Rieseberg L. H. (2017). The genetics and genomics of plant domestication. Bioscience 67 971–982. [Google Scholar]
- Kegode G. O. (1995). Germination ecology of shattercane (Sorghum bicolor (L.) Moench) and giant foxtail (Setaria faberi Herrm). [Ph.D dissertation]. Iowa,USA: Iowa State University. [Google Scholar]
- Konishi S., Izawa T., Lin S. Y., Ebana K., Fukuta Y., Sasaki T., et al. (2006). An SNP caused loss of seed shattering during rice domestication. Science 312 1392–1396. 10.1126/science.1126410 [DOI] [PubMed] [Google Scholar]
- Kord H., Shakib A. M., Daneshvar M. H., Azadi P., Bayat V., Mashayekhi M., et al. (2015). RNAi-mediated down-regulation of SHATTERPROOF gene in transgenic oilseed rape. 3 Biotech. 5 271–277. 10.1007/s13205-014-0226-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladizinsky G. (1985). Founder effect in crop-plant evolution. Econ. Bot. 39 191–199. 10.1007/bf02907844 [DOI] [Google Scholar]
- Lang H., He Y., Sun J., Li F., Ma D. (2021). Integrative hormone and transcriptome analysis underline the role of abscisic acid in seed shattering of weedy rice. Plant Growth Regul. 2021:e00714–18. 10.1007/s10725-021-00714-8 [DOI] [Google Scholar]
- Ł;angowski Ł, Goñi O., Quille P., Stephenson P., Carmody N., Feeney E., et al. (2019). A plant biostimulant from the seaweed Ascophyllum nodosum (Sealicit) reduces podshatter and yield loss in oilseed rape through modulation of IND expression. Sci. Rep. 9:16644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson S. R., Kellogg E. A. (2009). Genetic dissection of seed production traits and identification of major-effect seed retention QTL in hybrid Leymus (Triticeae) wildryes. Crop Sci. 49 29–40. 10.2135/cropsci2008.05.0277 [DOI] [Google Scholar]
- Lee G. H., Kang I. K., Kim K. M. (2016). Mapping of novel QTL regulating grain shattering using doubled haploid population in rice (Oryza sativa L.). Int. J. Genom. 2:128010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Yoon T. H., Lee J., Jeon S. Y., Lee J. H., Lee M. K., et al. (2018). A lignin molecular brace controls precision processing of cell walls critical for surface integrity in Arabidopsis. Cell 173 1468–1480. 10.1016/j.cell.2018.03.060 [DOI] [PubMed] [Google Scholar]
- Lemke B. M., Gibson L. R., Knapp A. D., Dixon P. M., Moore K. J., Hintz R. (2003). Maximizing seed production in eastern gamagrass. Agron. J. 95 863–869. 10.2134/agronj2003.0863 [DOI] [Google Scholar]
- Lenski R. E. (2017). What is adaptation by natural selection? Perspectives of an experimental microbiologist. PLoS Genet. 13:e1006668. 10.1371/journal.pgen.1006668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C. B., Zhou A. L., Sang T. (2006). Rice domestication by reducing shattering. Science 311 1936–1939. 10.1126/science.1123604 [DOI] [PubMed] [Google Scholar]
- Li F., Numa H., Hara N., Sentoku N., Ishii T., Fukuta Y., et al. (2019). Identification of a locus for seed shattering in rice (Oryza sativa L.) by combining bulked segregant analysis with whole-genome sequencing. Mol. Breed. 39:36. [Google Scholar]
- Liljegren S. J., Ditta G. S., Eshed Y., Savidge B., Bowman J., Yanofsky M. F. (2000). SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404 766–770. 10.1038/35008089 [DOI] [PubMed] [Google Scholar]
- Liljegren S. J., Roeder A. H., Kempin S. A., Gremski K., Østergaard L., Guimil S., et al. (2004). Control of fruit patterning in Arabidopsis by INDEHISCENT. Cell 116 843–853. 10.1016/s0092-8674(04)00217-x [DOI] [PubMed] [Google Scholar]
- Lin Z., Li X., Shannon L. M., Yeh C. T., Wang M. L., Bai G., et al. (2012). Parallel domestication of the Shattering1 genes in cereals. Nat. Genet. 44 720–724. 10.1038/ng.2281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Wang J., Wang H., Wang W., Zhou R., Mei D., et al. (2016). Multigenic control of pod shattering resistance in Chinese rapeseed germplasm revealed by genome-wide association and linkage analyses. Front. Plant. Sci. 7:1058. 10.3389/fpls.2016.01058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo S., Muñoz-Amatriaín M., Boukar O., Herniter I., Cisse N., Guo Y.-N., et al. (2018). Identification of QTL controlling domestication-related traits in cowpea (Vigna unguiculata L. Walp). Sci. Rep. 8:6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love H. H., Craig W. T. (1919). The synthetic production of wild wheat forms. J. Hered. 10 51–64. 10.1093/oxfordjournals.jhered.a101882 [DOI] [Google Scholar]
- Lv S., Wu W., Wang M., Meyer R. S., Ndjiondjop M. N., Tan L., et al. (2018). Genetic control of seed shattering during African rice domestication. Nat. Plants 4 331–337. 10.1038/s41477-018-0164-3 [DOI] [PubMed] [Google Scholar]
- Mahajan G., Chauhan B. S. (2021). Interference of wild oats (Avena fatua) and sterile oats [Avena sterilis ssp. ludoviciana (Durieu)] in wheat. Weed Sci. 2021 1–20. 10.1017/wsc.2021.25 [DOI] [Google Scholar]
- Maity A., Chakrabarty S. K., Yadav J. B. (2012). Standardisation of hybrid seed production technology of first Indian mustard (Brassica juncea) hybrid NRCHB 506. Indian J. Agric. Sci. 82 753–758. [Google Scholar]
- Maity A., Singh V., Martins M. B., Ferreira P. J., Smith G. R., Bagavathiannan M. (2021). Species identification and morphological trait diversity assessment in ryegrass (Lolium perenne ssp. multiflorum) populations from texas blacklands. Weed Sci. 2021 1–45. 10.1017/wsc.2021.18 [DOI] [Google Scholar]
- Maity A., Vijay D., Mukherjee A., Lamichaney A. (2016). “Potential Impacts of Climate Change on Quality Seed Production: A Perspective of Hill Agriculture,” in Conservation Agriculture: An approach to combat climate change in Indian Himalaya, eds Bisht J. K., Meena V. S., Mishra P. K., Pattanayak A. (Singapore: Springer Science + Business Media; ), 459–485. 10.1007/978-981-10-2558-7_18 [DOI] [Google Scholar]
- Marciniak K., Kucko A., Wilmowicz E., Swidzinski M., Przedniczek K., Kopcewicz J. (2018). Gibberellic acid affects the functioning of the flower abscission zone in Lupinus luteus via cooperation with the ethylene precursor independently of abscisic acid. J. Plant Physiol. 229 170–174. 10.1016/j.jplph.2018.07.014 [DOI] [PubMed] [Google Scholar]
- Margheim J. F., Baltensperger D. D., Wilson R. G., Lyon D. J., Hein G. L., Harveson R. M., et al. (2004). Chickpea Production in the High Plains. Lincoln, NE: University of Nebraska-Lincoln. [Google Scholar]
- Martinez-Anduijar C., Martin R. C., Nonogaki H. (2012). Seed traits and genes important for translational biology–highlights from recent discoveries. Plant Cell Physiol. 53 5–15. 10.1093/pcp/pcr112 [DOI] [PubMed] [Google Scholar]
- Martinez A. L., Vera A. (2009). “Arabidopsis fruit development,” in The Fruit Development and Seed Dispersal, ed. Ostergaard L. (Hoboken, NY: Blackwell Publishing Ltd; ), 172–203. 10.1002/9781444314557.ch5 [DOI] [Google Scholar]
- Massinga R. A., Al-Khatib K., Amand P. St, Miller P. S. (2003). Gene flow from imidazolinone-resistant domesticated sunflower to wildrelatives. Weed Sci. 51 854–862. 10.1614/ws-03-032r [DOI] [Google Scholar]
- Maxwell B. D., Smith R. G., Brelsford M. (2007). Wild oat (Avena fatua) seed bank dynamics in transition to organic wheat production systems. Weed Sci. 55 212–217. 10.1614/ws-06-179.1 [DOI] [Google Scholar]
- McWilliam J. R. (1980). “The development and significance of seed retention in grasses,” in Seed Production, ed. Hebblethwaite P. D. (London: Butterworths; ), 51–60. [Google Scholar]
- Mehmood M. Z., Qadir G., Afzal O., Ud Din A. M., Raza M. A., Khan I., et al. (2021). Paclobutrazol improves sesame yield by increasing dry matter accumulation and reducing seed shattering under rainfed conditions. Int. J. Plant Prod. 2021:132. 10.1007/s42106-021-00132-w [DOI] [Google Scholar]
- Meir S., Philosoph-Hadas S., Sundaresan S., Selvaraj K. S., Burd S., Ophir R., et al. (2010). Microarray analysis of the abscission-related transcriptome in the tomato flower abscission zone in response to auxin depletion. Plant Physiol. 154 1929–1956. 10.1104/pp.110.160697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz R. (1969). Causes of the increasing spread of wild oats (Avena fatua) and some field hygiene measures for destroying or eliminating wild oat seeds. NachBL. Dt. PflSchutzdienst. Berl. 24 85–88. [Google Scholar]
- Meyer R. S., DuVal A. E., Jensen H. R. (2012). Patterns and processes in crop domestication: an historical review and quantitative analysis of 203 global food crops. New Phytol. 196 29–48. 10.1111/j.1469-8137.2012.04253.x [DOI] [PubMed] [Google Scholar]
- Mitsuda N., Ohme-Takagi M. (2008). NAC transcription factors NST1 and NST3 regulate pod dehiscence in a partially redundant manner by promoting secondary wall formation after the establishment of tissue identity. Plant J. 56 768–778. 10.1111/j.1365-313x.2008.03633.x [DOI] [PubMed] [Google Scholar]
- Mongkolporn O., Kadkol G. P., Pang C. K., Taylor P. W. J. (2003). Identification of RAPD markers linked to recessive genes conferring siliqua shatter resistance in Brassica rapa. Plant Breed. 122 479–484. 10.1046/j.0179-9541.2003.00910.x [DOI] [Google Scholar]
- Murgia M. L., Attene G., Rodriguez M., Bitocchi E., Bellucci E., Fois D., et al. (2017). A comprehensive phenotypic investigation of the ‘pod-shattering syndrome’ in common bean. Front. Plant Sci. 8:215. 10.3389/fpls.2017.00251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadir S., Xiong H. B., Zhu Q., Zhang X. L., Xu H. Y., Li J., et al. (2017). Weedy rice in sustainable rice production. A review. Agron. Sust. Dev. 37:46. [Google Scholar]
- Nakano T., Ito Y. (2013). Molecular mechanisms controlling plant organ abscission. Plant Biotechnol. 30 209–216. 10.5511/plantbiotechnology.13.0318a [DOI] [Google Scholar]
- Niruntrayakul S., Rerkasem B., Jamjod S. (2009). Crossability between cultivated rice (Oryza sativa) and common wild rice (O. rufipogon) and characterization of F1 and F2 populations. Sci. Asia 35 161–169. [Google Scholar]
- Obraztsov V., Shchedrina D., Kadyrov S. (2018). Film agents as an effective means of reducing seed shattering in Festulolium. Agron. Res. 16 2130–2136. [Google Scholar]
- Ohadi S., Matthew L., Mohsen M., William R., Muthukumar B. (2018). Surveying the spatial distribution of feral sorghum (Sorghum bicolor L.) and its sympatry with johnsongrass (S. halepense) in South Texas. PLoS One 13:e0200984. 10.1371/journal.pone.0200984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen K. (2012). One gene’s shattering effects. Nat. Genet. 44 616–617. 10.1038/ng.2289 [DOI] [PubMed] [Google Scholar]
- Onishim K., Kyoko T., Manabu K., Takumi T., Yoshio S. (2007). Different patterns of genealogical relationships found in the two major QTLs causing reduction of seed shattering during rice domestication. Genome 50 757–766. 10.1139/g07-051 [DOI] [PubMed] [Google Scholar]
- Østergaard L., Kempin S. A., Bies D., Klee H. J., Yanofsky M. F. (2006). Pod shatter-resistant Brassica fruit produced by ectopic expression of the FRUITFULL gene. Plant Biotechnol. J. 4 45–51. 10.1111/j.1467-7652.2005.00156.x [DOI] [PubMed] [Google Scholar]
- Paterson A. H., Lin Y. R., Li Z., Schertz K. F., Doebley J. F., Pinson S. R. M., et al. (1995). Convergent domestication of cereal crops by independent mutations at corresponding genetic loci. Science 269 1714–1718. 10.1126/science.269.5231.1714 [DOI] [PubMed] [Google Scholar]
- Pfeiffer H. (1928). “Die Pflanzlichen Trennungsgewebe,” in Handbuch der Pflanzenanatomie, ed. Linsbauer K. (Berlin: Gebrüder Borntraeger; ), 236. [Google Scholar]
- Philbrook B., Oplinger E. S. (1989). Soybean field losses as influenced by harvest delays. Agron. J. 81 251–258. 10.2134/agronj1989.00021962008100020023x [DOI] [Google Scholar]
- Pickersgill B. (2007). Domestication of plants in the Americas: insights from Mendelian and molecular genetics. Ann. Bot. 100 925–940. 10.1093/aob/mcm193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinyopich A., Ditta G. S., Savidge B., Liljegren S. J., Baumann E., Wisman E., et al. (2003). Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424 85–88. 10.1038/nature01741 [DOI] [PubMed] [Google Scholar]
- Platt A. W., Wells S. A. (1949). Shattering, breaking and threshability in barley varieties. Sci. Agric. 292 453–464. [Google Scholar]
- Poncet V., Lamy F., Enjalbert J., Joly H., Sarr A., Robert T. (1998). Genetic analysis of the domestication syndrome in pearl millet (Pennisetum glaucum L., Poaceae): inheritance of the major characters. Heredity 81 648–658. 10.1046/j.1365-2540.1998.00445.x [DOI] [Google Scholar]
- Pope K. S., Dose V., Silva D. D., Brown P. H., Leslie C. A., Dejong T. M. (2013). Detecting nonlinear response of spring phenology to climate change by Bayesian analysis. Glob. Chang. Biol. 19 1518–1525. 10.1111/gcb.12130 [DOI] [PubMed] [Google Scholar]
- Price J. S., Hobson R. N., Neale M. A., Bruce D. M. (1996). Seed losses in commercial harvesting of oilseed rape. J. Agric. Eng. Res. 65 183–191. 10.1006/jaer.1996.0091 [DOI] [Google Scholar]
- Rajani S., Sundaresan V. (2001). The Arabidopsis myc/bHLH gene ALCATRAZ enables cell separation in fruit dehiscence. Curr. Biol. 11 1914–1922. 10.1016/s0960-9822(01)00593-0 [DOI] [PubMed] [Google Scholar]
- Raman H., Raman R., Kilian A., Detering F., Carling J., Coombes N., et al. (2014). Genome-wide delineation of natural variation for pod shatter resistance in Brassica napus. PLoS One 9:e101673. 10.1371/journal.pone.0101673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman R., Qiu Y., Coombes N., Song J., Kilian A., Raman H. (2017). Molecular diversity analysis and genetic mapping of pod shatter resistance loci in Brassica carinata L. Front. Plant Sci. 8:1765. 10.3389/fpls.2017.01765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reagon M., Snow A. (2006). Cultivated Helianthus annuus (Asteraceae) volunteers as a genetic “bridge” to weedy sunflower populations in north America. Am. J. Bot. 93 127–133. 10.3732/ajb.93.1.127 28924508 [DOI] [Google Scholar]
- Roberts J. A., Elliott K. A., Gonzalez-Carranza Z. H. (2002). Abscission, dehiscence, and other cell separation processes. Annu. Rev. Plant Biol. 53 131–158. [DOI] [PubMed] [Google Scholar]
- Roeder A. H. K., Ferrandiz C., Yanofsky M. F. (2003). The role of the REPLUMLESS homeodomain protein in patterning the Arabidopsis fruit. Curr. Biol. 13 1630–1635. 10.1016/j.cub.2003.08.027 [DOI] [PubMed] [Google Scholar]
- Roeth F., Martin A., Klein R. N. (1994). G94-1205 Shattercane and its control. Historical materials from University of Nebraska-Lincoln Extension. Lincoln, NE: University of Nebraska-Lincoln, 1503. [Google Scholar]
- Romkaew J., Nagaya Y., Goto M., Suzuki K., Umezaki T. (2008). Pod dehiscence in relation to chemical components of pod shell in soybean. Plant Prod. Sci. 11 278–282. 10.1626/pps.11.278 33707419 [DOI] [Google Scholar]
- Ros B. A. (1997). Lignification in plant cell walls. Int. Rev. Cytol. 176 87–132. 10.1016/s0074-7696(08)61609-5 [DOI] [PubMed] [Google Scholar]
- Saeidnia F., Majidi M. M., Mirlohi A. (2017). Analysis of seed production and its association with forage production and agronomic traits in orchardgrass (Dactylis glomerata) under different moisture conditions. Crop Pasture Sci. 68 657–669. 10.1071/cp17115 [DOI] [Google Scholar]
- Schwartz L. M., Norsworthy J. K., Young B. G., Bradley K. W., Kruger G. R., Davis V. M., et al. (2016). Tall waterhemp (Amaranthus tuberculatus) and Palmer amaranth (Amaranthus palmeri) seed production and retention at soybean maturity. Weed Technol. 30 284–290. 10.1614/wt-d-15-00130.1 [DOI] [Google Scholar]
- Schwartz-Lazaro L. M., Green J. K., Norsworthy J. K. (2017). Seed retention of Palmer amaranth (Amaranthus palmeri) and barnyardgrass (Echinochloa crusgalli) in soybean. Weed Technol. 31 617–622. 10.1017/wet.2017.25 [DOI] [Google Scholar]
- Senda T., Hiraoka Y., Tominaga T. (2006). Inheritance of seed shattering in Lolium temulentum and L. persicum hybrids. Genet. Resour. Crop Evol. 53 449–451. 10.1007/s10722-005-6096-6 [DOI] [Google Scholar]
- Serebrenik A. (2013). Genetic control of seed shattering in the model C4 grass, Setaria viridis, using transcription activator-like effector nucleases. Available online at: http://hdl.handle.net/11299/150158 (accessed January 20, 2021). [Google Scholar]
- Seymour G. B., Ostergaard L., Chapman N. H., Knapp S., Martin C. (2013). Fruit development and ripening. Annu. Rev. Plant Biol. 64 219–241. [DOI] [PubMed] [Google Scholar]
- Shaheb M., Islam M., Nessa A., Hossain M. (2015). Effect of harvest times on the yield and seed quality of French bean. SAARC J. Agric. 13 1–13. 10.3329/sja.v13i1.24175 [DOI] [Google Scholar]
- Sharma H. C., Waines J. G. (1980). Inheritance of tough rachis in crosses of Triticum monococcum and T. boeoticum. J. Hered. 71 214–216. 10.1093/oxfordjournals.jhered.a109352 [DOI] [Google Scholar]
- Shirtliffe S. J., Entz M. H., Acker R. C. V. (2000). Avena fatua development and seed shatter as related to thermal time. Weed Sci. 48 555–560. 10.1614/0043-1745(2000)048[0555:afdass]2.0.co;2 [DOI] [Google Scholar]
- Shirtliffe S. J., Kenkel N. C., Entz M. H. (2002). Fractal analysis of seed dispersal and spatial pattern in wild oats. Comm. Ecol. 3 101–107. 10.1556/comec.3.2002.1.12 [DOI] [Google Scholar]
- Shivrain V. K., Burgos N. R., Agrama H. A., Lawton-Raug H., Lu B., Sales M. A., et al. (2010). Genetic diversity of weedy red rice (Oryza sativa) in Arkansas, USA. Weed Res. 50 289–302. [Google Scholar]
- Silberstein T. B., Mark E. M., Thomas G. C., William C. Y. I. I. I. (2010). Using Seed Moisture as a Harvest Management Tool. Corvallis, OR: Oregan State University, 1–8. [Google Scholar]
- Simons K. J., Fellers J. P., Trick H. N., Zhang Z., Tai Y. S., Gill B. S., et al. (2006). Molecular characterization of the major wheat domestication gene Q. Genetics 172 547–555. 10.1534/genetics.105.044727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sintim H. Y., Zheljazkov V. D., Obour A. K., Garcia y Garcia A. (2016). Managing harvest time to control pod shattering in oilseed camelina. Agron. J. 108 656–661. 10.2134/agronj2015.0300 [DOI] [Google Scholar]
- Soni N., Nissen S. J., Westra P., Norsworthy J. K., Walsh M. J., Gaines T. A. (2020). Seed retention of winter annual grass weeds at winter wheat harvest maturity shows potential for harvest weed seed control. Weed Technol. 34 266–271. 10.1017/wet.2019.108 [DOI] [Google Scholar]
- Suanum W., Somta P., Kongjaimun A., Yimram T., Kaga A., Tomooka N., et al. (2016). Co-localization of QTLs for pod fiber content and pod shattering in F2 and backcross populations between yardlong bean and wild cowpea. Mol. Breed. 36 1–11. [Google Scholar]
- Subudhi P. K., Singh P. K., DeLeon T., Parco A., Karan R., Biradar H., et al. (2014). Mapping of seed shattering Loci provides insights into origin of weedy rice and rice domestication. J. Hered. 105 276–287. 10.1093/jhered/est089 [DOI] [PubMed] [Google Scholar]
- Summers J. E., Bruce D. M., Vancanneyt G., Redig P., Werner C. P., Morgan C., et al. (2003). Pod shatter resistance in the resynthesized Brassica napus line DK142. J. Agric. Sci. 140 43–52. 10.1017/s002185960200285x [DOI] [Google Scholar]
- Sun C., Ashworth M. B., Flower K., Vila-Aiub M. M., Rocha R. L., Beckie H. J. (2021). The adaptive value of flowering time in wild radish (Raphanus raphanistrum). Weed Sci. 69 203–209. 10.1017/wsc.2021.5 [DOI] [Google Scholar]
- Sun P., Zhang W., Wang Y., He Q., Shu F., Liu H., et al. (2016). OsGRF4 controls grain shape, panicle length and seed shattering in rice. J. Integr. Plant Bio. 58 836–847. 10.1111/jipb.12473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Cuevas H. E., Das S., Sezen U. U., Zhou C., Guo H., et al. (2013). Seed shattering in a wild sorghum is conferred by a locus unrelated to domestication. Proc. Natl. Acad. Sci. USA. 110 15824–15829. 10.1073/pnas.1305213110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L. H., Morishima H. (1989). Variation and inheritance of seed shedding in weed rice. Rice Genet. Newslett. 6 72–73. [Google Scholar]
- Thurber C. S. (2012). The evolutionary genetics of seed shattering and flowering time, two weed adaptive traits in US weedy rice. [dissertation/PhD thesis). Boston, MA: University of Massachusetts. [Google Scholar]
- Thurber C. S., Jia M. H., Jia Y., Caicedo A. L. (2013). Similar traits, different genes? Examining convergent evolution in related weedy rice populations. Mol. Eco. 22 685–698. 10.1111/mec.12147 [DOI] [PubMed] [Google Scholar]
- Thurber C. S., Reagon M., Gross B. L., Olsen K. M., Jia Y., Caicedo A. L. (2010). Molecular evolution of shattering loci in U.S. weedy rice. Mol. Eco. 19 3271–3284. 10.1111/j.1365-294x.2010.04708.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tidemann B. D., Hall L. M., Harker K. N., Beckie H. J., Johnson E. N., Stevenson F. C. (2017). Suitability of wild oat (Avena fatua), false cleavers (Galium spurium), and volunteer canola (Brassica napus) for harvest weed seed control in Western Canada. Weed Sci. 65 769–777. 10.1017/wsc.2017.58 [DOI] [Google Scholar]
- Tiwari S., Bhatnagar P. (1989). Minimizing pod shattering in soybean. Indian Farm. 39 23–24. [Google Scholar]
- Tiwari S., Bhatnagar P. (1991). Pod shattering as related to other agronomic attributes in soybean. Tropical Agri. 68 102–103. [Google Scholar]
- Tsuchiya T. (1987). Physiological and genetic analysis of pod shattering in soybeans. Japan Agric. Res. Q. 21 166–175. [Google Scholar]
- Tsujimura Y., Sugiyama S., Otsuka K., Htun T. M., Numaguchi K., Castillo C., et al. (2019). Detection of a novel locus involved in non-seed-shattering behaviour of Japonica rice cultivar, Oryzasativa ‘Nipponbare’. Theor. Appl. Genet. 132 2615–2623. 10.1007/s00122-019-03376-3 [DOI] [PubMed] [Google Scholar]
- Tukamuhabwa P. K. E., Dashiell P. R., Nabasirye M. (2002). Determination of field yield loss and effect of environment on pod shattering in soybean. Afr. Crop Sci. J. 10 203–209. [Google Scholar]
- Van Gastel A., Bishaw Z., Niane A., Gregg B., Gan Y. (2007). “Chickpea seed production,” in The Chickpea Breeding and Management, eds Yadav S. S., Chen W. (Oxford, UK: CABI; ), 417–444. 10.1079/9781845932138.020 [DOI] [Google Scholar]
- Vera C. L., Downey R. K., Woods S. M., Raney J. P., McGregor D. I., Elliott R. H., et al. (2007). Yield and quality of canola seed as affected by stage of maturity at swathing. Can. J. Plant Sci. 87 13–26. 10.4141/p05-077 [DOI] [Google Scholar]
- Vigueira C., Li W., Olsen K. M. (2013). The role of Bh4 in parallel evolution of hull colour in domesticated and weedy rice. J. Evol. Biol. 26 1738–1749. 10.1111/jeb.12171 [DOI] [PubMed] [Google Scholar]
- Vigueira C. C., Qi X., Song B. K., Li L. F., Caicedo A. L., Jia Y., et al. (2019). Call of the wild rice: Oryza rufipogon shapes weedy rice evolution in Southeast Asia. Evol. Appl. 12 93–104. 10.1111/eva.12581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivian-Smith A., Koltunow A. (1999). Genetic analysis of growth-regulator-induced parthenocarpy in Arabidopsis. Plant Physiol. 121 437–451. 10.1104/pp.121.2.437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh M., Newman P., Powles S. (2013). Targeting weed seeds in-crop: a new weed control paradigm for global agriculture. Weed Technol. 27 431–436. 10.1614/wt-d-12-00181.1 [DOI] [Google Scholar]
- Walsh M. J., John C. B., Lauren M. S., Jason K. N., Adam S. D., Breanne D. T., et al. (2018). Opportunities and challenges for harvest weed seed control in global cropping systems. Pest Manag. Sci. 74 2235–2245. 10.1002/ps.4802 [DOI] [PubMed] [Google Scholar]
- Walsh M. J., Powles S. B. (2014). High seed retention at maturity of annual weeds infesting crop fields highlights the potential for harvest weed seed control. Weed Technol. 28 486–493. 10.1614/wt-d-13-00183.1 [DOI] [Google Scholar]
- Wang H., Squina F., Segato F., Mort A., Lee D., Pappan K., et al. (2011). High-temperature enzymatic breakdown of cellulose. Appl. Environ. Microbiol. 77 5199–5206. 10.1128/aem.00199-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Scarth R., Campbell G. C. (2005). Inheritance of seed shattering in interspecific hybrids between Fagopyrum esculentum and F. homotropicum. Crop Sci. 45 693–697. 10.2135/cropsci2005.0693 [DOI] [Google Scholar]
- Widderick M. J., Keenan M., Walsh M. J. (2014). “Harvest weed seed control: is there a role in northern region farming systems?,” in The 19th Australasian Weeds Conference-Science, Community and Food Security: the Weed Challenge, ed. Baker M. T. (Hobart: Tasmanian Weed Society; ). [Google Scholar]
- Wiggans S., Metcalfe D., Thompson H. (1956). The use of desiccant sprays in harvesting birdsfoot trefoil for seed. Agron. J. 48 281–284. 10.2134/agronj1956.00021962004800070001x [DOI] [Google Scholar]
- Wilson B. J. (1970). Studies on the shedding of seed of Avena fatua in various cereal crops and the presence of the seed in the harvested matter, in proceedings of the 10th Brighton Weed Control Conference. Croydon, UK: British Crop Protection Council, 831–836. [Google Scholar]
- Wilson B. J., Cussans G. (1975). A study on the population dynamics of Avena fatua L. as influenced by straw burning, seed shedding and cultivations. Weed Res. 15 249–258. 10.1111/j.1365-3180.1975.tb01132.x [DOI] [Google Scholar]
- Wilson C. E., Castro K. L., Thurston G. B., Sissons A. (2016). “Pathway risk analysis of weed seeds in imported grain: A Canadian perspective,” in The Proceedings of 13th International EMAPi conference, eds van Kleunen D. C. M., Pysek P., Richardson D. M. (Waikoloa, Hawaii: NeoBiota; ), 49–74. [Google Scholar]
- Xangsayasane P., Vongxayya K., Phongchanmisai S., Mitchell J., Fukai S. (2019). Rice milling quality as affected by drying method and harvesting time during ripening in wet and dry seasons. Plant Prod. Sci. 22 98–106. 10.1080/1343943x.2018.1544463 [DOI] [Google Scholar]
- Yan H., Li L., Liu P., Jiang X., Wang L., Fang J., et al. (2017). Reduced weed seed shattering by silencing a cultivated rice gene: strategic mitigation for escaped transgenes. Transgenic Res. 26 465–475. 10.1007/s11248-017-0016-3 [DOI] [PubMed] [Google Scholar]
- Yoshida O., Nakagawa H., Ogura N., Sato T. (1984). Effect of heat treatment on the development of polygalacturonase activity in tomato fruit during ripening. Plant Cell Physiol. 25 505–509. [Google Scholar]
- Young B. L. (2021). Integration of harvest-time and post-harvest tactics for integrated management of johnsongrass (Sorghum halepense) in grain sorghum (Sorghum bicolor). M.S. Thesis. College Station: Texas A&M University. [Google Scholar]
- Zhao Y., Zhang J., Zhang Z., Xie W. (2019). Elymus nutans genes for seed shattering and candidate gene-derived EST-SSR markers for germplasm evaluation. BMC Plant Bio. 19:102. 10.1186/s12870-019-1691-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Sun G., Chen X. (2007). Characteristics of the short rachillae of rice from archaeological sites dating to 7000 years ago. Chinese Sci. Bull. 52 1654–1660. 10.1007/s11434-007-0258-1 [DOI] [Google Scholar]
- Zhou Y., Lu D., Li C., Luo J., Zhu B. F., Zhu J., et al. (2012). Genetic control of seed shattering in rice by the APETALA2 transcription factor shattering abortion1. Plant Cell 24 1034–1048. 10.1105/tpc.111.094383 [DOI] [PMC free article] [PubMed] [Google Scholar]