Abstract
The cytosolic sulfotransferases (SULTs) are Phase II detoxifying enzymes that mediate the sulfate conjugation of numerous xenobiotic molecules. While the research on the SULTs has lagged behind the research on Phase I cytochrome P-450 enzymes and other Phase II conjugating enzymes, it has gained more momentum in recent years. This review aims to summarize information obtained in several fronts of the research on the SULTs, including the range of the SULTs in different life forms, concerted actions of the SULTs and other Phase II enzymes, insights into the structure–function relationships of the SULTs, regulation of SULT expression and activity, developmental expression of SULTs, as well as the use of a zebrafish model for studying the developmental pharmacology/toxicology.
Keywords: sulfotransferase, 3′-phosphoadenosine-5′-phosphosulfate, Phase II, detoxifying enzymes
Graphical abstract
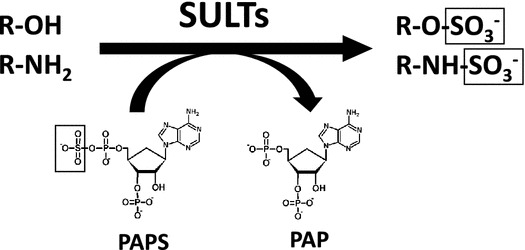
The cytosolic sulfotransferases (SULTs) are Phase II detoxifying enzymes that mediate the sulfate conjugation of numerous xenobiotic molecules.
The research on the cytosolic sulfotransferases (SULTs) and SULT-mediated sulfation of drugs and other xenobiotics, as well as a number of key endogenous compounds, has long lagged behind the research on cytochrome P-450s and other Phase II enzymes such as UDP-glucuronosyltransferases (UGTs) and catechol-O-methyltransferases (COMTs). In recent years, however, the research on SULTs has gained considerably more momentum. This review aims to summarize some recent advances concerning the SULT research.
I. Overview of the SULT-mediated sulfation
Biological sulfation was first discovered when phenyl sulfate was isolated from the urine of a patient being treated with phenol as an antiseptic.1) This finding had positioned the research on sulfation and sulfotransferase enzymes within the area of pharmacology/toxicology for over a century. Indeed, many studies using experimental animals or human subjects have demonstrated the metabolism of drugs through sulfation.2,3) The responsible enzymes, now called the SULTs, in vertebrates are hence viewed as detoxifying or Phase II drug-metabolizing enzymes.4–7) These enzymes catalyze the transfer of the sulfonate group from the active sulfate, 3′-phosphoadenosine 5′-phosphosulfate (PAPS), to substrate compounds containing hydroxyl or amino group(s).8) Sulfate conjugation may result in the inactivation of the substrate compounds or increase their water-solubility, thereby facilitating their removal from the body.4–7) Many of the SULT enzymes have also been shown to be involved in the sulfation of key endogenous compounds such as steroids and catecolamines.9) Fig. 1 shows typical SULT-mediated sulfation reactions.
Fig. 1.
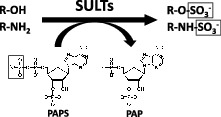
Typical SULT-mediated sulfation reactions.
Notes: Different SULT enzymes may catalyze the transfer of the sulfonate
(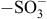 ) group from the donor compound, PAPS, to the
hydroxyl (−OH) or amino (−NH2) group of the substrate compound.
) group from the donor compound, PAPS, to the
hydroxyl (−OH) or amino (−NH2) group of the substrate compound.
II. Range of the SULTs in different life forms
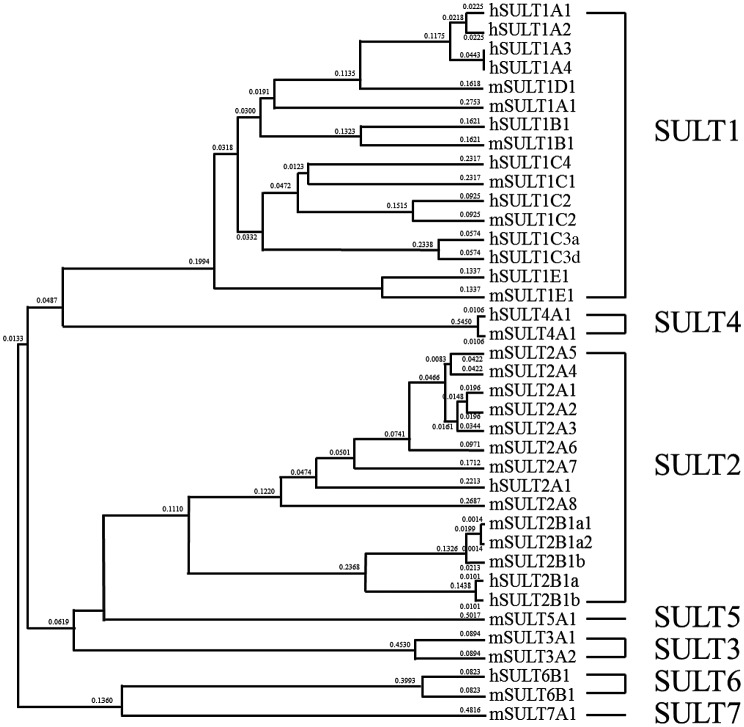
To distinguish them from the Golgi membrane-bound sulfotransferases that are responsible for the sulfation of proteins, proteoglycans, and glycolipids,10,11) the SULTs have been defined as cytosolic enzymes involved in the sulfation of low molecular weight compounds.4–7) Enzymes of this nature have been reported to be present in different life forms spanning both prokaryotes and eukaryotes. This is not surprising considering that a major function of these enzymes is the detoxification of numerous xenobiotic compounds that pose as a constant threat to intrude cells in all living forms. Evidence for the existence of sulfotransferases in bacteria first came from the detection of arylsulfotransferase activity in the feces of human and rat.12) It was later demonstrated that a novel type of arylsulfotransferase capable of mediating the sulfation of quercetin was present in a human intestinal bacterium.13) Following these initial findings, sulfotransferases from a number of bacteria, including Eubacterium, Klebsiella, Haemophilus, Citrobacter, and Mycobacteria were reported.14–17) Among eukaryotic micro-organisms, arylsulfotransferase activities had been detected in the filamentous fungus Cunninghamella elegans and the white rot fungus Pleurotus ostreatus.18,19) Among plants, the presence of sulfotransferase was first reported in spinach leaves.20) Subsequently, a variety of flavonol sulfotransferases were detected in different plant species.21) In a model plant organism, Arabidopsis thaliana, 21 distinct sulfotransferases have been identified.22) In the animal kingdom, the SULT can be found in Caenorhabditis elegans, which contains a sole SULT enzyme reported to be abundantly expressed at the dauer larvae stage during development.23) A family of SULTs has been detected in Drosophila melanogaster.24) Three of the four Drosophila SULTs were shown to be expressed during embryogenesis, with the fourth one being expressed post-embryonically.25) Among vertebrates, numerous SULTs have been reported.26,27) Based on their amino acid sequences, all SULTs found in vertebrates are proposed to constitute a gene superfamily.28) In humans, 15 distinct SULTs classified into 4 SULT gene families are present,28,29) whereas in mouse, 23 SULTs spread among 7 SULT gene families are known.28) As described below, 20 SULTs categorized into 6 SULT gene families have been identified in zebrafish. Fig. 2 shows a dendrogram comprising all human and mouse SULTs.
Fig. 2.
Classification of the human and mouse SULTs based on their amino acid sequences.
Note: The dendrogram was generated based on the degree of amino acid sequences among the human and mouse SULTs.
III. Concerted actions of the SULTs and other Phase II enzymes
It is generally known that the metabolism of drugs and other xenobiotics may proceed through two phases, with Phase I reactions involving the generation of functional groups that may subsequently be used in Phase II reactions.30) In the Phase II drug/xenobiotic metabolism, different groups of Phase II enzymes have also been shown to act in a concerted manner. For example, mono-conjugated metabolites, particularly methylated metabolites, have been reported to undergo sulfation reaction. Methylated catecholamines such as 3-O-methyldopamine and 3-O-methylepinephrine have been shown to be subjected to the sulfation mediated by SULT1A3, and methylated catechol estrogens (methoxyestrogens) have been shown to be subjected to sulfation by SULT1A1, SULT1A3, and SULT1E1.31–33) Methylated metabolites of quercetin have been also reported to be sulfated by the SULT enzyme(s) in rat liver lysates.34) These results implied a concerted action of the COMTs and the SULTs in the metabolism of catechol and polyphenol compounds. A number of glucuronide-sulfate double-conjugated metabolites of polyphenols, e.g. catechin and genistein, have been detected from human urine, implying the concerted action of UGTs and the SULTs in the metabolism of polyphenols.35–37) The enzymatic sulfation reaction of O-glucuronide polyphenols, however, has not yet been reported and remains to be investigated.
IV. Insights into the structure–function relationships of the SULTs and in silico studies identifying the substrate of SULTs
The majority of the SULT enzymes are known to be capable of catalyzing the sulfation of a range of substrate compounds, indicating their flexible active sites.38–40) Many SULT enzymes share the same substrates with differential kinetics of sulfation, making it difficult in projecting their physiological substrate spectra and functions. To better understand these latter aspects, structural analyses of the SULT enzymes have been undertaken in the past two decades. Starting from the early 1990s, cDNA encoding SULTs from human and other species have been cloned and sequenced.27) Deduced amino acid sequences of different SULTs provided useful information for not only the classification of the SULTs, but also the delineation of conserved sequences, particularly the so-called “signature sequences” that are involved in the binding of PAPS, a co-substrate and sulfonate donor involved in the SULT-mediated sulfation reaction.8) In 1997, the first crystal structure of SULT, that of the mouse estrogen sulfotransferase, was reported.41) In the following decade, crystal structures of a good number SULTs from different species have been determined and revealed details about the structures of the PAPS-binding and substrate-binding sites.38,40,42) Generally speaking, the overall structures of the SULTs, including the PAPS-binding regions, dimerization motif, and critical catalytic residues such as histidine and lysine, are conserved among SULT members. The substrate-binding sites, particularly the substrate-binding loops, however, showed considerable variations among SULT enzymes. Intriguingly, clustering analyses using sequences or structures in the substrate-binding sites have demonstrated that the clustering with substrate-binding site was relatively similar to that with enzymatic activity data rather than that with entire amino acid sequences used for the classification of the SULTs.38) This observation indicated that the enzymatic characteristics may correlate more with the substrate-binding site, which is formed by three substrate-binding loops, than the genetic distance, implying the feasibility of taking the in silico approach to predict the substrate compounds for different SULTs. Although the classic enzymatic assays remain useful for the identification of substrates, in silico studies are becoming an increasingly more useful methodology for identifying novel substrate compounds as well as for investigating the catalytic mechanisms at the molecular level. The first in silico substrate-screening study was performed in 2009, which virtually docked over 50,000 chemical compounds from World Drug Index into the substrate-binding pockets of SUULT1A3 and SULT1E1, respectively.43) The compounds with hydroxyl groups not correctly oriented at the sulfonate-transfer site were excluded. Interestingly, the final output showed that 10 of the top 12 high score compounds for SULT1A3 were dopaminergic compounds and 4 of the top 12 for SULT1E1 were estrogenic compounds. Although the status of these potential substrate compounds have not yet been verified, the results derived from this in silico study appeared compatible to the substrate specificity of SULT1A3 and SULT1E1 previously reported. It is noted that the structures used in the docking simulations included both the PAP(S)-bound structures and the rigid substrate-binding pockets derived from crystal structure data. It has been proposed that PAP(S) binding may alter the conformation of the substrate-binding pockets and that the substrate-binding pockets are flexible in response to the incoming substrates.40,44) A more recent in silico substrate-screening study utilizing structures without and with bound PAP(S) and taking into consideration the dynamics of substrate-binding pockets also yielded highly accurate predictions of substrate compounds.45) In short, in silico docking simulation is becoming a good method for screening candidate substrate compounds, and may be useful for finding substrates for orphan SULTs such as human SULT4A1.
V. Regulation of SULT expression and activity
A number of xenobiotics, e.g. polychlorinated biphenyls, drug compounds, and dietary polyphenol, have been shown to directly inhibit the SULT activity.46,47) The transcriptional regulation of SULTs has been a subject for investigation in the past two decades. Several nuclear receptors, e.g. constitutive androstane receptor, glucocorticoid receptor, and peroxisome proliferator-activated receptors, in conjunction with their ligand compounds, have been demonstrated to be involved in the regulation of the transcription of SULT genes.48–50) Xenobiotics such as 2,3,7,8-tetrachlorodibenzo-p-dioxin and dexamethasone have been reported to affect the expression of the SULTs through aromatic hydrocarbon receptor and pregnane X receptor.39,48) More recently, several studies revealed that a nuclear transcription factor, E2-related factor 2 (Nrf2), is likely involved in the regulation of SULT gene expression. Nrf2 is a transcription factor that binds to the antioxidant response elements (AREs) by responding the oxidative stress and controls the transcription of a number of Phase II drug-metabolizing/detoxifying enzymes.51,52) It has been reported that an antioxidant, caffeic acid, may upregulate the expression of SULT1A1 dependent on the phosphorylation of Nrf2 by the activation of p38 mitogen-activated protein kinase (MAPK) pathway.53) Nrf2 activators, such as butylated hydroxyanisole, oltiplaz, and ethoxyquin, have also been shown to affect the expression of many SULT genes, including SULT1A1, SULT1B1, SULT1C1, SULT1C2, SULT1D1, SULT1E1, SULT3A1, and SULT5A1 in mice.49) Indeed, Nrf2-null mice were found to manifest decreased expression of SULT3A1 and SULT5A1.54) However, Nrf2-null mice showed no change in the expression of SULT1A1, SULT1D1, and SULT1E1 regardless of induction by an Nrf2 activator, oltiplaz.54,55) In contrast, oxidative stress, caused by liver ischemia and reperfusion (I/R), was found to induce the expression of SULT1E1 through the binding of Nrf2 to ARE.56) Interestingly, accumulation of Nrf2 upon IR was more intense in female SULT1E1-/- mice than in wild-type mice,56) implying that estrogens may regulate the activation of Nrf2 and that the regulation of SULT1E1 expression by Nrf2 may be subjected to a negative feedback mechanism. It is worthwhile mentioning that Nrf2 activators are also known to modify or oxidize the cysteine residues of proteins, e.g. Keap 1, repressing Nrf2 action, and activate MAPK pathway, possibly through the generated reactive phenoxy radicals.52,57) Oxidative stress, therefore, may affect the expression of those SULT genes unresponsive to Nrf2 action, and perhaps genes coding for other detoxifying-enzymes as well. Previous studies have also demonstrated that Keap 1-knockdown mice displayed decreased acetaminophen-sulfating activity in the liver and oxidative stress treatment in liver cytosols or slices affected the sulfating activities of SULT1A1 and SULT1E1.58–60) Cellular antioxidant responses therefore may affect the regulation of expression and/or activity of the SULTs, although detailed mechanisms remain to be clarified. Other cellular responses, e.g. those to nitric oxide (NO) stress and cytokine stimulations, have also been shown to regulate the expression or activity of the SULTs. A study revealed that treatment with NO donors led to the decrease in estrogen-sulfating activity in MCF-7 human breast cancer cells, possibly due to the inactivation of SULT1E1.61) The mechanism underlying the decrease in estrogen-sulfating activity is not fully understood, but peroxynitrite treatment of SULT1E1 enzyme caused the tyrosine nitration of the SULT1E1 and a decrease in the sulfating activity of nitrated SULT1E1,61) indicating that nitrative stress may result in the inhibition of the activity of SULT1E1 through tyrosine nitration of SULT1E1. Treatment with NO donors has also been shown to downregulate the expression of SULT2A1 in HepG2 human hepatoma cells through the activation of MAPK pathway, leading to the decrease in hydroxysteroid-sulfating activity.62) SULT2A1 has also been shown to be downregulated in response to stimulation of lipopolysaccharide (LPS) and cytokines such as TNF and IL-1, implying that the steroid-sulfating activity may be suppressed under inflammatory conditions.63) Like SULT2A1, SULT2B1a has been also reported to be downregulated through NO signaling in response to the stimulation of LPS and TNF.64) These results implicated that NO signaling may regulate the steroid-sulfating SULTs, SULT1E1, SULT2A1, and SULT2B1a, in response to various stimulations. Like some other drug-metabolizing enzymes, recent studies have shown the induction of certain SULTs by their substrates, either directly or indirectly. For example, the expression of SULT1A3 was found to be induced by its prototype substrate, dopamine, via ERK pathway in SK-N-MC and SH-SY5Y human neuroblastoma cells.65) Ethanol has been shown to induce ethanol-sulfating SULTs,66,67) SULT1A1 and SULT1E1, in cultured HepG2 human hepatoma cells.68) Interestingly, 17β-estradiol, a substrate for SULT1A1 and SULT1E1, was demonstrated to be capable of inducing SULT2A1, implying that the sulfation of androgens, which are estrogen precursors, is regulated by 17β-estradiol.69) Taken together, the above-mentioned findings clearly indicated that the production and the functioning of the SULT enzymes are dynamic events that may undergo changes in response to cellular conditions.
VI. Developmental expressions of SULTs
Many of the Phase II drug-metabolizing/detoxifying enzymes are not strongly expressed in early life as they are in adulthood. The SULT enzymes are, however, unique in that they are strongly expressed early in life. Significant sulfating activities toward p-nitrophenol, β-naphthol, dopamine, and estrogens were detected in a variety of human fetal specimens prepared from the adrenals, gut, kidney, liver, and lung.70–72) In contrast, UGTs were found to be poorly expressed in fetal and early neonatal liver.73,74) Furthermore, comparative analyses of sulfation and glucuronidation activities between fetal and adult liver samples revealed that the fetal sulfation activities toward β-naphthol and ritodorine were stronger than those of glucuronidation, whereas adult sulfation activities were weaker than those of glucuronidation.75,76) Compared with other Phase II enzymes, the SULTs therefore are thought to play a more prominent role in the protection against xenobiotics as well as in the regulation of hormone functions during fetal, neonatal, and infant development. A number of ontogenic studies have been performed by investigating the sulfating activities of major SULT enzymes, including SULT1A1, SULT1A3, SULT1E1, and SULT2A1, using their prototype substrates such as p-nitrophenol, dopamine, estradiol, and dehydroepiandrosterone (DHEA).77–82) All these studies revealed high levels of expression of the SULT enzymes during the prenatal period, indicating the importance of these enzymes in fetus. Another SULT1 isoform, SULT1B1, has been shown to be extensively expressed in fetal intestine and continued to be expressed in adult intestine. In contrast to its low expression level in fetal liver, the expression of SULT1B1 was shown to be dramatically increased in adult liver.80) Since SULT1B1 has been shown to be capable of catalyzing the sulfation of thyroid hormones,83) it may play a role in their homeostasis. Of special interest are members of the SULT1C subfamily. While it remains to be fully investigated, a few studies have revealed the expression of SULT1C2 in fetal liver, kidney, and intestine.84) Moreover, the intestinal expression of SULT1C2 in the fetus was found to be much stronger than that in adult.80,84) SULT1C4 expression was detected in fetal kidney, lung, and heart, as well as in adult ovary and spinal cord, implying its specific expression in fetal and maternal tissues.85) Members of the SULT1C subfamily may, therefore, appear to play an important role in the protection against the exposure to xenobiotics during the fetal period. Of the two human SULT2B members, SULT2B1a was detected in fetal, but not adult, brain, whereas SULT2B1b was detected in adult, but not fetal, brain.86,87) SULT4A1, the sole member of the SULT4 subfamily,88,89) has been shown to be exclusively expressed in brain and other neuronal tissues, and has been linked to Schizophrenia.90,91) Although its ontogenic expression has not been fully characterized, the SULT4A1 mRNA expression has been confirmed in the fetal brain and the level of expression is in fact higher than that in adult brain.88,89) The ontogenic expression of the human SULTs and associated functional implications remained to be fully understood. In vivo studies using animal models are needed in order to elucidate the physiological functions of the SULTs in fetal life.
VII. Zebrafish as a model for SULT research
Zebrafish have been widely used as an animal model for developmental biology research.92,93) In recent years, zebrafish have also found their place in pharmacology/toxicology research.94,95) Our laboratories have recently embarked on the use of zebrafish as a model for investigating some fundamental aspects of the SULTs.96,97) The benefits of using zebrafish to explore the physiological role of the SULTs, especially in embryogenesis and organogenesis, will be discussed. To date, 20 distinct zebrafish SULTs, including 9 SULT1s, 3 SULT2s, 5 SULT3s, 1 SULT4, 1 SULT5, and 1 SULT6, have been cloned, expressed, purified, and characterized.98–109) All these zebrafish SULTs, similar to human and other mammalian SULTs, contain the so-called “signature sequences” (YPK(A/S)GTxW in the N-terminal region and RKGxxGDW(V/K)NxFT in the C-terminal region) characteristic of the SULT enzymes.27) Of the nine zebrafish SULT1 enzymes, SULT1 ST1 through ST4 as well as SULT1 ST7 through ST9 were capable of sulfating a variety of low-molecular weight xenobiotic compounds and therefore appeared to be involved in the general detoxification processes.99,102,105,106,108,109) Interestingly, SULT1 ST5 and ST6 displayed strict substrate specificity, respectively, for thyroid hormones and estrogens.98,100) These two enzymes, therefore, appear to be the zebrafish counterparts of the human thyroid hormone-sulfating SULT1B1 and estrogen-sulfating SULT1E1. Indeed, zebrafish SULT1 ST5 and ST6 exhibited the highest % homology values at the amino acid sequence level to human SULT1B1 and SULT1E1, respectively, among all 11 known human SULTs.83,110) The three zebrafish SULT2 STs, which appear to correspond to the three human SULT2 enzymes, exhibited differential sulfating activities toward various hydroxysteroids, with optimal activities for SULT2 ST1, ST2, and ST3 being toward pregnenolone, DHEA, and corticosterone, respectively.103,107) The five zebrafish SULT3 enzymes showed sulfating activities toward a number of endogenous compounds including 17β-estradiol, DHEA and pregnenolone, and xenobiotic compounds such as bisphenol A, 2-naphthol, and 2-naphthylamine.103) The enzymatic properties of both zebrafish SULT4 and SULT5, however, remain to be fully characterized. It is noted that zebrafish SULT4 shares an 89% amino acid sequence identity with the human SULT4A1.88,89) SULT4-knocked down zebrafish has recently been generated and shown to be associated with altered expression of 135 genes including phototransduction genes, circadian rhythm-related genes, and CREB signaling genes, whereas no morphological phenotypes were observed.111) It was subsequently reported that SULT4 frameshift mutant zebrafish manifested slightly less active locomotor behaviors during the daytime.112) Zebrafish SULT6, on the other hand, has been shown to exhibit differential sulfating activities toward various endogenous and xenobiotic compounds tested as substrates.104) It is to be noted that a human SULT6B1 has also been identified in human genome.29) It therefore appears that the zebrafish possesses many homologous SULT enzymes that are present in humans.
As mentioned above, previous studies have revealed the SULTs as predominant conjugation enzymes during early development, particularly the fetal and neonatal stages.70–72,75,76) Primarily due to the inconvenience and limitations involved in using mammalian animal models, zebrafish may serve as a useful model for systematically exploring the expression patterns and in vivo involvement of the SULTs during embryogenesis onto larval development. In view of the regulation of key endogenous compounds through the sulfation during the development, it is an interesting issue whether the expression of SULTs may correlate with the development of the endocrine or nervous system. The developmental expression of the zebrafish SULTs have been investigated using RT-PCR. Different SULTs exhibited distinct patterns of expression at different stages during embryogenesis as well as larval development.98–103,113,114) Fig. 3 shows the expression patterns of several SULTs during the zebrafish development. Of the four monoamine-sulfating SULTs, SULT1 ST1, SULT1 ST2, SULT1 ST3, and SULT1 ST6, the expression of SULT1 ST1 started during the pharyngula period and the expression of the other three did not start until 3 days into the larval stage. In both cases, the expression continued to increase until maturation. It has been reported that during zebrafish development, aminergic neurons first appear at diencephalon and locus coeruleus in the pharyngula period (24 hpf). Innervation to posterior tuberculum and spinal cord occurs by the hatching period (2–3 dpf), and the development of the aminergic neuron system completes shortly after the hatching time.115) Based on their developmental expression patterns, only SULT1 ST1 may, therefore, play an role in the regulation of monoamines at the beginning of aminergic system development and other three SULTs may get involved later toward the completion of the aminergic system. SULT1 ST5, a thyroid hormone-sulfating SULT, was found to appear at the beginning of the hatching period and gradually increase on to maturity.100) The thyroid hormones are essential for embryonic and larval development.116,117) Interestingly, fish eggs have been shown to contain high concentrations of thyroid hormones, which are probably of maternal origin, which decrease during the embryogenesis.116) The thyroid gland is formed during the hatching period and may start producing the thyroid hormones thereafter.117) Sulfation of thyroid hormones, as mediated by SULT1 ST5, may therefore regulate the action of thyroid hormones that are produced by the developing larva. Among the hydroxysteroid-sulfating SULTs, SULT3 ST1 and SULT3 ST5 have been shown to be expressed in unfertilized egg and upon fertilization, throughout the entire developmental process.103,113) Major hydroxysteroid-sulfating SULTs, SULT3 ST2, SULT3 ST3, and SULT3 ST4 have been shown to exhibit similar ontogenic expression patterns, first appearing during the blastula to neurula/segmentation period, decreasing temporarily, and re-emerging dramatically in the post-hatching embryos (at 72 hpf) where most organ rudiments are rapidly developing and fairly complete.103,113,114) CYP1A1, a first-step and rate-limiting enzyme in the biosynthesis of steroids, has been shown to be present from the time of unfertilized egg through the gastrula period and gradually descend on to maturity. CYP1A1 appears to be essential for embryogenesis, especially in epiboly process during blastula and gastrula periods.118,119) SULT3 ST1 and SULT3 ST5, which exhibited lower activities toward hydroxysteroids, may be involved in the homeostasis of basal level of embryonic steroids and major hydroxysteroid-sulfating SULTs may regulate the action of steroids during epiboly process. The expression pattern of SULT1 ST6, an estrogen-sulfating SULT, is in accordance with the appearance of aromatase, which is responsible for the conversion of androgens to estrogens. Both enzymes were detected in unfertilized eggs, dramatically decreased post fertilization, and re-emerged during the hatching periods.120) Sulfation of estrogens may be related to the organogenesis, including the development of nervous system, eye, and muscle.121,122) Considering the role of the SULTs in the detoxification and disposal of xenobiotics, it is important to explore to what extent the xenobiotic-sulfating SULTs may contribute toward the chemical defense of the developing zebrafish embryos/larvae. Interestingly, a recent report showed that different zebrafish SULTs displayed differential sulfating activities toward a panel of drugs tested as substrates.123) Moreover, a metabolic labeling study showed that cultured zebrafish liver cells displayed a similar pattern of sulfation of several tested drugs as that of HepG2 human hepatoma cells, implying that human and zebrafish liver cells share considerable similarities with regard to their constituent drug-sulfating SULT enzymes. It therefore appears that the mechanism of drug metabolism through sulfation, to a considerable extent, is conserved between the zebrafish and humans.
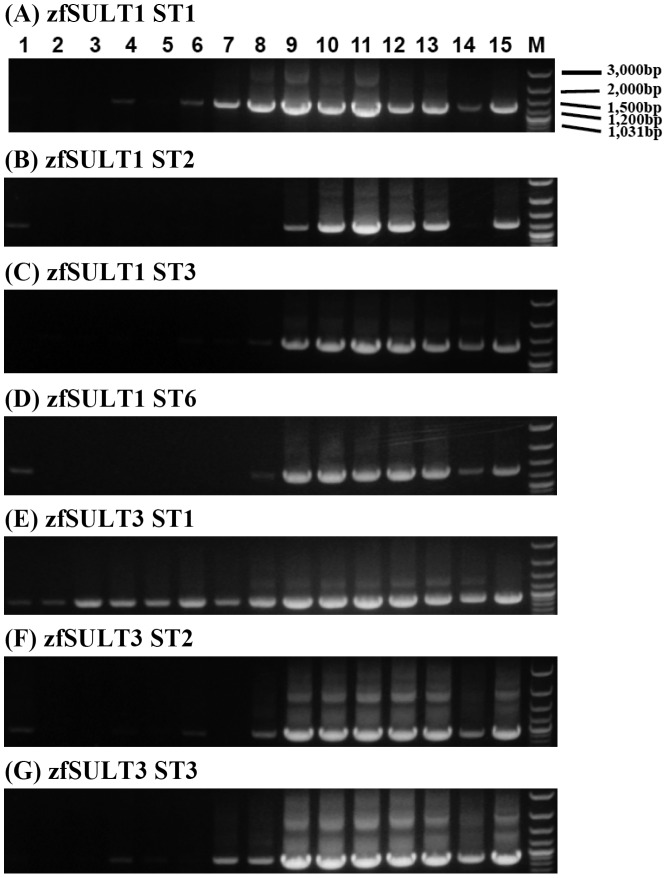
Fig. 3.
Developmental stage-dependent expression of representative zebrafish SULTs.
Notes: Lanes 1 through 15 correspond to unfertilized zebrafish eggs, 0-, 1-, 3-, 6-, 12-, 24-, 48-, and 72-h zebrafish embryos, 1-, 2-, 3-, and 4-week-old zebrafish larvae, and 3-month-old male and female zebrafish.
Overall, the information concerning the enzymatic characteristics and developmental expression of zebrafish SULTs may provide useful clues for designing in vivo studies to clarify the physiological and/or pharmacological roles of the SULTs using gene targeting strategies such as knock-down, knock-out, and transgenic manipulations. For example, since some SULT enzymes are highly expressed in the early developmental stages, the research using zebrafish embryos/larvae may reveal the role of sulfation, mediated by those SULTs, in the regulation of key endogenous compounds during embryogenesis and organogenesis. Further studies using knock-down, knock-out, and transgenic zebrafish will clarify the impact of inhibiting or potentiating a specific SULT on the endocrine disorders/diseases and provide information that may aid in the treatment of these endocrine disorders/diseases. Another application of using zebrafish is related to the developmental pharmacology, which may provide clues for the rational design of drugs that may be metabolized through sulfation, thereby alleviating potential adverse effects of obstetric and/or pediatric medications. The orthology and the enzymatic properties of the orthologous SULTs will need to be carefully scrutinized in order to connect the study using zebrafish to humans. In this regard, there may be some insurmountable difficulties. For example, SULT1A3 is the monoamine-sulfating SULT in humans and no orthologous SULT has been identified in zebrafish. Nevertheless, in zebrafish, there may be functionally related enzymes, such as SULT1S T1, SULT1 ST2, and SULT1 ST3, which have been shown to be capable of mediating the sulfation of monoamines. For the sulfation of thyroid hormones, SULT1 ST5 is likely the responsible sulfating enzyme in zebrafish, whereas in humans, SULT1A1 is accepted to be responsible for the sulfation instead of SULT1B1, the orthologous human SULT to zebrafish SULT1 ST5. On the other hand, zebrafish SULT1 ST6 and human SULT1E1 appear to be orthologous SULTs; the two share common enzymatic characteristics toward endogenous estrogens, although their expression patterns remain to be verified. Detailed comparative analyses of the expression pattern between zebrafish and human SULTs as well as functional characterizations therefore are needed. It should be pointed that zebrafish embryos/larvae are freely living in environments and no placenta is involved. This fact may pose a significant difference between zebrafish and humans, but nevertheless may provide an advantage to understand the biological function of the SULTs in embryo independent of the influence from maternal SULTs.
VIII. Concluding remarks and future directions
Although the sulfation of xenobiotics was first discovered back in the nineteenth century, the research on the responsible enzymes has been slow until the 1980s. The past three decades have witnessed significant progress made in the elucidation of the diversity of the SULT enzymes and their enzymatic characteristics, the phylogenetic relationships between the SULTs, the structural biology of the SULTs, the developmental expression of the SULTs, the mechanisms underlying the regulation of the SULT gene expression and the sulfating activity of the SULTs, as well as the development of the zebrafish as a model for use in SULT research. While continued efforts need to be made in all these latter aspects of the SULT research, additional fronts, particularly the implications of the polymorphisms of the SULT genes and the systems biology regarding the physiological involvement of the SULTs—not only in detoxification of xenobiotics but also in the homeostasis of key endogenous compounds such as thyroid/steroid hormones and catecholamine neurotransmitter/hormones—will need to be addressed.
Disclosure statement
No potential conflict of interest was reported by the authors.
Funding
This work was in part supported by a grant from National Institutes of Health [Grant # R03HD071146; to MCL]; a Grant-in-Aid for Scientific Research (C) [#26450130; to M.S.], (B) [#15H04502; to Y.S.]; a Grant-in Aid from Strategic Young Researcher Overseas Visits Program for Accelerating Brain Circulation [#S2511; to M.S.] from Japan Society for the Promotion of Science (JSPS).
Footnotes
This review was written in response to the author’s receipt of the Japan Society for Bioscience, Biotechnology, and Agrochemistry Senior Scientists Award in 2015.
Abbreviations: SULT, cytosolic sulfotransferase; PAP, 3′-phosphoadenosine-5′-phosphate; PAPS, 3′-phosphoadenosine-5′-phosphosulfate; TLC, thin-layer chromatography.
References
- Baumann E. Ueber Sulfosäuren im Harn, Ber. Dtsch. Chem.Ges. 1876; 9: 54–58. 10.1002/(ISSN)1099-0682 [DOI] [Google Scholar]
- Nelson SD, Gordon WP. Mammalian drug metabolism. J. Nat. Prod. 1983;46: 71–78. 10.1021/np50025a005 [DOI] [PubMed] [Google Scholar]
- Chen X, Zhong D, Blume H. Stereoselective pharmacokinetics of propafenone and its major metabolites in healthy Chinese volunteers. Eur. J. Pharm. Sci. 2000;10: 11–16. 10.1016/S0928-0987(99)00083-4 [DOI] [PubMed] [Google Scholar]
- Mulder GJ, Jakoby WB. Sulfation. In: Mulder GJ, Jakoby W, editors. Conjugation reactions in drug metabolism. London: Taylor and Francis; 1990. p. 107–161. [Google Scholar]
- Weinshilboum R, Otterness D. Sulfotransferase enzymes. In: Kaufmann FC, editor. Conjugation–deconjugation reactions in drug metabolism and toxicity. Berlin: Springer-Verlag; 1994. p. 45–78. 10.1007/978-3-642-78429-3 [DOI] [Google Scholar]
- Falany CN. Enzymology of human cytosolic sulfotransferases. FASEB J. 1997;11: 206–216. [DOI] [PubMed] [Google Scholar]
- Duffel MW. Sulfotransferases. In: Guengerich FP, editor. Comprehensive toxicology. Oxford: Elsevier Science Ltd.; 1997. p. 365–383. [Google Scholar]
- Lipmann F. Biological sulfate activation and transfer. Science. 1958;128: 575–580. 10.1126/science.128.3324.575 [DOI] [PubMed] [Google Scholar]
- Strott CA. Sulfonation and molecular action. Endocr. Rev. 2002;23: 703–732. 10.1210/er.2001-0040 [DOI] [PubMed] [Google Scholar]
- Niehrs C, Beisswanger R, Huttner WB. Protein tyrosine sulfation, 1993—an update. Chem. Biol. Interact. 1994;92: 257–271. 10.1016/0009-2797(94)90068-X [DOI] [PubMed] [Google Scholar]
- Thacker BE, Xu D, Lawrence R, et al. Heparan sulfate 3-O-sulfation: a rare modification in search of a function. Matrix Biol. 2014;35: 60–72. 10.1016/j.matbio.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Kobashi K. The role of intestinal flora in metabolism of phenolic sulfate esters. Biochem. Pharmacol. 1986;35: 3507–3510. 10.1016/0006-2952(86)90619-2 [DOI] [PubMed] [Google Scholar]
- Koizumi M, Shimizu M, Kobashi K. Enzymatic sulfation of quercetin by arylsulfotransferase from a human intestinal bacterium. Chem. Pharm. Bull. (Tokyo) 1990;38: 794–796. 10.1248/cpb.38.794 [DOI] [PubMed] [Google Scholar]
- Konishi-Imamura L, Kim DH, Kobashi K. Relationship between substrate activity and pKa value of phenols on sulfotransferase from Eubacterium A-44. Biochem. Int. 1992;28: 725–734. [PubMed] [Google Scholar]
- Baek MC, Kim SK, Kim DH, et al. Cloning and sequencing of the klebsiella K-36 astA gene, encoding an arylsulfate sulfotransferase. Microbiol. Immunol. 1996;40: 531–537. 10.1111/mim.1996.40.issue-8 [DOI] [PubMed] [Google Scholar]
- Kang JW, Jeong YJ, Kwon AR, et al. Cloning, sequence analysis, and characterization of the astA gene encoding an arylsulfate sulfotransferase from Citrobacter freundii. Arch. Pharm. Res. 2001;24: 316–322. 10.1007/BF02975099 [DOI] [PubMed] [Google Scholar]
- Tanaka S, Moriizumi Y, Kimura M, et al. Overproduction, purification and preliminary X-ray diffraction analysis of a sulfotransferase from Mycobacterium tuberculosis H37Rv. Acta Crystallogr. Sect F Struct. Biol. Cryst. Commun. 2005;61: 33–35. 10.1107/S1744309104022328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Yang Y, Leakey JE, et al. Phase I and phase II enzymes produced by Cunninghamella elegans for the metabolism of xenobiotics. FEMS Microbiol. Lett. 1996;138: 221–226. 10.1111/fml.1996.138.issue-2-3 [DOI] [PubMed] [Google Scholar]
- Bezalel L, Hadar Y, Cerniglia CE. Enzymatic mechanisms involved in phenanthrene degradation by the White Rot Fungus Pleurotus ostreatus. Appl. Environ. Microbiol. 1997;63: 2495–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A. A sulfotransferase from spinach leaves using adenosine-5′-phosphosulfate. Planta. 1975;124: 267–275. 10.1007/BF00388689 [DOI] [PubMed] [Google Scholar]
- Varin L, Marsolais F, Richard M, et al. Sulfation and sulfotransferases 6: biochemistry and molecular biology of plant sulfotransferases. FASEB J. 1997;11: 517–525. [DOI] [PubMed] [Google Scholar]
- Hirschmann F, Krause F, Papenbrock J. The multi-protein family of sulfotransferases in plants: composition, occurrence, substrate specificity, and functions. Front. Plant Sci. 2014;5: 556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori K, Inoue M, Inoue T, et al. A novel sulfotransferase abundantly expressed in the dauer larvae of Caenorhabditis elegans. J. Biochem. 2006;139: 355–362. 10.1093/jb/mvj041 [DOI] [PubMed] [Google Scholar]
- Hattori K, Motohashi N, Kobayashi I, et al. Cloning, expression, and characterization of cytosolic sulfotransferase isozymes from Drosophila melanogaster. Biosci. Biotechnol. Biochem. 2008;72: 540–547. 10.1271/bbb.70647 [DOI] [PubMed] [Google Scholar]
- Fahmy K, Baumgartner S. Expression analysis of a family of developmentally-regulated cytosolic sulfotransferases (SULTs) in Drosophila. Hereditas. 2013;150: 44–48. 10.1111/more.2013.150.issue-2-3 [DOI] [PubMed] [Google Scholar]
- Yamazoe Y, Nagata K, Ozawa S, et al. Structural similarity and diversity of sulfotransferases. Chem. Biol. Interact. 1994;92: 107–117. 10.1016/0009-2797(94)90057-4 [DOI] [PubMed] [Google Scholar]
- Weinshilboum RM, Otterness DM, Aksoy IA, et al. Sulfation and sulfotransferases 1: sulfotransferase molecular biology: cDNAs and genes. FASEB J. 1997;11: 3–14. [PubMed] [Google Scholar]
- Blanchard RL, Freimuth RR, Buck J, et al. A proposed nomenclature system for the cytosolic sulfotransferase (SULT) superfamily. Pharmacogenetics. 2004;14: 199–211. 10.1097/00008571-200403000-00009 [DOI] [PubMed] [Google Scholar]
- Freimuth RR, Wiepert M, Chute CG, et al. Human cytosolic sulfotransferase database mining: identification of seven novel genes and pseudogenes. Pharmacogenomics J. 2004;4: 54–65. 10.1038/sj.tpj.6500223 [DOI] [PubMed] [Google Scholar]
- Brunton LL, Chabner BA, Knollmann BC. Goodman & Gilman’s the pharmacological basis of therapeutics. 12th ed. New York (NY): McGraw-Hill; 2011. [Google Scholar]
- Hui Y, Yasuda S, Liu MY, et al. On the sulfation and methylation of catecholestrogens in human mammary epithelial cells and breast cancer cells. Biol. Pharm. Bull. 2008;31: 769–773. 10.1248/bpb.31.769 [DOI] [PubMed] [Google Scholar]
- Yasuda S, Yasuda T, Hui Y, et al. Concerted action of the cytosolic sulfotransferase, SULT1A3, and catechol-O-methyltransferase in the metabolism of dopamine in SK-N-MC human neuroblastoma cells. Neurosci. Res. 2009;64: 273–279. 10.1016/j.neures.2009.03.011 [DOI] [PubMed] [Google Scholar]
- Kurogi K, Alazizi A, Liu MY, et al. Concerted actions of the catechol O-methyltransferase and the cytosolic sulfotransferase SULT1A3 in the metabolism of catecholic drugs. Biochem. Pharmacol. 2012;84: 1186–1195. 10.1016/j.bcp.2012.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Woude H, Boersma MG, Vervoort J, et al. Identification of 14 quercetin Phase II mono- and mixed conjugates and their formation by rat and human Phase II in vitro model systems. Chem. Res. Toxicol. 2004;17: 1520–1530. 10.1021/tx049826v [DOI] [PubMed] [Google Scholar]
- Donovan JL, Kasim-Karakas S, German JB, et al. Urinary excretion of catechin metabolites by human subjects after red wine consumption. Br. J. Nutr. 2002;87: 31–37. 10.1079/BJN2001482 [DOI] [PubMed] [Google Scholar]
- Roura E, Andrés-Lacueva C, Jáuregui O, et al. Rapid liquid chromatography tandem mass spectrometry assay to quantify plasma (-)-epicatechin metabolites after ingestion of a standard portion of cocoa beverage in humans. J. Agric. Food Chem. 2005;53: 6190–6194. 10.1021/jf050377u [DOI] [PubMed] [Google Scholar]
- Del Rio D, Calani L, Cordero C, et al. Bioavailability and catabolism of green tea flavan-3-ols in humans. Nutrition. 2010;26: 1110–1116. 10.1016/j.nut.2009.09.021 [DOI] [PubMed] [Google Scholar]
- Allali-Hassani A, Pan PW, Dombrovski L, et al. Structural and chemical profiling of the human cytosolic sulfotransferases. PLoS Biol. 2007;5: e97. 10.1371/journal.pbio.0050097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamage N, Barnett A, Hempel N, et al. Human sulfotransferases and their role in chemical metabolism. Toxicol. Sci. 2006;90: 5–22. [DOI] [PubMed] [Google Scholar]
- Tibbs ZE, Rohn-Glowacki KJ, Crittenden F, et al. Structural plasticity in the human cytosolic sulfotransferase dimer and its role in substrate selectivity and catalysis. Drug Metab. Pharmacokinet. 2015;30: 3–20. 10.1016/j.dmpk.2014.10.004 [DOI] [PubMed] [Google Scholar]
- Kakuta Y, Pedersen LG, Carter CW, et al. Crystal structure of estrogen sulphotransferase. Nat. Struct. Biol. 1997;4: 904–908. 10.1038/nsb1197-904 [DOI] [PubMed] [Google Scholar]
- Dong D, Ako R, Wu B. Crystal structures of human sulfotransferases: insights into the mechanisms of action and substrate selectivity. Expert Opin. Drug Metab. Toxicol. 2012;8: 635–646. 10.1517/17425255.2012.677027 [DOI] [PubMed] [Google Scholar]
- Campagna-Slater V, Schapira M. Evaluation of virtual screening as a tool for chemical genetic applications. J. Chem. Inf. Model. 2009;49: 2082–2091. 10.1021/ci900219u [DOI] [PubMed] [Google Scholar]
- Leyh TS, Cook I, Wang T. Structure, dynamics and selectivity in the sulfotransferase family. Drug Metabo. Rev. 2013;45: 423–430. 10.3109/03602532.2013.835625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook I, Wang T, Falany CN, et al. High accuracy in silico sulfotransferase models. J. Biol. Chem. 2013;288: 34494–34501. 10.1074/jbc.M113.510974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hurk P, Wiley FE, Lavoie ET, et al. Activity patterns of biotransformation enzymes in juvenile chickens after in ovo dosage of PCB126. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2007;146: 301–307. 10.1016/j.cbpc.2007.03.004 [DOI] [PubMed] [Google Scholar]
- Wang LQ, James MO. Inhibition of sulfotransferases by xenobiotics. Curr. Drug Metab. 2006;7: 83–104. 10.2174/138920006774832596 [DOI] [PubMed] [Google Scholar]
- Runge-Morris M, Kocarek TA. Regulation of sulfotransferases by xenobiotic receptors. Curr. Drug Metab. 2005;6: 299–307. 10.2174/1389200054633871 [DOI] [PubMed] [Google Scholar]
- Alnouti Y, Klaassen CD. Regulation of sulfotransferase enzymes by prototypical microsomal enzyme inducers in mice. J. Pharmacol. Exp. Ther. 2008;324: 612–621. [DOI] [PubMed] [Google Scholar]
- Runge-Morris M, Kocarek TA. Regulation of sulfotransferase and UDP-glucuronosyltransferase gene expression by the PPARs. PPAR Res. 2009;2009: 728941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaassen CD, Reisman SA. Nrf2 the rescue: effects of the antioxidative/electrophilic response on the liver. Toxicol. Appl. Pharmacol. 2010;244: 57–65. 10.1016/j.taap.2010.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Li W, Su ZY, et al. The complexity of the Nrf2 pathway: beyond the antioxidant response. J. Nutr. Biochem. 2015;26: 1401–1413. 10.1016/j.jnutbio.2015.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh CT, Yen GC. Involvement of p38 MAPK and Nrf2 in phenolic acid-induced P-form phenol sulfotransferase expression in human hepatoma HepG2 cells. Carcinogenesis. 2006;27: 1008–1017. 10.1093/carcin/bgi281 [DOI] [PubMed] [Google Scholar]
- Aleksunes LM, Klaassen CD. Coordinated regulation of hepatic phase I and II drug-metabolizing genes and transporters using AhR-, CAR-, PXR-, PPARα-, and Nrf2-null mice. Drug Metab. Dispos. 2012;40: 1366–1379. 10.1124/dmd.112.045112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisman SA, Yeager RL, Yamamoto M, et al. Increased Nrf2 activation in livers from keap1-knockdown mice increases expression of cytoprotective genes that detoxify electrophiles more than those that detoxify reactive oxygen species. Toxicol. Sci. 2009;108: 35–47. 10.1093/toxsci/kfn267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Hu B, Huang H, et al. Estrogen sulfotransferase is an oxidative stress-responsive gene that gender-specifically affects liver ischemia/reperfusion injury. J. Biol. Chem. 2015;290: 14754–14764. 10.1074/jbc.M115.642124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu R, Tan TH, Kong AN. Butylated hydroxyanisole and its metabolite tert-butylhydroquinone differentially regulate mitogen-activated protein kinases. The role of oxidative stress in the activation of mitogen-activated protein kinases by phenolic antioxidants. J. Biol. Chem. 1997;272: 28962–28970. 10.1074/jbc.272.46.28962 [DOI] [PubMed] [Google Scholar]
- Reisman SA, Csanaky IL, Aleksunes LM, et al. Altered disposition of acetaminophen in Nrf2-null and Keap1-knockdown mice. Toxicol. Sci. 2009;109: 31–40. 10.1093/toxsci/kfp047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti S, Zhang J, Chen G. Redox regulation of human estrogen sulfotransferase (hSULT1E1). Biochem. Pharmacol. 2007;73: 1474–1481. 10.1016/j.bcp.2006.12.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammanahalli JK, Duffel MW. Oxidative modification of rat sulfotransferase 1A1 activity in hepatic tissue slices correlates with effects on the purified enzyme. Drug Metab. Dispos. 2012;40: 298–303. 10.1124/dmd.111.042044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui Y, Yasuda T, Yasuda S, et al. Inhibitory effects of nitrative stress on the sulfation of 17β-estradiol and 4-methoxyestradiol by human MCF 10A mammary Epithelial cells. Biol. Pharm. Bull. 2010;33: 1633–1637. 10.1248/bpb.33.1633 [DOI] [PubMed] [Google Scholar]
- Chandru HK, Chen G. Human hydroxysteroid sulfotransferase 2A1 is down regulated by nitric oxide in human HEP G2 cells. Int. J. Pharmacol. 2010;6: 631–637. [Google Scholar]
- Kim MS, Shigenaga J, Moser A, et al. Suppression of DHEA sulfotransferase (Sult2A1) during the acute-phase response. Am. J. Physiol. Endocrinol. Metab. 2004;287: E731–E738. 10.1152/ajpendo.00130.2004 [DOI] [PubMed] [Google Scholar]
- Kohjitani A, Fuda H, Hanyu O, et al. Regulation of SULT2B1a (pregnenolone sulfotransferase) expression in rat C6 glioma cells: relevance of AMPA receptor-mediated NO signaling. Neurosci. Lett. 2008;430: 75–80. 10.1016/j.neulet.2007.10.023 [DOI] [PubMed] [Google Scholar]
- Sidharthan NP, Minchin RF, Butcher NJ. Cytosolic sulfotransferase 1A3 is induced by dopamine and protects neuronal cells from dopamine toxicity: role of D1 receptor-N-methyl-D-aspartate receptor coupling. J. Biol. Chem. 2013;288: 34364–34374. 10.1074/jbc.M113.493239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurogi K, Davidson G, Mohammed YI, et al. Ethanol sulfation by the human cytosolic sulfotransferases: a systematic analysis. Biol. Pharm. Bull. 2012;35: 2180–2185. 10.1248/bpb.b12-00547 [DOI] [PubMed] [Google Scholar]
- Schneider H, Glatt H. Sulpho-conjugation of ethanol in humans in vivo and by individual sulphotransferase forms in vitro. Biochem. J. 2004;383: 543–549. 10.1042/BJ20040925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti S, Chen G. Ethanol up-regulates phenol sulfotransferase (SULT1A1) and hydroxysteroid sulfotransferase (SULT2A1) in rat liver and intestine. Arch. Physiol. Biochem. 2015;121: 68–74. 10.3109/13813455.2014.992440 [DOI] [PubMed] [Google Scholar]
- Li W, Ning M, Koh KH, et al. 17β-estradiol induces sulfotransferase 2A1 expression through estrogen receptor α. Drug Metab. Dispos. 2014;42: 796–802. 10.1124/dmd.113.055178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacifici GM, Franchi M, Colizzi C, et al. Sulfotransferase in humans: development and tissue distribution. Pharmacology. 1988;36: 411–419. 10.1159/000138330 [DOI] [PubMed] [Google Scholar]
- Cappiello M, Giuliani L, Rane A, et al. Dopamine sulphotransferase is better developed than p-nitrophenol sulphotransferase in the human fetus. Dev. Pharmacol. Ther. 1991;16: 83–88. [PubMed] [Google Scholar]
- Jones AL, Hume R, Bamforth KJ, et al. Estrogen and phenol sulfotransferase activities in human fetal lung. Early Hum. Dev. 1992;28: 65–77. 10.1016/0378-3782(92)90007-4 [DOI] [PubMed] [Google Scholar]
- Kawade N, Onishi S. The prenatal and postnatal development of UDP-glucuronyltransferase activity towards bilirubin and the effect of premature birth on this activity in the human liver. Biochem. J. 1981;196: 257–260. 10.1042/bj1960257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strassburg CP, Strassburg A, Kneip S, et al. Developmental aspects of human hepatic drug glucuronidation in young children and adults. Gut. 2002;50: 259–265. 10.1136/gut.50.2.259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacifici GM, Franchi M, Giuliani L, et al. Development of the glucuronyltransferase and sulphotransferase towards 2-naphthol in human fetus. Dev. Pharmacol. Ther. 1988;14: 108–114. [PubMed] [Google Scholar]
- Pacifici GM, Kubrich M, Giuliani L, et al. Sulphation and glucuronidation of ritodrine in human foetal and adult tissues. Eur. J. Clin. Pharmacol. 1993;44: 259–264. 10.1007/BF00271368 [DOI] [PubMed] [Google Scholar]
- Cappiello M, Giuliani L, Rane A, et al. Dopamine sulphotransferase is better developed than p-nitrophenol sulphotransferase in the human fetus. Dev. Pharmacol. Ther. 1990;16: 83–88. [PubMed] [Google Scholar]
- Barker EV, Hume R, Hallas A, et al. Dehydroepiandrosterone sulfotransferase in the developing human fetus: quantitative biochemical and immunological characterization of the hepatic, renal, and adrenal enzymes. Endocrinology. 1994;134: 982–989. [DOI] [PubMed] [Google Scholar]
- Richard K, Hume R, Kaptein E, et al. Sulfation of thyroid hormone and dopamine during human development: ontogeny of phenol sulfotransferases and arylsulfatase in liver, lung, and brain. J. Clin. Endocrinol. Metab. 2001;86: 2734–2742. [DOI] [PubMed] [Google Scholar]
- Stanley EL, Hume R, Coughtrie MW. Expression profiling of human fetal cytosolic sulfotransferases involved in steroid and thyroid hormone metabolism and in detoxification. Mol. Cell. Endocrinol. 2005;240: 32–42. 10.1016/j.mce.2005.06.003 [DOI] [PubMed] [Google Scholar]
- Duanmu Z, Weckle A, Koukouritaki SB, et al. Developmental expression of aryl, estrogen, and hydroxysteroid sulfotransferases in pre-and postnatal human liver. J. Pharmacol. Exp. Ther. 2006;316: 1310–1317. [DOI] [PubMed] [Google Scholar]
- Adjei AA, Gaedigk A, Simon SD, et al. Interindividual variability in acetaminophen sulfation by human fetal liver: implications for pharmacogenetic investigations of drug-induced birth defects. Birth Defects Res. A Clin. Mol. Teratol. 2008;82: 155–165. 10.1002/(ISSN)1542-0760 [DOI] [PubMed] [Google Scholar]
- Wang J, Falany JL, Falany CN. Expression and characterization of a novel thyroid hormone-sulfating form of cytosolic sulfotransferase from human liver. Mol. Pharmacol. 1998;53: 274–282. [DOI] [PubMed] [Google Scholar]
- Her C, Kaur GP, Athwal RS, et al. Human sulfotransferase SULT1C1: cDNA cloning, tissue-specific expression, and chromosomal localization. Genomics. 1997;41: 467–470. 10.1006/geno.1997.4683 [DOI] [PubMed] [Google Scholar]
- Sakakibara Y, Yanagisawa K, Katafuchi J, et al. Molecular cloning, expression, and characterization of novel human SULT1C sulfotransferases that catalyze the sulfonation of N-hydroxy-2-acetylaminofluorene. J. Biol. Chem. 1998;273: 33929–33935. 10.1074/jbc.273.51.33929 [DOI] [PubMed] [Google Scholar]
- Geese WJ, Raftogianis RB. Biochemical characterization and tissue distribution of human SULT2B1. Biochem. Biophys. Res. Commun. 2001;288: 280–289. 10.1006/bbrc.2001.5746 [DOI] [PubMed] [Google Scholar]
- Shimizu M, Tamura HO. Identification and localization of two hydroxysteroid sulfotransferases in the human brain. J. Health Sci. 2002;48: 467–472. 10.1248/jhs.48.467 [DOI] [Google Scholar]
- Falany CN, Xie X, Wang J, et al. Molecular cloning and expression of novel sulphotransferase-like cDNAs from human and rat brain. Biochem. J. 2000;346: 857–864. 10.1042/bj3460857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara Y, Suiko M, Pai TG, et al. Highly conserved mouse and human brain sulfotransferases: molecular cloning, expression, and functional characterization. Gene. 2002;285: 39–47. 10.1016/S0378-1119(02)00431-6 [DOI] [PubMed] [Google Scholar]
- Minchin RF, Lewis A, Mitchell D, et al. Sulfotransferase 4A1. Int. J. Biochem. Cell. Biol. 2008;40: 2686–2691. 10.1016/j.biocel.2007.11.010 [DOI] [PubMed] [Google Scholar]
- Meltzer HY, Brennan MD, Woodward ND, et al. Association of Sult4A1 SNPs with psychopathology and cognition in patients with schizophrenia or schizoaffective disorder. Schizophr. Res. 2008;106: 258–264. 10.1016/j.schres.2008.08.029 [DOI] [PubMed] [Google Scholar]
- Vascotto SG, Beckham Y, Kelly GM. The zebrafish’s swim to fame as an experimental model in biology. Biochem. Cell. Biol. 1997;75: 479–485. 10.1139/o97-081 [DOI] [PubMed] [Google Scholar]
- Westerfield M. The zebrafish book. Eugene: University of Oregon Press; 2000. [Google Scholar]
- Sipes NS, Padilla S, Knudsen TB. Zebrafish-as an integrative model for twenty-first century toxicity testing. Birth Defects Res. C Embryo Today. 2011;93: 256–267. 10.1002/bdrc.v93.3 [DOI] [PubMed] [Google Scholar]
- MacRae CA, Peterson RT. Zebrafish as tools for drug discovery. Nat. Rev. Drug Discov. 2015;14: 721–731. 10.1038/nrd4627 [DOI] [PubMed] [Google Scholar]
- Liu TA, Bhuiyan S, Liu MY, et al. Zebrafish as a model for the study of the phase II cytosolic sulfotransferases. Curr. Drug Metab. 2010;11: 538–546. 10.2174/138920010791636158 [DOI] [PubMed] [Google Scholar]
- Kurogi K, Liu TA, Sakakibara Y, et al. The use of zebrafish as a model system for investigating the role of the SULTs in the metabolism of endogenous compounds and xenobiotics. Drug Metab. Rev. 2013;45: 431–440. 10.3109/03602532.2013.835629 [DOI] [PubMed] [Google Scholar]
- Yasuda S, Liu CC, Takahashi S, et al. Identification of a novel estrogen-sulfating cytosolic SULT from zebrafish: molecular cloning, expression, characterization, and ontogeny study. Biochem. Biophys. Res. Commun. 2005;330: 219–225. 10.1016/j.bbrc.2005.02.152 [DOI] [PubMed] [Google Scholar]
- Liu MY, Sugahara T, Yasuda S, et al. Identification of a novel zebrafish SULT1 cytosolic sulfotransferase: cloning, expression, characterization, and developmental expression study. Arch. Biochem. Biophy. 2005;437: 10–19. 10.1016/j.abb.2005.02.029 [DOI] [PubMed] [Google Scholar]
- Yasuda S, Kumar AP, Liu MY, et al. Identification of a novel thyroid hormone-sulfating cytosolic sulfotransferase, SULT1 ST5, from zebrafish. FEBS J. 2005;272: 3828–3837. 10.1111/j.1742-4658.2005.04791.x [DOI] [PubMed] [Google Scholar]
- Yasuda S, Liu MY, Yang YS, et al. Identification of novel hydroxysteroid-sulfating cytosolic SULTs, SULT2 ST2 and SULT2 ST3, from zebrafish: cloning, expression, characterization, and developmental expression. Arch. Biochem. Biophys. 2006;455: 1–9. 10.1016/j.abb.2006.09.004 [DOI] [PubMed] [Google Scholar]
- Liu TA, Bhuiyan S, Snow R, et al. Identification and characterization of two novel cytosolic sulfotransferases, SULT1 ST7 and SULT1 ST8, from zebrafish. Aquat. Toxicol. 2008;89: 94–102. 10.1016/j.aquatox.2008.06.005 [DOI] [PubMed] [Google Scholar]
- Yasuda T, Yasuda S, Williams FE, et al. Characterization and ontogenic study of novel steroid-sulfating SULT3 sulfotransferases from zebrafish. Mol. Cell. Endocrinol. 2008;294: 29–36. 10.1016/j.mce.2008.06.014 [DOI] [PubMed] [Google Scholar]
- Sugahara T, Liu CC, Pai TG, et al. Molecular cloning, expression, and functional characterization of a novel zebrafish cytosolic sulfotransferase. Biochem. Biophys. Res. Commun. 2003;300: 725–730. 10.1016/S0006-291X(02)02915-7 [DOI] [PubMed] [Google Scholar]
- Sugahara T, Liu CC, Carter G, et al. cDNA cloning, expression, and functional characterization of a zebrafish SULT1 cytosolic sulfotransferase. Arch. Biochem. Biophys. 2003;414: 67–73. 10.1016/S0003-9861(03)00172-3 [DOI] [PubMed] [Google Scholar]
- Sugahara T, Liu CC, Pai TG, et al. Sulfation of hydroxychlorobiphenyls. Molecular cloning, expression, and functional characterization of zebrafish SULT1 sulfotransferases. Eur. J. Biochem. 2003;270: 2404–2411. 10.1046/j.1432-1033.2003.03608.x [DOI] [PubMed] [Google Scholar]
- Sugahara T, Liu CC, Pai TG, et al. Sulphonation of dehydroepiandrosterone and neurosteroids: molecular cloning, expression, and functional characterization of a novel zebrafish SULT2 cytosolic sulfotransferase. Biochem. J. 2003;375: 785–791. 10.1042/bj20031050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkimoto K, Sugahara T, Sakakibara Y, et al. Sulfonation of environmental estrogens by zebrafish cytosolic sulfotransferases. Biochem. Biophys. Res. Commun. 2003;309: 7–11. 10.1016/S0006-291X(03)01524-9 [DOI] [PubMed] [Google Scholar]
- Ohkimoto K, Liu MY, Suiko M, et al. Characterization of a zebrafish estrogen-sulfating cytosolic sulfotransferase: inhibitory effects and mechanism of action of phytoestrogens. Chem. Biol. Interact. 2004;147: 1–7. 10.1016/j.cbi.2003.09.001 [DOI] [PubMed] [Google Scholar]
- Aksoy IA, Wood TC, Weinshilboum R. Human liver estrogen sulfotransferase: identification by cDNA cloning and expression. Biochem. Biophys. Res. Commun. 1994;200: 1621–1629. 10.1006/bbrc.1994.1637 [DOI] [PubMed] [Google Scholar]
- Crittenden F, Thomas H, Ethen CM, et al. Inhibition of SULT4A1 expression induces up-regulation of phototransduction gene expression in 72-hour postfertilization zebrafish larvae. Drug Metab. Dispos. 2014;42: 947–953. 10.1124/dmd.114.057042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crittenden F, Thomas HR, Parant JM, et al. Activity suppression behavior phenotype in SULT4A1 frameshift mutant zebrafish. Drug Metab. Dispos. 2015;43: 1037–1044. 10.1124/dmd.115.064485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed YI, Kurogi K, Shaban AA, et al. Identification and characterization of zebrafish SULT1 ST9, SULT3 ST4, and SULT3 ST5. Aquat. Toxicol. 2012;112–113: 11–18. 10.1016/j.aquatox.2012.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda S, Burgess M, Yasuda T, et al. A novel hydroxysteroid-sulfating cytosolic sulfotransferase, SULT3 ST3, from zebrafish: identification, characterization, and ontogenic study. Drug Metab. Lett. 2009;3: 217–227. 10.2174/187231209790218154 [DOI] [PubMed] [Google Scholar]
- McLean DL, Fetcho JR. Ontogeny and innervation patterns of dopaminergic, noradrenergic, and serotonergic neurons in larval zebrafish. J. Comp. Neurol. 2004;480: 38–56. 10.1002/(ISSN)1096-9861 [DOI] [PubMed] [Google Scholar]
- Power DM, Llewellyn L, Faustino M, et al. Thyroid hormones in growth and development of fish. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2001;130: 447–459. 10.1016/S1532-0456(01)00271-X [DOI] [PubMed] [Google Scholar]
- Brown DD. The role of thyroid hormone in zebrafish and axolotl development. Proc. Natl. Acad. Sci. U.S.A. 1997;94: 13011–13016. 10.1073/pnas.94.24.13011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu HJ, Hsiao P, Kuo MW, et al. Expression of zebrafish cyp11a1 as a maternal transcript and in yolk syncytial layer. Gene Exp. Patterns. 2002;2: 219–222. 10.1016/S1567-133X(02)00059-5 [DOI] [PubMed] [Google Scholar]
- Hsu HJ, Hsu NC, Hu MC, et al. Steroidogenesis in zebrafish and mouse models. Mol. Cell Endocrinol. 2006;248: 160–163. 10.1016/j.mce.2005.10.011 [DOI] [PubMed] [Google Scholar]
- Sawyer SJ, Gerstner KA, Callard GV. Real-time PCR analysis of cytochrome P450 aromatase expression in zebrafish: gene specific tissue distribution, sex differences, developmental programming, and estrogen regulation. Gen. Comp. Endocrinol. 2006;147: 108–117. 10.1016/j.ygcen.2005.12.010 [DOI] [PubMed] [Google Scholar]
- Hamad A, Kluk M, Fox J, et al. The effects of aromatase inhibitors and selective estrogen receptor modulators on eye development in the zebrafish (Danio rerio). Curr. Eye Res. 2007;32: 819–827. 10.1080/02713680701573712 [DOI] [PubMed] [Google Scholar]
- Houser A, McNair C, Piccinini R, et al. Effects of estrogen on the neuromuscular system in the embryonic zebrafish (Danio rerio). Brain Res. 2011;1381: 106–116. 10.1016/j.brainres.2011.01.033 [DOI] [PubMed] [Google Scholar]
- Kurogi K, Dillon J, Nasser A, et al. Sulfation of drug compounds by the zebrafish cytosolic sulfotransferases (SULTs). Drug Metab. Lett. 2010;4: 62–68. 10.2174/187231210791292690 [DOI] [PubMed] [Google Scholar]