Abstract
Chemical control strategies are driving the evolution of pesticide resistance in pest populations. Understanding the genetic mechanisms of these evolutionary processes is of crucial importance to develop sustainable resistance management strategies. The acaricide pyflubumide is one of the most recently developed mitochondrial complex II inhibitors with a new mode of action that specifically targets spider mite pests. In this study, we characterize the molecular basis of pyflubumide resistance in a highly resistant population of the spider mite Tetranychus urticae. Classical genetic crosses indicated that pyflubumide resistance was incompletely recessive and controlled by more than one gene. To identify resistance loci, we crossed the resistant population to a highly susceptible T. urticae inbred strain and propagated resulting populations with and without pyflubumide exposure for multiple generations in an experimental evolution set-up. High-resolution genetic mapping by a bulked segregant analysis approach led to the identification of three quantitative trait loci (QTL) linked to pyflubumide resistance. Two QTLs were found on the first chromosome and centered on the cytochrome P450 CYP392A16 and a cluster of CYP392E6-8 genes. Comparative transcriptomics revealed a consistent overexpression of CYP392A16 and CYP392E8 in the experimental populations that were selected for pyflubumide resistance. We further corroborated the involvement of CYP392A16 in resistance by in vitro functional expression and metabolism studies. Collectively, these experiments uncovered that CYP392A16 N-demethylates the toxic carboxamide form of pyflubumide to a non-toxic compound. A third QTL coincided with cytochrome P450 reductase (CPR), a vital component of cytochrome P450 metabolism. We show here that the resistant population harbors three gene copies of CPR and that this copy number variation is associated with higher mRNA abundance. Together, we provide evidence for detoxification of pyflubumide by cytochrome P450s that is likely synergized by gene amplification of CPR.
Author summary
Our understanding of the causal genetic variants that drive the evolution of quantitative traits, such as polygenic pesticide resistance, remains very limited. Here, we followed a high-resolution genetic mapping approach to localize the genetic variants that cause pyflubumide resistance in the two-spotted spider mite Tetranychus urticae. Three well-supported QTL were uncovered and pointed towards a major role for cytochrome P450-mediated detoxification. Cis-regulatory variation for cytochrome P450s was observed, and in vitro cytochrome P450 experiments showed that pyflubumide was metabolized into a non-toxic derivate. A third QTL centered on cytochrome P450 reductase (CPR), which is required for cytochrome P450 activity, and is amplified in pyflubumide resistant populations. Our results indicate that pyflubumide resistance is mediated by cytochrome P450 detoxification that is enhanced by gene amplification at the CPR locus.
Introduction
Arthropod herbivores cause over US$450 billion in crop yield loss worldwide [1], and pesticides of various modes of action have been developed to control populations of these pests. Unfortunately, our past and continuing dependence on chemical control strategies is driving the evolution of pesticide resistance in pest populations [2]. Pesticide resistance can be achieved by genetic changes in the molecular target of pesticides (toxicodynamic resistance). These genetic changes are most often non-synonymous substitutions in coding sequences, but (dosage sensitive) gene amplification and differential transcriptional regulation have also been reported, all of which lead to decreased pesticide sensitivity [2,3]. Alternatively, genetic variants that alter the penetration, metabolism, sequestration, efflux, and excretion of pesticides can underlie pesticide resistance (toxicokinetic resistance) [2,4]. Multi-gene families such as cytochrome P450 monooxygenases (CYP), ABC transporters, glutathione S-transferases, and carboxyl/cholinesterases are often associated with the metabolic changes that lead to toxicokinetic resistance. Transcriptomic studies of major global pests have shown that heritable changes in the transcriptional regulation of detoxification genes often strongly associate with pesticide resistance [5–8]. However, our understanding of the causal genetic variation that regulates these transcriptional changes remains limited, and comes disproportionally from insect model species such as drosophilid fruit flies. Genetic changes in both cis and trans regulation can alter the transcription of detoxification genes [2]. For instance, in Drosophila melanogaster, a transposable element insertion in the 5’-UTR region of Cyp6g1 leads to higher transcript levels that underlie resistance to DDT [9,10]. Cis-regulatory variation underpinning the overexpression of cytochrome P450s have been found in a number of insects, including the African malaria vector Anopheles funestus [11,12]. Only infrequently, changes in detoxification gene dosage has been associated with resistance, as for resistance to organophosphates by esterase sequestration in insect pests [3,13]. Alternatively, genetic variation affecting the coding sequence of a detoxification gene can also underlie toxicokinetic resistance. A single point mutation in the GSTe2 gene of A. funestus results in pyrethroid and DDT resistance [14]. In dipterans, mutations in the αE7 esterase gene have been found to contribute to a broad resistance to organophosphates [15,16], whereas in the moth Helicoverpa armigera, a gene conversion event generated CYP337B3 which codes for a novel cytochrome P450 that is able to metabolize fenvalerate [17,18].
The genetic architecture of pesticide resistance can therefore be variable, and can have independent evolutionary origins across different pest populations [19–22]. While monogenic resistance is typically toxicodynamic, and can lead to very high levels of pesticide resistance [22–24], polygenic resistance is nonetheless very common, and has been more challenging to study. For this mode of resistance inheritance, neither the molecular mechanisms that are involved, nor their additive or synergistic effects, are well understood. However, the accelerating development of genetic and genomic resources in diverse arthropods enables high-resolution genetic mapping approaches to characterize complex genetic architectures of pesticide resistance [25,26]. One such approach is bulked segregant analysis (BSA) genetic mapping [25]. A BSA approach relies on crossing parental strains with contrasting phenotypes, bulking (pooling) the segregating progeny that display phenotypic extremes, and identifying genomic intervals with divergent allele frequencies across progeny bulks. BSA genetic mapping has been successfully applied in arthropods to uncover both monogenic and quantitative traits, including pigmentation and host plant use [23,27–33]. The haplodiploid spider mite Tetranychus urticae is a major global agricultural pest which rapidly develops resistance to pesticides of various modes of action [34]. As revealed by recent studies, T. urticae is highly amenable to BSA genetic mapping, with QTL, including for quantitative pesticide resistance, narrowed to small genomic intervals that have suggested specific candidate genes and causal alleles [29,33]. In some instances, known causal variants were recovered [35–38], confirming the validity of the narrow genomic intervals identified by BSA studies using large, highly replicated T. urticae experimental populations [25].
The development of pesticides with novel modes of action is crucial to maintain control over pest populations that have developed multi-resistance. Pyflubumide is a carboxanilide acaricide and inhibits mitochondrial complex II, or succinate dehydrogenase. This recently developed compound is highly selective to spider mite pest species [39,40] and acts as a pro-acaricide that requires bioactivation into a toxic derivative. Pyflubumide is converted within the spider mite body into its active deacylated metabolite (carboxamide form, NNI-0711-NH) that strongly inhibits the mitochondrial complex II through binding to the quinone-binding pocket [39,40]. In the current study, we characterize the genetic basis of resistance to this novel acaricide using two resistant T. urticae strains (JPR-R1 and JPR-R2). Target-site resistance is not involved in high-level pyflubumide resistance in these strains, and synergism/antagonism bioassays strongly suggested the involvement of cytochrome P450s [41]. In this study, we employed high-resolution BSA genetic mapping and uncovered three QTL associated with pyflubumide resistance. In parallel, transcriptomic analyses were conducted on these experimental populations to further characterize the molecular mechanisms of pyflubumide resistance. In vitro functional characterization of the cytochrome P450 CYP392A16, a QTL candidate, suggested a role for resistance to pyflubumide and its active (deacylated) carboxamide metabolite. Additionally, another QTL interval was centered on cytochrome P450 reductase (CPR), which is required for cytochrome P450 activity, and is amplified in the resistant populations. Together, our results indicate that the mechanism of pyflubumide detoxification involves cytochrome P450 activity enhanced by gene amplification at the CPR locus.
Results
Mode of inheritance of pyflubumide resistance
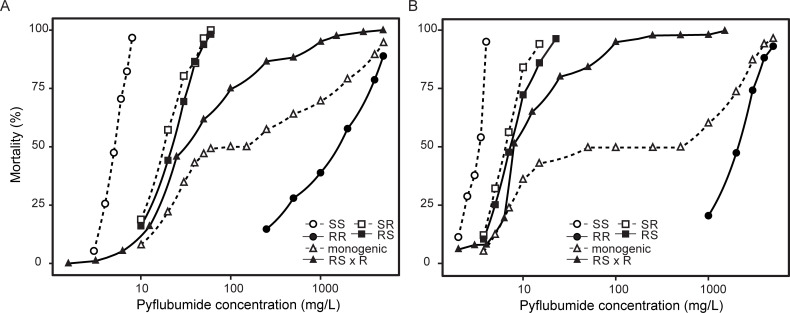
Previously, we selected a field-collected strain for higher resistance levels to pyflubumide and obtained two highly resistant strains, JPR-R1 and JPR-R2 [41]. In this study, we first confirmed the high levels of pyflubumide resistance and uncovered LC50 values higher than 1,000 mg/L of pyflubumide for both strains (S1 Table). Reciprocal crosses between resistant (JPR-R1 and JPR-R2) and susceptible (JP-S and Wasatch) strains revealed that pyflubumide resistance is not maternally inherited and has an incomplete recessive mode of inheritance (Fig 1 and S1 Table). The dose-response relationships of the two back-crossed populations were significantly different from the expected curves for a recessive monogenic mode of inheritance (p< 0.001) [42]. These results indicate that pyflubumide resistance in both JPR-R1 and JPR-R2 was determined by multiple loci.
Fig 1. Pyflubumide resistance has a polygenic basis in JPR-R1 and JPR-R2.
(A) Dose-response relationships of pyflubumide toxicity on JPR-R1, JP-S (circles), reciprocal crosses (squares), and back-crosses (triangles). (B) Dose-response relationships of pyflubumide toxicity on JPR-R2, Wasatch (circles), reciprocal crosses (squares), and back-crosses (triangles). Panels show that pyflubumide resistance has an incomplete recessive mode of inheritance in both resistant strains. After back-crossing, the observed dose-response relationships significantly differ from those calculated under the hypothesis of a recessive monogenic mode of inheritance (p< 0.001) (black versus white triangles, respectively), indicating that pyflubumide resistance is determined by multiple loci in both JPR-R1 and JPR-R2.
Response to pyflubumide selection pressure in an experimental evolution set-up
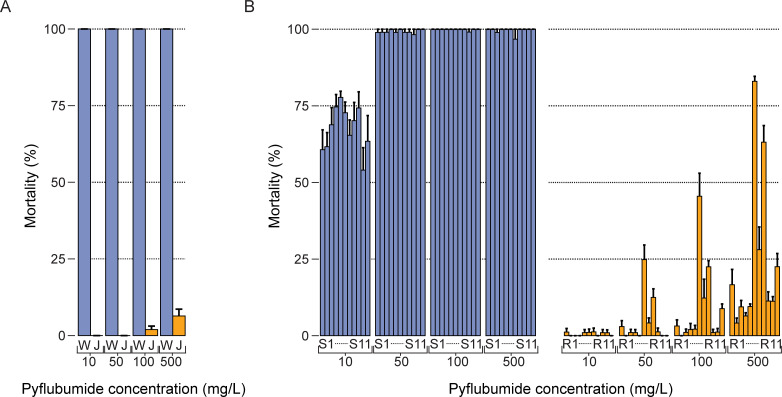
To further characterize the genetic architecture of pyflubumide resistance, we crossed a single haploid JPR-R1 male to diploid females of the susceptible inbred WasX strain. Mortality at 10, 50, 100, and 500 mg/L of pyflubumide was significantly different between JPR-R1 and WasX (X2 = 41.631, df = 1, p < 0.0001) (Fig 2). The segregating population resulting from the WasX × JPR-R1 cross was allowed to expand and develop for two months (approximately three-four generations), after which 11 pairs (22 populations) were set-up and allowed to propagate for approximately 35 generations. In this pairwise set-up, one sister population was exposed to pyflubumide on sprayed plants (a selected population), whereas the other was maintained on unsprayed plants (an unselected, control population). After a prolonged selection with increasing doses of pyflubumide, all populations were phenotyped, revealing that mortality at four pyflubumide doses was significantly lower for pyflubumide-selected populations as compared to the control populations (Fig 2) (X2 = 29.4361, df = 1, p<0.0001). Mortality was also significantly different across the four pyflubumide doses (each p-value < 0.0001).
Fig 2. Selection of segregating populations resulted in high levels of pyflubumide resistance.
(A) Mortality after exposure to 10, 50, 100, and 500 mg/L of pyflubumide of WasX (W) and JPR-R1 (J) (depicted in blue and orange, respectively), the two parental strains of the BSA cross. Error bars represent standard errors. Mortality was significantly different at every pyflubumide dose between JPR-R1 and WasX (p < 0.0001). (B) Mortality after exposure to 10, 50, 100, and 500 mg/L of pyflubumide of control (S1-S11) and pyflubumide-selected (R1-R11) populations (depicted in blue and orange, respectively). Error bars represent standard errors.
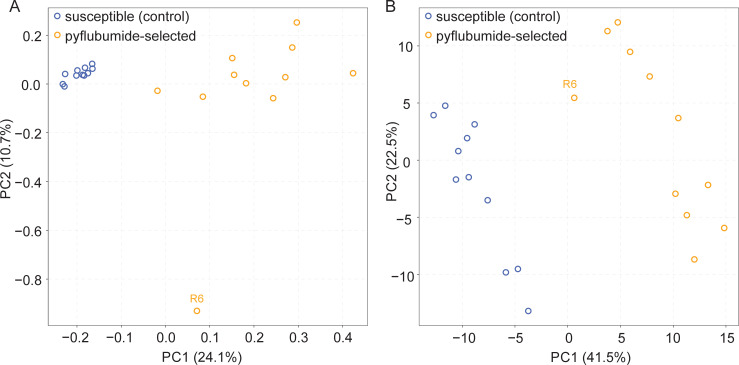
We subsequently sequenced DNA and RNA from the 22 experimental populations, and investigated both genomic and transcriptomic responses to pyflubumide selection by a principal component analysis (PCA). Using per sample allele frequencies at 214,502 high-quality SNP loci that distinguished JPR-R1 and WasX, control populations clustered closely together and the pyflubumide-selected populations were distinct from these controls along principal component 1 (PC1). Compared to the control populations, pyflubumide-selected populations were more dispersed; most notably, for an unknown reason, but that might have resulted from a population bottleneck that can occur in populations under pesticide selection pressure, population R6 stood out from the other selected populations along principal component 2 (PC2) (Fig 3A). Using our RNAseq data that consisted of a single RNA replicate per experimental population, the genome-wide transcript levels also clearly separated the control and pyflubumide-selected populations along PC1 (Fig 3B). Considering each experimental population as a replicate, a total of 413 genes were identified as differentially expressed between the pyflubumide-selected and control populations (S2 Data).
Fig 3. Selection for pyflubumide resistance is associated with a genomic and transcriptomic response.
(A) A principal component analysis of the eleven susceptible (control) and pyflubumide-selected populations using genome-wide allele frequency data at informative genomic loci. (B) A principal component analysis of the eleven susceptible (control) and pyflubumide-selected populations using genome-wide transcript levels. For both panels, each circle represents an individual population, the R6 population is labelled, treatment is color-coded as indicated (top left), and the percentage of explained variance is given in parentheses for each principal component (PC).
Genetic mapping uncovers multiple QTL associated with pyflubumide selection
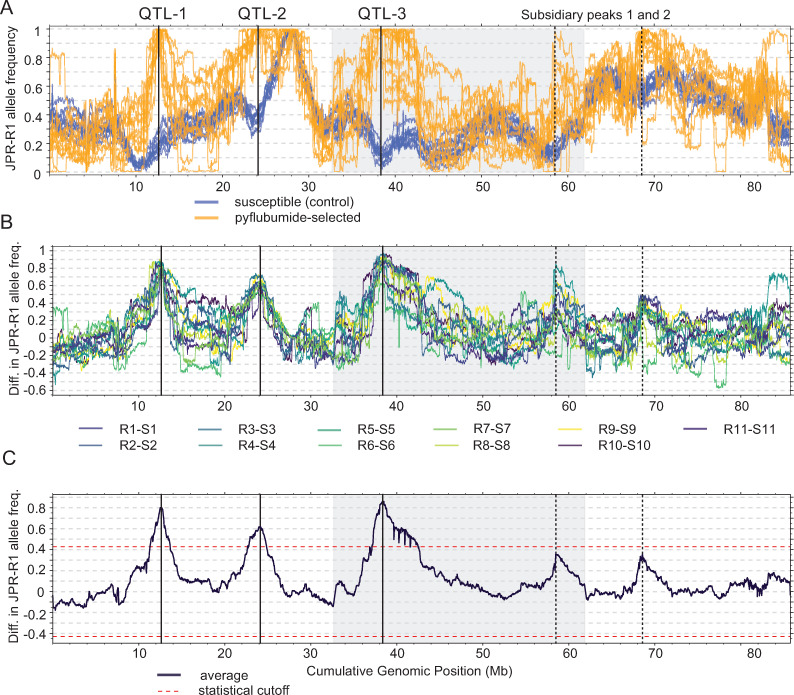
To locate the genomic regions that underlie pyflubumide resistance in the pyflubumide-selected populations, we employed a bulked segregant analysis (BSA) approach using high-quality SNP loci [25,29]. Genome-wide JPR-R1 allele frequencies between pyflubumide-selected and susceptible (control) populations (Fig 4A) and the matched pairs of populations (Fig 4B) revealed large deviations in allele frequencies on chromosomes 1 and 2. Using previously developed permutation-based methods for QTL detection with replicated paired selected and control populations [29], and with a false discovery rate (FDR) of 0.05, we identified two QTL on chromosome 1 (hereafter QTL-1 and QTL-2) and one on chromosome 2 (hereafter QTL-3) in the pyflubumide-selected populations (Figs 4C and S1). In all cases, alleles from the resistant JPR-R1 parent were selected, and for all pyflubumide-selected populations, near-fixation of the JPR-R1 haplotype was observed at the peaks of the three QTL regions (S1 Fig). Population R6 exhibited the lowest JPR-R1 allele frequency near the peaks of QTL-2 and QTL-3 (S1 Fig), a finding that is consistent with its lower resistance levels (Fig 2). We also noted two additional genomic regions where the allele frequency differences nearly exceeded the significance threshold (hereafter referred to as subsidiary peaks 1 and 2) (Fig 4C). Finally, a BSA mapping approach was also performed using informative SNP loci identified via the RNAseq data. This analysis further confirmed the three QTL, as well as the two subsidiary peaks (S2 Fig). Of the 413 differentially expressed genes (DEGs) detected between the selected and unselected populations, 123 (30.8%) were within the three top QTL genomic intervals defined as the regions with allele frequencies similar to those of the peaks (at least one of five consecutive windows having allele frequency values within 0.005 of the peak values). Specifically, 24 DEGs were located within the top 180 kb QTL-1 region, whereas 25 and 74 DEGs were within the 130 kb QTL-2 and 110 kb QTL-3 genomic intervals, respectively. The linkage information suggests that cis-regulation could play a dominant role in the different expression levels of these 123 DEGs (note that in the three QTL regions there are large differences in the frequency of the haplotypes from the two parents between the selected and unselected replicates, so cis-expression QTLs (eQTLs) are expected to be uncovered compared to distant genomic locations where haplotype frequencies are not significantly different). The RNA levels of the other (QTL-interval distant) 290 DEGs are presumably trans-regulated, with the different RNA levels controlled by genetic variants within the three QTL regions that genetically differentiate the selected and control populations.
Fig 4. BSA genetic mapping reveals three QTL for pyflubumide resistance.
(A) Frequency of JPR-R1 alleles in the susceptible control (blue lines) and pyflubumide-selected populations (orange lines) as assessed in a sliding window analysis. (B) Genome-wide differences in JPR-R1 allele frequencies between the eleven paired populations (R1-S1 to R11-S11), as assessed in a sliding window analysis. (C) Averaged genome-wide difference in JPR-R1 allele frequency using the eleven paired populations (R1-S1 to R11-S11). Dashed red lines delineate statistical significance for QTL detection as assessed by a permutation approach (FDR of 5%). In all panels, chromosomes are ordered by decreasing length and are indicated by alternating shading. Three QTL (QTL-1, QTL-2, and QTL-3) exceed the 5% FDR threshold; two sub-threshold peaks are also labeled (subsidiary peaks 1 and 2).
Candidate genes for pyflubumide resistance are linked to cytochrome P450-mediated detoxification
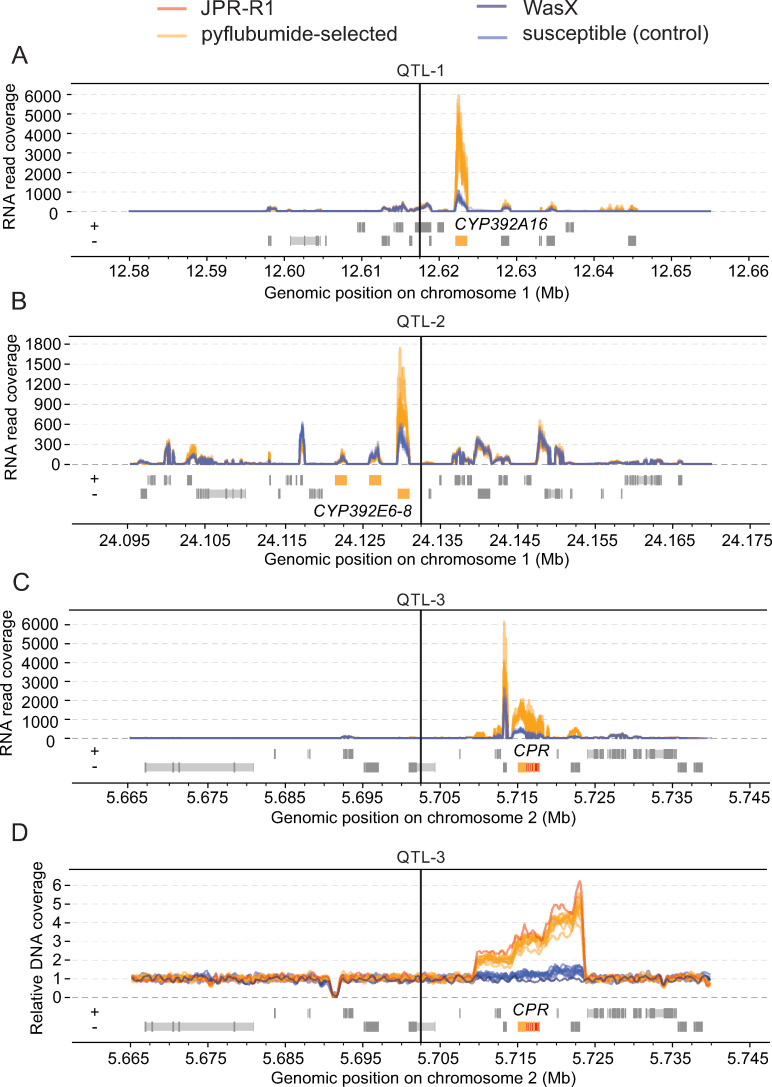
In Tetranychus spider mites, previous BSA studies of similar experimental design revealed that the average of peaks from replicated paired populations are often within several tens of kb from causal genes and variants [25,27,33]. Therefore, we examined ~75 kb genomic intervals that bracket the average of the BSA peaks to detect potential causal candidate genes (Fig 5 and S2 Data). For QTL-1, the cytochrome P450 CYP392A16 was the only gene in the interval with a predicted metabolic function that could be readily linked with xenobiotic metabolism of arthropods. CYP392A16 was within 10 kb of the averaged BSA peak (Fig 5A), and was one of three genes within the ~75 kb genomic interval with significantly higher expression in pyflubumide-selected populations compared to controls (S2 Data), a potential signal of cis-regulatory variation. Specifically, transcript levels of CYP392A16 were ~five-fold higher in pyflubumide-selected populations (Fig 5A, S3 Fig and S2 Data). DNA read coverage indicated that the segregating populations and the two parental strains had the same copy number of CYP392A16 (a single copy), ruling out copy number variation (CNV) as underpinning the higher transcript levels of CYP392A16 (S4 Fig). The ~75 kb genomic interval of QTL-1 also included tetur06g04470, a gene that codes for a protein that is classified as a member of the AIP/AIPL1 family (IPR039663), a group of proteins that include aryl hydrocarbon receptor (AhR)-interacting proteins. AhR participates in a signaling pathway that induces the transcription of detoxification genes in insects and changes in this pathway can promote pesticide resistance [43,44]. However, no significant differential transcription, nor non-synonymous single-nucleotide differences were observed for tetur06g04470, suggesting that tetur06g04470 likely does not contribute to pyflubumide resistance in our strains.
Fig 5. Genes at significant BSA peaks suggest a prominent role for cytochrome P450 activity in resistance to pyflubumide.
RNAseq read coverage and gene models in ~75kb genomic windows surrounding QTL-1 (A), QTL-2 (B), and QTL-3 (C). (D) Relative DNA coverage and gene models in a ~75kb genomic window surrounding QTL-3. For all panels, vertical lines indicate the locations of the averaged BSA peaks on the first (A and B) and second chromosome (C and D). Candidate genes CYP392A16 (A), CYP392E6-8 (B), and CPR (C and D) are highlighted in yellow and red (exons and introns, respectively). Otherwise, coding exons and introns are depicted as dark gray and lighter gray boxes, respectively. Symbols + and–denote forward and reverse gene orientations. Coverage is color-coded according to treatment and strain (legend, top).
A cluster of three cytochrome P450s, CYP392E6-8, was located very close to the BSA peak of QTL-2 (Fig 5B), and no other genes with obvious roles in arthropod detoxification were located in the interval. CYP392E8, which is located only 2 kb from the QTL-2 peak, was the only gene in the region that exhibited a significant increase in transcription in pyflubumide-selected populations (a fold change of ~2) (Fig 5 and S2 Data). Although substantial CNV for this cytochrome P450 gene cluster has been reported in other T. urticae strains [29], all CYP genes in the interval were single copy as revealed by genomic read coverage in the parental and segregating populations (S4 Fig).
No cytochrome P450s or genes encoding other detoxification enzymes were apparent at the QTL-3 peak region. However, NADPH cytochrome P450 reductase (tetur18g03390, CPR), the necessary electron donor for cytochrome P450 metabolism, was within 20 kb of the averaged BSA peak for QTL-3 (Fig 5). Previous work with T. urticae revealed an association between a CPR coding sequence variant and cytochrome P450-mediated detoxification of pesticides [29,33]; however, short-read DNA alignments revealed that CPR harbored no coding sequence differences between JPR-R1 and WasX. However, DNA read coverage revealed an ~14 kb genomic region that is copy number variable between the parental strains, and that harbors five genes including CPR (Fig 5D and S2 Data). Upstream of CPR, two genes coding for a hypothetical protein without clear homology to functionally characterized protein domains (tetur18g03400) and a protein that harbors a folate-binding domain (tetur18g03410, PF01571—cl33847) were within the copy number variable region of JPR-R1. Downstream of CPR, two genes coding for a ribosomal protein (tetur18g03380, PF00238—PTZ00054) and a archease protein (tetur18g03370, PF01951—MTH1598/TM1083) were also copy number variable. The short-read alignments revealed no variable sites within the copy number variable genomic region of JPR-R1, indicating that the different copies of this region are identical. Whereas WasX and the control populations were predicted to have a single copy of the region harboring CPR, for JPR-R1 and the resistant populations variable gene copies were predicted, with JPR-R1 having approximately three copies of CPR. For CPR, an additional copy number assay using an independent method (quantitative PCR) confirmed that CPR was present in an ~3:1 ratio in the genomes of pyflubumide-selected versus control populations (S3 Fig). This ratio was mirrored in the RNAseq read data where up-regulation of CPR transcription was ~3.66 in pyflubumide-selected versus control populations (Figs 5 and S3). Collectively, this indicates that CNV of CPR resulted in higher mRNA levels in JPR-R1 and pyflubumide-selected populations. It was also striking that in the control (unselected) populations, the JPR-R1 allele frequencies dipped to near zero coincident with the CPR-associated QTL-3 peak (Figs 4A and S1), potentially reflecting a negative pleiotropic effect on fitness that becomes apparent in absence of pyflubumide selection (see Discussion). This pattern was absent or less apparent for the other QTL peaks (for instance, a superficially similar pattern for QTL-1 is likely explained by linkage drag from a proximal locus at ~10 Mb on chromosome 1 that is unrelated to pyflubumide selection).
Consistent with previous work [41], our BSA genetic mapping with JPR-R1 excluded target-site insensitivity as a contributing mechanism to pyflubumide resistance. Only one of the genes encoding a mitochondrial complex II subunit, the known molecular target(s) of pyflubumide, was near a QTL peak (tetur08g03210, which is located ~640 kb from the BSA peak for QTL-2). Although the two subsidiary BSA peaks did not reach the significance threshold for QTL detection (Fig 4C), we nonetheless analyzed their genic content. For subsidiary peak 1, no obvious candidate detoxification genes were found within the peak ~75 kb region, nor were any genes differentially expressed. For subsidiary peak 2, two CYP genes, CYP392B2 and a predicted CYP pseudogene (tetur02g06640) in the London reference strain, were within the peak ~75 kb region, with the latter exhibiting lower RNA abundance in the pyflubumide-selected populations (a fold change of -2) (S2 Data). Several non-synonymous single-nucleotide differences were observed in both CYP genes between JPR-R1 and WasX. Despite the lack of statistical significance for subsidiary peak 2, CYP392B2 is of interest as it was previously associated with pyflubumide resistance [45] (and see Discussion).
CYP392A16 demethylates the toxic pyflubumide carboxamide
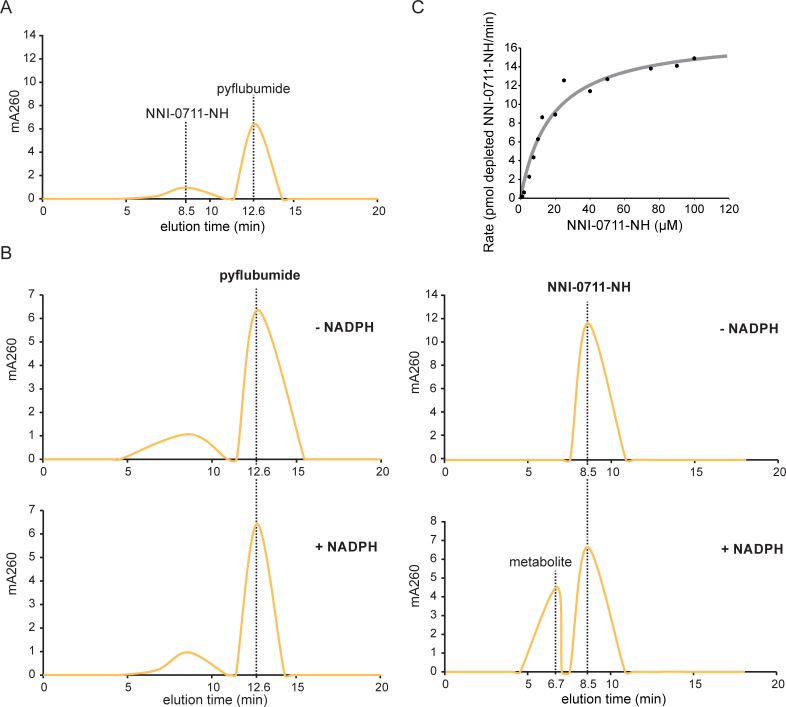
To test if CYP392A16 contributes to pyflubumide resistance in JPR-R1, we functionally expressed the gene to determine if the recombinant protein was active against pyflubumide and/or its toxic carboxamide metabolite (NNI-0711-NH) (Fig 6). The recombinant protein was well-folded (S5 Fig), exhibited activity towards the model substrate L-ME EGE (4 +/- 0.5 SE pmol D Luciferin/min/pmol P450), and membrane preparations exhibited CPR activity (2407 +/- 33.8 nmol/min/mg). By HPLC, substrate depletion was not observed when recombinant CYP392A16 was incubated with the proacaricide pyflubumide (Fig 6). In contrast, an NADPH-dependent depletion of its active, toxic NNI-0711-NH derivative was observed, along with the parallel formation of an unknown compound (eluting at 8.5 min and 6.7 min, respectively), after incubation with recombinant CYP392A16. Chromatograms showed no change in the amount of the carboxamide when incubations were performed in the absence of an NADPH regenerating system, which is necessary for cytochrome P450 activity (Fig 6). The ability of CYP392A16 to metabolize the pyflubumide carboxamide was further characterized by measuring substrate-dependent reaction rates. Its depletion rate as a function of substrate concentration revealed Michaelis-Menten kinetics (R2 of fitted curve = 0.96, Vmax = 17.42 (± 1.07 SE) pmol of depleted deacylated pyflubumide metabolite/min and Km = 17.79 (± 3.29 SE) μM) (Fig 6).
Fig 6. CYP392A16 is active towards the toxic deacylated pyflubumide metabolite, NNI-0711-NH, but not pyflubumide.
(A) HPLC chromatogram of pyflubumide and its toxic metabolite NNI-0711-NH, eluting at 12.6 and 8.5 min, respectively. (B) HPLC chromatograms show a NADPH-dependent depletion of the toxic NNI-0711-NH (eluting at 8.5 min) and the corresponding formation of a reaction product (eluting at 6.7 min). In contrast, reactions carried out in the absence and presence of a NADPH-regenerating system showed no change in the chromatogram profile with pyflubumide eluting at 12.6 min. Absorbance curves are plotted in yellow (A and B). (C) Michaelis–Menten kinetics of the depletion of NNI-0711-NH by CYP392A16. Plotted values represent the mean. The curve was calculated by non-linear regression.
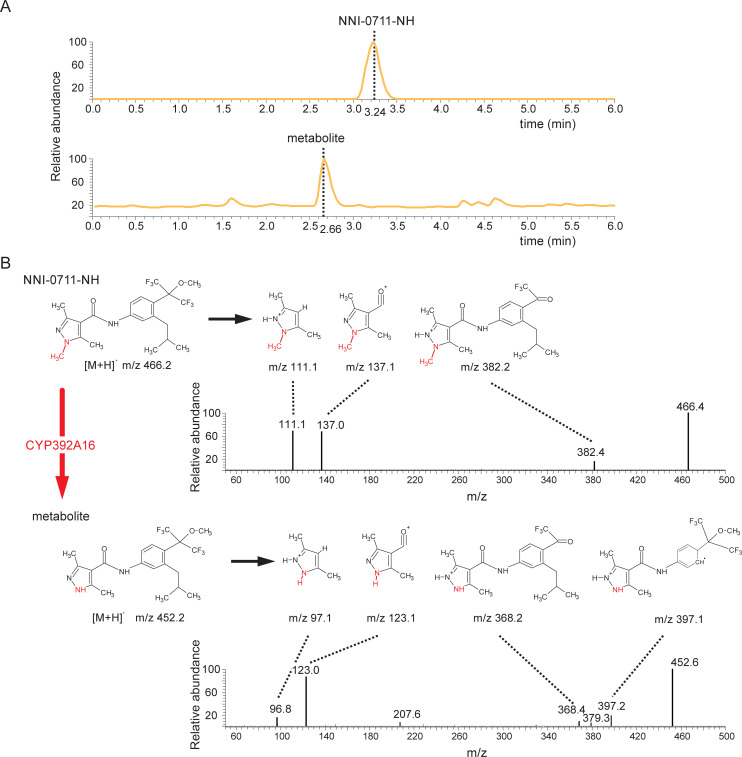
CYP392A16 metabolized 21% of the pyflubumide carboxamide NNI-0711-NH (with a concentration of 25 μM) after a 1-hour incubation. HPLC-MS/MS analysis of the reaction mixtures pointed to N-demethylation as the likely mechanism of the CYP392A16 reaction (Fig 7) [46]. Demethylation is expected to result in a mass difference of -14 Da between the NNI-0711-NH and reaction metabolite product ions. This was reflected in almost all substrate and reaction product ions formed by collision-induced dissociation (Fig 7). More specifically, the product ion MS data indicated that demethylation involves a pyrazole methyl group. We arrived at this conclusion because pyrazole-containing product ions originating from the reaction product are 14 Da lower than their corresponding substrate product ions of NNI-0711-NH (Fig 7).
Fig 7. The toxic pyflubumide metabolite NNI-0711-NH is demethylated by CYP392A16.
(A) Total ion chromatograms and product ion mass spectra obtained from the collision induced dissociation of NNI-0711-NH and the reaction metabolite molecular ions (m/z 466 and m/z 452 respectively) as obtained following their HPLC separation. TIC values for NNI-0711-NH and the reaction metabolite were 5.01E5 and 1.07E4, respectively. (B) Proposed product ions for NNI-0711-NH (m/z 466) and the reaction metabolite (m/z 452). Demethylation, which is expected to proceed via hydroxylation, results in a mass difference of -14 Da between NNI-0711-NH and the metabolite. The proposed demethylation site is indicated by red font. This scenario was confirmed in almost all product ions formed by collision induced dissociation.
Discussion
Pesticide resistance is a rapidly evolving trait within the arthropod phylum [19,47]. Despite its economic importance and its potential to contribute to our understanding of general evolutionary processes, several questions about the genetic architectures underlying pesticide resistance are still outstanding, especially in the case of polygenic resistance. Pyflubumide is a recently developed carboxanilide acaricide that is highly effective against plant-feeding spider mites [39–41,48]. Based on synergism/antagonism assays of previous work [41], the resistant T. urticae JPR-R1 and JPR-R2 strains most likely rely on cytochrome P450-mediated detoxification to overcome pyflubumide toxicity. The two strains also exhibit highly similar constitutive and plastic transcriptional patterns on a genome-wide level [41]. Here, we uncovered that pyflubumide resistance of both JPR-R1 and JPR-R2 is determined by multiple loci. Considering the two strains share a common ancestral genetic background with already moderate levels of pyflubumide resistance [41], these findings further suggest that JPR-R1 and JPR-R2 likely share (some) factors underlying pyflubumide resistance. Using JPR-R1 and susceptible WasX as parents, BSA genetic mapping revealed three major QTL that are associated with pyflubumide resistance based on the segregating genomes of 11 pyflubumide-selected and control population pairs. Interestingly, a BSA genetic mapping approach based on RNAseq data (instead of genomic DNA) uncovered highly similar allele frequency differences, mirroring the QTL pattern obtained with DNA. Less biological tissue is needed to obtain sufficient RNA, as compared to DNA, for high-quality sequencing, and our findings highlight the feasibility of using RNAseq data for BSA genetic mapping, a consideration that is relevant for microarthropods for which biological material is often limiting.
CYP392A16 was located at the center of QTL-1, was over-expressed in pyflubumide-selected populations and the JPR-R1 parent, and metabolized the toxic pyflubumide carboxamide in vitro. Incubation assays with membrane fractions of recombinant protein further uncovered that CYP392A16 demethylates the active carboxamide into a derivative that is not toxic to spider mites [48], confirming that CYP392A16 catalyzes a detoxification reaction. The CYP gene cluster of CYP392E6-8 was close to the peak of QTL-2 and CYP392E8 exhibited higher transcription in pyflubumide-selected populations and JPR-R1. In an earlier study, BSA genetic mapping uncovered a broad QTL (referred to as spiro-QTL 2) that included the CYP392E6-8 gene cluster when replicated populations were selected for resistance to spirodiclofen, a pesticide that targets lipid synthesis [29]. This raises the question of whether the same or a different CYP gene within this cluster was the target of selection and whether other CYP genes outside this cluster (partially) underpin pyflubumide and spirodiclofen resistance. The high susceptibility of the T. urticae SR-VP strain to pyflubumide, the spirodiclofen-resistant parental strain of the earlier BSA study, suggest that different CYP genes (or alleles) determine spirodiclofen and pyflubumide resistance [29,41]. CYP392E9-10 lie ~312 kb away from the CYP392E6-8 gene cluster but are still within the wider QTL-2 interval. CYP392E7 and CYP392E10 are overexpressed in spirodiclofen resistant strains, and previous work was able to show that CYP392E10 metabolizes spirodiclofen by hydroxylation [37,49]. Using the annotated genome of the London reference strain, QTL-2 is predicted to additionally hold several CYP gene fragments and pseudogenes [29,50]. As the CYP multi-gene family is characterized by highly dynamic birth and death rates [51,52], it is possible that these genes could be functional in JPR-R1. Further work is needed to fully characterize the functionality and enzymatic abilities of the multiple cytochrome P450s within QTL-2 and to understand why this genomic region has been a locus of selection for both spirodiclofen and pyflubumide resistance.
The causal genetic variants underpinning the over-expression of detoxification genes in resistant populations of agricultural pests has only been characterized in a limited number of cases [2,53–59]. We ruled out CNV of the coding sequences as causal for the over-expression of CYP392A16 and CYP392E8 in the pyflubumide resistant populations. Given the QTL peak locations, cis-regulatory variation appears to be at least partially responsible for the higher transcript levels. In insects, transcription of detoxification genes is controlled by a number of regulatory pathways, including the AhR/ARNT and CncC/Keap1 cascades, in which cis-acting transcription factor binding sites have been identified [2,43]. In contrast, transcriptional regulation remains poorly understood in mites, and promotor/enhancer analyses are currently not trivial to conduct or interpret. Future studies are therefore needed to test our hypothesis of cis-regulation, and to identify the potential cis-acting elements and their transcription factor binding partners, using suitable experimental set-ups [60,61].
QTL-3 centered on CPR, a necessary redox partner of cytochrome P450-mediated detoxification. Our analyses provide strong evidence for CNV at the CPR locus. Pyflubumide resistant populations were estimated to harbor three CPR copies by two methods, whereas susceptible populations have a single CPR copy. Neighboring genes on both sides of CPR also exhibit clear signs of CNV in JPR-R1, but with different copy numbers. The absence of CNV at this locus in other previously characterized T. urticae strains and the absence of genetic variation between the different genomic copies of JPR-R1 suggest a recent origin of CNV at the CPR locus. The JPR-R1 allele frequencies in the experimental control populations indicate that this copy number variable region could induce pleiotropic fitness penalties that lead to purging in the absence of strong selection pressure (i.e., continuous pyflubumide exposure). Potential fitness penalties may explain why CNV at the CPR locus has not yet been associated with other T. urticae resistance phenotypes that rely on cytochrome P450-mediated detoxification. The transcriptomic comparisons revealed that relative transcript levels were ~three-fold higher for CPR in the pyflubumide-selected populations compared to the controls. The nearly identical DNA and RNA coverage ratios strongly suggest that CNV determines the RNA levels of CPR in JPR-R1 by positive dosage effects. Previous studies have linked dosage-sensitive amplification of CYP genes to increased detoxification, and in turn, pesticide resistance [56–59]. We hypothesize that dosage-sensitive gene amplification of CPR in JPR-R1 strengthens cytochrome P450-mediated detoxification because P450 activity is dependent on the concentration of the CPR-P450 complex [62]. Indeed, reducing CPR transcription in T. urticae and other arthropod pests by RNA interference is known to increase susceptibility to pesticides [63,64]. Moreover, the combined up-regulation of CYP392A16, CYP392E8 and CPR could have additive or synergistic effects, resulting in higher levels of pyflubumide resistance. However, as the relationship between CNV, RNA levels, translation, and phenotypic traits can be complex [65], further work is needed to precisely quantify the impact of the dosage-sensitive CNV at the CPR locus on pyflubumide resistance. Using a BSA genetic mapping approach, CPR was also found at the center of a QTL associated with both spirodiclofen and Mitochondrial Electron Transport Inhibitors complex I (METI-I) resistance in T. urticae [29,33]. In both studies, the non-synonymous substitution D384Y was only observed in the pesticide-selected populations and the resistant parental strains SR-VP and MR-VP. Because METI-I and spirodiclofen resistance is mostly mediated by cytochrome P450s [36,37,66], it was hypothesized that D384Y might strengthen cytochrome P450 detoxification. Here, the coding sequences of the different CPR copies were identical and did not display D384Y or any other non-synonymous substitutions, compared to the genome of the reference London strain [29,50].
Previous BSA studies in T. urticae uncovered a genetic architecture that involves both toxicodynamic and toxicokinetic resistance to spirodiclofen and METI-Is [29,33]. QTL underlying spirodiclofen and METI-I resistance correspond with the molecular targets acetyl-CoA carboxylase and NADH:ubiquinone oxidoreductase, respectively [29,33,38]. None of the three QTLs of pyflubumide resistance were centered on the target of pyflubumide, strongly indicating that only toxicokinetic mechanisms were involved. Previously, Sugimoto et al. identified two QTLs that are associated with pyflubumide resistance in T. urticae using a microsatellite linkage map [45]. One QTL is located on the first T. urticae chromosome and centers on tetur01g15710, the gene that codes for succinate dehydrogenase subunit b (SdhB), the molecular target of pyflubumide [39,40]. Genotyping a panel of T. urticae strains revealed a strict association between the non-synonymous substitution I260V of SdhB and pyflubumide resistance. The I260V substitution is located at the I269 position of the fungus Zymoseptoria tritici (synonym Mycosphaerella graminicola) protein which impacts fungicide binding to succinate dehydrogenase and results in resistance in Z. tritici [67]. Our read data of the segregating populations did not reveal the I260V variant or other candidate target-site resistance mutations in SdhB. Additionally, Sugimoto et al. observed that their second broad QTL of ~4.1 Mb on the third T. urticae chromosome included three cytochrome P450 genes (CYP392A8, CYP392B1, and CYP392B2) [45]. These CYP genes were not differentially expressed between the pyflubumide-selected and control populations and were not included in any of the three well-supported QTL of this study. However, subsidiary peak 2 of the current study largely coincided with this QTL (its peak region also included CYP392B2). This could indicate that genetic variation within this genomic interval could have a minor effect on pyflubumide resistance in the segregating populations used in our study. Collectively, however, the two complementary genetic mapping approaches strongly suggest that pyflubumide resistance has been obtained by largely different molecular mechanisms between the strains of the current study and Sugimoto et al. Our work therefore adds to a growing body of evidence that populations of pest species can evolve different resistance mechanisms despite the highly specific selective pressures imposed by pesticides [19].
In summary, our work uncovered a complex genetic basis for pyflubumide resistance in the arthropod pest T. urticae that likely involves multiple genes with roles in cytochrome P450-mediated metabolism. Our in vitro findings suggest that CYP392A16 detoxifies the active form of the proacaricide pyflubumide, and that cis-regulatory variation at this gene contributes prominently to pyflubumide resistance. We also uncovered a potential novel molecular mechanism of toxicokinetic resistance–gene amplification of CPR–that may act in a synergistic manner.
Materials and methods
Mite strains and rearing
Five T. urticae strains were used for the genetic crosses: JP-S [68,69], Wasatch [28], WasX, JPR-R1, and JPR-R2. The JPR-R1 and JPR-R2 strains were selected for high levels of pyflubumide resistance from the JP-R strain with two different laboratory selection regimes. JPR-R1 was first selected by spraying pyflubumide on detached mite-infested bean leaves for two generations. Subsequently, both JPR-R1 and JPR-R2 were selected by feeding on pyflubumide sprayed potted bean plants (100 mg/L of pyflubumide) [41]. WasX was derived from the highly inbred Wasatch strain by two sequential rounds of mother-son mating. Unless otherwise stated, all T. urticae populations were kept under laboratory conditions (25°C, 65% relative humidity, and 16:8 hr light:dark photoperiod) and maintained on Phaseolus vulgaris cv. ‘Speedy’. JPR-R1 and JPR-R2 were kept on plants sprayed with 100 mg/L pyflubumide (commercial formulation, 20% SC (Danikong, Japan), kindly provided by Ralf Nauen).
Mode of inheritance of pyflubumide resistance
The mode of inheritance of pyflubumide resistance was determined by performing reciprocal crosses between JPR-R1 and JP-S, and JPR-R2 and Wasatch. For each cross, 80 teliochrysalid (virgin) females and 100 adult males were placed on leaf discs and allowed to mate. Females were transferred to new leaf discs every 2 days throughout their oviposition period. During development, 80 F1 teliochrysalid females were isolated for backcrosses to obtain F2 females. Dose-response bioassays were conducted for the parents and F1 and F2 females as previously described [41]. Briefly, 9 cm2 bean leaf discs were sprayed with 1 ml of acaricide solution at 1 bar pressure in a Potter spray tower (deionized water was used as the control). Depending on the population, 5–13 pyflubumide concentrations were tested using four replicates (20–35 females per replicate). Lethal concentrations and 95% confidence limits were determined using Probit analysis (POLO; LeORa Software). The degree of dominance (D) was calculated using Stone’s formula [70]. The expected dose-response relationships for F2 females for monogenic recessive resistance was calculated using c = 0.5 W (F1) + 0.5 W (JPR-R strain), where c is the expected response (mortality) at a given concentration and W is the observed response (mortality) of the parents at the respective concentration [42]. The hypothesis of a monogenic mode of inheritance was tested with χ2 goodness-of-fit tests.
Experimental evolution of pyflubumide resistance
A total of 26 teliochrysalid females of JPR-R1 were placed on individual leaf discs. Upon eclosion, unfertilized females were allowed to lay eggs for 2 days and were transferred to a 9 cm2 bean leaf disc. Resistance to pyflubumide was assessed by spraying 4,000 mg/L pyflubumide (1 bar pressure in a Potter spray tower with 2 mg aqueous deposit per cm2) and scoring mortality after 24 hr. Eggs of one of the resistant JPR-R1 females were allowed to hatch and develop into adult males. A single adult male (an isogenic genome, as males are haploid) was selected from this brood and was offered four female teliochrysalids of WasX on a daily basis. In total, 24 resulting females were allowed to produce an F1 generation of more than 500 female mites. The segregating population was expanded and allowed to propagate at a high population size (several hundreds of mites) for approximately two months. From this large population, we established 11 control populations on unsprayed potted bean plants using 650 founding females per control population. After approximately one month, sister populations were generated from these 11 control populations by transferring approximately 650 females to potted bean plants sprayed with 50 mg/L pyflubumide. Sprayed and unsprayed potted bean plants were offered to the 22 experimental populations every other week. During the experiment, selection pressure of pyflubumide was gradually increased until mites no longer exhibited acaricide-related mortality on beans sprayed until run-off with 100 mg/L of pyflubumide (selection was maintained for approximately a year). Dose-response bioassays were subsequently performed on the two parents and the 22 segregating populations as described above using four concentrations (10, 50, 100, and 500 mg/L pyflubumide). Differences in mortality percentages of the two parents were assessed using a generalized linear model with a binomial distribution. A generalized linear mixed model with a binomial distribution was used to analyze the mortality percentages of the pyflubumide-selected and control populations, with selection regime and pyflubumide dosage as fixed effects, and sister pair as a random effect. Percentage of mortality was the dependent variable in both models. Statistical analyses were performed in R using lme4 [71].
DNA sequencing and variant detection
Immediately after the dose-response bioassays were finalized, DNA was extracted from 1,000 adult females of the 22 segregating populations using a phenol-chloroform method as previously described [27,33]. Six months after the BSA cross, DNA was collected from the genetically diverse JPR-R1 strain and the inbred WasX strain. DNA samples were sequenced at the Huntsman Cancer Institute at the University of Utah to produce Illumina paired-end reads with lengths of 151 bp with mean insert sizes of 450 bp. DNA reads were aligned to the T. urticae three-chromosome assembly [29] using the default options of mem in the Burrows-Wheeler Aligner (BWA) version 0.7.17-r1188 [72]. Duplicate reads were marked using the MarkedDuplicates function of Picard version 2.18.11-SNAPSHOT (http://broadinstitute.github.io/picard/). Left-justified indel realignment was performed using the GATK version 4.0.7.0 [73]. Throughout these steps, SAMtools version 1.9 [74] was used to sort and index the reads in each BAM output. Two joint variant calls were performed using GATK version 4.0.7.0; one that included the two parents and the segregating populations, and one without the parents (the latter joint variant call was only used for the PCA). Each variant call was performed with HaplotypeCaller and then merged into a single gVCF file using the GVCFs function, after which the GenotypeGVCFs function was used to jointly predict SNPs and small indels.
RNA sequencing and differential expression analysis
Thirty-seven days after DNA was extracted from the segregating populations, the pyflubumide selection treatment was terminated by transferring mites from all experimental populations to new unsprayed bean plants where they were allowed to develop for one generation (eleven days). RNA was extracted from 110 adult females using the RNeasy minikit (Qiagen) for every segregating experimental population. Total RNA was sequenced with the Illumina method at NXTGNT (Ghent, Belgium) to give read lengths of 100 bp. The paired-end RNA reads were aligned to the T. urticae three-chromosome assembly [29,50] using STAR version 2.5.3a [75] with the 2-pass mode, the maximum intron size set to 20 kb, and without the use of the GFF annotation. SAMtools version 1.9 [74] was used for sorting and indexing the BAM files. Read counts were obtained with HTSeq version 0.11.2 [76] using the T. urticae annotation of Wybouw et al. [29]. Analysis of gene expression was performed using DESeq2 version 1.24.0 [77]. Differential expression was identified by applying a 2 and 0.05 cut-off for FC and adjusted p-value, respectively. Coverage from the BAM files for downstream analyses was obtained using the defaults of the pileup function within pysam version 0.15.1 (https://github.com/pysam-developers/pysam), and normalized using the estimated size factors generated by DESeq2 with “normalized = T”.
PCA of genomic and transcriptomic responses
PCA using variants (SNPs) ascertained from the DNA read data was performed with all the pyflubumide-selected and paired control populations using SNPRelate version 1.18.0 [78]. The VCF file was filtered as described in [33] and transformed to the gds [78,79] format using the snpgdsVCF2GDS function with the option "method = copy.num.of.ref", at which point the snpgdsPCA function was used to construct the PCA with the “autosome.only = FALSE” option. A PCA based on the RNAseq read data was performed using the regularized log transformed counts.
BSA genetic mapping
BSA genetic mapping was performed using methods adapted from previous studies [28,29,33], and specifically, using the defaults of the haplodiploid option of Kurlovs et al. [25]. Briefly, after quality control, JPR-R1 was specified as the haplodiploid male parent, and which alleles got passed on from its heterozygous sites (via the single haploid JPR-R1 male used in the BSA cross) was inferred using allele frequency information from the offspring; if the paired pyflubumide-selected and control populations had a frequency of 0.95 or higher, the site was considered non-segregating and not included in the final analysis. Sliding window analyses were performed using 75 kb windows with 5 kb offsets; at least 38 sites had to be present in a window for it to be included in the analysis. Significant responses to pyflubumide selection were detected using a permutation-based approach as described in Wybouw et al. [29]. The genome-wide allele frequency differences were permuted for the 11 pairs of selected/control populations 104 times and were averaged across all pairs per permutation. For these analyses, the three chromosomes were concatenated and made circular to maintain linkage information (the end of the third chromosome was connected with the beginning of the first chromosome). Using the 104 permutations, the distribution of the absolute values of the maximal deviations in the averaged allele frequency differences was calculated and QTLs were assigned using an FDR of 0.05. BSA genetic mapping was also performed on the RNA read data using each set of paired BAM files. For each biallelic position in a coding exon where RNA read coverage was at least 20 and the major allele frequencies were less than 0.95 in the control populations, we calculated the allele frequencies for the two bases. The absolute allele frequency differences were calculated per paired set of pyflubumide-selected and susceptible populations and were plotted in averages of 500 kb non-overlapping sliding windows across the T. urticae genome [29]. Finally, the allele frequency differences were averaged across the replicated pairs. Protein domains were identified using Pfam- and CDD-searches [80,81].
DNA quantitative PCR assay
RNA was reverse transcribed to cDNA using a RevertAid RT Reverse Transcription Kit (ThermoFisher Scientific). Primers 5’-CAAGGTGATCTCCAGCTTCA-3’ and 5’-ACCGTTCGTGTATGCATGTT-3’ were used to amplify CPR, yielding a 150 bp product. As a reference single-copy gene, we used the voltage-gated sodium channel (VGSC; tetur34g00970). Primers 5’-ATGATGCTTGGCAGTGAAAG-3’ and 5’-ATAAGCAGCAGCAGCAAGAA-3’ produced a 111 bp fragment of VGSC. PCR reactions were performed using Maxima SYBR Green/Rox qPCR Master Mix (Fermentas Life Sciences). All reactions were run in duplicate on an Agilent Technologies Stratagene Mx3005P thermocycler with the following conditions: an initial 10 min plateau at 95°C, followed by 40 cycles that included denaturation at 95°C for 15 sec, annealing at 55°C for 30 sec, and extension for 30 sec at 72°C. Melting curves were determined by an additional cycle of 95°C for 1 min, 65°C for 30 sec and 95°C for 30 sec. One duplicate was excluded from the VGSC and CPR standard curves due to pipetting errors. Efficiencies were calculated from these standard curves and were incorporated in further calculations. First, the number of VGSC copies was determined by 2*(efficiency(Ct-1)). Second, the Ct-values of CPR were corrected by log(efficiency)(VGSC copies/2)+1. The final estimate of CPR copy number was obtained with efficiency(corrected Ct–Ct).
Cytochrome P450 functional expression
We adopted a previously successful functional expression strategy for CYP392A16 [82]. Briefly, competent E. coli BL21STAR cells were co-transformed with the pCW_ompA-CYP392A16 and pACYQ_TuCPR plasmids [82]. Transformed cells were grown at 37°C in Terrific Broth under ampicillin and chloramphenicol selection until the optical density at 595 nm was higher than 0.95 cm-1. The heme precursor δ-aminolevulinic acid was added to a final concentration of 1 mM. Induction was initiated by adding 1 mM of isopropyl-1-thio-β-D-galactopyranoside. After growing at 25°C for 24 hours, cells were harvested and the spheroplasts were prepared. Membrane fractions were obtained from spheroplast solutions by ultracentrifugation at 180,000 g for 30 min at 4°C. CYP392A16 membranes were diluted in TSE buffer (0.1 M Tris-acetate, pH 7.6, 0.5 M sucrose, 0.5 mM EDTA) and stored in aliquots at -80°C. P450 content was measured by CO-difference spectra in reduced CYP392A16 membrane samples [83]. CPR activity was estimated by measurements of NADPH-dependent reduction of cytochrome c at 550 nm [84]. CYP activity was tested using the chemi-luminescent substrate Luciferin-ME EGE [82]. Total protein concentrations of CYP392A16 membranes were determined using the Bradford assay [85].
Cytochrome P450 HPLC-UV activity assay
Pyflubumide and its carboxamide metabolite (NNI-0711-NH) (both 99.6% purity) were provided by Nihon Nohyaku (Japan). Pyflubumide and the toxic carboxamide were incubated at a concentration of 25 μM with 25 pmol of recombinant CYP392A16 in 100 μl Tris-HCl buffer (0.2 M, pH 7.4), containing 0.25 mM MgCl2 and 2.5% acetonitrile (30°C and 1250 rpm shaking). The reaction was performed in the presence and absence of an NADPH generating system: 1 mM glucose-6-phosphate (Sigma Aldrich), 0.1 mM NADP+ (Sigma Aldrich), and 1 unit/ml glucose-6-phosphate dehydrogenase (G6PDH; Sigma Aldrich). Reactions were stopped after 0 and 1 hr using 100 μl acetonitrile and further stirred for an additional 30 min. Finally, the quenched reactions were centrifuged at 10,000 rpm for 10 min and 100 μl of the supernatant was transferred to glass vials for HPLC analysis. Acaricides were separated on a UniverSil HS C18 (250mm 5um) reverse phase analytical column (Fortis). Reactions with pyflubumide and its NNI-0711-NH metabolite were separated using an isocratic mobile phase of 20% H20 and 80% acetonitrile with a flow rate of 1 ml/min for 20 min. Reactions were monitored by changes in absorbance at 260 nm and quantified by peak integration (Chromeleon, Dionex). For enzyme reaction kinetics, different concentrations of the NNI-0711-NH pyflubumide metabolite were used. Rates of substrate turnover from two independent reactions were plotted versus substrate concentration. Km and Vmax were determined using SigmaPlot 12.0 (Systat Sofware, London, UK).
Identification of reaction products by HPLC-MS
Prior to HPLC-MS analysis, reaction mixtures were desalted by solid phase extraction (Bond Elute LRC-C18, 200 mg cartridges; Agilent, USA). Briefly, reaction mixtures were loaded to cartridges that were pre-conditioned with 3 mL 100% acetonitrile, followed by 3 mL 2.5% acetonitrile in water. Cartridges were subsequently washed with 1 mL water and samples were eluted with 1.5 mL acetonitrile. Eluents were transferred to HPLC autosampler vials and 500 μL of water was added to enhance chromatographic separation. Eluents were analyzed using an HPLC-MS/MS system. Sample injections (20 μl loop) were performed via a Surveyor Autosampler (Thermo Scientific, USA). Chromatographic separation was achieved using a Surveyor LCsystem (Thermo Scientific, USA), equipped with a Gemini C18 (3 mm, 100 mm x 2 mm) analytical column (Phenomenex, USA). An isocratic elution was applied with 80% acetonitrile-20% water and flow rate was set at 200 μL/min. Analyte detection was achieved using an electrospray ionization (ESI) triple quadrupole mass spectrometer (TSQ Quantum; Thermo Scientific, USA) operated in the positive ion mode. Mass spectrometry was operated both in full scan and product ion scan modes. The system was controlled by the Xcalibur software, which was also used for data acquisition and analysis. The optimum mass spectrometer parameters were set as follows: spray voltage at +4500 V, sheath gas pressure at 20 arbitrary units, auxiliary gas pressure at 10 arbitrary units, ion transfer capillary temperature at 300°C and source collision induced dissociation at 26 eV. In the product ion mode the collision cell contained Argon at 1.5 mTorr. Sheath/auxiliary gas was high purity nitrogen and collision gas was high purity argon. For MS/MS analysis, collision energy was set at 25 eV.
Supporting information
(A) Frequencies of JPR-R1 alleles in the susceptible (control) and pyflubumide-selected populations across the three QTL, as assessed in a sliding window analysis (see also Fig 4A). Chromosomes are ordered by decreasing length and are indicated by alternating shading. (B-D) Frequencies of JPR-R1 alleles in the susceptible (control) and pyflubumide-selected populations at QTL-1, QTL-2, and QTL-3, respectively. Pyflubumide-selected populations that do not show near-fixation levels of JPR-R1 allele frequencies at the averaged BSA peaks are indicated by their identifier. For all panels, coverage is color-coded according to treatment (legend, top). Vertical lines indicate the locations of the averaged BSA peaks.
(PDF)
Chromosomes are ordered by decreasing length and are indicated by alternating shading. Vertical lines indicate the locations of the three QTL, whereas vertical dashed lines indicate the two subsidiary peaks (Fig 4).
(PDF)
CYP392A16 (A) and CPR (B) expression in pyflubumide-selected populations relative to the mean RNA abundance in the susceptible control populations. (C) DNA coverage of CPR in pyflubumide-selected and control populations, relative to single-copy VGSC. DNA coverage was estimated by quantitative PCR analysis of the segregating populations, confirming estimates using short-read coverage (Fig 5). Panels are color-coded according to treatment.
(PDF)
Vertical lines indicate the locations of averaged BSA peaks. Candidate genes CYP392A16 (A) and CYP392E6-8 (B) are highlighted in yellow. Otherwise, coding exons and introns are depicted as dark gray and lighter gray boxes, respectively. Symbols + and–denote forward and reverse gene orientations. Coverage is color-coded according to treatment and strain (legend, top).
(PDF)
(PDF)
(DOCX)
(XLSX)
(XLSX)
Acknowledgments
We thank Ludek Tikovsky and Harold Lemereis for their assistance in plant rearing and maintenance of greenhouse rooms, and Robert Greenhalgh for assistance with bioinformatic analyses. We are grateful to René Feyereisen for helpful comments. We sincerely thank Nihon Nohyaku for providing technical standards of pyflubumide and its NH metabolite.
Data Availability
Sequence data has been deposited at the Sequence Read Archive (PRJNA596790). Phenotypic quantification data is available at S1 Data.
Funding Statement
This work was supported by the Research Foundation - Flanders (FWO) [grant G009312N and grant G053815N to T.V.L.] and the Research Council (ERC) under the European Union's Horizon 2020 research and innovation program [grant 772026-POLYADAPT to T.V.L. and 773902–SUPERPEST to J.V. and T.V.L.], and by the USA National Science Foundation (award 1457346 to R.M.C). N.W. was supported by a Research Foundation - Flanders (FWO) and a BOF (Ghent University) post-doctoral fellowship (12T9818N and 01P03420, respectively). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Arora R, Sandhu S. Breeding Insect Resistant Crops for Sustainable Agriculture. Singapore; 2017.
- 2.Feyereisen R, Dermauw W, Van Leeuwen T. Genotype to phenotype, the molecular and physiological dimensions of resistance in arthropods. Pesticide Biochemistry and Physiology. 2015;121: 61–77. doi: 10.1016/j.pestbp.2015.01.004 [DOI] [PubMed] [Google Scholar]
- 3.Bass C, Field LM. Gene amplification and insecticide resistance. Pest Manag Sci. 2011;67: 886–890. doi: 10.1002/ps.2189 [DOI] [PubMed] [Google Scholar]
- 4.Balabanidou V, Grigoraki L, Vontas J. Insect cuticle: a critical determinant of insecticide resistance. Current Opinion in Insect Science. 2018;27: 68–74. doi: 10.1016/j.cois.2018.03.001 [DOI] [PubMed] [Google Scholar]
- 5.Dermauw W, Wybouw N, Rombauts S, Menten B, Vontas J, Grbic M, et al. A link between host plant adaptation and pesticide resistance in the polyphagous spider mite Tetranychus urticae. Proceedings of the National Academy of Sciences. 2013;110: E113–E122. doi: 10.1073/pnas.1213214110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Marco L, Sassera D, Epis S, Mastrantonio V, Ferrari M, Ricci I, et al. The choreography of the chemical defensome response to insecticide stress: insights into the Anopheles stephensi transcriptome using RNA-Seq. Sci Rep. 2017;7: 41312. doi: 10.1038/srep41312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu F, Moural TW, Nelson DR, Palli SR. A specialist herbivore pest adaptation to xenobiotics through up-regulation of multiple Cytochrome P450s. Sci Rep. 2016;6: 20421. doi: 10.1038/srep20421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kalsi M, Palli SR. Transcription factor cap n collar C regulates multiple cytochrome P450 genes conferring adaptation to potato plant allelochemicals and resistance to imidacloprid in Leptinotarsa decemlineata (Say). Insect Biochemistry and Molecular Biology. 2017;83: 1–12. doi: 10.1016/j.ibmb.2017.02.002 [DOI] [PubMed] [Google Scholar]
- 9.Daborn PJ, Yen JL, Bogwitz MR, Le Goff G, Feil E, Jeffers S, et al. A Single P450 Allele Associated with Insecticide Resistance in Drosophila. Science. 2002;297: 2253–2256. doi: 10.1126/science.1074170 [DOI] [PubMed] [Google Scholar]
- 10.Chung H, Bogwitz MR, McCart C, Andrianopoulos A, ffrench-Constant RH, Batterham P, et al. Cis -Regulatory Elements in the Accord Retrotransposon Result in Tissue-Specific Expression of the Drosophila melanogaster Insecticide Resistance Gene Cyp6g1. Genetics. 2007;175: 1071–1077. doi: 10.1534/genetics.106.066597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weedall GD, Mugenzi LMJ, Menze BD, Tchouakui M, Ibrahim SS, Amvongo-Adjia N, et al. A cytochrome P450 allele confers pyrethroid resistance on a major African malaria vector, reducing insecticide-treated bednet efficacy. Sci Transl Med. 2019;11: eaat7386. doi: 10.1126/scitranslmed.aat7386 [DOI] [PubMed] [Google Scholar]
- 12.Mugenzi LMJ, Menze BD, Tchouakui M, Wondji MJ, Irving H, Tchoupo M, et al. Cis-regulatory CYP6P9b P450 variants associated with loss of insecticide-treated bed net efficacy against Anopheles funestus. Nat Commun. 2019;10: 4652. doi: 10.1038/s41467-019-12686-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Devonshire AL, Field LM. Gene Amplification and Insecticide Resistance. Annu Rev Entomol. 1991;36: 1–21. doi: 10.1146/annurev.en.36.010191.000245 [DOI] [PubMed] [Google Scholar]
- 14.Riveron JM, Yunta C, Ibrahim SS, Djouaka R, Irving H, Menze BD, et al. A single mutation in the GSTe2 gene allows tracking of metabolically based insecticide resistance in a major malaria vector. Genome Biol. 2014;15: R27. doi: 10.1186/gb-2014-15-2-r27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Newcomb RD, Campbell PM, Ollis DL, Cheah E, Russell RJ, Oakeshott JG. A single amino acid substitution converts a carboxylesterase to an organophosphorus hydrolase and confers insecticide resistance on a blowfly. Proceedings of the National Academy of Sciences. 1997;94: 7464–7468. doi: 10.1073/pnas.94.14.7464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Claudianos C, Russell RJ, Oakeshott JG. The same amino acid substitution in orthologous esterases confers organophosphate resistance on the house fly and a blowfly. Insect Biochemistry and Molecular Biology. 1999;29: 675–686. doi: 10.1016/s0965-1748(99)00035-1 [DOI] [PubMed] [Google Scholar]
- 17.Joussen N, Agnolet S, Lorenz S, Schone SE, Ellinger R, Schneider B, et al. Resistance of Australian Helicoverpa armigera to fenvalerate is due to the chimeric P450 enzyme CYP337B3. Proceedings of the National Academy of Sciences. 2012;109: 15206–15211. doi: 10.1073/pnas.1202047109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rasool A, Joußen N, Lorenz S, Ellinger R, Schneider B, Khan SA, et al. An independent occurrence of the chimeric P450 enzyme CYP337B3 of Helicoverpa armigera confers cypermethrin resistance in Pakistan. Insect Biochemistry and Molecular Biology. 2014;53: 54–65. doi: 10.1016/j.ibmb.2014.07.006 [DOI] [PubMed] [Google Scholar]
- 19.Hawkins NJ, Bass C, Dixon A, Neve P. The evolutionary origins of pesticide resistance. Biol Rev. 2019;94: 135–155. doi: 10.1111/brv.12440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Leeuwen T, Dermauw W. The Molecular Evolution of Xenobiotic Metabolism and Resistance in Chelicerate Mites. Annu Rev Entomol. 2016;61: 475–498. doi: 10.1146/annurev-ento-010715-023907 [DOI] [PubMed] [Google Scholar]
- 21.Li X, Schuler MA, Berenbaum MR. Molecular Mechanisms of Metabolic Resistance to Synthetic and Natural Xenobiotics. Annu Rev Entomol. 2007;52: 231–253. doi: 10.1146/annurev.ento.51.110104.151104 [DOI] [PubMed] [Google Scholar]
- 22.Roush RT, McKenzie JA. Ecological Genetics of Insecticide and Acaricide Resistance. Annu Rev Entomol. 1987;32: 361–380. doi: 10.1146/annurev.en.32.010187.002045 [DOI] [PubMed] [Google Scholar]
- 23.Van Leeuwen T, Demaeght P, Osborne EJ, Dermauw W, Gohlke S, Nauen R, et al. Population bulk segregant mapping uncovers resistance mutations and the mode of action of a chitin synthesis inhibitor in arthropods. Proceedings of the National Academy of Sciences. 2012;109: 4407–4412. doi: 10.1073/pnas.1200068109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riga M, Bajda S, Themistokleous C, Papadaki S, Palzewicz M, Dermauw W, et al. The relative contribution of target-site mutations in complex acaricide resistant phenotypes as assessed by marker assisted backcrossing in Tetranychus urticae. Sci Rep. 2017;7: 9202. doi: 10.1038/s41598-017-09054-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurlovs AH, Snoeck S, Kosterlitz O, Van Leeuwen T, Clark RM. Trait mapping in diverse arthropods by bulked segregant analysis. Current Opinion in Insect Science. 2019;36: 57–65. doi: 10.1016/j.cois.2019.08.004 [DOI] [PubMed] [Google Scholar]
- 26.Groen SC, Whiteman NK. Using Drosophila to study the evolution of herbivory and diet specialization. Current Opinion in Insect Science. 2016;14: 66–72. doi: 10.1016/j.cois.2016.01.004 [DOI] [PubMed] [Google Scholar]
- 27.Wybouw N, Kurlovs AH, Greenhalgh R, Bryon A, Kosterlitz O, Manabe Y, et al. Convergent evolution of cytochrome P450s underlies independent origins of keto-carotenoid pigmentation in animals. Proc R Soc B. 2019;286: 20191039. doi: 10.1098/rspb.2019.1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bryon A, Kurlovs AH, Dermauw W, Greenhalgh R, Riga M, Grbić M, et al. Disruption of a horizontally transferred phytoene desaturase abolishes carotenoid accumulation and diapause in Tetranychus urticae. PNAS. 2017;114: E5871–E5880. doi: 10.1073/pnas.1706865114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wybouw N, Kosterlitz O, Kurlovs AH, Bajda S, Greenhalgh R, Snoeck S, et al. Long-Term Population Studies Uncover the Genome Structure and Genetic Basis of Xenobiotic and Host Plant Adaptation in the Herbivore Tetranychus urticae. Genetics. 2019;211: 1409–1427. doi: 10.1534/genetics.118.301803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bastide H, Lange JD, Lack JB, Yassin A, Pool JE. A Variable Genetic Architecture of Melanic Evolution in Drosophila melanogaster. Genetics. 2016;204: 1307–1319. doi: 10.1534/genetics.116.192492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park Y, González-Martínez RM, Navarro-Cerrillo G, Chakroun M, Kim Y, Ziarsolo P, et al. ABCC transporters mediate insect resistance to multiple Bt toxins revealed by bulk segregant analysis. BMC Biol. 2014;12: 46. doi: 10.1186/1741-7007-12-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kunte K, Zhang W, Tenger-Trolander A, Palmer DH, Martin A, Reed RD, et al. doublesex is a mimicry supergene. Nature. 2014;507: 229–232. doi: 10.1038/nature13112 [DOI] [PubMed] [Google Scholar]
- 33.Snoeck S, Kurlovs AH, Bajda S, Feyereisen R, Greenhalgh R, Villacis-Perez E, et al. High-resolution QTL mapping in Tetranychus urticae reveals acaricide-specific responses and common target-site resistance after selection by different METI-I acaricides. Insect Biochemistry and Molecular Biology. 2019;110: 19–33. doi: 10.1016/j.ibmb.2019.04.011 [DOI] [PubMed] [Google Scholar]
- 34.Van Leeuwen T, Vontas J, Tsagkarakou A, Dermauw W, Tirry L. Acaricide resistance mechanisms in the two-spotted spider mite Tetranychus urticae and other important Acari: A review. Insect Biochemistry and Molecular Biology. 2010;40: 563–572. doi: 10.1016/j.ibmb.2010.05.008 [DOI] [PubMed] [Google Scholar]
- 35.Riga M, Myridakis A, Tsakireli D, Morou E, Stephanou EG, Nauen R, et al. Functional characterization of the Tetranychus urticae CYP392A11, a cytochrome P450 that hydroxylates the METI acaricides cyenopyrafen and fenpyroximate. Insect Biochemistry and Molecular Biology. 2015;65: 91–99. doi: 10.1016/j.ibmb.2015.09.004 [DOI] [PubMed] [Google Scholar]
- 36.Van Pottelberge S, Van Leeuwen T, Nauen R, Tirry L. Resistance mechanisms to mitochondrial electron transport inhibitors in a field-collected strain of Tetranychus urticae Koch (Acari: Tetranychidae). Bull Entomol Res. 2009;99: 23–31. doi: 10.1017/S0007485308006081 [DOI] [PubMed] [Google Scholar]
- 37.Van Pottelberge S, Van Leeuwen T, Khajehali J, Tirry L. Genetic and biochemical analysis of a laboratory-selected spirodiclofen-resistant strain of Tetranychus urticae Koch (Acari: Tetranychidae). Pest Manag Sci. 2009;65: 358–366. doi: 10.1002/ps.1698 [DOI] [PubMed] [Google Scholar]
- 38.Bajda S, Dermauw W, Panteleri R, Sugimoto N, Douris V, Tirry L, et al. A mutation in the PSST homologue of complex I (NADH:ubiquinone oxidoreductase) from Tetranychus urticae is associated with resistance to METI acaricides. Insect Biochemistry and Molecular Biology. 2017;80: 79–90. doi: 10.1016/j.ibmb.2016.11.010 [DOI] [PubMed] [Google Scholar]
- 39.Furuya T, Machiya K, Fujioka S, Nakano M, Inagaki K. Development of a novel acaricide, pyflubumide. J Pestic Sci. 2017;42: 132–136. doi: 10.1584/jpestics.J17-02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakano M, Yasokawa N, Suwa A, Fujioka S, Furuya T, Sakata K. Mode of action of novel acaricide pyflubumide: Effects on the mitochondrial respiratory chain. Journal of Pesticide Science. 2015;40: 19–24. doi: 10.1584/jpestics.D14-086 [DOI] [Google Scholar]
- 41.Fotoukkiaii SM, Mermans C, Wybouw N, Van Leeuwen T. Resistance risk assessment of the novel complex II inhibitor pyflubumide in the polyphagous pest Tetranychus urticae. J Pest Sci. 2020;93: 1085–1096. doi: 10.1007/s10340-020-01213-x [DOI] [Google Scholar]
- 42.Georghiou GP. Genetics of resistance to insecticides in houseflies and mosquitoes. Experimental Parasitology. 1969;26: 224–255. doi: 10.1016/0014-4894(69)90116-7 [DOI] [PubMed] [Google Scholar]
- 43.Amezian D, Nauen R, Le Goff G. Transcriptional regulation of xenobiotic detoxification genes in insects—An overview. Pesticide Biochemistry and Physiology. 2021;174. [DOI] [PubMed] [Google Scholar]
- 44.Hung CF, Holzmacher R, Connolly E, Berenbaum MR, Schuler MA. Conserved promoter elements in the CYP6B gene family suggest common ancestry for cytochrome P450 monooxygenases mediating furanocoumarin detoxification. Proceedings of the National Academy of Sciences. 1996;93: 12200–12205. doi: 10.1073/pnas.93.22.12200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sugimoto N, Takahashi A, Ihara R, Itoh Y, Jouraku A, Van Leeuwen T, et al. QTL mapping using microsatellite linkage reveals target-site mutations associated with high levels of resistance against three mitochondrial complex II inhibitors in Tetranychus urticae. Insect Biochemistry and Molecular Biology. 2020; 103410. doi: 10.1016/j.ibmb.2020.103410 [DOI] [PubMed] [Google Scholar]
- 46.Tao J, Kang Y, Xue Z, Wang Y, Zhang Y, Chen Q, et al. Theoretical study on the N-demethylation mechanism of theobromine catalyzed by P450 isoenzyme 1A2. Journal of Molecular Graphics and Modelling. 2015;61: 123–132. doi: 10.1016/j.jmgm.2015.06.017 [DOI] [PubMed] [Google Scholar]
- 47.Sparks TC, Nauen R. IRAC: Mode of action classification and insecticide resistance management. Pesticide Biochemistry and Physiology. 2015;121: 122–128. doi: 10.1016/j.pestbp.2014.11.014 [DOI] [PubMed] [Google Scholar]
- 48.Furuya T, Suwa A, Nakano M, Fujioka S, Yasokawa N, Machiya K. Synthesis and biological activity of a novel acaricide, pyflubumide. Journal of Pesticide Science. 2015;40: 38–43. doi: 10.1584/jpestics.D14-087 [DOI] [Google Scholar]
- 49.Demaeght P, Dermauw W, Tsakireli D, Khajehali J, Nauen R, Tirry L, et al. Molecular analysis of resistance to acaricidal spirocyclic tetronic acids in Tetranychus urticae: CYP392E10 metabolizes spirodiclofen, but not its corresponding enol. Insect Biochemistry and Molecular Biology. 2013;43: 544–554. doi: 10.1016/j.ibmb.2013.03.007 [DOI] [PubMed] [Google Scholar]
- 50.Grbić M, Van Leeuwen T, Clark RM, Rombauts S, Rouzé P, Grbić V, et al. The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature. 2011;479: 487–492. doi: 10.1038/nature10640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Feyereisen R. Insect CYP Genes and P450 Enzymes. Insect Molecular Biology and Biochemistry. Elsevier; 2012. pp. 236–316. doi: 10.1016/B978-0-12-384747-8.10008-X [DOI] [Google Scholar]
- 52.Dermauw W, Van Leeuwen T, Feyereisen R. Diversity and evolution of the P450 family in arthropods. Insect Biochemistry and Molecular Biology. 2020;127: 103490. doi: 10.1016/j.ibmb.2020.103490 [DOI] [PubMed] [Google Scholar]
- 53.Itokawa K, Komagata O, Kasai S, Masada M, Tomita T. Cis-acting mutation and duplication: History of molecular evolution in a P450 haplotype responsible for insecticide resistance in Culex quinquefasciatus. Insect Biochemistry and Molecular Biology. 2011;41: 503–512. doi: 10.1016/j.ibmb.2011.04.002 [DOI] [PubMed] [Google Scholar]
- 54.Pang R, Li Y, Dong Y, Liang Z, Zhang Y, Zhang W. Identification of promoter polymorphisms in the cytochrome P450 CYP6AY1 linked with insecticide resistance in the brown planthopper, N ilaparvata lugens: Promoter polymorphisms in P450 CYP6AY1. Insect Mol Biol. 2014;23: 768–778. doi: 10.1111/imb.12121 [DOI] [PubMed] [Google Scholar]
- 55.Bass C, Zimmer CT, Riveron JM, Wilding CS, Wondji CS, Kaussmann M, et al. Gene amplification and microsatellite polymorphism underlie a recent insect host shift. Proceedings of the National Academy of Sciences. 2013;110: 19460–19465. doi: 10.1073/pnas.1314122110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Puinean AM, Foster SP, Oliphant L, Denholm I, Field LM, Millar NS, et al. Amplification of a Cytochrome P450 Gene Is Associated with Resistance to Neonicotinoid Insecticides in the Aphid Myzus persicae. Stern DL, editor. PLoS Genet. 2010;6: e1000999. doi: 10.1371/journal.pgen.1000999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schmidt JM, Good RT, Appleton B, Sherrard J, Raymant GC, Bogwitz MR, et al. Copy Number Variation and Transposable Elements Feature in Recent, Ongoing Adaptation at the Cyp6g1 Locus. Begun DJ, editor. PLoS Genet. 2010;6: e1000998. doi: 10.1371/journal.pgen.1000998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Itokawa K, Komagata O, Kasai S, Okamura Y, Masada M, Tomita T. Genomic structures of Cyp9m10 in pyrethroid resistant and susceptible strains of Culex quinquefasciatus. Insect Biochemistry and Molecular Biology. 2010;40: 631–640. doi: 10.1016/j.ibmb.2010.06.001 [DOI] [PubMed] [Google Scholar]
- 59.Wondji CS, Irving H, Morgan J, Lobo NF, Collins FH, Hunt RH, et al. Two duplicated P450 genes are associated with pyrethroid resistance in Anopheles funestus, a major malaria vector. Genome Research. 2008;19: 452–459. doi: 10.1101/gr.087916.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wittkopp PJ, Haerum BK, Clark AG. Evolutionary changes in cis and trans gene regulation. Nature. 2004;430: 85–88. doi: 10.1038/nature02698 [DOI] [PubMed] [Google Scholar]
- 61.Wittkopp PJ, Kalay G. Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat Rev Genet. 2012;13: 59–69. doi: 10.1038/nrg3095 [DOI] [PubMed] [Google Scholar]
- 62.Murataliev M, Guzov V, Walker F, Feyereisen R. P450 reductase and cytochrome b5 interactions with cytochrome P450: Effects on house fly CYP6A1 catalysis. Insect Biochemistry and Molecular Biology. 2008;38: 1008–1015. doi: 10.1016/j.ibmb.2008.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Adesanya AW, Cardenas A, Lavine MD, Walsh DB, Lavine LC, Zhu F. RNA interference of NADPH-cytochrome P450 reductase increases susceptibilities to multiple acaricides in Tetranychus urticae. Pesticide Biochemistry and Physiology. 2020; 104550. doi: 10.1016/j.pestbp.2020.02.016 [DOI] [PubMed] [Google Scholar]
- 64.Zhang Y, Wang Y, Wang L, Yao J, Guo H, Fang J. Knockdown of NADPH-cytochrome P450 reductase results in reduced resistance to buprofezin in the small brown planthopper, Laodelphax striatellus (fallén). Pesticide Biochemistry and Physiology. 2016;127: 21–27. doi: 10.1016/j.pestbp.2015.08.006 [DOI] [PubMed] [Google Scholar]
- 65.Beckmann JS, Estivill X, Antonarakis SE. Copy number variants and genetic traits: closer to the resolution of phenotypic to genotypic variability. Nat Rev Genet. 2007;8: 639–646. doi: 10.1038/nrg2149 [DOI] [PubMed] [Google Scholar]
- 66.Devine GJ, Barber M, Denholm I. Incidence and inheritance of resistance to METI-acaricides in European strains of the two-spotted spider mite (Tetranychus urticae) (Acari: Tetranychidae). Pest Manag Sci. 2001;57: 443–448. doi: 10.1002/ps.307 [DOI] [PubMed] [Google Scholar]
- 67.Fraaije BA, Bayon C, Atkins S, Cools HJ, Lucas JA, Fraaije MW. Risk assessment studies on succinate dehydrogenase inhibitors, the new weapons in the battle to control Septoria leaf blotch in wheat: SDHI fungicides and Septoria leaf blotch control. Molecular Plant Pathology. 2012;13: 263–275. doi: 10.1111/j.1364-3703.2011.00746.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Asahara M, Uesugi R, Osakabe Mh. Linkage Between One of the Polygenic Hexythiazox Resistance Genes and an Etoxazole Resistance Gene in the Twospotted Spider Mite (Acari: Tetranychidae). Journal of Economic Entomology. 2008;101: 1704–1710. doi: 10.1603/0022-0493(2008)101[1704:lbootp]2.0.co;2 [DOI] [PubMed] [Google Scholar]
- 69.Khalighi M, Dermauw W, Wybouw N, Bajda S, Osakabe M, Tirry L, et al. Molecular analysis of cyenopyrafen resistance in the two-spotted spider mite Tetranychus urticae. Pest Manag Sci. 2016;72: 103–112. doi: 10.1002/ps.4071 [DOI] [PubMed] [Google Scholar]
- 70.Stone BF. A formula for determining degree of dominance in cases of monofactorial inheritance of resistance to chemicals. 1968. [PMC free article] [PubMed] [Google Scholar]
- 71.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL http://www.R-project.org. 2017. [Google Scholar]
- 72.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25: 1754–1760. doi: 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20: 1297–1303. doi: 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25: 2078–2079. doi: 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29: 15–21. doi: 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31: 166–169. doi: 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 2014;15: 550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zheng X, Levine D, Shen J, Gogarten SM, Laurie C, Weir BS. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics. 2012;28: 3326–3328. doi: 10.1093/bioinformatics/bts606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zheng X, Gogarten SM, Lawrence M, Stilp A, Conomos MP, Weir BS, et al. SeqArray—a storage-efficient high-performance data format for WGS variant calls. Bioinformatics. 2017;33: 2251–2257. doi: 10.1093/bioinformatics/btx145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lu S, Wang J, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, et al. CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Research. 2020;48: D265–D268. doi: 10.1093/nar/gkz991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bateman A. The Pfam protein families database. Nucleic Acids Research. 2004;32: 138D – 141. doi: 10.1093/nar/gkh121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Riga M, Tsakireli D, Ilias A, Morou E, Myridakis A, Stephanou EG, et al. Abamectin is metabolized by CYP392A16, a cytochrome P450 associated with high levels of acaricide resistance in Tetranychus urticae. Insect Biochemistry and Molecular Biology. 2014;46: 43–53. doi: 10.1016/j.ibmb.2014.01.006 [DOI] [PubMed] [Google Scholar]
- 83.Omura T, Sato R. The carbon monoxide-binding pigment of liver microsomes I. Evidence for its hemoprotein nature. Journal of Biological Chemistry. 1964; 2370–2378. [PubMed] [Google Scholar]
- 84.Pritchard MP, McLaughlin L, Friedberg T. Establishment of Functional Human Cytochrome P450 Monooxygenase Systems in Escherichia coli. Cytochrome P450 Protocols. New Jersey: Humana Press; 2005. pp. 19–30. doi: 10.1385/1-59259-998-2:19 [DOI] [PubMed] [Google Scholar]
- 85.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical biochemistry. 1976; 248–254. doi: 10.1006/abio.1976.9999 [DOI] [PubMed] [Google Scholar]