ABSTRACT
A confirmed African swine fever (ASF) outbreak in Nigeria was further investigated by partial sequencing of the B464L and E183L genes of ASF virus (ASFV). Results revealed the first-time presence of ASFV genotype II in Nigeria and West Africa. This finding has serious implications for control measures and food security.
ANNOUNCEMENT
African swine fever (ASF) is a highly fatal disease of pigs caused by ASF virus (ASFV), a double-stranded DNA virus of the genus Asfivirus and family Asfarviridae (1). Twenty-four genotypes of ASFV have been identified based on partial sequencing of the genes B646L, encoding the p72 capsid protein, and E183L, encoding the p54 envelope protein (1, 2). Only genotype 1 is known to be circulating in Nigeria and West Africa, while ASFV genotype II circulates in Europe and Asia (1, 3–5). In this study, investigation of an ASF outbreak that occurred in May 2020 on a pig farm in Oko Oba, Lagos, Nigeria, was carried out by characterizing the B646L (p72) and E183L (p54) genes of ASFV. Tissue samples (liver and spleen) were collected from dead pigs that showed clinical signs of ASF, and DNA was extracted using the QIAamp DNA minikit (Qiagen, Hilden, Germany).
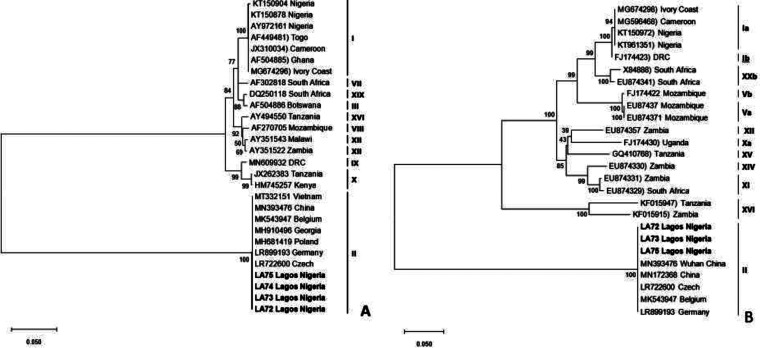
ASFV was detected in all tissue samples (4/4) collected from the pig farm using real-time PCR and analyzed at the National Veterinary Research Institute, Vom, Nigeria, as previously described (6). Genotyping was carried out by sequencing the B646L gene, which encodes the p72 protein, and E183L gene, which encodes the p54 protein, as previously described (7). The sequencing of the 4 ASFV-positive samples was carried out by the Sanger sequencing method at Macrogen Inc. (Netherlands). Chromatogram editing, assembly, and alignment of the raw sequences were done using the Staden package (http://staden.sourceforge.net/) and Bioedit (8). Two ASFV genes were sequenced, namely, B464L and E183L, but 7 sequences were obtained, including 4 sequences for B464L and 3 for E183L. They were compared with other sequences in the GenBank using the Basic Local Alignment Search Tool (BLAST) (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Twenty-four B646L (p72) and 21 E183L (p54) sequences representing the known genotypes were retrieved from GenBank for combined phylogenetic analysis with those generated from this study using MEGA X (9). Default parameters were applied for all software used. The minimum evolution method with 1,000 bootstraps was used for phylogenetic reconstruction. The comparison with ASFV sequences in GenBank revealed a similarity of 99% to 100% with ASFV genotype II from Vietnam (GenBank accession number MT332151) and China (MN393476). Phylogenetic analysis showed that sequences of both the p72 and p54 genes strongly clustered with ASFV genotype II (Fig. 1). The clustering of the sequences in our study with other genotype II sequences from different geographical areas had high bootstrap support. All of the p72 and p54 ASFV sequences that were obtained have been deposited in GenBank. In this report, sequencing and phylogenetic analysis revealed that ASFV genotype II was responsible for an ASF outbreak on a pig farm in Nigeria. Nucleotide sequences for p72 and p54 were used to describe the ASFV genotype in this study. Sequences from previous outbreaks in Nigeria and West Africa clustered only with genotype I. For the first time, ASFV genotype II was found and reported in West Africa. Introducing a new ASFV genotype into Nigeria presents complex epidemiology and will further complicate the already constrained control measures against the disease in the region.
FIG 1.
(A) Phylogenetic tree of partial B646L (p72) gene nucleotide sequences of Nigerian ASFV from a pig farm in Lagos, Nigeria. The tree was inferred using the minimum evolution method with 1,000 bootstrap replicates. The Nigerian ASFV isolates from this study are indicated in boldface and clustered with ASFV genotype II. (B) Phylogenetic tree based on the full-length E183L gene (p54). The phylogenetic tree was inferred using the minimum evolution method. The Nigerian ASFV sequences analyzed in this study are indicated by boldface and clustered within genotype II. The scale bar indicates the number of nucleotide substitutions per site.
Data availability.
The ASFV genome sequences in this report are available in GenBank under accession numbers MW852486 to MW852492.
ACKNOWLEDGMENT
This research was funded by the African Union Commission (AURG-II-1-196-2016).
Contributor Information
Adeyinka J. Adedeji, Email: yinkadeji@yahoo.com.
John J. Dennehy, Queens College CUNY
REFERENCES
- 1.Dixon LK, Stahl K, Jori F, Vial L, Pfeiffer DU. 2020. African swine fever epidemiology and control. Annu Rev Anim Biosci 8:221–246. doi: 10.1146/annurev-animal-021419-083741. [DOI] [PubMed] [Google Scholar]
- 2.Malogolovkin A, Burmakina G, Titov I, Sereda A, Gogin A, Baryshnikova E, Kolbasov D. 2015. Comparative analysis of African swine fever virus genotypes and serogroups. Emerg Infect Dis 21:312–315. doi: 10.3201/eid2102.140649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Penrith ML. 2020. Current status of African swine fever. CABI Agric Biosci 1:11. doi: 10.1186/s43170-020-00011-w. [DOI] [Google Scholar]
- 4.Costard S, Wieland B, de Glanville W, Jori F, Rowlands R, Vosloo W, Roger F, Pfeiffer DU, Dixon LK. 2009. African swine fever: how can global spread be prevented? Philos Trans R Soc Lond B Biol Sci 364:2683–2696. doi: 10.1098/rstb.2009.0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quembo CJ, Jori F, Vosloo W, Heath L. 2018. Genetic characterization of African swine fever virus isolates from soft ticks at the wildlife/domestic interface in Mozambique and identification of a novel genotype. Transbound Emerg Dis 65:420–431. doi: 10.1111/tbed.12700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.King DP, Reid SM, Hutchings GH, Grierson SS, Wilkinson PJ, Dixon LK, Bastos AD, Drew TW. 2003. Development of a TaqMan PCR assay with internal amplification control for the detection of African swine fever virus. J Virol Methods 107:53–61. doi: 10.1016/s0166-0934(02)00189-1. [DOI] [PubMed] [Google Scholar]
- 7.Bastos AD, Penrith ML, Cruciere C, Edrich JL, Hutchings G, Roger F, Couacy-Hymann E, Thomson GR. 2003. Genotyping field strains of African swine fever virus by partial p72 gene characterisation. Arch Virol 148:693–706. doi: 10.1007/s00705-002-0946-8. [DOI] [PubMed] [Google Scholar]
- 8.Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98. [Google Scholar]
- 9.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The ASFV genome sequences in this report are available in GenBank under accession numbers MW852486 to MW852492.