Abstract
Our understanding of the contribution of genetic risk factors to neuropsychiatric diseases is limited to abnormal neurodevelopment and neuronal dysfunction. Much less is known about the mechanisms whereby risk variants could affect the physiology of glial cells. Our prior studies have shown that a mutant (dominant-negative) form of a rare but highly penetrant psychiatric risk factor, Disrupted-In-Schizophrenia-1 (DISC1), impairs metabolic functions of astrocytes and leads to cognitive dysfunction. In order to overcome the limitations of the mutant DISC1 model and understand the putative regional properties of astrocyte DISC1, we assessed whether knockdown of Disc1 (Disc1-KD) in mature mouse astrocytes of the prefrontal cortex (PFC) or the hippocampus would produce behavioral abnormalities that could be attributed to astrocyte bioenergetics. We found that Disc1-KD in the hippocampus but not PFC impaired trace fear conditioning in adult mice. Using the innovative deep learning approach and convolutional deep neural networks (cDNNs), ResNet50 or ResNet18, and single cell-based analysis, we found that Disc1-KD decreased the spatial density of astrocytes associated with abnormal levels and distribution of the mitochondrial markers and the glutamate transporter, GLAST. Disc1-KD in astrocytes also led to decreased expression of the glutamatergic and increased expression of the GABA-ergic synaptic markers, possibly via non-apoptotic activation of caspase 3 in neurons located within the individual territories of Disc1-KD astrocytes. Our results indicate that altered expression of DISC1 in astrocytes could impair astrocyte bioenergetics, leading to abnormalities in synaptic neurotransmission and cognitive function in a region-dependent fashion.
Introduction
Astrocytes are the most abundant cells in the brain, and pathophysiological changes in astrocyte bioenergetics could lead to cognitive dysfunction (1,2). Recent progress in psychiatric genetics has advanced our knowledge of how genetic variants can affect neurodevelopment and adult brain function (3). Unfortunately, our understanding of the underlying molecular mechanisms is limited to neurons, and whether genetic variants could also affect the physiology of astrocytes remains understudied (4,5).
Disrupted-In-Schizophrenia-1 (DISC1) is a gene disrupted by balanced (1:11) (q42.1; q14.3) translocation, segregating in the Scottish family with several major psychiatric disorders (6). Despite its original name, DISC1 has not been associated with schizophrenia in the latest genome-wide association studies (7). Instead, owing to its extended protein–protein interaction network, DISC1 may be a key player in abnormal brain development and/or adult brain activities relevant to neuropsychiatric diseases (8–11). In this context, we use DISC1 and its protein products as valuable molecular tools to advance our understanding of the molecular pathobiology of different mental illnesses irrespective of their categorical classifications (11–14).
Multiple reports have demonstrated that DISC1 localizes to mitochondria to be involved in mitochondria trafficking and functions, including oxidative phosphorylation, Adenosine triphosphate (ATP) production and calcium buffering (15–22). However, the majority of these studies was performed on neuronal models (20,23), and whether the same partners underlying mechanisms and/or functional consequences of altered expression of DISC1 can be observed in other brain cells remains unknown. Our recent reports demonstrate that DISC1 is also expressed in astrocytes (24), and astrocyte-restricted expression of a mutant form of DISC1, dominant-negative DISC1 (DN-DISC1) in mice, produces cell-type-specific molecular changes, metabolic alterations and associated cognitive dysfunction (25,26). However, the DN-DISC1 model is confounded by several limitations, including possible gain-of-function effects unrelated to altered (i.e. decreased) expression of endogenous mouse DISC1. Thus, we selectively decreased Disc1 expression in astrocytes in vivo to assess whether Disc1 knockdown (Disc1-KD) would affect the mitochondrial markers in astrocytes and if Disc1-KD produced astrocyte dysfunction would alter levels of synaptic markers and cognitive function in a brain region-dependent fashion.
The recent advances in image recognition and classification have significantly expanded the application of the convolutional deep neural networks (cDNNs) in the biomedical fields, including pathology and oncology (27,28). In addition to the established software for image analysis such as Image-J/FIJI (imagej.nih.gov) and IMARIS (Bitplane AG, Zurich, Switzerland), cDNNs are increasingly being used for unbiased evaluation of images of different types of tissues (27,28). Here, using the innovative deep learning approach with transfer learning and cDNNs, ResNet50 or ResNet18, and single cell-based IMARIS-based analysis, we show that Disc1-KD in astrocytes of the hippocampus but not prefrontal cortex (PFC) impaired trace fear conditioning (TFC) in mice. This cognitive phenotype was associated with altered astrocyte morphology, expression of the mitochondrial markers and abnormal expression and cellular distribution of glutamate aspartate transporter (GLAST), possibly leading to decreased levels of glutamatergic and increased levels of GABAergic synaptic markers within the individual territories of affected astrocytes.
Results
Disc1-KD produced brain region-dependent cognitive impairment
Our prior studies have shown that expression of DN-DISC1 in astrocytes produced anxiety-like behavior and deficient performance in challenging cognitive tasks characterized by significant information load (25). As the DN-DISC1 transgenic model does not allow for evaluation of the regional effects and might have some untoward gain-of-function effects, we assessed the behavioral and brain effects of Disc1-KD in astrocytes in a brain region-dependent manner. We focused on female mice as most profound cognitive effects were noted in female DN-DISC1 mice (25). The schematic of the tests and post-mortem experiments is presented on Supplementary Material, Figure S1A. No significant effects of Disc1-KD were noted on general activity or anxiety in mice regardless of the brain area targeted (Supplementary Material, Fig. S1B and C). In TFC, similar to the DN-DISC1 model, Disc1-KD in hippocampal astrocytes did not affect the freezing behavior during the training session or the context-dependent freezing, but significantly decreased the cue-dependent freezing in mice (t = 2.33, P < 0.05, n = 7–11/group; Fig. 1A). In contrast to the hippocampus, Disc1-KD in astrocytes of the infralimbic area of the PFC produced no changes in the cue-dependent freezing (Fig. 1B). Based on the cognitive effects of Disc1-KD predominantly in the hippocampus, all subsequent studies focused on the mouse hippocampus.
Figure 1 .
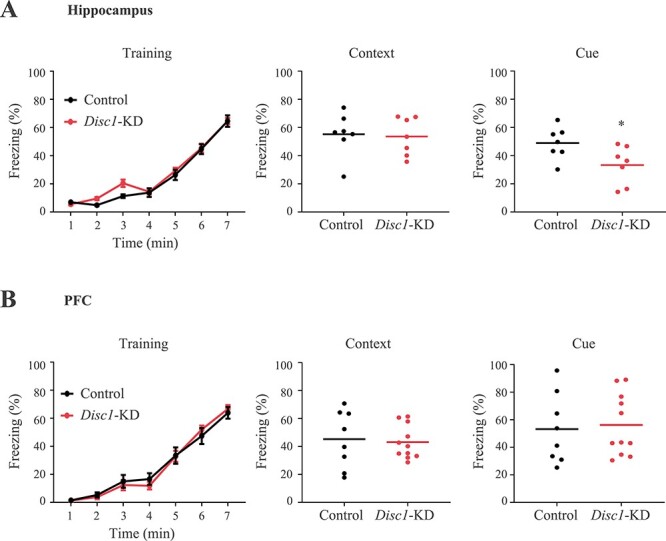
Disc1-KD in the hippocampus impairs trace fear conditioning. (A) Disc1-KD of hippocampal astrocytes decreased cue-dependent freezing without altering training or context-dependent freezing. (B) Disc1-KD of cortical astrocytes had no effects on training, context-dependent or cue-dependent freezing. * denotes P < 0.05, n = 7–11/group; Student two-tail t-test, t = 2.33
Disc1-KD changed astrocyte morphology
Consistent with the behavioral effects, Disc1-KD in astrocytes of the hippocampus significantly increased their spatial density (t = −3.168, P = 0.0034, N = 19–24 images/group; Fig. 2A), while we saw no significant effects of Disc1-KD on spatial density of cortical astrocytes (Supplementary Material, Fig. S2).
Figure 2 .
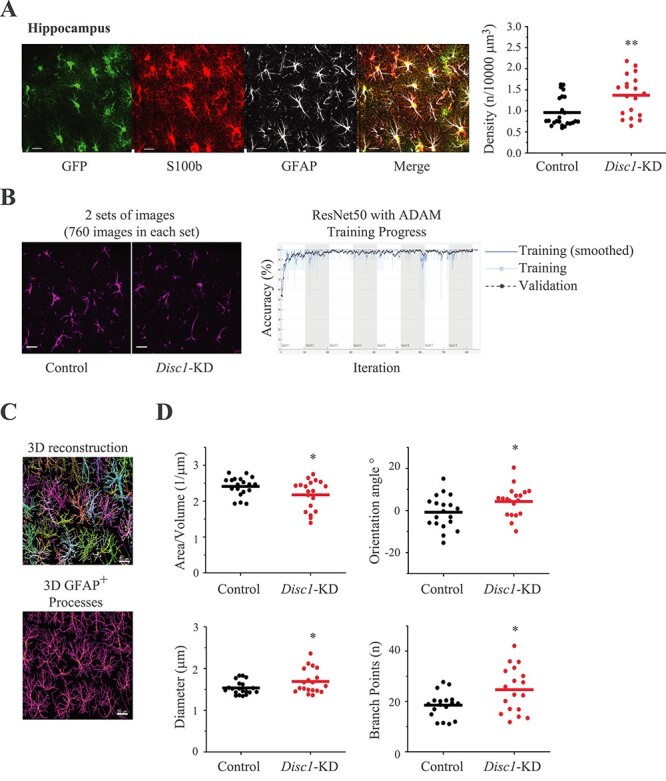
Disc1-KD affects astrocyte morphology. (A) Spatial density of astrocytes; the left panel—representative images of GFP+ (green) astrocytes co-stained with anti-S100b (red) and anti-GFAP (gray); scale bar—20 μm; the right panel—quantitative analysis of spatial density; Student two-tail t-test, t = −3.168, **- P = 0.0034, N = 19–24 images/2 sections per mouse/4 mice per group; (B) Transfer learning approach using cDNNs; the left panel—representative images of control and Disc1-KD GFAP+ astrocytes used for classification with pre-trained cDNNs ResNet50 and transfer learning, GFP channel is not shown, scale bar—20 μm; the right panel—representative histogram of training progress of the ResNet50 with ADAM optimizer; the X axis represents the number of epochs (total 8) with 103 iterations in each epoch. (C) 3D reconstruction of GFAP+ astrocytes with single astrocytes being colored with different colors and 3D visualization of GFAP+ main processes of astrocytes, scale bar—20 μm; (D) Quantitative analysis of area/volume ratio (Student two-tail t-test, t = 2.070, *- P = 0.046), processes orientation angle (Student two-tail t-test, t = −2.102, *- P = 0.042), diameter of GFAP+ processes (Student two-tail t-test, t = −2.278, *- P = 0.028) and number of branch points (Student two-tail t-test, t = −2.528, *- P = 0.018); N = 19 images/2 section per mouse/4 mouse per group.
To assess morphological changes in astrocytes with a different approach, we used transfer learning and cDNNs with either 50 deep layers (ResNet50) or 18 deep layers (ResNet18). cDNNs performed a classification of GFAP+ labeled images from the Disc1-KD and control image sets (Fig. 2B). Specifically, when cDNN ResNet50 was retrained to extract features of GFAP+ processes in both sets and classify images, accuracy of prediction during the classification was 99.77% with the ADAM optimizer (Fig. 2B) and 99.10% with the SGDM optimizer (Supplementary Material, Fig. S3A). When cDNN ResNet18 was used, it produced better accuracy of prediction—99.10% with the SGDM optimizer and 100% with the ADAM one. (Supplementary Material, Fig. S3B). As the ADAM optimizer demonstrated superior accuracy, we use it to test the quality of image set preparation. We retrained ResNet50 to discriminate the sets containing the same image sets, i.e. ‘Control’ versus ‘Control’ or ‘Disc1-KD’ versus ‘Disc1-KD’. We found no successful retraining and low accuracy of prediction, which was 15.13% for the control sets, and 22.56% for the Disc1-KD sets (Supplementary Material, Fig. S3C). The same procedure with ResNet18 produced accuracy of prediction 25.88% for the control sets and 39.77% for the Disc1-KD sets (data not shown).
We also compared the microstructure of the hippocampal GFP+ surface of astrocytes between the control and Disc1-KD groups using images extracted from confocal stacks taken with a higher magnification (100× objective and zoom = 1.5×). cDNN ResNet50 was retrained to classify images from the ‘Control’ and ‘Disc1-KD’ image sets. Accuracy of prediction was again better with the ADAM optimizer—98.43% (Supplementary Material, Fig. S4A) compared to the SGDM optimizer—95.13% (data not shown). Similarly, cDNN ResNet18 showed better accuracy of prediction with ADAM optimizer (98.49%) (Supplementary Material, Fig. S4AB) than with the SGDM one (94.55%). When cDNN ResNet50 was retrained with the same image sets (‘Control’ versus ‘Control’ and ‘Disc1-KD’ versus ‘Disc1-KD’), accuracy of prediction was 48.35% and 49.34%, respectively (Supplementary Material, Fig. S4C). ResNet18 showed 50.10% for the ‘Control’ versus ‘Control’ sets and 50.87% for the ‘Disc1-KD’ versus ‘Disc1-KD’ sets (Supplementary Material, Fig. S4D).
To validate the results of astrocyte processes classification with cDNN ResNet50 and ResNet18, we compared the same images of GFAP+ astrocytes using the Imaris software and found that Disc1-KD led to a significantly bigger diameter of GFAP+ processes (t = −2.278, P = 0.028), a decreased surface area/volume ratio (t = 2.070, P = 0.046), the significant changes in the orientation angle of secondary processes (t = −2.102, P = 0.042) and the significantly increased numbers of branch points (t = −2.528, P = 0.018; N = 19 images/group; Fig. 2C and D). Consistent with the results for GFP+ astrocyte surface generated with transfer learning and cDNN approaches, Imaris-based analyses also revealed a significant increase in the surface area/volume ratio (t = −2.363, P = 0.023) and a reduced vertices/astrocyte volume ratio following Disc1-KD (t = 2.414, P = 0.020, N = 22–23/group; Supplementary Material, Fig. S5). Collectively, our results demonstrate significant changes in astrocyte morphology after knockdown of Disc1 in the hippocampus.
Disc1-KD affected expression and distribution of GLAST
Glutamate re-uptake is a key function of astrocytes (29,30). We sought to evaluate whether Disc1-KD would alter expression and/or distribution of the glutamate transporters, glutamate transporter 1 (GLT-1) and GLAST.
Disc1-KD had no effects on GLT-1+ immunoreactivity (Supplementary Material, Fig. S6) but significantly decreased density (t = 6.4748, P = 1.3*10−7) and increased the size of GLAST+ spots (t = −4.957, P = 6.6*10−5) as well as reduced intensity (t = 3.4965, P = 0.0015) and increased the distance of GLAST+ spots to the surface of the astrocyte (t = −2.436, P = 0.021; Fig. 3). We also separately assessed GLAST+ spots in the astrocyte soma and found decreased spatial density of GLAST+ spots (t = 3.082, P = 0.0045, N = 19 images/group) with no changes in GLAST+ spots’ size (Supplementary Material, Fig. S7).
Figure 3 .
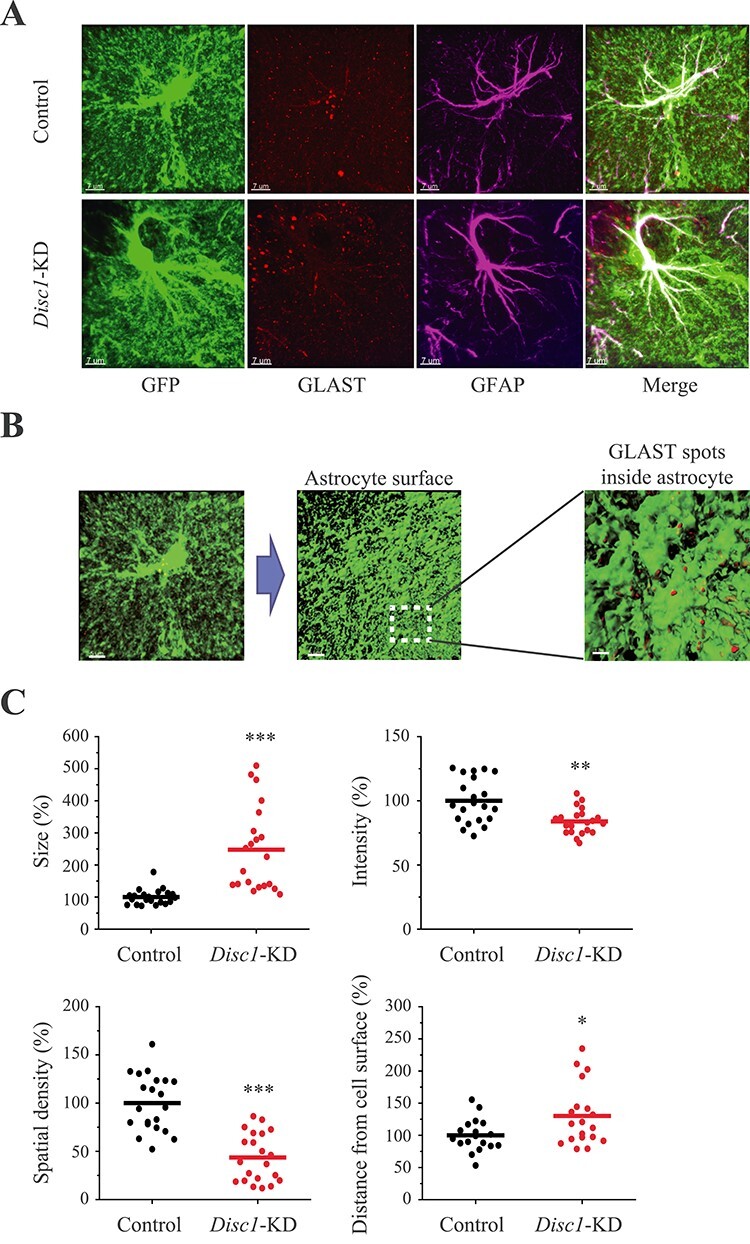
Disc1-KD alters expression and distribution of GLAST in astrocytes. (A) Representative images of control and Disc1-KD GFP+ (green) astrocytes co-stained with anti-GLAST (red) and anti-GFAP (magenta); scale bar—7 μm; (B) Assessing GLAST+ spots within individual GFP+ astrocytes; representative image of a GFP+ astrocyte, scale bar—5 μm; 3D surface visualization, scale bar—5 μm; a blown-up area outlined with GLAST+ spots within (brown) and on the surface of the GFP+ astrocyte (red); scale bar—1 μm; (C) Quantitative analyses of GLAST+ spots within individual astrocytes: size (Student two-tail t-test, t = −4.957, ***- P = 6.6*10−5), intensity (Student two-tail t-test, t = 3.4965, **- P = 0.0015), spatial density (Student two-tail t-test, t = 6.4748, ***- P = 1.3*10−7) and distance from astrocyte surface (Student two-tail t-test, t = −2.436, *- P = 0.021); N = 19–20 images/2 sections per mouse/4 mice per group.
Disc1-KD affected expression of the mitochondrial markers
Glutamate transport is an energy demanding process (31,32). Since DISC1 is a partner of mitofilin that is critical for protein transport in the mitochondrion and an important part of the MICOS complex (mitochondrial contact site and cristae organizing system) (20,33), we sought to evaluate the measures of mitochondria bioenergetics in Disc1-KD astrocytes that could contribute to abnormal expression and distribution of GLAST in transduced astrocytes. We assessed density, intensity and size of immunoreactive spots of TIM23 (translocase of the inner membrane), PDH (pyruvate dehydrogenase, one of the two component enzymes of a pyruvate dehydrogenase complex), MTCO1 (cytochrome c oxidase subunit I) and ATP5A (subunit of mitochondrial ATP synthase). Disc1-KD significantly increased density of TIM23+ and PDH+ spots (t = −4.0026, P = 0.00029 and t = −2.107, P = 0.045, respectively, N = 19–24 images/group) without altering those of MTCO1+ or ATP5+ (Fig. 4; Supplementary Material, Fig. S8). A prior study found that the distance between the glutamate transporters and mitochondria is critical for efficiency of glutamate uptake (31). Thus, we also measured the distance between the mitochondrial markers TIM23, PDH, MTCO1, ATP5A and GLAST+ spots. We found that Disc1-KD decreased distance between GLAST+ spots and those positive for TIM23 (t = 5.406; P = 8.2 × 10−6; Fig. 4), MTCO1 and ATP5A (t = 3.104, P = 0.0039 and t = 2.693, P = 0.011, respectively, N = 20–21 Images/group; Supplementary Material, Fig. S8).
Figure 4 .
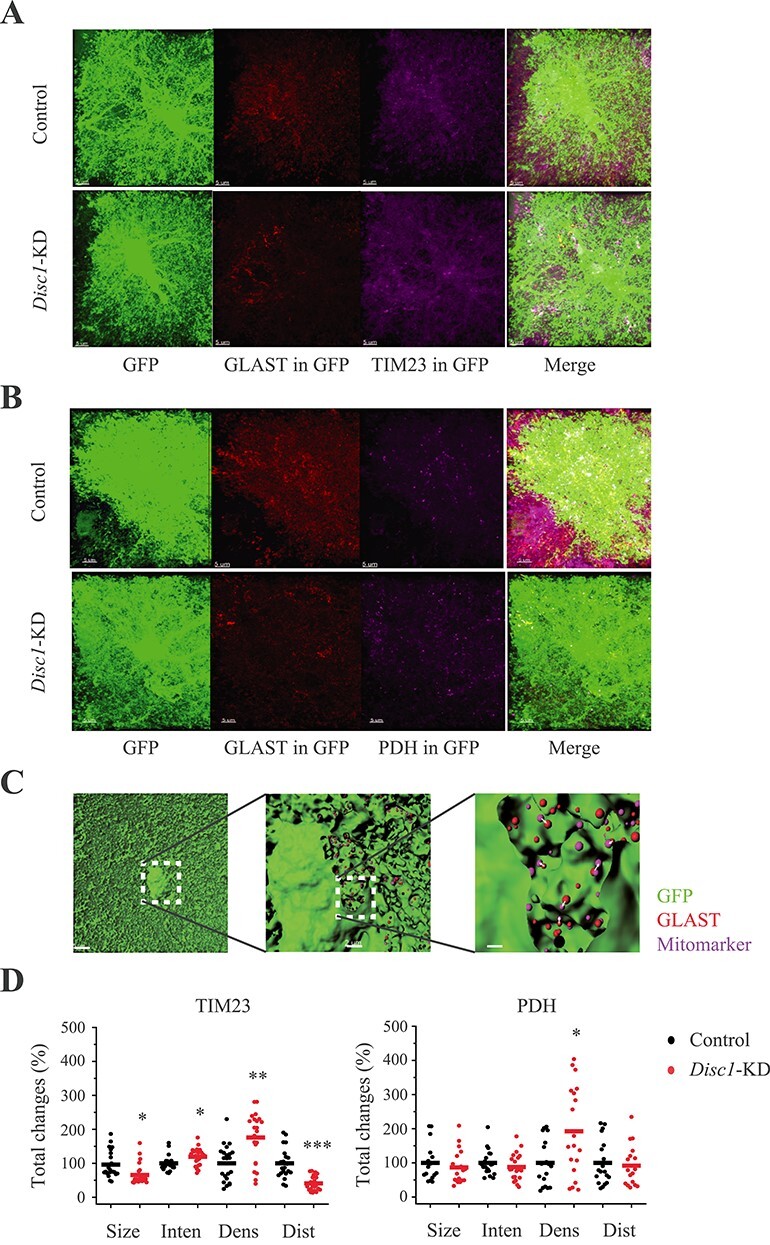
Disc1-KD affects expression of the mitochondrial markers in astrocytes. (A) Representative images of GFP+ control and Disc1-KD GFP+ (green) astrocytes stained with anti-GLAST (red) and anti-TIM23 (magenta); GLAST+ (GLAST in GFP) and TIM23+ (TIM23 in GFP) immunoreactivity inside GFP+ astrocyte are shown, scale bar—5 μm. (B) Representative images of GFP+ control and Disc1-KD astrocytes stained with anti-GLAST (red) and anti- anti-PDH (magenta); GLAST+ (GLAST in GFP) and PDH + (PDH in GFP) immunoreactivity inside GFP+ astrocyte are shown, scale bar—5 μm. (C) A workflow for analysis of mitomarkers and GLAST+ spots: 3D surface of the GFP+ mask (green) for astrocytes (the left panel), scale bar—5 μm; GLAST+ (red) and a mitochondrial marker (magenta) spot within an astrocyte (a clipping plane, the middle panel), scale bar—2 μm; a blown-up clipping plane with GLAST+ (red) and mitomarker (magenta) spots inside an astrocyte (the right panel); arrows—distance between GLAST and mitomarker; scale bar—1 μm. (D) Quantitative analyses of PDH+ and TIM23+ spots within individual astrocytes. For TIM23: Size (Student two-tail t-test, t = 3.2426, *- P = 0.0023), Inten—intensity (Student two-tail t-test, t = −2.659, *- P = 0.0111), Dens—spatial density (Student two-tail t-test, t = −4.0026, **- P = 0.00029), and Dist—distance (Student two-tail t-test, t = 5.406; ***- P = 8.2*10−6), between GLAST+ puncta and the mitochondrial marker inside individual astrocytes. For PDH: Dens—spatial density, Student two-tail t-test, t = −2.107, *- P = 0.045. N = 19–24 images/2–3 sections per mouse/4 mice per group.
Disc1-KD in astrocytes led to synaptic changes
As Disc1-KD in astrocytes affected the level and cellular distribution of GLAST that regulates glutamate uptake to influence glutamate synaptic transmission (29,30), we evaluated whether Disc1-KD would lead to altered expression of synaptic markers within the individual territories of Disc1-KD astrocytes. Specifically, we measured density of immune-positive puncta related to glutamatergic (VGLUT1+/PSD95+) or GABAergic (VGAT+/Gephyrin+) synapses located within individual territories of astrocytes. For analysis, we selected puncta of pre-synaptic and post-synaptic markers located less than 0.5 μm from each other (34). Disc1-KD significantly reduced density of VGLUT1+/PSD95+ puncta pairs (t = 2.177, P = 0.039, N = 15–18 images/group) and significantly increased density of VGAT+/Gephyrin+ puncta pairs (t = 2.381, P = 0.041, N = 8–9 images/group). The size of the co-labeled or individually labeled puncta, with the exception of PSD95+ puncta (t = −2.564, P = 0.017), was not altered (Fig. 5; Supplementary Material, Fig. S9). Compared to controls, Disc1-KD led to a greater number of VGLUT1+/PSD95+ puncta located farther away from the surface of the transduced astrocyte. In contrast, the distance between the astrocyte surface and VGAT+/Gephyrin+ puncta were unchanged (Fig. 5).
Figure 5 .
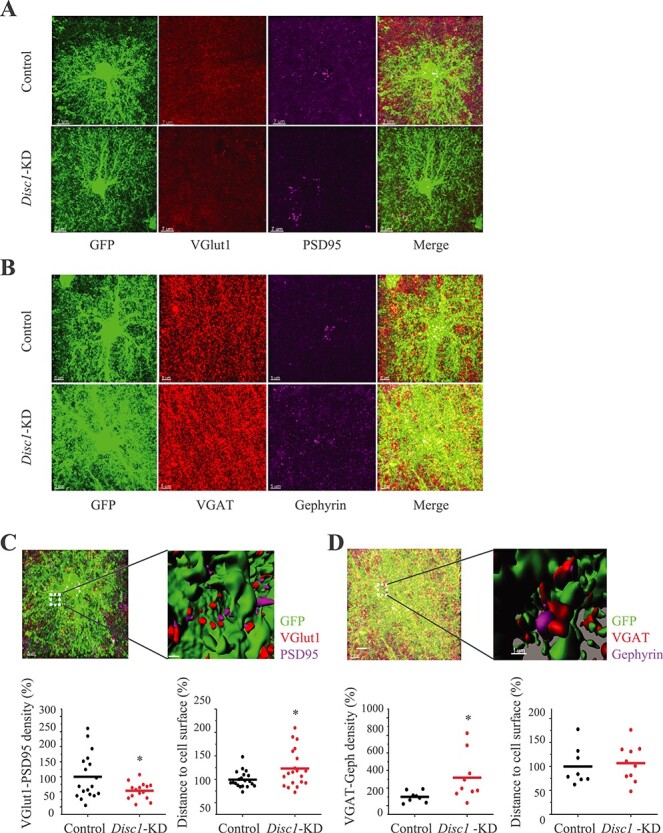
Disc1-KD alters expression of the synaptic markers within the astrocyte zone. (A) Representative images of GFP+ astrocytes co-stained with anti-VGLUT1 (red), anti-PSD95 (magenta), scale bar—7 μm; (B) Representative images of GFP+ astrocytes co-stained with anti-VGAT (red), anti-Gephyrin (magenta), scale bar—5 μm; (C) The upper panels: 3D visualization of glutamatergic puncta near the astrocyte surface within the individual territory of astrocyte; VGLUT1 (red) and PSD95 (magenta) puncta, scale bar—5 μm; and a blown-up 3D visualization of the co-stained area with VGLUT1 (red) and PSD95 (magenta) puncta, scale bar 1 μm. The bottom panels: only VGLUT1+/PSD95+ puncta with the between-markers distance less than 0.5 μm were selected for analysis. Decreased spatial density of VGLUT1+/PSD95+ puncta after Disc1-KD in astrocytes (Student two-tail t-test,t = 2.177, *- P = 0.039); increased distance from VGLUT1+ spots to the astrocyte surface (Student two-tail t-test, t = −2.491, *- P = 0.020), N = 15–18 images/2–3 sections per mouse/4 mice per group. (D) The upper panels: 3D visualization of GABAergic spots near the astrocyte surface within a single astrocyte zone; VGAT (red) and Gephyrin (magenta) spots puncta, scale bar—5 μm; a blown-up 3D visualization with VGAT (red) and Gephyrin (magenta) spots, scale bar 1 μm. The bottom panels: Only VGAT+/Gephyrin+ spots with the between-markers distance less than 0.5 μm were selected for analysis. Increased spatial density of VGAT+/Gephyrin+ puncta after Disc1-KD in astrocytes (Student two-tail t-test, t = 2.381, *- P = 0.041); no changes in distance from VGAT+ spots to the astrocyte surface; N = 8–9 images/2 sections per mouse/4 mice per group.
Caspase 3 activation in neurons within the territories of Disc1-KD astrocytes
Synaptic abnormalities in the hippocampus might result from abnormal glutamate metabolism because of an altered level and distribution of GLAST in Disc1-KD astrocytes. It is conceivable that decreased GLAST expression led to elevated levels of extracellular glutamate that could produce synaptic abnormalities in neurons by inducing apoptotic processes (35–37). To test that possibility, we assessed levels of two different markers of apoptosis, namely, the active subunit of Caspase3 p17 (Casp3–17), a marker of the ‘before no return point’, and DNA fragmentation, a marker of the ‘after no return point’ (38).
Disc1-KD in astrocytes significantly increased the spots’ size and the spatial density of Casp3–17+ spots outside PV+ neurons and outside GFP+ astrocytes, as well as increased the spatial density of Casp3–17+ spots inside PV+ neurons (Fig. 6). TUNEL assay for detecting apoptotic DNA fragmentation showed the absence or a very low signal intensity in the pyramidal layer of the CA1-CA2 areas of the hippocampus in Disc1-KD or control brains. As expected, 30-min incubation of the brain sections with DNase I produced a dramatic increase in TUNEL signal intensity (t = 6.797, P = 0.00053 for Control versus DNase I; t = 6.308, P = 0.00147 for Disc1-KD versus DNase I, N = 6–10 Images/group; Supplementary Material, Fig. S10).
Figure 6 .
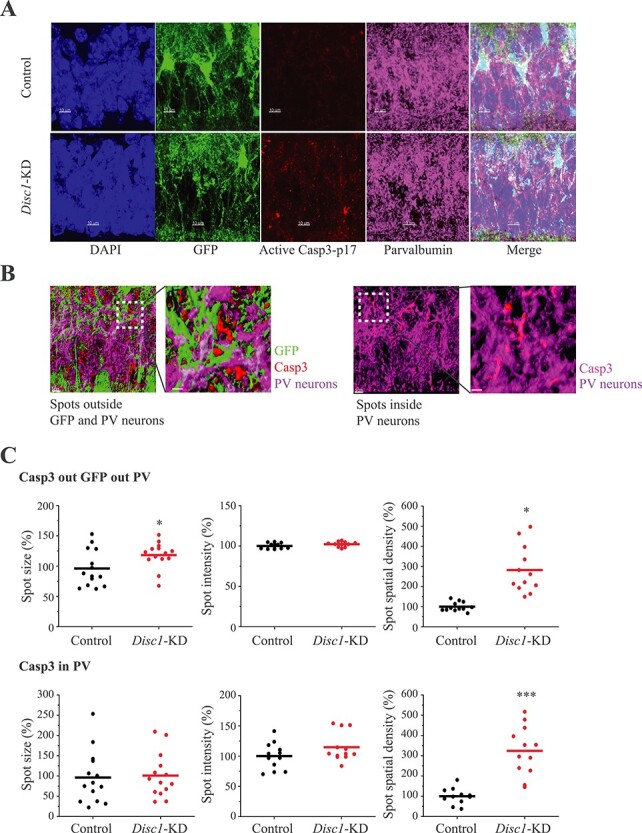
Caspase activation in neurons within the Disc1-KD astrocyte territories. (A) Representative images of GFP+ (green) astrocytes, DAPI+ (blue) nuclei, Caspase3 p17+ (red) and Parvalbumin+ (magenta) immunoreactivity, scale bar—10 μm; (B) 3D visualization of Caspase3 p17+ immunoreactivity in neurons; 3D visualization of Caspase3 p17+ immunoreactivity outside GFP+ and PV+ neurons (the left panel); 3D visualization of Caspase3 p17+ immunoreactivity inside PV+ neurons (the right panel); scale bar—5 μm; (C) Quantitative analyses of Caspase3 p17+ immunoreactivity outside GFP+ cells and PV+ neurons (the upper panels) and inside PV+ neurons (the bottom panels); For Casp3 out GFP out PV: spots size (Student two-tail t-test, t = −2.638, *- P = 0.0195); spot intensity; spot spatial density (t = −5.313, ***- P = 0.00018). For Casp3 in PV: spots size; spot intensity; spot spatial density (Student two-tail t-test, t = −4.115, ***- P = 0.00046), N = 12/2–3 sections per mouse/4 mice per group.
Discussion
Our findings indicate that knockdown of Disc1 expression selectively in astrocytes impairs TFC in a brain area-dependent manner. Using the innovative cDNN approaches, and single cell-based analyses, we found that deficient TFC was linked to decreased spatial density of astrocytes and reduced expression of the glutamate transporter, GLAST. It is tempting to speculate these astrocyte abnormalities might be, at least in part, due to Disc1-KD produced abnormal expression of the key mitochondrial proteins in astrocytes. These astrocyte anomalies could contribute to altered levels of the glutamatergic and GABA-ergic synaptic markers, possibly via activation of caspase 3 in neurons located within the individual territories of Disc1-KD astrocytes.
The present results concord with our prior studies using a mouse model of inducible expression of DN-DISC1 (25,39). Decreasing levels of DISC1 in astrocytes with either over-expression of DN-DISC1 or KD-Disc1 worsen TFC without significantly affecting general activity or anxiety-like behaviors. In both models, deficient TFC is associated with metabolic alterations in astrocytes (39). Similar phenotypic outcomes produced by DN-DISC1 and Disc1-KD in astrocytes provide additional validation for the past demonstrations of the cellular and behavioral effects reported for transgenic DISC1 mouse models.
Further, similar to the astrocyte DN-DISC1 model, our findings with Disc1-KD point to altered astrocyte bioenergetics as a possible mechanism underlying abnormal spatial density of astrocytes and decreased levels of GLAST. Several studies have implicated DISC1 in mitochondrial dynamics, regulation of mitochondrial quality control and function (16–23). We also reported that either DN-DISC1 or Disc1-KD decreased oxidative phosphorylation and glycolysis in primary astrocytes (39). In the current study, using an individual astrocyte-based analysis, we demonstrate that Disc1-KD increases density of the TIM23 and PDH-positive spots, the key mitochondria factors (40,41). These changes appear consistent with interaction of DISC1 with the mitochondrial protein, Mitophilin, which is involved in the import of mitochondrial proteins (20). It is conceivable that astrocyte DISC1 might also be involved in the protein import machinery by interacting with Mitofilin, and Disc1-KD disrupts this interaction leading to mitochondrial abnormalities.
In the context of possible abnormal astrocyte bioenergetics, it is worth mentioning that Disc1-KD decreased distance between GLAST-positive puncta and three mitochondria factors. Prior studies demonstrated the physical proximity of mitochondria and glutamate transporters at sites of glutamate uptake and neuronal influences on mitochondrial co-localization with glutamate transporters in astrocytes (31,42). One could propose that mitochondrial abnormalities following Disc1-KD might decrease the distance between GLAST and mitochondria as a compensatory mechanism of decreased spatial density of expression of GLAST-positive puncta in astrocytes. Although not evaluated directly in our study, all these changes might potentially affect the glutamate uptake capacity of transduced astrocytes. Future studies will shed light on the possible mechanisms whereby DISC1 is involved in mitochondria-glutamate transporters interplay in astrocytes and whether pathological changes in this interplay would affect glutamate uptake by astrocytes.
We found significant changes in expression of the pre- and postsynaptic markers of glutamatergic and GABAergic synapses. Notably, these alterations were noted within the individual territories of transduced astrocytes, suggesting that the synaptic abnormalities might result from altered GLAST expression that could lead to chronically elevated levels of extracellular glutamate (43–45). Decreased density of the markers for glutamatergic synapses and increased distance between the glutamatergic synaptic markers and the neuronal surface would seem to be consistent with this hypothesis. Interestingly, neurons located within the territories of individual transduced astrocytes exhibited elevated levels of Caspase 3. Although we cannot completely rule out that increased expression of Caspase 3 is a marker of pro-apoptotic processes, full-blown apoptosis appears less likely as no signs of cell death were detected by TUNEL assay. Instead, we would like to propose that Caspase 3 activation could be responsible for changing levels of the synaptic markers. For example, there were a few reports to indicate that elevated levels of caspases could contribute to postsynaptic remodeling and synaptic maturation (46). Future studies will evaluate this intriguing possibility in detail.
Our study also demonstrates for the first time a successful application of the innovative approach for assessment of morphological effects of Disc1-KD in astrocytes, namely cDNNs and transfer learning. Our results show that both cDNN models, ResNet50 and ResNet18, were successfully re-trained to extract the specific features of images in control and Disc1-KD groups and classified them with high accuracy of prediction. This was even possible using 2D images prepared from confocal stacks, with ADAM optimizer and cDNN ResNet18 demonstrating better results compared to those being generated with SGDM and cDNN ResNet50. Importantly, the outcomes produced with the cDNN approaches were validated using the popular Imaris software. Thus, our data demonstrate that cDNN and transfer learning can be used as a first line of a faster analysis of astrocyte morphology for future studies, with Imaris-based evaluation being used for subsequent assessments of more subtle changes if indicated.
Our study has some limitations. Although the main focus of the present work is to evaluate the in vivo role of DISC1 in astrocyte bioenergetics, the Adeno-associated virus (AAV) we used does not target GFP to the astrocyte membrane, limiting our ability to evaluate potential effects of Disc1-KD on fine astrocyte branchlets and processes and expression of the mitochondria markers in those fine structures. In addition, the titers of control and Disc1 were not exactly the same. However, given that it is control AAV that had a greater titer, one might expect that the effects produced by Disc1-KD might have been even more pronounced. Based on our initial characterization of the behavioral effects of Disc1-KD in astrocytes, we focused on the in-depth studies of the role of DISC1 in astrocytes of female mice. Still, our data do not rule out the possibility that using additional behavioral and cognitive tests and/or assessing other brain areas, one could have detected significant alterations in male mice as well. The future studies will address this possibility in greater detail.
In conclusion, we found that Disc1 knockdown in astrocytes of the CA1-CA2 areas of the hippocampus but not the infralimbic area of the PFC impairs TFC. Our data also suggest that this cognitive phenotype may be linked to dysfunction of glutamatergic and/or GABA synapses produced by abnormal expression of the glutamate transporter, GLAST, as a consequence of mitochondrial abnormalities in transduced astrocytes.
Materials and Methods
Animals
Female C57BL6/j mice, 8 weeks of age, were purchased from the Jackson Lab for this study. We focused on female mice because our prior studies with expression of DN-DISC1 in astrocytes showed the strongest behavioral changes in female mice (25). Mice were housed in sex-matched groups of five in standard mouse cages on a 12-h light/dark cycle at a room temperature (RT) of 23°C with free access to food and water. The animal protocol was approved by the Johns Hopkins University Animal Care and Use Committee.
AAV injections
To study the role of DISC1 in astrocyte bioenergetics, we knocked down expression of mouse Disc1 selectively in astrocytes using AAV vectors as previously described (26). Briefly, control (2.012 GC/ml) and Disc1-KD (2.011 GC/ml) AAV vectors were injected into the infralimbic area of the PFC (AP + 2.0, ML ± 1, DV-2.6, angle 17°), or in the CA1 area of the hippocampus (AP, ML, DV: −2.1; ±2.0; −1.5; and −2.1; ±1.4; −1.3). AAV vectors were delivered bilaterally in each target region in the amount of 0.5 μl at the rate of 50 nl/min. (Supplementary Material, Fig. S11) (47).
Behavioral tests
Four weeks after the injections, mice were tested in a series of behavioral tests (Supplementary Material, Fig. S1A).
Open field
Locomotor activity was assessed over 30 min in a 40 × 40 cm activity chamber with infrared beams (San Diego Instruments Inc., San Diego, CA, USA). Horizontal activity, as well as time spent in the center or periphery of the chamber, was automatically recorded.
Elevated Plus Maze
Anxiety-related behavior was assessed in the elevated plus maze. Mice were placed in the center of the maze consisting of two open arms and two closed arms 33 cm in length (San Diego Instruments Inc., San Diego, CA, USA) and allowed to explore the maze for 5 min. Distance travelled and time spent in the open arms were automatically recorded using Anymaze Tracking Software (Stoelting, Co., Wood Dale, IL, USA).
Trace Fear Conditioning
TFC was conducted as previously described (25). Briefly, TFC consisted of a habituation day, a training day and a test day over three consecutive days. On the habituation day, mice were exposed to the shock box (Coulbourn, Holliston, MA, USA) for 10 min. On the training day, mice were placed in the shock box and given a 2-min habituation, after which a 20-s white noise tone (80 db, 2000 Hz) was delivered. Twenty seconds following the termination of the tone, a scrambled 2-s 0.5 mA shock was delivered. The tone–shock pairing was repeated three additional times. On the test day, mice were placed in the shock box for 3 min to measure freezing in response to the context. Mice were then placed in a separate context and freezing in response to the 20-s white noise tone was measured. Freezing behavior was automatically scored using Cleversys Freezescan (Cleversys Inc., Reston, VA, USA).
Immunostaining
Immediately after completion of all behavioral tests, mice were sacrificed for the subsequent morphological studies (Supplementary Material, Fig. S1A).
Mice were deeply anesthetized with Forane (isofluran USP, NDC 10019-360-60, Baxter Healthcare Corporation, Deerfield, IL, USA) followed by transcardial perfusion with ice cold 0.1-M phosphate buffer (PBS; pH 7.4) with heparin (10 000 U/L) and 4.0% paraformaldehyde in 0.1-M PBS. The brains were dissected out and post-fixed in 4.0% paraformaldehyde in 0.1-M PBS for 24 h at 4°C. After cryoprotection in 30% sucrose in 0.1-M PBS for 48 h, the brains were cut into 40-μm thick parasagittal sections.
Transduced brain areas were visualized using expression of GFP. To test cell specificity of transduction, brain sections were stained with chicken anti-GFAP (1:1000), rabbit anti-S100 beta (1:1000), rabbit anti-Iba1 (1:1000) or guinea pig anti-NeuN (1:1000) antibodies as previously described (26,48). Briefly, after incubating brain sections in the blocking solution for 1 h at RT, the sections were incubated for 48 h at 4°C with the primary antibodies. Afterwards, the sections were incubated for 2 h at RT with the corresponding Alexa 488-, 568-, 633-labeled species-specific secondary antibodies (1:1000) followed by three 3 × 5-min PBS washes and DAPI 10 min staining (1:10000). After 3 × 5-min PBS washes and 30-min drying sections were mounted on Superfrost plus slides (Thermo Fisher Scientific, Waltham, MA; Cat # 22–037-246) with Aqua-Poly/Mount medium (Polysciences, Inc; Warrington, PA; Cat # 18606–20) and stored at 4°C until imaging. Images were collected with a Zeiss 880 confocal laser scanning microscope with GaAsP detector at the Johns Hopkins University Neuroscience Multiphoton/Electrophysiology Core Facility. Laser settings and optical parameters were kept constant between all images in Control and Disc1-KD groups.
To evaluate expression of glutamate transporters, rabbit anti-EAAT1 (1:1000) and anti-EAAT2 (1:1000) antibodies were used. To evaluate co-expression of pre- and postsynaptic markers of GABAergic or glutamatergic synapses, guinea pig anti-VGAT (1:1000), and monoclonal mouse anti-Gephyrin (1:1000) (for GABAergic synapses), or mouse anti-PSD95 (1:1000) and rabbit anti-VGLUT1 (1:1000) (for glutamatergic synapses) antibodies were used.
To assess cell apoptosis in AVV-transduced areas and evaluate co-localization of the apoptotic marker with GABAergic or glutamatergic neurons, mouse anti-Caspase3 p17 antibody were used. In addition, we used Click-iT™ Plus TUNEL Assay for in situ apoptosis detection (Thermo Fisher Scientific, Waltham, MA; Cat # C10618) to evaluate the final steps of apoptosis. Supplementary Material, Table S1 contains detailed information about the antibodies used.
Astrocyte morphology
In order to evaluate putative effects of Disc1-KD on the morphology of astrocytes, we used three complementary but independent approaches.
Spatial density
A prior study demonstrated no overlap between individual astrocyte territories in adult mice (49). Based on this finding, we assessed the effects of Disc1-KD on astrocyte spatial density by dividing the number of GFAP+/S100ß+ astrocytes within a confocal image by the total volume of the confocal image as previously described (49). Images were acquired with objective EC Plan-Neofluar 40×/1.30 Oil DIC and zoom = 1×. Frame size was set to 1024 × 1024 pixels, bit depths to 8-bit, averaging to 2× and z-step size to 0.5 μm (voxel dimension 208 nm × 208 nm × 500 nm, 35–45 optical sections per image) to perform precise 3D analysis and calculations.
Deep neural network
We also used a deep learning approach in the Matlab environment as described in Matlab with some minor modifications (Github). The training procedure with the cDNN takes time and requires multiple images. In order to improve learning feature accuracy and reduce the training time, we applied a transfer learning and used cDNN already trained with the ImageNet set with 1.2 million images (50). We used pre-trained cDNN ResNet50 (51) as it was already utilized for recognition and classification for images of breast cells (52) and pluripotent stem cells (53). In addition, we compared cDNN ResNet50 with cDNN ResNet18 with 18 deep layers (51) to examine if fewer number of layers, lower computational complexity and memory usage (54) would be sufficient and save time for cDNN retraining. We used cDNNs ResNet50 and ResNet18 with the following parameters and options: 1. Optimizer (optimization algorithm used for faster training and better performance of neural networks): SGDM (stochastic gradient descent with momentum; (55)) or ADAM (adaptive moment estimation; (56)); 2. Number of epochs = 8; 3. Learning rate: 0.0001; 4. Batch size: 10.
For both image sets, we captured images of GFP+/GFAP+ astrocytes using objective EC Plan-Neofluar 40x/1.30 Oil DIC and zoom = 1× as mentioned above. Then, we excluded GFP+ channel and extracted 2D images from GFAP+ channel of each confocal image (1024 × 1024, 24 bit), rotated each 2D image (‘90°’, ‘180°’, ‘270°’) to avoid orientation-dependent effects (57) and increase number of images in sets. Afterwards, we added those images to the original Control or Disc1-KD set. Each set was randomly divided on two groups—‘Training’ with 70% of images and ‘Validation’ with 30% of images (n = 716 per set, with 501 and 215 images being for training and validation, respectively).
We also used the transfer learning with pre-trained Resnet50 and ResNet18 to compare microstructure of the GFP+ astrocyte surface. For this experiment, we took confocal images with 1–2 astrocytes per image using alpha Plan-Apochromat 100×/1.46 Oil DIC M27 Elyra objective and zoom = 1.5×. For more accurate 3D reconstruction and analysis, frame size was set to 1024 × 1024 pixels, bit depths to 8-bit, averaging to 2×, and z-step size to 0.34 μm (voxel dimension 55 nm × 55 nm × 340 nm, 50–65 optical sections per image). Then, we extracted 2D images (1024 × 1024, 24 bit, n = 4840) from confocal stacks. The rest of image preparation and parameters/options for ResNet50 and ResNet18 training was the same as above. After retraining with the ‘Training’ group, the cDNN model classified images using ‘Validation’ group and calculated the ‘Accuracy of prediction’ (accuracy of prediction = number of correct predictions/total number of predictions). The cDNN training was performed utilizing GPU (NVIDIA GeForce GTX 1070 8GB) on system with processor Intel(R) Core(TM) i7-6850K CPU @ 3.60 GHz (6 Cores/12 Threads) and 64GB RAM memory.
Imaris-based approach
To validate the ability of the ResNet50 and ResNet18 to distinguish images of astrocytes between control and Disc1-KD groups, we used the commercially available software Imaris (Bitplane AG, Zurich, Switzerland). We reconstructed astrocyte arborization (‘Filament’ algorithm for 3D reconstruction of dendrites arborization) in the same GFAP+ confocal images used for deep learning. Visualization and measuring of GFAP+ processes in astrocytes were conducted as in (58) with minor improvements. To prevent influence of background on results of reconstruction and avoid manual correction, we created 3D mask with ‘GFAP’ channel (‘Smoothing with surface details’ = 0.1 μm, ‘Automatic’ threshold, ‘Number of Voxels’ above 100), adjusted the intensity of GFAP+ signal inside that mask to ‘255’ and generate new channel ‘GFAP255’. We then reconstructed astrocyte processes with ‘GFAP255’ channel (‘Dendrite starting Point Diameter’ = 10.0 μm; ‘Dendrite Seed Point Diameter’ = 0.5 μm; ‘Dendrite Starting Point Threshold’ = 50.0, ‘Dendrite Seed Point Threshold’ = 210.000) and automatically measured the basic parameters of these 3D processes. In addition, we assessed the surface area/volume ratio of the processes. We used ‘Dendrite Seed Point Diameter’ for GFAP+ processes of 0.5 μm because it was within the resolution limit provided by our microscope settings. This algorithm demonstrated good accuracy for reconstruction of three-dimensional GFAP+ processes.
Using the same confocal images of GFP+ astrocytes acquired with alpha Plan-Apochromat 100×/1.46 Oil DIC M27 Elyra objective and zoom = 1.5×, we examined astrocytic surface microstructure by creating a 3D surface with ‘GFP’ channel as previously described in (59,60) with a minor modification, including smoothing with the detail level of the surface area at 0.1 μm and automatic threshold. The ratio between the surface area and the volume of single astrocytes in a single confocal image and the number of vertices in the cell surface of astrocytes for Control and Disc1-KD samples were calculated and analyzed.
Individual astrocyte analysis of the glutamate transporters
As abnormal mitochondrial functioning could affect levels and trafficking of glutamate transporters in astrocytes (61,62), we simultaneously assessed the effects of Disc1-KD on expression of the glutamate transporters, glutamate transporter 1 (GLT1) or GLAST. Levels of GLT1 and GLAST were assessed within the individual zones of transduced astrocytes using the Imaris software combined with Matlab (MathWorks, MA, USA). Specifically, images of single astrocytes and their individual territories were taken with alpha Plan-Apochromat 100×/1.46 Oil DIC M27 Elyra objective and zoom = 1.5× (the other settings were kept as above to perform a precise 3D reconstruction and analysis).
We created a 3D cell surface of astrocytes as mentioned above and used this surface to mask the ‘GLT1in’ or ‘GLASTin’ channels and create a new channel of only GLT1+- or GLAST+-immunoreactivity inside the astrocyte surface. We then analyzed individual ‘GLT1in’ or ‘GLASTin’ channels to create GLT1+ or GLAST+ spots inside GFP+ astrocytes. We used the estimated diameter smoothing with 0.5 μm, quality 10 with local contrasting and region growing manual threshold =10 in the surface area detail level at 0.1 μm and the local contrast with diameter of largest sphere 0.5 μm and threshold 20. We have chosen ‘the estimated diameter of the spots’ of 0.5 μm because it was within resolution limit of microscope settings. The total number of GLT1+ or GLAST+ spots inside the astrocyte was divided by the total volume of the astrocyte to calculate the density (N/μm3), while size (μm3) and fluorescent intensity (optical units) of spots in the individual astrocyte were measured automatically. As Disc1-KD could impair internalization/externalization of glutamate transporters in astrocytes, we measured the distance between GLT1+ and GLAST+ spots and the cell surface using the ‘Distance Transformation’ option and creating a new channel with the value for each voxel equaling the shortest distance from a given spot to the astrocyte cell surface.
To evaluate a possible impairment in GLAST trafficking, we assessed the density and size of GLAST+ spots in the astrocyte soma using the same images. To select GLAST+ spots in the astrocyte soma, we created a 3D mask of GFP-labeled astrocyte soma with the estimated diameter smoothing 0.5 μm, absolute intensity threshold 220 and number of voxels >10 000. We also created a new channel, GLASTsoma, and analyzed the size (μm3) and spatial density (n/μm3) of GLAST+ spots in the astrocyte soma using the same algorithm and calculations as above.
Individual astrocyte analysis of the mitochondrial markers
Given the role of DISC1 in mitochondrial functions (16–21,23), we evaluated the effects of Disc1-KD on expression of the major mitochondrial markers, PDH, TIM23, cytochrome c oxidase subunit I (MTCO1) and ATP synthase F1 subunit alpha (ATP5A). We took confocal images with alpha Plan-Apochromat 100×/1.46 Oil DIC M27 Elyra objective and zoom = 1.5× (other settings as mentioned above). Then we created the GFP+ surface of transduced astrocytes as mentioned above and used this surface as 3D mask to select immunoreactivity for each mitochondrial marker inside. Using an individual channel, we created mitomarker+ spots inside GFP+ astrocyte surface with the estimated diameter 0.5 μm for spot detection, the background subtraction and the quality above 8 with the local contrasting and region growing manual threshold =10. We have chosen ‘estimated diameter of the spots’ 0.5 μm because it was within resolution limit of microscope settings. We then analyzed the size, mean intensity and spatial density (number of spots divided on volume of GFP+ surface, N/μm3).
A previous study has demonstrated that GLT1+ spots overlap with mitochondria along astrocytic processes, suggesting that mitochondria provide energy support for glutamate transport (31). Thus, we examined if Disc1-KD in astrocytes would alter the distance between GLAST+ spots and each mitochondrial marker. To measure the distance, we used the option ‘Distance Transformation’, and created a new channel with the value for each voxel equaling the shortest distance from GLAST+ spot to mitochondrial marker.
Analysis of the synaptic markers within the zones of transduced astrocytes
To assess the effects of Disc1-KD on the spatial density of the pre- and postsynaptic markers of excitatory and inhibitory synapses within the zones of transduced astrocytes we acquired confocal images with alpha Plan-Apochromat 100×/1.46 Oil DIC M27 Elyra objective and zoom = 1.5× with soma of astrocyte located in the center of image and processes that fits the whole image (frame size 1024 × 1024 pixels; voxel dimension 55 nm × 55 nm × 340 nm, 50–65 optical sections per image). GFP+ 3D cell surface was created using the same protocol as when we assessed GLT-1 and GLAST spots inside astrocyte (see above). We used PSD95+ or VGLUT1+ immunoreactivity to evaluate glutamatergic synaptic ‘puncta’ with the smoothing with surface area detail level at 0.1 μm and local contrast with diameter of largest sphere 0.5 μm and threshold 20 for both pre- and post-synaptic markers. For counting synapses, we selected pre- and postsynaptic markers localized within 0–0.5 μm from each other as previously described (34,59,63). Then using the ‘PSD95’ channel, we performed distance transformation and filtered all ‘VGLUT1’ to include only those that are localized with ‘PSD95’ within 0–0.5 μm from each other (34). We have selected ‘the estimated diameter of the spots’ of 0.5 μm and the distance between markers of 0–0.5 μm because it was within resolution limit of the microscope settings. The total number of PSD95+/VGLUT1+ pairs of puncta within zone of an astrocyte (density, N/μm3), and the size of PSD95+ and VGLUT1+ spots (μm3) were calculated. In addition, we determined the distance between synaptic puncta and the astrocyte surface using distance transformation. To evaluate the effects of Disc1-KD on GABA-ergic synapses, we used the same procedure and settings and evaluated density, the puncta size and the distance to astrocyte surface for VGAT+/Gephyrin+ pairs of puncta.
Expression of the markers of apoptosis and/or cell death
To elucidate possible effects of Disc1-KD on neuronal apoptosis in the transduced brain area, we measured expression of the p17 subunit of active Caspase-3 (Casp3+): (i) inside GABAergic parvalbumin neurons (PV+), or (ii) outside PV+ neurons and outside GFP-labeled transduced astrocytes. To assess expression of Casp3 in different brain cells, we created two separate 3D surfaces using the ‘PV’ or ‘GFP’ channel (algorithm was similar with those for glutamate transporters) in confocal images taken with EC Plan-Neofluar 40×/1.30 Oil DIC and zoom = 1×. Frame size was set to 1024 × 1024 pixels, bit depths to 8-bit, averaging to 2× and z-step size to 0.439 μm (voxel dimension 208 nm × 208 nm × 439 nm, 25 optical sections per image). For the ‘PV’ channel we used smoothing with surface area detail level at 0.1 μm, threshold 90 and the number of voxels for selected immunoreactivity was above 10 (to exclude possible background of staining and imaging). For the ‘GFP’ channel we applied the same parameters as mentioned above. With these two surfaces, we extracted ‘Casp3-PV’ and ‘Casp3 out PV+ out GFP+’ channels with Casp3+ immunoreactivity inside PV+ neurons or outside PV+ neurons and GFP+ astrocytes. Next, we analyzed individual ‘Casp3+ in PV+’ and ‘Casp3 out PV+ out GFP+’ channels to measure Casp3+ spots inside the two channels with the smoothing 0.1 μm and threshold 40. The size of Casp3+ spots, their intensity and spatial density (total number of Casp3+ spots dividing by volume of PV+ neurons or ‘out GFP+ out PV+’ volume, V = Vall − VGFP − VPV) were calculated.
TUNEL assay
TUNEL assay (Cat # C10618, Thermo Fisher Scientific) was performed according to the manufacturer’s instructions. As a positive control, adjacent brain sections were treated with one unit of DNase I (Cat # 108068–015) diluted in 1× DNase I Reaction Buffer (20 mM Tris-HCl, pH 8.4, 2 mM MgCl2, 50 mM KCl) for 30 min at RT. Images of brain sections were taken using Keyence BZ-X710 microscope with a 20× objective (frame size 960 × 720 pixels, bit depths 16-bit). To measure signal intensity, we created a 2D mask of pyramidal layers of hippocampus in the affected area and determined the mean fluorescence of ‘TUNEL’ positive signal in all sections using ImageJ-FIJI software (https://imagej.net/Fiji).
Statistical Analyses
Normality and equal variance tests were performed before the following statistical tests were applied to data. Data are expressed as means ± standard error of the mean (SEM). No statistical methods were used to estimate sample sizes. We determined animal numbers by considering our previous studies on the same mouse model and behavioral tests (25). Mice were randomly allocated to different treatment groups. No animals or data points were excluded from analyses. Although data collection and analysis were not performed with blinding to the experimental conditions, the same analysis and comparison criteria were applied to all mice.
Results are expressed as mean ± SEM or as normalized data (% of control values) throughout. The behavioral data were analyzed using analysis of variance or Student’s t-test when appropriate. Morphological data were analyzed by two-tailed Student’s t-test. Bonferroni correction was applied to multiple comparisons; the significance level was set to P < 0.05.
All raw data are available to the scientific community upon reasonable request.
Supplementary Material
Acknowledgments
The authors are grateful to Drs Daisuke Fukudome and Sun Hong Kim for their expert technical assistance and Drs Michele Pucak and James Show (Imaris, MA, USA) for their expert advice and suggestions for confocal microscopy and individual cell analysis.
Conflict of Interest statement. None declared.
Contributor Information
Alexey V Shevelkin, Departments of Psychiatry and Behavioral Sciences.
Chantelle E Terrillion, Departments of Psychiatry and Behavioral Sciences.
Yuto Hasegawa, Departments of Psychiatry and Behavioral Sciences.
Olga A Mychko, Departments of Psychiatry and Behavioral Sciences.
Yan Jouroukhin, Departments of Psychiatry and Behavioral Sciences.
Akira Sawa, Departments of Psychiatry and Behavioral Sciences; Solomon H. Snyder Department of Neuroscience; Department of Biomedical Engineering; Department of Genetic Medicine, Johns Hopkins University School of Medicine; Department of Mental Health, Johns Hopkins University Bloomberg School of Public Health, Baltimore, MD 21287, USA.
Atsushi Kamiya, Departments of Psychiatry and Behavioral Sciences; Solomon H. Snyder Department of Neuroscience.
Mikhail V Pletnikov, Departments of Psychiatry and Behavioral Sciences; Solomon H. Snyder Department of Neuroscience.
Funding
The study was supported by the MH-083728, R01DA041208, R01AG065168 and MH-094268 grants. The work at the JHU Neuroscience Multiphoton Imaging Core was supported by the NS050274 grant.
Financial Disclosures
The authors declare no biomedical financial interests or potential conflicts of interest.
References
- 1. Chung, W.S., Welsh, C.A., Barres, B.A. and Stevens, B. (2015) Do glia drive synaptic and cognitive impairment in disease? Nat. Neurosci., 18, 1539–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Molofsky, A.V., Krencik, R., Ullian, E.M., Tsai, H.H., Deneen, B., Richardson, W.D., Barres, B.A. and Rowitch, D.H. (2012). Astrocytes and disease: a neurodevelopmental perspective. Genes Dev., 26, 891–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sullivan, P.F., Daly, M.J. and O'Donovan, M. (2012) Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat. Rev. Genet., 13, 537–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kondziella, D., Brenner, E., Eyjolfsson, E.M. and Sonnewald, U. (2007) How do glial-neuronal interactions fit into current neurotransmitter hypotheses of schizophrenia? Neurochem. Int., 50, 291–301. [DOI] [PubMed] [Google Scholar]
- 5. Schnieder, T.P. and Dwork, A.J. (2011) Searching for neuropathology: gliosis in schizophrenia. Biol. Psychiatry, 69, 134–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Millar, J.K., Christie, S., Anderson, S., Lawson, D., Hsiao-Wei Loh, D., Devon, R.S., Arveiler, B., Muir, W.J., Blackwood, D.H. and Porteous, D.J. (2001) Genomic structure and localisation within a linkage hotspot of disrupted in schizophrenia 1, a gene disrupted by a translocation segregating with schizophrenia. Mol. Psychiatry, 6, 173–178. [DOI] [PubMed] [Google Scholar]
- 7. Schizophrenia Working Group of the Psychiatric Genomics, C (2014) Biological insights from 108 schizophrenia-associated genetic loci. Nature, 511, 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Facal, F. and Costas, J. (2019) Evidence of association of the DISC1 interactome gene set with schizophrenia from GWAS. Prog. Neuro-Psychopharmacol. Biol. Psychiatry, 95, 109729. [DOI] [PubMed] [Google Scholar]
- 9. Ryan, N.M., Lihm, J., Kramer, M., McCarthy, S., Morris, S.W., Arnau-Soler, A., Davies, G., Duff, B., Ghiban, E., Hayward, C. et al. (2018) DNA sequence-level analyses reveal potential phenotypic modifiers in a large family with psychiatric disorders. Mol. Psychiatry, 23, 2254–2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Malavasi, E.L.V., Economides, K.D., Grunewald, E., Makedonopoulou, P., Gautier, P., Mackie, S., Murphy, L.C., Murdoch, H., Crummie, D., Ogawa, F. et al. (2018) DISC1 regulates N-methyl-D-aspartate receptor dynamics: abnormalities induced by a Disc1 mutation modelling a translocation linked to major mental illness. Transl. Psychiatry, 8, 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Niwa, M., Cash-Padgett, T., Kubo, K.I., Saito, A., Ishii, K., Sumitomo, A., Taniguchi, Y., Ishizuka, K., Jaaro-Peled, H., Tomoda, T. et al. (2016) DISC1 a key molecular lead in psychiatry and neurodevelopment: no-more disrupted-in-schizophrenia 1. Mol. Psychiatry, 21, 1488–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Farrell, M.S., Werge, T., Sklar, P., Owen, M.J., Ophoff, R.A., O'Donovan, M.C., Corvin, A., Cichon, S. and Sullivan, P.F. (2015) Evaluating historical candidate genes for schizophrenia. Mol. Psychiatry, 20, 555–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cuthbert, B.N. and Insel, T.R. (2013) Toward the future of psychiatric diagnosis: the seven pillars of RDoC. BMC Med., 11, –126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sawa, A. (2019) DISC1 and its protein Interactomes for mental function. Biol. Psychiatry, 85, 283–284. [DOI] [PubMed] [Google Scholar]
- 15. Ji, B., Higa, K.K., Kim, M., Zhou, L., Young, J.W., Geyer, M.A. and Zhou, X. (2014) Inhibition of protein translation by the DISC1-Boymaw fusion gene from a Scottish family with major psychiatric disorders. Hum. Mol. Genet., 23, 5683–5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Millar, J.K., James, R., Christie, S. and Porteous, D.J. (2005) Disrupted in schizophrenia 1 (DISC1): subcellular targeting and induction of ring mitochondria. Mol. Cell. Neurosci., 30, 477–484. [DOI] [PubMed] [Google Scholar]
- 17. James, R., Adams, R.R., Christie, S., Buchanan, S.R., Porteous, D.J. and Millar, J.K. (2004) Disrupted in schizophrenia 1 (DISC1) is a multicompartmentalized protein that predominantly localizes to mitochondria. Mol. Cell. Neurosci., 26, 112–122. [DOI] [PubMed] [Google Scholar]
- 18. Norkett, R., Modi, S., Birsa, N., Atkin, T.A., Ivankovic, D., Pathania, M., Trossbach, S.V., Korth, C., Hirst, W.D. and Kittler, J.T. (2016) DISC1-dependent regulation of mitochondrial dynamics controls the morphogenesis of complex neuronal dendrites. J. Biol. Chem., 291, 613–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ogawa, F., Malavasi, E.L., Crummie, D.K., Eykelenboom, J.E., Soares, D.C., Mackie, S., Porteous, D.J. and Millar, J.K. (2014) DISC1 complexes with TRAK1 and Miro1 to modulate anterograde axonal mitochondrial trafficking. Hum. Mol. Genet., 23, 906–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pinero-Martos, E., Ortega-Vila, B., Pol-Fuster, J., Cisneros-Barroso, E., Ruiz-Guerra, L., Medina-Dols, A., Heine-Suner, D., Llado, J., Olmos, G. and Vives-Bauza, C. (2016) Disrupted in schizophrenia 1 (DISC1) is a constituent of the mammalian mitochondrial contact site and cristae organizing system (MICOS) complex, and is essential for oxidative phosphorylation. Hum. Mol. Genet., 25, 4157–4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Eykelenboom, J.E., Briggs, G.J., Bradshaw, N.J., Soares, D.C., Ogawa, F., Christie, S., Malavasi, E.L., Makedonopoulou, P., Mackie, S., Malloy, M.P. et al. (2012) A t(1;11) translocation linked to schizophrenia and affective disorders gives rise to aberrant chimeric DISC1 transcripts that encode structurally altered, deleterious mitochondrial proteins. Hum. Mol. Genet., 21, 3374–3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Park, S.J., Lee, S.B., Suh, Y., Kim, S.J., Lee, N., Hong, J.H., Park, C., Woo, Y., Ishizuka, K., Kim, J.H. et al. (2017) DISC1 modulates neuronal stress responses by gate-keeping ER-mitochondria Ca(2+) transfer through the MAM. Cell Rep., 21, 2748–2759. [DOI] [PubMed] [Google Scholar]
- 23. Park, C., Lee, S.A., Hong, J.H., Suh, Y., Park, S.J., Suh, B.K., Woo, Y., Choi, J., Huh, J.W., Kim, Y.M. et al. (2016) Disrupted-in-schizophrenia 1 (DISC1) and Syntaphilin collaborate to modulate axonal mitochondrial anchoring. Mol. Brain, 9, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Seshadri, S., Kamiya, A., Yokota, Y., Prikulis, I., Kano, S., Hayashi-Takagi, A., Stanco, A., Eom, T.Y., Rao, S., Ishizuka, K. et al. (2010) Disrupted-in-Schizophrenia-1 expression is regulated by beta-site amyloid precursor protein cleaving enzyme-1-neuregulin cascade. Proc. Natl. Acad. Sci. U. S. A., 107, 5622–5627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Terrillion, C.E., Abazyan, B., Yang, Z., Crawford, J., Shevelkin, A.V., Jouroukhin, Y., Yoo, K.H., Cho, C.H., Roychaudhuri, R., Snyder, S.H. et al. (2017) DISC1 in astrocytes influences adult neurogenesis and hippocampus-dependent behaviors in mice. Neuropsychopharmacology, 42, 2242–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jouroukhin, Y., Zhu, X., Shevelkin, A.V., Hasegawa, Y., Abazyan, B., Saito, A., Pevsner, J., Kamiya, A. and Pletnikov, M.V. (2019) Adolescent delta(9)-tetrahydrocannabinol exposure and astrocyte-specific genetic vulnerability converge on nuclear factor-kappaB-cyclooxygenase-2 signaling to impair memory in adulthood. Biol. Psychiatry, 85, 891–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sheikhzadeh, F., Ward, R.K., van Niekerk, D. and Guillaud, M. (2018) Automatic labeling of molecular biomarkers of immunohistochemistry images using fully convolutional networks. PLoS One, 13, e0190783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Md Zahangir Alom, T.A., Tarek M. Taha, Vijayan K. Asari, TJ Bowen, Dave Billiter, Simon Arkell. (2019) Advanced deep convolutional neural network approaches for digital pathology image analysis: a comprehensive evaluation with different use cases. arXiv, https://arxiv.org/abs/1904.09075.
- 29. Kanai, Y., Smith, C.P. and Hediger, M.A. (1993) A new family of neurotransmitter transporters: the high-affinity glutamate transporters. FASEB J., 7, 1450–1459. [DOI] [PubMed] [Google Scholar]
- 30. Danbolt, N.C. (1994) The high affinity uptake system for excitatory amino acids in the brain. Prog. Neurobiol., 44, 377–396. [DOI] [PubMed] [Google Scholar]
- 31. Jackson, J.G., O'Donnell, J.C., Takano, H., Coulter, D.A. and Robinson, M.B. (2014) Neuronal activity and glutamate uptake decrease mitochondrial mobility in astrocytes and position mitochondria near glutamate transporters. J. Neurosci., 34, 1613–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jackson, J.G. and Robinson, M.B. (2018) Regulation of mitochondrial dynamics in astrocytes: mechanisms, consequences, and unknowns. Glia, 66, 1213–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Park, Y.U., Jeong, J., Lee, H., Mun, J.Y., Kim, J.H., Lee, J.S., Nguyen, M.D., Han, S.S., Suh, P.G. and Park, S.K. (2010) Disrupted-in-schizophrenia 1 (DISC1) plays essential roles in mitochondria in collaboration with Mitofilin. Proc. Natl. Acad. Sci. U. S. A., 107, 17785–17790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fogarty, M.J., Hammond, L.A., Kanjhan, R., Bellingham, M.C. and Noakes, P.G. (2013) A method for the three-dimensional reconstruction of Neurobiotin-filled neurons and the location of their synaptic inputs. Front. Neural Circuits, 7, 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ikonomidou, C. and Turski, L. (1995) Excitotoxicity and neurodegenerative diseases. Curr. Opin. Neurol., 8, 487–497. [DOI] [PubMed] [Google Scholar]
- 36. Nicholls, D.G. (2004) Mitochondrial dysfunction and glutamate excitotoxicity studied in pri mary neuronal cultures. Curr. Mol. Med., 4, 149–177. [DOI] [PubMed] [Google Scholar]
- 37. Wang, Y. and Qin, Z.H. (2010) Molecular and cellular mechanisms of excitotoxic neuronal death. Apoptosis, 15, 1382–1402. [DOI] [PubMed] [Google Scholar]
- 38. Smyth, P.G., Berman, S.A. and Bursztajn, S. (2002) Markers of apoptosis: methods for elucidating the mechanism of apoptotic cell death from the nervous system. BioTechniques, 32, 648. [DOI] [PubMed] [Google Scholar]
- 39. Jouroukhin, Y., Kageyama, Y., Misheneva, V., Shevelkin, A., Andrabi, S., Prandovszky, E., Yolken, R.H., Dawson, V.L., Dawson, T.M., Aja, S. et al. (2018) DISC1 regulates lactate metabolism in astrocytes: implications for psychiatric disorders. Transl. Psychiatry, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Giorgi, C., Marchi, S. and Pinton, P. (2018) The machineries, regulation and cellular functions of mitochondrial calcium. Nat. Rev. Mol. Cell Biol., 19, 713–730. [DOI] [PubMed] [Google Scholar]
- 41.Spinelli, J.B., Haigis, M.C. (2018) The multifaceted contributions of mitochondria to cellular metabolism. Nat. Cell Biol., 20, 745–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ugbode, C.I., Hirst, W.D. and Rattray, M. (2014) Neuronal influences are necessary to produce mitochondrial co-localization with glutamate transporters in astrocytes. J. Neurochem., 130, 668–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rothstein, J.D. (1995) Excitotoxicity and neurodegeneration in amyotrophic lateral sclerosis. Clin. Neurosci., 3, 348–359. [PubMed] [Google Scholar]
- 44. Yi, J.H. and Hazell, A.S. (2006) Excitotoxic mechanisms and the role of astrocytic glutamate transporters in traumatic brain injury. Neurochem. Int., 48, 394–403. [DOI] [PubMed] [Google Scholar]
- 45. Pajarillo, E., Rizor, A., Lee, J., Aschner, M. and Lee, E. (2019) The role of astrocytic glutamate transporters GLT-1 and GLAST in neurological disorders: potential targets for neurotherapeutics. Neuropharmacology, 161, 107559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gibon, J., Unsain, N., Gamache, K., Thomas, R.A., De Leon, A., Johnstone, A., Nader, K., Seguela, P. and Barker, P.A. (2016) The X-linked inhibitor of apoptosis regulates long-term depression and learning rate. FASEB J., 30, 3083–3090. [DOI] [PubMed] [Google Scholar]
- 47. Pletnikov, M.V., Ayhan, Y., Nikolskaia, O., Xu, Y., Ovanesov, M.V., Huang, H., Mori, S., Moran, T.H. and Ross, C.A. (2008) Inducible expression of mutant human DISC1 in mice is associated with brain and behavioral abnormalities reminiscent of schizophrenia. Mol. Psychiatry, 13, 173–186, 115. [DOI] [PubMed] [Google Scholar]
- 48. Hoffman, G.E., Murphy, K.J. and Sita, L.V. (2016) The importance of titrating antibodies for Immunocytochemical methods. Curr. Protoc. Neurosci, 76, 2 12 11-12 12 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Grosche, A., Grosche, J., Tackenberg, M., Scheller, D., Gerstner, G., Gumprecht, A., Pannicke, T., Hirrlinger, P.G., Wilhelmsson, U., Huttmann, K. et al. (2013) Versatile and simple approach to determine astrocyte territories in mouse neocortex and hippocampus. PLoS One, 8, e69143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Russakovsky, O., Deng, J., Su, H., Krause, J., Satheesh, S., Ma, S., Huang, Z.H., Karpathy, A., Khosla, A., Bernstein, M. et al. (2015) ImageNet large scale visual recognition challenge. Int. J. Comput. Vis., 115, 211–252. [Google Scholar]
- 51. He, K.M., Zhang, X.Y., Ren, S.Q. and Sun, J. (2016) Deep residual learning for image recognition. arXiv, https://arxiv.org/abs/1512.03385.
- 52. Kensert, A., Harrison, P.J. and Spjuth, O. (2019) Transfer learning with deep convolutional neural networks for classifying cellular morphological changes. SLAS Discov., 24, 466–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Waisman, A., La Greca, A., Mobbs, A.M., Scarafia, M.A., Velazque, N.L.S., Neiman, G., Moro, L.N., Luzzani, C., Sevlever, G.E., Guberman, A.S. et al. (2019) Deep learning neural networks highly predict very early onset of pluripotent stem cell differentiation. Stem Cell Rep., 12, 845–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bianco, S., Cadene, R., Celona, L. and Napoletano, P. (2018) Benchmark analysis of representative deep neural network architectures. arXiv, https://arxiv.org/abs/1810.00736.
- 55. Qian, N. (1999) On the momentum term in gradient descent learning algorithms. Neural Netw., 12, 145–151. [DOI] [PubMed] [Google Scholar]
- 56. Kingma D.P. and Ba J. (2014) Adam: a method for stochastic optimization. arXiv, https://arxiv.org/abs/1412.6980.
- 57. Lanjakornsiripan, D., Pior, B.J., Kawaguchi, D., Furutachi, S., Tahara, T., Katsuyama, Y., Suzuki, Y., Fukazawa, Y. and Gotoh, Y. (2018) Layer-specific morphological and molecular differences in neocortical astrocytes and their dependence on neuronal layers. Nat. Commun., 9, 1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Siemsen, B.M., Reichel, C.M., Leong, K.C., Garcia-Keller, C., Gipson, C.D., Spencer, S., McFaddin, J.A., Hooker, K.N., Kalivas, P.W. and Scofield, M.D. (2019) Effects of methamphetamine self-administration and extinction on astrocyte structure and function in the nucleus accumbens core. Neuroscience, 406, 528–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Scofield, M.D., Li, H., Siemsen, B.M., Healey, K.L., Tran, P.K., Woronoff, N., Boger, H.A., Kalivas, P.W. and Reissner, K.J. (2016) Cocaine self-administration and extinction leads to reduced glial fibrillary acidic protein expression and morphometric features of astrocytes in the nucleus accumbens core. Biol. Psychiatry, 80, 207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Octeau, J.C., Chai, H., Jiang, R., Bonanno, S.L., Martin, K.C. and Khakh, B.S. (2018) An optical neuron-astrocyte proximity assay at synaptic distance scales. Neuron, 98, 49–66 e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Robinson, M.B. and Jackson, J.G. (2016) Astroglial glutamate transporters coordinate excitatory signaling and brain energetics. Neurochem. Int., 98, 56–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Robinson, M.B., Lee, M.L. and DaSilva, S. (2020) Glutamate transporters and mitochondria: signaling, co-compartmentalization, functional coupling, and future directions. Neurochem. Res., 45, 526–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Testen, A., Sepulveda-Orengo, M.T., Gaines, C.H. and Reissner, K.J. (2018) Region-specific reductions in morphometric properties and synaptic colocalization of astrocytes following cocaine self-administration and extinction. Front. Cell. Neurosci., 12, 246. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


