Abstract
Background
Precise detection of anaplastic lymphoma kinase (ALK) rearrangement guides the application of ALK-targeted tyrosine kinase inhibitors (ALK-TKIs) in patients with non-small-cell lung cancer (NSCLC). Next-generation sequencing (NGS) has been widely used in clinics, but DNA-based NGS used to detect fusion genes has delivered false-negative results. However, fusion genes can be successfully detected at the transcription level and with higher sensitivity using RNA-based reverse transcription polymerase chain reaction (RT-PCR).
Objective
This study compared the performance of RT-PCR and NGS in the detection of echinoderm microtubule-associated protein-like 4 (EML4)-ALK fusion in Chinese patients with NSCLC.
Methods
Formalin-fixed paraffin-embedded tissues from 153 patients who were pathologically diagnosed as having NSCLC were collected from November 2017 to October 2019. Both DNA/RNA-based NGS and RNA-based RT-PCR were used to detect EML4-ALK fusion. For samples with discordant ALK status results, fluorescence in situ hybridization (FISH) or Sanger sequencing was used to further confirm the ALK status.
Results
In total, 124 samples were successfully analyzed using both NGS and RT-PCR. For 118 samples, results were consistent between NGS and RT-PCR, with 25 reported as ALK fusion positive and 93 as ALK fusion negative, achieving a concordance rate of 95.16%. Among the six samples with disconcordant results, five were positive using RT-PCR but negative using NGS, and one was positive using NGS but negative using RT-PCR. Four of six cases with disconcordant results (three RT-PCR positive and one NGS positive) were successfully validated using either FISH or Sanger sequencing.
Conclusions
Compared with NGS, RT-PCR appears to be a reliable method of detecting EML4-ALK fusion in patients with NSCLC.
Supplementary Information
The online version contains supplementary material available at 10.1007/s40291-021-00532-8.
Key Points
| At the transcriptional level, reverse transcription polymerase chain reaction (RT-PCR) displays a reliable capacity to detect anaplastic lymphoma kinase (ALK) fusion in tissue samples from patients with non-small-cell lung cancer (NSCLC). |
| Compared with next-generation sequencing, RT-PCR appears to be a reliable method for the detection of ALK fusion in cases with a low abundance of ALK fusions. |
| Our research suggested that, for patients with newly diagnosed NSCLC, RT-PCR may be a better method for ALK testing because of its accuracy, short turnaround time, and low cost. |
Introduction
The echinoderm microtubule-associated protein-like 4 (EML4)–anaplastic lymphoma kinase (ALK) fusion constitutes a major subset of ALK fusions, whereby 2–7% of non-small-cell lung cancers (NSCLCs) can be directly targeted by ALK tyrosine kinase inhibitors (TKIs) [1–4]. ALK fusions can be identified using various techniques, including fluorescence in situ hybridization (FISH) [5], reverse transcription polymerase chain reaction (RT-PCR) [6], next-generation sequencing (NGS) [6], or immunohistochemistry (IHC) [7]. Lu et al. [8] reported that the incidence of EML4-ALK, as detected with IHC, RT-PCR, and NGS, was 9.51% (170/1787), 11.62% (33/284), and 5.84% (58/994), respectively, in patients with lung adenocarcinoma. Similar results were also observed in a cohort of 200 patients with NSCLC, in which RT-PCR yielded the highest EML4-ALK positivity (12.5%) (results with IHC or FISH analysis were 6.7 and 4.5%, respectively) [6]. Therefore, RT-PCR seems to be a sensitive, reliable, and economical approach to the detection of EML4-ALK [7, 9]. Although RT-PCR was the first published method for the determination of EML4-ALK fusion [4], NGS has gradually become widely available, providing high-throughput molecular analysis and genetic diagnostics, including fusion gene variation [10]. However, direct head-to-head comparison studies of the ability of RT-PCR and NGS to detect EML4-ALK fusions are scarce, especially RNA-based comparisons. Letovanec et al. [11] provided evidence that RT-PCR might be equivalent to RNA-based NGS in detecting ALK fusion. In this study, we investigated the concordance of EML4-ALK fusion status detection between RT-PCR and NGS in a cohort of NSCLC samples.
Materials and Methods
Patient Selection and Study Design
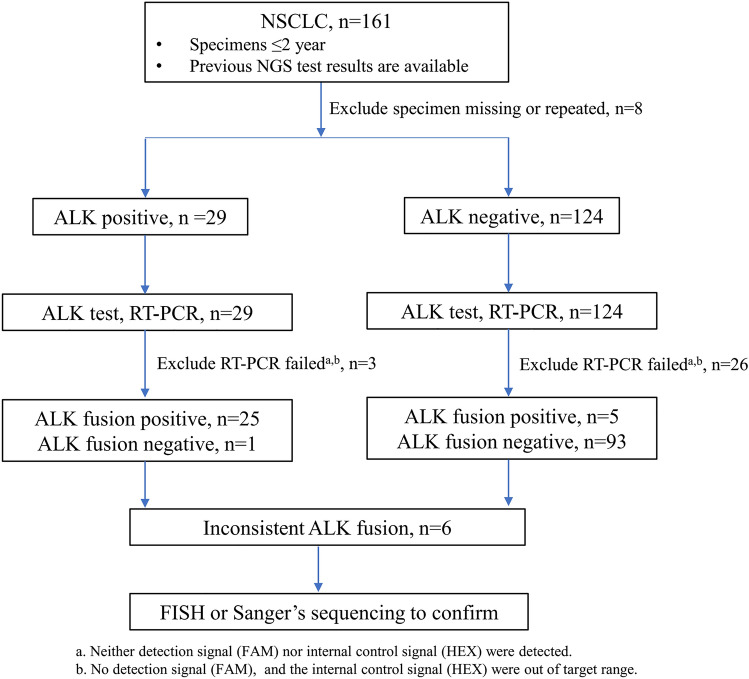
Eligible patients with pathologically confirmed NSCLC from November 2017 to October 2019 were reviewed based on previous ALK results determined using NGS (the DNA-sequencing library preparation used a commercially available 168-gene panel by Burning Rock Biotech [Guangzhou, China]; the DNA/RNA-sequencing library preparation used two commercially available gene panels [13 and 161 genes] per the protocol of Ion AmpliSeq™ Colon & Lung Cancer Research Panel and Oncomine™ Comprehensive Assay v3 [ThermoFisher, Waltham, MA, USA]). NGS method details were prepared as previously described [12, 13]. In total, 153 patients underwent NGS testing, with results showing 29 were ALK fusion positive and 124 were negative. Formalin-fixed paraffin-embedded (FFPE) tissues from 153 patients were accessible. As approved by the institutional review board of the First Affiliated Hospital, Sun Yat-sen University, RT-PCR was used to detect ALK rearrangements in these groups, and a total of 124 samples were successfully tested (of the 124 successful samples detected by RT-PCR, the NGS results of 119 cases were based on RNA library preparation, and the other five cases were based on DNA library preparation; Table 1. Samples with discordant results were validated using FISH or Sanger sequencing. Histology and stage were determined based on the 2015 World Health Organization classification (Table 1). All patients provided written informed consent before enrollment, and participation in this study was covered by this protocol (Fig. 1).
Table 1.
Demographic and clinical characteristics of the patients with samples tested using reverse transcription polymerase chain reaction
| Characteristics | Totals, N = 124 |
|---|---|
| Age, years | |
| Median | 60 |
| Range | 26–87 |
| Sex | |
| Male | 51 (41.3) |
| Female | 73 (58.7) |
| Smoking status | |
| Yes | 37 (29.8) |
| No/unknown | 87 (61.2) |
| Histologic type | |
| Adenocarcinoma | 101 (81.5) |
| Squamous cell carcinoma | 20 (16.1) |
| Other | 3 (2.4) |
| Stage | |
| I–IIIa | 60 (48.38) |
| IIIb–IV | 59 (47.58) |
| Unknown | 5 (4.03) |
| NGS detection | |
| DNA library preparation | 5 (4.03) |
| RNA library preparation | 119 (95.97) |
Data are presented as n (%) unless otherwise indicated
NGS next-generation sequencing
Fig. 1.
Flowchart showing the selection of study participants. ALK anaplastic lymphoma kinase, FISH fluorescence in situ hybridization, NGS next-generation sequencing, NSCLC non-small-cell lung cancer, RT-PCR reverse transcription polymerase chain reaction
Nucleic Acid Preparation
Genomic DNA and RNA was extracted from 4- to 5-μm FFPE sections using AmoyDx DNA, RNA Kits (Amoy Diagnostics Co., Xiamen, China) following the manufacturer’s instructions.
Detecting Anaplastic Lymphoma Kinase (ALK) Fusion Using Reverse Transcription Polymerase Chain Reaction (RT-PCR)
ALK fusion was detected using the AmoyDx EML4-ALK Fusion Gene Diagnostic Kit (Cat no. ADx-FF04; Amoy Diagnostics Co., Xiamen, China) within a range of 26 known transcript variants of EML4-ALK fusion (see Table 1 in the electronic supplementary material [ESM]) following the manufacturer’s instructions).
Fluorescence In Situ Hybridization (FISH) and Sanger Sequencing
A commercially available ALK probe (Vysis LSI ALK Dual Colour, Break Apart Rearrangement Probe; Abbott Molecular Inc., Abbott Park, IL, USA) was used according to the manufacturer’s instructions. Findings were defined as ALK positive and weakly positive, respectively, if > 15% of 50 (minimum) or 100 and 10–15% of 100 analyzed tumor cells displayed split red-green probes signals or isolated red signals. In addition, for cases with insufficient FFPE tissue, the RT-PCR product underwent Sanger sequencing to confirm ALK fusion.
Statistical Analysis
Analysis was performed using SPSS version 22.0 (IBM Corporation, Armonk, NY, USA). The concordance of results generated with RT-PCR and NGS was demonstrated using McNemar–Bowker’s test and kappa (κ) statistics. A relative level of EML4-ALK fusion was determined using the comparative cycle threshold (CT) method quantification (2−ΔCT method) [14, 15]. Student’s T test was applied to examine the statistical significance. A two-tailed P value of 0.05 was considered significant.
Results
Concordance of ALK Fusion Detected by RT-PCR and Next-Generation Sequencing (NGS)
In total, 124 samples were successfully analyzed using both NGS and RT-PCR. RT-PCR detected EML4-ALK fusion in five of the 98 ALK-negative patients with NSCLC (5.1%) defined using RNA-based NGS. Among 26 NGS-defined (five cases for DNA based, 21 cases for RNA based) ALK fusion-positive patients, only one patient with a rare fusion partner detected with RNA-based NGS displayed a negative result using RT-PCR. The consistency of the two methods for EML4-ALK fusion detection was 94.96% (95% confidence interval [CI] 89.89–97.93) (Table 2). No statistically significant difference in efficiency in detecting EML4-ALK fusion between RT-PCR and NGS was observed for RNA-based detection (kappa = 0.8386, P = 0.2188).
Table 2.
Consistency of the two methods for EML4-ALK fusion of RNA-based detection
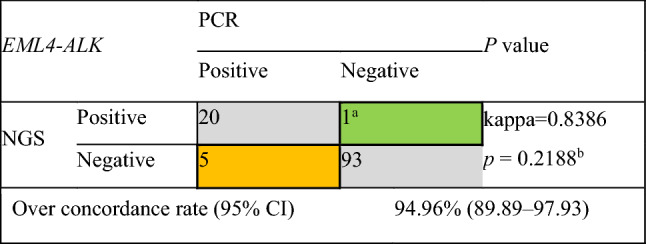
ALK anaplastic lymphoma kinase, ARMS amplification-refractory mutation system, CI confidence interval, EML4 echinoderm microtubule-associated protein-like 4, NGS next-generation sequencing, PCR polymerase chain reaction
aEML4-ALK (E6:A18) fusion that exceeded the detection scope of ARMS-PCR (see Table 1 in the electronic supplementary material)
bMcNemar’s test
Comparison of EML4-ALK Cycle Threshold Value between NGS-Negative and -Positive Samples
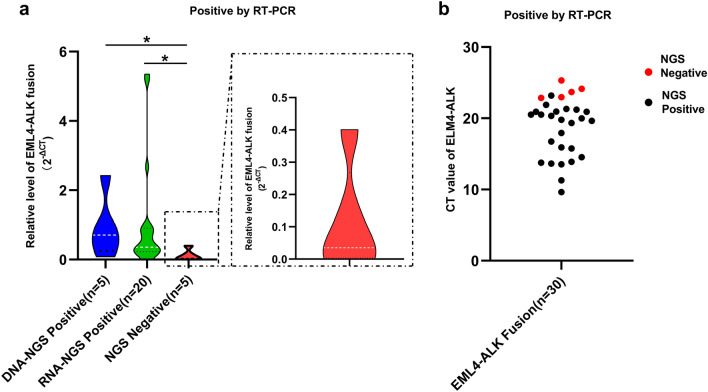
CT values for RT-PCR for all samples were divided into three groups according to their library construction approaches: DNA-NGS positive, RNA-NGS positive, and NGS negative. As shown in Fig. 2a, a relative level of EML4-ALK fusion was presented as 2−ΔCT for three group samples, the 2−ΔCT value of the samples with NGS-negative results was significantly lower than that of the samples with DNA-NGS-positive and RNA-NGS-positive results (P < 0.05). In addition, the five NGS-negative samples detected as positive using RT-PCR all had a low abundance of ALK fusion (Fig. 2b). These results suggested that RT-PCR can detect positive signals even if the level of ALK fusion in tumor tissue is low.
Fig. 2.
Consistency of the two methods of detecting EML4-ALK fusion. a The relative level of EML4-ALK fusion (2−ΔCT value) tested using RT-PCR in DNA-NGS-positive, RNA-NGS-positive, and NGS-negative samples was compared. b The CT value of EML4-ALK in NGS-positive and NGS-negative samples (red dots indicate high CT values). *P <0.05 unless specified otherwise. ALK anaplastic lymphoma kinase, EML4 echinoderm microtubule-associated protein-like 4, CT cycle threshold, NGS next-generation sequencing, RT-PCR reverse transcription polymerase chain reaction
Validation of Inconsistent RT-PCR or NGS-Yielded ALK Fusions by FISH or Sanger Sequencing
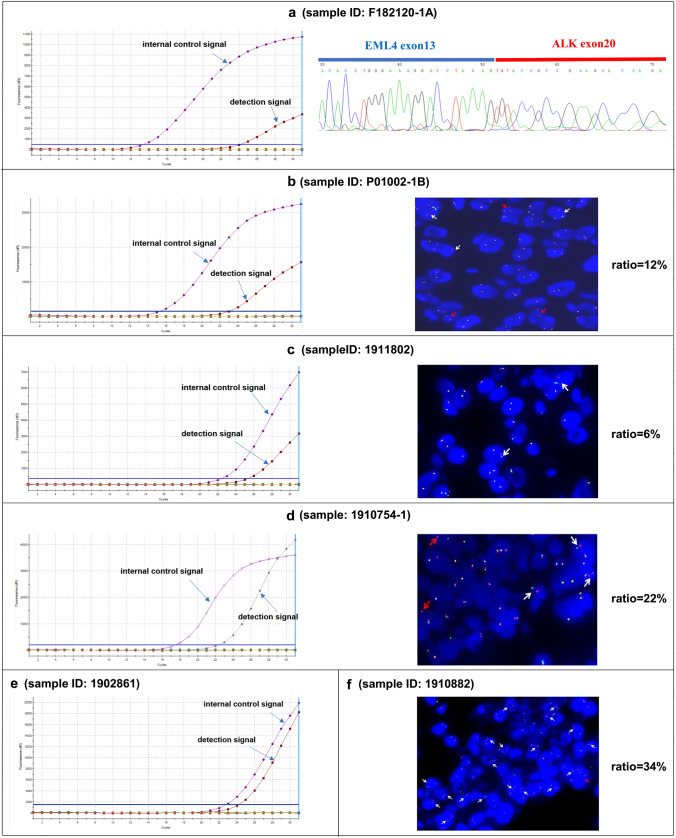
Results for six patients were inconsistent between RT-PCR and NGS, and the NGS results of these samples were all based on RNA library preparation. One PCR-positive/NGS-negative case was not validated because no sample was available. Among four RT-PCR-positive/NGS-negative samples, three samples were successfully validated using either Sanger sequencing (n = 1) or FISH (n = 2); the other sample displayed a 6% FISH-positive rate. Furthermore, one patient with RT-PCR had a rare EML4-ALK subtype, and FISH displayed a positive result (Table 3, Fig. 3).
Table 3.
Comparison of different methods to detect ALK fusion
| Sample ID | Percentage of tumor cells | ALK-NGS | ALK-RT-PCR | FISH/Sanger sequencing |
|---|---|---|---|---|
| f182120-1A | 50 | Negative | Positive | Positive (E13:A20) |
| 1902861 | 2 | Negative | Positive | NA |
| P01002-1B | 25 | Negative | Positive | Weakly positive (12%) |
| 1911802 | 6 | Negative | Positive | Negative (6%) |
| 1910754-1 | 3 | Negative | Positive | Positive (22%) |
| 1910882 | 15 | Positive (E6:A18) | Negative | Positive (34%) |
ALK anaplastic lymphoma kinase, FISH fluorescence in situ hybridization, ID identification, NA not available, NGS next-generation sequencing, RT-PCR reverse transcription polymerase chain reaction
Fig. 3.
Results of ALK status confirmed using fluorescence in situ hybridization (FISH) or Sanger sequencing. ALK status of a specimen F182120-1A tested using reverse transcription polymerase chain reaction (RT-PCR) and Sanger sequencing; b specimen P01002-1B tested using RT-PCR and FISH (magnification × 60); c specimen 1911802 tested using RT-PCR and FISH (magnification × 60); d specimen 1910754-1 tested using RT-PCR and FISH (magnification × 60); e specimen 1902861 tested using RT-PCR; and f specimen 1910882 tested using FISH (magnification × 60). White arrows indicate split red-green signal indicative of EML4-ALK fusion, red arrows indicate isolated red signals indicative of EML4-ALK fusion. ALK anaplastic lymphoma kinase
Discussion
A recent large-scale population-based study by Lu et al. [8] indicated that, when investigating the use of IHC, RT-PCR, and NGS, RT-PCR provided the highest EML4-ALK-positive rate. Although this conclusion was not obtained from the same sample set, it indicated a putative advantage of RT-PCR for the detection of ALK fusion in real-world clinical routine scenarios.
Detecting fusions at the transcriptional level is a better approach than DNA-based NGS. Targeted DNA-based NGS sought to detect all types of oncogenic alterations, including fusions [16]. However, the complexity of genomic rearrangements meant that breakpoints usually occurred at introns that could not be completely covered by DNA-based targeted sequencing [17, 18]. Detecting fusions at the transcriptional level is easier. Benayed et al. [19] reported that samples from patients with lung adenocarcinoma lacking oncogenic driver alterations given by DNA sequencing (MSK-IMPACT) underwent a clinically validated targeted RNA sequencing assay (MSK-Fusion). Among 232 successfully sequenced samples, 33 cases showed actionable in-frame fusions, including ALK fusions.
In this study, most samples (119 of 124) successfully detected by RT-PCR were previously tested using RNA-based NGS. Interestingly, RT-PCR found extra ALK fusion-positive patients among those with negative RNA-based NGS results. Among 30 patients with PCR-defined ALK rearrangements, the 2−ΔCT values of the samples with NGS-negative results were significantly lower than those of the samples with DNA and RNA-NGS-positive results. Most of these extra ALK-positive patients were further successfully validated using Sanger sequencing or FISH. These results suggested that, even at the transcriptional level, RT-PCR is more sensitive than RNA-based NGS and was able to find extra EML4-ALK fusions of low abundance.
For patients with ALK NGS-negative/RT-PCR-positive/FISH-negative results, the following reasons may contribute to the inconsistency. First, the original tissue was no longer available (ID: 1902861); second, the proportion of tumor cells in the FFPE sample was too low (6%, ID: 1902861) to obtain an adequate positive EML4-ALK signal. In clinical practice, it is generally recommended that samples used for molecular detection should have >20% tumor cells [20, 21], which may yield sufficient tumor cells for a reliable result. Additionally, the EML4-ALK subtype (E6:A18) detected by NGS exceeded the primers’ scope of RT-PCR (ID: 1910882). Information about the therapeutic response to ALK-TKIs of these patients was not included for analysis because of a lack of therapeutic information about ALK inhibition, but treatment information for these patients is provided in Table 2 in the ESM. It is reported that the median progression-free survival of osimertinib should be much longer than 3 months, even where EGFR and TP53 mutations co-exist [22]. Thus, for one patient (ID: 1902861), the resistance mechanism should be ALK fusion, which was not detected with NGS.
This study has several limitations. First, this was a retrospective study with selected patients, which could have induced a selection bias. Second, the number of patients included in the study was insufficient. Third, only FFPE samples were used for comparison, resulting in a lack of further validation by other specimen forms, such as cytological samples [23, 24]. Fourth, the two tests were performed with the same specimens at different times, whereas it would have been better to perform the two tests at the same time with the same specimens to prevent technical concerns related to discordant results. Fifth, EML4-ALK subtypes detected with NGS might exceeded the primers’ scope of RT-PCR. Furthermore, therapeutic response was not included for analysis because of the lack of therapeutic information about ALK inhibition (Table 2 in the ESM). Thus, further large-scale and prospective investigations are warranted to resolve some of these limitations.
Conclusions
This study found that, even at the transcriptional level, RT-PCR displays a reliable capacity to detect EML4-ALK fusion in tissue samples from patients with NSCLC, especially in those with low levels of ALK fusion. These results suggested that, for patients with newly diagnosed NSCLC, RT-PCR may be a better method of testing for ALK fusions because of its accuracy, short turnaround time, and low cost.
Supplementary Information
Below is the link to the electronic supplementary material.
Declarations
Funding
This work and the open access were supported by a grant from Guangzhou Science and Technology Planning Program (202002030023) and Natural Science Foundation of Guangdong Province (18zxxt17).
Conflict of interest
Fei Yao and Changbin Zhu are employed by Amoy Diagnostics Co., Ltd. Yukun Kuang, Peihang Xu, Jiyu Wang, Yifan Zheng, Xue Sun, Zimu Li, RunJing Gan, Huixia Li, Yubiao Guo, Zunfu Ke, Kejing Tang have no conflicts of interest that are directly relevant to the content of this article.
Ethics approval
This study was approved by the institutional review board of the First Affiliated Hospital, Sun Yat-sen University (no. Lunshen[2013]C-084).
Consent
All patients provided written informed consent before enrollment; participation in this study was covered by this protocol.
Author contributions
KT, ZK, YK, LB, PX, JW, and YZ designed the study and wrote the paper. XS, ZL, RG, HL, and YG performed the study by collecting cases with clinical data and performing the sample mutation test. FY and CZ analyzed the data and wrote the paper.
Data availability statement
The data that support the findings of this study are available on request from the corresponding author.
Footnotes
Yukun Kuang, Peihang Xu, Jiyu Wang, and Yifan Zheng have contributed equally to this work.
Contributor Information
Zunfu Ke, Email: kezunfu@mail.sysu.edu.cn.
Kejing Tang, Email: tangkj@mail.sysu.edu.cn.
References
- 1.Solomon BJ, Mok T, Kim DW, Wu YL, Nakagawa K, Mekhail T, et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med. 2014;371(23):2167–2177. doi: 10.1056/NEJMoa1408440. [DOI] [PubMed] [Google Scholar]
- 2.Tao H, Shi L, Zhou A, Li H, Gai F, Huang Z, et al. Distribution of EML4-ALK fusion variants and clinical outcomes in patients with resected non-small cell lung cancer. Lung Cancer. 2020;149:154–161. doi: 10.1016/j.lungcan.2020.09.012. [DOI] [PubMed] [Google Scholar]
- 3.Shaw AT, Engelman JA. Ceritinib in ALK-rearranged non-small-cell lung cancer. N Engl J Med. 2014;370(26):2537–2539. doi: 10.1056/NEJMc1404894. [DOI] [PubMed] [Google Scholar]
- 4.Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448(7153):561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 5.Rogers TM, Russell PA, Wright G, et al. Comparison of methods in the detection of ALK and ROS1 rearrangements in lung cancer. J Thorac Oncol. 2015;10(4):611–618. doi: 10.1097/JTO.0000000000000465. [DOI] [PubMed] [Google Scholar]
- 6.Teixidó C, Karachaliou N, Peg V, Gimenez-Capitan A, Rosell R. Concordance of IHC, FISH and RT-PCR for EML4-ALK rearrangements. Transl Lung Cancer Res. 2014;3(2):70–74. doi: 10.3978/j.issn.2218-6751.2014.02.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu YC, Chang IC, Wang CL, et al. Comparison of IHC, FISH and RT-PCR methods for detection of ALK rearrangements in 312 non-small cell lung cancer patients in Taiwan. PLoS ONE. 2013;8:e70839. doi: 10.1371/journal.pone.0070839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu S, Lu C, Xiao Y, et al. Comparison of EML4-ALK fusion gene positive rate in different detection methods and samples of non-small cell lung cancer. J Cancer. 2020;11(6):1525–1531. doi: 10.7150/jca.36580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, Zhang J, Gao G, et al. EML4-ALK fusion detected by RT-PCR confers similar response to crizotinib as detected by FISH in patients with advanced non-small-cell lung cancer. J Thorac Oncol. 2015;10:1546–1552. doi: 10.1097/JTO.0000000000000668. [DOI] [PubMed] [Google Scholar]
- 10.Kamps R, Brandão RD, Bosch BJ, et al. Next-generation sequencing in oncology: genetic diagnosis, risk prediction and cancer classification. Int J Mol Sci. 2017;18:308. doi: 10.3390/ijms18020308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Letovanec I, Finn S, Zygoura P, et al. Evaluation of NGS and RT-PCR methods for ALK rearrangement in European NSCLC patients: results from the European Thoracic Oncology Platform Lungscape Project. J Thorac Oncol. 2018;13(3):413–425. doi: 10.1016/j.jtho.2017.11.117. [DOI] [PubMed] [Google Scholar]
- 12.Lih CJ, Harrington RD, Sims DJ, Harper KN, Bouk CH, Datta V, Yau J, Singh RR, Routbort MJ, Luthra R, et al. Analytical validation of the next-generation sequencing assay for a nationwide signal-finding clinical trial: molecular analysis for Therapy Choice Clinical Trial. J Mol Diagn. 2017;19:313–327. doi: 10.1016/j.jmoldx.2016.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu J, Mu Z, Liu L, Li K, Jiang R, Chen P, Zhou Q, Jin M, Ma Y, Xie Y, et al. Frequency, clinical features and differential response to therapy of concurrent ALK/EGFR alterations in Chinese lung cancer patients. Drug Des Devel Ther. 2019;13:1809–1817. doi: 10.2147/DDDT.S196189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 15.Lee WI, Huang JL, Lin SJ, et al. Lower T regulatory and Th17 cell populations predicted by RT-PCR-amplified FOXP3 and RORγt genes are not rare in patients with primary immunodeficiency diseases. Front Immunol. 2020;11:1111. doi: 10.3389/fimmu.2020.01111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wen S, Dai L, Wang L, et al. Genomic signature of driver genes identified by target next-generation sequencing in Chinese non-small cell lung cancer. Oncologist. 2019;24(11):e1070–e1081. doi: 10.1634/theoncologist.2018-0572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li W, Liu Y, Li W, Chen L, Ying J. Intergenic breakpoints identified by DNA sequencing confound targetable kinase fusion detection in NSCLC. J Thorac Oncol. 2020;15(7):1223–1231. doi: 10.1016/j.jtho.2020.02.023. [DOI] [PubMed] [Google Scholar]
- 18.Davies KD, Lomboy A, Lawrence CA, et al. DNA-based versus RNA-based detection of MET Exon 14 skipping events in lung cancer. J Thorac Oncol. 2019;14(4):737–741. doi: 10.1016/j.jtho.2018.12.020. [DOI] [PubMed] [Google Scholar]
- 19.Benayed R, Offin M, Mullaney K, et al. High yield of RNA sequencing for targetable kinase fusions in lung adenocarcinomas with no mitogenic driver alteration detected by DNA sequencing and low tumor mutation burden. Clin Cancer Res. 2019;25(15):4712–4722. doi: 10.1158/1078-0432.CCR-19-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thunnissen E, Kerr KM, Herth FJ, et al. The challenge of NSCLC diagnosis and predictive analysis on small samples. Practical approach of a working group. Lung Cancer. 2012;76(1):1–18. doi: 10.1016/j.lungcan.2011.10.017. [DOI] [PubMed] [Google Scholar]
- 21.Smits AJ, Kummer JA, de Bruin PC, et al. The estimation of tumor cell percentage for molecular testing by pathologists is not accurate. Mod Pathol. 2014;27(2):168–174. doi: 10.1038/modpathol.2013.134. [DOI] [PubMed] [Google Scholar]
- 22.Zheng MM, Li YS, Tu HY, et al. Genotyping of cerebrospinal fluid associated with osimertinib response and resistance for leptomeningeal metastases in EGFR-mutated NSCLC. J Thorac Oncol. 2021;16(2):250–258. doi: 10.1016/j.jtho.2020.10.008. [DOI] [PubMed] [Google Scholar]
- 23.Heriyanto DS, Trisnawati I, Kumara EG, et al. The prevalence of the EML4-ALK fusion gene in cytology specimens from patients with lung adenocarcinoma. Pulm Med. 2020;2020:3578748. doi: 10.1155/2020/3578748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Liu Y, Zhao C, et al. Feasibility of cytological specimens for ALK fusion detection in patients with advanced NSCLC using the method of RT-PCR. Lung Cancer. 2016;94:28–34. doi: 10.1016/j.lungcan.2016.01.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author.