Abstract
Circulating cell-free DNA (ccfDNA) has emerged as a promising diagnostic tool in oncology. Identification of tumour-derived ccfDNA (i.e. circulating tumour DNA [ctDNA]) provides non-invasive access to a malignancy’s molecular landscape to diagnose, inform therapeutic strategies, and monitor treatment efficacy. Current applications of ccfDNA to detect somatic mutations, however, have been largely constrained to tumour-informed searches and identification of common mutations because of the interaction between ctDNA signal and next-generation sequencing (NGS) noise. Specifically, the low allele frequency of ctDNA associated with non-metastatic and early-stage lesions may be indistinguishable from artifacts that accrue during sample preparation and NGS. Thus, using ccfDNA to achieve non-invasive and personalized molecular profiling to optimize individual patient care is a highly sought goal that remains limited in clinical practice. There is growing evidence, however, that further advances in the field of ccfDNA diagnostics may be achieved by improving detection of somatic mutations through leveraging the inherently shorter fragment lengths of ctDNA compared to non-neoplastic ccfDNA. Here, the origins and rationale for seeking to improve the mutation-based detection of ctDNA by using ccfDNA size profiling are reviewed. Subsequently, in vitro and in silico methods to enrich for a target ccfDNA fragment length are detailed to identify current practices and provide perspective into the potential of using ccfDNA size profiling to impact clinical applications in oncology.
Key Findings/Implications
| Application of next-generation sequencing for the detection of somatic mutations in circulating cell-free DNA derived from solid tumour malignancies seeks to revolutionize precision medicine by using a simple blood draw to detect cancers, monitor response to therapies, and personalize treatment strategies. |
| Advancing cell-free DNA diagnostics to early-stage and non-metastatic cancers has been limited by challenges associated with distinguishing the true signal of low-frequency tumour-derived cell-free DNA from noise generated during next-generation sequencing. |
| Differences in the fragment length between tumour-derived cell-free DNA (circulating tumour DNA [ctDNA]) and cell-free DNA originating from healthy cells is a biologic phenomenon that can be leveraged to improve detection of ctDNA. |
| Both laboratory-based (in vitro) and computer-based (in silico) methods are being developed to use the fragment length profile of ctDNA to improve cell-free DNA diagnostics in cancer. |
| Further investigations into using size-based analyses in difficult to detect cancers are necessary to expand the role of cell-free DNA in non-invasive applications of precision oncology. |
Why is the Fragment Length of Circulating Tumour DNA (ctDNA) Important?
In 1948, the presence of DNA in plasma without a protective membrane, which has been subsequently termed circulating cell-free DNA (ccfDNA) (Fig. 1a), was described by Mandel and Métais [1]. In 1977, the first report of a difference in the ccfDNA concentration (ng/mL plasma) between cancer patients and healthy individuals hinted at the possibility of harnessing ccfDNA as a diagnostic tool in clinical oncology [2]. Unfortunately, applications based on ccfDNA concentration alone proved limited because of significant overlap between healthy individuals and patients with cancer, particularly patients with early-stage and non-metastatic disease [2–5]. In addition, factors such as age [6], weight [7], inflammation [8], infection [9], and exercise [10], among other clinical factors, have been shown to affect ccfDNA abundancy, likely undermining the use of ccfDNA concentration to discern and monitor malignancy [11]. Thus, the 1994 discovery of a KRAS p.G12D point mutation in ccfDNA derived from pancreatic cancer was a landmark occasion because the detection of tumour-derived ccfDNA (circulating tumour DNA [ctDNA]) provided incontrovertible molecular proof for the viability of the non-invasive detection of cancer [12].
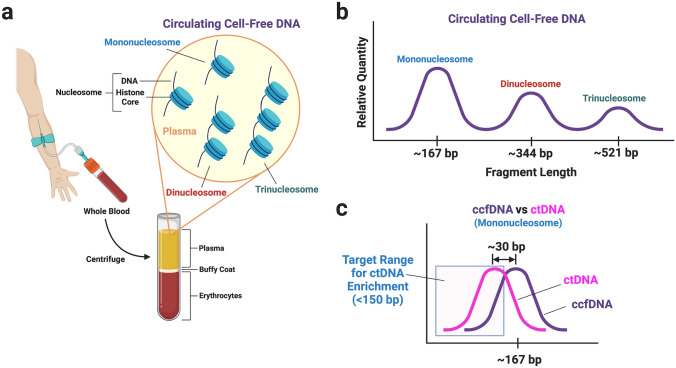
Fig. 1.
Acquisition and characteristics of circulating cell-free DNA (ccfDNA). Whole blood acquired through venipuncture is centrifuged to separate plasma from buffy coat and erythrocytes (a). ccfDNA derived from apoptosis is present in plasma as various multiples of nucleosomes—DNA wrapped around a histone core with a linker fragment of DNA (~10 bp) joining adjacent nucleosomes (a). The relative quantity and fragment length distribution of ccfDNA is shown in (b), where the most abundant fragment length corresponds to the length of the mononucleosome. The fragment lengths of circulating tumour DNA (ctDNA) tend to be shorter than ccfDNA (c); however, there is substantial overlap. Enrichment of ctDNA has generally focused on isolation of fragment lengths < 150 bp to improve the ratio between ctDNA and ccfDNA
Haematopoietic cells account for > 90% of the ccfDNA in healthy individuals [13–16]. The remaining portion of ccfDNA is principally derived from endothelial cells and hepatocytes [13, 14, 16]. In the presence of solid tumours, the proportion of ccfDNA composed of ctDNA is generally correlated to disease severity [3]. In advanced and metastatic disease, the somatic mutations present in ctDNA may overwhelm the wild-type alleles derived from haematopoietic cell DNA and appear with strikingly high allele frequencies (> 20% [17]). In turn, the allele frequency of ctDNA associated with early-stage or non-metastatic disease may be quite low (< 1%). Because a widely sought goal of ccfDNA diagnostics is detection of ctDNA before advanced disease occurs to potentially maximize therapeutic options, the field has been driven towards development of sensitive methods to identify ctDNA among the abundant ccfDNA from normal healthy cells—the proverbial “needle in a haystack”. From 1994 to 2012, research efforts were largely led by polymerase chain reaction (PCR)-based methods conducting narrow tumour-informed searches or analysing common tumourigenic loci (e.g. EGFR p.T790M) for evidence of ctDNA. During this period, ctDNA was detected from malignancies associated with a multitude of organs, such as the colon [3], breast [18], prostate [19], lung [20], skin [21], and pancreas [22] among others. In some instances, detection of ctDNA correlated with poor survival [23]. Other studies found that serial quantitative monitoring of ctDNA informed responses to therapy [24]. Overall, however, the successful detection of ctDNA based on mutation detection tended towards advanced disease where the ctDNA fraction was generally observed at > 1%.
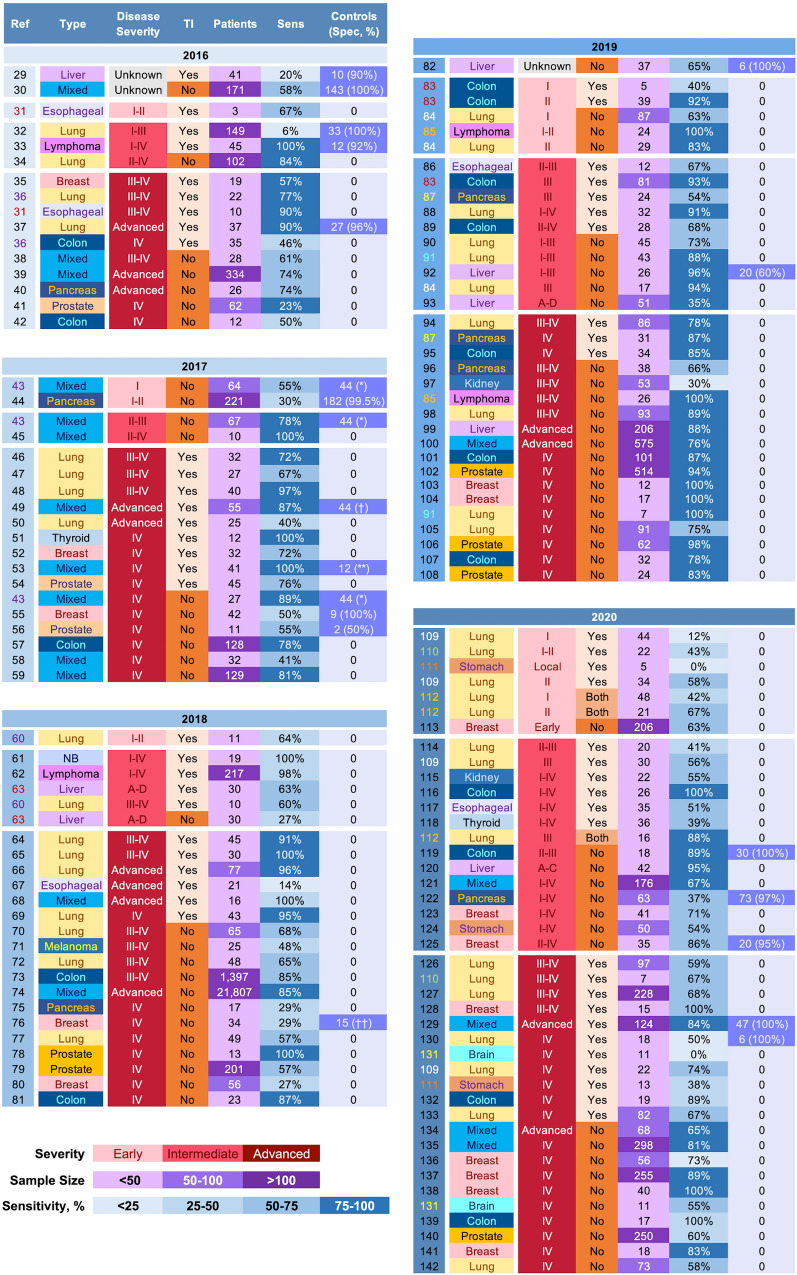
In 2012, the first uses of next-generation sequencing (NGS) to detect ctDNA were detailed [25, 26]. Integration of NGS into ccfDNA diagnostics promised the broader characterization of ctDNA because capture-enrichment panels used during NGS can be customized to target a wide range of molecular foci depending on the desired applications. Thus, detection of ctDNA could move beyond a narrow search to a broad surveillance of a multitude of genes enabling a more extensive genotyping through perfusion sampling rather than focal tissue analysis. Theoretically, NGS should enable substantial advances in precision oncology via (1) detection of both common and patient-specific mutations, (2) monitoring of molecular evolution, and (3) detection of ctDNA derived from metastatic lesions with a potentially separate molecular profile. Each of these mutation-based features is key in the advancement of personalized medicine through the enabling of treatment optimization for each patient’s genotypically unique primary and possibly distal lesions. Although NGS is on par with PCR-based methods in ability to detect ctDNA from a wide array of human cancers [27], further attainment of NGS’s potential has been constrained by assay noise generated during NGS because a portion of artifacts can occur randomly with an allele frequency similar to low-frequency ctDNA [28]. An overview of publications from 2016 to 2020 highlights several key effects of NGS-associated noise on the mutation-based detection of ctDNA (Fig. 2, [29–142]). First, the sensitivity of a mutation-based ctDNA detection strategy to identify cancer reveals a high degree of variability across all disease severities (Figs. 2, 3a, b). Although detection of advanced and metastatic disease of some cancers has improved, others remain challenging likely indicating a low ctDNA burden associated with some cancers regardless of clinical stage. As such, development of more sensitive techniques to discern ctDNA from noise may not only extend ctDNA applications to a broader range of cancer types and stages, but also improve characterization of genetic heterogeneity in advanced cancers. Second, a large portion of studies use a tumour-informed search to guide mutation-based ctDNA detection to distinguish true positives from false positive (Fig. 2). Thus, the ctDNA mutational landscape is constrained to somatic mutations present in a focal tissue sample that may not adequately represent genetic heterogeneity of the primary tumour and/or mutations unique to metastatic lesions. In addition, dependence on tumour DNA to detect ctDNA largely voids the concept of the non-invasive liquid biopsy, particularly applications pertaining to diagnosis and monitoring of molecular evolution. Finally, inclusion of healthy control ccfDNA is uncommon in studies searching for ctDNA with or without a priori knowledge of somatic mutations (Fig. 2). Because a principal component of NGS-associated error is stochastic [28, 143], investigations using a mutation-based detection of ctDNA without validating specificity pertinent to the experimental design may be challenging to interpret due to the confounding effects random noise may have on identification of low-frequency ctDNA, especially when tumour DNA is not used as a guide. Collectively, the data presented in Fig. 2 and 3 show positive trends in the overall progress achieved in the past 5 years, but also provide compelling evidence that the true potential of NGS-based ccfDNA diagnostics in precision oncology remains unrealized. As such, laboratory methods, technical innovations, or both to improve the signal-to-noise ratio during NGS are needed to translate mutation-based ctDNA detection into impactful clinical applications.
Fig. 2.
Profile of publications from 2016 to 2020 using NGS for mutation-based ctDNA detection. A PubMed querya was used to initiate a search in each year for publications with a total sample size ≥ 10. Publications that sought to detect ctDNA associated with minimal residual disease were excluded. The list is first grouped by year (Yr), then disease severity, and finally by whether or not the mutation-based search for ctDNA was tumour informed. The reference (Ref) column identifies the citation. When feasible, data associated with different disease severities are presented separately and the reference number is non-black to support matching of data from the same source. In one study (Ref. [112]), data obtained with and without a tumour-informed search were merged and separation was not possible (marked ‘Both’). Sensitivity (Sens) represents a study value corresponding to the mutation-based detection of ctDNA to determine presence/absence of a malignancy. The associated sample size is presented in the adjacent column (Patients). Specificity (Spec) is reported only if obtained from healthy control data. Sensitivity and specificity values should be interpreted cautiously as calculations can vary substantially within and between publications. For example, in Ref. [131] the authors report detection of ctDNA in 55% of patients. However, no mutations identified in solid tumour DNA were present in ccfDNA, so sensitivity is also shown at 0% for a tumour-informed search. To gain adequate contextual understanding of values, reviewing the publication’s supplemental data may be necessary. ccfDNA circulating cell-free DNA, ctDNA circulating tumour DNA, mixed = cancers from different organs, NB neuroblastoma. *Specificity not calculated, controls used to determine error rate (< 3.3 × 10−7 false positive mutation calls per base); †controls used for error modelling; **specificity not calculated, median error rate of 0.03 non-silent single nucleotide variants per Mb; ††specificity not calculated, 307 of 342 targeted positions error free. aPubMed query: (cell-free DNA[Title]) AND (cancer) AND (("YEAR/01/01"[Date - Publication] : "YEAR/12/31"[Date - Publication]) AND (next-generation sequencing) AND (circulating))
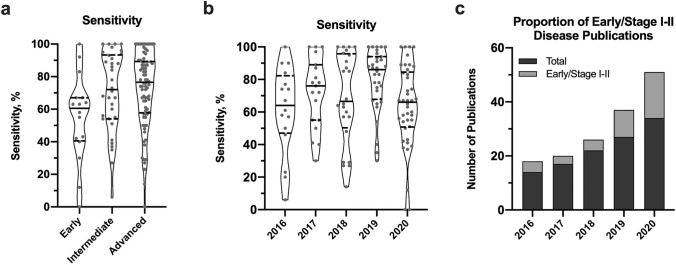
Fig. 3.
Characteristics of publications from 2016 to 2020 using NGS for mutation-based ctDNA detection. In a, the violin plots show the sensitivity of mutation-based ctDNA detection relative to disease severity. In general, there was a trend towards increased detection with more severe disease. In b, sensitivity is relatively similar across years likely because more difficult to detect tumours are being included in later years, which may adversely affect sensitivity but also indicates detection strategies are working towards inclusion of more challenging cancers. In c, the total number of publications profiled for each year from Fig. 2 is shown (dark gray) and the number of those publications that include early/stage I–II disease is also depicted (light gray). In more recent years, a larger proportion of publications are seeking to detect ctDNA in early-stage and non-metastatic cancers. ctDNA circulating tumour DNA, NGS next-generation sequencing
One route to improving detection of low-frequency variants is using a larger DNA input so that the signal exceeds the level of noise. Unfortunately, the amount of ccfDNA in plasma is finite. In healthy individuals, the concentration of ccfDNA ranges between 5 and 15 ng/mL plasma and the quantity of ccfDNA in patients with early-stage or non-metastatic disease is similar or only marginally higher. As a reference point, a single human cell (diploid) contains 7 pg of DNA. In 1 ng of human DNA, each genomic position associated with the non-sex chromosomes is represented ~ 286 times (i.e. genomic equivalents). Thus, a 30-ng ccfDNA library input provides ~ 8580 unique copies of DNA at each base position assuming a lossless library preparation and absence of gene amplifications and deletions. Using NGS, a ctDNA variant with an allele frequency of 0.05% may be indistinguishable from noise because fewer than five unique copies of the mutation under ideal conditions are present in the sequencing data. Because of losses that occur during sample handling for NGS in combination with the low quantity of cell-free DNA in plasma, achieving the necessary boost in signal at such low allele frequencies to overcome NGS-associated noise requires large volumes of blood (i.e. > 100 mL [121]). However, a distinctive characteristic of ctDNA may help overcome these signal-to-noise obstacles. There is growing evidence that the predominance of ctDNA occurs with a fragment length shorter than ccfDNA from healthy cells. Given the challenges associated with detecting ctDNA derived from early-stage and non-metastatic cancers, differences in fragment length size between ctDNA and ccfDNA may represent an exploitable phenomenon to further move ccfDNA beyond research development and towards a non-invasive diagnostic tool in clinical oncology.
The Fragment Length of ctDNA
The fragment length of ctDNA was originally used to discern the type of cell death driving the release of cell-free DNA into the circulation—apoptosis versus necrosis. DNA derived from apoptosis has a characteristic ladder-like distribution with the principal peak at the length of the mononucleosome (~167 bp) and subsequent peaks of decreasing magnitude corresponding to di- and trinucleosomes (Fig. 1b) [144]. Necrosis produces genomic DNA with lengths > 10,000 bp. Initially, there was evidence supporting the origins of ccfDNA from both pathways [145]. Terms such as apoptosis index and DNA integrity index subsequently emerged to characterize PCR-based analyses using ratios of short and long amplicon lengths [146–148]. By 2011, there was a preponderance of evidence that ccfDNA originated from apoptosis. The first application of NGS to better characterize ctDNA fragment length occurred in 2015, when Jiang et al. used copy number aberrations in hepatocellular carcinoma to infer the difference in size distributions between ccfDNA and ctDNA [149]. A positive correlation was identified between the tumour DNA fraction in plasma and the proportion of DNA fragments less than 150 bp, suggesting a shorter size length profile for ctDNA compared to the mononucleosome’s ~ 167-bp length associated with normal ccfDNA.
In 2016, two technical aspects of NGS were used to characterize the specific fragment length of ctDNA. First, during library preparation of ccfDNA, mechanical and enzymatic shearing is not used because the predominant component of ccfDNA is found in association with the mononucleosome, which has a median length of ~167 bp (Fig. 1b). Therefore, to prevent generation of short fragments that are difficult to sequence, ccfDNA NGS workflows do not include a DNA shearing step, which is commonly used during preparation of genomic DNA extracted from white blood cells and tumour cells. Second, NGS generally employs a constrained sequencing length (e.g. 100 bp, 125 bp, 150 bp), which means the entire length of the ccfDNA molecule may not be sequenced. Thus, many NGS applications use paired-end sequencing to read the sequence from both ends of the molecule. Once paired reads are aligned to a reference genome, the length of the original DNA molecule is measured. These two components of NGS were combined with rat xenograft models of a brain tumour and hepatocellular carcinoma to precisely measure fragment lengths of normal ccfDNA and ctDNA, which were parsed based on species (rat DNA vs. human DNA, respectively) [150]. Overall, the median fragment length of ctDNA was 134–144 bp, which was ~ 20–30 bp shorter than the median length of 167 bp for rat ccfDNA (Fig. 1c). In addition, smaller peaks along the overall fragment distribution profile of the mononucleosome were present at regular intervals (i.e. ~ 10-bp periodicity). Observations regarding differences in fragment length size and, to a lesser extent, periodicity translated to a patient with widely metastatic melanoma where the principal peak associated with the BRAF p.V600E mutation was ~20 bp shorter than the wild type distribution. Similar differences in fragment size were also observed in lung cancer patients harbouring the common EGFR p.T790M and p.L858R mutations. These findings not only solidified observations by Jiang et al. [149], but also echoed findings from several earlier reports. First, Giacona et al. applied a microspreading technique in 1998 to measure double-stranded DNA with transmission electron microscopy to compare strand length between three controls and three patients with adenocarcinoma of the pancreas [151]. The median ccfDNA strand length in cancer patients was 185 nm compared to 273 nm in controls, which is a ~ 30-bp difference (assuming 0.34 nm/bp). Second, in 2010, Lo et al. used NGS to describe characteristics of foetal ccfDNA in the maternal circulation [152]. The principal fragment length of foetal ccfDNA in the maternal circulation was 143 bp, shorter than the principal fragment length of maternal ccfDNA at 166 bp. In addition, the fragment distribution for both foetal and maternal ccfDNA exhibited a 10-bp periodicity. Finally, Zheng et al. showed in 2012 that ccfDNA derived from a transplanted liver exhibited a 10-bp periodicity consisting of significantly shorter fragments than that of the recipient-specific ones [153]. Collectively, these studies established and provided orthogonal support for the narrow difference in fragment size length between ctDNA and ccfDNA. Importantly, multiple studies using a mixture of xenograft models and point mutations have subsequently confirmed the discrete size disparity between ctDNA and ccfDNA [68, 112, 121, 154, 155], which has also been the topic of previous reviews [156]. In addition, the overall size profile in patients with metastatic disease has also been shown to be shorter compared to healthy controls [154], but substantial overlap between patients and controls limits size profiling alone as a clinical diagnostic tool, particularly for early-stage and non-metastatic cancers. Although the origins of the 10-bp periodicity seen in the fragmentation pattern of ccfDNA has been linked to both intra- and extracellular processes [157, 158], the origins of the size difference between ctDNA and ccfDNA is unclear. However, there is compelling evidence that chromatin structure or epigenetic changes may have a role [15, 159–161] (see [162] for a review of epigenetic modifications associated with ccfDNA). Regardless of aetiology, the focus of this review is exploring prior use of ctDNA fragment length to improve the non-invasive detection of somatic mutations in ccfDNA associated with malignant disease. Both in vitro and in silico methods to isolate subfractions of ccfDNA are reviewed to identify strengths and weaknesses associated with different approaches. In so doing, the goal if this review is to establish the current state of the art in ccfDNA size-selection technology to better understand the necessary steps forward to potentially bring ccfDNA diagnostics to earlier stages of disease and enable precision oncology through detection of patient-specific somatic mutations regardless of disease severity.
In Vitro Enrichment of ctDNA
In vitro refers to a mechanical or wet laboratory-based technique. Gel-based methods have been the principal in vitro approach for extracting a narrow size profile range. The first description of this occurred in 2016 and used polyacrylamide gel electrophoresis to achieve sufficient resolution to obtain six fractions from the ccfDNA mononucleosome peak [150]. In four lung cancer patients harbouring the EGFR p.T790M mutation, isolation of the ccfDNA fraction 20–50 bp shorter than the principal peak increased the mutant allele frequency (MAF) measured by droplet digital PCR by 2.8-fold to 9.1-fold in 75% of samples (Table 1). Although the size-selection methodology was not scalable to broader applications, the potential for using the size difference to enrich for ctDNA was successfully demonstrated.
Table 1.
In vitro size selection for ctDNA detection
| Publication | Methods | Cancer (sample size) | Baseline detection characteristics | Results |
|---|---|---|---|---|
| Underhill et al., 2016 [150] |
PAGE with manual excision to extract six fractions from the mononucleosome after library generation and PCR amplification ddPCR used to measure MAF |
Lung (N = 4) | EGFR T790M MAF range in ccfDNA 0.33–7.1% |
1.1- to 9.1-fold enrichment of EGFR p.T790M mutations in ccfDNA fraction 20–50 bp shorter than the mononucleosome peak The sample with a MAF of 0.33% showed strongest enrichment at 9.1-fold |
| Mouliere et al., 2018 [68] |
3.0% agarose cassette targeting a single ccfDNA fraction (90–150 bp) prior to library generation and PCR amplification sWGS to calculate tMAD scores |
HGSOC (N = 35; 48 samples) Healthy controls (N = 46; 65 samples) |
Median tMAD score of 0.015 in patients (mean of 0.05; range 0.005–0.30) Median tMAD score of 0.01 in controls (range 0.004–0.015) 50% of patients had an increased tMAD score compared to controls |
0.9- to 6.4-fold enrichment of tMAD scores (mean of 2.5-fold and median of 2.1-fold enrichment) After size selection in 48 patient samples and 18 control samples, 82% of samples had an increased tMAD score compared to controls tMAD scores increased ≥ 2-fold after size selection in 54.3% (19/35) and 22.2% (4/18) of pre-treatment patients and controls, respectively |
|
3.0% agarose cassette targeting a single ccfDNA fraction (90–150 bp) prior to library generation and PCR amplification WES for point mutations |
HGSOC (N = 6; pre-treatment) | 821 mutations identified in 6 patients, with mean MAF of 12.0% (median of 9.5%, range 0.53–100%) |
Mean increase in MAF of 4.19-fold (median 4.27-fold increase) In 5 of 6 patients, size selection identified an additional 171 mutations with mean MAF of 27.9% (median of 25%, range 8.3–75%) |
|
| HGSOC (N = 6; post-treatment) | 202 mutations identified in 6 patients, with mean MAF of 8.1% (median of 6.5%, range 1.2–47.6%) |
Mean increase of 2.1-fold (median 1.5-fold, range 0.9- to 11-fold) In 4 of 6 patients, size selection identified an additional 89 mutations, with mean MAF of 39.3% (median 37.5%, range 15.4–100%) |
||
| Hellwig et al., 2018 [155] |
Automated liquid handler using a 3.0% agarose matrix in a 12-channel cassette to extract 3 fractions after library generation and PCR amplification ddPCR Capture-enrichment panel |
Melanoma (N = 8), colorectal adenocarcinoma (N = 3), pancreatic adenocarcinoma (N = 2) | All samples were positive for point mutations in KRAS or BRAF by ddPCR, with a mean MAF of 2.4% (median of 1.1%, range 0.39–12.3%) |
At a median insert size of 141 bp, MAFs were enriched on average by 2.9-fold (median 2.5-fold, range 0.2–10.3) as measured by ddPCR At a median insert size of 141 bp, MAFs were enriched on average by 2.0-fold (median of 2.0-fold, range 0.8–3.3) as measured by NGS At median insert size of 167 bp, MAFs were reduced by a median of 0.8 and 0.7-fold using ddPCR and NGS, respectively MAF measured by ddPCR and NGS were strongly correlated (Pearson’s r = 0.97, P < 0.001) |
| Ishida et al., 2020 [116] |
SPRIselect beads to obtain fractionated small ccfDNA (100–400 bp) dPCR Hotspot amplicon-based sequencing in 50 genes |
Colorectal cancer (N = 26; stage I–II, N = 6; stage III–IV, N = 20) | KRAS, BRAF, and TP53 mutations present in tumour DNA were detected in serum by dPCR in 24 of 26 patients (92.3%), with a mean MAF of 11.7% (median of 1.2%, range 0.1–63.4%) |
Size selection increased mean MAFs of driver genes by dPCR from 6.8% to 10.7% Size selection increased mean MAFs by NGS from 16.3% to 18.8% For MAFs > 1%, NGS detected a higher average number of mutations in size-selected small ccfDNA compared to unselected ccfDNA (1.8 vs. 1.0 per case, respectively) |
ccfDNA circulating cell-free DNA, ctDNA circulating tumour DNA, dPCR digital PCR, ddPCR droplet dPCR, HGSOC high-grade serous ovarian carcinoma, MAF mutant allele frequency, NGS next-generation sequencing, PAGE polyacrylamide gel electrophoresis, PCR polymerase chain reaction, sWGS shallow whole genome sequencing, tMAD trimmed median absolute deviation from copy number neutrality, WES whole exome sequencing
In 2018, in vitro size selection was applied to larger sample sizes using benchtop microfluidic devices. Mouliere et al. loaded 8–20 ng of pre-library ccfDNA (i.e. ccfDNA prior to library generation and PCR amplification) into a 3% agarose cassette (HTC3010, Sage Bioscience) and targeted the ccfDNA fraction between 90 and 150 bp for extraction on a PippinHT (Sage Bioscience; Table 1) [68]. Using shallow whole genome sequencing (sWGS) (read depth of 0.5 ×), the trimmed median absolute deviation from copy number neutrality (tMAD; a measure of copy number alterations) increased by a median of 2.1-fold in 47 of 48 plasma samples from 35 patients with high-grade serous ovarian cancer. In a subset of six patients with ctDNA variants detected by whole exome sequencing (WES) (median MAF 9.5%, range 0.5–100%), in vitro size selection increased MAF by a median factor of 4.27-fold. Three weeks after therapy when MAFs were reduced (median MAF 6.5%, range 1.2–48%), the median increase in MAF via in vitro size selection was less, at 1.5-fold, suggesting that in vitro size selection may have reduced benefits at lower MAFs.
Instead of using ccfDNA during size selection prior to library preparation, Hellwig et al. used 1 µg of ccfDNA obtained after library generation and PCR amplification as input into an automated liquid handler (NIMBUS Select, Hamilton, Reno, NV) that incorporated Ranger Technology (Coastal Genomics, Burnaby, BC) for the monitoring and real-time manipulation of electrophoretic mobilities through a 3.0% agarose matrix in a 12-channel cassette (Table 1) [155]. Three ccfDNA fractions centred at ~ 141 bp, ~ 155 bp, and ~ 167 bp were obtained from 13 patients harbouring mutations in a BRAF p.V600, KRAS p.G12, or KRAS p.G13 codon associated with colorectal cancer, melanoma, and pancreatic cancer for comparison to unselected ccfDNA. The median MAF in unselected ccfDNA was 1.1% (range 0.39–12.3%), with MAF ≤ 0.81% in 46.2% of the mutations. Using NGS, an average 2.0-fold increase in MAF was observed in the fraction centred at 141 bp compared to unselected ccfDNA, which was significantly higher than the 1.3-fold gain in the 155-bp fraction and the 0.7-fold reduction in the 167-bp fraction. All NGS results were cross-validated with droplet digital PCR, which similarly confirmed enrichment of ctDNA through in vitro size selection. In addition, the observed enrichment of ctDNA associated with a low MAF (< 1%) suggests that size selection conducted on ccfDNA libraries rather than pre-library ccfDNA may be advantageous by mitigating loss of rare unique molecules through PCR amplification to generate more copies for acquisition.
Most recently, Ishida et al. used solid-phase reversible immobilization (SPRI) beads (SPRIselect beads, Beckman Coulter, CA) to fractionate pre-library ccfDNA to obtain DNA sizes of 100–400 bp (Table 1) [116]. In so doing, the proportion of ccfDNA in the range of 90–150 bp increased from 3.0% in unselected ccfDNA to 25.9% in the size-selected ccfDNA. In 20 patients with colorectal cancer (63% with distant metastasis), the mean MAFs of driver genes as measured by digital PCR increased from 6.8% in unselected ccfDNA to 10.7% in size-selected ccfDNA. Using NGS, a similar increase in mean MAFs was observed. In addition, the average number of mutations with an MAF > 1% detected in size-selected ccfDNA compared to unselected ccfDNA increased from 1.0 to 1.8 per patient. In contrast to gel-based techniques that sought to isolate a subfraction of the mononucleosome, the bead-based approach yielded a size-selected fraction of ccfDNA inclusive of both the mono- and dinucleosomes. Although the extent of ctDNA enrichment was less than the gel-based methods, the study demonstrated that even a marginal shift in the overall size profile of ccfDNA towards shorter fragment lengths has the potential to improve ctDNA detection, which may have implications for high-throughput labs using bead-based methods as part of automated protocols.
Additional publications that use in vitro size selection to enrich for ctDNA were not identified in the literature. However, in vitro size selection has been more extensively investigated in prenatal diagnostics, where foetal ccfDNA in the maternal circulation has a similar fragment length shortening relative to maternal ccfDNA [152]. Thus, several papers are highlighted herein to substantiate the observations described above and to also provide a broader scope of in vitro methods associated with ccfDNA size selection that may have potential ctDNA applications. The in vitro methodologies to enrich for foetal ccfDNA from maternal ccfDNA broadly organize into two categories: (1) isolation of the entire mononucleosome or (2) extraction of the shorter fragments associated with the mononucleosome. Regarding the former category, Hahn et al. described a microsystem to preconcentrate ccfDNA using electrokinetic trapping followed by separation of fragments using ionic mobility to effectively parse ccfDNA with a length < 300 bp from fragments > 500 bp [163]. Yang et al. established a PCR-based enrichment protocol by modifying the amplification reaction conditions [164]. Using an optimized lower denaturing temperature that biased PCR towards amplification of shorter fragments, the foetal fraction was comparably increased to that of removing fragments longer than 300 bp from the maternal ccfDNA sample. A potential advantage of this method is that existing workflows do not require alterations beyond optimization of thermal cycling temperatures. To isolate the short fraction (< 160 bp) of cell-free DNA from the mononucleosome, Hu et al. used custom-designed magnetic beads with an average particle size of 1 µm (for reference, the average size of the industry standard SPRI beads is similarly 1 µm [165]) to enrich for the foetal fraction of ccfDNA [166]. Using a bead ratio of 1.5 × for size selection after end-repair and prior to adapter ligation increased the proportion of ccfDNA < 160 bp from ~ 25% in unfractionated ccfDNA to ~85% in fractionated ccfDNA. Foetal ccfDNA was enriched in the fractionated maternal ccfDNA by 1.5- to 4-fold, which improved detection of missed foetal trisomies by 45.5%. Compared to the approach by Ishida et al. described above [116], the method employed by Hu et al. appears to more strongly select for the shorter fraction of the mononucleosome, suggesting that a bead-based approach may be able to more strongly enrich for ctDNA once optimized. Most recently, Welker et al. used gel electrophoresis on 2% agarose cassettes (BluePippin, Sage Bioscience) to isolate short ccfDNA fragments from the mononucleosome with an average length of 140 bp (not including adapters) [167]. Of the 2401 maternal ccfDNA samples tested, 2395 (99.8%) showed an increase in the foetal fraction, with an average increase of 2.3-fold. Samples with a foetal fraction < 4% showed the largest gain, with an average increase of 3.9-fold. Because the general methodology was similar to that used by Mouliere et al. [68] and Hellwig et al. [155] for ctDNA enrichment, an important aspect to recognize in the Welker et al. study [167] is the feasibility of scaling gel-based techniques to meet high-throughput demands.
Overall, there is compelling evidence that in vitro size selection enriches for ctDNA. The brief overview of a few foetal ccfDNA studies supports this conjecture, while also identifying additional in vitro strategies that may facilitate ccfDNA diagnostics in cancer. A key caveat to recognize, however, is the limited enrichment data for MAF < 1%. Moreover, data have not been previously reported for in vitro enrichment of ctDNA associated with an MAF < 0.1%. Thus, additional studies are necessary to determine if the strengths of in vitro fragment size selection to enrich for ctDNA extend to the lower allele frequencies that are more representative of early-stage and non-metastatic disease.
In Silico Applications of ctDNA Fragment Length
In silico refers to the computer-based or bioinformatic curation of NGS data. Because paired-end sequencing associated with NGS enables strand length measurements, reads can be sorted based on the length of the original DNA molecules used during library preparations. In 2018, Mouliere et al. applied an in silico size-selection analysis to synthetically enrich for ctDNA by selecting for ccfDNA fragments with lengths of 90–150 bp (Table 2) [68]. In 189 cancer patients and 65 healthy controls, in silico size selection of sWGS data improved the area under the curve (AUC) for discerning cancer patients based on tMAD scores from 0.69 to 0.90. In silico size selection also increased MAF as measured by WES by 2.2-fold in six patients with high-grade serous ovarian carcinoma. In a separate cohort of 16 cancer patients that underwent WES, in silico size selection increased the MAF of ctDNA on average by 1.7-fold and additional mutations were detected in 13 of 16 patients. In 2020, Smith et al. used in silico size selection for ccfDNA fragments between 90 and 150 bp to improve detection of ctDNA associated with renal tumours (Table 2) [115]. In silico size profiling improved somatic copy number alteration (SCNA) detection in ctDNA from 6.3 to 22.9% in 48 patients with renal tumours ranging from benign tumours to metastatic malignancies. In a separate cohort of 43 patients with renal cell carcinoma (41 with metastatic disease) and most with the primary tumour removed (35/43 patients), in silico size selection identified SCNA-based ctDNA in six additional patients, increasing the detection rate from 18.6 to 32.6%. The original MAFs of SCNAs ranged from 4 to 17% and increased on average by 2.2-fold (range 0.9–5.7). Also in 2020, Nygard et al. similarly used in silico size selection for ccfDNA fragments between 90 and 150 bp to improve SCNA-based detection of ctDNA derived from stage III non-small cell lung cancer (Table 2) [168]. In 23 samples obtained from six patients undergoing radiation therapy, in silico size selection improved SCNA detection from 22 to 70% of samples. Although it is important to recognize that these three studies largely identified ctDNA associated with MAFs > 1% [68, 115, 168], this MAF range was governed by study design rather than technical limitations of in silico size selection. The studies were either targeting SCNA via sWGS or point mutations detected by WES. Both approaches are associated with read depths that are not amenable to detection of MAFs < 1%. Thus, the use of in silico size selection to improve detection of specific variants in ctDNA at low MAFs remains uncertain. However, these studies collectively provide compelling evidence that integration of size profiling into bioinformatic applications of ctDNA detection may be advantageous.
Table 2.
In silico size-based filtering of insert size for ctDNA detection
| Publication | Methods | Cancer (sample size) | Baseline detection characteristics | Results |
|---|---|---|---|---|
| Mouliere et al., 2018 [68] |
sWGS to calculate tMAD scores Size selection for inserts with lengths of 90–150 bp |
“High ctDNA” cancers from melanoma, ovarian, lung, colorectal, cholangiocarcinoma, and other (N = 189) Healthy controls (N = 65) |
AUC = 0.69 | After size selection, AUC increase to 0.90 |
|
WES for point mutations Size selection for inserts with lengths of 90–150 bp |
HGSOC (N = 6; pre-treatment) | 821 mutations identified in 6 patients, with mean MAF of 12.0% (median of 9.5%, range 0.53–100%) |
Mean increase in MAF of 2.2-fold (median 2.25-fold increase) In 6 of 6 patients, size selection identified an additional 188 mutations, with mean MAF of 21.5% (median of 16.9%, range 3.0–88.9%) |
|
| HGSOC (N = 6; post-treatment) | 202 mutations identified in 6 patients, with mean MAF of 8.1% (median of 6.5%, range 1.2–47.6%) | In 6 of 6 patients, size selection identified an additional 122 mutations, with mean MAF of 30.3% (median of 25.0%, range 7.3–85.7%) | ||
| Colorectal, cholangiocarcinoma pancreatic, and prostate (N = 16) | 2133 mutations in plasma with matched mutations in tumour DNA (MAF range ~1% to ~70%) |
Size selection increased mean MAF by 1.7-fold in 97% of mutations In 13 of 16 patients, size selection identified additional mutations |
||
| Smith et al., 2020 [115] |
sWGS to detect SCNAs based on tMAD score Size selection for inserts with lengths of 90–150 bp |
Renal tumours (benign to metastatic; N = 48) | SCNA detected in 4 of 48 (6.3%) samples |
After size selection, 41 of 48 samples met criteria for tMAD analysis (> 2 million reads) Average tMAD score increased 2.2-fold (range 1.25–4.83) SCNA-based ctDNA detected in 8 additional patients (11/48, 22.9%) |
| RCC, N = 43, 41 patients with metastatic disease, 35 patients with primary tumour removed | SCNA detected in 8 of 43 samples (18.6%), with median MAF of 7% (range 4–17%) |
SCNA detection increased to 14 of 43 samples (32.6%) MAF increased by a mean of 2.2-fold (range 0.9–5.7) to a median of 8% (range 4–23%) |
||
| Nygard et al., 2020 [168] |
sWGS to detect SCNAs based on tMAD score Size selection for inserts with lengths of 90–150 bp |
Inoperable, stage III NSCLC (N = 6; 23 samples) | SCNA detection in 5 (22%) samples from 3 of 6 patients | SCNA detection increased to 16 of 23 samples (70%) from 6 of 6 patients |
AUC area under the curve, ctDNA circulating tumour DNA, HGSOC high-grade serous ovarian carcinoma, MAF mutant allele frequency, NSCLC non-small cell lung cancer, RCC renal cell carcinoma, SCNA somatic copy number alteration, sWGS shallow whole genome sequencing, tMAD trimmed median absolute deviation from copy number neutrality, WES whole exome sequencing
As an alternative to the explicit exclusion of fragments outside of a size profile window, in silico approaches have also assigned a weight based on fragment length as part of a probability model for cancer detection. Using sWGS data and tMAD scores, Mouliere et al. applied a random forest algorithm that included the proportion of fragments in defined size ranges to enhance the detection of tumours [68] (Table 3). In 68 patients with high ctDNA cancers, the model was able to distinguish cancer patients from 26 healthy controls with an AUC of 0.994. In 57 patients with low ctDNA cancers, the AUC was 0.914. Notably, using fragmentation features without the tMAD score yielded AUCs of 0.989 and 0.891, respectively, suggesting that fragment size was the principal component of the original model. Wan et al. used fragment length as part of an in silico error reduction strategy named INtegration of VAriant Reads (INVAR), which weights mutant reads across all patient-specific mutation loci based on the empirical distribution of mutant fragments in all other samples in the cohort being studied to give a size range enriched in cancer greater weight (Table 3) [121]. Using a tumour-informed search to guide ctDNA detection, INVAR achieved a median AUC of 0.80 (range 0.64–0.92) in early-stage disease and previously difficult to detect cancers such as stage I–III non-small cell lung cancer, stage I–II breast cancer, and renal and brain tumours, and after surgery for stage II–III melanoma. INVAR has also been applied by Smith et al. to detect renal tumours (Table 3) [115]. Using a tumour-informed search, ctDNA was detected in 12 of 22 patients (54.5%) with renal tumours ranging from benign to metastatic. In a search without a priori knowledge of somatic tumour mutations, INVAR detected ctDNA in 18.6% of patients with largely advanced renal cell carcinoma. Finally, Chabon et al. similarly integrated fragment length into a noise reduction strategy, an enhanced version of CAPP-Seq [37], to detect ctDNA derived from stage I–III non-small cell lung cancer (Table 3) [112]. Using a tumour-informed search, ctDNA was detected in 45% (38 of 85) of patients. In a subset of the 85-patient cohort where ctDNA was not detected, patient-specific panels derived from WES of the tumour were generated, which led to new ctDNA detection in 10 of 17 patients (59%). In a subsequent step, ccfDNA fragment length was integrated into a machine learning algorithm to screen for ctDNA indicative of non-small cell lung cancer in high-risk patients. At 98% specificity, sensitivity for detecting stage I–II non-small cell lung cancer via ctDNA in 41 patients was ~30%. Collectively, these studies demonstrate the promise of integrating ctDNA fragment length into probability-based models to detect early-stage and difficult to detect cancers. Importantly, there are two key aspects of these studies that merit additional consideration within the context of ccfDNA fragment length. First, ccfDNA fragment length was only one component in the overall ctDNA detection strategy. For example, Chabon et al. used a multi-tiered approach to estimate the probability that a ccfDNA mutation was tumour derived using background frequencies, ccfDNA fragment size, the gene affected, and likelihood of clonal haematopoiesis of indeterminate potential [112]. Second, the studies used fragment length to enhance tumour detection rather than mutation detection. For example, Wan et al. observed in nine of the 11 stage II–III melanoma patients where INVAR detected ctDNA that the integrated MAF determined from multiple loci was below the 95% limit of detection for a “perfect” single-locus assay based on the ccfDNA input [121], indicating that insufficient read depth and signal were available to confidently determine presence/absence of each individually targeted mutation in ctDNA. As such, additional studies are needed to determine if machine-learning algorithms using fragment length profiling can be further harnessed to improve detection of individual somatic mutations in ctDNA.
Table 3.
In silico size-based weighting of potential somatic mutations for ctDNA and tumour detection
| Publication | Methods | Cancer (sample size) | Results |
|---|---|---|---|
| Mouliere et al., 2018 [68] |
sWGS RF algorithm that included proportion of fragments in defined size ranges and tMAD score |
“High ctDNA” cancers from melanoma, ovarian, lung, colorectal, cholangiocarcinoma, and other (N = 68) “Low ctDNA” cancers from renal, brain, bladder, and pancreas (N = 57) Healthy controls (N = 26) |
In high ctDNA cancers, an RF model yielded an AUC of 0.994 for distinguishing cancer patients from controls In low ctDNA cancers, an RF model yielded an AUC of 0.914 for distinguishing cancer patients from controls An RF model using fragmentation features alone (leaving out tMAD score) yielded AUCs of 0.989 and 0.891 for cancer types with a high and low amount of ctDNA, respectively |
| Wan et al., 2020 [121] |
Tumour-informed, patient-specific, custom-capture panels INVAR weights mutant reads across all patient-specific mutation loci based on the empirical distribution of mutant fragments in all other samples in the cohort being studied to give a size range enriched in cancer greater weight |
Stage II–III melanoma after complete resection (N = 38) Stage IV melanoma (N = 9; 52 samples) |
ctDNA detected in 11 stage II–III patients (28.9%, specificity at 98.6%), and the integrated MAF in 9 of the 11 patients was below the 95% LOD for a “perfect” single-locus assay based on ccfDNA input amount (AUC = 0.64) ctDNA detected in the baseline samples of 9 stage IV patients (100%) ctDNA detected in 50 of 52 treatment and follow-up samples from 9 stage IV patients where the integrated MAF in 15 of the 50 samples was below the 95% LOD for a “perfect” single-locus assay based on ccfDNA input amount |
|
Stage I–IV breast cancer (N = 7) NSCLC (N = 19) Renal tumours (N = 24) Brain tumours (N = 8) |
16 samples from stage I–II breast cancer, sensitivity of 62.5% at specificity of 90% (AUC = 0.81) 19 samples from stage IV breast cancer, sensitivity of 100% at specificity of 100% (AUC = 1.00) 8 patients with grade II–IV brain tumours, sensitivity of 75% at specificity of 90% (AUC = 0.92) 24 patients with stage I–IV renal tumours, sensitivity of 41.7% at specificity of 90% (AUC = 0.66) 19 patients with stage I–III NSCLC, sensitivity of 63.1% at specificity of 98% (AUC = 0.80) |
||
| Smith et al., 2020 [115] |
Custom-panel with patient-specific mutations detected in tumour DNA by WES and 109 genes commonly mutated in RCC INVAR |
Renal tumours (benign to metastatic; N = 22) | ctDNA was detected in 12 patients (54.5%) |
|
Custom-panel targeting 10 genes in renal cancers INVAR |
RCC (N = 43, 41 patients with metastatic disease, 35 patients with primary tumour removed) |
ctDNA was detected in 8 patients (18.6%) Mean MAF was 8.3% (range 3.5–18%) |
|
| Chabon et al., 2020 [112] |
Tumour-informed search using a 355-kb panel of 255 genes recurrently mutated in NSCLC Lung-CLiP – a multi-tiered machine-learning approach that includes fragment size to estimate the probability that a ccfDNA mutation is tumour derived |
Stage I–II NSCLC (N = 41) Stage III NSCLC (N = 5) Risk-matched controls (N = 48) |
For stage I–II patients: Sensitivity of ~30% at 98% specificity Sensitivity of ~56% at 80% specificity For stage III patients: Sensitivity of ~60% at 98% specificity Sensitivity of ~100% at 80% specificity |
AUC area under the curve, ccfDNA circulating cell-free DNA, ctDNA circulating tumour DNA, INVAR INtegration of VAriant Reads, LOD limit of detection, Lung-CLiP lung cancer likelihood in plasma, MAF mutant allele frequency, NSCLC non-small cell lung cancer, RCC renal cell carcinoma, RF random forest, sWGS shallow whole genome sequencing, tMAD trimmed median absolute deviation from copy number neutrality, WES whole exome sequencing
In silico size selection offers a multitude of opportunities to analyse ccfDNA using hard filters, probability modelling, or both. An advantage of in silico size analysis is the integration of fragment length with other features of ccfDNA and/or other biomarkers of cancer [169] in a lossless manner to optimize ctDNA and tumour detection. Relative to ctDNA enrichment for somatic mutation detection, however, the caveats associated with in silico size selection are identical to those introduced with in vitro enrichment—there is limited data available for MAFs < 1%. Although in silico size selection has been used during detection of foetal ccfDNA in the maternal circulation [170, 171], the approaches are similar to that described for ctDNA, and the foetal fraction is generally > 1%, with few exceptions. As such, additional insights are unlikely to be gleaned from reviewing the foetal ccfDNA literature. Accordingly, future studies devoted to using in silico size selection for enrichment of ctDNA associated with early-stage and non-metastatic cancers are needed to better understand the potential impact on mutation-based ctDNA detection to further improve the non-invasive genotyping of tumours.
In Vitro Versus in Silico ccfDNA Size Selection, Which Is Better?
Both in vitro and in silico approaches to enrich for ctDNA present strengths and challenges. In a direct comparison between methods, Mouliere et al. found that in vitro size selection afforded a higher degree of ctDNA enrichment compared to in silico size selection (mean increase in MAF of 4.19-fold vs. 2.2-fold, respectively) [68]. However, several factors regarding ctDNA fragment size within the context of in vitro size selection warrant careful review. Although ctDNA generally aggregates around a shorter distribution of fragment lengths (< 150 bp), longer fragments outside the targeted range for enrichment may also be present. For example, Hellwig et al. found the fraction of isolated ccfDNA fragments with a median length of ~ 167 bp to be associated with a reduced MAF rather than absent ctDNA indicating that ctDNA is not exclusive to fragments < 150 bp [155]. In addition, the size difference between ctDNA and normal ccfDNA has been shown to extend beyond the mononucleosome to the dinucleosome, where a second cluster of enriched ctDNA may occur at 230–310 bp [112, 121, 172]. Finally, Chabon et al. found in silico hard filtering based on ctDNA-enriched size windows disproportionately favoured variants with higher pre-enrichment MAFs [112]. As such, using in vitro size selection to exclude molecules > 150 bp has the potential to adversely affect sensitivity [173], particularly for ctDNA with a low allele frequency where accrual of DNA losses during sample preparation for NGS may be compounded by the in vitro size-selection process.
Whereas in vitro size selection may lead to irretrievable loss of rare molecules, in silico size selection allows a lossless size-based analysis. In addition, a multitude of size analyses using hard filters and/or probability modelling can be conducted iteratively to optimize ctDNA detection that may be specific to each type and severity of cancer. In combination with bioinformatic strategies to reduce error [174], the additional potential boost in ctDNA signal via in vitro size selection to overcome NGS-associated noise may not be necessary. A potential weakness of in silico size selection is that application of algorithms may lead to fewer samples available for analysis and/or reduced read depth. For example, Smith et al. found that seven of 29 samples (24.1%) were excluded as technical failures because there were insufficient reads after size selection to meet the requirement of the INVAR algorithm [115]. However, these effects can likely be anticipated and mitigated by using a higher number of total paired reads for each sample during sequencing.
Heretofore, in vitro size selection has been discussed within the context of ctDNA enrichment. However, an in vitro approach may also have beneficial bioinformatic effects that merit consideration. Specifically, Hellwig et al. showed that in vitro size selection reduced sample complexity, leading to the generation of larger family sizes without compromising ctDNA detection [155]. During library preparation, amplicons of each unique DNA molecule are generated. Subsequently, molecular barcodes are used to identify amplicons derived from the same original DNA molecule. During in silico analysis of NGS data, amplicons with the same molecular barcode are grouped into a family and used to generate a single consensus sequence that represents the original unique DNA molecule. Family size is the number of amplicons used to generate a consensus sequence and larger family sizes correspond to larger error reduction. Thus, in vitro size selection has the potential to enrich for ctDNA, while concomitantly reducing NGS error.
Currently, there is not strong evidence that supports using one approach over the other. In vitro size selection may enable enrichment of ctDNA while also yielding fewer NGS-associated errors, but at the cost of losing rare ctDNA molecules that may mitigate the desired enrichment effect. In silico size selection allows a multitude of lossless bioinformatic analyses, but at the cost of potentially reduced ctDNA enrichment that may be insufficient to overcome NGS-associated noise. Finally, and perhaps most importantly, there are insufficient data at very low allele frequencies to adequately determine which approach will best position ccfDNA diagnostics to improve non-invasive precision oncology. Until further data become available, study goals will need to continue to dictate integration of in vitro or in silico approaches into experimental designs.
Additional Observations in ctDNA Fragmentation
Although this review has largely focused on the discrete size difference between ctDNA and ccfDNA relative to the mono- and dinucleosomes, other studies have identified additional size-based differences between ctDNA and ccfDNA that should be considered to provide a more complete overview of ctDNA fragment length. In 2016, single-stranded DNA library preparation methods found that ccfDNA shorter than 100 nt was more accessible compared to the more commonly used double-stranded DNA library generation approach because of the latter’s requirement to use size-selective steps to remove adapter dimers, which tends to also eliminate shorter ccfDNA fragments [15, 175]. In 2018, Sanchez et al. used single-stranded DNA library preparation in a small cohort of cancer patients to show highly fragmented ccfDNA with a 10 nt periodicity and a size range of 41 to 167 nt. In contrast, double-stranded DNA library preparation yielded the same periodicity, but was observed only from 81 to 166 bp [176]. Subsequently, Liu et al. used single-stranded library preparation to detect ctDNA present in stage II–III pancreatic ductal adenocarcinoma patients with paired tumour DNA and ccfDNA [177]. In 13 patients, they found evidence of corresponding ctDNA in ~ 69% of patients that were either the KRAS p.G12D or p.G12C variants (median allele frequency of 0.12%; range 0.05–0.56%). Overall, ~ 30% of mutations present in tumour tissue with an MAF > 1% were present in ccfDNA, which were all associated with common KRAS mutations. However, the proportion of pancreatic patients with detected ctDNA was similar to previous reports [44, 45]. In addition, Moser et al. reported that single-stranded DNA library preparation enriched for smaller DNA fragments, but did not preferentially enrich for ctDNA [178]. As such, more direct comparisons between single- and double-stranded DNA library generation inclusive of different preparation methods for both are necessary to determine if the additional steps involved in single-stranded library generation are warranted.
In contrast to previous studies that have shown ctDNA is shorter than ccfDNA, a report in 2019 found that ctDNA fragment length may include both shorter and longer fragments relative to ccfDNA. Cristiano et al. used whole genome sequencing at 1–2 × coverage in 208 cancer patients and analysed the ratio of small (100–150 bp) to large (151–220 bp) ccfDNA fragments to evaluate the fragmentation profile in 504 windows of 5 Mb, covering approximately 2.6 Gb of the genome [160]. They found the ratio indicated both longer and shorter fractions associated with cancer. Moreover, the size profile indicated tissue of origin. Across a range of stages and cancer types, detection of cancer ranged between 68 and 79% at a 98% specificity. The origins of the variation of fragment lengths within the same patient are unclear. The authors hypothesized that the fragmentation pattern in patients with cancer likely resulted from mixtures of nucleosomal DNA from blood and neoplastic cells that reflected both epigenomic and genomic alterations. Although genomic monitoring may not identify patient-specific somatic mutations, broad surveillance of cancer across multiple types of malignancy may be feasible by profiling the fragmentation profile across the entire genome.
Finally, viral genomes appear to have an alternate size profile after integration into somatic cells that may enable viral-driven malignancies to be discerned from an active or concurrent viral infection. Lam et al. studied the size profile of Epstein-Barr virus (EBV) with and without the occurrence of nasopharyngeal carcinoma (NPC) [179]. They observed that EBV DNA in plasma from non-NPC patients were distributed over shorter fragment lengths compared to NPC patients, leading to a higher proportion of EBV DNA molecules shorter than 110 bp in non-NPC patients. Subsequently, the proportion of EBV DNA within 80–110 bp relative to autosomal DNA within 80–110 bp was used to discern EBV-positive patients at a higher risk of harbouring NPC. These observations regarding the viral genome fragmentation pattern may have significant implications for detecting a variety of virus-associated cancers such as human papilloma virus-associated squamous cell carcinoma [180].
Conclusions
Over the past decade, tremendous growth in the development of ccfDNA as a diagnostic tool in oncology has occurred. Challenges associated with the amount of ccfDNA available in plasma (i.e. signal) and errors associated with NGS (i.e. noise) have driven significant advances in the state of the art. Leveraging differences in fragment size between ctDNA and healthy ccfDNA may be one avenue of innovation that further advances clinical applications in oncology. Both in vitro and in silico data for ctDNA enrichment and detection are encouraging. Overall, however, there is a paucity of data to support definitive conclusions regarding utility in clinical diagnostics, particularly for early-stage and non-metastatic cancers. Many key questions persist. For example, does in vitro enrichment for ctDNA adversely affect detection of ultra-low-frequency ctDNA because of losses that may occur during sample recovery? Can in silico probability estimates of ctDNA based on fragment size be used to discover patient-specific somatic mutations without a tumour-informed search? Is a combination of in vitro and in silico methods warranted? Future studies that address these questions, among others, would provide the necessary evidence to establish fragment length as a key component in ccfDNA diagnostics. Additionally, greater inclusion of healthy control data into experimental designs may provide better support for determination of assay specificity, especially as ccfDNA diagnostics advances towards harder to detect cancers. Finally, investigations into the mechanisms that govern the size difference between ctDNA and ccfDNA may prove meaningful not only for detection, but also lead to new therapeutic strategies. Regardless, ctDNA’s fragmentation pattern is an exploitable biologic phenomenon with the potential to enhance ctDNA detection in clinical oncology that may lead to an advanced era of non-invasive, personalized cancer diagnostics to improve patient outcomes.
Acknowledgements
BioRender.com was used for figure generation.
Declarations
Funding
National Cancer Institute of the National Institutes of Health (HRU; R37CA246183).
Conflict of interest
The author (HRU) has a US patent pending (US-2019106737-A1) for using size selection of cell-free DNA to increase family size during next-generation sequencing.
Ethics approval
Not applicable.
Author contributions
HRU is the sole author and is responsible for all content.
References
- 1.Mandel P, Métais P. Les acides nucléiques du plasma sanguin chez l'homme. C R Seances Soc Biol Fil. 1948;142(3–4):241–243. [PubMed] [Google Scholar]
- 2.Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Can Res. 1977;37(3):646–650. [PubMed] [Google Scholar]
- 3.Diehl F, Li M, Dressman D, He Y, Shen D, Szabo S, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci USA. 2005;102(45):16368–16373. doi: 10.1073/pnas.0507904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sozzi G, Conte D, Leon M, Ciricione R, Roz L, Ratcliffe C, et al. Quantification of free circulating DNA as a diagnostic marker in lung cancer. J Clin Oncol. 2003;21(21):3902–3908. doi: 10.1200/JCO.2003.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Huang ZH, Li LH, Hua D. Quantitative analysis of plasma circulating DNA at diagnosis and during follow-up of breast cancer patients. Cancer Lett. 2006;243(1):64–70. doi: 10.1016/j.canlet.2005.11.027. [DOI] [PubMed] [Google Scholar]
- 6.Jylhava J, Kotipelto T, Raitala A, Jylha M, Hervonen A, Hurme M. Aging is associated with quantitative and qualitative changes in circulating cell-free DNA: the vitality 90+ study. Mech Ageing Dev. 2011;132(1–2):20–26. doi: 10.1016/j.mad.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 7.Haghiac M, Vora NL, Basu S, Johnson KL, Presley L, Bianchi DW, et al. Increased death of adipose cells, a path to release cell-free DNA into systemic circulation of obese women. Obesity (Silver Spring) 2012;20(11):2213–2219. doi: 10.1038/oby.2012.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan EM, Schur PH, Carr RI, Kunkel HG. Deoxybonucleic acid (DNA) and antibodies to DNA in the serum of patients with systemic lupus erythematosus. J Clin Investig. 1966;45(11):1732–1740. doi: 10.1172/JCI105479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boender PJ, Heijtink RA, Hellings JA. Nucleosomal fragments in serum may directly reflect cell-mediated cytotoxic activity in vivo. Clin Immunol Immunopathol. 1989;53(1):87–98. doi: 10.1016/0090-1229(89)90104-9. [DOI] [PubMed] [Google Scholar]
- 10.Stawski R, Walczak K, Kosielski P, Meissner P, Budlewski T, Padula G, et al. Repeated bouts of exhaustive exercise increase circulating cell free nuclear and mitochondrial DNA without development of tolerance in healthy men. PLoS One. 2017;12(5):e0178216. doi: 10.1371/journal.pone.0178216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dawson SJ, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin SF, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368(13):1199–1209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- 12.Sorenson GD, Pribish DM, Valone FH, Memoli VA, Bzik DJ, Yao SL. Soluble normal and mutated DNA sequences from single-copy genes in human blood. Cancer Epidemiol Biomark Prev. 1994;3(1):67–71. [PubMed] [Google Scholar]
- 13.Sun K, Jiang P, Chan KC, Wong J, Cheng YK, Liang RH, et al. Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc Natl Acad Sci USA. 2015;112(40):E5503–E5512. doi: 10.1073/pnas.1508736112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moss J, Magenheim J, Neiman D, Zemmour H, Loyfer N, Korach A, et al. Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat Commun. 2018;9(1):5068. doi: 10.1038/s41467-018-07466-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Snyder MW, Kircher M, Hill AJ, Daza RM, Shendure J. Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell. 2016;164(1–2):57–68. doi: 10.1016/j.cell.2015.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laurent D, Semple F, Starkey Lewis PJ, Rose E, Black HA, Coe J, et al. Absolute measurement of the tissue origins of cell-free DNA in the healthy state and following paracetamol overdose. BMC Med Genom. 2020;13(1):60. doi: 10.1186/s12920-020-0705-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Long-Mira E, Ilie M, Chamorey E, Leduff-Blanc F, Montaudie H, Tanga V, et al. Monitoring BRAF and NRAS mutations with cell-free circulating tumor DNA from metastatic melanoma patients. Oncotarget. 2018;9(90):36238–36249. doi: 10.18632/oncotarget.26343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Board RE, Wardley AM, Dixon JM, Armstrong AC, Howell S, Renshaw L, et al. Detection of PIK3CA mutations in circulating free DNA in patients with breast cancer. Breast Cancer Res Treat. 2010;120(2):461–467. doi: 10.1007/s10549-010-0747-9. [DOI] [PubMed] [Google Scholar]
- 19.Schwarzenbach H, Alix-Panabieres C, Muller I, Letang N, Vendrell JP, Rebillard X, et al. Cell-free tumor DNA in blood plasma as a marker for circulating tumor cells in prostate cancer. Clin Cancer Res. 2009;15(3):1032–1038. doi: 10.1158/1078-0432.CCR-08-1910. [DOI] [PubMed] [Google Scholar]
- 20.Taniguchi K, Uchida J, Nishino K, Kumagai T, Okuyama T, Okami J, et al. Quantitative detection of EGFR mutations in circulating tumor DNA derived from lung adenocarcinomas. Clin Cancer Res. 2011;17(24):7808–7815. doi: 10.1158/1078-0432.CCR-11-1712. [DOI] [PubMed] [Google Scholar]
- 21.Panka DJ, Sullivan RJ, Mier JW. An inexpensive, specific and highly sensitive protocol to detect the BrafV600E mutation in melanoma tumor biopsies and blood. Melanoma Res. 2010;20(5):401–407. doi: 10.1097/CMR.0b013e32833d8d48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mulcahy HE, Lyautey J, Lederrey C, Qi CX, Anker P, Alstead EM, et al. A prospective study of K-ras mutations in the plasma of pancreatic cancer patients. Clin Cancer Res. 1998;4(2):271–275. [PubMed] [Google Scholar]
- 23.Castells A, Puig P, Mora J, Boadas J, Boix L, Urgell E, et al. K-ras mutations in DNA extracted from the plasma of patients with pancreatic carcinoma: diagnostic utility and prognostic significance. J Clin Oncol. 1999;17(2):578–584. doi: 10.1200/JCO.1999.17.2.578. [DOI] [PubMed] [Google Scholar]
- 24.Shinozaki M, O'Day SJ, Kitago M, Amersi F, Kuo C, Kim J, et al. Utility of circulating B-RAF DNA mutation in serum for monitoring melanoma patients receiving biochemotherapy. Clin Cancer Res. 2007;13(7):2068–2074. doi: 10.1158/1078-0432.CCR-06-2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forshew T, Murtaza M, Parkinson C, Gale D, Tsui DW, Kaper F, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med. 2012;4(136):136ra68. doi: 10.1126/scitranslmed.3003726. [DOI] [PubMed] [Google Scholar]
- 26.Narayan A, Carriero NJ, Gettinger SN, Kluytenaar J, Kozak KR, Yock TI, et al. Ultrasensitive measurement of hotspot mutations in tumor DNA in blood using error-suppressed multiplexed deep sequencing. Can Res. 2012;72(14):3492–3498. doi: 10.1158/0008-5472.CAN-11-4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6(224):224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nix DA, Hellwig S, Conley C, Thomas A, Fuertes CL, Hamil CL, et al. The stochastic nature of errors in next-generation sequencing of circulating cell-free DNA. PLoS One. 2020;15(2):e0229063. doi: 10.1371/journal.pone.0229063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liao W, Yang H, Xu H, Wang Y, Ge P, Ren J, et al. Noninvasive detection of tumor-associated mutations from circulating cell-free DNA in hepatocellular carcinoma patients by targeted deep sequencing. Oncotarget. 2016;7(26):40481–40490. doi: 10.18632/oncotarget.9629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwaederle M, Husain H, Fanta PT, Piccioni DE, Kesari S, Schwab RB, et al. Detection rate of actionable mutations in diverse cancers using a biopsy-free (blood) circulating tumor cell DNA assay. Oncotarget. 2016;7(9):9707–9717. doi: 10.18632/oncotarget.7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ueda M, Iguchi T, Masuda T, Nakahara Y, Hirata H, Uchi R, et al. Somatic mutations in plasma cell-free DNA are diagnostic markers for esophageal squamous cell carcinoma recurrence. Oncotarget. 2016;7(38):62280–62291. doi: 10.18632/oncotarget.11409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohira T, Sakai K, Matsubayashi J, Kajiwara N, Kakihana M, Hagiwara M, et al. Tumor volume determines the feasibility of cell-free DNA sequencing for mutation detection in non-small cell lung cancer. Cancer Sci. 2016;107(11):1660–1666. doi: 10.1111/cas.13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scherer F, Kurtz DM, Newman AM, Stehr H, Craig AF, Esfahani MS, et al. Distinct biological subtypes and patterns of genome evolution in lymphoma revealed by circulating tumor DNA. Sci Transl Med. 2016;8(364):364ra155. doi: 10.1126/scitranslmed.aai8545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson JC, Yee SS, Troxel AB, Savitch SL, Fan R, Balli D, et al. Detection of therapeutically targetable driver and resistance mutations in lung cancer patients by next-generation sequencing of cell-free circulating tumor DNA. Clin Cancer Res. 2016;22(23):5772–5782. doi: 10.1158/1078-0432.CCR-16-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang DH, Ensor JE, Liu ZB, Patel A, Patel TA, Chang JC, et al. Cell-free DNA as a molecular tool for monitoring disease progression and response to therapy in breast cancer patients. Breast Cancer Res Treat. 2016;155(1):139–149. doi: 10.1007/s10549-015-3635-5. [DOI] [PubMed] [Google Scholar]
- 36.Rachiglio AM, Esposito Abate R, Sacco A, Pasquale R, Fenizia F, Lambiase M, et al. Limits and potential of targeted sequencing analysis of liquid biopsy in patients with lung and colon carcinoma. Oncotarget. 2016;7(41):66595–66605. doi: 10.18632/oncotarget.10704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Newman AM, Lovejoy AF, Klass DM, Kurtz DM, Chabon JJ, Scherer F, et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol. 2016;34(5):547–555. doi: 10.1038/nbt.3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chae YK, Davis AA, Carneiro BA, Chandra S, Mohindra N, Kalyan A, et al. Concordance between genomic alterations assessed by next-generation sequencing in tumor tissue or circulating cell-free DNA. Oncotarget. 2016;7(40):65364–65373. doi: 10.18632/oncotarget.11692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jovelet C, Ileana E, Le Deley MC, Motte N, Rosellini S, Romero A, et al. Circulating cell-free tumor DNA analysis of 50 genes by next-generation sequencing in the prospective MOSCATO trial. Clin Cancer Res. 2016;22(12):2960–2968. doi: 10.1158/1078-0432.CCR-15-2470. [DOI] [PubMed] [Google Scholar]
- 40.Pishvaian MJ, Joseph Bender R, Matrisian LM, Rahib L, Hendifar A, Hoos WA, et al. A pilot study evaluating concordance between blood-based and patient-matched tumor molecular testing within pancreatic cancer patients participating in the Know Your Tumor (KYT) initiative. Oncotarget. 2017;8(48):83446–83456. doi: 10.18632/oncotarget.13225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wyatt AW, Azad AA, Volik SV, Annala M, Beja K, McConeghy B, et al. Genomic alterations in cell-free DNA and enzalutamide resistance in castration-resistant prostate cancer. JAMA Oncol. 2016;2(12):1598–1606. doi: 10.1001/jamaoncol.2016.0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beije N, Helmijr JC, Weerts MJA, Beaufort CM, Wiggin M, Marziali A, et al. Somatic mutation detection using various targeted detection assays in paired samples of circulating tumor DNA, primary tumor and metastases from patients undergoing resection of colorectal liver metastases. Mol Oncol. 2016;10(10):1575–1584. doi: 10.1016/j.molonc.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Phallen J, Sausen M, Adleff V, Leal A, Hruban C, White J, et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med. 2017;9(403):eaan2415. [DOI] [PMC free article] [PubMed]
- 44.Cohen JD, Javed AA, Thoburn C, Wong F, Tie J, Gibbs P, et al. Combined circulating tumor DNA and protein biomarker-based liquid biopsy for the earlier detection of pancreatic cancers. Proc Natl Acad Sci USA. 2017;114(38):10202–10207. doi: 10.1073/pnas.1704961114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vietsch EE, Graham GT, McCutcheon JN, Javaid A, Giaccone G, Marshall JL, et al. Circulating cell-free DNA mutation patterns in early and late stage colon and pancreatic cancer. Cancer Genet. 2017;218–219:39–50. doi: 10.1016/j.cancergen.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iwama E, Sakai K, Azuma K, Harada T, Harada D, Nosaki K, et al. Monitoring of somatic mutations in circulating cell-free DNA by digital PCR and next-generation sequencing during afatinib treatment in patients with lung adenocarcinoma positive for EGFR activating mutations. Ann Oncol. 2017;28(1):136–141. doi: 10.1093/annonc/mdw531. [DOI] [PubMed] [Google Scholar]
- 47.Yao Y, Liu J, Li L, Yuan Y, Nan K, Wu X, et al. Detection of circulating tumor DNA in patients with advanced non-small cell lung cancer. Oncotarget. 2017;8(2):2130–2140. doi: 10.18632/oncotarget.12883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mao X, Zhang Z, Zheng X, Xie F, Duan F, Jiang L, et al. Capture-based targeted ultradeep sequencing in paired tissue and plasma samples demonstrates differential subclonal ctDNA-releasing capability in advanced lung cancer. J Thorac Oncol. 2017;12(4):663–672. doi: 10.1016/j.jtho.2016.11.2235. [DOI] [PubMed] [Google Scholar]
- 49.Janku F, Zhang S, Waters J, Liu L, Huang HJ, Subbiah V, et al. Development and validation of an ultradeep next-generation sequencing assay for testing of plasma cell-free DNA from patients with advanced cancer. Clin Cancer Res. 2017;23(18):5648–5656. doi: 10.1158/1078-0432.CCR-17-0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bartels S, Persing S, Hasemeier B, Schipper E, Kreipe H, Lehmann U. Molecular analysis of circulating cell-free DNA from lung cancer patients in routine laboratory practice: a cross-platform comparison of three different molecular methods for mutation detection. J Mol Diagn. 2017;19(5):722–732. doi: 10.1016/j.jmoldx.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 51.Sandulache VC, Williams MD, Lai SY, Lu C, William WN, Busaidy NL, et al. Real-time genomic characterization utilizing circulating cell-free DNA in patients with anaplastic thyroid carcinoma. Thyroid. 2017;27(1):81–87. doi: 10.1089/thy.2016.0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maxwell KN, Soucier-Ernst D, Tahirovic E, Troxel AB, Clark C, Feldman M, et al. Comparative clinical utility of tumor genomic testing and cell-free DNA in metastatic breast cancer. Breast Cancer Res Treat. 2017;164(3):627–638. doi: 10.1007/s10549-017-4257-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adalsteinsson VA, Ha G, Freeman SS, Choudhury AD, Stover DG, Parsons HA, et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat Commun. 2017;8(1):1324. doi: 10.1038/s41467-017-00965-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wyatt AW, Annala M, Aggarwal R, Beja K, Feng F, Youngren J, et al. Concordance of circulating tumor DNA and matched metastatic tissue biopsy in prostate cancer. J Natl Cancer Inst. 2017;109(12):djx118. [DOI] [PMC free article] [PubMed]
- 55.Page K, Guttery DS, Fernandez-Garcia D, Hills A, Hastings RK, Luo J, et al. Next generation sequencing of circulating cell-free DNA for evaluating mutations and gene amplification in metastatic breast cancer. Clin Chem. 2017;63(2):532–541. doi: 10.1373/clinchem.2016.261834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goldstein A, Toro PV, Lee J, Silberstein JL, Nakazawa M, Waters I, et al. Detection fidelity of AR mutations in plasma derived cell-free DNA. Oncotarget. 2017;8(9):15651–15662. doi: 10.18632/oncotarget.14926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pereira AAL, Morelli MP, Overman M, Kee B, Fogelman D, Vilar E, et al. Clinical utility of circulating cell-free DNA in advanced colorectal cancer. PLoS One. 2017;12(8):e0183949. doi: 10.1371/journal.pone.0183949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koeppel F, Blanchard S, Jovelet C, Genin B, Marcaillou C, Martin E, et al. Whole exome sequencing for determination of tumor mutation load in liquid biopsy from advanced cancer patients. PLoS One. 2017;12(11):e0188174. doi: 10.1371/journal.pone.0188174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim ST, Banks KC, Lee SH, Kim K, Park JO, Park SH, et al. Prospective feasibility study for using cell-free circulating tumor DNA-guided therapy in refractory metastatic solid cancers: an interim analysis. JCO Precis Oncol. 2017;1:1–15. doi: 10.1200/PO.16.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guo Q, Wang J, Xiao J, Wang L, Hu X, Yu W, et al. Heterogeneous mutation pattern in tumor tissue and circulating tumor DNA warrants parallel NGS panel testing. Mol Cancer. 2018;17(1):131. doi: 10.1186/s12943-018-0875-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chicard M, Colmet-Daage L, Clement N, Danzon A, Bohec M, Bernard V, et al. Whole-exome sequencing of cell-free DNA reveals temporo-spatial heterogeneity and identifies treatment-resistant clones in neuroblastoma. Clin Cancer Res. 2018;24(4):939–949. doi: 10.1158/1078-0432.CCR-17-1586. [DOI] [PubMed] [Google Scholar]
- 62.Kurtz DM, Scherer F, Jin MC, Soo J, Craig AFM, Esfahani MS, et al. Circulating tumor DNA measurements as early outcome predictors in diffuse large B-cell lymphoma. J Clin Oncol. 2018;36(28):2845–2853. doi: 10.1200/JCO.2018.78.5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ng CKY, Di Costanzo GG, Tosti N, Paradiso V, Coto-Llerena M, Roscigno G, et al. Genetic profiling using plasma-derived cell-free DNA in therapy-naive hepatocellular carcinoma patients: a pilot study. Ann Oncol. 2018;29(5):1286–1291. doi: 10.1093/annonc/mdy083. [DOI] [PubMed] [Google Scholar]
- 64.Veldore VH, Choughule A, Routhu T, Mandloi N, Noronha V, Joshi A, et al. Validation of liquid biopsy: plasma cell-free DNA testing in clinical management of advanced non-small cell lung cancer. Lung Cancer (Auckl). 2018;9:1–11. doi: 10.2147/LCTT.S147841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guibert N, Hu Y, Feeney N, Kuang Y, Plagnol V, Jones G, et al. Amplicon-based next-generation sequencing of plasma cell-free DNA for detection of driver and resistance mutations in advanced non-small cell lung cancer. Ann Oncol. 2018;29(4):1049–1055. doi: 10.1093/annonc/mdy005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Helman E, Nguyen M, Karlovich CA, Despain D, Choquette AK, Spira AI, et al. Cell-free DNA next-generation sequencing prediction of response and resistance to third-generation EGFR inhibitor. Clin Lung Cancer. 2018;19(6):518–530. doi: 10.1016/j.cllc.2018.07.008. [DOI] [PubMed] [Google Scholar]
- 67.Pasternack H, Fassunke J, Plum PS, Chon SH, Hescheler DA, Gassa A, et al. Somatic alterations in circulating cell-free DNA of oesophageal carcinoma patients during primary staging are indicative for post-surgical tumour recurrence. Sci Rep. 2018;8(1):14941. doi: 10.1038/s41598-018-33027-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mouliere F, Chandrananda D, Piskorz AM, Moore EK, Morris J, Ahlborn LB, et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci Transl Med. 2018;10(466):eaat4921. [DOI] [PMC free article] [PubMed]
- 69.Tsui DWY, Murtaza M, Wong ASC, Rueda OM, Smith CG, Chandrananda D, et al. Dynamics of multiple resistance mechanisms in plasma DNA during EGFR-targeted therapies in non-small cell lung cancer. EMBO Mol Med. 2018;10(6):e7945. doi: 10.15252/emmm.201707945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu L, Liu H, Shao D, Liu Z, Wang J, Deng Q, et al. Development and clinical validation of a circulating tumor DNA test for the identification of clinically actionable mutations in nonsmall cell lung cancer. Genes Chromosomes Cancer. 2018;57(4):211–220. doi: 10.1002/gcc.22522. [DOI] [PubMed] [Google Scholar]
- 71.Gangadhar TC, Savitch SL, Yee SS, Xu W, Huang AC, Harmon S, et al. Feasibility of monitoring advanced melanoma patients using cell-free DNA from plasma. Pigment Cell Melanoma Res. 2018;31(1):73–81. doi: 10.1111/pcmr.12623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Laufer-Geva S, Rozenblum AB, Twito T, Grinberg R, Dvir A, Soussan-Gutman L, et al. The clinical impact of comprehensive genomic testing of circulating cell-free DNA in advanced lung cancer. J Thorac Oncol. 2018;13(11):1705–1716. doi: 10.1016/j.jtho.2018.07.101. [DOI] [PubMed] [Google Scholar]
- 73.Strickler JH, Loree JM, Ahronian LG, Parikh AR, Niedzwiecki D, Pereira AAL, et al. Genomic landscape of cell-free DNA in patients with colorectal cancer. Cancer Discov. 2018;8(2):164–173. doi: 10.1158/2159-8290.CD-17-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zill OA, Banks KC, Fairclough SR, Mortimer SA, Vowles JV, Mokhtari R, et al. The Landscape Of Actionable Genomic Alterations In Cell-Free Circulating Tumor DNA from 21,807 advanced cancer patients. Clin Cancer Res. 2018;24(15):3528–3538. doi: 10.1158/1078-0432.CCR-17-3837. [DOI] [PubMed] [Google Scholar]
- 75.Perets R, Greenberg O, Shentzer T, Semenisty V, Epelbaum R, Bick T, et al. Mutant KRAS circulating tumor DNA is an accurate tool for pancreatic cancer monitoring. Oncologist. 2018;23(5):566–572. doi: 10.1634/theoncologist.2017-0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Masunaga N, Kagara N, Motooka D, Nakamura S, Miyake T, Tanei T, et al. Highly sensitive detection of ESR1 mutations in cell-free DNA from patients with metastatic breast cancer using molecular barcode sequencing. Breast Cancer Res Treat. 2018;167(1):49–58. doi: 10.1007/s10549-017-4487-y. [DOI] [PubMed] [Google Scholar]
- 77.Goldberg SB, Narayan A, Kole AJ, Decker RH, Teysir J, Carriero NJ, et al. Early assessment of lung cancer immunotherapy response via circulating tumor DNA. Clin Cancer Res. 2018;24(8):1872–1880. doi: 10.1158/1078-0432.CCR-17-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Belic J, Graf R, Bauernhofer T, Cherkas Y, Ulz P, Waldispuehl-Geigl J, et al. Genomic alterations in plasma DNA from patients with metastasized prostate cancer receiving abiraterone or enzalutamide. Int J Cancer. 2018;143(5):1236–1248. doi: 10.1002/ijc.31397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Annala M, Vandekerkhove G, Khalaf D, Taavitsainen S, Beja K, Warner EW, et al. Circulating tumor DNA genomics correlate with resistance to abiraterone and enzalutamide in prostate cancer. Cancer Discov. 2018;8(4):444–457. doi: 10.1158/2159-8290.CD-17-0937. [DOI] [PubMed] [Google Scholar]
- 80.Lupini L, Moretti A, Bassi C, Schirone A, Pedriali M, Querzoli P, et al. High-sensitivity assay for monitoring ESR1 mutations in circulating cell-free DNA of breast cancer patients receiving endocrine therapy. Sci Rep. 2018;8(1):4371. doi: 10.1038/s41598-018-22312-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mansukhani S, Barber LJ, Kleftogiannis D, Moorcraft SY, Davidson M, Woolston A, et al. Ultra-sensitive mutation detection and genome-wide DNA copy number reconstruction by error-corrected circulating tumor DNA sequencing. Clin Chem. 2018;64(11):1626–1635. doi: 10.1373/clinchem.2018.289629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xiong Y, Xie CR, Zhang S, Chen J, Yin ZY. Detection of a novel panel of somatic mutations in plasma cell-free DNA and its diagnostic value in hepatocellular carcinoma. Cancer Manag Res. 2019;11:5745–5756. doi: 10.2147/CMAR.S197455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Reinert T, Henriksen TV, Christensen E, Sharma S, Salari R, Sethi H, et al. Analysis of plasma cell-free DNA by ultradeep sequencing in patients with stages I to III colorectal cancer. JAMA Oncol. 2019;5(8):1124–1131. doi: 10.1001/jamaoncol.2019.0528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Peng M, Xie Y, Li X, Qian Y, Tu X, Yao X, et al. Resectable lung lesions malignancy assessment and cancer detection by ultra-deep sequencing of targeted gene mutations in plasma cell-free DNA. J Med Genet. 2019;56(10):647–653. doi: 10.1136/jmedgenet-2018-105825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sun P, Chen C, Xia Y, Wang Y, Liu PP, Bi XW, et al. Mutation profiling of malignant lymphoma by next-generation sequencing of circulating cell-free DNA. J Cancer. 2019;10(2):323–331. doi: 10.7150/jca.27615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Meng P, Wei J, Geng Y, Chen S, Terpstra MM, Huang Q, et al. Targeted sequencing of circulating cell-free DNA in stage II–III resectable oesophageal squamous cell carcinoma patients. BMC Cancer. 2019;19(1):818. doi: 10.1186/s12885-019-6025-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mohan S, Ayub M, Rothwell DG, Gulati S, Kilerci B, Hollebecque A, et al. Analysis of circulating cell-free DNA identifies KRAS copy number gain and mutation as a novel prognostic marker in pancreatic cancer. Sci Rep. 2019;9(1):11610. doi: 10.1038/s41598-019-47489-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang Y, Chang L, Yang Y, Fang W, Guan Y, Wu A, et al. Intratumor heterogeneity comparison among different subtypes of non-small-cell lung cancer through multi-region tissue and matched ctDNA sequencing. Mol Cancer. 2019;18(1):7. doi: 10.1186/s12943-019-0939-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Takeda K, Yamada T, Takahashi G, Iwai T, Ueda K, Kuriyama S, et al. Analysis of colorectal cancer-related mutations by liquid biopsy: Utility of circulating cell-free DNA and circulating tumor cells. Cancer Sci. 2019;110(11):3497–3509. doi: 10.1111/cas.14186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang B, Niu X, Zhang Q, Wang C, Liu B, Yue D, et al. Circulating tumor DNA detection is correlated to histologic types in patients with early-stage non-small-cell lung cancer. Lung Cancer. 2019;134:108–116. doi: 10.1016/j.lungcan.2019.05.034. [DOI] [PubMed] [Google Scholar]
- 91.Chen Y, Han T, Zhou Y, Mao B, Zhuang W. Comparing the efficacy of targeted next-generation sequencing in the identification of somatic mutations in circulating tumor DNA from different stages of lung cancer. Neoplasma. 2019;66(4):652–660. doi: 10.4149/neo_2018_181130N910. [DOI] [PubMed] [Google Scholar]
- 92.An Y, Guan Y, Xu Y, Han Y, Wu C, Bao C, et al. The diagnostic and prognostic usage of circulating tumor DNA in operable hepatocellular carcinoma. Am J Transl Res. 2019;11(10):6462–6474. [PMC free article] [PubMed] [Google Scholar]
- 93.Howell J, Atkinson SR, Pinato DJ, Knapp S, Ward C, Minisini R, et al. Identification of mutations in circulating cell-free tumour DNA as a biomarker in hepatocellular carcinoma. Eur J Cancer. 2019;116:56–66. doi: 10.1016/j.ejca.2019.04.014. [DOI] [PubMed] [Google Scholar]
- 94.Guibert N, Jones G, Beeler JF, Plagnol V, Morris C, Mourlanette J, et al. Targeted sequencing of plasma cell-free DNA to predict response to PD1 inhibitors in advanced non-small cell lung cancer. Lung Cancer. 2019;137:1–6. doi: 10.1016/j.lungcan.2019.09.005. [DOI] [PubMed] [Google Scholar]
- 95.Shi X, Duose DY, Mehrotra M, Harmon MA, Hu P, Wistuba II, et al. Non-invasive genotyping of metastatic colorectal cancer using circulating cell free DNA. Cancer Genet. 2019;237:82–89. doi: 10.1016/j.cancergen.2019.06.004. [DOI] [PubMed] [Google Scholar]
- 96.Wei T, Zhang Q, Li X, Su W, Li G, Ma T, et al. Monitoring tumor burden in response to FOLFIRINOX chemotherapy via profiling circulating cell-free DNA in pancreatic cancer. Mol Cancer Ther. 2019;18(1):196–203. doi: 10.1158/1535-7163.MCT-17-1298. [DOI] [PubMed] [Google Scholar]
- 97.Yamamoto Y, Uemura M, Fujita M, Maejima K, Koh Y, Matsushita M, et al. Clinical significance of the mutational landscape and fragmentation of circulating tumor DNA in renal cell carcinoma. Cancer Sci. 2019;110(2):617–628. doi: 10.1111/cas.13906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zugazagoitia J, Ramos I, Trigo JM, Palka M, Gomez-Rueda A, Jantus-Lewintre E, et al. Clinical utility of plasma-based digital next-generation sequencing in patients with advance-stage lung adenocarcinomas with insufficient tumor samples for tissue genotyping. Ann Oncol. 2019;30(2):290–296. doi: 10.1093/annonc/mdy512. [DOI] [PubMed] [Google Scholar]
- 99.Kaseb AO, Sanchez NS, Sen S, Kelley RK, Tan B, Bocobo AG, et al. Molecular profiling of hepatocellular carcinoma using circulating cell-free DNA. Clin Cancer Res. 2019;25(20):6107–6118. doi: 10.1158/1078-0432.CCR-18-3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sanchez NS, Kahle MP, Bailey AM, Wathoo C, Balaji K, Demirhan ME, et al. Identification of actionable genomic alterations using circulating cell-free DNA. JCO Precis Oncol. 2019;3:1–10. doi: 10.1200/PO.19.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Osumi H, Shinozaki E, Takeda Y, Wakatsuki T, Ichimura T, Saiura A, et al. Clinical relevance of circulating tumor DNA assessed through deep sequencing in patients with metastatic colorectal cancer. Cancer Med. 2019;8(1):408–417. doi: 10.1002/cam4.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sonpavde G, Agarwal N, Pond GR, Nagy RJ, Nussenzveig RH, Hahn AW, et al. Circulating tumor DNA alterations in patients with metastatic castration-resistant prostate cancer. Cancer. 2019;125(9):1459–1469. doi: 10.1002/cncr.31959. [DOI] [PubMed] [Google Scholar]
- 103.Keup C, Storbeck M, Hauch S, Hahn P, Sprenger-Haussels M, Tewes M, et al. Cell-free DNA variant sequencing using CTC-depleted blood for comprehensive liquid biopsy testing in metastatic breast cancer. Cancers (Basel) 2019;11(2):238. doi: 10.3390/cancers11020238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Li S, Wang X, Li Y, Lai H, Liu Y, Jin L. Non-invasive analysis of tumor mutation profiles and druggable mutations by sequencing of cell free DNA of Chinese metastatic breast cancer patients. Thorac Cancer. 2019;10(4):807–814. doi: 10.1111/1759-7714.13002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li BT, Janku F, Jung B, Hou C, Madwani K, Alden R, et al. Ultra-deep next-generation sequencing of plasma cell-free DNA in patients with advanced lung cancers: results from the Actionable Genome Consortium. Ann Oncol. 2019;30(4):597–603. doi: 10.1093/annonc/mdz046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Torquato S, Pallavajjala A, Goldstein A, Toro PV, Silberstein JL, Lee J, et al. Genetic alterations detected in cell-free DNA are associated with enzalutamide and abiraterone resistance in castration-resistant prostate cancer. JCO Precis Oncol. 2019;3:1–14. doi: 10.1200/PO.18.00227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.van Helden EJ, Angus L, van der Houven M, van Oordt CW, Heideman DAM, Boon E, van Es SC, et al. RAS and BRAF mutations in cell-free DNA are predictive for outcome of cetuximab monotherapy in patients with tissue-tested RAS wild-type advanced colorectal cancer. Mol Oncol. 2019;13(11):2361–2374. doi: 10.1002/1878-0261.12550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Taavitsainen S, Annala M, Ledet E, Beja K, Miller PJ, Moses M, et al. Evaluation of commercial circulating tumor DNA test in metastatic prostate cancer. JCO Precis Oncol. 2019;3:1–9. doi: 10.1200/PO.19.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jiang J, Adams HP, Yao L, Yaung S, Lal P, Balasubramanyam A, et al. Concordance of genomic alterations by next-generation sequencing in tumor tissue versus cell-free DNA in stage I–IV non-small cell lung cancer. J Mol Diagn. 2020;22(2):228–235. doi: 10.1016/j.jmoldx.2019.10.013. [DOI] [PubMed] [Google Scholar]
- 110.Ottestad AL, Wahl SGF, Gronberg BH, Skorpen F, Dai HY. The relevance of tumor mutation profiling in interpretation of NGS data from cell-free DNA in non-small cell lung cancer patients. Exp Mol Pathol. 2020;112:104347. doi: 10.1016/j.yexmp.2019.104347. [DOI] [PubMed] [Google Scholar]
- 111.Serrano C, Vivancos A, Lopez-Pousa A, Matito J, Mancuso FM, Valverde C, et al. Clinical value of next generation sequencing of plasma cell-free DNA in gastrointestinal stromal tumors. BMC Cancer. 2020;20(1):99. doi: 10.1186/s12885-020-6597-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chabon JJ, Hamilton EG, Kurtz DM, Esfahani MS, Moding EJ, Stehr H, et al. Integrating genomic features for non-invasive early lung cancer detection. Nature. 2020;580(7802):245–251. doi: 10.1038/s41586-020-2140-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Radovich M, Jiang G, Hancock BA, Chitambar C, Nanda R, Falkson C, et al. Association of circulating tumor DNA and circulating tumor cells after neoadjuvant chemotherapy with disease recurrence in patients with triple-negative breast cancer: preplanned secondary analysis of the BRE12-158 randomized clinical trial. JAMA Oncol. 2020;6(9):1410–1415. doi: 10.1001/jamaoncol.2020.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ohara S, Suda K, Sakai K, Nishino M, Chiba M, Shimoji M, et al. Prognostic implications of preoperative versus postoperative circulating tumor DNA in surgically resected lung cancer patients: a pilot study. Transl Lung Cancer Res. 2020;9(5):1915–1923. doi: 10.21037/tlcr-20-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Smith CG, Moser T, Mouliere F, Field-Rayner J, Eldridge M, Riediger AL, et al. Comprehensive characterization of cell-free tumor DNA in plasma and urine of patients with renal tumors. Genome Med. 2020;12(1):23. doi: 10.1186/s13073-020-00723-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ishida Y, Takano S, Maekawa S, Yamaguchi T, Yoshida T, Kobayashi S, et al. Fractionated small cell-free DNA increases possibility to detect cancer-related gene mutations in advanced colorectal cancer. JGH Open. 2020;4(5):978–986. doi: 10.1002/jgh3.12379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Openshaw MR, Mohamed AA, Ottolini B, Fernandez-Garcia D, Richards CJ, Page K, et al. Longitudinal monitoring of circulating tumour DNA improves prognostication and relapse detection in gastroesophageal adenocarcinoma. Br J Cancer. 2020;123(8):1271–1279. doi: 10.1038/s41416-020-1002-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lan X, Bao H, Ge X, Cao J, Fan X, Zhang Q, et al. Genomic landscape of metastatic papillary thyroid carcinoma and novel biomarkers for predicting distant metastasis. Cancer Sci. 2020;111(6):2163–2173. doi: 10.1111/cas.14389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zviran A, Schulman RC, Shah M, Hill STK, Deochand S, Khamnei CC, et al. Genome-wide cell-free DNA mutational integration enables ultra-sensitive cancer monitoring. Nat Med. 2020;26(7):1114–1124. doi: 10.1038/s41591-020-0915-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhao W, Qiu L, Liu H, Xu Y, Zhan M, Zhang W, et al. Circulating tumor DNA as a potential prognostic and predictive biomarker during interventional therapy of unresectable primary liver cancer. J Gastrointest Oncol. 2020;11(5):1065–1077. doi: 10.21037/jgo-20-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wan JCM, Heider K, Gale D, Murphy S, Fisher E, Mouliere F, et al. ctDNA monitoring using patient-specific sequencing and integration of variant reads. Sci Transl Med. 2020;12(548):eaaz8084. [DOI] [PubMed]
- 122.Macgregor-Das A, Yu J, Tamura K, Abe T, Suenaga M, Shindo K, et al. Detection of circulating tumor DNA in patients with pancreatic cancer using digital next-generation sequencing. J Mol Diagn. 2020;22(6):748–756. doi: 10.1016/j.jmoldx.2020.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Xu B, Shan G, Wu Q, Li W, Wang H, Li H, et al. Concordance of genomic alterations between circulating tumor DNA and matched tumor tissue in Chinese patients with breast cancer. J Oncol. 2020;2020:4259293. doi: 10.1155/2020/4259293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Leal A, van Grieken NCT, Palsgrove DN, Phallen J, Medina JE, Hruban C, et al. White blood cell and cell-free DNA analyses for detection of residual disease in gastric cancer. Nat Commun. 2020;11(1):525. doi: 10.1038/s41467-020-14310-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kleftogiannis D, Ho D, Liew JX, Poon PSY, Gan A, Ng RC, et al. Detection of genomic alterations in breast cancer with circulating tumour DNA sequencing. Sci Rep. 2020;10(1):16774. doi: 10.1038/s41598-020-72818-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Iwama E, Sakai K, Hidaka N, Inoue K, Fujii A, Nakagaki N, et al. Longitudinal monitoring of somatic genetic alterations in circulating cell-free DNA during treatment with epidermal growth factor receptor-tyrosine kinase inhibitors. Cancer. 2020;126(1):219–227. doi: 10.1002/cncr.32481. [DOI] [PubMed] [Google Scholar]
- 127.Fukuhara T, Saito H, Furuya N, Watanabe K, Sugawara S, Iwasawa S, et al. Evaluation of plasma EGFR mutation as an early predictor of response of erlotinib plus bevacizumab treatment in the NEJ026 study. EBioMedicine. 2020;57:102861. doi: 10.1016/j.ebiom.2020.102861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Winn JS, Hasse Z, Slifker M, Pei J, Arisi-Fernandez SM, Talarchek JN, et al. Genetic variants detected using cell-free DNA from blood and tumor samples in patients with inflammatory breast cancer. Int J Mol Sci. 2020;21(4):1290. doi: 10.3390/ijms21041290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Razavi P, Li BT, Brown DN, Jung B, Hubbell E, Shen R, et al. High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat Med. 2019;25(12):1928–1937. doi: 10.1038/s41591-019-0652-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Liu HE, Vuppalapaty M, Wilkerson C, Renier C, Chiu M, Lemaire C, et al. Detection of EGFR mutations in cfDNA and CTCs, and comparison to tumor tissue in non-small-cell-lung-cancer (NSCLC) patients. Front Oncol. 2020;10:572895. doi: 10.3389/fonc.2020.572895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bagley SJ, Nabavizadeh SA, Mays JJ, Till JE, Ware JB, Levy S, et al. Clinical utility of plasma cell-free DNA in adult patients with newly diagnosed glioblastoma: a pilot prospective study. Clin Cancer Res. 2020;26(2):397–407. doi: 10.1158/1078-0432.CCR-19-2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.He Y, Ma X, Chen K, Liu F, Cai S, Han-Zhang H, et al. Perioperative circulating tumor DNA in colorectal liver metastases: concordance with metastatic tissue and predictive value for tumor burden and prognosis. Cancer Manag Res. 2020;12:1621–1630. doi: 10.2147/CMAR.S240869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bustamante Alvarez JG, Janse S, Owen DH, Kiourtsis S, Bertino EM, He K, et al. Treatment of non-small-cell lung cancer based on circulating cell-free DNA and impact of variation allele frequency. Clin Lung Cancer. 2020. 10.1016/j.cllc.2020.11.007. [DOI] [PubMed]
- 134.Palmieri M, Baldassarri M, Fava F, Fabbiani A, Gelli E, Tita R, et al. Two-point-NGS analysis of cancer genes in cell-free DNA of metastatic cancer patients. Cancer Med. 2020;9(6):2052–2061. doi: 10.1002/cam4.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Pairawan S, Hess KR, Janku F, Sanchez NS, Mills Shaw KR, Eng C, et al. Cell-free circulating tumor DNA variant allele frequency associates with survival in metastatic cancer. Clin Cancer Res. 2020;26(8):1924–1931. doi: 10.1158/1078-0432.CCR-19-0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Shibayama T, Low SK, Ono M, Kobayashi T, Kobayashi K, Fukada I, et al. Clinical significance of gene mutation in ctDNA analysis for hormone receptor-positive metastatic breast cancer. Breast Cancer Res Treat. 2020;180(2):331–341. doi: 10.1007/s10549-019-05512-5. [DOI] [PubMed] [Google Scholar]
- 137.Davis AA, Jacob S, Gerratana L, Shah AN, Wehbe F, Katam N, et al. Landscape of circulating tumour DNA in metastatic breast cancer. EBioMedicine. 2020;58:102914. doi: 10.1016/j.ebiom.2020.102914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Keup C, Benyaa K, Hauch S, Sprenger-Haussels M, Tewes M, Mach P, et al. Targeted deep sequencing revealed variants in cell-free DNA of hormone receptor-positive metastatic breast cancer patients. Cell Mol Life Sci. 2020;77(3):497–509. doi: 10.1007/s00018-019-03189-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Gupta R, Othman T, Chen C, Sandhu J, Ouyang C, Fakih M. Guardant360 circulating tumor DNA assay is concordant with FoundationOne next-generation sequencing in detecting actionable driver mutations in anti-EGFR Naive metastatic colorectal cancer. Oncologist. 2020;25(3):235–243. doi: 10.1634/theoncologist.2019-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kohli M, Tan W, Zheng T, Wang A, Montesinos C, Wong C, et al. Clinical and genomic insights into circulating tumor DNA-based alterations across the spectrum of metastatic hormone-sensitive and castrate-resistant prostate cancer. EBioMedicine. 2020;54:102728. doi: 10.1016/j.ebiom.2020.102728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Keup C, Storbeck M, Hauch S, Hahn P, Sprenger-Haussels M, Hoffmann O, et al. Multimodal targeted deep sequencing of circulating tumor cells and matched cell-free DNA provides a more comprehensive tool to identify therapeutic targets in metastatic breast cancer patients. Cancers (Basel). 2020;12(5):1084. doi: 10.3390/cancers12051084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Dietz S, Christopoulos P, Yuan Z, Angeles AK, Gu L, Volckmar AL, et al. Longitudinal therapy monitoring of ALK-positive lung cancer by combined copy number and targeted mutation profiling of cell-free DNA. EBioMedicine. 2020;62:103103. doi: 10.1016/j.ebiom.2020.103103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Newman AM, Lovejoy AF, Klass DM, Kurtz DM, Chabon JJ, Scherer F, et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol. 2016;34(5):547–555. doi: 10.1038/nbt.3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Jiang P, Lo YM. The long and short of circulating cell-free DNA and the ins and outs of molecular diagnostics. Trends Genet. 2016;32(6):360–371. doi: 10.1016/j.tig.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 145.Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD, et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Can Res. 2001;61(4):1659–1665. [PubMed] [Google Scholar]
- 146.Thierry AR, Mouliere F, Gongora C, Ollier J, Robert B, Ychou M, et al. Origin and quantification of circulating DNA in mice with human colorectal cancer xenografts. Nucleic Acids Res. 2010;38(18):6159–6175. doi: 10.1093/nar/gkq421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Mouliere F, Robert B, Arnau Peyrotte E, Del Rio M, Ychou M, Molina F, et al. High fragmentation characterizes tumour-derived circulating DNA. PLoS One. 2011;6(9):e23418. doi: 10.1371/journal.pone.0023418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ellinger J, Bastian PJ, Ellinger N, Kahl P, Perabo FG, Buttner R, et al. Apoptotic DNA fragments in serum of patients with muscle invasive bladder cancer: a prognostic entity. Cancer Lett. 2008;264(2):274–280. doi: 10.1016/j.canlet.2008.01.038. [DOI] [PubMed] [Google Scholar]
- 149.Jiang P, Chan CW, Chan KC, Cheng SH, Wong J, Wong VW, et al. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc Natl Acad Sci USA. 2015;112(11):E1317–E1325. doi: 10.1073/pnas.1500076112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Underhill HR, Kitzman JO, Hellwig S, Welker NC, Daza R, Baker DN, et al. Fragment length of circulating tumor DNA. PLoS Genet. 2016;12(7):e1006162. doi: 10.1371/journal.pgen.1006162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Giacona MB, Ruben GC, Iczkowski KA, Roos TB, Porter DM, Sorenson GD. Cell-free DNA in human blood plasma: length measurements in patients with pancreatic cancer and healthy controls. Pancreas. 1998;17(1):89–97. doi: 10.1097/00006676-199807000-00012. [DOI] [PubMed] [Google Scholar]
- 152.Lo YM, Chan KC, Sun H, Chen EZ, Jiang P, Lun FM, et al. Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci Transl Med. 2010;2(61):61ra91. doi: 10.1126/scitranslmed.3001720. [DOI] [PubMed] [Google Scholar]
- 153.Zheng YW, Chan KC, Sun H, Jiang P, Su X, Chen EZ, et al. Nonhematopoietically derived DNA is shorter than hematopoietically derived DNA in plasma: a transplantation model. Clin Chem. 2012;58(3):549–558. doi: 10.1373/clinchem.2011.169318. [DOI] [PubMed] [Google Scholar]
- 154.Lapin M, Oltedal S, Tjensvoll K, Buhl T, Smaaland R, Garresori H, et al. Fragment size and level of cell-free DNA provide prognostic information in patients with advanced pancreatic cancer. J Transl Med. 2018;16(1):300. doi: 10.1186/s12967-018-1677-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hellwig S, Nix DA, Gligorich KM, O'Shea JM, Thomas A, Fuertes CL, et al. Automated size selection for short cell-free DNA fragments enriches for circulating tumor DNA and improves error correction during next generation sequencing. PLoS One. 2018;13(7):e0197333. doi: 10.1371/journal.pone.0197333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Shi J, Zhang R, Li J, Zhang R. Size profile of cell-free DNA: a beacon guiding the practice and innovation of clinical testing. Theranostics. 2020;10(11):4737–4748. doi: 10.7150/thno.42565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Cheng THT, Lui KO, Peng XL, Cheng SH, Jiang P, Chan KCA, et al. DNase1 does not appear to play a major role in the fragmentation of plasma DNA in a knockout mouse model. Clin Chem. 2018;64(2):406–408. doi: 10.1373/clinchem.2017.280446. [DOI] [PubMed] [Google Scholar]
- 158.Han DSC, Ni M, Chan RWY, Chan VWH, Lui KO, Chiu RWK, et al. The biology of cell-free DNA fragmentation and the roles of DNASE1, DNASE1L3, and DFFB. Am J Hum Genet. 2020;106(2):202–214. doi: 10.1016/j.ajhg.2020.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Guo J, Ma K, Bao H, Ma X, Xu Y, Wu X, et al. Quantitative characterization of tumor cell-free DNA shortening. BMC Genom. 2020;21(1):473. doi: 10.1186/s12864-020-06848-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Cristiano S, Leal A, Phallen J, Fiksel J, Adleff V, Bruhm DC, et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature. 2019;570(7761):385–389. doi: 10.1038/s41586-019-1272-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ivanov M, Baranova A, Butler T, Spellman P, Mileyko V. Non-random fragmentation patterns in circulating cell-free DNA reflect epigenetic regulation. BMC Genom. 2015;16(Suppl 13):S1. doi: 10.1186/1471-2164-16-S13-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.van der Pol Y, Mouliere F. Toward the early detection of cancer by decoding the epigenetic and environmental fingerprints of cell-free DNA. Cancer Cell. 2019;36(4):350–368. doi: 10.1016/j.ccell.2019.09.003. [DOI] [PubMed] [Google Scholar]
- 163.Hahn T, Drese KS, O'Sullivan CK. Microsystem for isolation of fetal DNA from maternal plasma by preparative size separation. Clin Chem. 2009;55(12):2144–2152. doi: 10.1373/clinchem.2009.127480. [DOI] [PubMed] [Google Scholar]
- 164.Yang Q, Du Z, Song Y, Gao S, Yu S, Zhu H, et al. Size-selective separation and overall-amplification of cell-free fetal DNA fragments using PCR-based enrichment. Sci Rep. 2017;19(7):40936. doi: 10.1038/srep40936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.DeAngelis MM, Wang DG, Hawkins TL. Solid-phase reversible immobilization for the isolation of PCR products. Nucleic Acids Res. 1995;23(22):4742–4743. doi: 10.1093/nar/23.22.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Hu P, Liang D, Chen Y, Lin Y, Qiao F, Li H, et al. An enrichment method to increase cell-free fetal DNA fraction and significantly reduce false negatives and test failures for non-invasive prenatal screening: a feasibility study. J Transl Med. 2019;17(1):124. doi: 10.1186/s12967-019-1871-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Welker NC, Lee AK, Kjolby RAS, Wan HY, Theilmann MR, Jeon D, et al. High-throughput fetal fraction amplification increases analytical performance of noninvasive prenatal screening. Genet Med. 2020;23(3):443–450. doi: 10.1038/s41436-020-01009-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Nygard L, Ahlborn LB, Persson GF, Chandrananda D, Langer JW, Fischer BM, et al. Circulating cell free DNA during definitive chemo-radiotherapy in non-small cell lung cancer patients—initial observations. PLoS One. 2020;15(4):e0231884. doi: 10.1371/journal.pone.0231884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Cohen JD, Li L, Wang Y, Thoburn C, Afsari B, Danilova L, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359(6378):926–930. doi: 10.1126/science.aar3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Chan KC, Jiang P, Sun K, Cheng YK, Tong YK, Cheng SH, et al. Second generation noninvasive fetal genome analysis reveals de novo mutations, single-base parental inheritance, and preferred DNA ends. Proc Natl Acad Sci USA. 2016;113(50):E8159–E8168. doi: 10.1073/pnas.1615800113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Cirigliano V, Ordonez E, Rueda L, Syngelaki A, Nicolaides KH. Performance of the neoBona test: a new paired-end massively parallel shotgun sequencing approach for cell-free DNA-based aneuploidy screening. Ultrasound Obstet Gynecol. 2017;49(4):460–464. doi: 10.1002/uog.17386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Heitzer E, Auer M, Hoffmann EM, Pichler M, Gasch C, Ulz P, et al. Establishment of tumor-specific copy number alterations from plasma DNA of patients with cancer. Int J Cancer. 2013;133(2):346–356. doi: 10.1002/ijc.28030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Liu X, Lang J, Li S, Wang Y, Peng L, Wang W, et al. Fragment enrichment of circulating tumor DNA with low-frequency mutations. Front Genet. 2020;11:147. doi: 10.3389/fgene.2020.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Esposito Abate R, Frezzetti D, Maiello MR, Gallo M, Camerlingo R, De Luca A, et al. Next generation sequencing-based profiling of cell free DNA in patients with advanced non-small cell lung cancer: advantages and pitfalls. Cancers (Basel). 2020;12(12):3804. doi: 10.3390/cancers12123804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Burnham P, Kim MS, Agbor-Enoh S, Luikart H, Valantine HA, Khush KK, et al. Single-stranded DNA library preparation uncovers the origin and diversity of ultrashort cell-free DNA in plasma. Sci Rep. 2016;14(6):27859. doi: 10.1038/srep27859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Sanchez C, Snyder MW, Tanos R, Shendure J, Thierry AR. New insights into structural features and optimal detection of circulating tumor DNA determined by single-strand DNA analysis. NPJ Genom Med. 2018;3:31. doi: 10.1038/s41525-018-0069-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Liu X, Liu L, Ji Y, Li C, Wei T, Yang X, et al. Enrichment of short mutant cell-free DNA fragments enhanced detection of pancreatic cancer. EBioMedicine. 2019;41:345–356. doi: 10.1016/j.ebiom.2019.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Moser T, Ulz P, Zhou Q, Perakis S, Geigl JB, Speicher MR, et al. Single-stranded DNA library preparation does not preferentially enrich circulating tumor DNA. Clin Chem. 2017;63(10):1656–1659. doi: 10.1373/clinchem.2017.277988. [DOI] [PubMed] [Google Scholar]
- 179.Lam WKJ, Jiang P, Chan KCA, Cheng SH, Zhang H, Peng W, et al. Sequencing-based counting and size profiling of plasma Epstein–Barr virus DNA enhance population screening of nasopharyngeal carcinoma. Proc Natl Acad Sci USA. 2018;115(22):E5115–E5124. doi: 10.1073/pnas.1804184115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Mazurek AM, Rutkowski T, Fiszer-Kierzkowska A, Malusecka E, Skladowski K. Assessment of the total cfDNA and HPV16/18 detection in plasma samples of head and neck squamous cell carcinoma patients. Oral Oncol. 2016;54:36–41. doi: 10.1016/j.oraloncology.2015.12.002. [DOI] [PubMed] [Google Scholar]