To the Editor:
Of the many immune components involved in asthma pathobiology, recent studies have focused on the potential roles of receptor for advanced glycation end products (RAGE).1 RAGE is a member of the immunoglobulin superfamily, which serves as a pattern-recognition receptor for many glycated and non-glycated ligands.1 Membrane-bound RAGE (mRAGE), through binding with ligands, activates various downstream inflammatory pathways.1 By contrast, the cleaved form—soluble RAGE (sRAGE)—functions as a decoy receptor through binding proinflammatory ligands destined for mRAGE, and hence offers anti-inflammatory effects.1 These RAGE pathways are involved in a range of lung diseases (e.g., acute respiratory distress syndrome) and are a candidate for targeted therapies.1
Epidemiological studies have reported associations of lower circulating sRAGE levels with incident asthma2 and higher disease severity.1 For example, a multicenter cohort analysis of bronchiolitis has shown that infants with lower sRAGE levels have a higher risk of developing recurrent wheeze and asthma in childhood.2 Genome-wide association studies (GWAS) have also found that polymorphisms in RAGE-coding gene (AGER) are associated with natural variations of lung function in Europeans.1 Additionally, its missense variant (rs2070600) has been associated with lower levels of sRAGE, more airflow limitation, and higher asthma severity.1,3 Notwithstanding these epidemiological associations, the causal role of sRAGE in asthma (and the role of potential RAGE-targeted therapies) remains unclear. To address the knowledge gap, we performed a Mendelian randomisation study to examine the causal relationship of sRAGE with asthma, its major phenotypes, and risk factors.
This is a two-sample Mendelian randomisation study using GWAS summary statistics from two large studies—the SCALLOP consortium4 and UK Biobank.5-8 Details of the Methods are available in the Online Repository. Briefly, Mendelian randomisation can provide unbiased causal estimates from observational data because the genetic polymorphisms associated with the exposure (sRAGE) are allocated randomly at conception and its causal inference is less susceptible to confounding and reverse causation.9 Of 15 cohorts that comprise the SCALLOP consortium, 13 cohorts conducted a genome-wide protein quantitative trait loci (pQTL) analysis of plasma proteins in 21,758 individuals of European ancestry.4 The UK Biobank is a prospective cohort study that enrolled approximately 500,000 individuals, and longitudinally measured comprehensive phenotypic data and performed genome-wide genotyping. The current analysis restricted the sample to subjects of European ancestry (46,799 cases with asthma and 347,457 controls) to minimize population stratification bias. The current analysis has complied with all ethical regulations according to UK Biobank policy5 and was approved by the institutional review board of Massachusetts General Hospital.
As the genetic instruments, we identified independent cis-acting variants strongly associated with plasma sRAGE levels (PGWAS<1×10−5, r2<0.001, 500kb from AGER; Table S1) in the SCALLOP consortium.4 To ensure no confounding for variants, we investigated Bonferroni-corrected associations with potential confounders—alcohol intake, physical activity and smoking status—in the UK Biobank. We also assured that variants affect outcomes solely through sRAGE by examining pleiotropy in Ensembl, GWAS catalog, and PhenoScanner. Separately, using the UK Biobank data, we computed the GWAS statistics for asthma (primary outcome) and six major asthma phenotypes (secondary outcomes): 1) childhood, 2) adult-onset, 3) allergic, 4) non-allergic, 5) obese, and 6) non-obese asthma, as previously described.6-8 We weighted the magnitude of association of each variant with outcomes by that with sRAGE, and estimated the combined effect of sRAGE on each outcome by inverse-variance weighted method with a fixed-effects model. In the sensitivity analysis, we repeated the analysis by selecting a variant only from AGER coding-region (PGWAS<1×10−5, r2<0.001). We also examined the role of potential asthma risk factors (atopic dermatitis, total IgE level, blood eosinophil and neutrophil counts, body mass index [BMI]) using 2-step Mendelian randomisation method.
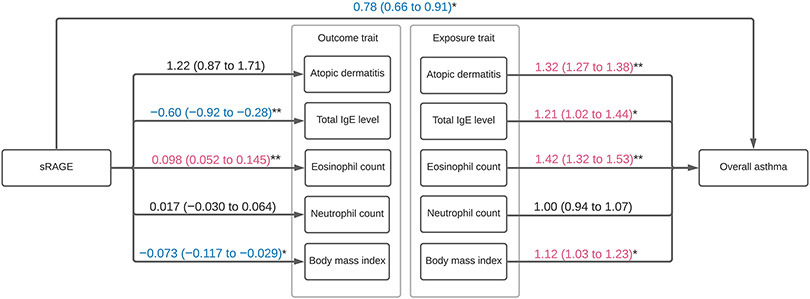
The analysis identified two genetic instruments (rs204993, rs9267536) strongly associated with plasma sRAGE levels. These variants had an F-statistic of >10, without significant association with the potential confounders (Table S1). Higher genetically-instrumented sRAGE levels were associated with a significantly lower risk of overall asthma (OR per one standard deviation [SD] increment in inverse-rank normalized sRAGE level, 0.78; 95%CI, 0.66-0.91; P=0.002; Figures 1 and S1). In the stratified analysis, with limited statistical power, the association appeared consistent across the asthma phenotypes (Figure 1)—e.g., allergic (OR, 0.75; 95%CI, 0.60-0.93; P=0.008) and obese (OR, 0.69; 95%CI, 0.52-0.92; P=0.01) asthma. The sensitivity analysis using a variant in AGER coding-region (rs3134940) also demonstrated a consistent result—a negative relationship between sRAGE and overall asthma risk (OR, 0.74; 95%CI, 0.58-0.95; P=0.02; Figure S2). In the 2-step Mendelian randomisation analysis (Figure 2 and Table S2), higher sRAGE levels were associated with a significantly lower total IgE level (SD difference, −0.60; 95%CI, −0.92 to −0.28; P<0.001) and BMI (SD difference, −0.073; 95%CI, −0.12 to −0.029; P=0.001). Both of these risk factors were also associated with a significantly higher risk of overall asthma. Similarly, a higher total IgE level was significantly associated with an increased risk of allergic asthma (OR, 1.27; 95%CI, 1.01-1.62; P=0.04), while it was not associated with non-allergic asthma (OR, 1.16; 95% CI 0.92-1.47; P=0.21; Table S3 and Figure S3).
Figure 1. Mendelian randomisation estimates for the effect of soluble receptor for advanced glycation end products (sRAGE) on asthma and its phenotypes.
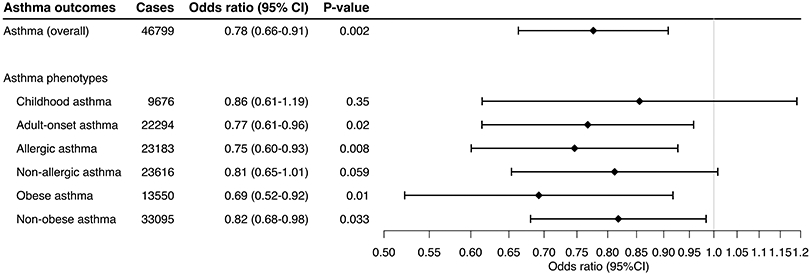
By using the inverse variance weighted method, the combined causal effect of sRAGE on asthma (overall) and six asthma phenotypes was estimated. The odds ratios were estimated per one standard deviation increment in the inverse-rank normalized sRAGE level.
Abbreviation: CI, confidence interval; sRAGE, soluble receptor for advanced glycation end products
Figure 2. Two-step Mendelian randomisation estimates for the effect of soluble receptor for advanced glycation end products (sRAGE) on risk factors and their effect on asthma.
The numbers indicate estimated odds ratios (ORs) or regression coefficients with 95% CIs from the Mendelian randomisation analyses. For the effect of the plasma sRAGE level on the binary risk factor (atopic dermatitis), an OR per one standard deviation (SD) increment in the inverse-rank normalized sRAGE level was estimated. For the effect of sRAGE on the continuous risk factors (plasma total IgE level, blood eosinophil and neutrophil count, and body mass index), coefficients in their SD scale per one SD increment in the inverse-rank normalized sRAGE level were estimated
For the effect of the binary risk factor on asthma risk, an OR was estimated by comparing the patients with the risk factor to those without. For the effect of the continuous risk factors, ORs per one SD increment in the corresponding risk factors were estimated. Arrows indicate causal directions in the 2-step Mendelian randomisation analyses. *, P<0.05; **, P<0.001.
Abbreviations: CI, confidence interval; IgE, immunoglobulin E; sRAGE, soluble receptor for advanced glycation end products
The current Mendelian randomisation analysis demonstrated that the genetically-instrumented sRAGE level is negatively associated with the asthma risk and that the causal association is consistent across a wide range of asthma phenotypes. Our results are concordant with previous epidemiological findings that lower circulating sRAGE levels are associated with greater risks of incident asthma2 and higher disease severity, both in children and adults.1 Prior research also has shown that higher levels of sRAGE ligands (e.g., danger-associated molecular patterns [DAMPs], including HMGB1) are associated with increased blood IgE level and asthma severity.1 In the present analysis, the validity is buttressed by the use of Mendelian randomisation approach. This approach has the major advantage that it can address unmeasured confounding and reverse causation that occur in traditional observational studies.9 Our analysis meets the three assumptions of the Mendelian randomisation design:9 1) the genetic instruments are strongly correlated with sRAGE (the relevance assumption); 2) they do not share common causes with the outcome (the independence assumption); and 3) they affect the outcome only though their effect on sRAGE (the exclusion restriction assumption). The present analysis, using large cohort data, builds on earlier reports, and extends them by robustly examining causal effects of sRAGE on asthma, its phenotypes, and risk factors.
Our observations suggest that sRAGE plays a fundamental role in the pathobiology of asthma and its phenotypes. The exact mechanism(s) underlying the observed relationships is beyond the scope of current study and merits further investigations. Lower levels of sRAGE (as a decoy receptor) reduce binding of numerous RAGE ligands, thereby leading to upregulated signalling of mRAGE—which are most abundantly-expressed in type I alveolar cells—and downstream inflammatory (e.g., NFκB, JAK/STAT) pathways in the lungs.1 Additionally, experimental models have shown that environmental allergen exposures induce alarmins in an mRAGE-dependent fashion, inducing interleukin (IL)-4/IL-13 production and T2 inflammation.1,10 Furthermore, research also reported that administration of sRAGE diminishes LPS-induced T1 and T17 cytokines.11 Taken together, there is a growing rationale for targeting the RAGE signalling pathways in asthma.
Several potential limitations should be noted. Firstly, misclassification of risk factors and asthma outcomes in the GWAS datasets is possible. Yet, it is unrelated to the measured sRAGE levels in the current two-sample design; thus, this independent nondifferential misclassification would have biased the inferences toward the null. Secondly, the effects of genetic instruments may be mitigated by developmental processes. Yet, such mechanisms would have decreased the genetic effects, also biasing the inferences toward the null. Finally, to minimize the population stratification bias, we restricted the study samples to individuals of European ancestry. Therefore, the inferences should be cautiously generalized to other racial/ethnic populations.
To conclude, the current Mendelian randomisation analysis demonstrated that a higher genetically-instrumented sRAGE level is causally associated with a lower risk of asthma. The observation was consistent across major asthma phenotypes. Our inferences should encourage further research into identifying patients with specific molecular phenotype(s) in which the RAGE pathways play major pathobiological roles.
Supplementary Material
Acknowledgments
Funding Statement This study was supported by grants (R01 AI-134940 and R01 AI-148338) from the National Institutes of Health (Bethesda, MD). The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The funding organization was not involved in the collection, management, or analysis of the data; preparation or approval of the manuscript; or decision to submit the manuscript for publication.
Footnotes
Disclosure Statement: Dr. Camargo has participated in asthma-related scientific advisory boards for AstraZeneca, GSK, and Sanofi. Dr. Hasegawa has received an asthma-related research grant from Novartis. The authors have no conflicts of interest relevant to this article to disclose.
REFERENCES
- 1.Perkins TN, Donnell ML, Oury TD. The axis of the receptor for advanced glycation endproducts in asthma and allergic airway disease. Allergy. Published online September 25, 2020. doi: 10.1111/all.14600 [DOI] [PubMed] [Google Scholar]
- 2.Patregnani JT, Fujiogi M, Camargo CA, et al. Serum soluble receptor for advanced glycation end-products (sRAGE) in infants with bronchiolitis: Associations with acute severity and recurrent wheeze. Clin Infect Dis. Published online November 10, 2020. doi: 10.1093/cid/ciaa1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lyu Y, Zhao H, Ye Y, et al. Decreased soluble RAGE in neutrophilic asthma is correlated with disease severity and RAGE G82S variants. Mol Med Rep. 2018;17(3):4131–4137. doi: 10.3892/mmr.2017.8302 [DOI] [PubMed] [Google Scholar]
- 4.Folkersen L, Gustafsson S, Wang Q, et al. Genomic and drug target evaluation of 90 cardiovascular proteins in 30,931 individuals. Nat Metab. 2020;2(10):1135–1148. doi: 10.1038/s42255-020-00287-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sudlow C, Gallacher J, Allen N, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12(3):e1001779. doi: 10.1371/journal.pmed.1001779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu Z, Lee PH, Chaffin MD, et al. A genome-wide cross-trait analysis from UK Biobank highlights the shared genetic architecture of asthma and allergic diseases. Nat Genet. 2018;50(6):857–864. doi: 10.1038/s41588-018-0121-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu Z, Guo Y, Shi H, et al. Shared genetic and experimental links between obesity-related traits and asthma subtypes in UK Biobank. J Allergy Clin Immunol. 2020;145(2):537–549. doi: 10.1016/j.jaci.2019.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu Z, Zhu X, Liu C-L, et al. Shared genetics of asthma and mental health disorders: a large-scale genome-wide cross-trait analysis. Eur Respir J. 2019;54(6). doi: 10.1183/13993003.01507-2019 [DOI] [PubMed] [Google Scholar]
- 9.Davies NM, Holmes MV, Davey Smith G. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ. 2018;362. doi: 10.1136/bmj.k601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oczypok EA, Milutinovic PS, Alcorn JF, et al. Pulmonary receptor for advanced glycation end-products promotes asthma pathogenesis through IL-33 and accumulation of group 2 innate lymphoid cells. J Allergy Clin Immunol. 2015;136(3):747–756.e4. doi: 10.1016/j.jaci.2015.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang F, Su X, Huang G, et al. sRAGE alleviates neutrophilic asthma by blocking HMGB1/RAGE signalling in airway dendritic cells. Sci Rep. 2017;7(1):14268. doi: 10.1038/s41598-017-14667-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.